Abstract
The human herpes simplex virus type 1 regulatory protein ICP4 binds DNA as a dimer and forms a single protein-DNA complex (A complex) with short DNA probes. ICP4 oligomerized in a DNA-dependent manner, forming two or more protein-DNA complexes with longer DNA fragments containing a single DNA binding site. When resolved electrophoretically, one or more low-mobility DNA-protein complexes follow the fast-moving A complex. The major protein-DNA complex (B complex) formed by ICP4 with long DNA probes migrates just behind the A complex in the electric field, implying the oligomerization of ICP4 on the DNA. Binding experiments with circularly permutated DNA probes containing one ICP4 binding site revealed that about 70 bp of nonspecific DNA downstream of the cognate ICP4 binding site was required for efficient B complex formation. In addition, the C-terminal domain of ICP4 was found to be required for DNA-dependent oligomerization and B complex formation. Gel mobility shift analysis of protein-DNA complexes, combined with supershift analysis using different monoclonal antibodies, indicated that the B complex contained two ICP4 dimers. DNase I footprinting of ICP4-DNA complexes showed that one ICP4 dimer contacts the specific binding site and another ICP4 dimer contacts nonspecific DNA in the B complex. DNA-dependent oligomerization increased the affinity of ICP4 for relatively weak binding sites on large DNA molecules. The results of this study suggest how ICP4 may use multiple weak binding sites to aid in transcription activation.
Herpes simplex virus type 1 (HSV-1) encodes a 175-kDa nuclear phosphoprotein (11) referred to as infected-cell polypeptide 4 (ICP4) that both positively and negatively regulates RNA polymerase II-dependent transcription of the viral genes. ICP4 negatively regulates the ICP4 promoter (12, 41), latency-associated promoter (3), and L/ST promoter (5, 59) and activates most of the early and late genes of the virus (12, 16, 22, 41, 45, 57, 60). Genetic and biochemical analyses revealed that ICP4 contains discrete domains responsible for DNA binding, nuclear localization, and transcriptional regulation (6, 14, 43, 44, 49, 51). ICP4 is a dimer in solution (39) and probably functions as a dimer in DNA binding and transcriptional regulation (40, 50). A region of the protein close to the DNA binding domain has been shown to be sufficient for ICP4 dimerization (50). ICP4 is a DNA binding protein (21) with preferences for specific DNA sequences (18, 19, 34). Analysis of different known ICP4 binding sites yielded a relatively degenerate consensus, RTCGTCNNYNYSG, where R is purine, Y is pyrimidine, S is C or G, and N is any base (15).
It has been shown that ICP4-mediated repression requires specifically located, appropriately oriented, and relatively strong ICP4 binding sites (24, 35, 40, 46). Biochemical studies showed that purified ICP4, TATA binding protein (TBP), and transcription factor IIB (TFIIB) cooperatively interact on ICP4 and L/ST promoters (47, 54) and that the degree of cooperative TBP-DNA-TFIIB-ICP4 complex formation correlates positively with the degree of repression of the promoters (35). ICP4 and L/ST promoters contain a TATA box and a functional ICP4 binding site. Biochemical analyses also showed that at least part of the N-terminal region of ICP4 is required for efficient cooperativity in the TBP-TFIIB-DNA-ICP4 complex formation and repression in vitro. The C-terminal region of ICP4 is dispensable for these two functions (26, 54). Less is known about how ICP4 as a DNA binding protein activates transcription. Most genetic and biochemical studies indicate that DNA binding is essential for the activating functions of ICP4 (1, 44, 51). However, mutational alteration of the binding sites in and around the promoters of activated genes appears to have little effect (25, 53). At present, it is unclear how the specific DNA binding of ICP4 is involved in ICP4-mediated activation. There could be mechanisms that increase the affinity of ICP4 for DNA and/or allow ICP4 to work over a long distance. Cellular DNA binding proteins, such as high-mobility-group proteins, TATA box binding proteins, and initiator binding proteins, may be involved in ICP4-mediated gene activation in this way (8, 33, 35, 42). In the present study, we observed that ICP4 forms two (or more) protein-DNA complexes with longer DNA probes containing a single effective ICP4 binding site. Our results showed that ICP4 forms tetramers and higher-order complexes over long DNA fragments through multiple protein-protein and protein-DNA contacts. Such binding may affect the affinity of ICP4 for DNA, particularly DNA containing relatively weak binding sites. Our results may explain how ICP4 activates early and late genes that may not have strong and indispensable ICP4 binding sites.
MATERIALS AND METHODS
Proteins.
Wild-type (WT) ICP4 and mutant ICP4 proteins were purified from Vero cells infected with HSV strain Kos or specific mutant viruses as previously described (29, 51). The mutant viruses d810, n208, and nd810 express ICP4 proteins corresponding to amino acids 1 to 142 and 210 to 1,298, 1 to 774, and 1 to 142 and 210 to 774, respectively. ICP4 mutant protein X25 (amino acids 1 to 30 and 274 to 774) was purified from the nuclei of X25-expressing Vero cells as described previously (50, 54). ICP4 mutants EICP4 and En208 are similar to WT ICP4 and n208, respectively, except that EICP4 and En208 contain the influenza virus hemagglutinin (HA) epitope (7).
Plasmids and DNA probes.
The plasmids pk1-2, containing the ICP4 gene starting from position −330 relative to the start site (13), and pBend4, containing a fragment of the ICP4 promoter from positions −40 to +27 relative to the start site, have previously been described (35). The plasmid pBend4R is similar to pBend4 except that the ICP4 binding site is reversed in orientation. Double-stranded oligonucleotides equivalent to the gC promoter region spanning positions +75 to +125 (including the ICP4 binding site) were cloned in plasmid pBend (34). The resulting recombinant plasmid, pBendG, was cut with the appropriate restriction endonucleases to generate DNA probes GR and GH with ICP4 binding sites located very close to the left and right ends, respectively, of the DNA probes. All the newly constructed plasmids were sequenced to confirm the integrity of the constructs. To make DNA probes, the plasmids were digested with the appropriate restriction endonuclease and then dephosphorylated by treating them with alkaline phosphatase. Alkaline phosphatase was inactivated by phenol extraction. DNA was ethanol precipitated, air dried, and then phosphorylated using T4 polynucleotide kinase in the presence of [γ-32P]ATP. To prepare strand-specific labeled DNA probes, a small fragment of DNA from one or the other end of the labeled probes was removed by digesting the probes with a restriction endonuclease. The labeled DNA fragments were resolved on 5% polyacrylamide gels, the proper bands were located by exposing the gels to photographic films, and the bands were excised. The DNA present in the gel slice was electroeluted or purified by filtration using Spin-X cartridges (Costar, Cambridge, MA). Electroeluted DNA was further purified using Elutip-d mini columns (Schleicher and Schuell, Keene, NH) and Sephadex G50 spin columns (Roche, Indianapolis, IN).
Electrophoretic mobility shift assays (EMSA).
DNA binding reactions and the subsequent gel shift analyses were performed as previously described (54) with slight modifications. Unless otherwise indicated, 40 ng of ICP4 or mutant ICP4 was incubated with 1 ng of DNA probe in a buffer containing 10 mM HEPES-KOH (pH 7.9), 5 mM ammonium sulfate, 8% (vol/vol) glycerol, 2% (wt/vol) polyethylene glycol 8000, 80 mM KCl, 5 mM β-mercaptoethanol, 0.2 mM EDTA, and 25 μg/ml poly(dG-dC)·poly(dG-dC) in a total volume of 30 μl for 40 min at 30°C. For monoclonal antibody supershift experiments, diluted monoclonal antibodies were added to the binding reactions 30 min after initial incubation. The mixtures were incubated for an additional 10 minutes prior to being loaded onto the gel. The binding mixtures were electrophoretically resolved under constant voltage (12.5 V/cm) on a native 3% polyacrylamide gel containing 0.5× TBE (45 mM Tris, 45 mM borate, and 1 mM EDTA [pH 8.0]) buffer. The gels were dried and exposed to Hyperfilm MP (Amersham, Arlington Heights, IL). For quantitative analyses, the dried gels were scanned using the Ambis radioanalytic imaging system (Ambis Inc., San Diego, CA).
DNase I footprinting.
ICP4 or mutant ICP4 (40 ng) was incubated with 1 ng of DNA end-labeled probe in the binding buffer (described above). The mixture was incubated at 30°C for 40 min, subjected to DNase I digestion, and then resolved on a 3% polyacrylamide gel by electrophoresis as described previously (54). The DNA probes were labeled at the 5′ end of the top or the bottom strand. The bound and free DNA bands were located by exposing the wet gel (covered with Saran Wrap) to photographic film. The bands were excised, and the DNA was eluted by soaking the gel slice overnight in a buffer (0.5 ml) containing 10 mM Tris-HCl (pH 8.0), 1 mM EDTA, and 0.5 M ammonium acetate. The eluted DNA was purified using an Elutip-d mini column (Schleicher and Schuell) and resolved on a 6% polyacrylamide sequencing gel containing 7.3 M urea. A Maxam-Gilbert sequencing reaction (38) for the same probe was resolved in the same gel as that of the marker. After electrophoresis, the gel was dried and then exposed to photographic film.
Calculation of KD.
Constant amounts of ICP4 were incubated with increasing concentrations of radiolabeled DNA ligand under standard binding conditions except that the poly(dG-dC)·poly(dG-dC)·poly(dG-dC) concentration was 10 μg/ml (to obtain measurable amounts of ICP4-DNA complexes with weaker binding sites). The reaction products were resolved on native polyacrylamide gels and the net radioactivities corresponding to the bound and free DNA bands were measured from the dried gels by using the Ambis radioanalytic imager. The values were then used to calculate the apparent equilibrium dissociation constant (KD) from the Scatchard equation, where a plot of DNAbound/DNAfree versus DNAbound is a straight line with a slope of 1/KD (48).
RESULTS
ICP4 forms multiple protein-DNA complexes with long DNA molecules.
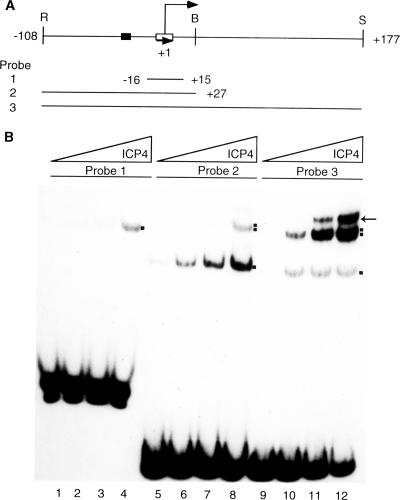
To understand the nature of ICP4-DNA interactions, purified ICP4 protein was incubated with DNA probes of various sizes, all containing a single ICP4 binding site, and then run on native polyacrylamide gels (Fig. 1A). Purified ICP4 formed predominantly a single protein-DNA complex (A complex) with the shorter DNA probe (probe 1) (Fig. 1B, lanes 2 to 4). ICP4 formed an A complex alone with probe 2 at lower protein concentrations (Fig. 1B, lanes 6 to 7) but two complexes at higher protein concentrations (Fig. 1B, lane 8). With probe 3, ICP4 formed two or more complexes with all protein concentrations tested, with B complex as the major species, and one or more slower-moving species (Fig. 1B, lanes 10 to 12). We examined the nature of the B complex in further detail.
FIG. 1.
ICP4-DNA complexes with ICP4 promoter and start site DNA. (A) DNA probes generated from the ICP4 promoter (drawn to scale). The probes were generated from plasmid pk1-2 by digestion with the restriction endonucleases shown above (R, EcoRI; B, BamHI; S, SalI). The TATA box and the ICP4 binding sites are indicated by filled and empty boxes, respectively. The numbers indicate nucleotide positions relative to the start site of transcription. The start site is indicated by a bent arrow, and the orientation of the ICP4 binding site is indicated by a straight arrow. (B) Results of an EMSA of ICP4-DNA complexes formed with probes 1, 2, and 3. The mixtures of the reactions corresponding to lanes 2, 6, and 10 contained 20 ng ICP4. Those corresponding to lanes 3, 7, and 11 contained 70 ng ICP4. Those corresponding to lanes 4, 8, and 12 contained 200 ng ICP4. The binding reaction mixtures loaded in lanes 1, 5, and 9 lacked ICP4. The mixtures for reactions 1 to 4, 5 to 8, and 9 to 12 were loaded at three different times (probe 1 was loaded first, followed by probe 2 and then probe 3) to resolve all lanes in the same gel. The A and B complexes are indicated by single and double dots, respectively. The additional complex formed with probe 3 is indicated by an arrow.
A long stretch of DNA downstream of the ICP4 binding site is required for effective B complex formation.
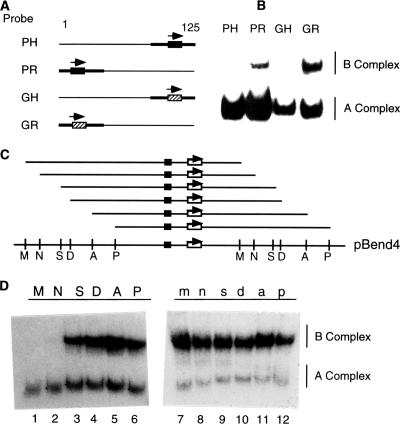
To further examine the DNA requirements for B complex formation, we tested four additional probes containing a single ICP4 binding site located at either the left or the right end of the probes (Fig. 2A). The PH and PR probes contain the ICP4 binding site that spans the start site of ICP4 transcription, while the GH and GR probes contain an ICP4 binding site from the region downstream of the gC transcription start site (positions +75 to +125). As shown in Fig. 2B, ICP4 formed only an A complex with probes PH and GH (both probes contained ∼10 bp of DNA downstream of the ICP4 binding site) but both A and B complexes with probes PR and GR (both probes contained ∼100 bp of DNA downstream of the ICP4 binding site). To find out the minimal length of DNA needed for effective B complex formation, we inserted a fragment of the ICP4 promoter containing the ICP4 binding site (positions −40 to +27 relative to the start site) into the pBend2 vector (30) and made a series of circularly permutated DNA probes (Fig. 2C). The DNA probes are identical in DNA sequence, but the ICP4 binding sites are positioned at different distances from the ends of the probes. The probes were incubated with a fixed concentration of ICP4, and the resulting complexes were separated electrophoretically. ICP4 formed only an A complex with probes M and N (Fig. 2D). These two probes have a short stretch of DNA (<45 bp) downstream of the ICP4 binding site. However, ICP4 formed both A and B complexes (Fig. 2D, lanes S, D, A, and P) with the rest of the probes. These probes contained >70 bp of DNA downstream of the ICP4 binding site. We constructed another set of circularly permutated probes identical to those shown in Fig. 2C except that the orientation of the ICP4 binding site was reversed. ICP4 formed both A and B complexes with all of the probes (Fig. 2D, right). The reverse orientation of the ICP4 binding site in these probes extended the length of DNA downstream of the ICP4 binding site by >70 bp.
FIG. 2.
A long stretch of DNA downstream of the ICP4 binding site is required for B complex formation. (A) Maps of the four DNA probes (all ∼125 bp) containing one ICP4 promoter (black boxes) or gC (hashed boxes) downstream of the ICP4 binding site. The orientation of the ICP4 binding site is indicated by an arrow. (B) Results of an EMSA of ICP4 bound to probes in panel A. A and B complexes are indicated. The unbound probe is not shown. (C) Map of ICP4 binding site-containing probes. The plasmid pBend4 was digested with different restriction enzymes (M, MluI; N, NheI; S, SpeI; D, DraI; A, SmaI; P, SspI) to generate the probes (M, N, S, D, A, and P). The TATA box (black box) and the ICP4 binding site (white box with an arrow indicating the orientation of the binding site) are indicated. The location of the ICP4 binding site relative to the ends of the probe is drawn to scale. The probes m, n, s, d, a, and p generated from pBend4R are identical to probes M, N, S, D, A, and P, respectively, except that the orientation of the ICP4 binding site is reversed. (D) Results of an EMSA of ICP4 bound to probes M, N, S, D, A, and P and m, n, s, d, a, and p. The EMSA was conducted with the probes in panel C as described in the legend to Fig. 1. The different mobilities of the A complexes are due to ICP4-induced DNA bends as reported earlier (35).
The C-terminal region of ICP4 is required for effective B complex formation.
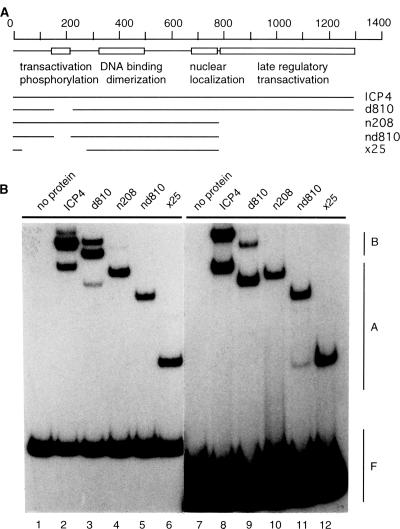
To identify the domains of ICP4 (other than the DNA binding domain) required for B complex formation, we used several mutants of ICP4 (Fig. 3A) in DNA binding studies. Purified ICP4 or the mutants were separately incubated with two different DNA probes. Both probes contained >70 bp of DNA downstream of the single ICP4 binding site. ICP4 molecules containing the DNA binding domain and the C-terminal region (WT ICP4 and d810) formed both A and B complexes with both of the probes (Fig. 3B, lanes 2, 3, 8, and 9). All three ICP4 mutants lacking the C-terminal region (n208, nd810, and ×25) failed to form the B complex (Fig. 3B, lanes 4 to 6 and 10 to 12).
FIG. 3.
ICP4 domains required for B complex formation. (A) Schematic diagrams of WT ICP4 and different deletion mutants (drawn to scale). The numbers on the top indicate the amino acid positions. The map of the known domains of ICP4 (indicated by empty boxes) was adapted from references 14 and 50. (B) Results of an EMSA of mutant and WT ICP4 DNA complexes. Purified WT ICP4 and the mutant proteins were incubated with DNA probes derived from plasmids pk1-2 (lanes 2 to 6) and pBend4 (probe D) (lanes 8 to 12). Lanes 1 and 7 contained reaction mixtures lacking protein.
Protein-protein interactions in the B complex.
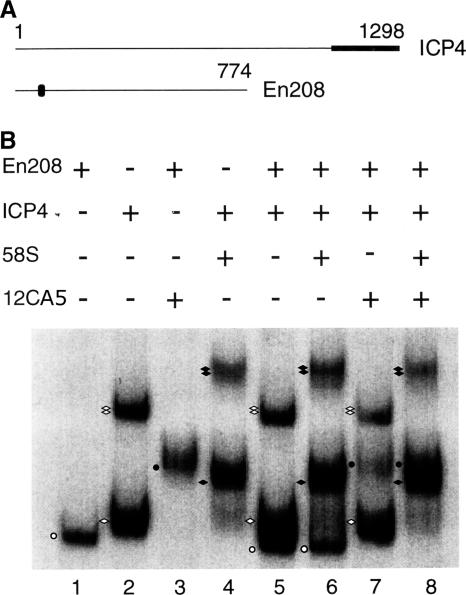
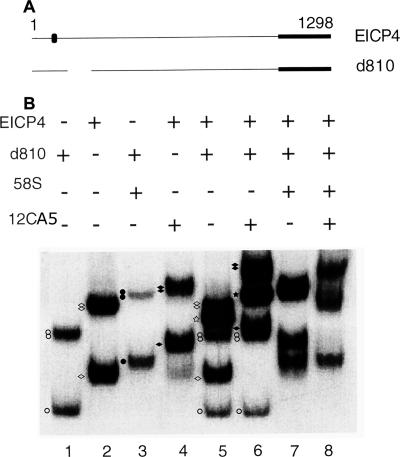
To further investigate the role of the C-terminal region of ICP4 in B complex formation, we mixed WT ICP4 and mutant ICP4 lacking the C-terminal region (En208) in DNA binding experiments. The mutant En208 is similar to n208 except that the former is tagged with an influenza virus HA epitope close to the N terminus of the protein (Fig. 4A). Two monoclonal antibodies, 58S and 12CA5, were used to identify the protein-DNA complexes formed by ICP4 and En208, respectively. 58S recognizes the C-terminal region of ICP4 (52), and 12CA5 recognizes the HA epitope (20) present in En208. The probe used was pBend4D (Fig. 2C). As expected, En208 formed only an A complex with the probe (Fig. 4B, lane 1), which was supershifted by 12CA5 (Fig. 4B, lane 3). ICP4 formed both A and B complexes (Fig. 4B, lane 2), and both complexes were supershifted by 58S (Fig. B, lane 4). When ICP4 and En208 were incubated together with the probe, three different complexes were observed (Fig. 4B, lane 5). Considering the motilities of the complexes formed individually by En208 and ICP4, these three complexes were an A complex formed by En208 and A and B complexes formed by ICP4 (Fig. 4B, compare lane 5 with lanes 1 and 2). When the monoclonal antibody 58S was added to the binding reaction mixture containing ICP4, En208, and the probe (Fig. 4B, lane 6), the A and B complexes formed by ICP4 supershifted but the mobility of the A complex formed by En208 remained unaffected (Fig. 4B, compare lanes 5 and 6). When the monoclonal antibody 12CA5 was added to a binding reaction mixture containing ICP4, En208, and the probe (Fig. 4B, lane 7), the A complex formed by En208 supershifted but the mobilities of the A and B complexes formed by ICP4 remained unaffected (Fig. 4B, compare lanes 5 and 7). These data suggest that En208 formed an A complex only and failed to form a heterogeneous DNA-protein complex with ICP4. When both 58S and 12CA5 were added to a binding reaction mixture containing ICP4, En208, and the probe, both A and B complexes supershifted (Fig. 4B, lane 8) but there was no complex with a mobility lower than that of the supershifted B complex formed by ICP4 (Fig. 4B, compare lane 8 with lanes 6 and 7). These data further suggest that a heterogeneous protein-DNA complex containing En208 and ICP4 was not formed. Together, these data indicate that B complex formation is dependent on protein-protein interactions through the C-terminal regions of ICP4 molecules.
FIG. 4.
B complex formation involves protein-protein interactions. (A) Schematic diagrams of ICP4 and En208. The locations of the epitopes recognized by the monoclonal antibodies 58S (black box) and 12CA5 (black oval) are indicated. The numbers on the top indicate the amino acid positions. (B) ICP4 and/or En208 was incubated with probe pBend4D (Fig. 2C) for mobility shift assays. Monoclonal antibodies 58S and 12CA5 were added to the indicated reactions. En208 formed only an A complex (white circle), which was supershifted by 12CA5 (black circle). ICP4 formed both A (single white diamond) and B (double white diamonds) complexes (lane 2), which were supershifted by 58S (corresponding black diamonds) (lane 4). ICP4 and En208 did not form a mixed complex (lane 5) to be supershifted by 58S (lane 6) or 12CA5 (lane 7) or both 12CA5 and 58S (lane 8).
Two ICP4 dimers are present in the B complex.
To investigate the number of ICP4 dimers present in the B complex, we used two variants of ICP4 capable of forming the B complex. For this series of experiments, we used d810 and EICP4 (7). EICP4 is equivalent to WT ICP4 but contains an HA epitope tag close to the N terminus (Fig. 5A). The monoclonal antibodies 12CA5 and 58S were used to identify protein-DNA complexes formed by EICP4 and d810. Again, pBend4D (Fig. 2C) served as the probe. As expected, both d810 and EICP4 formed A and B complexes (Fig. 5B, lanes 1 and 2, respectively). Both the A and B complexes formed by d810 were supershifted by 58S (Fig. 5B, lane 3), and both the A and B complexes formed by EICP4 were supershifted by 12CA5 (Fig. 5B, lane 4). When EICP4 and d810 were incubated together with the probe (Fig. 5B, lane 5), a heterogeneous B complex in addition to A and B complexes formed by EICP4 and d810 appeared (Fig. 5B, compare lane 5 with lanes 1 and 2). When the monoclonal antibody 12CA5 was added to a binding reaction mixture containing EICP4, d810, and the probe (Fig. 5B, lane 6), the mobilities of the A and B complexes formed by d810 remained unchanged but the A and B complexes formed by EICP4 and the unique B complex supershifted (Fig. 5B, compare lane 6 with lanes 4 and 5). When the monoclonal antibody 58S alone or along with the monoclonal antibody 2CA5 was added to binding reaction mixtures containing EICP4, d810, and the probe, the A and B complexes formed by both EICP4 and d810 supershifted (Fig. 5B, lanes 7 and 8). Since 58S recognizes both d810 and EICP4, the nature of the protein-DNA complexes formed in Fig. 5B, lanes 7 and 8, could not be definitively interpreted. The mobility of the unique DNA-protein complex formed in lane 5 (Fig. 5B) was faster than the mobility of the B complex formed by EICP4 alone (and slower than the mobility of the B complex formed by d810) and that of the unique complex supershifted by the monoclonal antibody 12CA5 (Fig. 5B, lane 6). This observation is not easily reconciled with the conclusion that the unique complex is an octomer. If octomers were forming these complexes, then one would expect multiple unique complexes, which were not seen. Therefore, these data indicate that the unique protein-DNA complex was formed by one dimer of EICP4 and one dimer of d810.
FIG. 5.
Number of ICP4 dimers present in the B complex. (A) Schematic diagram showing EICP4 and d810. The locations of the epitopes recognized by 58S (black box) and 12CA5 (black oval) are indicated. The numbers on the top indicate the amino acid positions. (B) ICP4 DNA complexes formed between EICP4 and d810 with the probe pBend4D (Fig. 2C). The indicated proteins and monoclonal antibodies, 58S and 12CA5, were added to the indicated reactions. EICP4 and d810 formed a unique B complex (lane 5, indicated by a white star) in addition to the A and B complexes formed by EICP4 and d810. This additional B complex was supershifted by 12CA5 (lane 6, indicated by a black star). The A complexes are indicated by a white circle and a white diamond for d810 and EICP4, respectively; the B complexes are indicated by double white circles and double white diamonds for d810 and EICP4, respectively. The supershifted A and B complexes are indicated by the corresponding black circle or black diamonds. The lack of adequate resolution prevented the identification of the complexes formed in lanes 7 and 8.
ICP4 contacts DNA outside its primary binding site in the B complex.
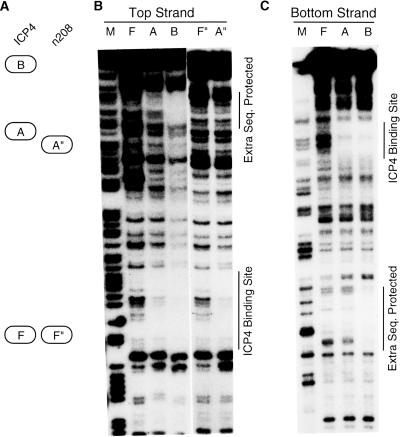
To find out whether ICP4 physically contacts DNA outside the primary binding site in the B complex, we performed DNase I footprinting of isolated ICP4-DNA complexes. Figure 6A shows a schematic diagram of the protein-DNA complexes used for this analysis. ICP4 protected only the ICP4 binding site in the A complex (Fig. 6B, lane A). ICP4 protected the cognate ICP4 binding site and an extra stretch of DNA about 50 bp downstream of the ICP4 binding site in the B complex (Fig. 6B, lane B). A similar pattern of DNA protection was observed for DNA probes labeled uniquely at the top or the bottom strand (Fig. 5C). This result suggests that ICP4 makes contact with DNA outside and downstream of its primary binding site in the B complex. The mutant n208 formed only an A complex with the probe and protected only the cognate ICP4 binding site (Fig. 6B, lane A″).
FIG. 6.
ICP4 contacts DNA outside its cognate binding site in the B complex. (A) Schematic diagram identifying the ICP4-DNA and n208-DNA complexes. (B) DNase I protection for the top strand of the probe by ICP4 and n208. (C) DNase I protection for the bottom strand of the probe by ICP4. A, ICP4-DNA A complex; A″, n208-DNA A complex; B, ICP4-DNA B complex; F, free DNA in ICP4-DNA binding reaction; F″, free DNA in n208-DNA binding reaction; M, CT- or G-specific sequencing ladder of the probe. The specific ICP4 binding site and extra DNA sequence (Seq.) protected are shown by vertical lines.
B complex formation and kinetics of ICP4-DNA interaction.
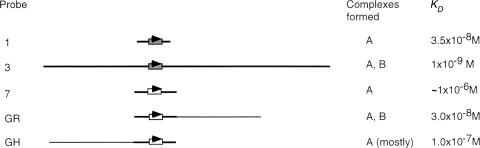
We determined the KD of DNA-protein complexes formed by ICP4 with short and long DNA probes containing different ICP4 binding sites. Under the binding condition used, ICP4 had a higher affinity for the long DNA probes, with >70 bp of nonspecific DNA downstream of the ICP4 binding site, than for the short (35 to 50 bp) DNA probes containing the corresponding ICP4 binding sites (Fig. 7). ICP4 has a much greater affinity for the binding site of the gC start site of transcription than for the binding site located downstream of the gC start site. The addition of extra DNA downstream of the stronger binding site increased affinity only threefold. However, the addition of extra DNA downstream of the weaker ICP4 binding site increased affinity >300-fold.
FIG. 7.
Probes used for determining KD of ICP4-DNA complexes. The probes are drawn to scale. Probe 1, equivalent to ICP4 promoter positions −16 to +15; probe 3, equivalent to ICP4 promoter positions −108 to +177; probe 7, equivalent to gC promoter positions +75 to +124, all relative to the transcription start site; probe GR, probe 7 trailed by 75 bp of plasmid DNA; probe GH, probe 7 appended downstream of 75 bp of plasmid DNA. The KD value for probe 7 was estimated to be 1 × 10−6 to 2 × 10−6 M; the value was difficult to determine due to the high degree of dissociation of the DNA-protein complex.
DISCUSSION
The oligomerization of regulatory factors plays a key role in the growth and differentiation of cells. Ligand-dependent dimerization of epidermal growth factor receptor is a prerequisite for normal receptor signaling (37). In contrast, ligand-independent oligomerization of (mutated) erythropoietin receptors induces cell transformation (56). DNA binding proteins, such as TBP, dimerize in solution or as a part of TFIID when not bound to DNA (55). A cellular transcription factor known as germ cell nuclear factor binds DNA as a dimer, oligomerizes in a DNA-dependent manner, and affects the expression of various factors involved in cell differentiation (27). Similar to germ cell nuclear factor, dimeric ICP4 oligomerizes in a DNA-dependent manner and presumably affects viral gene transcription. Here, we investigated the cis and trans requirements of the process. Our investigation revealed that DNA-dependent oligomerization of ICP4 involves multiple protein-DNA interactions and specific ICP4 dimer-ICP4 dimer interactions.
Data from previous reports (3, 15) indicated DNA-dependent oligomerization of ICP4. It was reported that ICP4 formed both low-mobility and high-mobility complexes with a latency-associated promoter but only a high-mobility complex with an ICP4 promoter (3). The low-mobility complexes with an ICP4 promoter were not obtained presumably because of the small size of the DNA probes used. It was also reported earlier that ICP4 formed higher-order complexes with the ICP4 promoter but contacted DNA only in the consensus binding site (15). The low-mobility complexes were presumably formed through protein-protein interactions alone, or the additional complexes were not stable enough to withstand the highly active and low-molecular-weight DNA cleaving reagent (1,10-phenanthroline-copper) employed for the protection experiments (15). We observed that DNA-dependent oligomerization of ICP4 involved DNA contacts in the specific binding site and another stretch of DNA about 50 bp downstream. We found that the DNA sequence of the additional protected patch was 5′-GCTAGCATCGATCCATGGA-3′. This sequence does not constitute an ICP4 binding site. The sequence is present upstream of the ICP4 binding site (in pBend4 probes M and N) (Fig. 2C), but the sequence did not cause the formation of the B complex nor was it protected from DNase I in the A complex (Fig. 6B).
Structure of oligomerized ICP4.
ICP4 is an obligatory dimer in solution (39). The results presented here indicate that B complex formation involves protein-protein interactions requiring the C termini of the interacting ICP4 dimers. Several lines of evidence attest this point: (i) ICP4 mutants lacking the C-terminal region were unable to form the B complex, (ii) deletion within the N-terminal region of the protein did not affect B complex formation, and (iii) an N-terminal domain mutant formed heterotetramers with WT ICP4. In addition, a specific ICP4 binding site and a downstream nonspecific stretch of DNA were required for oligomerization. We conclude that the B complex we analyzed in this study consists of two dimers of ICP4 interacting with each other. One of the interacting dimers interacts with a specific binding site, and another interacts with nonspecific DNA downstream of the specific binding. The requirement for DNA on only one side of the ICP4 binding site attests to the asymmetric nature of the ICP4 binding site and possibly ICP4 itself. However, it should be noted that higher-order complexes appeared in the EMSA when the conditions were right for B complex formation (Fig. 1 and 3). This suggests that higher-order oligomers can form. It has previously been reported that ICP4 may almost coat the viral genome early in infection (17). The interactions described in this study may be the basis for this phenomenon. The experiments conducted in this study were performed by EMSA, in which a large excess of nonspecific competitor was used. An intended outcome of this was the visualization of complexes requiring some specific protein-DNA interactions. Therefore, it cannot be ruled out that the DNA-dependent multimerization of ICP4 may not require specific protein-DNA interactions in virus-infected cells. Considering this possibility, using reconstituted in vitro transcription and gC promoter templates of defined composition and length downstream of the TATA box, we have previously observed that the requirement for DNA-protein contacts could be met by the presence of an ICP4 binding site in the leader, by the presence of a site more than 150 nucleotides further downstream, by the presence of an inserted site that normally acts to repress transcription, or by the addition of sufficient non-site-containing DNA (25).
Function of oligomerized ICP4.
Our results show that ICP4 has relatively high affinity (KD = 35 nM) for a binding site at the start of P4 transcription and that oligomerization does not dramatically increase the stability of binding (only threefold). For a weaker ICP4 binding site, the affinities of ICP4 for short and long DNA fragments differed by more than 2 orders of magnitude. Such an increase in affinity may play a significant role in ICP4-mediated transcriptional activation under physiological conditions. These data suggest that ICP4 may interact with long DNA molecules with multiple weak binding sites as effectively as with its DNA containing one strong binding site. A somewhat similar finding was reported previously, where it was observed that a slow-moving complex formed by ICP4 with a latency-associated prompter binding site dissociated much slower than the fast-moving complex formed by an ICP4 binding site probe (3). ICP4 binding sites located at gC and gD promoters are dispensable for the activation of the promoters (25, 53). If B complex formation significantly increases the affinity of ICP4 for DNA, a relatively weak binding site located in the appended DNA may serve as a docking site for ICP4 for subsequent promoter activation, particularly if ICP4 can extend its reach, so to speak, by oligomerizing on the viral genome. This in turn could further promote the assembly of transcription preinitiation complexes through protein-protein interactions between ICP4 and TFIID (7, 23).
n208 (which is unable to form a B complex) activates transcription very poorly from supercoiled DNA templates in vitro and activates only a subset of viral genes during infection (7, 14). The burst size of n208 is over 600-fold lower than that of WT HSV (14). The lack of activation by C-terminal domain mutants indirectly indicates that ICP4-ICP4 interactions involved in B complex formation may be important in ICP4-mediated activation. B complex formation, however, is apparently not essential for ICP4-mediated repression in vitro. For example, n208 efficiently represses transcription from promoters containing a single relatively strong ICP4 binding site located at the start site of transcription (26). ICP4-mediated repression from such promoters may depend more on ICP4-TBP-TFIIB cooperative interactions (26, 35, 54) and DNA distortion by TBP (28, 31, 32, 36) and ICP4 (18, 35). However, B complex formation may be important for repression as well when the ICP4 binding site is not strong.
The C-terminal region of ICP4 is involved in other protein-protein interactions. ICP4 and analogs in other herpesviruses interact with TFIID in vitro (2, 7, 23). Biochemical studies have shown that the C-terminal region of ICP4 and TAF250 of TFIID are directly involved in ICP4-TFIID interaction (7). Biochemical studies have also shown that the HSV regulatory protein ICP0 interacts with the C-terminal region of ICP4 (58). The C-terminal region of ICP4 is a large domain and may provide interfaces for many different interacting factors. The C-terminal regions of ICP4 analogs are highly conserved among different alphaherpesviruses (9, 57). The available data indicate that even small mutations in the C-terminal domain of ICP4 significantly alter the biological activities of the protein (6, 10). Studies are under way to determine the region within the C-terminal domain of ICP4 involved in DNA-dependent oligomerization to explore its contribution activation independent of other functions in the C-terminal region of ICP4.
Acknowledgments
This work was supported by NIH grants AI30612 and AI27431 to N.A.D.
Footnotes
Published ahead of print on 20 June 2007.
REFERENCES
- 1.Allen, K. E., and R. D. Everett. 1997. Mutations which alter the DNA binding properties of the herpes simplex virus type 1 transactivating protein Vmw175 also affect its ability to support virus replication. J. Gen. Virol. 78:2913-2922. [DOI] [PubMed] [Google Scholar]
- 2.Abmayr, S. M., J. L. Workman, and R. G. Roeder. 1988. The pseudorabies virus immediate early protein stimulates in vitro transcription by facilitating TFIID:promoter interactions. Genes Dev. 2:542-553. [DOI] [PubMed] [Google Scholar]
- 3.Batchelor, A. H., K. W. Wilcox, and P. O'Hare. 1994. Binding and repression of the latency-associated promoter of herpes simplex virus by the immediate early 175K protein. J. Gen. Virol. 75:753-767. [DOI] [PubMed] [Google Scholar]
- 4.Bates, P. A., and N. A. DeLuca. 1998. The polyserine tract of herpes simplex virus ICP4 is required for normal viral gene expression and growth in murine trigeminal ganglia. J. Virol. 72:7115-7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bohenzky, R. A., A. G. Papavassiliou, I. H. Gelman, and S. Silverstein. 1993. Identification of a promoter mapping within the reiterated sequences that flank the herpes simplex virus type 1 UL region. J. Virol. 67:632-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruce, J. W., and K. W. Wilcox. 2002. Identification of a motif in the C terminus of herpes simplex virus regulatory protein ICP4 that contributes to activation of transcription. J. Virol. 76:195-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carrozza, M. J., and N. A. DeLuca. 1996. Interaction of the viral activator protein ICP4 with TFIID through TAF250. Mol. Cell. Biol. 16:3085-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrozza, M. J., and N. A. DeLuca. 1998. The high mobility group protein 1 is a coactivator of herpes simplex virus ICP4 in vitro. J. Virol. 72:6752-6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung, A. K. 1989. DNA nucleotide sequence analysis of the immediate-early gene of pseudorabies virus. Nucleic Acids Res. 17:4637-4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Compel, P., and N. A. DeLuca. 2003. Temperature-dependent conformational changes in herpes simplex virus ICP4 that affect transcription activation. J. Virol. 77:3257-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Courtney, R. J., and M. Benyesh-Melnick. 1974. Isolation and characterization of a large molecular weight polypeptide of herpes simplex virus type 1. Virology 62:539-551. [DOI] [PubMed] [Google Scholar]
- 12.DeLuca, N. A., and P. A. Schaffer. 1985. Activation of immediate-early, early, and late promoters by temperature-sensitive and wild-type forms of herpes simplex virus type 1 protein ICP4. Mol. Cell. Biol. 5:1997-2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeLuca, N. A., and P. A. Schaffer. 1987. Activities of herpes simplex virus type 1 (HSV-1) ICP4 genes specifying nonsense peptides. Nucleic Acids Res. 15:4491-4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeLuca, N. A., and P. A. Schaffer. 1988. Physical and functional domains of the herpes simplex virus transcriptional regulatory protein ICP4. J. Virol. 62:732-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DiDonato, J. A., and M. T. Muller. 1989. DNA binding and gene regulation by herpes simplex virus type 1 protein ICP4 and involvement of the TATA element. J. Virol. 63:3737-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Everett, R. D. 1984. Trans-activation of transcription by herpes virus product: requirements for two HSV-1 immediate-early polypeptides for maximum activity. EMBO J. 3:3135-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Everett, R. D., G. Sourvinos, and A. Orr. 2003. Recruitment of herpes simplex virus type 1 transcriptional regulatory protein ICP4 into foci juxtaposed to ND10 in live, infected cells. J. Virol. 77:3680-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faber, S. W., and K. W. Wilcox. 1986. Association of the herpes simplex virus regulatory protein ICP4 with specific nucleotide sequences in DNA. Nucleic Acids Res. 14:6067-6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faber, S. W., and K. W. Wilcox. 1988. Association of the herpes simplex virus regulatory protein ICP4 with sequences spanning the ICP4 gene transcription initiation site. Nucleic Acids Res. 16:555-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Field, J., J.-I. Nikawa, D. Broek, B. MacDonald, L. Rodgers, I. A. Wilson, R. A. Lerner, and M. Wigler. 1988. Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol. Cell. Biol. 8:2159-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freeman, M. J., and K. L. Powell. 1982. DNA-binding properties of a herpes simplex virus immediate early protein. J. Virol. 44:1084-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Godowski, P. J., and D. M. Knipe. 1986. Transcriptional control of herpes virus gene expression: gene functions required for positive and negative regulation. Proc. Natl. Acad. Sci. USA 83:256-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grondin, B., and N. DeLuca. 2000. Herpes simplex virus type 1 ICP4 promotes transcription preinitiation complex formation by enhancing the binding of TFIID to DNA. J. Virol. 74:11504-11510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gu, B., R. Rivera-Gonzalez, C. A. Smith, and N. A. DeLuca. 1993. Herpes simplex virus infected cell polypeptide 4 preferentially represses Sp1-activated over basal transcription from its own promoter. Proc. Natl. Acad. Sci. USA 90:9528-9532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu, B., and N. A. DeLuca. 1994. Requirements for activation of the herpes simplex virus glycoprotein C promoter in vitro by the viral regulatory protein ICP4. J. Virol. 68:7953-7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu, B., R. Kuddus, and N. A. DeLuca. 1995. Repression of activator-mediated transcription by herpes simplex virus ICP4 via a mechanism involving interactions with the basal transcription factors TATA-binding factor and TFIIB. Mol. Cell. Biol. 15:3618-3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gu, P., D. H. Morgan, M. Sattar, X. Xu, R. Wagner, M. Ravisconi, O. Lichtarge, and A. J. Cooney. 2005. Evolutionary trace-based peptides identify a novel asymmetric interaction that mediates oligomerization in nuclear receptors. J. Biol. Chem. 250:31818-31829. [DOI] [PubMed] [Google Scholar]
- 28.Horikoshi, M., C. Bertuccioli, R. Takada, J. Wang, T. Yamamoto, and R. G. Roeder. 1992. Transcription factor IID induces DNA bending upon binding to the TATA element. Proc. Natl. Acad. Sci. USA 89:1060-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Imbalzano, A. N., A. A. Shepard, and N. A. DeLuca. 1990. Functional relevance of specific interactions between herpes simplex virus type 1 ICP4 and sequences from the promoter-regulatory domain of the viral thymidine kinase gene. J. Virol. 64:2620-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim, J., C. Zwieb, C. Wu, and S. Adhya. 1989. Bending of DNA by gene regulatory proteins: construction and use of a DNA bending vector. Gene 85:16-23. [DOI] [PubMed] [Google Scholar]
- 31.Kim, J. L., D. B. Nikolov, and S. K. Burley. 1993. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature (London) 365:520-527. [DOI] [PubMed] [Google Scholar]
- 32.Kim, Y., J. H. Geiger, S. Hahn, and P. B. Sigler. 1993. Crystal structure of a yeast TBP/TATA box complex. Nature (London) 365:512-520. [DOI] [PubMed] [Google Scholar]
- 33.Kim, D.-B., S. Zabierowski, and N. A. DeLuca. 2002. The initiator element in a herpes simplex virus type 1 late-gene promoter enhances activation by ICP4, resulting in abundant late-gene expression. J. Virol. 76:1548-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kristie, T. M., and B. Roizman. 1986. DNA-binding site of major regulatory protein a4 specifically associated with promoter-regulatory domains of alpha genes of herpes simplex virus type 1. Proc. Natl. Acad. Sci. USA 83:4700-4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuddus, R., B. Gu, and N. A. DeLuca. 1995. Relationship between TATA-binding protein and herpes simplex virus type 1 ICP4 DNA-binding sites in complex formation and repression of transcription. J. Virol. 69:5568-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuddus, R., and M. C. Schmidt. 1993. Effect of the non-conserved N-terminus on the DNA binding activity of the yeast TATA binding protein. Nucleic Acids Res. 21:1789-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lemmon, M. A., and J. Schlessinger. 1994. Regulation of signal transduction and signal diversity by receptor oligomerization. Trends Biochem. Sci. 19:459-463. [DOI] [PubMed] [Google Scholar]
- 38.Maxam, A. M., and W. Gilbert. 1980. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 65:499-559. [DOI] [PubMed] [Google Scholar]
- 39.Metzler, D. W., and K. W. Wilcox. 1985. Isolation of herpes simplex virus regulatory protein ICP4 as a homodimeric complex. J. Virol. 55:329-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Michael, N., and B. Roizman. 1993. Repression of the herpes simplex virus 1 a4 gene by its gene product occurs within the context of the viral genome and is associated with all three identified cognate sites. Proc. Natl. Acad. Sci. USA 90:2286-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Hare, P., and G. S. Hayward. 1985. Evidence for a direct role for both the 175,000- and 110,000-molecular-weight immediate-early proteins of herpes simplex virus in the transactivation of delayed-early promoters. J. Virol. 53:751-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Panagiotidis, C. A., and S. J. Silverstein. 1999. The host-cell architectural protein HMG I(Y) modulates binding of herpes simplex virus type 1 ICP4 to its cognate promoter. Virology 256:64-74. [DOI] [PubMed] [Google Scholar]
- 43.Paterson, T., and R. D. Everett. 1988. The regions of the herpes simplex virus type 1 immediate early protein Vmw175 required for site specific DNA binding closely correspond to those involved in transcriptional regulation. Nucleic Acids Res. 15:11005-11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paterson, T., and R. D. Everett. 1988. Mutational dissection of the HSV-1 immediate-early protein Vmw175 involved in transcriptional transactivation and repression. Virology 166:186-196. [DOI] [PubMed] [Google Scholar]
- 45.Quinlan, M., and D. Knipe. 1985. Stimulation of expression of a herpes simplex virus DNA-binding protein by two viral functions. Mol. Cell. Biol. 5:957-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roberts, M. S., A. Boundy, P. O'Hare, M. C. Pizzorno, D. M. Ciufo, and G. S. Hayward. 1988. Direct correlation between a negative autoregulatory response element at the cap site of the herpes simplex virus type 1 IE175 (α4) promoter and a specific binding site for the IE175 (ICP4) protein. J. Virol. 62:4307-4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Samaniego, L. A., A. L. Webb, and N. A. DeLuca. 1995. Functional interactions between herpes simplex virus immediate-early proteins during infection: gene expression as a consequence of ICP27 and different domains of ICP4. J. Virol. 69:5705-5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Segel, I. H. 1968. Biochemical calculations. John Wiley & Sons, New York, NY.
- 49.Shepard, A. A., and N. A. DeLuca. 1991. Activities of heterodimers composed of DNA-binding- and transactivation-deficient subunits of the herpes simplex virus regulatory protein ICP4. J. Virol. 65:299-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shepard, A. A., P. Tolentino, and N. A. DeLuca. 1990. Transdominant inhibition of the herpes simplex virus transcriptional regulatory protein ICP4 by heterodimer formation. J. Virol. 64:3916-3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shepard, A. A., A. N. Imbalzano, and N. A. DeLuca. 1989. Separation of primary structural components conferring autoregulation, transactivation, and DNA-binding properties to the herpes simplex virus transcriptional regulatory protein ICP4. J. Virol. 63:3714-3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Showalter, S. D., M. Zweig, and B. Hampar. 1981. Monoclonal antibodies to herpes simplex virus type 1 proteins, including the immediate-early protein ICP 4. Infect. Immun. 34:684-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smiley, J. R., D. C. Johnson, L. I. Pizer, and R. D. Everett. 1992. The ICP4 binding sites in the herpes simplex virus type 1 glycoprotein (gD) promoter are not essential for efficient gD transcription during virus infection. J. Virol. 66:623-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith, C. A., P. Bates, R. Rivera-Gonzalez, B. Gu, and N. A. DeLuca. 1993. ICP4, the major transcriptional regulatory protein of herpes simplex virus type 1, forms a tripartite complex with TATA-binding protein and TFIIB. J. Virol. 67:4676-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taggart, A. K. P., and F. Pugh. 1996. Dimerization of TFIID when not bound to DNA. Science 272:1331-1333. [DOI] [PubMed] [Google Scholar]
- 56.Watowich, S. S., A. Yoshimura, G. D. Longmore, D. J. Hilton, Y. Yoshimura, and H. F. Lodish. 1992. Homodimerization and constitutive activation of the erythropoietin receptor. Proc. Natl. Acad. Sci. USA 89:2140-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Watson, R., and J. B. Clements. 1980. A herpes simplex virus type 1 function continuously required for early and late virus RNA synthesis. Nature 285:329-330. [DOI] [PubMed] [Google Scholar]
- 58.Yao, F., and P. A. Schaffer. 1994. Physical interaction between the herpes simplex virus type 1 immediate-early regulatory proteins ICP0 and ICP4. J. Virol. 68:8158-8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yeh, L., and P. A. Schaffer. 1993. A novel class of transcripts expressed with late kinetics in the absence of ICP4 spans the junction between the long and short segments of the herpes simplex virus type 1 genome. J. Virol. 67:7373-7382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zabierowski, S., and N. A. DeLuca. 2004. Differential cellular requirements for activation of herpes simplex virus type 1 early (tk) and late (gC) promoters by ICP4. J. Virol. 78:6162-6170. [DOI] [PMC free article] [PubMed] [Google Scholar]