Abstract
The bovine papillomavirus type 1 (BPV-1) E7 oncoprotein is required for the full transformation activity of the virus. Although BPV-1 E7 by itself is not sufficient to induce cellular transformation, it enhances the abilities of the other BPV-1 oncogenes to induce anchorage independence. We have been exploring the mechanisms by which E7 might affect the transformation efficiency of other viral oncoproteins and in particular whether it might protect cells from apoptosis. We report here that BPV-1 E6 and E7 can each independently inhibit anoikis, a type of apoptosis that is induced upon cell detachment. Using site-directed mutagenesis, we determined regions of the E7 protein that were essential for its antiapoptotic activity. The ability of E7 to inhibit anoikis did partially correlate with an ability to enhance anchorage independence of BPV-1 E6-transformed cells. In addition, the antiapoptotic activity of E7 also only partially correlated with its ability to bind p600, a cellular protein that has previously been reported to play a role in anoikis. We conclude that the contribution of E7 to BPV-induced cellular transformation may involve its ability to inhibit anoikis but that additional functional activities must also be involved.
Bovine papillomavirus type 1 (BPV-1) has been well characterized for its ability to induce cellular transformation and has served as an excellent model for studying the many molecular biological aspects of the papillomaviruses (14). BPV-1 induces fibropapillomas in its natural host and is also able to transform a variety of rodent cell lines in tissue culture. This transformation activity has provided a robust assay to define the viral genes involved in cellular transformation and to study the molecular mechanisms of papillomavirus-mediated transformation (6, 16). BPV-1-transformed mouse cells have a fully transformed phenotype as measured by anchorage independence and are tumorigenic in nude mice.
Genetic analyses have mapped the transforming activities of BPV-1 to three viral genes: E5, E6, and E7 (14). The E5 gene encodes a hydrophobic membrane protein of 44 amino acids that binds and dimerizes the platelet-derived growth factor-β receptor, thereby stimulating its tyrosine kinase activity and inducing cellular proliferation (5, 13, 23, 24). E5 also binds the 16-kDa subunit of the vacuolar H+-ATPase complex (10) and has been reported to decrease cell surface expression of major histocompatibility complex class I by causing its retention within the cell (17). The BPV-1 E6 gene encodes an oncoprotein of 137 amino acids that disrupts the actin cytoskeleton and binds a number of cellular proteins including ERC-55, paxillin, the E3 ubiquitin ligase E6AP, and the clathrin adaptor complex AP-1 (3, 29-32). Studies with BPV-1 E6 mutants have revealed a strong correlation between E6 binding to the focal adhesion protein paxillin and its transformation capacity (31). Either E5 or E6 alone is sufficient to transform cells, and mutation of either, in the context of the full BPV-1 genome, decreases its transforming activity (5, 26).
The BPV-1 E7 gene encodes a 127-amino-acid zinc-binding protein that on its own does not induce colony formation or anchorage independence but does enhance transformation mediated by other BPV-1 oncogenes. E7 expression leads to an increase in both the number and size of anchorage-independent colonies induced by E5 or E6 expression (2, 4). In addition, mutation of E7, in the context of the entire viral genome, results in a reduction in its transformation potential (22, 25).
We have previously reported that BPV-1 E7 interacts with p600, a 600-kDa protein that has been reported to play a role in cell survival and morphogenesis (20). Mutations in BPV-1 E7 that abrogate its interaction with p600 also compromise its ability to stimulate E6-mediated transformation, suggesting that the E7-p600 interaction could be important for the E7 oncogenic function. It should be noted that BPV-1 E7 does not bind to the retinoblastoma (pRB) tumor suppressor family of proteins, nor does it contain the LXCXE motif found in human papillomavirus (HPV) E7 proteins that mediates the binding of the HPV E7 proteins to the pRB family of proteins (19). The ability to bind p600 may be a general property of all papillomavirus E7 proteins and has been shown for HPV16 and HPV6 E7 as well as BPV-1 E7 (4, 15), suggesting a conserved role for the E7-p600 interaction that may be important in some aspect of the virus-host cell interaction.
Since p600 has been implicated in playing a role in cell survival, we examined whether BPV-1 E7 also affected the apoptotic program of the cell and found that BPV-1 E7 expression can indeed inhibit cell detachment-induced apoptosis. This type of apoptosis, known as “anoikis” (Greek for “homelessness”), is commonly inhibited in cancer cells, which allows cells to survive in the absence of normal cell-matrix interactions (8, 9). Our data indicate that BPV-1 E7 expression can inhibit anoikis of C127 murine cells and that the ability of BPV-1 E7 to inhibit anoikis correlated in part with an ability to bind p600.
MATERIALS AND METHODS
Cell lines.
Monolayer cultures of murine C127 cells and transformed C127 cells were maintained at 37°C with 5% CO2 in Dulbecco modified Eagle medium (DMEM) (GIBCO-BRL, Gaithersburg, MD) containing 10% fetal bovine serum (FBS) and supplemented with penicillin (10 U/ml) and streptomycin (100 μg/ml). BPV-1-transformed C127 cells (clone H2), containing a stable episome of the BPV-1 genome, were kindly provided by Carl Baker (National Cancer Institute, Bethesda, MD).
Mutagenesis of BPV-1 E7 and generation of cell lines.
Site-directed mutations in the E7 open reading frame (ORF) were constructed using the QuikChange XL site-directed mutagenesis kit (Stratagene, LaJolla, CA) according to the manufacturer's instructions. Briefly, mutagenic oligonucleotides were used to amplify E7 by PCR, and the resulting ORF was cloned into the retroviral expression vector pOZ-C, a plasmid which placed the E7 protein in frame with a C-terminal FLAG/hemagglutinin (HA) tag (21). This retroviral vector expressed, under the control of a long terminal repeat promoter, a bicistronic mRNA containing the FLAG/HA-E7 protein followed by the interleukin-2 receptor-α, used as a selectable cell surface marker. All versions of E7 generated in this report involved changing conserved amino acids to alanine (as shown in Fig. 3), including E7-C PNTHR (plasmid 5921), E7-C LLIL (plasmid 5922), E7-C RR (plasmid 5923), E7-C TSSTS (plasmid 5924), E7-C DLD (plasmid 5925), and E7-C CPRC (plasmid 5926). E7 constructs previously published (4) include wild-type E7-C (plasmid 5087); E7 proteins with a deletion of amino acids 5 to 9 (E7-C Δ5-9; plasmid 5090), a deletion of amino acids 90 to 94 (E7-C Δ90-94; plasmid 5091), and a proline-to-alanine substitution at residue 5 (E7-C P5A; plasmid 5092); and an untagged E7 protein (plasmid 5927). E7-N (plasmid 5088), also previously published, placed the E7 protein in frame with an N-terminal FLAG/HA tag.
FIG. 3.
Sequence alignment of papillomavirus E7 proteins. The E7 proteins from the deltapapillomaviruses BPV-1, DPV-1, and EPV-1 were aligned using the ClustalW algorithm. These three viruses share the ability to transform rodent cells. Site-directed mutagenesis of BPV-1 E7 was performed, and the constructs used in this report are shown above the amino acid sequence. With the exception of the deletion constructs (E7-CΔ5-9 and Δ90-94), all constructs involved changing the boxed residues to alanine.
Using standard transfection and retroviral production procedures, C127 cells were generated that stably expressed FLAG/HA-tagged E7 proteins. The transduced subpopulation was purified by repeated cycles of affinity cell sorting for the interleukin-2 receptor-α surface marker (21).
For expression of BPV-1 E6, C127 cells were transduced with a retrovirus generated using pBabe-puro-E6 (3). Cells expressing E6 were selected for by passage in 1 μg/ml puromycin.
Induction of anoikis.
Cells were normally propagated on coated tissue culture plasticware. In order to stimulate cell detachment-induced apoptosis, cells were trypsinized, washed with DMEM-10% FBS, and placed in ultra low-attachment tissue culture plates (Corning Inc., Corning, NY). Typically, cells were suspended at 1 × 105 cells per 2 ml medium. Plates were then placed in tissue culture incubators and cells harvested at various times postplating, as indicated.
DNA fragmentation assay.
Cells were assayed for apoptosis using the cell death detection enzyme-linked immunosorbent assay (ELISA) kit (Roche, Germany) according to the manufacturer's instructions. Briefly, 1 × 105 cells were plated on either normal tissue culture dishes (attached) or ultra low-attachment dishes (suspended) and incubated at 37°C for 18 h. Attached cells were scraped from the plates, while cells plated in the low-attachment dishes were collected by pipetting. Both samples were then pelleted, lysed, and incubated for 2 h with the provided anti-histone biotin-anti-DNA peroxidase immunoreagent in strepavidin-coated microtiter plates. The wells were washed, and fragmented DNA was detected using a colorimetric substrate measured at 405 nm on a Victor Wallac plate reader (PerkinElmer, Waltham, MA). All samples were processed in triplicate and data averaged. Readings were normalized as relative DNA fragmentation levels using the maximum values observed as 100%.
Caspase activity assay.
The caspase profiling assay (BD Biosciences, Mountain View, CA) was performed according to the manufacturer's instructions. Cells (2 ×105) were plated in 4 ml on either normal tissue culture dishes (attached) or ultra low-attachment dishes (suspended) and incubated at 37°C. At various times after plating, attached cells were scraped from the plates, while cells plated in the low-attachment dishes were collected by pipetting. Both samples were then pelleted, lysed, and incubated in the provided reaction buffer with caspase-specific, fluorescently labeled substrates. The reaction mixture was incubated at 37°C for 2 h and then measured on a Victor Wallac reader with excitation at 355 nm and emission at 460 nm. When indicated, lysates were incubated with caspase-specific inhibitors included in the kit. All samples were processed in duplicate and data averaged. Readings were normalized as relative caspase activity using values observed for attached cells as 1.
Immunoprecipitation and immunoblotting.
Cells were lysed in FLAG lysis buffer (1% Triton X-100, 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 0.2 mM phenylmethylsulfonyl fluoride, and protease inhibitors) at 4°C for 20 min. Insoluble proteins were pelleted by centrifugation at 15,000 × g for 10 min at 4°C, and soluble lysates were incubated with anti-FLAG M2 agarose (Sigma) for 1 h at 4°C. Beads were washed four times with FLAG lysis buffer and bound proteins eluted with 0.1 M glycine, pH 2.5, at room temperature for 10 min. Samples were then adjusted back to a neutral pH and boiled in sodium dodecyl sulfate sample buffer. Samples were separated on NuPAGE 4 to 12% Bis-Tris gels (Invitrogen, Carlsbad, CA) run in 1× morpholineethanesulfonic acid buffer. After polyacrylamide gel electrophoresis, proteins were transferred to polyvinylidene difluoride membranes and dried overnight at 4°C. The membrane was then rehydrated and probed with 1:500 anti-HA-horseradish peroxidase (Roche) to detect tagged E7 protein and visualized by development in Western Lightning chemiluminescence reagent (PerkinElmer, Waltham, MA). p600 was detected with 1:2000 rabbit polyclonal anti-p600, clone 5F7 (20), and 1:2000 anti-rabbit Alexa Fluor 680, visualized on a LI-COR imaging machine.
Anchorage-independent growth assays.
Thirty-five-millimeter dishes were prepared with a bottom layer of 0.5% Noble agar (Difco, Detroit, MI), DMEM, and 10% FBS and a top layer of 0.3% Noble agar, DMEM, and 10% FBS seeded with cells (either 5 × 102 or 1 × 103 cells). Plates were incubated at 37°C and 5% CO2, and colony formation was quantified after 14 days. All samples were plated in duplicate, and colony numbers were averaged.
RESULTS
C127 cells are anoikis sensitive.
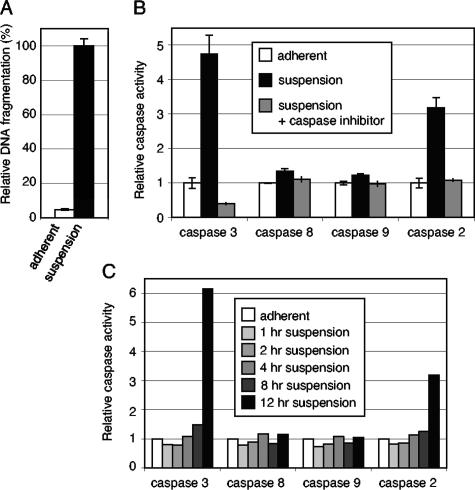
Cellular transformation studies with BPV-1 have often utilized murine C127 cells in a focus assay first described by Dvoretzky et al. (6). In order to investigate whether the ability of BPV-1 E7 to enhance the transformation phenotype might be due to an ability to inhibit anoikis, we examined whether C127 cells were anoikis sensitive. To do so, we plated equivalent numbers of C127 cells on two different substrates: normal tissue culture dishes or low-attachment tissue culture dishes, the surfaces of which are coated with a hydrogel layer that is neutrally charged, thus inhibiting cell attachment. As expected, C127 cells plated on normal tissue culture dishes attached readily and formed monolayers. In contrast, C127 cells plated on low-attachment plates did not attach to the plate and instead remained free floating in the medium. Eighteen hours postplating, we determined the apoptotic status of the cells by measuring the levels of DNA fragmentation using an ELISA-based colorimetric assay. C127 cells plated on the low-attachment dishes consistently had an approximately 20-fold-higher level of DNA fragmentation than cells plated on normal tissue culture dishes (Fig. 1A). These results are similar to results reported for other immortalized, nontransformed cell lines, such as MDCK and SCP-2, which undergo apoptosis upon loss of cell attachment (1, 7).
FIG. 1.
C127 cells are anoikis sensitive and induce caspase-2 and caspase-3 activity upon loss of cell attachment. (A) C127 cells were plated in triplicate on either normal tissue culture dishes (adherent) or low-attachment dishes (suspended) for 18 h. Cells were then harvested, and apoptosis was measured via DNA fragmentation using an ELISA-based colorimetric assay as described in Materials and Methods. Results were normalized to C127 cells in suspension. (B) C127 cells were plated in duplicate on either normal tissue culture dishes (adherent) or low-attachment dishes (suspended) for 18 h. Cells were then harvested, and apoptosis was measured via caspase activation using a fluorimetric assay as described in Materials and Methods. Results were normalized to the amount of caspase activity measured in adherent cells. The gray bars represent caspase activity after the addition of specific caspase inhibitors to cell lysates. (C) Cells were plated on either normal tissue culture dishes (adherent) or low-attachment dishes (suspended). At the indicated hours postplating, cells were harvested, and caspase activation was measured.
To examine whether the DNA fragmentation observed in C127 cells in suspension correlated with other indicators of apoptosis such as caspase activation, we utilized fluorogenic substrates specific for different caspases to analyze caspase activation. We observed a 4.5-fold induction of caspase-3 activity and a threefold induction of caspase-2 activity in C127 cells plated in suspension for 18 h, relative to attached cells (Fig. 1B). There was no increase of caspase-8 activity or caspase-9 activity in these cells at 18 h.
We next performed a time course analysis to characterize when the apoptotic signaling due to cell detachment was manifested by caspase-2 and caspase-3 activation. We observed no significant increase in caspase activity up to 8 h after cells were plated in suspension compared to caspase activity in cells plated on normal plates (Fig. 1C). However, at 12 h postplating, there was an increase in caspase-3 activity (sixfold) and caspase-2 activity (threefold). Again, no increase in caspase-8 activity or caspase-9 activity was observed at these times.
We concluded, from the observed increase in DNA fragmentation, the increase in caspase-2 and caspase-3 activities, and the cleavage of the PARP protein (another indicator of apoptosis) (data not shown), that C127 cells are anoikis sensitive.
Expression of BPV-1 E6 or E7 inhibits anoikis.
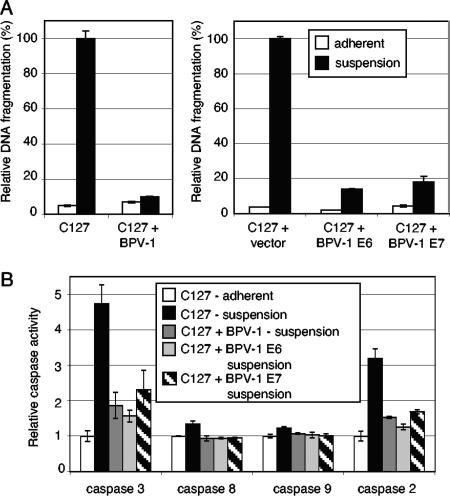
Anoikis-sensitive cells that have undergone cellular transformation typically become anoikis resistant (7). Since BPV-1-transformed C127 cells are anchorage independent, we asked whether BPV-1-transformed cells were anoikis resistant using the DNA fragmentation assay described above for normal C127 cells. As expected, BPV-1-transformed C127 cells survived when plated in suspension for 18 h with no increase in DNA fragmentation compared to attached cells (Fig. 2A, left panel), and with little increase in caspase-3 and caspase-2 activity compared to control cells (Fig. 2B). We also examined whether cells transformed by either BPV-1 E6 or BPV-1 E7 were anoikis resistant. As might be anticipated, BPV-1 E6-transformed C127 cells showed no increase in DNA fragmentation compared to attached cells (Fig. 2A, right panel) and showed little increase in caspase-3 and caspase-2 activity compared to control cells (Fig. 2B), indicating that BPV-1 E6 expression alone induces anoikis resistance. This result was not unexpected, since many transforming genes are able to inhibit anoikis.
FIG. 2.
BPV-1 E7 and BPV-1 E6 protect C127 cells from anoikis. (A) C127 cells were plated in triplicate on either normal tissue culture dishes (adherent) or low-attachment dishes (suspended) for 18 h. Cells were then harvested, and DNA fragmentation was determined as a measure of apoptosis. Samples included C127 parental cells, BPV-1-transformed C127 cells (H2), vector-transduced C127 cells, and C127 cells transduced with a retrovirus expressing BPV-1 E6 or BPV-1 E7. Results were normalized to C127 cells grown in suspension. (B) The indicated cells were plated in duplicate on either normal tissue culture dishes (adherent) or low-attachment dishes (suspended) for 18 h. Cells were harvested, and caspase activation was determined. Results were normalized to the amount of caspase activity measured in adherent C127 cells.
The question of whether or not BPV-1 E7 expression, which by itself does not transform cells, could confer anoikis resistance was less predictable. To examine this, we generated C127 cells expressing BPV-1 E7 alone using a Moloney murine leukemia virus-derived expression vector in which E7 was under the control of the retroviral long terminal repeat promoter. E7-expressing cells plated in suspension had significantly lower levels of DNA fragmentation than cells transduced with the empty retrovirus vector (Fig. 2A, right panel). There was also a smaller increase in caspase-3 and caspase-2 activity in E7-expressing cells plated in suspension than in control cells (Fig. 2B). Taken together, these data indicate that BPV-1 E7 itself has antiapoptotic activity and can inhibit apoptosis induced during cell detachment.
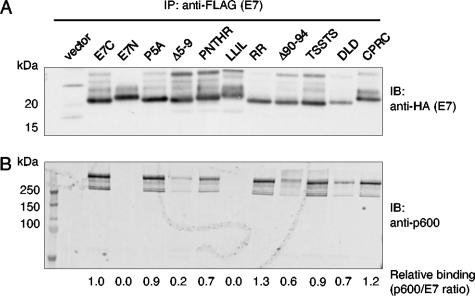
To determine the regions of E7 required for its antiapoptotic activity, we generated cell lines stably expressing various E7 mutant proteins (listed in Fig. 3 and Table 1). To guide our mutagenesis strategy, we compared the protein sequence of BPV-1 E7 with the E7 proteins encoded by other transforming members of the deltapapillomaviruses (14), including the deer papillomavirus (DPV) and the elk papillomavirus (EPV). BPV-1 E7 shares extensive homology with DPV-1 E7 and EPV-1 E7, and, like BPV-1, these viruses readily transform rodent cells in tissue culture (11, 27). Thus, we chose a number of conserved regions for targeted mutagenesis. Expression of the various E7 mutant proteins was validated by immunoprecipitation using an anti-FLAG antibody and immunoblotting using an anti-HA antibody, as all constructs contained a FLAG/HA tag at their C terminus (with the exception of E7-N, which contains a FLAG/HA tag at its N terminus) (Fig. 4A).
TABLE 1.
Summary of BPV-1 E7 constructs used in this study
| BPV-1 E7 construct | Anoikis inhibitiona | Stimulates E6 transformationb | p600 bindingc |
|---|---|---|---|
| E7 | + | + | + |
| E7-C (FLAG-HA) | + | + | + |
| E7-N (FLAG-HA) | − | − | − |
| Δ5-9 | ± | − | ± |
| P5A | − | + | + |
| Δ90-94 | + | + | + |
| PNTHR (5-9) AAAAA | + | + | + |
| LLIL (19-22) AAAA | − | − | − |
| RR (73-74) AA | − | + | + |
| TSSTS (95-99) AAAAA | + | + | + |
| DLD (110-112) AAA | + | + | + |
| CPRC (115-118) APRA | + | + | + |
The ability of E7 protein to inhibit anoikis was assayed (shown in Fig. 5). +, 0-25% relative DNA fragmentation; ±, 26-50% relative DNA fragmentation; −, 50-100% relative DNA fragmentation.
The ability of E7 protein to stimulate anchorage-independent growth was assayed (shown in Table 2). +, greater than 30 colonies; −, less than 30 colonies.
Binding of BPV-1 E7 to p600 was assayed by coimmunoprecipitation (shown in Fig. 4). −, a binding ratio (p600/E7) of zero; ±, a binding ratio of 0.1-0.5; +, a relative binding ratio of greater than 0.5.
FIG. 4.
Expression of BPV-1 E7 mutants and interaction with p600 during anoikis induction. C127 cells stably expressing E7 constructs, or vector alone, were plated in suspension for 18 h. Cells were harvested and lysed under nondenaturing conditions, and E7 was immunoprecipitated using anti-FLAG antibody. Protein complexes were resolved on a NuPAGE 4 to 12% Bis-Tris gel and probed by immunoblotting with anti-HA antibody to detect the E7 proteins (panel A) or probed with anti-p600 antibody (panel B). All E7 constructs have a FLAG/HA epitope tag at the C terminus, with the exception of E7-N, which has a FLAG/HA epitope tag at the N terminus. Signal intensities were measured using ImageQuant software, and the ratio of the amount of p600 detected to the amount of E7 detected is shown.
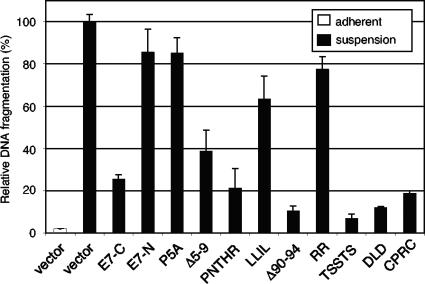
Cells expressing the mutant E7 proteins grew normally under attachment conditions and manifested a nontransformed, contact-inhibited phenotype (data not shown). We next asked which E7 constructs retained the ability to inhibit apoptosis when cells were grown in suspension by measuring DNA fragmentation. Cell lines stably expressing E7-C or E7-C PNTHR, Δ90-94, TSSTS, DLD, or CPRC grown in suspension exhibited significantly less DNA fragmentation than cells containing the empty pOZ vector (Fig. 5), indicating that these mutations did not disrupt the antiapoptotic activity of E7. In contrast, cell lines stably expressing E7-N and E7-C P5A, LLIL, and RR grown in suspension manifested high levels of DNA fragmentation, similar to C127 cells transduced with the empty pOZ vector, indicating that these mutations abrogated the antiapoptotic activity of E7. The E7 construct E7-C Δ5-9 showed an intermediate phenotype in this assay.
FIG. 5.
Mutations in BPV-1 E7 affect its ability to protect cells from anoikis. C127 cells stably expressing E7 constructs, or vector alone, were plated in triplicate on either normal tissue culture dishes (adherent) or low-attachment dishes (suspended) for 18 h. Cells were harvested, and apoptosis was measured via DNA fragmentation as described in the text. Results were normalized to C127 cells transduced with empty pOZ vector, grown in suspension.
Stimulation of anchorage independence by BPV-1 E7.
We next examined whether the ability of BPV-1 E7 to protect cells from anoikis correlated with its ability to enhance transformation by BPV-1 E6. BPV-1 E7 has previously been shown to stimulate the anchorage-independent growth of BPV-1 E6- and BPV-1 E5-transformed cells when suspended in soft agar (2, 4). The expression of E7 results in an increase in both the number and size of the colonies formed. We therefore examined whether the ability of the BPV-1 E7 mutants to stimulate anchorage independence of BPV-1 E6-expressing C127 cells correlated with their ability to induce anoikis resistance in low-attachment dishes. In agreement with previous results, expression of wild-type E7 increased the number of colonies formed in E6-transformed cells as measured by growth in soft agar over the course of 2 weeks (Table 2). Of the various E7 constructs tested, E7-C and E7-C P5A, PNTHR, Δ90-94, TSSTS, DLD, RR, and CPRC consistently stimulated the anchorage-independent growth of E6-expressing cells, whereas the expression of E7-N and E7-C Δ5-9 and LLIL failed to significantly enhance the anchorage-independent phenotype. The E7 Δ5-9 mutant had an intermediate phenotype in the anoikis DNA fragmentation assay (Fig. 5), and the P5A and RR mutants enhanced the E6 transforming activity but did not protect C127 cells from anoikis.
TABLE 2.
Anchorage-independent growth of cells expressing BPV-1 E6 and BPV-1 E7
| Cells | Number of anchorage-independent coloniesa
|
||
|---|---|---|---|
| Expt. 1 | Expt. 2 | Expt. 3 | |
| C127 | 0 | 0 | 0 |
| E6 + vector | 13 | 9 | 8 |
| E6 + E7-C (FLAG-HA) | 102 | 119 | 118 |
| E6 + E7-N (FLAG-HA) | 14 | 19 | 27 |
| E6 + Δ5-9 | 16 | 13 | 21 |
| E6 + P5A | 79 | 70 | 76 |
| E6 + Δ90-94 | 98 | 106 | 90 |
| E6 + PNTHR (5-9) AAAAA | 90 | 91 | 101 |
| E6 + LLIL (19-22) AAAA | 20 | 9 | 22 |
| E6 + RR (73-74) AA | 81 | 86 | 79 |
| E6 + TSSTS (95-99) AAAAA | 88 | 92 | 80 |
| E6 + DLD (110-112) AAA | 98 | 90 | 92 |
| E6 + CPRC (115-118) APRA | 80 | 76 | 76 |
Cells were assayed for anchorage-independent growth with transduced BPV-1 oncogenes: C127 control cells, cells expressing BPV-1 E6 alone, and cells expressing BPV-1 E6 and BPV-1 E7. Cells (5 × 102 cells) were suspended in 0.3% Noble agar, DMEM, and 10% FBS and grown for 14 days. Cells were plated in duplicate, and the number of anchorage-independent colonies were counted after 14 days and averaged. Results are shown from three separate experiments.
Interaction of BPV-1 E7 and p600 during anoikis induction.
We have previously identified the cellular p600 as an E7-interacting partner for BPV-1 E7 (4). This E7-p600 interaction is not limited to BPV, since HPV6 and HPV16 E7 proteins can also complex with p600 (15). In our previous studies, we found a correlation between the ability of various BPV-1 E7 mutants and constructs to bind p600 and to stimulate anchorage independence. Therefore, we next performed coimmunoprecipitation experiments with the additional E7 mutants generated in this study to determine whether the ability to inhibit anoikis correlated with an ability to bind p600.
C127 cell lines expressing E7 were plated on low-attachment plates for 18 h, harvested, and assayed for the E7-p600 binding by immunoprecipitation for E7 followed by immunoblotting for p600 (Fig. 4B). As previously shown, the E7-C-tagged protein binds p600, and the E7-N-tagged protein does not (4). Most of the E7 mutants retained an ability to interact with p600. The amount of p600 immunoprecipitated with E7-C Δ5-9 or LLIL was significantly less than that observed for the other E7 mutant proteins. It is interesting to note that the constructs E7-N and E7-C LLIL were each deficient in their abilities to inhibit anoikis, to stimulate transformation, and to interact with p600. The Δ5-9 mutant that displayed lower binding to p600 also had an intermediate phenotype in being able to protect C127 cells from anoikis. The P5A and RR mutants, however, bound to p600 but did not protect C127 cells from anoikis (Table 1 and Fig. 5).
Taken together, these data show a good correlation between the ability of BPV-1 E7 mutants to bind p600 strongly and to enhance anchorage independence of BPV-1 E6-expressing cells. The E7 mutants that were deficient in p600 binding were unable to protect cells from anoikis, and the Δ5-9 mutant that had reduced binding to p600 had an intermediate phenotype in providing anoikis resistance. However, some of the mutants (E7-C P5A and RR) that retained p600 binding were unable to protect C127 cells from anoikis as assayed by DNA fragmentation. These data are consistent with a role for p600 binding being necessary but not sufficient for E7 to inhibit anoikis.
DISCUSSION
In this study, we have identified a novel activity for BPV-1 E7, namely, the inhibition of anoikis. BPV-1-transformed C127 cells are resistant to suspension-induced apoptosis and, importantly, expression of E7 by itself can elicit this phenotype. Resistance to apoptosis in these cells was accompanied by decreased activity of the apoptotic-specific caspase-3 and -2. Furthermore, interaction between E7 and the previously identified cellular protein, p600, may be important for this effect, since E7 mutants deficient in p600 did not provide C127 cells resistance to anoikis. However, several E7 mutants that retained the ability to bind p600 were unable to protect C127 cells from anoikis, consistent with a model in which E7 binding to p600 might be necessary but not sufficient for protection of cells from anoikis.
Anchorage dependence is a property of nontumorigenic cells that is required for regulated cell growth, proliferation, migration, and differentiation. Proper cell attachment is critical for these events, and failure of cells to correctly attach impairs these processes and results in a specific type of apoptosis termed anoikis (1, 12). These anchorage-dependent functions are achieved by the process of cell adhesion, which is the physical interaction of integrin receptors with substrates of the extracellular matrix. Integrin engagement leads to the activation of signal transduction cascades that regulate cell growth, proliferation, differentiation, and migration. One such pathway, which is activated by engagement of β1 or β3 integrins, is the phosphatidyl inositol-protein kinase B/Akt pathway (PI3K-PKB/Akt). Activation of this pathway represents a potent survival signal, and cell detachment results in a cessation of this signal, leading to anoikis. Elevated PKB/Akt activity has been documented in many cancer cell lines and primary tumors and may represent the predominant mechanism of resistance to anoikis (9). Preliminary data in our laboratory suggest that the PI3K-PKB/Akt pathway may be so activated in BPV-1-transformed cells.
We have shown previously that BPV-1 E7 binding to p600 correlates with its transformation phenotype and that mutants of E7 unable to bind p600 were defective for their ability to enhance BPV-1 E6-dependent anchorage independence (4). This is in general agreement with the findings presented in the current study, in which we have extended the correlation to a series of additional E7 mutants. Furthermore, we have examined whether E7 binding to p600 was also important for anoikis resistance, since both anchorage-independent colony growth and resistance to anoikis are tumorigenic events that are somewhat related. We found that E7 mutant proteins unable to bind p600 were also unable to protect cells from anoikis. However, several E7 mutants that were able to bind p600 could not inhibit anoikis, although they were still capable of enhancing the transforming phenotype in E6-expressing C127 cells.
A number of reports on p600 suggest it may have a variety of cellular functions. A role for p600 in regulating cell adhesion signaling was initially suggested by Nakatani and colleagues, who observed decreased activity of the focal adhesion kinase in adherent human cells in which p600 protein expression was suppressed by small interfering RNA (20). However, since cell adhesion assays and integrin blocking experiments were not performed in their analysis, a direct role for p600 in regulating integrin function was not directly demonstrated. In another study, murine p600, also known as UBR4 or Zubr1 (NCBI GeneID, 69116), was identified in an affinity assay for proteins that bind to the destabilizing N-terminal residues responsible for targeting proteins for ubiquitin-mediated proteolysis via the N-end rule pathway (28). Indeed, p600/UBR4 is a member of the UBR family, a group of mammalian E3 ubiquitin ligases that contain a zinc finger-like domain termed the UBR box involved in the recognition of N-degrons. Bona fide cellular substrates of p600/UBR4 have yet to be identified. Human p600 (also known as ZURB1; NCBI GeneID, 23352) and the Drosophila homolog (Calossin/CalO/pushover) have been reported to bind calmodulin (20, 34), suggesting a role in calcium-mediated signaling pathways. As p600 and its homologs are enormous (with a length of over 5,000 amino acids), p600 is likely to play multiple and complex roles within the cell and could very well function as a scaffold.
Our finding that the interaction between E7 and p600 appears to be required for the inhibition of anoikis in suspension suggests a role for p600 in regulating cell survival, possibly through its proposed role in cell adhesion signaling. Indeed, in DNA microarray profiling experiments, p600 has been shown to be upregulated after transformation of mouse embryonic fibroblasts, as well as upregulated in metastatic tumor cell lines (33, 18). These data suggest that p600 may play a role in cancer progression and metastasis, possibly by promoting cell survival after transformation and loss of attachment. Our results would support a hypothesis that p600 promotes cell survival during transformation, which may be exploited by E7 during papillomavirus-mediated transformation in order to enhance resistance to anoikis. The papillomavirus E7 proteins could prove to be useful tools in further analyzing and dissecting these functions of p600.
While we have shown that E7 expression in C127 cells does inhibit cell detachment-induced apoptosis in tissue culture, the relevance of this antiapoptotic activity in the viral life cycle requires further elucidation. In previous experiments, viral genomes in which the E7 ORF has been disrupted have no defect in transformation a determined using the soft-agar assay (22). Since these E7 knockout mutants are not defective in anchorage independence, other viral genes must also be able to protect cells from this type of apoptosis. Indeed, as we have shown in this report, E6 also induces anoikis resistance, indicating that this E7 function is redundant with that of E6. It is possible that the antiapoptotic activities of E7 and E6 might manifest themselves at different stages in the life cycle of the virus, possibly through different molecular mechanisms or at different stages of cell differentiation.
Acknowledgments
This work has been supported by a grant from the National Cancer Institute to P.M.H. (PO1 CA050661) and an American Cancer Society fellowship to J.D. (PF-0414701).
Footnotes
Published ahead of print on 27 June 2007.
REFERENCES
- 1.Attwell, S., C. Roskelley, and S. Dedhar. 2000. The integrin-linked kinase (ILK) suppresses anoikis. Oncogene 19:3811-3815. [DOI] [PubMed] [Google Scholar]
- 2.Bohl, J., B. Hull, and S. B. Vande Pol. 2001. Cooperative transformation and coexpression of bovine papillomavirus type 1 E5 and E7 proteins. J. Virol. 75:513-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, J. J., C. E. Reid, V. Band, and E. J. Androphy. 1995. Interaction of papillomavirus E6 oncoproteins with a putative calcium-binding protein. Science 269:529-531. [DOI] [PubMed] [Google Scholar]
- 4.DeMasi, J., K. W. Huh, Y. Nakatani, K. Munger, and P. M. Howley. 2005. Bovine papillomavirus E7 transformation function correlates with cellular p600 protein binding. Proc. Natl. Acad. Sci. USA 102:11486-11491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DiMaio, D., D. Guralski, and J. T. Schiller. 1986. Translation of open reading frame E5 of bovine papillomavirus is required for its transforming activity. Proc. Natl. Acad. Sci. USA 83:1797-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dvoretzky, I., R. Shober, S. K. Chattopadhyay, and D. R. Lowy. 1980. A quantitative in vitro focus assay for bovine papilloma virus. Virology 103:369-375. [DOI] [PubMed] [Google Scholar]
- 7.Frisch, S. M., and H. Francis. 1994. Disruption of epithelial cell-matrix interactions induces apoptosis. J. Cell Biol. 124:619-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frisch, S. M., and E. Ruoslahti. 1997. Integrins and anoikis. Curr. Opin. Cell Biol. 9:701-706. [DOI] [PubMed] [Google Scholar]
- 9.Frisch, S. M., and R. A. Screaton. 2001. Anoikis mechanisms. Curr. Opin. Cell Biol. 13:555-562. [DOI] [PubMed] [Google Scholar]
- 10.Goldstein, D. J., M. E. Finbow, T. Andersson, P. McLean, K. Smith, V. Bubb, and R. Schlegel. 1991. Bovine papillomavirus E5 oncoprotein binds to the 16K component of the vacuolar H+-ATPase. Nature 352:347-349. [DOI] [PubMed] [Google Scholar]
- 11.Groff, D. E., J. P. Sundberg, and W. D. Lancaster. 1983. Extrachromosomal deer fibromavirus DNA in deer fibromas and virus-transformed mouse cells. Virology 131:546-550. [DOI] [PubMed] [Google Scholar]
- 12.Grossmann, J. 2002. Molecular mechanisms of “detachment-induced apoptosis-anoikis.” Apoptosis 7:247-260. [DOI] [PubMed] [Google Scholar]
- 13.Horwitz, B. H., A. L. Burkardt, R. Schlegel, and D. DiMaio. 1988. The 44-amino-acid E5 transforming protein of bovine papillomavirus requires a hydrophobic core and specific carboxyl-terminal amino acids. Mol. Cell. Biol. 8:4071-4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howley, P. M., and D. R. Lowy. 2007. Papillomaviruses, p. 2299-2354. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed. Lippincott, Williams and Wilkins, Philadelphia, PA.
- 15.Huh, K. W., J. DeMasi, H. Ogawa, Y. Nakatani, P. M. Howley, and K. Munger. 2005. Association of the human papillomavirus type 16 E7 oncoprotein with the 600-kDa retinoblastoma protein-associated factor, p600. Proc. Natl. Acad. Sci. USA 102:11492-11497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lowy, D. R., I. Dvoretzky, R. Shober, M. F. Law, L. Engel, and P. M. Howley. 1980. In vitro tumorigenic transformation by a defined subgenomic fragment of bovine papillomavirus DNA. Nature 287:72-74. [DOI] [PubMed] [Google Scholar]
- 17.Marchetti, B., G. H. Ashrafi, E. Tsirimonaki, P. M. O'Brien, and M. S. Campo. 2002. The bovine papillomavirus oncoprotein E5 retains MHC class I molecules in the Golgi apparatus and prevents their transport to the cell surface. Oncogene 21:7808-7816. [DOI] [PubMed] [Google Scholar]
- 18.Montel, V., T. Y. Huang, E. Mose, K. Pestonjamasp, and D. Tarin. 2005. Expression profiling of primary tumors and matched lymphatic and lung metastases in a xenogeneic breast cancer model. Am. J. Pathol. 166:1565-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Münger, K., B. A. Werness, N. Dyson, W. C. Phelps, and P. M. Howley. 1989. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 8:4099-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakatani, Y., H. Konishi, A. Vassilev, H. Kurooka, K. Ishiguro, J. Sawada, T. Ikura, S. J. Korsmeyer, J. Qin, and A. M. Herlitz. 2005. p600, a unique protein required for membrane morphogenesis and cell survival. Proc. Natl. Acad. Sci. USA 102:15093-15098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakatani, Y., and V. Ogryzko. 2003. Immunoaffinity purification of mammalian protein complexes. Methods Enzymol. 370:430-444. [DOI] [PubMed] [Google Scholar]
- 22.Neary, K., and D. DiMaio. 1989. Open reading frames E6 and E7 of bovine papillomavirus type 1 are both required for full transformation of mouse C127 cells. J. Virol. 63:259-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petti, L., and D. DiMaio. 1992. Stable association between the bovine papillomavirus E5 transforming protein and activated platelet-derived growth factor receptor in transformed mouse cells. Proc. Natl. Acad. Sci. USA 89:6736-6740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petti, L., L. Nilson, and D. DiMaio. 1991. Activation of the platelet-derived growth factor receptor by the bovine papillomavirus E5 protein. EMBO J. 10:845-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sarver, N., M. S. Rabson, Y. C. Yang, J. C. Byrne, and P. M. Howley. 1984. Localization and analysis of bovine papillomavirus type 1 transforming functions. J. Virol. 52:377-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schiller, J. T., W. C. Vass, K. H. Vousdan, and D. R. Lowy. 1986. The E5 open reading frame of bovine papillomavirus type 1 encodes a transforming gene. J. Virol. 57:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stenlund, A., J. Moreno-Lopez, H. Ahola, and U. Pettersson. 1983. European elk papillomavirus: characterization of the genome, induction of tumors in animals, and transformation in vitro. J. Virol. 48:370-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tasaki, T., L. C. Mulder, A. Iwamatsu, M. J. Lee, I. V. Davydov, A. Varshavsky, M. Muesing, and Y. T. Kwon. 2005. A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons. Mol. Cell. Biol. 25:7120-7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tong, X., W. Boll, T. Kirschhausen, and P. M. Howley. 1998. Interaction of the bovine papillomavirus E6 protein with the clathrin adaptor complex AP-1. J. Virol. 72:476-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tong, X., and P. M. Howley. 1997. The bovine papillomavirus E6 oncoprotein interacts with paxillin and disrupts the actin cytoskeleton. Proc. Natl. Acad. Sci. USA 94:4412-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tong, X., R. Salgia, J.-L. Li, J. D. Griffin, and P. M. Howley. 1997. The bovine papillomavirus E6 protein binds to the LD motif repeats of paxillin and blocks its interaction with vinculin and the focal adhesion kinase. J. Biol. Chem. 272:33373-33376. [DOI] [PubMed] [Google Scholar]
- 32.Vande Pol, S. B., M. C. Brown, and C. E. Turner. 1998. Association of bovine papillomavirus type 1 E6 oncoprotein with the focal adhesion protein paxillin through a conserved protein interaction motif. Oncogene 16:43-52. [DOI] [PubMed] [Google Scholar]
- 33.Vasseur, S., C. Malicet, E. L. Calvo, J. C. Dagorn, and J. L. Iovanna. 2005. Gene expression profiling of tumours derived from rasV12/E1A-transformed mouse embryonic fibroblasts to identify genes required for tumour development. Mol. Cancer 4:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu, X. Z., P. D. Wes, H. Chen, H. S. Li, M. Yu, S. Morgan, Y. Liu, and C. Montell. 1998. Retinal targets for calmodulin include proteins implicated in synaptic transmission. J. Biol. Chem. 273:31297-31307. [DOI] [PubMed] [Google Scholar]