Abstract
Infection of cells with flaviviruses in vitro is reduced by pretreatment with small amounts of type I interferon (IFN-α/β). Similarly, pretreatment of animals with IFN and experiments using mice defective in IFN signaling have indicated a role for IFN in controlling flavivirus disease in vivo. These data, along with findings that flavivirus-infected cells block IFN signaling, suggest that flavivirus infection can trigger an IFN response. To investigate IFN gene induction by the very first cells infected during in vivo infection with the flavivirus West Nile virus (WNV), we infected mice with high-titer preparations of WNV virus-like particles (VLPs), which initiate viral genome replication in cells but fail to spread. These studies demonstrated a brisk production of IFN in vivo, with peak levels of over 1,000 units/ml detected in sera between 8 and 24 h after inoculation by either the intraperitoneal or footpad route. The IFN response was dependent on genome replication, and WNV genomes and WNV antigen-positive cells were readily detected in the popliteal lymph nodes (pLN) of VLP-inoculated mice. High levels of IFN mRNA transcripts and functional IFN were also produced in VLP-inoculated IFN regulatory factor 3 null (IRF3−/−) mice, indicating that IFN production was independent of the IRF3 pathways to IFN gene transcription, consistent with the IFN type produced (predominantly α).
West Nile virus (WNV) is a member of the Flavivirus genus of the family Flaviviridae. This genus contains a number of arthropod-borne human pathogens, including dengue virus (DV), Japanese encephalitis virus, yellow fever virus, and tick-borne encephalitis virus (26). WNV was associated with fever and infrequent encephalitis cases in humans in Africa, the Middle East, and Europe from its discovery in 1938 through the 1990s. In 1999, WNV was first detected in New York City, and from there it rapidly spread across the United States, some regions of Canada, Mexico, and Central America. The majority of WNV infections are asymptomatic; however, a portion of infections result in West Nile fever, and a subset of infections lead to viral invasion of the central nervous system that results in encephalitis, paralysis, and meningitis, outcomes which are especially prevalent in immunocompromised and aged individuals (4).
The mechanisms by which WNV causes disease are not completely understood, but studies with a hamster model of the disease suggest that following a brief peripheral replication cycle, the virus crosses the blood-brain barrier, where it infects neurons, causing cell death (47). The direct effect of WNV on neurons or an immune response to their infection may also be responsible for encephalitis and meningoencephalitis in humans (13). Pretreatment of animals with type I interferons (IFN-α/IFN-β) has been shown to block flavivirus disease (3, 23), and animals defective in the IFN response have been shown to be more susceptible to flavivirus infection (19, 28, 39). Although the precise IFN-stimulated genes (ISGs) that are responsible for preventing or controlling flavivirus infections are unknown, a number of studies have demonstrated that flavivirus-infected cells prevent IFN-induced phosphorylation of STAT molecules, which carry the signal from the ligated IFN receptor to the nucleus to activate the transcription of ISGs (1, 11, 20, 24, 25, 32, 37, 44). Interestingly, the precise mechanism by which flaviviruses alter STAT phosphorylation appears to differ among members of the genus (1, 24, 32).
IFN is produced by most eukaryotic cells in response to viral infection and/or recognition of virus-associated macromolecules (known as pathogen-associated molecular patterns [PAMPs]), such as single- or double-stranded RNA (ssRNA and dsRNA, respectively). In many mammalian cell types, ligation of Toll-like receptor 3 (TLR3) to extracellular dsRNA or recognition of intracellular dsRNA by the intracellular helicase mda5 or RIG-I induces the phosphorylation and activation of the constitutively expressed IFN regulatory factor 3 (IRF3), leading to nuclear translocation and activation of transcription of the genes for IFN-β and IFN-α subtype 4 (14). Recent studies have indicated that in cell cultures, a subset of viruses appear to activate the mda5 pathway, whereas others, including the flavivirus Japanese encephalitis virus, activate the RIG-I pathway (21). In some cases, IFN-α expression has been linked to protein kinase R activation via binding of intracellular dsRNA (9). Furthermore, multiple IFN-α subtypes can also be induced by PAMP-stimulated signal transduction pathways that lead to the phosphorylation of IRF7, which can be triggered by binding of ssRNA to TLR7/8. Interestingly, IRF7 is constitutively expressed in only a subset of cells, but the IRF7 gene is an ISG, so following IFN binding, many cells can produce IRF7, permitting them to amplify the IFN signal if stimulated by ssRNA (12).
For many infections, a subset of cells known as plasmacytoid dendritic cells (pDCs), which express IRF7 constitutively, have been implicated as key IFN producers (16). These cells can produce extremely high levels of IFN in response to stimulation with infectious disease agents or components thereof. Activated pDCs also produce other cytokines, notably interleukin-12 (IL-12), tumor necrosis factor alpha, granulocyte-macrophage colony-stimulating factor, and IL-3, and chemokines, such as CCL3, CCL4, CCL5, CCL22, CCL19, and CXCL13 (2, 5, 33). However, pDCs are not the only cell type that express IRF7 constitutively. Other lymphocytes, including monocytes, B cells, and dendritic cell precursors (pDC2) (18), also express IRF7 in the resting state and are hence able to induce synthesis of IFN-α through the IRF7 pathway (reviewed in reference 22).
Virus-like particles (VLPs) have been used as tools to study RNA virus infection in vitro and in vivo. VLPs consist of subgenomic replicating genomes lacking structural protein genes (replicons) that have been encapsidated by the missing structural proteins provided in trans by packaging cells. VLPs are thus able to infect cells and initiate genome replication in a manner that mimics that of normal virus, but unlike infections with normal virus, VLP infections cannot spread in the absence of trans-expressed structural proteins. In the case of both alphaviruses (35) and flaviviruses (43), VLPs have been used to identify the first cells that are infected in insect vectors of these viruses. Furthermore, for the alphavirus Venezuelan equine encephalitis virus, VLPs have been used to identify the first cells that are infected in animal models (29), and recent studies with Venezuelan equine encephalitis virus VLPs have shown that these VLPs (referred to as VRPs in the previous study) can strongly stimulate antiviral responses (46).
In the studies described in this paper, we used WNV VLPs to study the early events in WNV infection in mice, demonstrating that WNV VLPs induce a rapid IFN response, resulting in very high levels of IFN-α in the serum between 8 and 24 h after either intraperitoneal (i.p.) inoculation or subcutaneous inoculation in the footpad (f.p. inoculation). Since VLPs can undergo only a single round of infection, these studies have demonstrated that the very first cells infected in an animal are capable of stimulating the production of high levels of IFN. IFN production was dependent on the replicative capacity of VLPs, since mice inoculated with UV-inactivated VLPs did not produce IFN. Immunohistochemical (IHC) detection of WNV antigen-positive cells in the popliteal lymph nodes (pLN) after f.p. inoculation with VLPs and the demonstration of high levels of IFN mRNA in pLN collected from VLP-inoculated animals implicate this lymphoid organ as a prominent source of IFN production. Finally, the finding that IRF3−/− animals produced levels of IFN similar to those of wild-type animals suggests that neither the RIG-I/mda5 nor the TLR3 pathway, known to be important for IFN-β induction, is important for this early response to WNV infection.
MATERIALS AND METHODS
Cell lines.
Baby hamster kidney (BHK) and Vero cell lines have been described previously (37). The mouse embryo fibroblast (MEF) cell line was obtained from I. Frolov (University of Texas Medical Branch [UTMB]).
WNV and WNV VLP replicons.
A genetically defined WNV derived from an infectious cDNA clone developed from a 2002 human isolate (37) was used for all studies. WNV replicon (WNR) C-NS1-5 (carrying the 5′ untranslated region [UTR], a complete C coding region, the entire nonstructural protein-encoding region, and the 3′ UTR) was produced from WNR NS1-5 (37) by the addition of the complete C-encoding region (R. Fayzulin and P. W. Mason, unpublished data). WNR C-Luc2A NS1-5, carrying the firefly luciferase reporter gene, has been described previously (10). VLPs were produced by electroporation of BHK VEErep/C*-E/Pac packaging cells (10) with in vitro transcripts of WNR C-NS1-5 (VLPs used for all animal studies were derived from a single electroporation with this WNR) or WNR C-Luc2A NS1-5 (used for IFN bioassays [see below]). In both cases, the packaging cells were maintained in medium with 1% fetal bovine serum (FBS; a low serum concentration was used to prevent cell overgrowth) and VLP-containing supernatants were collected at various times and frozen for subsequent analysis. In some cases, VLPs were concentrated by precipitation in the cold in 10% polyethylene glycol (molecular weight, 8,000) and 0.64 M NaCl (final concentration), followed by collection by centrifugation and solubilization in tissue culture medium. VLP titers were determined by enumerating WNV antigen-positive cells detected in IHC-stained Vero cell monolayers 16 to 24 h after infection with dilutions of VLPs (37). All titers are expressed in terms of infectious units (IU) on Vero cells. VLPs were inactivated by exposure to UV light from a 4-W, 254-nm source placed at a distance of 10 cm for 1 min. This dose of UV, which yielded reductions in titer of >99.99%, was used to minimize the physical effects of UV exposure on RNA and protein structure.
Animals.
Outbred Swiss Webster mice were obtained from Harlan (Houston, TX). C57BL/6J (BL6) mice were obtained from Jackson Laboratory (Bar Harbor, ME). IRF3−/− mice (41) on a C57BL/6J background (kindly provided by T. Moran and T. Taniguchi) were maintained in a breeding colony at UTMB. All mice were housed in sterile microisolator cages under specific-pathogen-free conditions in the AAALAC-approved UTMB animal facility. All procedures were conducted in accordance with the UTMB Institutional Animal Care and Use Committee.
IFN bioassay.
IFN levels were measured using a bioassay. Briefly, samples were diluted fourfold with Dulbecco's modified Eagle's medium containing 1% FBS and 10 mM HEPES and used to treat MEF monolayers grown in 96-well black-walled plates at 37°C. After an overnight incubation, the samples were aspirated from the monolayers and 50 μl of VLPs containing WNR C-luc-NS1-5 expressing firefly luciferase (Luc) was added to each well. Twenty-four hours after VLP infection, cells were lysed by the addition of lysis buffer containing d-luciferin and ATP (SteadyGlo; Promega), and after a 5-minute incubation, the plates were read on a TR717 microplate luminometer (Applied Biosystems). Percent inhibition was determined by normalizing the value (photons/s) for each sample to that for a sample that had been pretreated with Dulbecco's modified Eagle's medium containing only FBS and HEPES. All bioassay data were standardized by side-by-side comparisons to a murine IFN-β (an NIH standard produced from L cells obtained from the NIAID Reference Reagent Repository, which is operated by Braton Biotech, Gaithersburg, MD) or a commercial murine IFN-β (Sigma) calibrated to the NIAID standard. Using the amount of the standard required to inhibit Luc activity by 50% (between 1.5 and 5 U/ml of the NIAID IFN-β standard), the amount of IFN in each sample was determined by interpolation to the sample dilution yielding a 50% inhibition of Luc activity.
IFN ELISA.
IFN-α and -β were measured in serum samples by an enzyme-linked immunosorbent assay (ELISA), using a commercial murine IFN ELISA kit (PBL Biomedical Laboratories, New Brunswick, NJ). Sera were diluted 5- to 10-fold before analysis to overcome nonspecific interference with the assay. At these dilutions, normal sera produced optical densities slightly lower than those obtained with sample diluent, so standard curves used to quantitate IFN levels were generated by adding IFN standards to normal sera at the same concentrations used in the test samples (20% for IFN-β ELISA and 10% for IFN-α ELISA). The limits of detection with these assays were 100 pg/ml for IFN-β in sera (due to the 5-fold dilution factor and assay sensitivity) and 100 pg/ml for IFN-α in sera (due to the 10-fold dilution factor and assay sensitivity).
Bead protein array analysis of cytokines and chemokines in mouse sera.
Serum samples were analyzed for cytokines and chemokines by use of a Bio-Plex bead array (Bio-Rad, Hercules, CA) according to the manufacturer's recommendations. Samples were analyzed in parallel to known cytokine reference standards (2 to 5,000 pg/ml) provided by the manufacturer. A multiplex premixed panel of 23 mouse cytokines and chemokines was analyzed in all samples.
Animal inoculations.
Mice were inoculated with WNV or WNV VLPs by i.p. injection or by subcutaneous rear f.p. injection. For intermediate points in time course studies, animals were anesthetized and sera were collected by retro-orbital bleeds. At the end of studies, the mice were sacrificed and sera were collected postmortem by cardiac puncture. In some studies, peritoneal lavage samples were collected and pLN were harvested following sacrifice.
Real-time RT-PCR analysis.
BL6 and IRF3−/− mice were inoculated with 1 × 107 IU of WNV VLPs by either the subcutaneous rear left f.p. route in a volume of 17 μl or the i.p. route in a volume of 100 μl or were mock inoculated with medium by both routes. Twenty-four hours after inoculation, the rear left pLN were harvested from all animals and stored overnight in RNAlater (Ambion). The next day, total RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer's specifications. Extracts were DNase I (Ambion) treated to remove contaminating genomic DNA, precipitated, and resuspended in nuclease-free water. Reverse transcription (RT) was carried out with an ImProm II RT kit (Promega), with random hexamers as primers. Real-time PCR analysis was carried out using an iQ SYBR green supermix kit (Bio-Rad), using the primers listed in Table 1. Reaction mixes were set up in 96-well PCR plates (Eppendorf). Amplifications were carried out for 50 cycles, followed by a melting curve analysis of resulting products to confirm the specificity of the reactions.
TABLE 1.
Primers for real-time analysis
| Target | Primer sequence (5′-3′)
|
Product size (bp) | Reference | |
|---|---|---|---|---|
| Forward | Reverse | |||
| GAPDH | ATGTCAGATCCACAACGGATACAT | ACTCCCTCAAGATTGTCAGCAAT | 307 | 42 |
| IFN-β | GGAGATGACGGAGAAGATGC | CCCAGTGCTGGAGAAATTGT | 107 | 45 |
| IFN-α | ATGGCTAGGCYCTGTGCTTTC | TCTGAYCACCTCCCAGGCACA | 492 | 8 |
| IRF7 | CAGCGAGTGCTGTTTGGAGAC | AAGTTCGTACACCTTATGCGG | 351 | 7 |
| MCP-1 | ATCCCAATGAGTAGGCTGGAGAGC | GGTGGTTGTGGAAAAGGTAGTGG | 279 | 34 |
| WNV NS5 | CGGTCGGAAAAGTGATTGACC | GCCCTTTGTGTACCCTCTGACTTC | 100 | This study |
The following cells and treatments were used for the preparation of PCR standards: for IRF7, macrophage chemoattractant protein 1 (MCP-1), IFN-α, and IFN-β, Raw 264.7 mouse macrophages infected with Sendai virus for 16 h; for the WNV genome, Vero cells infected with WNR NS1-5 VLPs; and for glyceraldehyde-3-phosphate dehydrogenase (GAPDH), unstimulated Raw 264.7 cells. Total RNA was isolated from the cells, and 300- to 600-bp fragments of the gene(s) of interest were amplified by RT-PCR using appropriate primer sets. PCR fragments were gel purified and quantitated, and the copy number was calculated. Serial 10-fold dilutions were then prepared for use as templates to create standard curves by real-time PCRs. All samples were normalized to GAPDH. To control for plate-to-plate variation, GAPDH reactions were run for all samples on the same plate as the respective real-time PCR for the gene of interest. All data are expressed as the copy number of the gene of interest per 106 copies of GAPDH.
IHC staining of pLN.
pLN collected from mice inoculated in the f.p. with WNV VLPs or from control animals as described above were snap frozen in OCT medium in cryomolds. Five-micrometer-thick cryostat sections were prepared, mounted on Superfrost Plus slides, and stored at −80°C until needed. Before being stained, the slides were brought to room temperature, fixed in ice-cold acetone for 5 min, and air dried for 30 min. After being washed in phosphate-buffered saline, slides were treated with 3% hydrogen peroxide for 10 min. Following sequential 15-min incubations with 0.1% avidin and 0.01% biotin (Vector Laboratories, Inc., Burlingame, CA) to block endogenous reactivity, slides were incubated in blocking solution (Histomouse-SP kit; Zymed Laboratories Inc., South San Francisco, CA) for 30 min, and then a biotinylated anti-WNV immunoglobulin G (IgG) fraction prepared by biotin-N-hydroxysuccinimide conjugation to protein A-purified IgG from hyperimmune mouse ascitic fluid was applied to the sections for 60 min at 37°C. Following washing, the bound biotin was detected using a Histomouse-SP kit according to the manufacturer's instructions.
RESULTS
Mice produce high titers of IFN following inoculation with WNV VLPs.
In initial studies, we showed that i.p. inoculation of BL6 mice with a high titer of WNV VLPs elicited a robust IFN response at 24 h postinoculation, as determined in a bioassay standardized with an NIH standard (Fig. 1). IFN was present in these animals at similar concentrations in both the serum and peritoneal fluid, and IFN was undetectable in both of these fluids in animals inoculated with medium alone (Fig. 1A and B). The similarity of IFN levels in these two compartments led us to conclude that the cytokine was freely moving between these compartments, and hence, subsequent data are only reported from serum bioassays, which have an enhanced level of sensitivity since the serum samples are not diluted at the time of collection, as is necessary for peritoneal lavage samples. Interestingly, IFN levels were lower at 48 h postinoculation, indicating a transient response to the VLP treatment. Figure 1C, which was obtained for a second experiment, shows that large amounts of IFN were detectable in as little as 8 h and that high levels of IFN were also detectable when animals were inoculated with a lower dose of VLPs. In addition, Fig. 1C shows that infectious VLPs were needed to elicit IFN production, since animals inoculated with UV-inactivated VLPs did not produce detectable levels of IFN.
FIG. 1.
Detection of IFN in sera and peritoneal fluids from mice inoculated with WNV VLPs. Pairs of BL6 mice were injected i.p. with the indicated doses (in IU) of WNV VLPs. Serum and peritoneal lavage samples were collected at the indicated times postinoculation and tested for IFN activity by a bioassay. (A) IFN levels detected in sera 24 and 48 h after inoculation. (B) IFN levels detected in the peritoneal fluids from the same animals. (C) IFN levels detected in serum samples from a second experiment targeting earlier times of collection, with demonstration that UV inactivation (4 W at 254 nm for 1 min at 10 cm, yielding an inactivation rate of >99.9985%) prevented IFN production. (D) IFN levels detected in serum samples collected at 24 h post-i.p. inoculation with the indicated amounts of VLPs, determined by an IFN-α ELISA (open bars) or an IFN activity bioassay (closed bars). Graphs in panels A, B, and C are truncated at the level of detection for the individual assays, and the filled and open bars in these panels represent data obtained for two mice. <, the values displayed were at the limits of detection of these assays (D).
To learn more about the IFN response to VLP injection, groups of five mice were inoculated with sequentially lower doses of VLPs by the i.p. route. Sera from these mice were collected at 24 h postinoculation and tested for IFN levels by bioassay and an IFN-α ELISA (Fig. 1D). The data in Fig. 1D demonstrate some variability in the responses of individual animals to VLP inoculation, but they display a clear dose-dependent response and demonstrate a concordance between these assays. IFN-β ELISAs performed on sera pooled for the highest dose shown in Fig. 1D (107 VLPs) failed to detect the presence of any IFN-β. Since this assay has a limit of detection of 100 pg/ml and the same pool contained IFN-α levels of over 5,000 pg/ml, the IFN-α levels detected by ELISA were at least 50-fold greater than the levels of IFN-β detected by ELISA.
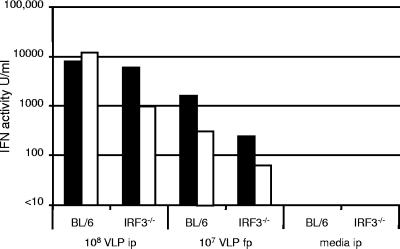
IFN production in response to WNV VLPs and WNV is independent of IRF3.
To help us to determine the mechanism by which WNV replication elicits this strong IFN response, we compared IFN production in BL6 and IRF3−/− mice. These studies showed that both BL6 and IRF3−/− animals produced a strong IFN response following VLP inoculation (Fig. 2). In addition, high levels of IFN were also detected following f.p. inoculation, indicating that multiple routes of inoculation could produce high levels of IFN (Fig. 2). Interestingly, IRF3−/− mice have been reported to produce about 20 times less IFN than wild-type mice when inoculated with Newcastle disease virus (41), suggesting that the mechanism of activation of the IFN response to the negative-strand RNA virus Newcastle disease virus is substantially different from that used in response to the positive-strand RNA virus WNV. The IFN produced in WNV VLP-injected mice was completely neutralized with a high dilution (1:10,000) of a polyclonal anti-murine IFN reference serum (obtained from the NIAID Reference Reagent Repository, operated by Braton Biotech, Gaithersburg, MD) but was not inhibited by the control serum (same source), indicating that this antiviral activity was type I IFN (results not shown), consistent with the ELISA data shown in Fig. 1D.
FIG. 2.
Comparison of levels of IFN produced by BL6 and IRF3−/− mice following inoculation with WNV VLPs by different routes. Pairs of mice were injected by the i.p. or f.p. route with the indicated doses of WNV VLPs. After 24 h, sera were collected and IFN levels were determined by bioassay (filled and open bars represent the values obtained for the two mice in each group).
Since IRF3 plays a key role in IFN production in cells which have been exposed to PAMPs from multiple viruses, including WNV, we sought to further examine the phenotype of our IRF3−/− animals in response to inoculation with WNV compared to that in response to inoculation with the dsRNA mimetic poly(IC:LC) (Hiltonal; generously supplied by A. M. Salazar, Oncovir, Inc.). Table 2 shows that in these studies the poly(IC:LC) rapidly induced an IFN response that began to wane at 24 h, whereas the WNV induction was slower, peaking at 24 h before diminishing at 48 h, similar but not identical to the kinetics shown for VLP inoculation (Fig. 1A and C). Interestingly, inoculation with 107 VLPs produced more IFN than inoculation with 107 “live” WNV virions. This interesting finding could be due to a variety of factors, the most intriguing of which is the possibility that the spreading infection of the live virus could actually interfere with the innate response to infection. Interestingly, the IRF3−/− mice produced a high response to poly(IC:LC), suggesting either that this dsRNA mimetic acts through a non-TLR3 pathway or that an intermediate pathway other than IRF3 transduces the TLR3 signal for IFN synthesis.
TABLE 2.
IFN response following poly(IC:LC) or WNV inoculation of mice
| Mouse genotype | Treatmenta | Serum IFN concn (U/ml)b
|
||
|---|---|---|---|---|
| 8 h | 24 h | 48 h | ||
| IRF3−/− | Poly(IC:LC) | >28,000 | 1,300 | <10 |
| WNV | 11 | 139 | 15 | |
| WNV | 14 | 520 | 11 | |
| WNV | 39 | 821 | 21 | |
| WNV | 14 | 325 | 15 | |
| BL6 | Poly(IC:LC) | >28,000 | 2,600 | <10 |
| WNV | 20 | 433 | 15 | |
| WNV | 27 | 780 | 23 | |
| WNV | 30 | 419 | 15 | |
| WNV | 15 | 173 | 14 | |
Mice were administered 40 μg poly(IC:LC) or 1 × 107 focus-forming units of WNV per animal i.p.
Determined by bioassay.
Analyses of pLN reveal a role in IFN production and the sites of VLP replication.
To analyze the role of lymphoid tissue in the generation of the early IFN response, groups of three BL6 and IRF3−/− animals were inoculated in the left rear f.p. or inoculated i.p. with 107 IU VLPs or VLP diluent. Sera were collected at 8 h postinoculation, and at 24 h postinoculation the animals were euthanized and the sera as well as the left rear pLN were collected. As shown in Fig. 3A and B, most animals responded to the inoculations, producing readily detectable IFN in both serum samples. However, one of three IRF3−/− animals inoculated by the i.p. route did not produce detectable levels of IFN at either time point. Detection of WNV RNA by real-time RT-PCR revealed high levels of WNV RNA in the pLN of animals inoculated with VLPs by the f.p. route, very low levels of WNV RNA in the pLN of i.p. inoculated animals, and no detectable WNV RNA in the pLN of medium-inoculated mice (Fig. 3C). These findings suggest that the f.p.-inoculated VLPs (or cells infected with the VLPs) ultimately trafficked to the pLN, where they replicated, producing high levels of viral RNA. The absence of WNV genomes and the lower levels of IFN-α RNA (see below; Fig. 3D) in the pLN of mice inoculated i.p. suggest that VLPs delivered i.p. trafficked to different secondary lymphoid organs, where they likely replicated and induced IFN production. Analyses of pLN mRNAs from these animals also demonstrated a modest elevation in the level of mRNA for IFN-β (up to several hundred copies per 1 million copies of GAPDH mRNA), whereas the IFN-α level showed a remarkable elevation (in the range of 50,000 to 650,000 copies per 1 million copies of GAPDH mRNA) (Fig. 3D and E). The very high levels of IFN-α mRNA and low levels of IFN-β mRNA detected in the draining LN of f.p.-inoculated mice are consistent with the results shown in Fig. 1D that revealed high levels of IFN-α in sera of VLP-inoculated animals but no detectable IFN-β in the same sera (see above). The presence of IFN-α in the sera of the IRF3−/− animals used for the study in Fig. 3 was corroborated by performing ELISAs on 24-hour sera from two animals of each genotype injected i.p. with VLPs. These assays failed to detect IFN-β at levels above the limit of detection of the assay, whereas IFN-α levels were found to be over 7,000 pg/ml in these four samples. Despite the low levels of IFN-β mRNA in the pLN, it is intriguing that there were higher levels in the pLN of f.p.-inoculated mice, suggesting that IFN-β production at this site could contribute to the production of IFN-α at this site and the resulting systemic IFN-α response. Analyses of the mRNA levels for IRF7 in the pLN showed high responses in all inoculated animals, including the animals inoculated i.p. (Fig. 3F). These findings are consistent with the expected transcriptional activation of the IRF7 gene by the systemic IFN produced from either a local (f.p.) or distant (i.p.) route of inoculation. As expected, the IRF3−/− animal with undetectable levels of serum IFN following i.p. inoculation with the VLPs (see above; Fig. 3A and B) was the one showing the lowest level of pLN IRF7 mRNA (Fig. 3F).
FIG. 3.
Comparison of levels of serum IFN and levels of pLN mRNAs for WNV, IFN-α, IFN-β, and IRF7 following WNV VLP (or mock) inoculation into BL6 and IRF3−/− mice. (A) IFN levels, determined by bioassay, in sera recovered 8 h after inoculation by the indicated routes. (B) IFN levels, determined by bioassay, in sera recovered 24 h after inoculation by the indicated routes. (C) WNV mRNA levels determined from RNAs isolated from pLN obtained from the left rear foot of animals 24 h after inoculation by the indicated routes. (D) IFN-α mRNA levels determined from RNAs isolated from pLN obtained from the left rear foot of animals 24 h after inoculation by the indicated routes. (E) IFN-β mRNA levels determined from RNAs isolated from pLN obtained from the left rear foot of animals 24 h after inoculation by the indicated routes. (F) IRF7 mRNA levels determined from RNAs isolated from pLN obtained from the left rear foot of animals 24 h after inoculation by the indicated routes. Bars represent three individual mice of each genotype, arranged in the same order in each panel.
IHC staining of pLN from VLP-injected animals.
To further evaluate the events following inoculation of VLPs, pLN were dissected from VLP- or medium-inoculated animals at 24 h postinjection, cryopreserved, sectioned, and stained with a biotin-labeled polyclonal antibody preparation from mice hyperimmunized with WNV (see Materials and Methods). Figure 4 shows representative sections from these studies, demonstrating the presence of a large number of WNV antigen-positive cells in the cortex of the pLN from a VLP-inoculated animal and the absence of staining of a similarly prepared section from a medium-inoculated animal. These antigen-positive cells almost certainly represent cells undergoing active infection (rather than cells that have taken up viral antigen) due to the intensity of their staining and the fact that β-galactosidase (β-Gal) was also detected in a similar region of the pLN in animals inoculated with β-Gal-expressing VLPs (results not shown).
FIG. 4.
IHC staining of pLN from VLP-inoculated mice. Mice were inoculated with VLPs or medium by f.p. injection, and the pLN were dissected and processed for IHC 24 h later, as described in Materials and Methods.
Analyses of selected cytokines and chemokines demonstrate that VLP inoculation results in a broad antiviral response.
To further define the potential cellular target(s) for WNV infection, serum cytokines and chemokines were measured at two times (8 or 24 h) postinfection in mice inoculated with VLPs by two different routes (i.p. and f.p. inoculation). Table 3 shows a comparison of IFN activities in these samples as well as the levels of six type 1 proinflammatory cytokines and chemokines selected from the 23 potential analytes detected in the BioPlex assay we utilized. β-Chemokines (RANTES, macrophage inflammatory protein 1α [MIP-1α], and MCP-1) were up regulated approximately twofold and a proinflammatory cytokine (IL-12p70) was up regulated marginally relative to the levels found in medium-inoculated animals following i.p. inoculation with WNV VLPs. Modest increases in IFN-γ production were noted from VLP inoculation in mice via the i.p. route (up to 1.5-fold over baseline by 24 h postinfection). Combined, these effects could be mediated via TLR activation (TLR-3, TLR-7, or TLR-9) of cells at the site of infection. However, several of these molecules (MIP-1α [38], MCP-1 [15, 17], MCP-3 [31], MIP-1β, and RANTES [6]) are known to be up regulated by type I IFN treatment and alterations in cellular recruitment/infiltrates, so we cannot exclude the possibility that these factors were produced in response to the systemic effects of type I IFN produced by VLP infection.
TABLE 3.
Temporal serum cytokine and chemokine responses following inoculation with WNV VLPs
| Groupa | Cytokine or chemokine concn (fold increase)e
|
Amt of MCP-1 RNAd (no. of copies/106 GAPDH copies [fold increase]) | ||||||
|---|---|---|---|---|---|---|---|---|
| IFN-α/βb (U/ml) | IFN-γc (pg/ml) | TNF-α (pg/ml) | IL-12 (pg/ml) | RANTES (pg/ml) | MIP-1α (pg/ml) | MCP-1 (pg/ml) | ||
| Control | <10 | 319 | 1,822 | 813 | 447 | 60 | 543 | 51 |
| f.p. inoculation | ||||||||
| 8 h | 203 (>20.3) | 318 (1.0) | 1,167 (0.6) | 674 (0.8) | 426 (1.0) | 160 (2.7) | 636 (1.2) | ND |
| 24 h | 527 (>52.7) | 356 (1.1) | 1,838 (1.0) | 764 (0.9) | 564 (1.3) | 131 (2.2) | 850 (1.6) | 965 (18.9) |
| i.p. inoculation | ||||||||
| 8 h | 857 (>85.7) | 418 (1.3) | 1,739 (1.0) | 919 (1.1) | 534 (1.2) | 355 (5.9) | 1,409 (2.6) | ND |
| 24 h | 5,816 (>581.6) | 487 (1.5) | 2,208 (1.2) | 1,053 (1.3) | 860 (1.9) | 193 (3.2) | 2,094 (3.9) | 436 (8.5) |
Groups of three BL6 mice were inoculated with WNV VLPs by the f.p. or i.p. route or with medium by both routes to serve as controls. Eight and 24 h after inoculation, sera were collected and pooled for all analyses.
Determined by bioassay.
Determined by Bio-Plex bead array analysis.
Determined by real-time RT-PCR analysis of pLN-derived RNA samples. ND, not done.
Increase values are for comparisons to the control animal values.
DISCUSSION
The outcome of a viral infection involves a complex interaction between the host's (and host cell's) countermeasures and the virus's ability to circumvent these. In acute viral infections, the countermeasures put in place by the host involve both short-term and long-term responses that are intended to result in pathogen elimination/reduction of disease and the production of memory responses that prevent or ameliorate subsequent infections, respectively. In the case of arboviruses, the vast majority of infections are acute and rate limiting, but the pathological outcome of infection can vary greatly both among different host species and between individuals of the same species.
In the case of WNV, there is ample evidence that the immune status of the host is an important determinant of the pathological outcome of infection (see the introduction). Here we have shown that WNV VLPs induce a rapid production of IFN upon inoculation into animals by either the subcutaneous f.p. or the i.p. route. Although the former would appear to mimic a natural infection by a virus-infected mosquito, the latter provides for more direct access to the circulatory system, the target of mosquito blood feeding and the site where much of the saliva-carried virus is likely deposited. The levels of IFN that we detected in peritoneal fluid and sera harvested from these animals at 8 h postinoculation are 20- to 100-fold higher than the dose that prevents WNV infection in vitro (44), indicating that these values are biologically relevant.
We also observed IFN production in animals inoculated with WNV, a finding that has been reported in other studies (27, 36, 40); however, we focused most of our studies on WNV VLPs. The reason for this emphasis is due to the properties of these particles, namely, their inability to spread from cell to cell, which indicates that the IFN observed was produced by the cells that were infected with the inoculating dose or stimulated by the products of these primary infected cells. As stated in the introduction, alphavirus VLPs were used by Johnston and coworkers to track the earliest sites of infection in studies that also demonstrated early infection of lymphoid tissues by VLPs derived from this family of arboviruses (29). In the case of WNV infection, the rapid amplification of the virus in certain tissues confounds these types of analyses, since the IFN production reported by us and others (see above) could result in part from amplification of the virus and its ability to spread to other cells. Although our studies do not preclude IFN production by other cells later in the course of infection with the natural virus, they do clearly demonstrate that the primary inoculation is capable of activating this important aspect of the immune response.
The kinetics of IFN production in animals inoculated with WNV VLPs and poly(IC:LC) were dramatically different. Poly(IC:LC) elicited IFN much more quickly, but the response also began to wane very rapidly. These properties are consistent with our demonstration that VLPs need to be able to replicate to produce the PAMP responsible for the induction of IFN production, whereas poly(IC:LC) is injected at high doses and is directly active.
The finding that production of this early IFN response required VLP replication and was accompanied in f.p.-inoculated animals by high levels of IFN-α mRNA in the pLN implies that the site of IFN production is lymphoid tissue. Furthermore, the IFN activity was neutralized by a polyclonal antiserum that recognizes IFN-α/β, and direct ELISA demonstrated the presence of IFN-α but not IFN-β in the sera of VLP-infected animals. These findings, as well as our findings that IFN was produced in similar quantities by IRF3−/− mice, argue for a role of lymphoid cells that constitutively express high levels of IRF7 as the source of IFN. Although the cells producing IFN could be DCs, a cell type that has been implicated in IFN production in infection with DV and yellow fever virus, to our knowledge there are no reports demonstrating that infection of these cells is important for IFN production in WNV-infected animals. Unfortunately, our analysis of 23 cytokines and chemokines by Bioplex array did not allow us to pinpoint expression to a single cell type given the rather broad array of potential expressing cell types for molecules that were up regulated at least twofold over baseline in the circulation. This will be the focus of future studies with sorted cells from treated animals.
The early production (and recognition) of IFN by immunocompetent hosts could readily explain why such hosts are much more likely to survive infection. Likewise, this early production of IFN could have interesting ramifications in dose escalation experiments designed to determine the virulence of viral isolates. In particular, we have noted that as arbovirus isolates become more attenuated, there is often a threshold in virulence, such that viruses with a predicted large difference in virulence show identical inabilities to produce disease and mortality in animals, even when administered at very high doses. In light of the data presented here, these experiments could indicate that the early IFN responses to very high doses of the virus could block replication of viruses of various levels of virulence, confounding measurement of attenuation when high doses are administered.
The IFN responses we have observed here also have important ramifications for vaccine design and delivery. For the reasons cited above, escalating doses of live attenuated flavivirus vaccines, especially in the case of multiserotype vaccines (as needed, for example, in the case of DV), could result in IFN responses that would dampen virus spread and, consequently, potency/efficacy. However, this effect would likely be offset by the autoadjuvant effect of IFN production. Thus, the balance between IFN production that blocks attenuated vaccine virus spread and the need to attenuate viral replication to prevent disease could help to explain why live attenuated vaccines for some flaviviruses have been so difficult to produce. On the other hand, the autoadjuvant effect would have a less severe effect on the immune responses engendered by VLP or VLP-like vaccines, such as our recently reported pseudo-infectious flavivirus vaccines (30). Interestingly, recent reports on VLPs from alphaviruses have also reported that they can stimulate strong immune responses, likely through a similar mechanism of activation (46).
Multiple important aspects of WNV VLP-induced IFN production remain to be elucidated. These include the precise nature of the cells that are infected in the lymphoid tissues and the mechanism of infection. In particular, we are interested in determining if the VLPs traffic to the lymphoid tissues and infect lymphoid cells in these organs or if they infect cells in the periphery which then traffic to the draining LN. Further studies to explore the expected adjuvant activity that this IFN production can have are also warranted, as this may have important implications for the design of effective vaccines.
Acknowledgments
We thank A. M. Salazar of Oncovir, Inc., for providing poly(IC:LC) (Hiltonol) for our studies; I. Frolov, UTMB, for supplying the MEF cells; and T. Moran of Mt. Sinai School of Medicine for facilitating transfer of IRF3−/− mice from T. Taniguchi at the University of Tokyo.
S.L.R. was supported by a James McLaughlin Fellowship, and this work was supported by a grant from NIAID to P.W.M. through the Western Regional Center of Excellence for Biodefense and Emerging Infectious Disease Research (NIH grant number U54 AI057156), AI061441, and by the Sealy Center for Vaccine Development.
Footnotes
Published ahead of print on 13 June 2007.
REFERENCES
- 1.Best, S. M., K. L. Morris, J. G. Shannon, S. J. Robertson, D. N. Mitzel, G. S. Park, E. Boer, J. B. Wolfinbarger, and M. E. Bloom. 2005. Inhibition of interferon-stimulated JAK-STAT signaling by a tick-borne flavivirus and identification of NS5 as an interferon antagonist. J. Virol. 79:12828-12839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boonstra, A., R. Rajsbaum, M. Holman, R. Marques, C. Asselin-Paturel, J. P. Pereira, E. E. Bates, S. Akira, P. Vieira, Y. J. Liu, G. Trinchieri, and A. O'Garra. 2006. Macrophages and myeloid dendritic cells, but not plasmacytoid dendritic cells, produce IL-10 in response to MyD88- and TRIF-dependent TLR signals, and TLR-independent signals. J. Immunol. 177:7551-7558. [DOI] [PubMed] [Google Scholar]
- 3.Brooks, T. J., and R. J. Phillpotts. 1999. Interferon-alpha protects mice against lethal infection with St. Louis encephalitis virus delivered by the aerosol and subcutaneous routes. Antivir. Res. 41:57-64. [DOI] [PubMed] [Google Scholar]
- 4.Campbell, G. L., A. A. Marfin, R. S. Lanciotti, and D. J. Gubler. 2002. West Nile virus. Lancet Infect. Dis. 2:519-529. [DOI] [PubMed] [Google Scholar]
- 5.Cao, W., and Y. J. Liu. 2006. Innate immune functions of plasmacytoid dendritic cells. Curr. Opin. Immunol. 19:24-30. [DOI] [PubMed] [Google Scholar]
- 6.Cremer, I., V. Vieillard, and E. De Maeyer. 2000. Retrovirally mediated IFN-beta transduction of macrophages induces resistance to HIV, correlated with up-regulation of RANTES production and down-regulation of C-C chemokine receptor-5 expression. J. Immunol. 164:1582-1587. [DOI] [PubMed] [Google Scholar]
- 7.Delhaye, S., S. Paul, G. Blakqori, M. Minet, F. Weber, P. Staeheli, and T. Michiels. 2006. Neurons produce type I interferon during viral encephalitis. Proc. Natl. Acad. Sci. USA 103:7835-7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deonarain, R., A. Alcami, M. Alexiou, M. J. Dallman, D. R. Gewert, and A. C. Porter. 2000. Impaired antiviral response and alpha/beta interferon induction in mice lacking beta interferon. J. Virol. 74:3404-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diebold, S. S., M. Montoya, H. Unger, L. Alexopoulou, P. Roy, L. E. Haswell, A. Al-Shamkhani, R. Flavell, P. Borrow, and C. Reis e Sousa. 2003. Viral infection switches non-plasmacytoid dendritic cells into high interferon producers. Nature 424:324-328. [DOI] [PubMed] [Google Scholar]
- 10.Fayzulin, R., F. Scholle, O. Petrakova, I. Frolov, and P. W. Mason. 2006. Evaluation of replicative capacity and genetic stability of West Nile virus replicons using highly efficient packaging cell lines. Virology 351:196-209. [DOI] [PubMed] [Google Scholar]
- 11.Guo, J. T., J. Hayashi, and C. Seeger. 2005. West Nile virus inhibits the signal transduction pathway of alpha interferon. J. Virol. 79:1343-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haller, O., G. Kochs, and F. Weber. 2006. The interferon response circuit: induction and suppression by pathogenic viruses. Virology 344:119-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayes, E. B., and D. J. Gubler. 2006. West Nile virus: epidemiology and clinical features of an emerging epidemic in the United States. Annu. Rev. Med. 57:181-194. [DOI] [PubMed] [Google Scholar]
- 14.Hiscott, J., R. Lin, P. Nakhaei, and S. Paz. 2006. MasterCARD: a priceless link to innate immunity. Trends Mol. Med. 12:53-56. [DOI] [PubMed] [Google Scholar]
- 15.Hokeness, K. L., W. A. Kuziel, C. A. Biron, and T. P. Salazar-Mather. 2005. Monocyte chemoattractant protein-1 and CCR2 interactions are required for IFN-alpha/beta-induced inflammatory responses and antiviral defense in liver. J. Immunol. 174:1549-1556. [DOI] [PubMed] [Google Scholar]
- 16.Honda, K., H. Yanai, H. Negishi, M. Asagiri, M. Sato, T. Mizutani, N. Shimada, Y. Ohba, A. Takaoka, N. Yoshida, and T. Taniguchi. 2005. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434:772-777. [DOI] [PubMed] [Google Scholar]
- 17.Ishikawa, R., and C. A. Biron. 1993. IFN induction and associated changes in splenic leukocyte distribution. J. Immunol. 150:3713-3727. [PubMed] [Google Scholar]
- 18.Izaguirre, A., B. J. Barnes, S. Amrute, W. S. Yeow, N. Megjugorac, J. Dai, D. Feng, E. Chung, P. M. Pitha, and P. Fitzgerald-Bocarsly. 2003. Comparative analysis of IRF and IFN-alpha expression in human plasmacytoid and monocyte-derived dendritic cells. J. Leukoc. Biol. 74:1125-1138. [DOI] [PubMed] [Google Scholar]
- 19.Johnson, A. J., and J. T. Roehrig. 1999. New mouse model for dengue virus vaccine testing. J. Virol. 73:783-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones, M., A. Davidson, L. Hibbert, P. Gruenwald, J. Schlaak, S. Ball, G. R. Foster, and M. Jacobs. 2005. Dengue virus inhibits alpha interferon signaling by reducing STAT2 expression. J. Virol. 79:5414-5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato, H., O. Takeuchi, S. Sato, M. Yoneyama, M. Yamamoto, K. Matsui, S. Uematsu, A. Jung, T. Kawai, K. J. Ishii, O. Yamaguchi, K. Otsu, T. Tsujimura, C. S. Koh, C. Reis e Sousa, Y. Matsuura, T. Fujita, and S. Akira. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101-105. [DOI] [PubMed] [Google Scholar]
- 22.Kawai, T., and S. Akira. 2006. Innate immune recognition of viral infection. Nat. Immunol. 7:131-137. [DOI] [PubMed] [Google Scholar]
- 23.Leyssen, P., C. Drosten, M. Paning, N. Charlier, J. Paeshuyse, E. De Clercq, and J. Neyts. 2003. Interferons, interferon inducers, and interferon-ribavirin in treatment of flavivirus-induced encephalitis in mice. Antimicrob. Agents Chemother. 47:777-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin, R. J., B. L. Chang, H. P. Yu, C. L. Liao, and Y. L. Lin. 2006. Blocking of interferon-induced Jak-Stat signaling by Japanese encephalitis virus NS5 through a protein tyrosine phosphatase-mediated mechanism. J. Virol. 80:5908-5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin, R. J., C. L. Liao, E. Lin, and Y. L. Lin. 2004. Blocking of the alpha interferon-induced Jak-Stat signaling pathway by Japanese encephalitis virus infection. J. Virol. 78:9285-9294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1041. In D. M. Knipe, P. M. Howley, D. G. Griffin, R. A. Lamb, M. A. Martin, and B. Roizman (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 27.Liu, W. J., X. J. Wang, D. C. Clark, M. Lobigs, R. A. Hall, and A. A. Khromykh. 2006. A single amino acid substitution in the West Nile virus nonstructural protein NS2A disables its ability to inhibit alpha/beta interferon induction and attenuates virus virulence in mice. J. Virol. 80:2396-2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lobigs, M., A. Mullbacher, Y. Wang, M. Pavy, and E. Lee. 2003. Role of type I and type II interferon responses in recovery from infection with an encephalitic flavivirus. J. Gen. Virol. 84:567-572. [DOI] [PubMed] [Google Scholar]
- 29.MacDonald, G. H., and R. E. Johnston. 2000. Role of dendritic cell targeting in Venezuelan equine encephalitis virus pathogenesis. J. Virol. 74:914-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mason, P. W., A. V. Shustov, and I. Frolov. 2006. Production and characterization of vaccines based on flaviviruses defective in replication. Virology 351:432-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Menten, P., P. Proost, S. Struyf, E. Van Coillie, W. Put, J. P. Lenaerts, R. Conings, J. M. Jaspar, D. De Groote, A. Billiau, G. Opdenakker, and J. Van Damme. 1999. Differential induction of monocyte chemotactic protein-3 in mononuclear leukocytes and fibroblasts by interferon-alpha/beta and interferon-gamma reveals MCP-3 heterogeneity. Eur. J. Immunol. 29:678-685. [DOI] [PubMed] [Google Scholar]
- 32.Munoz-Jordan, J. L., G. G. Sanchez-Burgos, M. Laurent-Rolle, and A. Garcia-Sastre. 2003. Inhibition of interferon signaling by dengue virus. Proc. Natl. Acad. Sci. USA 100:14333-14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piqueras, B., J. Connolly, H. Freitas, A. K. Palucka, and J. Banchereau. 2006. Upon viral exposure, myeloid and plasmacytoid dendritic cells produce 3 waves of distinct chemokines to recruit immune effectors. Blood 107:2613-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ransohoff, R. M., T. Wei, K. D. Pavelko, J. C. Lee, P. D. Murray, and M. Rodriguez. 2002. Chemokine expression in the central nervous system of mice with a viral disease resembling multiple sclerosis: roles of CD4+ and CD8+ T cells and viral persistence. J. Virol. 76:2217-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Romoser, W. S., L. P. Wasieloski, Jr., P. Pushko, J. P. Kondig, K. Lerdthusnee, M. Neira, and G. V. Ludwig. 2004. Evidence for arbovirus dissemination conduits from the mosquito (Diptera: Culicidae) midgut. J. Med. Entomol. 41:467-475. [DOI] [PubMed] [Google Scholar]
- 36.Rossi, S. L., R. Fayzulin, N. Dewsbury, N. Bourne, and P. W. Mason. 2007. Mutations in West Nile virus nonstructural proteins that facilitate replicon persistence in vitro attenuate virus replication in vitro and in vivo. Virology 364:184-195. [DOI] [PubMed] [Google Scholar]
- 37.Rossi, S. L., Q. Zhao, V. K. O'Donnell, and P. W. Mason. 2005. Adaptation of West Nile virus replicons to cells in culture and use of replicon-bearing cells to probe antiviral action. Virology 331:457-470. [DOI] [PubMed] [Google Scholar]
- 38.Salazar-Mather, T. P., C. A. Lewis, and C. A. Biron. 2002. Type I interferons regulate inflammatory cell trafficking and macrophage inflammatory protein 1alpha delivery to the liver. J. Clin. Investig. 110:321-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Samuel, M. A., and M. S. Diamond. 2005. Alpha/beta interferon protects against lethal West Nile virus infection by restricting cellular tropism and enhancing neuronal survival. J. Virol. 79:13350-13361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Samuel, M. A., K. Whitby, B. C. Keller, A. Marri, W. Barchet, B. R. Williams, R. H. Silverman, M. Gale, Jr., and M. S. Diamond. 2006. PKR and RNase L contribute to protection against lethal West Nile virus infection by controlling early viral spread in the periphery and replication in neurons. J. Virol. 80:7009-7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sato, M., H. Suemori, N. Hata, M. Asagiri, K. Ogasawara, K. Nakao, T. Nakaya, M. Katsuki, S. Noguchi, N. Tanaka, and T. Taniguchi. 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity 13:539-548. [DOI] [PubMed] [Google Scholar]
- 42.Savard, C. E., T. A. Blinman, H. S. Choi, S. K. Lee, S. J. Pandol, and S. P. Lee. 2002. Expression of cytokine and chemokine mRNA and secretion of tumor necrosis factor-alpha by gallbladder epithelial cells: response to bacterial lipopolysaccharides. BMC Gastroenterol. 2:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scholle, F., Y. A. Girard, Q. Zhao, S. Higgs, and P. W. Mason. 2004. trans-Packaged West Nile virus-like particles: infectious properties in vitro and in infected mosquito vectors. J. Virol. 78:11605-11614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scholle, F., and P. W. Mason. 2005. West Nile virus replication interferes with both poly(I:C)-induced interferon gene transcription and response to interferon treatment. Virology 342:77-87. [DOI] [PubMed] [Google Scholar]
- 45.Stewart, M. J., K. Smoak, M. A. Blum, and B. Sherry. 2005. Basal and reovirus-induced beta interferon (IFN-beta) and IFN-beta-stimulated gene expression are cell type specific in the cardiac protective response. J. Virol. 79:2979-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson, J. M., A. C. Whitmore, J. L. Konopka, M. L. Collier, E. M. Richmond, N. L. Davis, H. F. Staats, and R. E. Johnston. 2006. Mucosal and systemic adjuvant activity of alphavirus replicon particles. Proc. Natl. Acad. Sci. USA 103:3722-3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao, S. Y., H. Guzman, H. Zhang, A. P. Travassos da Rosa, and R. B. Tesh. 2001. West Nile virus infection in the golden hamster (Mesocricetus auratus): a model for West Nile encephalitis. Emerg. Infect. Dis. 7:714-721. [DOI] [PMC free article] [PubMed] [Google Scholar]