Abstract
Herpesviruses require membrane-associated glycoproteins gB, gH, and gL for entry into host cells. Epstein-Barr virus (EBV) gp42 is a unique protein also required for viral entry into B cells. Key interactions between EBV gp42 and the EBV gH/gL complex were investigated to further elucidate their roles in membrane fusion. Deletion and point mutants within the N-terminal region of gp42 revealed residues important for gH/gL binding and membrane fusion. Many five-residue deletion mutants in the N-terminal region of gp42 that exhibit reduced membrane fusion activity retain binding with gH/gL but map out two functional stretches between residues 36 and 96. Synthetic peptides derived from the gp42 N-terminal region were studied in in vitro binding experiments with purified gH/gL and in cell-cell fusion assays. A peptide spanning gp42 residues 36 to 81 (peptide 36-81) binds gH/gL with nanomolar affinity, comparable to full-length gp42. Peptide 36-81 efficiently inhibits epithelial cell membrane fusion and competes with soluble gp42 to inhibit B-cell fusion. Additionally, this peptide at low nanomolar concentrations inhibits epithelial cell infection by intact virus. Shorter gp42 peptides spanning the two functional regions identified by deletion mutagenesis had little or no binding to soluble gH/gL and were also unable to inhibit epithelial cell fusion, nor could they complement gp42 deletion mutants in B-cell fusion. These studies identify key residues of gp42 that are essential for gH/gL binding and membrane fusion activation, providing a nanomolar inhibitor of EBV-mediated membrane fusion.
Epstein-Barr virus (EBV) is a member of the human herpesviruses, all of which are membrane-enveloped viruses that utilize multiple glycoproteins to enter cells (8, 43). More than 90% of the adult population is seropositive for EBV, which efficiently infects both epithelial and B cells, the latter providing the latency reservoir (2, 40). Acute EBV infection acquired in adolescence or adulthood can cause infectious mononucleosis, accompanied by a proliferation of B cells (15). In addition, both cell types can develop tumors that are directly linked to EBV, such as nasopharyngeal carcinoma and Burkitt's lymphoma (4, 16, 18, 50, 56). Other disorders also connected to EBV include posttransplant lymphoproliferative disorder as well as oral hairy leukoplakia and B-cell lymphomas of the central nervous system prevalent in AIDS and immunosuppressed individuals (17, 53). The medical relevance of EBV makes it an important subject of study, especially understanding the mechanism by which the virus enters its two target cell types.
The minimum requirement for membrane fusion to occur with epithelial cells is the coexpression of EBV envelope glycoproteins gH, gL, and gB. EBV additionally requires the gp42 protein for entry into B cells (11, 27, 28). EBV gp42 binds major histocompatibility complex class II (MHC-II) proteins expressed on B cells to trigger viral-cell membrane fusion. One requirement for this interaction is the presence of a glutamic acid at the MHC beta-chain residue 46, which is present in all HLA-DR and HLA-DP but only some HLA-DQ alleles (10, 12, 19, 26, 29, 32, 45, 46, 54). A soluble gp42-Fc chimeric protein can stimulate the entry of a gp42-null virus into B cells, and, likewise, the addition of baculovirus-produced, soluble gp42 to cells transfected with gH, gL, and gB allows membrane fusion to occur with B cells (23, 55). However, membrane fusion with epithelial cells is actually inhibited by the presence of gp42 for both virus infection and a cell-cell fusion assay (23, 55). Increasing levels of inhibition occur as exogenous soluble gp42 is added in a cell-cell fusion assay, beginning in the low-nanomolar-concentration range (23). This is likely due to the formation of three-part gH/gL/gp42 complexes that are unable to mediate membrane fusion with epithelial cells, possibly due to steric hindrance of gH/gL receptor binding or direct insertion of gH/gL into the target membrane (3, 19). These studies are consistent with the proposal that levels of gp42 in the virus dictate EBV tropism in vivo (55).
Initial studies suggested that gL might bind gp42 in the heterotrimeric complex, but more recent data implicate gH as a primary determinant of the interaction (27, 36, 55, 59). The N-terminal region of gp42 was demonstrated to be important for interactions with gH/gL, initially using a truncated soluble gp42-Fc chimeric protein lacking the first 58 gp42 residues. This mutant maintained class II binding, and although it did not detectably coprecipitate gH/gL, it did partially inhibit epithelial cell infection, which was attributed to a reduced ability to interact with gH/gL (55). Further studies of purified, soluble proteins revealed that gp42 stably binds gH/gL with 1:1:1 stoichiometry, with residues 33 through 85 being critical for this complex formation. A 30-residue gp42-derived peptide spanning residues 36 to 65 was sufficient to inhibit membrane fusion with epithelial cells with micromolar affinity, partially mimicking the ability of gp42 to bind gH/gL (23). This present study further elucidates the interaction between gH/gL and the N-terminal region of gp42 critical for binding and fusion.
Nearly all herpesviruses contain a gH/gL complex, which serves an indispensable role in membrane fusion and infection (13, 30, 35, 43, 52, 55, 58). EBV gH requires the presence of gL in order for EBV gH to fold properly and be transported to the cell membrane, and both EBV gL and the related varicella-zoster virus gL proteins function effectively in mediating the folding and expression of EBV gH protein (25, 27, 60). It has also been established that EBV gH/gL exists as a noncovalently associated heterodimer complex in a 1:1 subunit ratio (23). Three lines of evidence suggest that EBV infection of epithelial cells requires the interaction of gH/gL with a specific receptor. First, EBV virions lacking gH are unable to attach to epithelial cells. Second, soluble gH/gL has been shown to bind to epithelial cells permissive in membrane fusion and infection. Third, viruses using gH/gL for both adhesion and fusion are compromised in infection. Despite all the data, this putative epithelial cell receptor remains elusive (3, 31, 33, 55).
EBV and other herpesviruses use a multicomponent system for membrane fusion, which is different from other known viral fusion systems that utilize a single fusion protein (21). It remains unclear as to how these proteins interact to accomplish their entry function, although crystal structures of the EBV gp42-HLA-DR1 MHC-II complex and the herpes simplex virus type 1 (HSV-1) functional homolog gD bound to the herpesvirus entry mediator receptor have been determined (5, 32). The unliganded HSV-1 gD crystals revealed a flexible hairpin loop, the conformation of which is clearly altered after receptor binding, something which might occur similarly in gp42 after MHC-II engagement (24, 41, 44). HSV-1 gD has been shown to form complexes sequentially with both gH and gB (7, 22, 37, 49). Subramanian and Geraghty developed an exquisite assay to detect hemifusion as well as complete fusion and demonstrated the sufficiency of gD with gH/gL for hemifusion. Interestingly, gB was not required for hemifusion but was necessary for complete fusion, providing an elegant model of sequential glycoprotein involvement in HSV-1 membrane fusion. The HSV-1 gB structure has also recently been determined, revealing characteristics resembling both class I and class II viral fusion proteins, such as residues positioned as possible fusion loops and coiled-coil helices in a trimeric form (14, 39). Nevertheless, the requirement for gH/gL in herpesviruses suggests that the membrane fusion mechanism may be different from the known single-component systems in which trimeric fusion proteins assemble into hairpin-like conformations that bring the viral and cellular membranes together (39, 47). Since the presence or absence of EBV gp42 appears to act as a switch for virus entry into B cells or epithelial cells, respectively, its interaction with gH/gL is a fundamental feature in EBV-mediated membrane fusion (2).
In order to better understand the interactions between EBV gH/gL and gp42 in the virus entry mechanism, we studied the gp42 residues involved in gH/gL binding. We examined the effects of point mutations and deletions in the gp42 N-terminal region on membrane fusion and interaction with gH/gL. Peptides derived from gp42 spanning the N-terminal region were observed to directly bind to gH/gL, inhibit epithelial cell fusion, and compete for gp42 binding in B-cell fusion. A synthetic 46-mer peptide from gp42 could bind gH/gL with similar nanomolar affinity as the intact gp42 protein, proving a potent antagonist of EBV-mediated membrane fusion.
MATERIALS AND METHODS
Cells and proteins.
High Five insect cells (BTI-TN-5B1-4; Invitrogen) were grown in shaker flasks in Excell-405 medium (JRH Biosciences). Sf9 insect cells (Invitrogen) were grown in shaker flasks in Sf900 medium (Gibco).
All media for mammalian cell growth were made complete by supplementing with 10% FetalPlex animal serum complex (Gemini Bio-Products) and 1% penicillin-streptomycin (BioWhittaker), and all mammalian cells were grown in 75-cm2 cell culture flasks (Corning). Mammalian epithelial cells were human embryonic kidney 293 cells that express simian virus 40 large T antigen (293T; ATCC, Manassas, VA) and modified to stably express T7 RNA polymerase under selection of 100 μg/ml zeocin in complete Dulbecco's modified Eagle medium (BioWhittaker), which is known as line 14, as described previously (35). Mammalian B cells were Daudi B lymphocytes that are EBV positive and express HLA class II and CD21 (ATCC) and that are modified to stably express T7 RNA polymerase under selection of G418 (700 to 900 μg/ml) in complete RPMI 1640 medium (BioWhittaker) (41). Chinese hamster ovary cells (CHO-K1) were kindly provided by Nanette Susmarski and were grown in complete Ham's F-12 medium (BioWhittaker). Versene (1 mM EDTA in phosphate-buffered saline [PBS]) or trypsin-versene (BioWhittaker) was used to detach adherent cells.
Monoclonal antibodies E1D1 (anti-gH/gL) and F-2-1 (anti-gp42) were gifts generously provided by L. Hutt-Fletcher (Louisiana State University Health Sciences Center, Shreveport, LA) (1, 48). Monoclonal antibody 3H3 (anti-gp42) was obtained as previously described (23). Polyclonal anti-gp42 antibody serum PB1114 was used as previously described (29, 41). Milligram quantities of antibodies were produced by the Northwestern Monoclonal Antibody Facility and the National Cell Culture Center. For cell enzyme-linked immunosorbent assay (CELISA) experiments with soluble gH/gL, monoclonal antibodies 3H3 and E1D1 were purified by a HiTrap protein G agarose column (Amersham Biosciences), eluted with 0.2 M glycine-HCl, pH 2.5, immediately neutralized to pH ∼7 by adding 1/10 volume of 1 M Tris, pH 9.0. Purity was >95% as demonstrated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis. Purified antibodies were exchanged into PBS, and concentration was determined by absorbance at 280 nm using a theoretical extinction coefficient (0.1%) of 1.4 for immunoglobulin G (IgG).
Soluble EBV gp42 and soluble EBV gH/gL were produced and purified as described elsewhere (23). Briefly, gp42 and gH/gL proteins were obtained using the baculovirus expression system in High Five and Sf9 insect cells, respectively. Soluble gp42 was purified by cobalt metal affinity chromatography, and soluble gH/gL was purified by affinity purification using monoclonal anti-gH/gL antibody E1D1. Gel filtration using a Superdex-200 HR 10/30 analytical column (Amersham Biosciences) provided the final purification in running buffers consisting of PBS or 25 mM Tris (pH 7.4)-150 mM NaCl (TBS).
Peptides.
Synthetic peptides were obtained commercially (EZBiolab) at 95% purity, as determined by high-performance liquid chromatography and mass spectrometry, and used without further purification. Lyophilized peptides were reconstituted in TBS or PBS. Peptide concentration was measured by absorbance at 280 nm using the theoretical 0.1% value. For peptide labeled with fluorescein isothiocyanate (FITC), the absorbance maximum at 495 nm for FITC was additionally used to confirm peptide concentration. Peptide 36-81 is N-terminally labeled with FITC and corresponds to gp42 residues 36 to 81 (RVAAAAITWVPKPNVEVWPVDPPPPVNFNKTAEQEYGDKEVKLPHW). Peptide 36-65 corresponds to gp42 residues 36 to 65 (RVAAAAITWVPKPNVEVWPVDPPPPVNFNK) (23). Peptide 36-56 corresponds to gp42 residues 36 to 56 (RVAAAAITWVPKPNVEVWPVD). Peptide 42-56 corresponds to gp42 residues 42 to 56 (ITWVPKPNVEVWPVD). Peptide 67-81 corresponds to gp42 residues 67 to 81 (AEQEYGDKEVKLPHW).
Mutants.
Mutants were generated using a double-arm PCR approach. Mutant primers were generated to incorporate either five-residue deletions or single-residue changes as well as silent restriction enzyme cutting sites. The first PCR used primers matched to flank the EBV gp42 gene in the pCAGGS.mcs vector (5′ primer p-1, TCCTGGGCAACGTGCTGGTTGTTG; 3′ primer p-2, GCCAGAAGTCAGATGCTCAAGGGG) paired with their complementary directional mutant primer on a wild-type EBV gp42 plasmid template. The PCR Tails program was used as follows: (i) 94°C for 2 min, (ii) 94°C for 15 s, (iii) 58°C for 1 min, (iv) 72°C for 90 s (40 cycles of steps ii to iv), (v) 72°C for 15 min, and (vi) 4°C for an unlimited period. PCR products were confirmed by gel electrophoresis and then used as templates with the flanking primers in the second PCR to generate full-length mutant gp42, which was confirmed by gel electrophoresis, cut with EcoR1 and BglII, and ligated overnight at 16°C with vector that had been digested under the same conditions. Ligated products were transformed and selected on ampicillin plates. Colonies were picked and grown overnight to generate mini-preparations, which were double digested to confirm the introduction of the mutation. Mini-preparations were sequenced, and positive clones were then grown in large quantities and isolated in cesium chloride density gradients by ultracentrifugation and sequenced again. Double mutants were generated similarly except mutant gp42 plasmids were used for the first PCR template. The first deletion mutant was used as a template with a different pair of mutant primers to generate the second deletion; e.g., d37-41/67-71 (where d37-41 is a deletion mutant lacking gp42 residues 37 to 41) was created using d37-41 plasmid template with d67-71 mutant primers for the first PCR. Two mutants studied had incorporated inconsequential extra point mutations: mutant K47A gained P48S and d67-71 gained T13I. These mutations were detected after sequencing of CsCl preparations, so new mutants were generated and confirmed to lack any incidental mutations introduced by PCR. Table 1 identifies mutants used in this study. Mutant W44A was generated as previously described (41).
TABLE 1.
Activity summary of gp42 deletion mutantsa
| Category and mutantb | Surface expression | Fusion | gH/gL binding
|
|
|---|---|---|---|---|
| Immunoprecipitation | CELISA | |||
| Category 1 | ||||
| R30A | + | + | Y | ND |
| R32A | + | + | Y | ND |
| R36A | + | + | Y | ND |
| W44A | + | + | Y | ND |
| K47A | + | + | Y | ND |
| K47A/P48S | + | + | Y | ND |
| E51A | + | + | Y | ND |
| d32-36 | + | + | Y | + |
| d57-61c | + | + | Y | − |
| d62-66 | + | + | Y | + |
| Category 2 | ||||
| NT1 | ||||
| d37-41 | + | − | Y | + |
| d42-46 | + | − | Y | + |
| NT2 | ||||
| d82-86 | + | − | Y | + |
| d87-91 | + | − | Y | + |
| d92-96 | + | − | Y | + |
| Category 3 | ||||
| NT1 | ||||
| d47-51 | + | − | Y | +/− |
| d52-56 | + | − | Y | − |
| NT2 | ||||
| d67-71 | + | +/− | Y | − |
| d72-76 | + | − | Y | − |
| d77-81 | + | − | Y | +/− |
| NT1 and NT2 | ||||
| d45-89 | + | − | ND | − |
| d37-41/d67-71 | + | − | Y | − |
| d42-46/d72-76 | + | − | N | − |
| d47-51/d77-81 | + | − | N | ND |
| d52-56/d77-81 | + | − | ND | ND |
| d77-81/d47-51 | + | − | ND | − |
Activity is defined as 100% wild-type to 50% (+), between 50% and 25% (+/−), or less than 25% (−). ND; not done; Y, observed by immunoprecipitation with Western blotting; N, greatly reduced or not observed by immunoprecipitation with Western blotting.
Category 1, functional; category 2, fusion impaired, not due to gH/gL binding; category 3, fusion impaired due to altered gH/gL binding.
Mutant d57-61 exhibits nearly wild-type fusion but reduced interactions with soluble gH/gL.
Transfection.
CHO-K1 cells were transfected in Opti-MEM I medium (Gibco) by a uniform protocol using Lipofectamine 2000 (Invitrogen) as previously described (41). Cells were plated in a six-well format, and after 24 h each well received 5 μl of Lipofectamine 2000, and various combinations of pCAGGS expression vector containing the gene of interest were used in the following amounts: 0.5 μg for gH, 0.5 μg for gL, 0.8 μg for gB, 0.8 μg for luciferase, and 1.7 μg for the pCAGGS vector control (11, 34). In the case of gp42-transfected CHO cells, pCAGGS vector control plasmid was replaced by 1.7 μg of gp42 vector. For fusion assays (see Fig. 1), amounts of DNA were changed to 0.5 μg for gB, 2.0 μg of wild-type or mutant gp42, and 1.0 μg each of pCAGGS and enhanced green fluorescent protein (GFP) to allow for negative control and visual confirmation of transfection. For immunoprecipitations, gB and luciferase plasmids were excluded, and 1.0 μg of gH and gL was transfected with 2.0 μg of wild-type or mutant gp42. For negative control and visualization, 2.0 μg each of pCAGGS and enhanced GFP was included with no other glycoprotein.
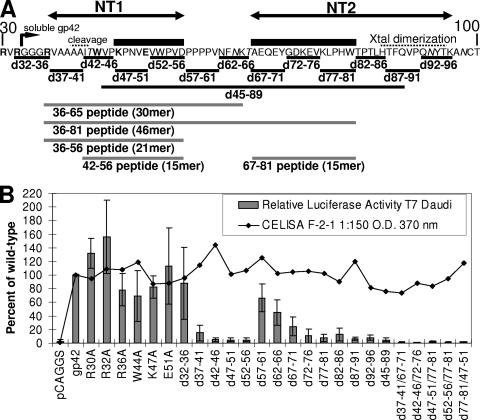
FIG. 1.
Schematic of gp42 mutants, fusion assay, and CELISA data. (A) Map of EBV gp42 amino-terminal ectodomain residues 30 to 100 depicting mutant constructs of either five-residue and longer deletions or single-residue substitutions. Individual residues that were mutated to alanine are shown in bold. Italics highlight predicted glycosylation sites. Putative cleavage and crystal (Xtal) dimerization sites are indicated by dotted lines from above. Soluble gp42 starts at residue 33 and is indicated by an angled arrow. Important functional domains designated NT1 and NT2 are represented by double arrows, and peptides derived from gp42 are shown below. The core binding residues based on concordance between activity in fusion and binding soluble gH/gL in CELISA are below black boxes. Gray lines depict the peptides used in this study. (B) Relative luciferase activity measured in a cell-cell fusion assay using CHO cells transfected with gH, gL, gB, and wild-type or mutant gp42 and overlaid with B cells (bar graph). Surface expression of gp42 is measured by CELISA using monoclonal F-2-1 antibody, secondary biotinylated-anti-mouse-IgG antibody, tertiary streptavidin-horseradish peroxidase, and TMB substrate (line graph). Color development was measured by absorbance at 370 nm. Data shown are averages of at least three independent experiments with wild-type gp42 luciferase activity and expression set at 100%, and standard deviations are represented by the error bars.
Fusion assay.
CHO-K1 cells were transiently transfected as described above. At 12 h posttransfection, the cells were detached by versene, counted either by hand with a hemocytometer or using a Beckman Coulter Z1 particle counter, and 2.0 × 105 to 2.5 × 105 cells/well in 0.5 ml were transferred to a 24-well format, incubated for about 10 min after the addition of peptide and/or soluble proteins (in TBS or PBS without azide), and subsequently overlaid with 0.5 ml of target cells, either Daudi B cells or 293T epithelial cells. For simultaneous surface expression readings by CELISA, 4.0 × 104 to 5.0 × 104 CHO cells were also plated in 96-well plates (described below). To induce fusion, equal or greater numbers of Daudi or 293T cells were overlaid on CHO cells, and total volume was 1 ml or more per well. Both of the target cell types, Daudi and 293T, used in the fusion assay stably expressed T7 RNA polymerase and were under selection by G418 and zeocin, respectively (35, 41, 57). After a 24-h overlay, cells were washed with PBS and lysed with 100 μl of passive lysis buffer (Promega) per well. Luciferase activity was quantified by transferring triplicate 20-μl aliquots of lysed cells to 96-well opaque plates with clear bottoms (Wallac), and luminescence was measured on a Perkin-Elmer Victor plate reader immediately after adding 100 μl of luciferase assay reagent (Promega).
CELISA.
For testing gp42 mutant binding to soluble gH/gL, 1.7 μg of gp42 (wild type or mutant) DNA was added with 5 μl of Lipofectamine 2000 per well in the six-well format. After 12 h posttransfection, CHO cells were replated in the 96-well format. After a 24-h incubation, the medium was removed, cells were washed with PBS-ABC (PBS with 0.89 g of CaCl2 and 0.89 g of MgCl2·H2O per 8 liters), and soluble gH/gL was added in PBS-ABC with 3% bovine serum albumin (BSA) (Sigma). After a 30-min incubation, cells were washed, and primary antibody at a 50 nM final concentration was added in PBS-ABC with 3% BSA. Cells were fixed for 10 min in PBS with 2% formaldehyde and 0.2% glutaraldehyde and then blocked with 3% BSA in PBS-ABC. Biotinylated anti-mouse-IgG at 1:500 was added as the secondary antibody, followed by streptavidin-horseradish peroxidase at 1:20,000. TMB (3,3′,5,5′-tetramethylbenzidine; Sigma) substrate was added, and quantitative colorimetric measurement was performed at 370 nm using a Victor plate reader, typically achieving a high signal over background measurement by 30 to 60 min. Background measurements were obtained from negative control wells that omitted the transfected glycoprotein expression plasmid and/or omitted primary antibody. Surface expression of transfected gp42 was measured using 50 nM 3H3 antibody, and binding of soluble gH/gL to transfected gp42 was measured using 50 nM E1D1 antibody. Percent soluble gH/gL binding to gp42 was determined by dividing the background-subtracted binding of soluble gH/gL by the background-subtracted surface expression for each gp42 mutant. The wild-type gp42 sample binding to soluble gH/gL was then set to 100%, and gp42 mutants were normalized to that value. For measurement of cell surface expression simultaneous with fusion assays, CHO cells transfected for fusion were also plated in the 96-well format 12 h posttransfection, and CELISA was performed 24 h later. The primary antibody was F-2-1 hybridoma supernatant used at a dilution of 1:150.
Immunoprecipitation.
CHO cells were transfected with plasmids encoding EBV gH, gL, and either wild-type or mutant gp42. Opti-MEM medium was removed 12 h posttransfection and replaced with complete Ham's F-12 medium. After 24 h of incubation, cells were detached with versene, counted, washed twice in PBS, and lysed as previously described (41). Equal volumes of lysates were either added to 5× sample buffer or incubated with no antibody, anti-gH/gL E1D1 monoclonal antibody, or anti-gp42 F-2-1 monoclonal antibody and rotated at 4°C for at least 1 h. Agarose beads cross-linked to protein G (Amersham Biosciences) were incubated with lysates at 4°C for at least 1 h and used to pull down IgG complexes by centrifugation. Samples were washed at least three times with complete lysis buffer, and 100 μl of 2× SDS sample buffer was added. Samples of whole-cell lysate and immunoprecipitate were run on 12.5% or 10% Criterion Tris-HCl gels (Bio-Rad) at 120 V for 110 min, followed by transfer at 90 V for 90 min to Immobilon-P transfer membranes (Millipore). Western blotting immunodetection of gp42 was carried out using anti-gp42 antibody serum PB1114 as previously described (41). All the gp42 mutants with the exception of the d45-89 mutant were readily detected by Western blotting. The absence of the detection of the d45-89 mutant may be a result of the extensive deletion compared to the other mutants.
Fluorescence polarization.
A Beacon 2000 instrument (Invitrogen) was used to measure the fluorescence polarization signal that occurs when FITC-labeled gp42-derived peptide binds to soluble gH/gL. Samples were made in parallel in 100-μl volumes using 10-fold serial dilutions of protein and peptides, with subdivisions in between. To reduce nonspecific interactions and reduce protein losses at low concentrations, samples contained 0.1% Tween-20 detergent. To reduce pipetting errors, parallel samples were mixed in individual tubes (Kimble Glass) and prepared identically using the same volumes for each dilution. Samples were allowed to incubate at room temperature for at least 20 min before measurement. For competition assays, the two competitors were mixed together (FITC-labeled peptide and competitor protein or peptide), and then soluble gH/gL was added last. Data were collected at 25°C.
Cell-free virus infection assay.
GFP-positive EBV was produced by Akata cells (a gift from L. Hutt-Fletcher), as previously described (20, 31, 51). Briefly, Akata cells were grown in RPMI 1640 medium (BioWhittaker) with 10% fetal bovine serum (HyClone), 1% penicillin-streptomycin, and 500 μg/ml neomycin (Gemini Bio-Products). At a high cell concentration, 4 × 107 cells were induced with 50 μg/ml anti-human Ig (MP Biomedicals) for 5 days. After the cells were removed, 100 μg/ml bacitracin (Sigma Aldrich) was added, and virus was pelleted by centrifugation at 16,000 × g at 4°C for 90 min. Virus was resuspended in 1 ml of RPMI medium containing bacitracin and passed through a 0.8-μm-pore-size filter.
Virus aliquots were preincubated for 24 h at 37°C under conditions for testing viral inhibition, during which time human embryonic kidney 293 epithelial cells were plated in the 96-well format nearly at confluence. The infection assay was performed using 50 μl of cell-free virus per well that was centrifuged at 2,500 rpm for 1 h at 33°C. Subsequently, virus was removed by aspiration, cells were washed with 200 μl of RPMI complete medium, and cells were left with 200 μl of fresh RPMI complete medium to incubate for 72 h. Infected cells could be observed producing GFP after 24 h. Fluorescence-activated cell sorting (FACS) with a BD LSR II 488-nm laser was used to quantify GFP-expressing cells at 72 h postinfection. The percentage of infected cells was measured by collecting 30,000 total events and gating on the live cell population.
RESULTS
Mutants of the gp42 N-terminal region.
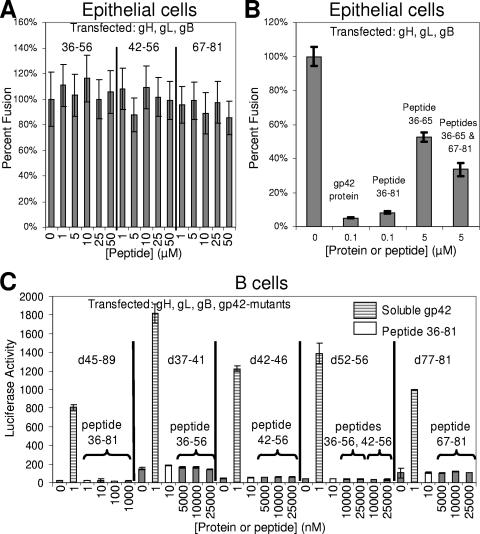
Mutants of gp42 were engineered to span the N-terminal region from residues 30 to 96 (Fig. 1A). Initially, six point mutants were made to substitute alanine for charged residues or tryptophan (Fig. 1A, bold amino acids). Since none of these mutants dramatically altered fusion, a more systematic approach was employed to remove five contiguous residues throughout the N-terminal region from amino acids 32 to 96 (Fig. 1A). Cell surface expression of gp42 mutants was similar to wild-type gp42 in transfected CHO cells as shown by CELISA (Fig. 1B, line graph). The mutants were tested for their ability to mediate membrane fusion using CHO cells transfected with gH, gL, gB, and mutant or wild-type gp42 and overlaid with B cells. Mutants with five-residue deletions between residues 37 to 56 or 72 to 96 showed significantly reduced membrane fusion activity, less than 25% of wild type (Fig. 1B, bar graph). In contrast, deletion of residues 57 to 61 or 62 to 66 showed only moderate reduction in membrane fusion activity to ∼50% of wild-type gp42. Mutant d67-71 averaged between 25% and 50% of wild-type fusion. These data suggest that there are two functional regions within the gp42 N terminus that we designate N-terminal region 1 (NT1) for residues 37 to 56 and N-terminal region 2 (NT2) for residues 67 to 96.
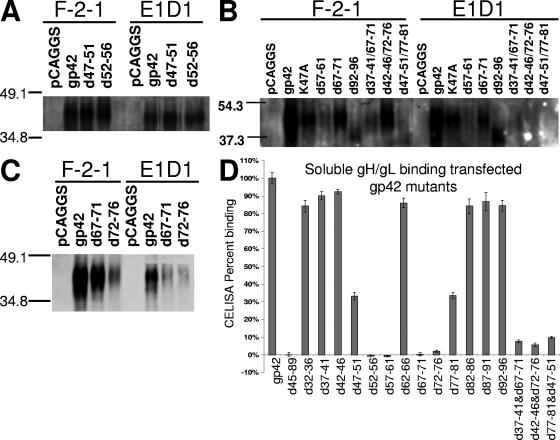
Representative Western blotting of lysates immunoprecipitated with anti-gp42 mouse monoclonal antibody demonstrated that the expression and size of mutant gp42 protein are nearly the same as wild-type gp42, except for d62-66 and d92-96, where two and one, respectively, potential glycosylation sites were removed (Fig. 2B and data not shown). Loss of the glycosylation sites did not affect the ability of the d62-66 mutant to trigger fusion, although the d92-96 mutant was unable to mediate fusion (Fig. 1B). This may indicate that glycosylation, at least at the second site, may be important for gp42 function. This would be in contrast to HSV-1 gD in which glycosylation is not essential for function (42). The observation with the d92-96 mutant may also indicate another role of this region in gp42 function such as dimerization, which was observed in the crystal structure (32). Immunoprecipitation with anti-gH/gL E1D1 antibody followed by Western blotting for gp42 was performed to monitor gH/gL binding with gp42 (Fig. 2A to C). Surprisingly, none of the deletion mutants with the exception of d45-89 was deficient in gH/gL binding as assessed by immunoprecipitation (representative data in Fig. 2A to C and summarized in Table 1). The d45-89 mutant is not readily detected by Western blotting even though surface expression was confirmed by CELISA (Fig. 1B).
FIG. 2.
Binding of gp42 mutants with transfected gH/gL in coimmunoprecipitations and soluble gH/gL in CELISA. (A to C) SDS-polyacrylamide gel electrophoresis and Western blotting of wild-type and mutant gp42 expressed in CHO cells, using rabbit polyclonal PB1114 anti-gp42 antibody at 1:1,000 for detection, reveal gp42 levels in immunoprecipitations using either F-2-1 anti-gp42 antibody or E1D1 anti-gH/gL antibody as labeled. Samples are from the same transfection and were simultaneously run in nonadjacent lanes. (D) CELISA-based quantitative measurements of soluble gH/gL binding to transfected gp42. Detection of bound gH/gL is carried out with E1D1 anti-gH/gL antibody, as in the CELISA described in the legend of Fig. 1B. Data shown are representative of three or more independent experiments, and error bars represent standard deviations across triplicate measurements within an experiment. Data for all mutants are summarized in Table 1.
As an alternative approach to further understand gp42 domains required for interaction with gH/gL, more quantitative analysis was performed using CELISA experiments with 50 nM soluble gH/gL to determine gp42 mutants that retain near wild-type binding, bind at a reduced level, and completely lose binding function (Fig. 2D). Interestingly, these results did not entirely agree with the immunoprecipitation data (Fig. 2 and summarized in Table 1). Whereas all of the single five-residue deletions in either NT1 or NT2 were able to bind to gH/gL in immunoprecipitation, one deletion mutant in NT1 and two deletions in NT2 were completely deficient in binding soluble gH/gL, while one in each category showed approximately half of wild-type binding. These results appeared to more quantitatively characterize gH/gL binding, demonstrating that some mutants at the beginning of NT1 and the end of NT2 were deficient for fusion but were still able to bind soluble gH/gL (Fig. 2D and Table 1). Most surprising was the d57-61 mutant that bound gH/gL in immunoprecipitation but did not appear to bind gH/gL in CELISA. Double deletion gp42 mutants were constructed to completely disrupt gH/gL binding since the immunoprecipitation results with the single deletion mutants suggested that both NT1 and NT2 are important for gp42 binding to gH/gL. Many of the double deletion mutants were similar to d45-89 in that they appeared to express well on the surface of transfected cells, they were unable to mediate membrane fusion (even lower than single deletion mutants), and some were difficult to detect in Western blots (Fig. 1A and 2B). However, d42-46/72-76 was easily detected by Western blotting, and binding to gH/gL was greatly reduced (Fig. 2B). Although d37-41/67-71 was not detected well by Western blotting, gp42 levels in F-2-1 and E1D1 immunoprecipitation were often nearly similar, implying that there was more gH/gL binding than in other double mutants (Fig. 2B and data not shown). However, soluble gH/gL binding to these double mutants showed little to no binding (Fig. 2D).
Peptide from gp42 residues 36 to 81 binds to soluble gH/gL with strong affinity.
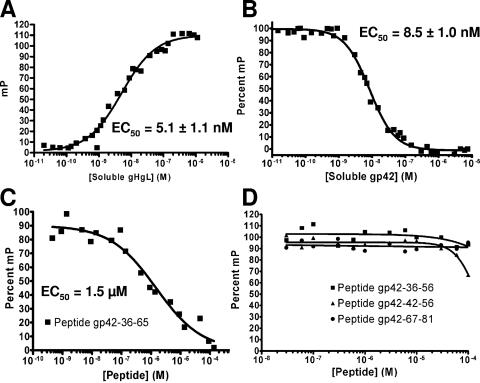
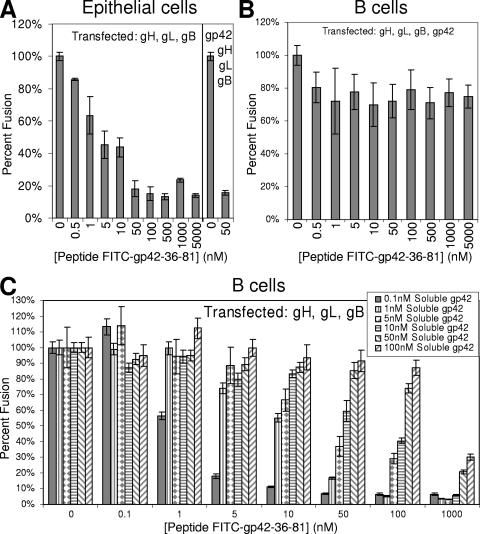
The gp42 mutants investigated here demonstrate that much of the N-terminal region of gp42 is critical for membrane fusion and interactions with gH/gL. We have previously demonstrated that a peptide corresponding to residues 36 to 65 binds to gH/gL with micromolar affinity, but this peptide appears to cover only one of the two N-terminal regions important for gH/gL binding (23). In particular the gH/gL CELISA data indicate that residues between 67 and 81, but not 82 and 96, are important for gH/gL binding (Fig. 2D). An FITC-labeled peptide containing residues 36 to 81 was therefore synthesized to determine in fluorescence polarization experiments the strength of binding to soluble gH/gL. This peptide was found to bind with low nanomolar affinity, with 50% binding calculated to be 5.1 nM by a nonlinear least squares fit (Fig. 3A). Compared to the gp42 peptide consisting of residues 36 to 65, which binds with a low micromolar Kd, the longer 46-mer peptide gains about 500-fold affinity by extending the 30-mer by 16 C-terminal amino acids to residue 81 (23). These binding data are consistent with the gp42 five-residue deletion mutants, demonstrating that mutants lacking residues between 72 and 81 were noticeably impaired in their ability to bind soluble gH/gL (Fig. 2D). Key interaction residues exist in the N-terminal region from residues 36 to 65 (based on activity of the peptide 36-65) and in the extended region from residues 66 and 81. These interaction regions were further investigated by studying shorter peptides.
FIG. 3.
Equilibrium binding of gp42 peptides to soluble gH/gL measured by fluorescence polarization. (A) Binding between soluble gH/gL and FITC-labeled gp42-peptide 36-81 shows increased fluorescence polarization signal as the concentration of soluble gH/gL is increased. The concentration of peptide was kept constant at 10 nM, and 50% of binding occurred at 5.1 ± 1.1 nM gH/gL. (B to D) Fluorescence polarization measurements using constant 10 nM concentrations of soluble gH/gL and 10 nM FITC-labeled gp42-peptide 36-81 with added competitor (100% corresponds to no competitor): soluble gp42 protein, EC50 at 8.5 ± 1.0 nM (B); peptide 36-65, EC50 at 1.5 μM (C). For peptides 36-56, 42-56, and 67-81, the extrapolated theoretical EC50 for peptide 42-56 is 150 μM (D). Competitor was premixed with labeled peptide 36-81, and soluble gH/gL was added to initiate binding.
Fluorescence polarization experiments with soluble gp42, an N-terminally truncated form of gp42 beginning at residue 86 (gp42-ΔN86), and short gp42-derived peptides 36-65, 36-56, 42-56, and 67-81.
In order to further map the gp42 regions involved in binding to gH/gL, competition fluorescence polarization experiments were performed using 10 nM soluble gH/gL and 10 nM FITC-labeled gp42-peptide 36-81 with the addition of a competitor gp42 protein (Fig. 3B). Following the competitive reduction of the peptide polarization signal, titrations of soluble gp42 demonstrated a binding affinity for gH/gL that is very similar to that of the FITC-labeled 46-mer peptide. A nonlinear least squares fit calculated 50% inhibition of binding to be at 8.5 nM soluble gp42. Addition of peptides 36-65, 36-56, 42-56, and 67-81 revealed their relative abilities to compete with peptide 36-81 for binding to gH/gL (Fig. 3C and 3D). Only peptides 36-65 and 42-56 were able to diminish the polarization signal, albeit at significantly higher concentrations, with 50% binding inhibition at 1.2 μM for peptide 36-65 and at an extrapolated value of approximately 150 μΜ for peptide 42-56. Peptides 36-56 and 67-81 did not show significant binding to soluble gH/gL up to the highest concentration tested (100 μM). Compared to the low nanomolar range for soluble gp42, these short peptides have much lower affinity for gH/gL (Table 2).
TABLE 2.
Activity summary of gp42-derived peptides
| Soluble EBV gp42 residues | Peptide concn (nM)
|
||
|---|---|---|---|
| 50% binding to gH/gLa | 50% inhibition of B-cell fusionb | 50% inhibition of epithelial cell fusionc | |
| 33-223 | 8.5 | 5 | |
| 36-81 | 5.1 | ∼2d | 5 |
| 36-65 | 1,500 | >10,000 | 5,000 |
| 36-56 | >100,000 | >50,000 | >50,000 |
| 42-56 | ∼150,000e | >50,000 | >50,000 |
| 67-81 | >100,000 | >50,000 | >50,000 |
Measured by fluorescence polarization competition experiments with 10 nM purified soluble gH/gL and 10 nM FITC-labeled gp42-peptide 36-81.
Measured by luciferase activity in fusion assay using CHO cells transfected with gH, gL, and gB and addition of known concentrations of soluble gp42, overlaid with Daudi B cells.
Measured by luciferase activity in fusion assay using CHO cells transfected with gH, gL, and gB and overlaid with 293T epithelial cells.
Measured using lowest gp42 concentration tested.
Theoretical extrapolated value based on trend of measured data.
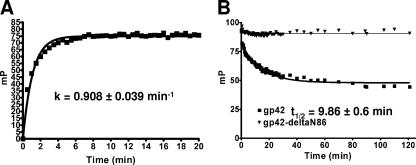
The binding kinetics for FITC-labeled gp42-derived peptide 36-81 and soluble gH/gL were also studied. Fluorescence polarization measurements were taken at 30-s time points after mixing 10 nM peptide and 10 nM soluble gH/gL (Fig. 4A). Maximum binding signal was achieved by 10 min, and the nonlinear least squares exponential fit for one-site binding shows a binding rate of 0.908 ± 0.039 min−1. For dissociation experiments, excess soluble gp42 was added to preincubated 10 nM peptide and 10 nM soluble gH/gL. Time points measured over 2 h were fit to an exponential dissociation curve with a calculated half-life of the gH/gL/peptide complex at 9.86 ± 0.6 min (Fig. 4B). A control sample with 1 μM soluble gp42-ΔN86 added to 10 nM peptide and 10 nM soluble gH/gL did not show any peptide dissociation (Fig. 4B).
FIG. 4.
Kinetics of FITC-labeled gp42 peptide 36-81 binding soluble gH/gL as determined by fluorescence polarization. The fluorescence polarization signal from FITC-labeled gp42 peptide 36-81 was monitored at time points immediately after samples were mixed. (A) Peptide 36-81 binding to soluble gH/gL, each present at a 10 nM concentration. Binding was nearly complete by 10 min with a rate constant of 0.908 ± 0.039 min−1. (B) Dissociation of premixed 10 nM gH/gL and 10 nM FITC-labeled gp42-peptide 36-81 is observed by adding soluble gp42 at 1 μM. The half-life (t1/2) of the gH/gL/peptide complex is 9.86 ± 0.6 min.
Peptide from gp42 residues 36 to 81 is an efficient inhibitor of EBV-mediated membrane fusion.
Previous work with peptide 36-65 revealed that it acted as an inhibitor of EBV-mediated epithelial cell membrane fusion in a dose-dependent manner in the low micromolar range (23). This finding is consistent with the ability of peptide 36-65 to inhibit binding of peptide 36-81 in the polarization assay. Since the longer peptide 36-81 bound gH/gL with much higher affinity than peptide 36-65, peptide 36-81 was also tested for inhibition of cell fusion. As shown in Fig. 5A, peptide 36-81 is a significantly stronger inhibitor of epithelial cell fusion, with an apparent 50% effective concentration (EC50) of approximately 5 nM. Both soluble gp42 and peptide 36-81 had a very similar inhibitory effect over comparable concentration ranges on epithelial cell membrane fusion (see Fig. 5A in reference 23). Further testing of the peptide in fusion assays with B cells revealed that the long peptide is also capable of inhibiting membrane fusion in the presence of gp42. With CHO cells transfected with gH, gL, gB, and gp42, the degree of inhibition is ∼25% of the control, which is a significant amount (P value of < 0.05) as determined by a Student's t test (Fig. 5B). This decrease in fusion is modest and likely attributable to difficulties in competing the membrane-bound gH/gL/gp42 complexes, which form over a 12-h transfection incubation. A significant amount of preformed gH/gL/gp42 complexes likely remain intact after the peptide is incubated with the CHO cells for only 10 min before it is overlaid with target B cells, especially as the half-life for dissociation is approximately 10 min, based on our peptide polarization results. Even greater inhibition of B-cell fusion was observed when the peptide was allowed to incubate for longer times (data not shown). To examine direct competition between gp42 and the peptide for binding cell-associated gH/gL, the B-cell fusion assay was performed using known concentrations of soluble gp42 added to CHO cells transfected with gH, gL, and gB. A clear dose-dependent inhibitory effect on B-cell fusion is observed when peptide and soluble gp42 compete for binding gH/gL on the same timescale (Fig. 5C). The data show a logical trend that as more soluble gp42 is added, more peptide is needed to inhibit fusion. Soluble gp42 and the 46-mer peptide are competitive in the same concentration ranges, suggesting that their binding affinities for gH/gL are similar.
FIG. 5.
Peptide from gp42 residues 36 to 81 inhibits epithelial and B-cell membrane fusion. Fusion assay graphs showing concentration-dependent effects of peptide 36-81 on the cell-cell fusion assay. (A) Epithelial cell fusion. (B and C) B-cell fusion. CHO cells were transfected with gH, gL, and gB (A and C) and additionally transfected with gp42 (B). (The data at the far right of graph A are for cells also transfected with gp42.) For panel C, soluble gp42 was added to trigger B-cell fusion and demonstrate efficient competition between the peptide and soluble protein. Data shown are the mean of triplicate measurements from at least three independent experiments, and error bars indicate standard deviations.
Complementation assays with peptides and gp42 deletion mutants.
The membrane fusion assay was used to test if B-cell fusion with nonfunctional gp42 deletion mutants could be rescued by adding gp42-derived peptides in trans. First, the soluble truncated gp42-ΔN86 protein was added along with the longest gp42-derived peptide (residues 36 to 81), but no membrane fusion occurred (data not shown). This lack of fusion was observed with or without transfected gp350 that might provide more favorable contacts between the fusion proteins and the target B cell (data not shown).
Next, the short gp42-derived peptides spanning residues 36 to 56, 42 to 56, and 67 to 81 were tested for the ability to inhibit epithelial cell fusion. Based on their inability to block peptide 36-81 binding in the polarization assay, we did not expect the peptides to block fusion, and indeed no inhibitory activity was detected up to a 50 μM final peptide concentration (Fig. 6A). The peptides were also added in combination to see if their inhibitory effects would become apparent at micromolar concentrations, but none of these three peptides exhibited any activity when added simultaneously. However, using the peptide derived from residues 36 to 65, which has an inhibitory effect on epithelial cell fusion in the low micromolar range in combination with the peptide from residues 67 to 81, we did observe consistently enhanced inhibition, although the effect was still moderate and required peptide concentrations in the low micromolar range (Fig. 6B). Simultaneous addition of peptides spanning residues 36 to 65 and 67 to 81 in the low micromolar range did not inhibit as strongly as the single peptide spanning residues 36 to 81 used in the nanomolar range, indicating that their covalent linkage is important for full binding and inhibition activity.
FIG. 6.
Shorter gp42-derived peptides tested in epithelial and B-cell fusion assays. (A) Fusion assay graphs showing no inhibitory effect on epithelial cell fusion with individual gp42-derived peptides tested up to 50 μM final concentration. (B) Enhanced inhibition of epithelial cell fusion using simultaneous addition of gp42 peptides 36-65 and 67-81 compared to peptide 36-65 alone. (C) There was no restoration of B-cell fusion using transfected gp42 deletion mutants deficient in membrane fusion activity with added peptides, although wild-type soluble gp42 restored fusion. Brackets above the solid bars indicate the peptides added. Data shown are representative from at least two independent experiments, and error bars indicate standard deviations.
Subsequently, the reverse experiments were performed to see if the short peptides could complement gp42 deletion mutants deficient in membrane fusion activity. Data were collected from the cell-cell fusion assay using CHO cells transfected with gH, gL, gB, and mutant gp42 (five-residue and longer deletions), and micromolar concentrations of short peptides were added immediately before overlay with B cells (Fig. 6C). Fusion assay data revealed that these short N-terminal gp42 peptides added in trans could not restore the function of gp42 deletion mutants that lost the ability to trigger fusion with B cells. The longest peptide from gp42 residues 36 to 81, which has nanomolar binding to gH/gL, was unable to rescue membrane fusion with any of the gp42 mutants, including the gp42 mutant lacking residues 45 to 89. These data indicate that the contiguity of the gp42 C-terminal lectin domain with N-terminal gp42/gH/gL complexes is important for full fusogenic activity.
EBV infection assay with gp42-derived peptide 36-81.
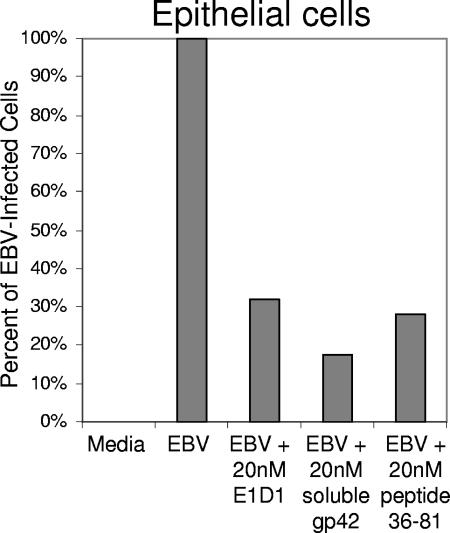
In order to test the effect of peptide 36-81 on entry of the whole virus, epithelial cells were infected by GFP-expressing EBV, which was produced by Akata cells. Virus was preincubated with a 20 nM concentration of anti-gH/gL antibody E1D1, soluble gp42, or peptide 36-81 for 24 h at 37°C prior to a 1-h spin infection of cells in a 96-well format. The effect of peptide 36-81 on the infection of epithelial cells by EBV was measured by FACS analysis. Each of the three experimental conditions showed a reduction of infection to approximately 25% of the uninhibited EBV control (Fig. 7). These data demonstrate that a low nanomolar concentration of peptide 36-81 is able to inhibit the intact virus from entering epithelial cells, consistent with observations made in the cell-cell fusion assays.
FIG. 7.
Peptide 36-81 inhibits EBV entry into epithelial cells. A graph of a representative EBV infection assay shows the inhibitory effect of gp42-derived peptide 36-81. Virus was incubated with anti-gH/gL antibody E1D1, soluble gp42, or peptide 36-81 at 20 nM. FACS analysis was used to measure the GFP-expressing cells, and data were transformed to set the medium-only control to 0% infected and the EBV-only control to 100% infected so that each experimental condition demonstrates a fraction of the uninhibited 100% infection. The data shown are representative of at least three independent experiments.
DISCUSSION
N-terminal deletion mutants of gp42 lose the ability to function in membrane fusion.
The deletion of five-residue segments within the N terminus of gp42 did not substantially affect protein expression levels compared to wild type. Membrane fusion experiments with B cells demonstrated that some mutants retain nearly wild-type fusion function, some allow moderate levels of fusion, and some are inactive in fusion. These deletion mutants reveal that residues 37 to 56 and 72 to 96 are absolutely required functional regions of the N terminus of gp42 to trigger fusion and suggest that some of the residues within residues 67 to 71 are also required (Fig. 1B). The gp42 mutants defective in membrane fusion can be rescued by the addition of soluble gp42 in the fusion assay but not with any gp42-derived peptides that span the regions deleted (Fig. 6C). Thus, there is a requirement for direct linkage between a gp42 lectin domain (residues 111 to 217) and the N-terminal gH/gL binding site to be within the same molecule.
CELISA with gp42 five-residue deletion mutants quantitatively measures binding to soluble gH/gL.
The loss of membrane fusion activity for some gp42 mutants may be due to a lack of binding to gH/gL or to deletion of gp42 residues otherwise important for fusion activation. To investigate this hypothesis, a gH/gL-based CELISA was employed in addition to immunoprecipitation, and the data revealed that some defective gp42 mutants are similar to wild-type gp42 in binding gH/gL, some show moderately diminished binding, and some completely lose any interaction with gH/gL. Correlating the ability of mutants to function in membrane fusion with the ability to bind soluble gH/gL distinguishes three categories of mutants (Table 1): category 1, inconsequential mutants remaining fully functional; category 2, loss of fusion but maintenance of wild-type gH/gL binding; and category 3, reduced or complete loss of fusion due to altered binding to gH/gL. Category 1 mutants are innocuous deletions and/or single substitutions that have no major effect on gp42 or gH/gL function. The d57-61 mutant appears to lack gH/gL binding in the CELISA but could mediate fusion. However, there are a few discrepancies between the immunoprecipitation data and CELISA measurements for the binding of gp42 mutants to gH/gL. These differences may be due to interactions gp42 may have with transfected gH/gL during protein expression and folding, possibly within the proteins' membrane spanning domains.
Mutant d57-61 is able to promote near wild-type membrane fusion but completely loses binding to soluble gH/gL as detected in CELISA. This is a peculiar mutant that was difficult to detect in Western blotting even though surface expression levels were consistently higher than wild-type gp42 (Fig. 1B and 2B). There may be a significant contribution by the unusual tetra-proline structure (residues 57 to 60) in binding gH/gL, but a nearly wild-type degree of fusion is maintained in its absence. For HSV gD, which is a functional homolog for gp42, it is thought that a flexible proline-rich region becomes exposed upon receptor binding, thus activating gH/gL and gB for membrane fusion (5, 6, 9, 24). We reasoned that the N-terminal region of gp42 might play an analogous role and that for gp42 the presence of the tetra-proline motif might enable a gH/gL-dependent regulation of the gp42-N-terminal peptide. In this scenario, the d57-61 mutant might be capable of activating gB-dependent fusion in the absence of gH/gL. However, fusion assays revealed that the d57-61 mutant is not capable of membrane fusion without gH/gL, i.e., transfected gB and the d57-61 mutant do not cause membrane fusion (data not shown). Furthermore, epithelial cell membrane fusion was inhibited in the presence of the d57-61 gp42 mutant, although d57-61 gp42 was somewhat less effective than wild-type gp42 (data not shown). These data suggest that the d57-61 mutant still binds gH/gL but with a lower affinity and significantly increased dissociation rate. As a preliminary test for altered binding kinetics, preformed E1D1-gH/gL complexes (500 nM E1D1 and 50 nM gH/gL) were tested in CELISA for binding d57-61, but no binding was detected even though the antibody complex is bivalent and thereby might be expected to interact more strongly with the mutant (data not shown). However, when the gp42 d57-61 mutant is transfected instead of wild-type gp42 in membrane fusion assays with B cells, peptide 36-81 does inhibit fusion somewhat more efficiently (data not shown). These data also suggest that the d57-61 mutant forms complexes with gH/gL with a lower binding affinity than wild-type gp42 and that this complex can be dissociated by adding peptide 36-81.
Interestingly, category 2 mutants that are at the beginning of NT1 and the end of NT2 are able to bind soluble gH/gL in the gH/gL CELISA but are still unable to mediate membrane fusion (Fig. 1B and 2; Table 1). At the N terminus of NT1, mutants d37-41 and d42-46 both appear to bind gH/gL well but are impaired in membrane fusion. The NT1 mutant d37-41 removes the cleavage site of residues 40 to 42 and does not secrete soluble gp42 (data not shown), potentially indicating that soluble gp42 plays an important role in membrane fusion (38). However, it is not clear why d42-46, which can bind soluble gH/gL, does not mediate fusion. Perhaps these missing residues are essential for triggering gH/gL function, even though binding to gH/gL occurs at wild-type levels. The NT2 mutants d82-86, d87-91, and d92-96 were also able to bind gH/gL but not mediate fusion. These mutants fall in or near the dimerization domain that was seen in the crystal structure of gp42-HLA-DR1 that includes residues 86 to 95 (32). The dimerization site might therefore be important for membrane fusion; alternatively, conceivably a linker region between the gH/gL binding site and the lectin domain is required. A five-residue insertion mutant previously studied (LI93) at this site does not affect membrane fusion, consistent with the idea that a minimal, but perhaps sequence-independent, linker region is important rather than dimerization (41). The d82-86 mutant had very low levels of fusion, and these residues link the end of the gH/gL binding domain to the putative dimerization site. Further studies are required to determine whether specific sequences, dimerization, or simply linker residues are required for gp42 function in these regions of NT1 and NT2. For example, it is possible that five-residue deletions between residues 82 to 96 primarily result in a loss of fusion function due to altered geometry of the lectin domain relative to gH/gL in the complex. These residues may act as a linker between the gH/gL binding site and the lectin domain, providing needed flexibility for the fusion process to proceed. Nevertheless, category 3 mutants reveal the “core” gp42 residues that interact with gH/gL. These residues are important for binding soluble gH/gL and essential for membrane fusion function (Fig. 1A).
The gp42-derived peptides directly bind to gH/gL.
Fluorescence polarization experiments reveal that the FITC-labeled gp42-derived peptide from residues 36 to 81 binds to gH/gL with high affinity. This direct measurement allows quantitative data to be gathered on the binding. Both binding at equilibrium and kinetics of binding were studied. The measured values for the on-rate and off-rate of FITC-labeled peptide 36-81 binding to soluble gH/gL are consistent with values obtained in equilibrium binding experiments. The affinity of peptide 36-81 for gH/gL was found to be nearly equivalent to the binding affinity of soluble gp42 protein. Thus, the residues sufficient for wild-type level binding to gH/gL appear to be present in the 46-mer peptide. Additional competition experiments with shorter peptides showed significantly weaker binding affinities. Peptide from residues 36 to 65 competes in the low-micromolar-concentration range, while the peptide from residues 42 to 56 just begins binding soluble gH/gL in the mid-micromolar range, confirming that the minimum residues required for high-affinity gH/gL binding cover a large residue range (Table 2).
Peptides derived from gp42 inhibit epithelial cell fusion.
Membrane fusion studies investigating the effect of soluble gp42 and peptides derived from the N-terminal region of gp42 revealed that they can effectively inhibit epithelial cell fusion. The soluble gp42 protein and the long peptide 36-81 inhibit in the low nanomolar range, the peptide 36-65 inhibits in the low micromolar range, and the shorter peptides do not show inhibition up to 50 μM. Nevertheless, a synergistic effect was found when adding peptides 36-65 and 67-81 in the low micromolar range, somewhat enhancing the inhibitory effect compared to peptide 36-65 alone. The small synergistic effect of peptides 36-65 and 67-81 suggests that these segments bind with an extended peptide binding site on gH/gL, which exhibits only mild cooperativity in binding. The fact that the longer peptide 36-81 gains nearly 1,000-fold binding affinity and concentration for 50% inhibition compared to the shorter peptides demonstrates the importance of having these two binding regions within a single peptide chain. Given that gp42 and gp42-derived peptides function by binding to gH/gL, it can be inferred that the inhibitory mechanism is either blockage of a critical binding site on gH/gL, such as an interaction of gH/gL with a gH/gL receptor, or inhibition or induction of a conformational change in gH/gL rendering it unable to mediate fusion.
Peptide from gp42 residues 36 to 81 inhibits B-cell fusion when competing with soluble gp42.
The long peptide from gp42 residues 36 to 81 binds soluble gH/gL with approximately the same affinity as the soluble protein gp42. This suggests that their activities in membrane fusion may be in the same functional range, namely, the low nanomolar concentration. Indeed, when tested in direct competition with soluble gp42, peptide 36-81 is able to inhibit B-cell fusion when its concentration is above that of soluble gp42. However, membrane fusion with B cells proceeds only when the N-terminal region of gp42 is connected to the lectin domain, which engages the MHC-II receptor. B-cell fusion did not occur in experiments with peptide 36-81 and soluble gp42-ΔN86 added together (data not shown). Therefore, membrane fusion requires the combination of the N-terminal region of gp42 occupying the gH/gL binding site and the same protein's proximal C-type lectin domain engaging the MHC-II receptor. In addition, none of the short gp42-derived peptides (peptides 36-56, 42-56, and 67-81) has any inhibitory effect on B-cell or epithelial cell fusion, testing even at 50 μM peptide. Moreover, none of the gp42-derived peptides can rescue B-cell fusion with transfected five-residue gp42 deletion mutants. Nevertheless, the gp42 residues in this region are critical for binding to gH/gL and triggering membrane fusion. It remains unclear why the presence of these residues is insufficient to stimulate membrane fusion, even when the high-affinity peptide 36-81 and the gp42-ΔN86 protein are present.
Peptide 36-81 inhibits epithelial cell infection by whole virus.
Since peptide 36-81 inhibited EBV-mediated epithelial cell fusion in the low nanomolar range, its ability to inhibit intact virus infection of epithelial and B cells was examined. For epithelial cell infections, virus was produced by Akata cells, which are thought to make virus with lower levels of gp42 that is therefore more efficient at infecting epithelial cells. At a concentration of 20 nM, which is approximately four times the EC50 for inhibiting epithelial cell fusion, the peptide acts similarly to anti-gH/gL antibody E1D1 and soluble gp42, reducing the infection to nearly one-quarter of the uninhibited control (Fig. 7). The peptide inhibits intact virus as effectively as it inhibits epithelial cell membrane fusion in the virus-free cell-cell fusion assay. Since the peptide is known to bind directly to gH/gL in competition for gp42, the infection assay data strongly suggest that the peptide is capable of binding gH/gL on the intact virus surface to inhibit its ability to enter epithelial cells.
In contrast, when peptide 36-81 was examined in a B-cell infection assay, using virus produced by B95-8 cells, it did not demonstrate inhibition (data not shown). This was likely due to the presence of higher concentrations of viral gp42 forming tight membrane-bound gH/gL/gp42 complexes, in which gp42 could not be displaced by the peptide. This finding is consistent with the results observed for peptide inhibition of B-cell membrane fusion in which even high concentrations of peptide could achieve only a modest decrease in cell-cell fusion when transfected gp42 was present (Fig. 5B). Design of other peptides or peptidomimetics might reveal a better inhibitor of viral entry into B cells.
Characterization of the gp42 residues important for gH/gL-mediated membrane fusion.
The gp42 residues found to primarily contribute to binding gH/gL (47 to 61 and 67 to 81) contain seven prolines, 10 charged residues, and three aromatic residues. Other residues important for triggering fusion function, are hydrophobic (residues 37 to 46) and mostly neutral (residues 82 to 96). However, there are putative glycosylation sites that suggest that these regions are not likely to mediate direct protein-protein interactions. In addition, gp42 has a cleavage site from residues 40 to 42 and a possible dimerization or linker region from residues 86 to 95. We must await a crystal structure of the N-terminal region of gp42 and the structure of the gH/gL complex to complete our understanding of this herpesvirus membrane fusion mechanism.
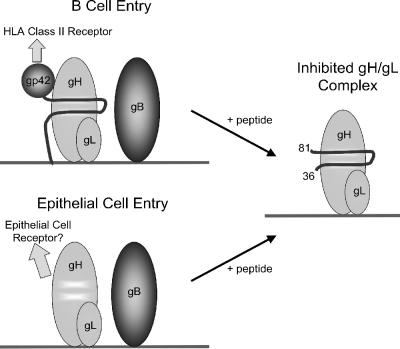
Based on the data gathered using the gp42-derived peptide 36-81 in vitro and in cell-based assays, a model of its inhibition of EBV-mediated membrane fusion is proposed (Fig. 8). Contacts mimicking gp42 are retained, and membrane fusion is inhibited when peptide 36-81 is bound to gH/gL.
FIG. 8.
Model of peptide inhibition of EBV membrane fusion apparatus. The N-terminal region of gp42 binds gH/gL with high affinity, and the peptide from gp42 residues 36 to 81 mimics that interaction. Epithelial cell fusion is inhibited by nanomolar concentrations of gp42, and the peptide 36-81 is sufficient to inhibit at those concentrations as well. B-cell membrane fusion only proceeds when a gp42 lectin domain is supplied in cis with the N-terminal region. Thus, peptide 36-81 is a strong inhibitor of both epithelial and B-cell membrane fusion.
The peptide derived from residues 36 to 81 acts as a low nanomolar inhibitor of B-cell membrane fusion, and it is as potent as gp42 at inhibiting epithelial cell membrane fusion and viral entry. This peptide proves useful for future drug development to treat EBV-mediated diseases of both B-cell and epithelial cell origin. The gp42-derived peptides, and others like them, provide new tools for studying herpesvirus entry and designing effective therapeutics.
Acknowledgments
We thank Hisae Matsuura for providing gp42-ΔN86, Jasmina Omerović for providing baculovirus encoding soluble gH/gL and for assistance with Western blotting, and Aileen Baldwin for providing GFP-expressing EBV and for assistance with the infection assay and FACS analysis. We thank Lindsey Hutt-Fletcher for providing key reagents. We acknowledge the use of the fluorescence polarization instrument at the Keck Biophysics Facility at Northwestern University [http://www.biochem.northwestern.edu/Keck/Keckmain.html]. We appreciate help and support provided by the members of the Jardetzky and Longnecker laboratories.
This research was supported by Public Health Service grants CA93444 (R.L. and T.S.J.) and CA117794 (R.L. and T.S.J.) from the National Cancer Institute. This work is supported in part by a predoctoral fellowship from Northwestern's Biotechnology Training Program from the National Institutes of Health (A.N.K.) and Northwestern's training program in the Cellular and Molecular Basis of Disease (T32 GM08061) from the National Institutes of Health (A.S.L.).
Footnotes
Published ahead of print on 20 June 2007.
REFERENCES
- 1.Balachandran, N., D. E. Oba, and L. M. Hutt-Fletcher. 1987. Antigenic cross-reactions among herpes simplex virus types 1 and 2, Epstein-Barr virus, and cytomegalovirus. J. Virol. 61:1125-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borza, C. M., and L. Hutt-Fletcher. 2002. Alternate Replication in B cells and epithelial cells switches tropism of Epstein-Barr virus. Nat. Med. 8:594-599. [DOI] [PubMed] [Google Scholar]
- 3.Borza, C. M., A. J. Morgan, S. M. Turk, and L. M. Hutt-Fletcher. 2004. Use of gHgL for attachment of Epstein-Barr virus to epithelial cells compromises infection. J. Virol. 78:5007-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burkitt, D. 1962. A tumour syndrome affecting children in tropical Africa. Postgrad. Med. J. 38:71-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carfi, A., H. Gong, H. Lou, S. H. Willis, G. H. Cohen, R. J. Eisenberg, and D. C. Wiley. 2002. Crystallization and preliminary diffraction studies of the ectodomain of the envelope glycoprotein D from herpes simplex virus 1 alone and in complex with the ectodomain of the human receptor HveA. Acta Crystallogr. D 58:836-838. [DOI] [PubMed] [Google Scholar]
- 6.Cocchi, F., D. Fusco, L. Menotti, T. Gianni, R. J. Eisenberg, G. H. Cohen, and G. Campadelli-Fiume. 2004. The soluble ectodomain of herpes simplex virus gD contains a membrane-proximal pro-fusion domain and suffices to mediate virus entry. Proc. Natl. Acad. Sci. USA 101:7445-7450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diakidi-Kosta, A., G. Michailidou, G. Kontogounis, A. Sivropoulou, and M. Arsenakis. 2003. A single amino acid substitution in the cytoplasmic tail of the glycoprotein B of herpes simplex virus 1 affects both syncytium formation and binding to intracellular heparan sulfate. Virus Res. 93:99-108. [DOI] [PubMed] [Google Scholar]
- 8.Epstein, M. A., B. G. Achong, and Y. M. Barr. 1964. Virus particles in cultured lymphoblasts from Burkitt's lymphoma. Lancet 15:702-703. [DOI] [PubMed] [Google Scholar]
- 9.Fusco, D., C. Forghieri, and G. Campadelli-Fiume. 2005. The pro-fusion domain of herpes simplex virus glycoprotein D (gD) interacts with the gD N terminus and is displaced by soluble forms of viral receptors. Proc. Natl. Acad. Sci. USA 102:9323-9328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haan, K. M., W. W. Kwok, R. Longnecker, and P. Speck. 2000. Epstein-Barr virus entry utilizing HLA-DP or HLA-DQ as a coreceptor. J. Virol. 74:2451-2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haan, K. M., S. K. Lee, and R. Longnecker. 2001. Different functional domains in the cytoplasmic tail of glycoprotein B are involved in Epstein-Barr virus-induced membrane fusion. Virology 290:106-114. [DOI] [PubMed] [Google Scholar]
- 12.Haan, K. M., and R. Longnecker. 2000. Coreceptor restriction within the HLA-DQ locus for Epstein-Barr virus infection. Proc. Natl. Acad. Sci. USA 97:9252-9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haddad, R. S., and L. M. Hutt-Fletcher. 1989. Depletion of glycoprotein gp85 from virosomes made with Epstein-Barr virus proteins abolishes their ability to fuse with virus receptor-bearing cells. J. Virol. 63:4998-5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heldwein, E. E., H. Lou, F. C. Bender, G. H. Cohen, R. J. Eisenberg, and S. C. Harrison. 2006. Crystal structure of glycoprotein B from herpes simplex virus 1. Science 313:217-220. [DOI] [PubMed] [Google Scholar]
- 15.Henle, G., W. Henle, and V. Diehl. 1968. Relation of Burkitt's tumor-associated herpes-type virus to infectious mononucleosis. Proc. Natl. Acad. Sci. USA 59:94-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henle, W., and G. Henle. 1970. Evidence for a relation of Epstein-Barr virus to Burkitt's lymphoma and nasopharyngeal carcinoma. Bibl. Haematol. 1970:706-713. [DOI] [PubMed] [Google Scholar]
- 17.Henle, W., and G. Henle. 1977. Evidence for an etiologic relation of the Epstein-Barr virus to human malignancies. Laryngoscope 87:467-473. [DOI] [PubMed] [Google Scholar]
- 18.Henle, W., and G. Henle. 1969. The relation between the Epstein-Barr virus and infectious mononucleosis, Burkitt's lymphoma and cancer of the postnasal space. East Afr. Med. J. 46:402-406. [PubMed] [Google Scholar]
- 19.Hutt-Fletcher, L. M., and C. M. Lake. 2001. Two Epstein-Barr virus glycoprotein complexes. Curr. Top. Microbiol. Immunol. 258:51-64. [DOI] [PubMed] [Google Scholar]
- 20.Hutt-Fletcher, L. M., and S. M. Turk. 2001. Virus isolation. Methods Mol. Biol. 174:119-123. [DOI] [PubMed] [Google Scholar]
- 21.Jardetzky, T. S., and R. A. Lamb. 2004. Virology: a class act. Nature 427:307-308. [DOI] [PubMed] [Google Scholar]
- 22.Jones, N. A., and R. J. Geraghty. 2004. Fusion activity of lipid-anchored envelope glycoproteins of herpes simplex virus type 1. Virology 324:213-228. [DOI] [PubMed] [Google Scholar]
- 23.Kirschner, A. N., J. Omerovic, B. Popov, R. Longnecker, and T. S. Jardetzky. 2006. Soluble Epstein-Barr virus glycoproteins gH, gL, and gp42 form a 1:1:1 stable complex that acts like soluble gp42 in B-cell fusion but not in epithelial cell fusion. J. Virol. 80:9444-9454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krummenacher, C., V. M. Supekar, J. C. Whitbeck, E. Lazear, S. A. Connolly, R. J. Eisenberg, G. H. Cohen, D. C. Wiley, and A. Carfi. 2005. Structure of unliganded HSV gD reveals a mechanism for receptor-mediated activation of virus entry. EMBO J. 24:4144-4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, Q., C. Buranathai, C. Grose, and L. M. Hutt-Fletcher. 1997. Chaperone functions common to nonhomologous Epstein-Barr virus gL and varicella-zoster virus gL proteins. J. Virol. 71:1667-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, Q., M. K. Spriggs, S. Kovats, S. M. Turk, M. R. Comeau, B. Nepom, and L. M. Hutt-Fletcher. 1997. Epstein-Barr virus uses HLA class II as a cofactor for infection of B lymphocytes. J. Virol. 71:4657-4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, Q., S. M. Turk, and L. M. Hutt-Fletcher. 1995. The Epstein-Barr virus (EBV) BZLF2 gene product associates with the gH and gL homologs of EBV and carries an epitope critical to infection of B cells but not of epithelial cells. J. Virol. 69:3987-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McShane, M. P., and R. Longnecker. 2004. Cell-surface expression of a mutated Epstein-Barr virus glycoprotein B allows fusion independent of other viral proteins. Proc. Natl. Acad. Sci. USA 101:17474-17479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McShane, M. P., M. M. Mullen, K. M. Haan, T. S. Jardetzky, and R. Longnecker. 2003. Mutational analysis of the HLA class II interaction with Epstein-Barr virus glycoprotein 42. J. Virol. 77:7655-7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller, N., and L. M. Hutt-Fletcher. 1988. A monoclonal antibody to glycoprotein gp85 inhibits fusion but not attachment of Epstein-Barr virus. J. Virol. 62:2366-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molesworth, S. J., C. M. Lake, C. M. Borza, S. M. Turk, and L. M. Hutt-Fletcher. 2000. Epstein-Barr virus gH is essential for penetration of B cells but also plays a role in attachment of virus to epithelial cells. J. Virol. 74:6324-6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mullen, M. M., K. M. Haan, R. Longnecker, and T. S. Jardetzky. 2002. Structure of the Epstein-Barr virus gp42 protein bound to the MHC class II receptor HLA-DR1. Mol. Cell 9:375-385. [DOI] [PubMed] [Google Scholar]
- 33.Oda, T., S. Imai, S. Chiba, and K. Takada. 2000. Epstein-Barr virus lacking glycoprotein gp85 cannot infect B cells and epithelial cells. Virology 276:52-58. [DOI] [PubMed] [Google Scholar]
- 34.Okuma, K. M., S. Nakamura, S. Nakano, Y. Niho, and Y. Matsuura. 1999. Host range of human T-cell leukemia virus type I analyzed by a cell fusion-dependent reporter gene activation assay. Virology 254:235-244. [DOI] [PubMed] [Google Scholar]
- 35.Omerovic, J., L. Lev, and R. Longnecker. 2005. The amino terminus of Epstein-Barr virus glycoprotein gH is important for fusion with epithelial and B cells. J. Virol. 79:12408-12415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Omerovic, J., and R. Longnecker. 1 May 2007, posting date. Functional homology of gHs and gLs from EBV-related γ-herpesviruses for EBV-induced membrane fusion. Virology 365:157-165. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perez-Romero, P., A. Perez, A. Capul, R. Montgomery, and A. O. Fuller. 2005. Herpes simplex virus entry mediator associates in infected cells in a complex with viral proteins gD and at least gH. J. Virol. 79:4540-4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ressing, M. E., D. van Leeuwen, F. A. Verreck, S. Keating, R. Gomez, K. L. Franken, T. H. Ottenhoff, M. Spriggs, T. N. Schumacher, L. M. Hutt-Fletcher, M. Rowe, and E. J. Wiertz. 2005. Epstein-Barr virus gp42 is posttranslationally modified to produce soluble gp42 that mediates HLA class II immune evasion. J. Virol. 79:841-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rey, F. A. 2006. Molecular gymnastics at the herpesvirus surface. EMBO Rep. 7:1000-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rickinson, A. B., and E. Kieff. 1996. Epstein-Barr virus, p. 2397-2446. In B. N. Fields, D. M. Knipe, P. M. Howley, R. M. Chanock, J. L. Melnick, T. P. Monath, B. Roizman, and S. E. Straus (ed.), Virology, 3rd ed. Raven Press, Ltd., New York, NY.
- 41.Silva, A. L., J. Omerovic, T. S. Jardetzky, and R. Longnecker. 2004. Mutational analyses of Epstein-Barr virus glycoprotein 42 reveal functional domains not involved in receptor binding but required for membrane fusion. J. Virol. 78:5946-5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sodora, D. L., G. H. Cohen, M. I. Muggeridge, and R. J. Eisenberg. 1991. Absence of asparagine-linked oligosaccharides from glycoprotein D of herpes simplex virus type 1 results in a structurally altered but biologically active protein. J. Virol. 65:4424-4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spear, P. G., and R. Longnecker. 2003. Herpesvirus entry: an update. J. Virol. 77:10179-10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spear, P. G., S. Manoj, M. Yoon, C. R. Jogger, A. Zago, and D. Myscofski. 2006. Different receptors binding to distinct interfaces on herpes simplex virus gD can trigger events leading to cell fusion and viral entry. Virology 344:17-24. [DOI] [PubMed] [Google Scholar]
- 45.Speck, P., K. M. Haan, and R. Longnecker. 2000. Epstein-Barr virus entry into cells. Virology 277:1-5. [DOI] [PubMed] [Google Scholar]
- 46.Spriggs, M. K., R. J. Armitage, M. R. Comeau, L. Strockbine, T. Farrah, B. Macduff, D. Ulrich, M. R. Alderson, J. Mullberg, and J. I. Cohen. 1996. The extracellular domain of the Epstein-Barr virus BZLF2 protein binds the HLA-DR beta chain and inhibits antigen presentation. J. Virol. 70:5557-5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steven, A. C., and P. G. Spear. 2006. Biochemistry. Viral glycoproteins and an evolutionary conundrum. Science 313:177-178. [DOI] [PubMed] [Google Scholar]
- 48.Strnad, B. C., T. Schuster, R. Klein, R. F. Hopkins III, T. Witmer, R. H. Neubauer, and H. Rabin. 1982. Production and characterization of monoclonal antibodies against the Epstein-Barr virus membrane antigen. J. Virol. 41:258-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Subramanian, R. P., and R. J. Geraghty. 2007. Herpes simplex virus type 1 mediates fusion through a hemifusion intermediate by sequential activity of glycoproteins D, H, L, and B. Proc. Natl. Acad. Sci. USA 104:2903-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takada, K. 2001. Role of Epstein-Barr virus in Burkitt's lymphoma. Curr. Top. Microbiol. Immunol. 258:141-151. [DOI] [PubMed] [Google Scholar]
- 51.Takada, K., K. Horinouchi, Y. Ono, T. Aya, T. Osato, M. Takahashi, and S. Hayasaka. 1991. An Epstein-Barr virus-producer line Akata: establishment of the cell line and analysis of viral DNA. Virus Genes 5:147-156. [DOI] [PubMed] [Google Scholar]
- 52.Urquiza, M., J. Suarez, R. Lopez, E. Vega, H. Patino, J. Garcia, M. A. Patarroyo, F. Guzman, and M. E. Patarroyo. 2004. Identifying gp85-regions involved in Epstein-Barr virus binding to B-lymphocytes. Biochem. Biophys. Res. Commun. 319:221-229. [DOI] [PubMed] [Google Scholar]
- 53.Volpi, A. 2004. Epstein-Barr virus and human herpesvirus type 8 infections of the central nervous system. Herpes 11(Suppl. 2):120A-127A. [PubMed] [Google Scholar]
- 54.Wang, X., and L. M. Hutt-Fletcher. 1998. Epstein-Barr virus lacking glycoprotein gp42 can bind to B cells but is not able to infect. J. Virol. 72:158-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang, X., W. J. Kenyon, Q. Li, J. Mullberg, and L. M. Hutt-Fletcher. 1998. Epstein-Barr virus uses different complexes of glycoproteins gH and gL to infect B lymphocytes and epithelial cells. J. Virol. 72:5552-5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wei, W. I., and J. S. Sham. 2005. Nasopharyngeal carcinoma. Lancet 365:2041-2054. [DOI] [PubMed] [Google Scholar]
- 57.Whetter, L. E., S. P. Day, E. A. Brown, O. Elroy-Stein, and S. M. Lemon. 1994. Analysis of hepatitis A virus translation in a T7 polymerase-expressing cell line. Arch. Virol. Suppl. 9:291-298. [DOI] [PubMed] [Google Scholar]
- 58.Wu, L., C. M. Borza, and L. M. Hutt-Fletcher. 2005. Mutations of Epstein-Barr virus gH that are differentially able to support fusion with B cells or epithelial cells. J. Virol. 79:10923-10930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu, L., and L. M. Hutt-Fletcher. 2007. Point mutations in EBV gH that abrogate or differentially affect B cell and epithelial cell fusion. Virology. [DOI] [PMC free article] [PubMed]
- 60.Yaswen, L. R., E. B. Stephens, L. C. Davenport, and L. M. Hutt-Fletcher. 1993. Epstein-Barr virus glycoprotein gp85 associates with the BKRF2 gene product and is incompletely processed as a recombinant protein. Virology 195:387-396. [DOI] [PubMed] [Google Scholar]