Abstract
Sylvatic dengue viruses (DENV) are transmitted in an enzootic cycle between nonhuman primates and arboreal Aedes mosquitoes in Southeast Asia and West Africa. Although previous analyses have revealed the evolutionary processes among endemic (human) DENV, little is known about viral evolution in the sylvatic cycle. Through an analysis of 14 complete coding regions of sylvatic Dengue type 2 virus sampled over a 33-year period, we show that both the rate of evolutionary change and the pattern of natural selection are similar among endemic and sylvatic DENV, although the latter have a uniquely high frequency of positive selection in the NS4B protein gene. Our findings support a recent cross-species transmission event and suggest the possibility of future DENV reemergence from the sylvatic cycle.
Dengue is an emerging disease that has spread widely in tropical and subtropical regions due to recent changes in human ecology. The causative positive-sense RNA viruses (dengue viruses [DENV]) are classified within the family Flaviviridae, genus Flavivirus, and comprise four serotypes (DENV type 1 [DENV-1] to DENV-4) that exhibit complex immunological interactions (1). The epidemiology of DENV involves both a sylvatic enzootic cycle, most likely among nonhuman primates and arboreal Aedes sp. mosquitoes, and an endemic cycle involving humans and the domestic mosquito Aedes aegypti (2, 3, 19). Sylvatic cycles of DENV have been demonstrated in Asia, where serologic and virus isolation data suggest an association between Macaca and Presbytis monkeys and zoonotic DENV-1, -2, and -4, with Aedes niveus mosquitoes as the primary vectors (18). Sylvatic DENV cycles have also been documented in West Africa, where only DENV-2 has been found to circulate among Erythrocebus patas monkeys and various sylvatic Aedes species, including Aedes taylori, Aedes furcifer, and Aedes luteocephalus (4, 5, 16, 20). Although many DENV strains isolated in Africa can be genetically defined as sylvatic (15, 16, 26), some were in reality isolated from humans who contacted sylvatic cycles in eastern Senegal (20, 29). Conversely, other viruses from Somalia and Burkina Faso have been classified genetically as endemic (25).
Although phylogenetic analyses suggest that endemic DENV strains have their ancestry in the sylvatic viruses (26), little is known about the evolutionary processes that characterize sylvatic DENV. However, revealing the extent and structure of the genetic diversity of sylvatic DENV is central to understanding how this virus crossed species barriers and emerged in humans. In particular, it is not clear whether sylvatic DENV evolve more slowly in their reservoir hosts than endemic isolates in humans because of a lower rate of replication, a decreased intensity of transmission, or greater purifying selection against amino acid change in viruses that may have reached a fitness peak. Recent experimental evidence suggests that the emergence of endemic DENV-2 from sylvatic progenitors may not have required adaptation in order to replicate efficiently in humans, implying that the potential of sylvatic DENV-2 to reemerge is high (25). Conversely, although the transmission of sylvatic DENV-2 to humans has been documented (20, 29), there is no evidence that these sylvatic genotypes are involved in outbreaks of epidemic dengue, probably due to their confinement to forest habitats (5).
Phylogenetic analysis provides a powerful means to reveal the rates of evolutionary change, the times of viral origin, and the nature of selection pressures acting across the DENV genome. Previous estimates of the rate of nucleotide substitution and age of genetic diversity in endemic DENV have been generally consistent; the four DENV serotypes have a mean evolutionary rate of ∼10−3 nucleotide substitutions per site per year (7, 9, 10, 12, 23, 30), and the genetic diversity within each serotype dates to ∼100 to 300 years ago (23, 26-28). These dates likely correspond either to the time of cross-species transmission from nonhuman primates to humans or to the emergence of Aedes aegypti as the principal vector for DENV transmission in humans. Evolutionary analyses have also revealed that the major selective pressure acting on the endemic DENV genomes is purifying (negative) selection, reflected in a low ratio of nonsynonymous (dN) to synonymous (dS) substitutions per nucleotide site (dN/dS ≪ 1), with only sporadic occurrences of positive selection (8, 24). Our determination of the complete genomic sequences of sylvatic DENV-2 isolates allows a detailed analysis of the patterns and process of evolution in these viruses for the first time.
We sequenced 14 complete open reading frames of sylvatic DENV-2 isolates sampled over a 33-year period from 1966 to 1999 (Table 1). Viral RNA was extracted using the QIAamp viral RNA minikit (QIAGEN), overlapping PCR amplicons spanning the entire length of the genome were purified from 1% agarose gels, and both strands were sequenced directly using ABI protocols with both the PCR and internal primers to derive a consensus sequence. A phylogenetic tree including 54 endemic isolates of DENV-2 and three representatives each of DENV-1 (including sylvatic isolate P72-1244), DENV-3 (sylvatic DENV-3 have not been isolated to date but are believed to exist in Malaysia based on the seroconversion of sentinel monkeys [17]), and DENV-4 (including sylvatic isolate P75-215) (total data set, 77 isolates, 10,185 nucleotides in length) was inferred using the maximum likelihood (ML) method available in the PAUP* package (22), employing the best-fit general time-reversible (GTR) + I + Γ4 model of nucleotide substitution as determined by MODELTEST (14) (Fig. 1). To assess the reliability of major groupings, a neighbor-joining bootstrap analysis (1,000 replications) was used with the ML substitution model. The rates of nucleotide substitution, model of population growth, and age of the most recent common ancestor (MRCA) of the 14 sylvatic isolates were estimated using a Bayesian Markov chain Monte Carlo method as implemented in the BEAST program (http://beast.bio.ed.ac.uk/) (6). Two models of demographic history were compared: (i) constant population size and (ii) exponential population growth. To account for any differences in substitution rates among lineages, we employed both strict and relaxed (uncorrelated exponential) molecular clocks. All analyses were run again using the GTR + I + Γ4 substitution model. Akaike's information criterion was used to determine which model best fit the data in hand, with uncertainty in parameter estimates (taken over millions of sampled trees) reflected in the 95% highest probability density (HPD) values, and all chains were run for a sufficient time to ensure convergence.
TABLE 1.
Passage history of DENV-2 isolatesa
| Isolate | Hostb | Passage history (host, no. of passages)c | Location | Yr | GenBank accession no. |
|---|---|---|---|---|---|
| PM33974 | Aedes africanus | Toxorhynchites amboinensis, 1; C6/36, 2 | Guinea | 1981 | EF105378 |
| P8-1407 | Sentinel monkey | SM, 3; C6/36, 2 | Malaysia | 1970 | EF105379 |
| Dak Ar 578 | Aedes taylori | SM, 8; C6/36, 2 | Ivory Coast | 1980 | EF105380 |
| Dak Ar 510 | Aedes taylori | SM, 4; C6/36, 2 | Ivory Coast | 1980 | EF105381 |
| Dak Ar 2039 | Aedes luteocephalus | SM, 6; C6/36, 2 | Burkina Faso | 1980 | EF105382 |
| Dak Ar A1247 | Aedes taylori | SM, 5; C6/36, 2 | Ivory Coast | 1980 | EF105383 |
| Dak HD10674 | Human | SM, 25; mosquito, 1; C6/36, 1 | Senegal | 1970 | EF105384 |
| Dak Ar D20761 | Aedes luteocephalus | SM, 8; C6/36, 1 | Senegal | 1974 | EF105385 |
| Dak Ar A2022 | Aedes africanus | SM, 6; C6/36, 1 | Burkina Faso | 1980 | EF105386 |
| IBH11206 | Human | SM, 30; C6/36, 1 | Nigeria | 1966 | EF105387 |
| IBH11664 | Human | SM, 5; C6/36, 1 | Nigeria | 1966 | EF105388 |
| Dak Ar 141069 | Aedes luteocephalus | AP61, 1; C6/36, 3 | Senegal | 1999 | EF105389 |
| Dak Ar 141070 | Aedes luteocephalus | AP61, 5; C6/36, 1 | Senegal | 1999 | EF105390 |
| Dak Ar D75505 | Aedes luteocephalus | AP61, 5; C6/36, 1 | Senegal | 1991 | EF457904 |
DENV isolates were obtained from the UTMB World Reference Center for Emerging Viruses and Arboviruses and from Institut Pasteur de Dakar, Dakar, Senegal.
Source of virus isolation.
C6/36, Aedes albopictus cell line; SM, suckling mouse; AP61, Aedes pseudoscutellaris cell line.
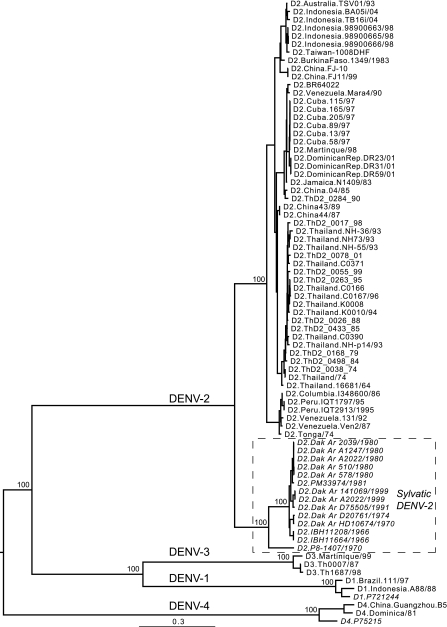
FIG. 1.
Phylogenetic relationships of 14 complete coding regions of sylvatic DENV-2 genomes compared to human isolates of DENV-2 and representatives of DENV-1, DENV-3, and DENV-4. The phylogeny was inferred using an ML procedure, and all horizontal branches are scaled according to the number of substitutions per site. All sylvatic isolates (from DENV-1, DENV-2, and DENV-4) are italicized, and DENV-2 sylvatic isolates are boxed. The genome sequences of the DENV-1 sylvatic isolate P72-1244 and the DENV-4 sylvatic isolate P75-215 were determined in this study. Bootstrap values (>95%) are shown for key nodes.
To determine the gene- and site-specific selection pressures acting on sylvatic DENV, we estimated dN/dS using two likelihood procedures available in the HYPHY package and accessed through the Datamonkey facility (11): the single-likelihood ancestor counting (SLAC) method and the more powerful random-effects (REL) methods, both incorporating the GTR model of nucleotide substitution, with phylogenetic trees inferred using the neighbor-joining method.
Due to the small sample of sylvatic DENV sequences, we estimated evolutionary rates for the entire coding region rather than for individual genes. For these data, a model of constant population size was best-fit to the data. The mean evolutionary rates estimated ranged from 0.374 × 10−3 substitutions per site per year (95% HPD, 0.178 × 10−3 to 0.550 × 10−3 substitutions/site/year) for the strict molecular clock to 2.350 × 10−3 substitutions/site/year (95% HPD, 0.752 × 10−3 to 4.038 × 10−3 substitutions/site/year) for the relaxed molecular clock. Since these rates varied considerably, evidently as a function of sample size, we suggest that the true rate lies between these estimates. More notably, the rates obtained were within the range of those previously estimated for endemic DENV-2 strains (isolated from humans and Aedes aegypti) (7, 23, 30). Hence, this analysis suggests that sylvatic DENV strains do not evolve more slowly than endemic strains. Further evidence that this is the case is that the branch lengths of evolutionary trees are similar among endemic and sylvatic strains in large-scale phylogenetic analyses (Fig. 1).
We were also able to estimate the time of the MRCA of the sylvatic DENV-2 isolates studied. Using our Bayesian Markov chain Monte Carlo procedure, the MRCA of the sylvatic DENV-2 strains sampled existed between 385 years ago (95% HPD, 214 to 612 years) assuming a strict clock and 72 years ago (95% HPD, 39 to 126 years) allowing for a relaxed clock. Despite this variation, the time scale of evolution for sylvatic DENV-2 is similar to that for human epidemic DENV (23).
The sylvatic DENV-2 were also broadly similar to their endemic counterparts in terms of overall selective pressure (Table 2) (30). Gene-specific dN/dS ratios presented a picture of strong selective constraints (dN/dS ≪ 1). This was especially true of NS2B, which was found to be far more conserved at nonsynonymous sites in sylvatic DENV-2 than in endemic DENV-2. While no positive selection was detected at any genomic site in sylvatic DENV-2 by the SLAC method, 16 amino acid sites showed evidence of positive selection by REL. These putatively selected codons were unevenly distributed across the DENV genome, being located only in the E gene (2 codon sites), the NS2A gene (1 site), and, most notably, the NS4B gene (13 sites). For all these sites, the functional importance of the changes observed remains undetermined, and none have previously been shown to exhibit adaptive evolution in DENV (24). Strikingly, there was no evidence of positive selection on NS4B in endemic DENV-2 strains (Table 2), and 11 positively selected sites remained in the sylvatic lineage analysis when the divergent sylvatic DENV-2 isolate P8-1407, from Malaysia, was excluded. Notably, no positive selection was detected in the sylvatic lineage at E390, a key virulence determinant (21) that has previously been reported to exhibit positive selection in endemic DENV-2 (24). In the future, reverse genetic techniques may allow the interpretation of the functional importance of these positively selected sites. Of particular interest is the concentration of putative positive selection in NS4B, a gene functioning as an interferon-signaling inhibitor, and the question of whether or not these mutations distinguish endemic and sylvatic DENV phenotypes.
TABLE 2.
Evolutionary processes among the proteins of DENV-2
| Protein | Length (codons) | Endemic DENV-2a
|
Sylvatic DENV-2
|
|||
|---|---|---|---|---|---|---|
| Total tree lengthb | dN/dS | Total tree length | dN/dS | Positively selected site(s) | ||
| Capsid | 114 | 0.380 | 0.104 | 0.174 | 0.075 | |
| Membrane | 166 | 0.582 | 0.097 | 0.206 | 0.048 | |
| Envelope | 495 | 0.473 | 0.055 | 0.238 | 0.075 | 177, 329 |
| NS1 | 352 | 0.514 | 0.085 | 0.235 | 0.060 | |
| NS2A | 218 | 0.626 | 0.074 | 0.307 | 0.084 | 195 |
| NS2B | 130 | 0.494 | 0.045 | 0.206 | 0.005 | |
| NS3 | 618 | 0.535 | 0.042 | 0.210 | 0.036 | |
| NS4A | 149 | 0.586 | 0.038 | 0.269 | 0.025 | |
| NS4B | 249 | 0.549 | 0.037c | 0.273 | 0.052 | 12, 19, 21, 24, 26, 49, 96, 113, 116, 123, 197, 242, 246 |
| NS5 | 900 | 0.525 | 0.066 | 0.222 | 0.048 | |
Estimated from a data set of 54 DENV-2 complete coding region sequences.
Expressed in substitutions per site.
No evidence for positive selection was observed by the SLAC and fixed-effects likelihood methods for human NS4B.
Overall, our data support previous studies that suggest that the recent emergence of all endemic DENV serotypes from sylvatic progenitors occurred concomitantly with the appearance of human populations large enough to support continuous viral transmission (28). Emergence in humans may also have been facilitated by the accumulation of adaptive mutations enabling DENV to replicate efficiently in the domestic Aedes vectors (13), although later analyses using a larger number and diversity of isolates have not supported this conclusion (K. Hanley, personal communication). Moreover, the ability of domestic Aedes mosquitoes to serve as vectors for sylvatic DENV transmission (5), as well as the lack of evidence that any adaptation of sylvatic DENV is needed for efficient replication in humans (as is demonstrated by the similarity of the mean replication titers of human and sylvatic DENV isolates in two surrogate human model hosts of DENV replication [25]), suggests that there is a relatively high potential for sylvatic DENV to reemerge in human populations.
Several key issues in the evolutionary ecology of DENV remain unanswered, including the role of other vertebrate hosts in the maintenance of sylvatic DENV, the degree of ecological contact between humans and sylvatic DENV, and the replication and immunological dynamics of sylvatic DENV in nonhuman primates. Our analyses indicate that sylvatic DENV evolves in a manner similar to that of endemic DENV, suggesting that the dynamics of mutation, replication, and selection are broadly equivalent for DENV-2 across its host range.
Nucleotide sequence accession numbers.
The genome sequences of the sylvatic DENV-1 isolate P72-1244 and the sylvatic DENV-4 isolate P75-215 have been assigned GenBank accession numbers EF457905 and EF457906, respectively.
Acknowledgments
We thank Robert Tesh for kindly providing DENV strains and K. A. Hanley for critical comments on the manuscript. Three anonymous referees also provided valuable comments.
N. Vasilakis was supported by the Centers for Disease Control and Prevention Fellowship Training Program in Vector-Borne Infectious Diseases, T01/CCT622892.
The authors have no conflicting financial interests.
Footnotes
Published ahead of print on 6 June 2007.
REFERENCES
- 1.Adams, B., E. C. Holmes, C. Zhang, M. P. Mammen, Jr., S. Nimmannitya, S. Kalayanarooj, and M. Boots. 2006. Cross-protective immunity can account for the alternating epidemic pattern of dengue virus serotypes circulating in Bangkok. Proc. Natl. Acad. Sci. USA 103:14234-14239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheong, W. H. 1986. The vectors of dengue and dengue hemorrhagic fevers in Malaysia. Inst. Med. Res. Malays. Bull. 23:155-167. [Google Scholar]
- 3.Cornet, M. 1993. Dengue in Africa, p. 39-47. In P. Thongcharoen (ed.), Monograph on dengue/dengue haemorrhagic fever. WHO regional publication, South-East Asia series no. 22. World Health Organization, Regional Office for South-East Asia, New Delhi, India.
- 4.Diallo, M., Y. Ba, A. A. Sall, O. M. Diop, J. A. Ndione, M. Mondo, L. Girault, and C. Mathiot. 2003. Amplification of the sylvatic cycle of dengue virus type 2, Senegal, 1999-2000: entomologic findings and epidemiologic considerations. Emerg. Infect. Dis. 9:362-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diallo, M., A. A. Sall, A. C. Moncayo, Y. Ba, Z. Fernandez, D. Ortiz, L. L. Coffey, C. Mathiot, R. B. Tesh, and S. C. Weaver. 2005. Potential role of sylvatic and domestic African mosquito species in dengue emergence. Am. J. Trop. Med. Hyg. 73:445-449. [PubMed] [Google Scholar]
- 6.Drummond, A. J., and A. Rambaut. 2003. Bayesian evolutionary analysis sampling trees: BEAST v1.4. http://beast.bio.ed.ac.uk/. [DOI] [PMC free article] [PubMed]
- 7.Dunham, E. J., and E. C. Holmes. 29 May 2007. Inferring the timescale of dengue virus evolution under realistic models of DNA substitution. J. Mol. Evol. doi: 10.1007/s00239-006-0278-5. [DOI] [PubMed]
- 8.Holmes, E. C. 2003. Patterns of intra- and interhost nonsynonymous variation reveal strong purifying selection in dengue virus. J. Virol. 77:11296-11298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jenkins, G. M., A. Rambaut, O. G. Pybus, and E. C. Holmes. 2002. Rates of molecular evolution in RNA viruses: a quantitative phylogenetic analysis. J. Mol. Evol. 54:152-161. [DOI] [PubMed] [Google Scholar]
- 10.Klungthong, C., C. Zhang, M. P. Mammen, Jr., S. Ubol, and E. C. Holmes. 2004. The molecular epidemiology of dengue virus serotype 4 in Bangkok, Thailand. Virology 329:168-179. [DOI] [PubMed] [Google Scholar]
- 11.Kosakovsky Pond, S. L., and S. D. W. Frost. 2005. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21:2531-2533. [DOI] [PubMed] [Google Scholar]
- 12.Lanciotti, R. S., D. J. Gubler, and D. W. Trent. 1997. Molecular evolution and phylogeny of dengue-4 viruses. J. Gen. Virol. 78:2279-2286. [DOI] [PubMed] [Google Scholar]
- 13.Moncayo, A. C., Z. Fernandez, D. Ortiz, M. Diallo, A. Sall, S. Hartman, C. T. Davis, L. Coffey, C. C. Mathiot, R. B. Tesh, and S. C. Weaver. 2004. Dengue emergence and adaptation to peridomestic mosquitoes. Emerg. Infect. Dis. 10:1790-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Posada, D., and K. A. Crandall. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 15.Rico-Hesse, R. 1990. Molecular evolution and distribution of dengue viruses type 1 and 2 in nature. Virology 174:479-493. [DOI] [PubMed] [Google Scholar]
- 16.Rodhain, F. 1991. The role of monkeys in the biology of dengue and yellow fever. Comp. Immunol. Microbiol. Infect. Dis. 14:9-19. [DOI] [PubMed] [Google Scholar]
- 17.Rudnick, A. 1984. The ecology of the dengue virus complex in peninsular Malaysia, p. 7. In T. Pang and R. Pathmanatan (ed.), Proceedings of the International Conference on Dengue/DHF. University of Malaysia Press, Kuala Lumpur, Malaysia.
- 18.Rudnick, A. 1986. Dengue virus ecology in Malaysia. Inst. Med. Res. Malays. Bull. 23:51-152. [Google Scholar]
- 19.Saluzzo, J. F., M. Cornet, P. Castagnet, C. Rey, and J. P. Digoutte. 1986. Isolation of dengue 2 and dengue 4 viruses from patients in Senegal. Trans. R. Soc. Trop. Med. Hyg. 80:5. [DOI] [PubMed] [Google Scholar]
- 20.Saluzzo, J. F., M. Cornet, C. Adam, M. Eyraud, and J. P. Digoutte. 1986. Dengue 2 in eastern Senegal: serologic survey in simian and human populations, 1974-85. Bull. Soc. Pathol. Exot. Filiales 79:313-322. (In French.) [PubMed] [Google Scholar]
- 21.Sanchez, I. J., and B. H. Ruiz. 1996. A single nucleotide change in the E protein gene of dengue virus 2 Mexican strain affects neurovirulence in mice. J. Gen. Virol. 77:2541-2545. [DOI] [PubMed] [Google Scholar]
- 22.Swofford, D. L. 2003. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sinauer Associates, Sunderland, Mass.
- 23.Twiddy, S. S., E. C. Holmes, and A. Rambaut. 2003. Inferring the rate and time-scale of dengue virus evolution. Mol. Biol. Evol. 20:122-129. [DOI] [PubMed] [Google Scholar]
- 24.Twiddy, S. S., C. H. Woelk, and E. C. Holmes. 2002. Phylogenetic evidence for adaptive evolution of dengue viruses in nature. J. Gen. Virol. 83:1679-1689. [DOI] [PubMed] [Google Scholar]
- 25.Vasilakis, N., E. J. Shell, E. B. Fokam, P. W. Mason, K. A. Hanley, D. M. Estes, and S. C. Weaver. 2007. Potential of ancestral sylvatic dengue-2 viruses to re-emerge. Virology 358:402-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang, E., H. Ni, R. Xu, A. D. T. Barrett, S. J. Watowich, D. J. Gubler, and S. C. Weaver. 2000. Evolutionary relationships of endemic/epidemic and sylvatic dengue viruses. J. Virol. 74:3227-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weaver, S. C., and A. D. Barrett. 2004. Transmission cycles, host range, evolution and emergence of arboviral disease. Nat. Rev. Microbiol. 2:789-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zanotto, P. M., E. A. Gould, G. F. Gao, P. H. Harvey, and E. C. Holmes. 1996. Population dynamics of flaviviruses revealed by molecular phylogenies. Proc. Natl. Acad. Sci. USA 93:548-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeller, H. G., M. Traore-Lamizana, E. Monlun, J. P. Hervy, M. Mondo, and J. P. Digoutte. 1992. Dengue-2 virus isolation from humans during an epizootic in southeastern Senegal in November, 1990. Res. Virol. 143:101-102. [DOI] [PubMed] [Google Scholar]
- 30.Zhang, C., M. P. Mammen, Jr., P. Chinnawirotpisan, C. Klungthong, P. Rodpradit, A. Nisalak, S. Nimmannitya, S. Kalayanarooj, D. W. Vaughn, and E. C. Holmes. 2006. Structure and age of genetic diversity of dengue virus type 2 in Thailand. J. Gen. Virol. 87:873-883. [DOI] [PubMed] [Google Scholar]