Abstract
The rate of insertion and deletion mutations of the replicase of Cucumber mosaic virus (CMV) was determined in planta by using a parasitic satellite RNA (satRNA) as a reporter. We found that the CMV replicase had different fidelity in different environments, with important implications in viral disease evolution. Insertions were very rare events, irrespective of the region of the satRNA genome assayed and independent of the hosts tested. On the other hand, deletion events were more frequent but were restricted to a highly structured region of the reporter. Deletion mutation rates were different for the two hosts tested, although the mutation distribution was not influenced by the hosts. Moreover, hot spots with high mutation rates were identified on the satRNA genome.
RNA viruses are characterized by extreme evolutionary capacities that allow them to successfully expand their host ranges and adapt to new environments, leading to emerging viral diseases such as severe acute respiratory syndrome and AIDS (4-7, 9, 15). This capacity for rapid evolution is powered by their replication by error-prone, self-encoded replicases that may lack proofreading functions (1, 2, 9, 26). A number of factors affect virus population diversity, including selection and genetic bottlenecks, but the low fidelity of the replicase is the underlying source for most variation. Although misincorporation is very important in RNA virus evolution, there is a lack of information about the polymerase fidelity of RNA viruses, especially in intact hosts. A few reports exist estimating the substitution mutation rates of RNA virus replicases in vitro or in tissue culture cells (5). However, no studies have characterized the rates of insertions and deletions that can lead to more profound evolutionary changes. Previous work indicated that the variation detected in a stable Cucumber mosaic virus (CMV) population is dependent on both the host and the virus (25). Here, we measured the polymerase fidelity of an RNA virus in planta. We analyzed the rate of insertion and deletion mutations (indels) introduced by the replicase of CMV in two hosts: pepper and tobacco.
CMV, genus Cucumovirus, family Bromoviridae, is related to several other plant and animal viruses, such as Sindbis virus, and is a well-established model system for RNA virus evolution studies (21). It sometimes harbors molecular parasites known as satellite RNAs (satRNAs). The satRNAs are noncoding, highly structured small RNAs that are completely dependent on the virus for replication and dissemination. They make excellent reporters for evolution studies (21). Since the structure of the plus strand of D4 satRNA is well established (20), we used this satellite as a reporter to measure CMV polymerase fidelity, and we correlated mutations with RNA structure.
MATERIALS AND METHODS
Viruses.
Plasmid pDsat4 was described previously (11). To obtain a D4 satRNA template that can be replicated in only one direction, we deleted 25 nucleotides (nt) from the 5′ end of pDsat4, including its presumed replication recognition sequences for CMV RNA polymerase, and added an SP6 RNA polymerase promoter for plus-strand transcript synthesized as follows. Primers CCGGGGGAATTCATTTAGGTGACACTATAGAGGGGTTATATCTACG and CCGGGGGATCCCTCGAGGGTCCTGCAGAGG (D4 satRNA sequences in are in boldface, the SP6 promoter sequence is italicized, and restriction sites are underlined [see below]) were used in a thermal cycling reaction using pDsat4 as a template to generate Δ5′ D4 satRNA. The amplified products were digested with restriction enzymes EcoRI and BamHI (underlined in the respective primers) and cloned into the analogous sites in pBluescript KS(+) (Stratagene, La Jolla, CA), resulting in Δ5′ pDsat4. Linearization of this plasmid with XhoI followed by transcription with bacteriophage SP6 polymerase (Ambion, Austin, TX) generated a precise Δ5′ D4 satRNA transcript. The transcript was then digested with RNase-free DNase (RQ1; Promega) to remove the plasmid template and purified from a 6% denaturing polyacrylamide gel (24). Synthesis of infectious transcripts of the Fny strain of CMV (helper virus) was described previously (22).
Plants and plant inoculations.
Plant hosts used were tobacco (Nicotiana tabacum cv. Xanthi nc) and pepper (Capsicum annuum cv. Morengo). Plants were maintained in a greenhouse with daytime temperatures of 28°C, nighttime temperatures of 22°C, and a 16-h day length. Fny CMV infectious transcripts were inoculated onto 3-week-old pepper and tobacco plants. As soon as CMV symptoms appeared (3 and 6 days postinoculation for tobacco and pepper, respectively), we inoculated the plants with Δ5′ D4 satRNA transcript. Control plants were healthy plants inoculated with the buffer only (50 mM Na2HPO4, pH 9) or with Δ5′ D4 satRNA transcript only.
Total RNA extraction and HiFi RT-PCR.
Twenty-four hours after inoculation of the Δ5′ D4 satRNA reporter, total RNA was isolated from inoculated or systemically infected leaves (upper leaves above satRNA-inoculated leaves) using Tri-Reagent, according to the manufacturer's protocol (Molecular Research Center Inc., Cincinnati, OH). One-fifth of the total RNA extraction was then used in a strand-specific high-fidelity reverse transcriptase (HiFi RT) thermal cycling reaction designed to prevent amplification of any self-primed cDNAs.
For the RT reaction, we used Superscript RT as recommended by the manufacturer (Invitrogen) with the first-strand primer, GCAGACCGAATTCGGGTTATATCTACG, which is specific for the 3′ end of the minus strand of Δ5′ D4 satRNA and has a 5′ sequence tag (underlined). After a 30-min incubation at 37°C, the excess of the first-strand primer was removed using a purification column (QIAGEN) to avoid any carryover to the thermal cycling reaction. The purified cDNAs were used as templates for the thermal cycling reactions with a primer specific for the sequence tag (underlined), CGCCGATATAGCAGACCGAAT, and primer CGGGCTGCAGCATAAGCCTTAGC, specific for the 5′ end of the Δ5′ D4 satRNA. The thermal cycling reactions were carried out in capillary tubes with an Idaho Technology Rapid Cycler for 15 cycles (94°C denaturation for 6 s, 50°C annealing for 6 s, and 72°C extension for 20 s). Products from this HiFi RT-PCR were purified from a 6% nondenaturing polyacrylamide gel (24) and used as the template for the indel assay.
Two regions of the reporter named nonstop (a region without an in-frame stop codon) region A and nonstop region B were identified for this assay. Nonstop region A contains 96 nt (nt 88 to 183), and nonstop region B contains 84 nt (nt 178 to 261). The indel assay primers were designed such that the entire sequence inserted in the multiple cloning site renders the lacZ gene out of frame unless an insertion or a deletion (except those in multiples of 3 nt) has occurred (Fig. 1 and data not shown).
FIG. 1.
The cDNA sequence of D4 satRNA plus-strand. Stop codons in the plus strand are shown in boldface, and stop codons in the minus strand are underlined. Primer sites for the nonstop A region are shown above the sequence line (AF, A region forward; AR, A region reverse). The forward primer contains two changes from the wild-type sequence to eliminate stop codons. The TGA in the reverse primer is in the same frame as the engineered stop codon and will be shifted out of frame in the event of an insertion or a deletion. Following the same strategy, primers BF (forward) and BR (reverse) were designed for the nonstop region B. The forward and reverse primers contain a PstI site and an EcoRI site (respectively) for directional cloning. The regions assayed include the sequences between (exclusive of) the primers.
To minimize artifacts due to misincorporation in the reactions, a DNA polymerase with proofreading capability (Pfu; Stratagene) was mixed with Taq DNA polymerase (Roche) at a ratio of 1:8. A maximum number 15 cycles was used for all thermal cycling steps.
Cloning and sequence analysis of mutant progenies.
Products from the second HiFi PCR step (amplifying the nonstop A and nonstop B regions) were digested with restriction enzymes PstI and EcoRI and ligated into the LacZ open reading frame (ORF) of pUC 19 such that they contain an ORF disrupted with a stop codon. DH5α competent cells were then transformed with ligated PCR products and plated onto LB agar containing IPTG (isopropyl-β-d-thiogalactopyranoside) and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). The total number of colonies was counted with a gel documentation system (Ultra-Lum, Inc.).
All the blue colonies were transferred to a broth culture, grown overnight, and used for a small-scale plasmid preparation. The sequence of the inserts of the resulting plasmids was determined using the DNA analyzer ABI 3730. The controls for the white and blue colony color screening consisted of the circularized pUC19 without insert (positive control, blue colony) and the pUC19 containing an insert of known size in the multiple cloning site that disrupted the lacZ gene (negative control, white colony). The error rate of the HiFi RT-PCR was determined by a control reaction done simultaneously, using in vitro generated transcripts (Table 1). For comparison to mutation frequencies of the CMV replicase, the incidence of indels in pepper and tobacco were calculated from experiments described previously (25) (Table 2).
TABLE 1.
Analysis of background mutations from SP6 transcription, RT-PCR, and cDNA cloning
| Expt no. | Background mutation rate (no. of blue colonies/total no. of colonies)a
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Control
|
Nonstop region A
|
Nonstop region B
|
||||||||
| Insertion
|
Deletion
|
Insertion
|
Deletion
|
|||||||
| Positive | Negative | Minus strand | Plus strand | Minus strand | Plus strand | Minus strand | Plus strand | Minus strand | Plus strand | |
| 1 | 0/1,997 | 2,010/2,010 | NA | NA | NA | NA | NA | NA | NA | NA |
| 2 | 0/3,052 | 3,132/3,132 | NA | NA | NA | NA | NA | NA | NA | NA |
| 3 | 0/2,500 | 2,143/2,143 | 0/2,024 | 0/1,843 | 0/1,625 | 0/1,774 | 0/1,832 | 0/2,280 | 0/1,770 | 0/2,345 |
| 4 | 0/2,241 | 2,031/2,031 | 0/1,735 | 0/1,822 | 0/1,530 | 0/1,724 | 0/1,529 | 0/1,840 | 0/2,025 | 0/2,322 |
| Total no. of colonies | 0/9,790 | 9,316/9,316 | 0/3,759 | 0/3,664 | 0/3,155 | 0/3,498 | 0/3,361 | 0/4,120 | 0/3,795 | 0/4,667 |
| Total no. of nucleotides analyzed | NA | NA | 360,864 | 351,840 | 302,880 | 335,808 | 278,963 | 341,960 | 314,985 | 387,361 |
NA, not applicable.
TABLE 2.
Indel incidence in pepper and tobacco populations of CMVa
| Plant | No. of insertions | No. of deletions | Total no. of indels | Description of mutationb
|
|
|---|---|---|---|---|---|
| Insertion(s) | Deletion(s) | ||||
| Pepper | 2 | 3 | 5 | 2006T, 1222C | 2006T, 1441A, 1088T/1089Tc |
| Tobacco | 1 | 0 | 1 | 2085T | |
Summary of the indels found in the CMV populations described in reference 25.
Mutations are described in terms of the nucleotide number and residue; e.g., the insertion 2006T is the insertion of a T residue after nt 2006, and the deletion 2006T is the deletion of a T residue after nt 2006. The nucleotide number is arbitrary when mutations occurred in runs of a single nucleotide.
Two nucleotides were deleted in one mutant. All other mutations were of single nucleotides.
Bootstrap program.
A description of the bootstrap program is available upon request.
Alignment of satRNA sequences.
Seventy-eight satRNA sequences were obtained from the GenBank database (accession numbers available upon request) and aligned with D4 satRNA (accession number M30584) using ClustalW.
RESULTS
Unidirectional replication and strand-specific HiFi RT-PCR.
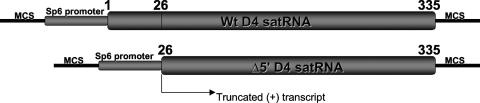
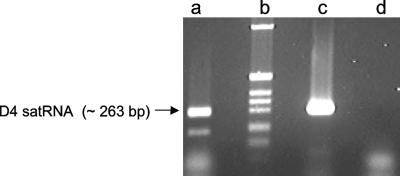
As defined by Domingo and Holland (5), the mutation rate refers to nucleotide misincorporation by the polymerase (including indels), and mutation frequency refers to the detectable occurrence of mutations in a population, after natural selection and genetic drift have acted on the mutant spectrum produced by the polymerase. To assess the CMV polymerase fidelity, we constructed a mutant cDNA clone of the CMV D4 satRNA in which the plus-strand promoter was deleted (Fig. 2). Transcript of positive polarity synthesized from this truncated satRNA clone was inoculated onto tobacco or pepper plants systemically infected with CMV (the helper virus). Since the CMV replicase had only the minus-strand satRNA promoter available, only half a round of replication, from plus strand to minus strand, could occur. The replicase error rate was therefore equal to the frequency of mutations in the minus-strand progeny satRNA. We recovered minus-strand progeny from plants systemically infected with CMV using a nested primer strategy that allows only primer-derived cDNAs (and not self-primed cDNAs) to be amplified, thereby preventing any amplification of the original plus-strand transcript inoculum recovered together with minus-strand progeny satRNA, which could bias the deletion rate estimates (Fig. 3). To eliminate any concern about self-replication of the reporter, we conducted in vivo control assays in which we used plants that were not infected with CMV for inoculation with transcript satRNA. The plus-strand inoculum was recovered, but no minus-strand progeny satRNA was recovered in these plants, confirming that the truncated satRNA transcript cannot replicate in the absence of its helper virus. To determine the background indel level due to SP6 transcription, RT-PCR, and cDNA cloning, plus- and minus-strand satRNA transcripts were used as in vitro controls. We found no background insertions or deletions in 2,674,661 nt of satRNA transcript analyzed (Table 1). This confirmed the high fidelity of our indel assay.
FIG. 2.
D4 satRNA cDNA and Δ5′ deletion mutant. The putative plus-strand promoter was deleted to allow only half a round of replication. MCS, multiple cloning site.
FIG. 3.
Strand specificity. Lane a, positive control reaction using plus-strand-specific RT-PCR and the plus-strand transcript used in lane d; lane b, 1-kb ladder marker; lane c, minus-strand-specific RT-PCR on minus-strand transcript; lane d, minus-strand-specific RT-PCR on plus-strand transcript. The size of the targeted fragment is indicated with the black arrow.
Insertion versus deletion.
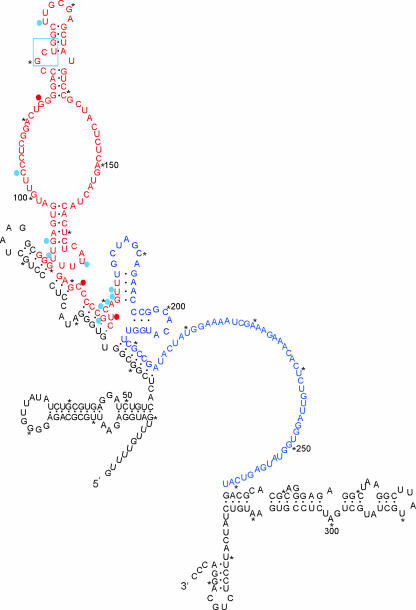
Using a modification of the method described for human immunodeficiency virus polymerase fidelity (16), mutations were measured by reversion of stop codons upstream of a LacZ gene, and the deletion-insertion rates of the replicase were calculated by measuring the ratio of blue to white colonies. The vast majority of the growing colonies were white as they contained a disrupted LacZ gene. However, the rare deletion or insertion mutations that occurred within the satRNA sequence reestablished the LacZ ORF and resulted in a blue colony. We measured CMV replicase indel rates using two regions of the D4 satRNA that are free of stop codons (Fig. 1). Nonstop region A is highly structured, containing nine helices, whereas nonstop region B consists of a long, single-stranded region (Fig. 4) (20). The indel assay was used with pepper and tobacco plants infected with CMV. We found that insertion events were very rare in both of the hosts tested. Only one insertion, a duplication of the 4-nt GCUG (position 120 to 123) was observed out of 4,283,270 bp assayed, compared to 49 deletion events from 5,624,531 bp assayed (Table 3). This does not allow us to estimate a rate of insertions, but it is likely near the limit of detection in this system.
FIG. 4.
Distribution of indels observed in planta and relationship to D4 satRNA secondary structure (revised from reference 18). The nonstop regions A and B are shown in red and blue, respectively. Deletions are indicated with blue circles or red circles (for hot spots). The nucleotide number is arbitrary when deletions occurred in runs of a single nucleotide. The box marks the nucleotides involved in the 4-base insertion event.
TABLE 3.
CMV replicase indel rates in planta
| Host | Plant no. | Leaf typea | Incidence of indel events (no. of events/total no. of nucleotides)b
|
Nonstop A deletion ratec | No. of deletions at the position indicated in nonstop region A
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nonstop A
|
Nonstop B
|
|||||||||||||||||||
| I | D | I | D | U-91 | G-93 | C-103 | G-113 | U-126 | U-165 | C-171 | G-176 | U-178 | C-179 | A-180 | G-181 | U-182 | ||||
| Pepper | Ct | 0/86,592 | 0/77,088 | 0/69,305 | 0/139,440 | NA | ||||||||||||||
| Pepper | 1 | In | 0/187,776 | 10/77,280 | 0/53,950 | 0/53,120 | 1.29 × 10−04 | 1 | 1 | 3 | 1 | 3 | 1 | |||||||
| 2 | In | 0/48,384 | 6/39,360 | 0/50,049 | 0/65,985 | 1.24 × 10−04 | 3 | 1 | 2 | |||||||||||
| 3 | In | 1/41,088 | 3/22368 | 0/29050 | 0/89,391 | 1.34 ×10−04 | 1 | 1 | 1 | |||||||||||
| Tobacco | 1 | In | 0/343,392 | 6/348,000 | 0/341,296 | 0/394,914 | 1.72 × 10−05 | 1 | 1 | 1 | 1 | 1 | 1 | |||||||
| 2 | In | 0/227,616 | 6/236,160 | 0/521,240 | 0/339,885 | 2.54 × 10−05 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||
| Sy | 0/386,496 | 1/517,536 | 0/390,515 | 0/500,158 | 1.93 × 10−06 | 1 | ||||||||||||||
| 3 | In | 0/115,200 | 8/116,640 | 0/124,500 | 0/108,315 | 6.85 × 10−05 | 1 | 4 | 1 | 2 | ||||||||||
| Sy | 0/240,288 | 1/310,176 | 0/390,764 | 0/500,324 | 3.22 × 10−06 | 1 | ||||||||||||||
| 4 | In | 0/96,480 | 2/98,400 | 0/101,592 | 0/89,225 | 2.03 × 10−05 | 1 | 1 | ||||||||||||
| Sy | 0/212,352 | 1/318,240 | 0/225,345 | 0/208,579 | 3.14 × 10−06 | 1 | ||||||||||||||
| Total | 1/1,985,664 | 44/2,161,248 | 0/2,297,606 | 0/2,489,336 | 2 | 2 | 2 | 8 | 1 | 2 | 7 | 4 | 7 | 1 | 4 | 3 | 1 | |||
Source of half-replicated, progeny Δ5′D4 satRNA reporter molecules. Ct, = control (healthy plant infected with reporter only); In, leaf of CMV-infected plant inoculated with Δ5′D4 satRNA reporter; Sy, leaf of CMV-infected plant systemically infected by Δ5′D4 satRNA reporter.
I, insertion; D, deletion.
NA, not applicable.
Template structure and host factors affect deletion mutation rates.
The deletions observed were all located in the highly structured nonstop A region, while the nonstop B region that is largely open did not contain any mutations (Fig. 4). This provides compelling evidence that CMV replicase fidelity is strongly dependent on template structure. CMV replicase deletion rates were calculated as a ratio of the number of deletions observed to the number of nucleotides assayed. The deletion rates were significantly higher for templates replicated in pepper versus tobacco (Table 3).
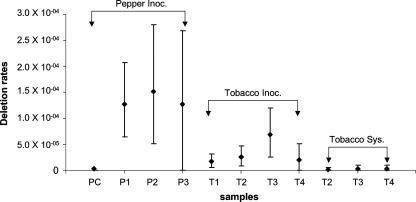
We used Octave, version 2.1.73 (J. W. Eaton; http://www.octave.org), to calculate 95% confidence intervals for the deletion rates using a bootstrap program that takes into account variation present in the data (Fig. 5). Using these intervals, the deletion rates from the in planta controls and those from systemically infected leaves were not significantly different from 0 or from each other. For inoculated leaves, most deletion rate estimates were significantly different from 0 (Fig. 5), although two samples (pepper plant 3 and tobacco plant 4) had relatively low numbers of colonies recovered during blue/white screening, which adversely affected the confidence intervals for these estimates. The deletion rate estimates were higher for the pepper-derived rates than for tobacco-derived rates, confirming trends detected in the raw data. It is difficult to precisely measure deletion rates because of the rare occurrence of indel events. Nevertheless, it was possible to estimate a deletion rate of CMV replicase in planta (Table 3).
FIG. 5.
Estimates of deletion rates and 95% confidence interval for each experiment. Samples are labeled according to the following key: P, pepper plant; T, tobacco plant; C, control. Samples were from inoculated leaves (Inoc.) or systemic leaves (Sys.). A description of the statistical analysis is available on request.
Selection effects on deletion mutant population.
To draw a parallel between our study and naturally occurring deletions that have survived selection and genetic bottlenecks, we compared 78 CMV satRNA sequences from the GenBank database and the D4 satRNA sequence. Only one of the deletions observed in our system, deletion U165, was fixed in 31 of the 79 natural isolates of CMV satRNA, suggesting that most deletions found here are probably not tolerated. We also measured deletion rates from the upper leaves of tobacco plants (systemically infected). Progeny satRNA that had moved out of the inoculated leaves lost most of the deletions (Table 3), most likely due to strong selective forces imposed during movement.
DISCUSSION
RNA viruses are known to generate high levels of genetic variation. We examined the extent to which host factors and template structure could influence viral RNA diversity and virus evolution by focusing our study on indels that can lead to profound evolutionary changes. We found that insertion events are very rare irrespective of the host tested. In nonstop region A, deletion mutations outnumbered insertion mutations at a ratio of 1 to 49, confirming results from previous studies (3, 18, 23). In long-term evolution, this dramatic difference in deletion and insertion rates may explain why most extant RNA viruses seem to be of minimal length; in the short term it illustrates the strength of selection in maintaining virus genomes, since reversions of deletion mutations are such rare events.
Since CMV satRNAs are noncoding, all of their biological activity resides in the primary sequence or the RNA structure. All of the deletions were found in the nonstop A region, which is highly structured, and most deletions were found more than once in these experiments (Table 3), with three occurring as often as seven or eight times, indicating potential hot spots. Pathak and Temin (19) demonstrated that the introduction of an artificial 34-bp stem-loop structure in an RT template triggered a threefold increase in the mutation rate, with 59% of the mutations consisting of deletions associated with the added stem-loop. Template secondary structure has also been shown to influence the generation of internal deletions in the genomic RNAs of Flock house virus and Tomato bushy stunt virus (14, 27). However, the mechanisms underlying RNA deletion mutation are unknown. Hence, structure seems to be important in the generation of deletion mutations, although we cannot discount the possibility that in our system there is something else that is unique about the nonstop A region that increases deletion rates.
Nagy and Bujarsky (17) proposed that pausing by Brome mosaic virus replicase within or close to an A-U-rich region of Brome mosaic virus RNA 3 induced deletions and imprecise homologous recombination. In vitro experiments suggested that viral RT pausing near the base of an extended hairpin triggered deletion events that truncated the structure (10), which correlates with recombination in vivo. Pausing during synthesis of RNA templates has also been attributed to stretches of G or C (28) and is more frequent when the template 6 to 10 nt ahead of the enzyme is base paired (8). Likewise, pausing diminishes when single unpaired nucleotides are introduced (12). Replicase pausing may explain why two deletions, G-113 and C-171, occurred so frequently (seven or eight times) (Table 3). C-171 is within a stretch of five Cs that were never methylated in in vivo analyses of satRNA structure (20), indicating that they were almost certainly involved in base pairing. G-113 is located within a stretch of four G residues; however, because G methylation by dimethyl sulfoxide is undetectable, their status (base paired or not) is uncertain. The other hypermutated nucleotide, U-178, is in the same highly structured region as the other nucleotides but is not in a homopolymer region. Since U-residue methylation by dimethyl sulfoxide is also undetectable, the base-pairing status of this nucleotide is also uncertain.
From analysis of systemically infected leaves in tobacco, we found that most of the deletions observed on the inoculated leaves do not move systemically. Several scenarios including a negative selection during the packaging process, a bottleneck during systemic movement (13), or the instability of the deleted molecules could explain this observation.
In previous studies, mutation frequencies of CMV populations were higher in pepper than in tobacco (25) (Table 2). These populations had undergone an unknown number of rounds of replication and were subjected to a variety of selective forces; thus, a role for replicase fidelity under these conditions is only one of several possibilities. Here, we clearly established a difference in replicase fidelity between pepper and tobacco, the first report of a viral replicase exhibiting variable fidelity in different hosts. The identification and manipulation of factors that regulate viral replicase fidelity in these hosts may offer a new set of tools to predict or control emerging diseases.
Acknowledgments
We thank F. Coker and V. Chandler for technical assistance, T. Feldman for help with statistical analyses, and K. Mysore and E. Urbanczyk-Wochniak for careful reviews of the manuscript.
This work was supported by the Samuel Roberts Noble Foundation.
Footnotes
Published ahead of print on 6 June 2007.
REFERENCES
- 1.Agol, V. I. 2006. Molecular mechanisms of poliovirus variation and evolution, p. 211-259. In E. Domingo (ed.), Quasispecies: concepts and implications for virology. Current topics in microbiology and immunology, vol. 299. Springer, Heidelberg, Germany. [DOI] [PubMed] [Google Scholar]
- 2.Castro, C., J. J. Arnold, and C. E. Cameron. 2005. Incorporation fidelity of the viral RNA-dependent RNA polymerase: a kinetic, thermodynamic and structural perspective. Virus Res. 107:141-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.deJong, W. W., and L. Rydén. 1981. Causes of more frequent deletions and insertions in mutations and protein evolution. Nature 290:157-159. [DOI] [PubMed] [Google Scholar]
- 4.Domingo, E., C. K. Biebricher, J. J. Holland, and M. Eigen. 2001. Quasispecies and RNA virus evolution, principles and consequences, vol. 14. Landes Bioscience, Austin, TX.
- 5.Domingo, E., and J. J. Holland. 1994. Mutation rates and rapid evolution of RNA viruses, p. 161-184. In S. S. Morse (ed.), The evolutionary biology of viruses. Raven Press, Ltd., New York, NY.
- 6.Eigen, M. 1993. Viral quasispecies. Sci. Am. 269:42-49. [DOI] [PubMed] [Google Scholar]
- 7.Gao, G., E. Bailes, D. L. Robertson, Y. Chen, C. M. Rodenburg, S. F. Michael, L. B. Cummins, L. O. Arthur, M. Peeters, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436-441. [DOI] [PubMed] [Google Scholar]
- 8.Harrison, G. P., M. S. Mayo, E. Hunter, and A. M. L. Lever. 1998. Pausing of reverse transcriptase on retroviral RNA templates is influenced by secondary structures both 5′ and 3′ of the catalytic site. Nucleic Acids Res. 26:3433-3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holland, J., K. Spindler, F. Horodyski, E. Grabau, S. Nichol, and S. VandePol. 1982. Rapid evolution of RNA genomes. Science 215:1577-1585. [DOI] [PubMed] [Google Scholar]
- 10.Konstantinova, P., P. de Haan, A. T. Das, and B. Berkhout. 2006. Haripin-induced tRNA-mediated (HITME) recombination in HIV-1. Nucleic Acids Res. 34:2206-2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurath, G., and P. Palukaitis. 1989. Satellite RNAs of cucumber mosaic virus: recombinants constructed in vitro reveal independent functional domains for chlorosis and necrosis in tomato. Mol. Plant-Microbe Interact. 2:91-96. [Google Scholar]
- 12.Lanciault, C., and J. J. Champoux. 2005. Effects of unpaired nucleotides within HIV-1 genomic secondary structures on pausing and strand transfer. J. Biol. Chem. 280:2413-2423. [DOI] [PubMed] [Google Scholar]
- 13.Li, H., and M. J. Roossinck. 2004. Genetic bottlenecks reduce population variation in an experimental RNA virus population. J. Virol. 78:10582-10587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, Y., and L. A. Ball. 1993. Nonhomologous RNA recombination during negative-strand synthesis of flock house virus RNA. J. Virol. 67:3854-3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lipsitch, M., T. Cohen, B. Cooper, J. M. Robins, S. Ma, L. James, G. Gopalakrishna, S. K. Chew, C. C. Tan, M. H. Samore, D. Fisman, and M. Murray. 2003. Transmission dynamics and control of severe acute respiratory syndrome. Science 300:1966-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mansky, L. M., and H. M. Temin. 1995. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J. Virol. 69:5087-5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagy, P. D., and J. Bujarski. 1996. Homologous RNA recombination in brome mosaic virus: AU-rich sequences decrease the accuracy of crossovers. J. Virol. 70:415-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ophir, R., and D. Graur. 1997. Patterns and rates of indel evolution in processed pseudogenes from humans and murids. Gene 205:191-202. [DOI] [PubMed] [Google Scholar]
- 19.Pathak, V. K., and H. M. Temin. 1992. 5-Azacytidine and RNA secondary structure increase the retrovirus mutation rate. J. Virol. 66:3093-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodríguez-Alvarado, G., and M. J. Roossinck. 1997. Structural analysis of a necrogenic strain of cucumber mosaic cucumovirus satellite RNA in planta. Virology 236:155-166. [DOI] [PubMed] [Google Scholar]
- 21.Roossinck, M. J. 2001. Cucumber mosaic virus, a model for RNA virus evolution. Mol. Plant Pathol. 2:59-63. [DOI] [PubMed] [Google Scholar]
- 22.Roossinck, M. J., I. Kaplan, and P. Palukaitis. 1997. Support of a cucumber mosaic virus satellite RNA maps to a single amino acid proximal to the helicase domain of the helper virus. J. Virol. 71:608-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saitou, N., and S. Ueda. 1994. Evolutionary rates of insertion and deletion in noncoding nucleotide sequences of primates. Mol. Biol. Evol. 11:504-512. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 25.Schneider, W. L., and M. J. Roossinck. 2001. Genetic diversity in RNA viral quasispecies is controlled by host-virus interactions. J. Virol. 75:6566-6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steinhauer, D. A., and J. J. Holland. 1986. Direct method for quantification of extreme polymerase error frequencies at selected single base sites in viral RNA. J. Virol. 57:219-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.White, K. A., and T. J. Morris. 1994. Nonhomologous RNA recombination in tombusviruses: generation and evolution of defective interfering RNAs by stepwise deletions. J. Virol. 68:14-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu, W., B. M. Blumberg, P. J. Fay, and R. A. Bambara. 1995. Strand transfer mediated by human immunodeficiency virus reverse transcriptase in vitro is promoted by pausing and results in misincorporation. J. Biol. Chem. 270:325-332. [DOI] [PubMed] [Google Scholar]