Abstract
The herpes simplex virus type 1 UL6 protein forms a 12-subunit ring structure at a unique capsid vertex which functions as a conduit for encapsidation of the viral genome. To characterize UL6 protein domains that are involved in intersubunit interactions and interactions with other capsid proteins, we engineered a set of deletion mutants spanning the entire gene. Three deletion constructs, D-5 (Δ198-295), D-6 (Δ322-416), and D-LZ (Δ409-473, in which a putative leucine zipper was removed), were introduced into the viral genome. All three mutant viruses produced only B capsids, indicating a defect in encapsidation. Western blot analysis showed that the UL6 protein was present in the capsids isolated from two mutants, D-6 and D-LZ. The protein encoded by D-5, on the other hand, was not associated with capsids and was instead localized in the cytoplasm of the infected cells, indicating that this deletion affected the nuclear transport of the portal protein. The UL6 protein from the KOS strain (wild type) and the D-6 mutant were purified from insect cells infected with recombinant baculoviruses and shown to form ring structures as assessed by sucrose gradient centrifugation and electron microscopy. In contrast, the D-LZ mutant protein formed aggregates that sedimented throughout the sucrose gradient as a heterogeneous mixture and did not yield stable ring structures. A mutant (L429E L436E) in which two of the heptad leucines of the putative zipper were replaced with glutamate residues also failed to form stable rings. Our results suggest that the integrity of the leucine zipper region is important for oligomer interactions and stable ring formation, which in turn are required for genome encapsidation.
The formation of mature capsids containing the linear double-stranded herpes simplex virus type 1 (HSV-1) genome is a complex process involving a preassembled protein shell (procapsid), concatemeric viral DNA, and seven other viral proteins believed to play a role in cleavage and packaging of the genome (1, 10). The procapsid contains three shell proteins, VP5, VP19C, and VP23, and the viral scaffolding proteins (encoded by UL26 and UL26.5). During wild-type (WT) infection, three capsid forms are observed: “A” capsids, which have initiated the encapsidation process and have lost both scaffold and DNA; “B” capsids, which contain the scaffold but no DNA; and DNA-containing mature “C” capsids, which have lost scaffold during the process of taking up DNA. The cleavage and packaging machinery consists of the UL6 portal ring, a two-subunit terminase (UL15 and UL28), a protein which seals the capsid after DNA uptake has occurred (UL25), and three other proteins which play less well defined roles in the process (UL17, UL32, and UL33). Defects in any of these genes except the UL25 gene result in the accumulation of B capsids.
The protein encoded by the UL6 gene is present in all three forms of viral capsids (19) and is believed to form a ring structure at a unique vertex which serves as the portal for the entry of viral DNA into the HSV-1 capsid. The UL6 protein is essential for cleavage of the replicated genome, and it can localize to the nucleus in the absence of other viral proteins (19, 20). The UL6 protein is also required for the efficient association of the terminase subunit, UL15, with the capsid (21, 33). Coimmunoprecipitation experiments with recombinant proteins showed that UL6 forms a complex with UL15 and UL28 (30). Newcomb et al. purified the UL6 protein from recombinant baculovirus-infected insect cells and showed in vitro that it forms oligomeric rings (17). The ring (average of 12 subunits) encloses a channel approximately 5 nm in diameter and resembles portals from bacteriophages such as T4, P22, SPP1, and Φ29. Trus et al. determined a low-resolution structure of the HSV-1 portal from cryoelectron microscopy images of UL6 rings (29). The rings average 15 nm in diameter and show narrow flanges on the outside, resembling the shape of a conical propeller. Using electron tomography of HSV-1 capsids from which the pentons were removed, Chang et al. showed the portal to be partly buried at a unique fivefold axis within the capsid shell (5). Recently, Cardone et al. confirmed the location of the HSV-1 portal at a unique capsid vertex; their tomographic analysis of A capsids, however, suggested that a major portion of the portal is present outside the capsid floor (3). The apparent discrepancy between these studies may be due to differences in the type of capsids used and the treatment of capsids with guanidine in the former study. The results from all these studies indicate that UL6 forms the portal ring and probably acts as the docking site for the terminase, which would be expected to bring the viral genome to the capsids prior to cleavage and encapsidation.
Structural studies have provided information about other portal rings. For instance, the three-dimensional structure of the Φ29 portal (connector, gp10) was determined by X-ray crystallography (24) and shown to consist of a cone-shaped dodecamer with a central channel lined by long α-helices and hydrophobic residues on the exterior. The Φ29 connector is associated with five to six copies of a 174-nucleotide prohead RNA (pRNA) which forms a ring structure at the portal vertex (8). Packaging of the DNA is believed to occur by the concerted action of the connector and the pRNA complex, causing a rotary motion that pumps the DNA into the procapsid (24). The terminase enzyme facilitates this process by hydrolyzing ATP and acting as a translocase. This model has subsequently been supported by several lines of evidence and serves as a paradigm for the packaging of genomic DNA into capsids of bacteriophages and herpesviruses (6, 26).
In HSV-1, the assembly of portal-containing capsid shells has been shown to require the capsid shell proteins and the UL26.5 scaffolding protein. Interaction between scaffold and portal proteins was demonstrated using an in vitro capsid assembly reaction with purified shell proteins and the UL6 protein, and it has been suggested that the scaffold acts to present the portal ring to the growing shell during assembly (18). The amino acids in the scaffold protein required for its interaction with the portal were recently identified by Singer et al. (25). UL6 may also interact with the major capsid protein, as suggested by the genetic analysis of a temperature-sensitive mutant, tsF18, which is defective in portal incorporation into capsids. A second site suppressor of tsF18 has been isolated which maps to the VP5 gene (Burch and Weller, unpublished data). The interaction of the portal protein with itself during ring formation and with the scaffold and major capsid proteins is likely to require dedicated domains within the UL6 gene. In the present study, we have engineered a set of deletion mutants within the UL6 protein in an attempt to map these domains. Three in-frame deletions (D-5, D-6, and D-LZ) were introduced into the viral genome and were found to be defective in cleavage and packaging. In cells infected with one of these mutants, D-5, the UL6 protein was localized to the cytoplasm, thereby preventing its incorporation into the capsids. The portal proteins of the D-6 and D-LZ mutants were present within the nuclear replication compartments and also were associated with B capsids in infected cells. Using purified proteins, we showed that the D-6 mutant produced rings in vitro, while the D-LZ mutant, in which a putative leucine zipper domain was deleted, was defective in the assembly of the portal ring. Site-directed mutagenesis of two of the conserved heptad leucines within the leucine zipper resulted in viral progeny unable to cleave and package the genome. Furthermore, the mutant protein when expressed in insect cells did not yield stable portal rings. Based on these results, we have proposed that the predicted leucine zipper region within the UL6 protein may play an essential role in portal assembly.
MATERIALS AND METHODS
Viruses, cells, antibodies, and other reagents.
The KOS strain of HSV-1 was used as the WT virus and as the parental strain for the generation of recombinant viruses. The UL6 null virus, hr74, and the UL15 null virus, hr81-1, contain insertions of the Escherichia coli lacZ gene under the control of the HSV-1 ICP6 promoter and were described previously (13, 32). African green monkey kidney cells (Vero) obtained from the ATCC were used to propagate the WT virus. A UL6-complementing cell line, the 31 cell line (13), and a UL15-complementing cell line, C2 (32), were used to propagate the respective mutant viruses. Polyclonal antibodies against the UL6 protein (anti-UL6PC) were custom made (HRP Inc., Denver, PA) against purified, glutathione S-transferase-tagged protein (13), and the anti-UL15 antibody (AS9) was a gift from Dan Tenney (Bristol-Myers Squibb, Wallingford, CT). The monoclonal antibody 1C9, directed against the UL6 ring, was a gift from Jay Brown, University of Virginia, Charlottesville. Anti-VP5 antibody (HSV-ICP5) was purchased from East Coast Bio, ME. All other reagents were of analytical grade.
Generation of UL6 deletion clones.
The coding sequence corresponding to the UL6 gene in frame with an N-terminal epitope tag (AU1) was cloned into the pcDNA1 plasmid vector. This plasmid, pAU1UL6, under the control of the cytomegalovirus promoter, was shown to express the full-length UL6 protein and complement the hr74 null virus in transient transfection assays; pAU1UL6 was used as the parental plasmid to engineer all the other mutations. A series of in-frame deletion mutants was constructed by deleting fragments of the UL6 gene using suitable restriction enzyme sites. The D-5 construct was made by deleting the region of the UL6 gene between an MscI site at nucleotide (nt) 593 and an FspI site at nt 884 within the UL6 open reading frame. D-6 contains a deletion of the DNA between two HpaI sites at nt 964 and 1243, respectively. The D-LZ plasmid was constructed with a deletion between the NarI site (1206) and an AgeI site (1429). All the recombinant plasmids were verified by restriction digestion analysis; the D-5, D-6, and D-LZ clones were further confirmed by DNA sequencing. A large EcoRI-BamHI fragment of HSV-1 strain KOS (nt 12592 to 21653 of the genomic sequence) was cloned into pUC119 to generate pBam15 with homologous sequences on either side of the UL6 gene to promote recombination during marker transfer. The D-5, D-6, and D-LZ deletions were then introduced into this plasmid by replacing a unique BsrGI/BsmI fragment within the UL6 gene with the corresponding fragment from the pcDNA clones to generate pBam15D-5, pBam15D-6, and pBam15D-LZ, respectively.
Site-directed mutagenesis.
The following oligonucleotide primers were synthesized for introducing two point mutations (underlined) within the UL6 leucine zipper region. Leucine codons 429 and 436 were replaced with glutamic acid codons (underlined): UL6_F1, 5′-CG GGA TCC ATG ACC GCA CCA CGC TCG CGG C3′; UL6_LZR, 5′-G CCG GCG CTC GAT GGT TCC AAA CCG GTT ATT TAT ATA GCC CTC-3′; UL6_LZF, 5′-C CGG TTT GGA ACC ATC GAG CGC CGG CGC GAG ACC AAC GCG G-3′; UL6_R1, 5′-G GAA TTC TCA TCG TCG GCC GTC GCG GCG GCC ATC-3′.
HSV-1 genomic DNA was used as the template in a two-step PCR-based mutagenesis protocol (9). The PCR product was gel purified and cloned into the pcDNA vector to generate the pcD6-L429E/L436E plasmid. The WT UL6 gene was also cloned in this vector to generate pCD6FL.
Marker transfer.
Monolayers of the 31 cell line (70% confluent) were transfected with plasmid DNA (1 μg) and hr74 virus DNA (1 μg) using the Lipofectamine reagent (Invitrogen Corporation), incubated in Dulbecco's modified Eagle medium (DMEM) containing 5% fetal bovine serum for 24 h, and harvested along with the culture supernatant. Viral stocks were titered on the 31 cell line and replated at a suitable dilution to yield approximately 100 plaques on a 10-cm dish. The dishes were overlaid with 2% methyl cellulose in DMEM, and recombinant plaques were identified by overlaying with methyl cellulose solution containing 5-bromo-4-chloro-3-indolyl-â-d-galactopyranoside (50 μg/ml) overnight. The parental hr74 plaques stain blue, while the recombinants that contain the mutant UL6 gene and hence have lost the lacZ gene are colorless. The colorless plaques were transferred individually to microcentrifuge tubes, resuspended in 100 μl of DMEM, freeze-thawed to release the virions, and plated again on indicator plates. This process was repeated three times to yield a pure clone for each mutant. Stocks were made from each clone, and the presence of the desired mutation was confirmed by Southern blot analysis.
Virus yield and plaque assays.
Virus yields were determined by infecting Vero cells at a multiplicity of infection (MOI) of 0.1 and collecting samples at various times after infection. Serial dilutions were plated on the 31 cell line, overlaid with methyl cellulose, incubated for 72 h, fixed in 4% formaldehyde, and stained with 1% crystal violet solution. Viral plaques were counted. All experiments were performed in triplicate and the mean of each sample plotted.
Transient expression and complementation assays.
Vero cells (70% confluent in a 35-mm dish) were transfected with plasmid DNA (1 μg) using the Lipofectamine reagent (Invitrogen Corporation), and cells were incubated overnight at 37°C in 5% CO2. One set of dishes was collected for Western blotting experiments and another set was superinfected with hr74 virus at an MOI of 1 PFU/cell. Twenty-four hours later, the cells were harvested along with the culture supernatant, and the amount of progeny virus was determined by titration on the 31 cell line. Mock-infected cells and transfection with the pcDNA1 vector alone were used as controls. The results were expressed as the ratio of the number of plaques obtained from each mutant plasmid divided by the plaque number for the WT plasmid, expressed as a percentage.
Sucrose density gradient purification of the capsids.
Vero cell monolayers (five 225-cm2 flasks) were infected with either WT or mutant virus at an MOI of 5 PFU/cell. Eighteen hours postinfection, the cells were harvested using a cell scraper, transferred into a centrifuge tube, pooled, and centrifuged at 2,000 rpm for 10 min. The supernatant was discarded, and the pellet was washed once with 10 ml of phosphate-buffered saline (PBS) and centrifuged again at 2,000 rpm for 10 min. The final pellet was resuspended in 10 ml of lysis buffer (20 mM Tris, 0.5 M NaCl, 1 mM EDTA, pH 7.4, 1% Triton X-100, plus one tablet of the protease inhibitor [Roche]), frozen, and thawed three times to release the capsids. This lysate was centrifuged at 10,000 × g for 30 min to remove the cell debris; capsids in the supernatant were separated by sedimentation through a cushion of 35% sucrose in TNE (20 mM Tris, 0.5 M NaCl, 1 mM EDTA, pH 7.4) in an SW41 ultracentrifuge tube at 100,000 × g for 1 h. The capsid pellet was resuspended in 0.5 ml TNE plus 1 mM dithiothreitol, loaded on a 20%-to-50% sucrose gradient in TNE, and centrifuged at 100,000 × g for 1 h. The gradients were fractionated (0.3 ml each) using a piston gradient fraction collector (BioComp Instruments Inc., New Brunswick, Canada), and the samples were processed for Western blotting experiments.
Western blot analysis.
Proteins were precipitated from the sucrose gradient fractions with 10% trichloroacetic acid, washed with ethanol, resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer, boiled for 3 min, and separated by 9% PAGE. The gel was blotted onto a nitrocellulose membrane and probed with antibodies against the UL6, UL15, or VP5 protein. Monoclonal IC9 (anti-UL6) was used at a dilution of 1:10,000, polyclonal AS9 (anti-UL15) at a 1:2,000 dilution, and the anti-VP5 monoclonal at a 1:2,000 dilution. Proteins were detected using secondary antibodies conjugated to either horseradish peroxidase or alkaline phosphatase and their appropriate substrates as described in the manufacturer's protocol.
Southern blot analysis.
Genomic DNA samples (1 μg) from infected cells or from virions were digested with restriction enzymes (BamHI for SQ termini and XhoI for analysis of recombinants), resolved on an 0.8% agarose gel, transferred to a nitrocellulose membrane, UV cross-linked, and hybridized with radiolabeled probes at 65°C. The blots were washed successively with 2×, 1×, and 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at 65°C, exposed to X-ray film overnight, and developed.
Recombinant baculoviruses and protein expression.
The WT, D-6, D-LZ, and L429E L436E gene constructs were subcloned into the pFastBac shuttle vector (Invitrogen) and transformed into DH10bac competent E. coli. Recombinant bacmids (bacterial artificial chromosomes) were isolated, and the presence of the mutation was confirmed by PCR sequencing of amplified DNAs. Plasmid DNAs isolated from the positive clones were then transfected into Sf9 insect cells (Novagen) to generate the baculovirus stocks. The recombinant proteins were expressed in Sf9 insect cells and purified according to the method of Newcomb et al. (18) with minor modifications. Insect cells were infected with recombinant baculoviruses at an MOI of 10 PFU/cell and incubated at 28°C for 48 h. The cells were harvested by centrifugation at 1,000 × g for 10 min at 4°C, and the pellet was washed once with PBS and spun again to collect the final cell pellet. The cell pellet (∼108 cells) was resuspended in 5 ml of PBS containing one tablet of the protease inhibitor cocktail (Roche), frozen and thawed three times to lyse the cells, and centrifuged at 10,000 × g for 10 min at 4°C. The supernatant was discarded, and the pellet containing the inclusion bodies was resuspended in 5 ml of TNE containing 2% Triton X-100 and 10 mM dithiothreitol plus one protease inhibitor tablet (Roche), kept on ice for 30 min to solubilize the membranes, and centrifuged again at 10,000 × g for 10 min, at 4°C. The supernatant was discarded, and the pellet containing the UL6 protein was resuspended in 2.5 ml of TNE plus 50 μl of 1.0 M MgCl2 with DNase being added to a final concentration of 0.5 mg/ml and incubated at room temperature for 5 min. The solution was then centrifuged for 10 min at 10,000 rpm, 4°C, and the supernatant discarded. The pellet was resuspended in 0.5 ml of arginine (1.0 M, pH 7.5), kept on ice for 15 min with occasional mixing, and centrifuged at 30,000 × g in a tabletop ultracentrifuge, 4°C, for 15 min. The supernatant which contained the solubilized UL6 protein was further purified by density gradient centrifugation.
Purification of UL6 rings and EM analysis.
The solubilized UL6 protein (0.25 ml) was loaded onto the top of a 10-to-30% sucrose gradient containing 1.0 M arginine (pH 7.4) in an SW 55Ti ultracentrifuge tube and spun at 45,000 rpm for 18 h at 4°C. Gradient fractions (250 μl each) were collected using a piston-type fraction collector (BioComp Instruments) and analyzed by SDS-PAGE and Western blotting. The rings band in the middle of the gradient and can be visualized by a light source. The fractions corresponding to the protein peaks were analyzed by electron microscopy (EM). For EM analysis, the samples were adsorbed onto formvar-carbon-coated grids, negatively stained with uranyl acetate, and visualized in a Philips CM10 transmission electron microscope operating at 60 kV (×52 magnification).
Immunofluorescence microscopy.
Vero cells attached to coverslips (70% confluent) were infected with WT or mutant viruses at an MOI of 5 PFU/cell; at 6 h postinfection, the coverslips were washed with PBS and cells were fixed with 4% paraformaldehyde, washed again with PBS, permeabilized with 0.5% Triton X-100, and finally washed with PBS to remove any residual detergent. The coverslips were incubated in a 5% dilution of normal goat serum in PBS as a blocking agent and treated with a 1:200 dilution of the respective antibody for 1 h. The coverslips were washed with PBS and incubated with secondary antibodies conjugated to Alexa Fluor 488 (green fluorescence) or Alexa Fluor 594 (red fluorescence) for 30 min. They were then washed and mounted on glass slides using glycerol-gelatin containing 1,4-diazabicyclo(2,2,2)octane to retard photo bleaching. The slides were examined at 60 × 10 magnification using a Carl Zeiss Axiovert A410 confocal microscope and images collected by scanning at two wavelengths (channels). The images were exported to Photoshop 6.0 to generate the final figures.
RESULTS
Deletions within the UL6 gene abrogate portal function.
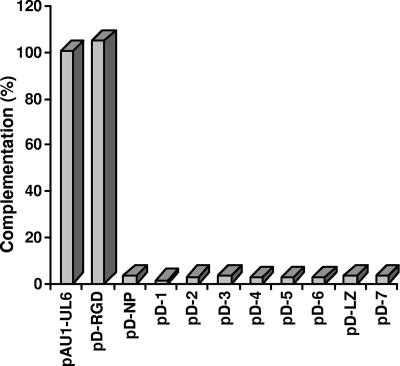
The UL6 protein takes part in many aspects of viral assembly and encapsidation, including ring formation, incorporation into the growing procapsid shell, binding of the terminase, and acting as a conduit for the packaging of viral DNA into capsids. In order to map functional domains of UL6 involved in its many activities, a series of deletions spanning the UL6 gene (Fig. 1) was generated and cloned into the pcDNA1 vector backbone containing a cytomegalovirus promoter and an N-terminal AU1 epitope tag. Vero cells were transfected with WT and mutant plasmids, and protein expression was analyzed by Western blot analysis. All the plasmids produced proteins of the expected sizes (data not shown), indicating that most of the deletions do not cause global protein instability. A transient complementation assay was performed to determine the ability of the deletion constructs to complement the growth of a UL6 null mutant. Figure 2 shows that with the exception of the RGD construct, none of the deletion mutants was able to complement the UL6 null virus, hr74. The ability of the RGD deletion plasmid to complement hr74 suggests that this region is not essential for portal function. These data suggest, however, that the UL6 gene in general is very intolerant to mutations.
FIG. 1.
UL6 gene deletion mutants generated for this study. Schematic representation of the UL6 gene showing the predicted leucine zipper region (amino acids 422 to 445) and the various deletion mutants engineered for this study. The numbers correspond to the positions of the amino acids within the UL6 protein; -RGD, -NP, and -LZ denote deletions of a putative RGD motif, a predicted nuclear pore binding region, and a leucine zipper motif, respectively.
FIG. 2.
Transient complementation assay. Vero cells were transfected with plasmid DNAs of deletion mutants and superinfected with hr74 virus, and the resulting progeny virus was titered on 31 cells. The percent complementation was calculated by dividing the titer obtained for the mutant plasmid with the titer of the WT plasmid and multiplying by 100.
Mutant UL6 viruses produce only B capsids and are deficient in DNA cleavage.
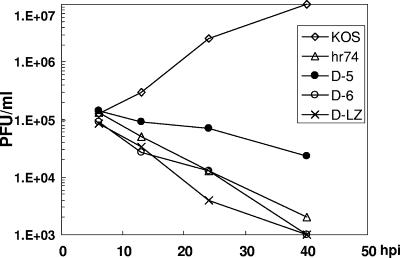
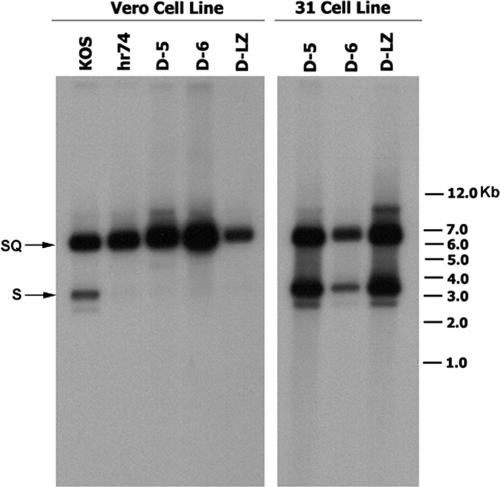
Three of these mutations, D-5, D-6, and D-LZ, were introduced into the viral genome by marker transfer, and recombinant viruses were plaque purified on a complementing cell line (31 cell line). Initially, we chose to study these three deletions for two reasons: (i) they affect conserved helices and a coiled-coil domain within UL6, and (ii) they span the middle portion of the UL6 gene and hence are unlikely to disrupt the cis-acting elements of the flanking UL5 and UL7 genes. The presence of the respective mutations within D-5, D-6, and D-LZ virus was confirmed by Southern blot analysis using 32P-labeled UL6 gene fragment as a probe (data not shown). All three mutant viruses formed plaques in a UL6-complementing cell line but not in Vero cells, supporting the conclusion that the growth defects were due to the mutations introduced into the UL6 gene. The growth of these mutant viruses on Vero cells was monitored using a viral yield assay. All three mutants exhibited severe defects in their ability to produce infectious virus (Fig. 3). Furthermore, all three viruses produced only B capsids when infected-cell lysates were subjected to sucrose gradient analysis, indicating a defect in DNA packaging (data not shown). Southern blot analysis of DNA samples from the virus-infected cells indicated that all three mutants synthesized viral DNA. The inability to cleave the replicated DNA in these mutants was confirmed by Southern hybridization using a 32P-labeled DNA fragment which detects the 6-kb internal junction repeat sequence (BamHI SQ fragment) and a 3.5-kb band corresponding to the left end of the genomic terminus (BamHI S fragment). In the BamHI-digested DNA from KOS-infected Vero cells, this probe detected both the junction fragment (SQ) and the expected terminal fragment (S) (Fig. 4, left panel). DNA from cells infected with the null mutant and the three deletion mutants generated only the junction fragment. The absence of the S terminus indicates that all three mutant viruses are defective in DNA cleavage. When the DNA from these mutants grown on the complementing cell line was analyzed, a band corresponding to the S terminus was detected (Fig. 4, right panel), indicating normal cleavage and packaging.
FIG. 3.
Mutant viruses show reduced virus yields. Graph showing the growth curves of UL6 WT and mutant viruses obtained following infection of Vero cells with the WT (KOS), the UL6 null (hr74), or one of the three mutant (D-5, D-6, or D-LZ) viruses harvested at different time points (h) after infection (hpi).
FIG. 4.
UL6 mutant viruses are defective in DNA cleavage. Southern blot of DNA from cells infected with WT (KOS), hr74 (null), or UL6 mutant virus (D-5, D-6, or D-LZ) probed with a terminal “b” repeat sequence. The probe was derived from the pNAR plasmid by digestion with NcoI and DraI, and it contains the “b” repeat sequences that detect the S termini and the internal SQ junction (16). The left panel shows DNA obtained from infected Vero cells, and the right panel shows DNA samples from the infected 31 cell line. The positions of the molecular weight markers are indicated on the right.
D-6 and D-LZ mutant UL6 proteins are associated with B capsids.
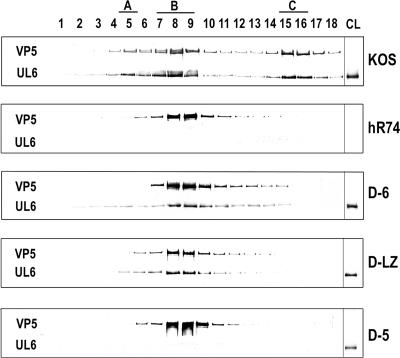
The inability to package DNA could be due to a failure to form the portal ring, its failure to be incorporated into the capsids, or the loss of association of the mutant protein with other components of the packaging machinery, for example, the putative terminase complex UL15/28. In order to distinguish between these possibilities, the capsids made by the D-5, D-6, and D-LZ viruses were fractionated using sucrose density gradient centrifugation and analyzed by Western blotting. The virus obtained from KOS (WT)-infected cell extracts resolved into three bands, corresponding to the A, B, and C capsids, while the mutants generated only a single band, corresponding to the B capsids. The identities of these bands were confirmed by probing Western blots of the fractions with antiscaffold antibodies which detect only the B capsids (data not shown). The capsid fractions were subjected to SDS-PAGE, transferred to a polyvinylidene difluoride membrane, and probed with monoclonal anti-UL6 and anti-VP5 antibodies. Capsid preparations from cells infected with the UL6 null virus (hr74) served as a control. A Western blot analysis of the capsid fractions is shown in Fig. 5. The VP5 antibody detected a 150-kDa band peaking at positions corresponding to A, B, and C capsids from cells infected with WT virus, while in the mutant virus-infected cell extracts, only the B-capsid peak is detected. Probing these blots with the UL6 antibody showed that the D-6 and D-LZ mutant proteins were present in B capsids (Fig. 5, panels D-6 and D-LZ), while the D-5 protein was not detectable in capsid fractions (Fig. 5, panel D-5). As expected from previous studies, the WT UL6 protein (KOS panel) was present in all three capsid types. We also probed the blot with antibodies to detect UL15, a subunit of the viral terminase, and observed that UL15 was present in capsids from WT, D-6, and D-LZ viruses (data not shown). These results indicate that the domains deleted in both D-6 and D-LZ mutants are essential for neither capsid nor terminase association.
FIG. 5.
The D-6 and D-LZ mutant proteins are associated with capsids. Western blot of fractions (1, top; 18, bottom) obtained by sucrose density gradient sedimentation of capsids from cell lysates of Vero cells infected with KOS (WT), hr74 (null), or the D-6, D-LZ, or D-5 mutant. Each blot was divided into two strips and probed separately with anti-UL6 and anti-VP5 antibodies. A, B, and C represent the positions of the three types of capsids in the sucrose gradient. The last lane, CL, contained a sample of the infected-cell lysate, used as a control to monitor the expression of the mutant proteins.
The D-5 mutant protein does not localize to the nucleus.
The absence of the D-5 mutant protein in the capsids was not due to a lack of protein expression, since the mutant protein was detected in infected-cell lysates (Fig. 5, panel D-5, lane CL), albeit at slightly lower levels than WT UL6. The inability to detect the D-5 protein in capsids raised the possibility that the protein may be defective in portal ring formation or capsid binding. Alternatively, the incorporation defect could be due to improper folding or abnormal localization of the mutant protein, making it unavailable for incorporation into the packing machinery.
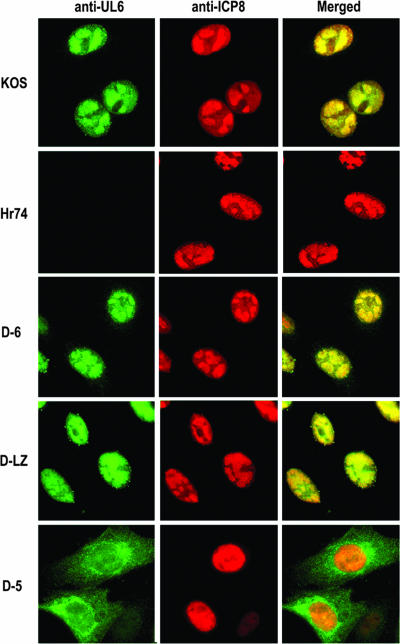
Since assembly and encapsidation are thought to occur in large globular domains in the nucleus called replication compartments, it is expected that in order for UL6 to be incorporated into growing capsids, it must also localize to replication compartments. The full-length UL6 protein has been shown to localize to the replication compartments in cells infected with WT virus (20). Vero cells infected with the WT or each of the three mutant viruses were immunostained with anti-UL6 and anti-ICP8, which is used as a marker for replication compartments. Figure 6 shows the dual immunofluorescence images collected in the green (UL6) and red (ICP8) channels and their merged signals. As expected from previous studies, the WT UL6 and ICP8 were both detected in replication compartments. In cells infected with hr74, replication compartments can be observed, but no UL6 staining could be detected. In cells infected with D-6 and D-LZ viruses, the mutant proteins colocalized with ICP8 in replication compartments. The D-5 protein, on the other hand, was detected predominantly in the cytoplasm of the infected cells (Fig. 6, bottom panels), while ICP8 staining was observed in the nucleus. Thus, the D-5 protein is defective for nuclear transport, which may indicate either that the nuclear localization signal (NLS) has been deleted or that the protein is misfolded. In either case, this result explains the absence of the UL6 protein in capsid fractions isolated from the D-5 mutant virus.
FIG. 6.
The D-5 protein localizes to the cytoplasm of infected cells. Immunofluorescence images of Vero cells infected with KOS, hr74 (null), or D-6, D-LZ, or D-5 mutant UL6 viruses double labeled with anti-UL6 and ant-ICP8 antibodies and detected by Alexa Fluor 488- and 594-conjugated secondary antibodies, respectively. The panels in each row show an image from the green channel (UL6) and the red channel (ICP8) and the merged image. The yellow color denotes colocalization of UL6 with ICP8 in the replication compartments.
The UL6 gene contains a putative leucine zipper.
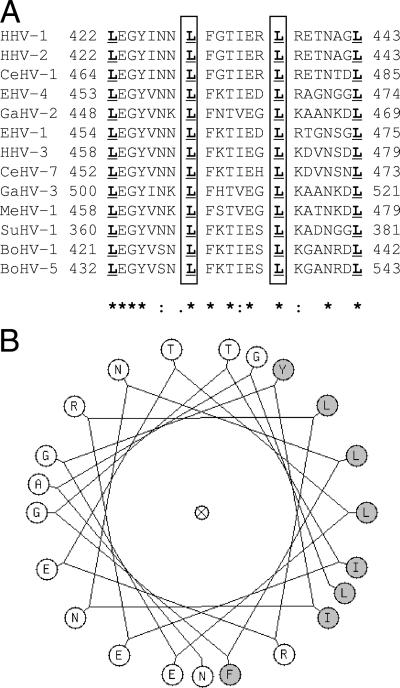
Sequence analysis of the UL6 protein and comparison with those of other herpesviruses predicts that the region spanning amino acids 422 to 443 has a high potential to form a leucine zipper (LZ domain). The heptad leucines within this region are conserved in all of the alphaherpesviruses shown in Fig. 7A. The COILS 2 program (15) predicts a strong likelihood of a leucine zipper in this region. It can be seen from the helical representation of the UL6-LZ domain that the invariant leucines line up on one side of the helix, which is predicted to have a high propensity to form a coiled-coil configuration (Fig. 7B). Thus, the LZ domain may represent a structurally conserved motif in the portal proteins of alphaherpesviruses. Leucine zipper motifs are generally involved in protein-protein and/or protein-DNA interactions. They have been shown to mediate intersubunit interactions in several multimeric proteins, including those that form ring structures (7, 23). The region deleted in the D-LZ mutant contains the putative leucine zipper, which we propose is necessary for stable portal formation.
FIG. 7.
A putative leucine zipper domain is present within the UL6 gene. Multiple sequence alignment of the predicted leucine zipper regions of the following alphaherpesvirus portal proteins: human herpesvirus 1 (HHV-1) gp011, human herpesvirus 2 (HHV-2) gp08, simian herpesvirus 1 (CeHV-1) gp07, equine herpesvirus 4 (EHV-4) gp55, chicken herpesvirus 2 (GaHV-2) gp018, equine herpesvirus 1 (EHV-1) orf56, human herpesvirus 3 (HHV-3) gp55, simian herpesvirus 7 (CeHV-7) gp55, chicken herpesvirus 3 (GaHV-3) gp019, meleagrid herpesvirus 1(MeHV-1) gp013, pseudorabies virus (SuHV-1) gp52, bovine herpesvirus 1 (BoHV-1) gp30, and bovine herpesvirus 5 (BoHV-5) gp51. The numbering on the left denotes the amino acid position of the first leucine of the zipper motif. The conserved heptad leucines are underlined. The stars in the bottom row represent identical residues, and the dots denote amino acid similarities at the respective positions. The boxed residues of the HSV-1 sequence were changed to glutamic acid to generate the L429E L436E mutant. The sequences were obtained from the NCBI viral genome database, accession numbers NC_001806, NC_001798, NC_004812, NC_001844, NC_002229, NC_001491, NC_001348, NC_002686, NC_002577, NC_002641, NC_006151, NC_001847, and NC_005261, respectively. Panel B shows a helical wheel diagram of the HSV1-LZ motif generated using the GCG Wisconsin package (31). The hydrophobic side chains, most of which face one side of the helix, are shaded gray.
Mutations within the putative leucine zipper abrogate portal function.
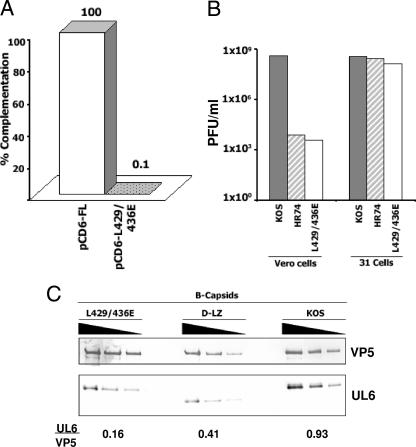
In order to test the significance of the predicted leucine zipper in UL6, we introduced two point mutations within this region, resulting in the substitution of glutamic acid residues for L429 and L436. The replacement of the heptad leucines with charged residues is expected to disrupt the coiled-coil interactions, thereby preventing zipper formation. These two leucine residues are positioned in the center of the predicted coiled-coil region and hence are a better target for disruption of the zipper domain. A two-step PCR-based strategy using mutagenic primer sets was used to amplify the UL6 gene and clone it into the pcDNA vector (pcD6-L429E/L436E). This construct, when transfected into Vero cells, expressed the mutant protein as determined by Western blot analysis of the cell extracts and localized to the nucleus as observed by using immunofluorescence microscopy (data not shown). In transient complementation experiments, this mutant failed to support growth of the UL6 null virus (Fig. 8 A), suggesting the importance of the two heptad leucines in UL6. We then introduced this mutation into the HSV-1 genome by marker transfer and generated a recombinant virus (UL6-L429E/L436E). This recombinant virus does not grow on Vero cells (Fig. 8B) and produces only B capsids (data not shown). With the complementing cell line (31 cell line), the L429E L436E mutant grew to levels similar to those of the WT (KOS) (Fig. 8B). Western blot analysis of B capsids isolated from cells infected with the UL6-L429E/L436E mutant virus showed that UL6 was present in the B capsids. It appears that the amount of UL6 associated with capsids is smaller for the mutant than for the WT (Fig. 8C). In order to determine the relative amounts of the UL6 protein in these capsids, the intensities of the UL6 and VP5 signals were quantified using NIH Image J software and expressed as a ratio (UL6/VP5). It should be noted that this ratio does not compare the actual amounts of the two proteins because of differences in the affinities of the VP5 and UL6 antibodies used; however, it can be used to determine relative differences in the amounts of UL6 associated with capsids from WT and mutant viruses. The capsids from D-LZ and L429E L436E mutant viruses showed lower levels of UL6 (0.16 and 0.41, respectively) than the WT (KOS) capsids (0.93), suggesting that the stoichiometry of portal incorporation into capsids is also affected in these mutants. The WT UL6 protein forms a dodecameric ring at a unique capsid vertex, which accounts for a defined stoichiometry of UL6 to VP5 observed in HSV-1 capsids (2). Thus, the smaller amounts of UL6 present in the leucine zipper mutants may indicate a defect in normal ring formation. These results indicate that the phenotype of the point mutant is similar to that of the leucine zipper deletion mutant and that the conserved leucines 429 and 436 within the zipper region are important for the production of mature C capsids.
FIG. 8.
The L429E L436E mutant virus does not grow on Vero cells and produces B capsids containing reduced amounts of the UL6 protein. (A) The WT (pCD6-FL) and the mutant (pCD6-L429E/L436E) plasmids were transfected into Vero cells, which were incubated for 18 h and then superinfected with the null virus (hr74), and the yield of resulting progeny viruses was determined by plating on the complementing cell line. The graph shows the percent complementation relative to results for the WT, normalized to 100%. (B) The Vero and 31 cell lines were infected with KOS (WT), hr74 (null), or the L429E L436E mutant (leucine zipper mutant) virus at an MOI of 0.5 PFU/cell; 24 h postinfection, progeny viruses were collected and assayed for yield on the complementing cell line. (C) B-capsid fractions from the sucrose density gradient-purified L429E L436E mutant (leucine zipper point mutant), D-LZ (leucine zipper deletion), and KOS (WT) viruses were serially diluted (1:1, 1:2, and 1:4) and analyzed by Western blotting using anti-VP5 and anti-UL6 antibodies. The images were quantified using Image J software, and results were expressed as a ratio of the relative intensities of the UL6 and VP5 protein bands.
The predicted leucine zipper region of the UL6 protein may be involved in the assembly of stable portal rings.
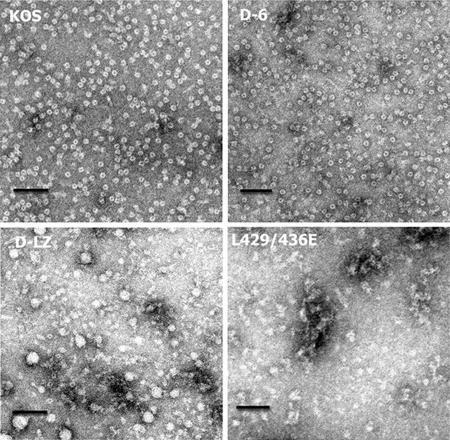
The purified UL6 protein expressed in insect cells forms ring structures in vitro, similar to the portal proteins of bacteriophages. We next asked whether the mutant UL6 proteins could form rings in vitro. WT and D-6, D-LZ, and L429E L436E mutant proteins were expressed in insect cells using the baculovirus expression system. The recombinant proteins were purified from inclusion bodies using 1 M arginine for solubilization and sucrose density gradient centrifugation. The purity of the fractions was determined to be >95% by SDS-PAGE and Coomassie staining. Aliquots from these fractions were adsorbed onto EM grids, stained with uranyl acetate, and examined by transmission EM. As expected, the WT UL6 protein formed ring structures in vitro (Fig. 9). The dimensions of these structures were similar to those observed previously (17). Interestingly, the D-6 mutant protein also formed ring structures, indicating that the amino acid residues deleted in this mutant were not essential for portal ring assembly. The D-LZ and L429E L436E mutants, on the other hand, did not form ring structures; instead the mutant proteins appeared to form polymorphic aggregates (Fig. 9). This may indicate that the amino acid residues within the leucine zipper domain are involved in portal ring formation.
FIG. 9.
The D-LZ mutant protein does not form rings in vitro. Electron micrographs show purified preparations of the WT and three mutant (D-6 and D-LZ deletions and the site-specific L429E L436E mutant) UL6 proteins made in insect cells using a recombinant baculovirus expression system. The photographs show negatively stained rings with dark central channels in the WT and the D-6 mutant. The D-LZ and L429 L436E mutant proteins show amorphous particles of variable dimensions and without a distinct central channel. The scale bar in each panel corresponds to 100 nm.
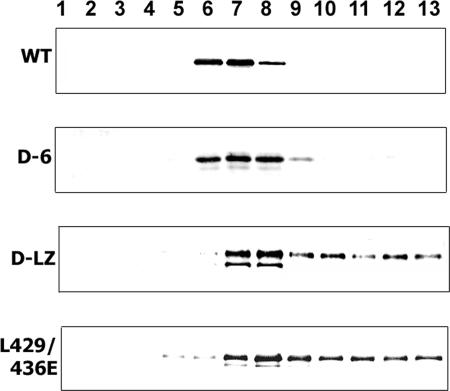
The oligomeric states of the purified WT and mutant proteins were analyzed using density gradient centrifugation. UL6 proteins purified from insect cells were subjected to sucrose density gradient (10% to 30%) centrifugation. The gradients were fractionated and analyzed by SDS-PAGE and Western blotting using anti-UL6 antibodies (Fig. 10). The WT protein migrated as a sharp peak (fractions 6 to 8), consistent with the formation of a homogeneous multimer. The D-6 protein exhibited a similarly sharp peak in fractions 6 to 8, indicating that it forms a homogeneous protein assembly. On the other hand, the D-LZ and L429E L436E mutant proteins were distributed throughout fractions 7 to 13 of the gradient, consistent with the formation of heterogeneous aggregates. The lower-molecular-weight bands seen in the D-LZ mutant could result from a small amount of degradation of the mutant protein.
FIG. 10.
Sedimentation profile of purified WT, D-6, D-LZ, and L429 436E recombinant UL6 proteins. Western blot image of the fractions (1, top; 13, bottom) from sucrose gradient centrifugation of the WT, D-6, D-LZ, and L429E L436E recombinant UL6 proteins probed with anti-UL6 antibody. The WT protein forms a sharp peak in the middle of the gradient, while the D-6 mutant migrates in a similar mode; the D-LZ and L429E L436E mutant proteins are distributed in a range of sizes throughout the gradient.
These results suggest that the leucine-rich region of UL6 may be specifically required for ring formation. Alternatively, the LZ domain mutants may produce misfolded proteins. We think this is unlikely in view of the results presented above demonstrating that the D-LZ protein can localize appropriately to the nucleus within replication compartments and can be incorporated into capsids; thus, the LZ deletion does not appear to lead to gross conformational changes. Based on the results presented here, we suggest that the putative leucine zipper domain within the UL6 protein plays a role in the formation and/or stability of the portal rings.
DISCUSSION
Encapsidation of the newly replicated genome is an important step in the maturation and assembly of all viruses. The fundamental aspects of this macromolecular assembly process have been conserved in organisms ranging from bacteriophages to viruses infecting mammalian cells. Assembly involves interactions between the preformed capsids, the viral nucleic acid, and the packaging machinery. The packaging machinery is believed to generate the required forces and direct the genome encapsidation. The processes of genome maturation and cleavage and packaging have been studied extensively with model systems including bacteriophage lambda (4), Φ29 (6, 28), and P22 (27), which serve as prototypes for the study of the complex molecular mechanisms in viruses which infect eukaryotes.
By analogy with the better-studied bacteriophages, the HSV-1 portal protein has an important role in the encapsidation process, serving both as a channel for DNA insertion and as a docking site for the other components of the packing machinery (17, 21, 33). The experiments described in this paper were designed to map regions of the UL6 protein that mediate its interaction within the ring and with the other cleavage and packaging proteins. Nine of the ten deletion mutants failed to complement the null virus despite detectable levels of protein expression. The portal protein is involved in interactions with multiple partners during cleavage and packaging of the HSV-1 genome, and the deletions may thus affect one or more of these interaction domains, rendering the UL6 gene relatively intolerant to mutations. The high degree of sequence similarity seen in the portal proteins from different herpesviruses is also consistent with this interpretation. We cannot rule out, however, that some deletions may have caused conformational changes in the UL6 protein. The complementing RGD mutant contains a seven-amino-acid deletion eliminating a putative integrin binding motif, indicating that this motif is not essential for UL6 function. A second RGD sequence (238 to 240) is also present within the UL6 protein; this motif is located within the sequences deleted in the D-5 mutant. In the phage Φ29 terminal protein, the RGD motif is required for its interaction with the viral DNA polymerase (11). It is possible that the second RGD motif in UL6 (deleted in the D-5 mutant) is involved in protein-protein interactions.
Three deletion mutants, D-5, D-6, and D-LZ, and a double point mutant, the L429E L436E mutant, were introduced into the viral genome for phenotypic analysis, and all four were shown to produce only immature B capsids. This is consistent with the phenotype of the previously described UL6 null mutant and of mutants in other components of the cleavage/packaging machinery. Most mutations isolated to date in the UL6, UL15, and UL28 genes show this “all or none” phenotype. It has been hypothesized that DNA packaging and capsid maturation are coupled processes, hence the difficulty in separating these events.
The D-5 protein was localized to the cytoplasm of infected cells and was not incorporated into capsids; furthermore, it could not be expressed stably in insect cells from recombinant baculoviruses. Interestingly, Preston and colleagues reported a UL6 insertion mutant (UL6 in269) in which the UL6 protein failed to localize to the nucleus; moreover, this mutant UL6 protein also appeared to retain both UL15 and UL28 in the cytoplasm (30). The UL6 amino acid sequence does not reveal any of the canonical nuclear localization sequence motifs; however, since the deletion of amino acids 198 to 295 in the D-5 mutant and an insertion at residue 269 prevented nuclear import, it is tempting to speculate that this region may contain an unrecognized NLS or domain of interaction with another NLS-containing protein. The ability of the UL6 in269 mutant to retain UL15 and UL28 in the cytoplasm suggests that the region of UL6 required for interaction with UL15 and UL28 does not reside in the region disrupted by this insertion. Taken together, these results suggest that the D-5 mutant may lack an NLS, but we cannot rule out that this protein is mislocalized due to a folding defect which may also contribute to its instability. Whether the D-5 protein can still interact with the terminase is not clear because of difficulties expressing this protein in insect cells.
The deletion mutants D-6 and D-LZ and the L429E L436E double point mutant are able to enter the nucleus, localize to the replication compartments, and are incorporated into B capsids. UL15 can still associate specifically with B capsids formed in cells infected with these mutants (data not shown); however, these mutants exhibit a severe defect in cleavage and packaging. The D-6 protein, however, was found to be competent to form oligomeric ring structures in vitro. This result suggests that this mutant is deficient in DNA packaging, perhaps due to changes in the properties of the channel which controls the entry and/or exit of DNA into or out of the capsid (12). Within the deleted region of D-6, sequence analysis of the UL6 protein predicts a potential alpha-helix and a sequence similar to the DNA binding region of integrases. The DNA channels of portal proteins for which atomic structures are known contain alpha-helices and are known to interact with the DNA. The D-6 mutant could be defective in these interactions, which may explain its inability to package DNA. It will be necessary to introduce point mutations within this region and test them for DNA interactions to address this possibility. Another possible reason for the defective DNA packaging of this mutant may be due to the loss of interaction with other components of the packaging machinery. The UL15 protein has been shown to be transiently associated with procapsids (22) and with B capsids (21, 33). The observation that UL15 does not specifically associate with capsids isolated from cells infected with the UL6 null mutant virus implies an interaction between UL15 and UL6 (33). Using immunoprecipitation assays, a direct interaction between UL6 and UL15 and UL28 was also observed in recombinant baculovirus-infected insect cells (30). However, we did not see any difference in levels of UL15 in the capsids isolated from the D-6 mutant virus, indicating that terminase association is not affected in this mutant (data not shown). Thus, this mutant may be reminiscent of DNA channel mutants described for the Salmonella phage SPP1 portal (12).
The results described in this article indicate that the leucine zipper in UL6 is required for assembly of portal-containing HSV-1 capsids which are competent for encapsidation. There is precedent for the use of leucine zippers in proteins that form rings. For example, the leucine zipper in phospholamban (a protein which regulates the levels of intracellular calcium) appears to play an important role in stabilizing the pentamers of this protein to form a coiled-coiled pore (23). Based on our finding that the D-LZ and L429E L436E mutants fail to form rings in vitro, we suggest that this region is important for the formation and/or stability of the HSV-1 portal ring. Interestingly, despite the inability of the recombinant proteins to form rings in vitro, the D-LZ and L429E L436E mutant proteins were associated with B capsids from infected cells. The association with capsids may indicate that the interaction domain between mutant UL6 and capsid proteins is not affected. The amount of UL6 present in mutant capsids was less than that observed in WT capsids, suggesting that the D-LZ and L429E L436E mutant proteins are not efficiently incorporated into capsids during assembly. It is possible that the leucine zipper mutant proteins form polymorphic aggregates that can initiate assembly, resulting in capsids with a lower ratio of UL6 to VP5. Aggregate formation might be expected if additional protein-protein interactions contribute to the formation of rings. The forces which stabilize UL6 rings are not known but could include electrostatic or hydrophobic interactions, disulfide linkages, and/or other structural motifs. In fact, we have preliminary data showing that intersubunit disulfide bonds involving two cysteines of the UL6 protein are also important in portal ring formation (J. Nellissery and S. Weller, unpublished data). Intersubunit disulfide bond formation may result in the formation of aggregates which still associate with capsids but are defective for encapsidation. This aggregation may explain the sedimentation behavior of the D-LZ and L429E L436E mutant proteins and the fact that monomers are not observed.
Recently the three-dimensional structure of the bacteriophage SPP1 portal protein was determined at 3.4 Å (14). Each subunit consists of a crown, wing, stem, and clip domains. Twelve to thirteen of these monomers are arranged in a ring around a central channel, similar to the portals of other bacteriophages and herpesviruses. The channel is lined by loops which protrude inward and are positioned so as to contact the major groove of the DNA; this model suggests that the movement of these loops is coupled to DNA translocation. The portal proteins of bacteriophages and herpesviruses show little sequence similarity, but their secondary structures display significant conservation of helices, sheets, and loop regions. When we aligned the predicted secondary structure of the UL6 protein to that of the SPP1 portal protein, we observed that our deletion mutants mapped to the wing (D-5), stem and channel (D-6), and major helix α6 (D-LZ and L429E L436E) domains. The D-6 mutant is particularly interesting since it lacks the channel loops that are thought to play a dynamic role during DNA packaging. The leucine zipper mutants (D-LZ and L429E L436E mutants) map to the conserved helix α6; the leucine zipper which is present within this region in alphaherpesviruses may be important in interactions that affect the stability of the portal ring.
In summary, we have generated four HSV-1 portal protein mutants that are defective in cleavage and packaging of the genome and have determined the localization of their gene products within infected cells and within viral capsids. We have also shown that the WT and one mutant (D-6) protein formed oligomeric rings in vitro, while the two other mutants (D-LZ and L429E L436E mutants), in which the putative leucine zipper is disrupted, did not form stable rings. This is the first report defining a region within the UL6 protein that is essential for the formation of stable portal rings.
Acknowledgments
We thank Art Hand and John Aghajanian of the Central Electron Microscopy facility at the University of Connecticut Health Center for analysis of the UL6 protein rings. We especially thank Ping Bai for help with protein purification. The UL6-specific monoclonal antibodies used in this study were obtained from Jay Brown, University of Virginia. We also thank the members of the Weller lab for helpful discussions and suggestions throughout the course of this study.
This work was supported by a grant from the NIH, AI37549, to S. K. Weller.
Footnotes
Published ahead of print on 20 June 2007.
REFERENCES
- 1.Baines, J. D., and S. K. Weller. 2005. Cleavage and packaging of herpes simplex virus 1 DNA, p. 135-145. In C. E. Catalano (ed.), Viral genome packaging machines: genetics, structure, and mechanism. Kluwer Academic/Plenum, New York, NY.
- 2.Beard, P. M., C. Duffy, and J. D. Baines. 2004. Quantification of the DNA cleavage and packaging proteins U(L)15 and U(L)28 in A and B capsids of herpes simplex virus type 1. J. Virol. 78:1367-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cardone, G., D. C. Winkler, B. L. Trus, N. Cheng, J. E. Heuser, W. W. Newcomb, J. C. Brown, and A. C. Steven. 2007. Visualization of the herpes simplex virus portal in situ by cryo-electron tomography. Virology 361:426-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Catalano, C. E., D. Cue, and M. Feiss. 1995. Virus DNA packaging: the strategy used by phage lambda. Mol. Microbiol. 16:1075-1086. [DOI] [PubMed] [Google Scholar]
- 5.Chang, J. T., M. F. Schmid, F. J. Rixon, and W. Chiu. 2007. Electron cryotomography reveals the portal in the herpesvirus capsid. J. Virol. 81:2065-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chemla, Y. R., K. Aathavan, J. Michaelis, S. Grimes, P. J. Jardine, D. L. Anderson, and C. Bustamante. 2005. Mechanism of force generation of a viral DNA packaging motor. Cell 122:683-692. [DOI] [PubMed] [Google Scholar]
- 7.Cornillez-Ty, C. T., and D. W. Lazinski. 2003. Determination of the multimerization state of the hepatitis delta virus antigens in vivo. J. Virol. 77:10314-10326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo, P., C. Zhang, C. Chen, K. Garver, and M. Trottier. 1998. Inter-RNA interaction of phage phi29 pRNA to form a hexameric complex for viral DNA transportation. Mol. Cell 2:149-155. [DOI] [PubMed] [Google Scholar]
- 9.Higuchi, R., B. Krummel, and R. K. Saiki. 1988. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 16:7351-7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Homa, F. L., and J. C. Brown. 1997. Capsid assembly and DNA packaging in herpes simplex virus. Rev. Med. Virol. 7:107-122. [DOI] [PubMed] [Google Scholar]
- 11.Illana, B., A. Zaballos, L. Blanco, and M. Salas. 1998. The RGD sequence in phage phi29 terminal protein is required for interaction with phi29 DNA polymerase. Virology 248:12-19. [DOI] [PubMed] [Google Scholar]
- 12.Isidro, A., A. O. Henriques, and P. Tavares. 2004. The portal protein plays essential roles at different steps of the SPP1 DNA packaging process. Virology 322:253-263. [DOI] [PubMed] [Google Scholar]
- 13.Lamberti, C., and S. K. Weller. 1996. The herpes simplex virus type 1 UL6 protein is essential for cleavage and packaging but not for genomic inversion. Virology 226:403-407. [DOI] [PubMed] [Google Scholar]
- 14.Lebedev, A. A., M. H. Krause, A. L. Isidro, A. A. Vagin, E. V. Orlova, J. Turner, E. J. Dodson, P. Tavares, and A. A. Antson. 2007. Structural framework for DNA translocation via the viral portal protein. EMBO J. 26:1984-1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lupas, A. 1996. Prediction and analysis of coiled-coil structures. Methods Enzymol. 266:513-525. [DOI] [PubMed] [Google Scholar]
- 16.Martinez, R., R. T. Sarisky, P. C. Weber, and S. K. Weller. 1996. Herpes simplex virus type 1 alkaline nuclease is required for efficient processing of viral DNA replication intermediates. J. Virol. 70:2075-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newcomb, W. W., R. M. Juhas, D. R. Thomsen, F. L. Homa, A. D. Burch, S. K. Weller, and J. C. Brown. 2001. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J. Virol. 75:10923-10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newcomb, W. W., D. R. Thomsen, F. L. Homa, and J. C. Brown. 2003. Assembly of the herpes simplex virus capsid: identification of soluble scaffold-portal complexes and their role in formation of portal-containing capsids. J. Virol. 77:9862-9871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patel, A. H., and J. B. MacLean. 1995. The product of the UL6 gene of herpes simplex virus type 1 is associated with virus capsids. Virology 206:465-478. [DOI] [PubMed] [Google Scholar]
- 20.Patel, A. H., F. J. Rixon, C. Cunningham, and A. J. Davison. 1996. Isolation and characterization of herpes simplex virus type 1 mutants defective in the UL6 gene. Virology 217:111-123. [DOI] [PubMed] [Google Scholar]
- 21.Salmon, B., and J. D. Baines. 1998. Herpes simplex virus DNA cleavage and packaging: association of multiple forms of UL15-encoded proteins with B capsids requires at least the UL6, UL17, and UL28 genes. J. Virol. 72:3045-3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheaffer, A. K., W. W. Newcomb, M. Gao, D. Yu, S. K. Weller, J. C. Brown, and D. J. Tenney. 2001. Herpes simplex virus DNA cleavage and packaging proteins associate with the procapsid prior to its maturation. J. Virol. 75:687-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simmerman, H. K., Y. M. Kobayashi, J. M. Autry, and L. R. Jones. 1996. A leucine zipper stabilizes the pentameric membrane domain of phospholamban and forms a coiled-coil pore structure. J. Biol. Chem. 271:5941-5946. [DOI] [PubMed] [Google Scholar]
- 24.Simpson, A. A., Y. Tao, P. G. Leiman, M. O. Badasso, Y. He, P. J. Jardine, N. H. Olson, M. C. Morais, S. Grimes, D. L. Anderson, T. S. Baker, and M. G. Rossmann. 2000. Structure of the bacteriophage phi29 DNA packaging motor. Nature 408:745-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singer, G. P., W. W. Newcomb, D. R. Thomsen, F. L. Homa, and J. C. Brown. 2005. Identification of a region in the herpes simplex virus scaffolding protein required for interaction with the portal. J. Virol. 79:132-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith, D. E., S. J. Tans, S. B. Smith, S. Grimes, D. L. Anderson, and C. Bustamante. 2001. The bacteriophage straight phi29 portal motor can package DNA against a large internal force. Nature 413:748-752. [DOI] [PubMed] [Google Scholar]
- 27.Strobel, E., W. Behnisch, and H. Schmieger. 1984. In vitro packaging of mature phage DNA by Salmonella phage P22. Virology 133:158-165. [DOI] [PubMed] [Google Scholar]
- 28.Tao, Y., N. H. Olson, W. Xu, D. L. Anderson, M. G. Rossmann, and T. S. Baker. 1998. Assembly of a tailed bacterial virus and its genome release studied in three dimensions. Cell 95:431-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trus, B. L., N. Cheng, W. W. Newcomb, F. L. Homa, J. C. Brown, and A. C. Steven. 2004. Structure and polymorphism of the UL6 portal protein of herpes simplex virus type 1. J. Virol. 78:12668-12671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White, C. A., N. D. Stow, A. H. Patel, M. Hughes, and V. G. Preston. 2003. Herpes simplex virus type 1 portal protein UL6 interacts with the putative terminase subunits UL15 and UL28. J. Virol. 77:6351-6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Womble, D. D. 2000. GCG: The Wisconsin Package of sequence analysis programs. Methods Mol. Biol. 132:3-22. [DOI] [PubMed] [Google Scholar]
- 32.Yu, D., A. K. Sheaffer, D. J. Tenney, and S. K. Weller. 1997. Characterization of ICP6::lacZ insertion mutants of the UL15 gene of herpes simplex virus type 1 reveals the translation of two proteins. J. Virol. 71:2656-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu, D., and S. K. Weller. 1998. Herpes simplex virus type 1 cleavage and packaging proteins UL15 and UL28 are associated with B but not C capsids during packaging. J. Virol. 72:7428-7439. [DOI] [PMC free article] [PubMed] [Google Scholar]