Abstract
Ty1 reverse transcriptase/RNase H (RT/RH) is exquisitely sensitive to manganese concentrations. Elevated intracellular free Mn2+ inhibits Ty1 retrotransposition and in vitro Ty1 RT-polymerizing activity. Furthermore, Mn2+ inhibition is not limited to the Ty1 RT, as this ion similarly inhibits the activities of both avian myeloblastosis virus and human immunodeficiency virus type 1 RTs. To further characterize Mn2+ inhibition, we generated RT/RH suppressor mutants capable of increased Ty1 transposition in pmr1Δ cells. PMR1 codes for a P-type ATPase that regulates intracellular calcium and manganese ion homeostasis, and pmr1 mutants accumulate elevated intracellular manganese levels and display 100-fold less transposition than PMR1+ cells. Mapping of these suppressor mutations revealed, surprisingly, that suppressor point mutations localize not to the RT itself but to the RH domain of the protein. Furthermore, Mn2+ inhibition of in vitro RT activity is greatly reduced in all the suppressor mutants, whereas RH activity and cleavage specificity remain largely unchanged. These intriguing results reveal that the effect of these suppressor mutations is transmitted to the polymerase domain and suggest biochemical communication between these two domains during reverse transcription.
The yeast Saccharomyces cerevisiae harbors long terminal repeat (LTR) retrotransposons known as Ty elements, the most abundant being Ty1. Transcription of the 6-kb Ty1 element produces a terminally redundant RNA molecule from which the structural precursor protein, Gag, and the precursor to the enzymes required for retrotransposition, Gag-Pol, are translated. Specifically, gag encodes the capsid protein, and pol encodes the protease, integrase, and reverse transcriptase/RNase H (RT/RH) (4, 12, 20). As in the case of retroviruses, pol is initially expressed as a Gag-Pol polyprotein from a programmed ribosomal frameshift (3). Upon transcription of the Ty1 RNA, its life cycle can be broken down into three stages; assembly, reverse transcription, and integration. Assembly consists of the formation of pre-virus-like particles (pre-VLPs) from Gag and Gag-Pol proteins with Ty1 RNA in the cytoplasm; subsequent proteolytic cleavage leads to a mature VLP in which reverse transcription is thought to occur (22), and upon completion, the DNA-containing VLPs are transported to the nucleus, where integration into the host genome can occur.
Generation of full-length, double stranded cDNA from Ty1 RNA is a complicated process that includes two strand transfer events. Briefly, reverse transcription is initiated from a host-encoded Meti-tRNA that anneals to the complementary primer binding site, immediately downstream of the 5′ LTR (2, 6). Elongation from this primer produces minus-strand strong-stop DNA and subsequent strand transfer to complementary regions of the 3′ LTR, and further elongation results in bulk minus-strand synthesis. Concomitant with this elongation, RH degrades the RNA template, with the exception of the polypurine tract (PPT) sequence, immediately upstream of U3, which serves as a primer for plus-strand synthesis (11, 16). As in the case with minus-stand synthesis, elongation from the PPT primer requires a strand transfer event before further elongation generates the full-length Ty1 cDNA. This entire process can be carried out in vitro and requires divalent cations, either Mg2+ or Mn2+ (12, 27), as well as the necessary primers, templates, and deoxynucleoside triphosphates (dNTPs) for active RT and RH activities.
It has been previously demonstrated that disruption of the PMR1 gene blocks the Ty1 life cycle at the reverse transcription stage (5). PMR1 codes for a Golgi-localized P-type ATPase that regulates intracellular calcium and manganese ion homeostasis (15), and pmr1 mutants accumulate fivefold-higher total cellular manganese levels and display 100-fold-reduced retrotransposition relative to PMR1 cells. Specifically, there is a defect in cDNA formation in VLPs, a defect explained by inhibition of the RT-polymerizing activity of Ty1 RT/RH by elevated Mn2+ concentrations (5).
We report here the selection of Mn2+ suppressor mutants capable of elevated retrotransposition in pmr1Δ cells. Surprisingly, these suppressor mutants contain mutations which map not to the RT itself but to the RH domain of the protein. Furthermore, Mn2+ inhibition of in vitro RT activity is greatly reduced in all the suppressor mutants, whereas RH activity and cleavage specificity remain largely unchanged. Taken together, these intriguing results reveal that the effect of these suppressor mutations is transmitted to the polymerase domain and thus suggest biochemical communication between these two domains during reverse transcription. However, the nature of this communication is still unknown.
MATERIALS AND METHODS
Strains and media.
The yeast strains used in this study were JB740 (MATα his3Δ200 leu2Δ1 ura3-167) and yEB104A (JB740 with pmr1Δ::hphMX4). Media were prepared as described previously (26).
Plasmid constructions.
Plasmids pECB9, a GAL-Ty1-HIS3 plasmid, and pGEX-4T-3-115A, with recombinant Ty1 RT subcloned into the BamHI and XhoI sites of pGEX-4T-3, were previously described (5). For glutathione S-transferase (GST)-tagged protein purification of Mn2+ suppressor mutants, suppressor point mutations were excised from mutagenized pECB9 and cloned into pGEX-4T-3-115A.
Mutagenic PCR and gapped plasmid repair.
Mutagenic amplification of the ClaI-NheI region of pECB9 with approximately 300 bp of flanking sequence at each end was performed using primers JB4036 (5′-CCCAGAGAAGTTGACCCCA-3′) and JB4037 (5′-CCGGGGGATCCTCTAGAGT-3′). Ninety-six mutagenic PCR mixtures (50 μl each) contained 1.5 mM MgCl2, 100 μM dNTPs, 13 ng pECB3K, 0.2 mM each primer, and 2.5 units of AmpliTaq DNA polymerase (Roche Molecular Biochemicals, Mannheim, Germany). Reactions were performed in a Perkin-Elmer 9600 PCR machine (Perkin-Elmer, Boston, MA) using the following method: 95°C for 5 min; 95°C for 1 min, 53°C for 1 min, and 72°C for 1 min (30 cycles); and a final 5-min extension at 72°C. The conditions described above generated one misincorporation per 1,800 bp polymerized or approximately one misincorporation per PCR product.
Transposition assay.
Transformants containing the URA3-marked Ty1 donor plasmid, pECB9, and mutant variants were patched onto SC-Leu-Ura with 2% glucose (10). After 2 days at 30°C, yeast patches were replica plated to similar medium with 2% galactose and incubated at 22°C for 3 days. After this period, plates were replica plated to YPD, incubated at 30°C for 1 day, and then replica plated again the following day to SC-His for an additional day at 30°C. Transposition was recorded quantitatively by the ability of patches to grow on SC-His plates. Quantitative assays were performed similarly by scraping cells from a 1-inch square patch grown on YPD at 30°C overnight into 1 ml H2O and diluted appropriately, with titers determined on YPD and SC-His plates.
Protein purification.
Recombinant wild-type (WT) and mutant Ty1 RTs (27) containing a cleavable N-terminal GST tag were expressed in Escherichia coli and purified by affinity chromatography, similar to the method described previously for human immunodeficiency virus type 1 (HIV-1) RT (17). The GST-tagged protein was eluted from a glutathione Sepharose 4B (Amersham Pharmacia Biotech, Piscataway, NJ) gravity column with buffer (50 mM HEPES-KOH [pH 7.8], 200 mM KCl, 10% glycerol, and 2 mM dithiothreitol [DTT]) containing 15 mM reduced glutathione. Samples were then dialyzed overnight into the storage buffer (50 mM HEPES-KOH [pH 7.8], 100 mM KCl, 50% glycerol, and 1 mM DTT) and stored at −80°C in 1-ml and 50-μl aliquots.
RT assay.
Assays of recombinant WT and mutant Ty1 RT proteins were performed in 30-μl reaction mixtures [50 mM HEPES-KOH (pH 7.8), 3 mM DTT, 0.2 μM dGTP, 0.5 μCi of [α-32P]dGTP, 1 μg/ml oligo(dG)12-18, 10 μg/ml poly(rC)n, and the indicated concentrations of MgCl2 and/or MnCl2]. Reactions were set up on ice, carried out for 1 h at 22°C, and kept on ice while being spotted onto DE81 anion-exchange paper (Whatman International Ltd., Maidstone, England) (5). In reactions involving heparin, enzyme and template were incubated together for 10 min before heparin (Sigma; 1 mg/ml) and divalent metal cations were added.
RH assays.
To analyze a generic RNA-DNA hybrid template (adapted from reference 18), RH activity was evaluated on a 5′-end-labeled 40-nucleotide RNA template (Dharmacon Research) annealed to a 30-nucleotide DNA primer (Integrated DNA Technologies), kindly provided by Stuart Le Grice. To prepare the RNA/DNA duplexes, a threefold molar excess of DNA was hybridized with the RNA in a buffer containing 10 mM Tris-HCl (pH 7.8) and 2.5 mM MgCl2. The mixture was heated at 90°C for 1 min and slowly cooled to room temperature. A reaction mixture containing 50 nM template-primer was prepared in a buffer of 10 mM Tris-HCl (pH 7.8), 80 mM NaCl, 5 mM DTT, 5 mM MgCl2, and various concentrations of MnCl2 as indicated in the figures. Hydrolysis was initiated by the addition of equal activities (normalized by RT activity) of WT or mutant enzymes at 22°C for 1 h in a final volume of 30 μl. The reaction was stopped by addition of an equal volume of formamide sample buffer (70% formamide, 6.7% formaldehyde, 10 mM EDTA, 0.1% bromophenol blue, 1× MOPS [morpholinepropanesulfonic acid] buffer).
In analyzing a PPT-containing RNA-DNA hybrid template (adapted from reference 28), RH activity was assayed on a radiolabeled 32P-labeled RNA 28-nucleotide (5′-AUUACAUUAUGGGUGGUAUGUUGGAAUA-3′; Dharmacon Research)-DNA 28-nucleotide (JB6856, 5′-TATTCCAACATACCACCCATAATGTAAT-3′) sequence consisting of the Ty1 PPT with flanking upstream sequence from the Ty1 3′ untranslated region and downstream sequence from the 3′ U3 segment. To prepare the RNA/DNA duplexes, a threefold molar excess of DNA was hybridized with the RNA in a buffer containing 50 mM Tris-HCl (pH 7.8), 15 mM NaCl, 8 mM β-mercaptoethanol, and 5 mM MgCl2. The mixture was heated at 90°C for 1 min and slowly cooled to room temperature. Hydrolysis was initiated at 22°C for 1 h as described above with the MnCl2 concentrations indicated in the figures. Reactions were stopped by addition of formamide sample buffer. In reactions involving heparin, enzyme and template were incubated together for 10 min before heparin (Sigma; 1 mg/ml) and divalent metal cations were added to initiate the RH cleavage.
RESULTS
Isolation of “Mn2+ suppressor” mutants.
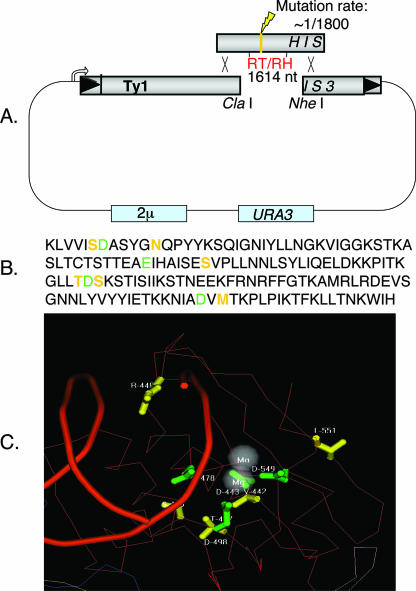
To further characterize the effects of elevated Mn2+ on Ty1 transposition, we mutagenized a pGal-Ty1-HIS3 plasmid and selected for reversion of the retrotransposition-defective phenotype. The entire RT/RH gene on the pGal-Ty1-HIS3 donor plasmid was mutagenized by a PCR-based method in which mutagenized PCR products were incorporated into the RT/RH (Fig. 1A) region of the Ty1-HIS3 element by in vivo gapped plasmid repair (5, 21). Colonies containing mutagenized pGal-Ty1-HIS3 donor plasmids were screened for elevated Ty1 transposition in a pmr1Δ yeast strain. The retrotransposition assay consists of a series of replica-plating steps, in which retrotransposition is induced on plates containing galactose and transposition is scored by the number of His+ papillae on plates lacking histidine (see Materials and Methods). Candidate His+ colonies were patched, and retrotransposition frequency was retested. Donor plasmids demonstrating elevated Ty1 retrotransposition levels in the pmr1Δ yeast strain were recovered, and the point mutations in recovered pGal-Ty1-HIS3 plasmids were identified by sequencing (Table 1).
FIG. 1.
Generation and positions of “Mn2+ suppressor” mutations. (A) Generation of Mn2+ suppressor mutations using mutagenic PCR and gapped repair. Conditions were such that there was one misincorporation per 1,800 bp polymerized, or approximately one misincorporation per PCR product. (B) Amino acid sequences of RT and RH. Green residues indicate metal-chelating amino acids, and yellow residues indicate Mn2+ suppressor mutations discovered in this study. (C) Ty1 Mn2+ suppressor mutations mapped onto the HIV-1 crystal structure (19, 27, 30). Green residues indicate metal-chelating amino acids, and yellow residues indicate Ty1 Mn2+ suppressor mutants on a red polypeptide backbone.
TABLE 1.
“Mn2+ suppressor” mutations
Suppressor point mutations lie in the RH domain.
Nearly all Mn2+ suppressor mutations identified in this study are present in the RH domain (Table 1); exceptions include mutant 1 (which has a missense mutation in the connector domain lying between the RT and RH domains) and mutant 5 (which has missense mutations in both the polymerase and RH domains). Isolate 5 was not further examined because we know that the N398D mutation in the RH domain alone (i.e., mutation 4) is sufficient to explain its phenotype. The location of these suppressor mutations is especially surprising given that it has been previously demonstrated that Mn2+ inhibits in vitro RT activity (5). Mapping these mutations onto the HIV-1 RT/RH crystal structure (Fig. 1C) (19, 27, 30) shows that most of these mutations cluster in three dimensions around the metal binding pocket of the RH domain and that four of these mutations lie adjacent to the residues that chelate divalent metal ions in RH.
HIS3 mobilization by manganese suppressor mutants.
The strength of the various Mn2+ suppressor mutants was determined by a quantitative transposition assay (see Materials and Methods). HIS3 marker mobilization, a reflection of Ty1 transposition frequency, of the suppressor mutants in the pmr1Δ strain ranged from 2.5- to 23.5-fold above WT Ty1 levels (Table 2). Since the Mn2+ suppressor mutants were selected in cells lacking PMR1, it was possible that certain Ty1 mutants were actually dependent on the elevated intracellular Mn2+ level in these cells. To test this possibility, quantitative transposition assays were also performed with PMR1 cells (Table 3). Indeed, transposition in PMR+ cells was decreased in all Mn2+ suppressors by 1.5- to 27-fold. The most dramatic losses in Ty1 transposition occurred in the two mutants containing the S469G mutation (9-fold and 27-fold in PMR1 cells). The manganese dependence of these two mutants is not altogether surprising given that the S469G and S392C substitutions occur within one residue of the metal ion-chelating residues.
TABLE 2.
HIS3 mobilization of suppressor mutants in the pmr1Δ strain
| Protein | Transposition frequency (mean ± SEM)a | Fold increase relative to WT Ty1 elementb |
|---|---|---|
| WT | (8.9 ± 0.71) × 10−5 | 1 |
| Y299C | (4.1 ± 0.47) × 10−4 | 4.65 |
| M520I | (2.1 ± 0.10) × 10−3 | 23.52 |
| S442P | (2.2 ± 0.17) × 10−4 | 2.54 |
| N398D | (4.5 ± 0.73) × 10−4 | 5.05 |
| S469G S392C | (7.6 ± 1.1) × 10−4 | 8.62 |
| S469G | (4.7 ± 1.0) × 10−4 | 5.32 |
Retroransposition frequencies of the WT element and suppressor mutants in the pmr1Δ strain were calculated as the number of His+ CFU/total CFU after 2 days on SC-Ura Gal medium. Results are from three measurements.
The relative increase in transposition-gained suppressor mutations was calculated as transposition frequency of suppressor mutant/transposition frequency of WT.
TABLE 3.
HIS3 mobilization of suppressor mutants in the PMR1 strain
| Protein | Transposition frequency (mean ± SEM)a | Fold decrease relative to WT Ty1 elementb |
|---|---|---|
| WT | (1.5 ± 0.50) × 10−2 | 1 |
| Y299C | (6.7 ± 0.07) × 10−3 | 2.31 |
| M520I | (9.7 ± 1.8) × 10−3 | 1.58 |
| S442P | (9.2 ± 0.70) × 10−3 | 1.68 |
| N398D | (3.6 ± 0.64) × 10−3 | 4.31 |
| S469G S392C | (1.6 ± 0.4) × 10−3 | 9.43 |
| S469G | (5.8 ± 1.1) × 10−4 | 27.0 |
Retroransposition frequencies of the WT element and suppressor mutants in the PMR1 strain were calculated as the number of His+ CFU/total CFU after 2 days on SC-Ura Gal medium. Results are from three measurements.
The relative decrease in retrotransposition frequency, or manganese dependence, was calculated as retrotransposition frequency of WT/retrotransposition frequency of suppressor mutant.
Suppressor mutant RTs are resistant to Mn2+ in vitro.
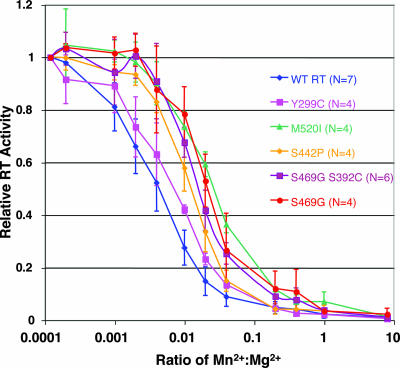
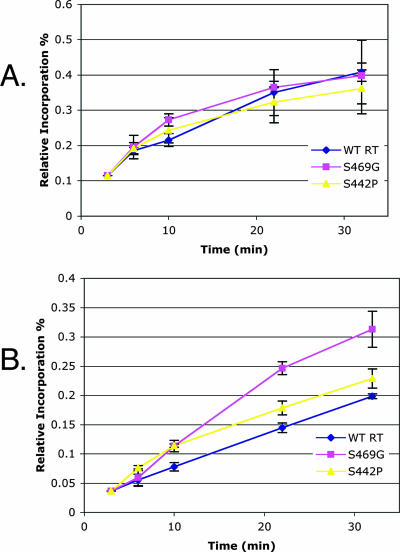
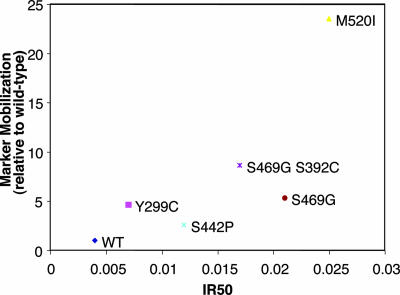
As it has been previously documented that trace amounts of Mn2+ could inhibit Ty1 in vitro RT activity (5), we wanted to ascertain whether such inhibition had been reduced or negated in the manganese suppressors. To determine whether manganese could indeed inhibit the Mn2+ suppressor mutants, in vitro RT activity of GST tag-purified mutant RT/RH (27) was assayed by incorporation of radiolabeled dGTP into the dG(12-18)-poly(rC) primer-template at a constant Mg2+ level and with increasing levels of Mn2+ (Fig. 2). Absolute levels of DNA synthesis for the suppressor mutants were comparable to that for the WT enzyme (data not shown), and as in the case of the WT enzyme, Mn2+ inhibition was observed for all of the mutants. However, every Mn2+ suppressor mutant examined displayed some significant quantitative increase in the degree of resistance to manganese-dependent inhibition of RT activity. In addition, given that the estimated ratio of free intracellular Mn2+ to Mg2+ in yeast cells (1 μM/1 mM = 0.001 [5]) falls along the steepest part of the inhibition curve, the observed quantitative differences in manganese resistance are predicted to have a substantial impact on the RT-polymerizing activity in vivo, and thus on Ty1 retrotransposition levels, in the presence of elevated levels of intracellular free Mn2+. Furthermore, in agreement with previous results, when representative mutants were analyzed in a time course, the suppressor mutants displayed an elevated polymerization rate compared to WT RT in the presence of manganese (Fig. 3). This result suggests that the WT RT polymerization rate is impaired in the presence of elevated manganese levels and that this inhibition is “rescued” in the suppressor mutants. Whereas the correlation between in vitro Mn2+ inhibition and suppressor activity is not directly proportional, the correlation is very good; the weaker suppressors show relatively small effects on Mn+2 inhibition (Fig. 4).
FIG. 2.
“Manganese suppressor” mutants display resistance to elevated Mn2+ in in vitro RT assays. Assays of recombinant WT and mutant Ty1 RT proteins were performed in 30-μl reaction mixtures [50 mM HEPES-KOH (pH 7.8), 3 mM DTT, 0.2 μM dGTP, 0.5 μCi of [α-32P]dGTP, 1 μg/ml oligonucleotide (dG)12-18, 10 μg/ml poly(rC)n, and the indicated concentrations of MgCl2 and/or MnCl2]. Reactions were set up on ice and carried out for 1 h at 22°C, and products were kept on ice while being spotted onto DE81 anion-exchange paper. The RT activity of manganese suppressor mutant proteins was normalized to WT RT activity, and N represents the number of times that each RT protein was assayed.
FIG. 3.
“Manganese suppressor” mutants display an increased polymerization rate in the presence of elevated manganese levels. (A) Time course assay with the WT and representative mutants in the absence of manganese. (B) Time course assay with the WT and representative mutants in the presence of a 0.1 Mn2+/Mg2+ ratio. Assays of recombinant WT and mutant Ty1 RT proteins were performed in 30-μl reaction mixtures [50 mM HEPES-KOH (pH 7.8), 3 mM DTT, 0.2 μM dGTP, 0.5 μCi of [α-32P]dGTP, 1 μg/ml oligonucleotide (dG)12-18, 10 μg/ml poly (rC)n]. Reactions were set up on ice and carried out at the indicated times (3, 6, 10, 22, and 32 min) at 22°C, and products were kept on ice while being spotted onto DE81 anion-exchange paper. The RT activity of manganese suppressor mutant proteins was normalized to WT RT activity.
FIG. 4.
Plot of HIS3 marker mobilization (Table 2) against IR50 (Mn2+/Mg2+ ratio at which WT and suppressor mutant RTs display 50% activity). The 50% RT activities are calculated from logarithmic regressions of the steepest part of the WT and suppressor mutant Mn2+ inhibition curves from Fig. 2.
Is RH activity required for Mn2+ inhibition of RT?
Based on the unexpected location of the suppressor mutations in the RH domain, we examined whether RH activity was required for RT activity and its inhibition by Mn2+. To address this issue, we made use of a D468S RNase catalytically dead mutant (28). We assayed D468S RT incorporation of radiolabeled dGTP into the dG(12-18)-poly(rC) primer-template at a constant Mg2+ level and increasing levels of Mn2+, and this RH mutant still displayed in vitro Mn2+ inhibition similar to that of the WT (data not shown). RH activity per se thus has no impact on RT-dependent Mn2+ inhibition.
Most Mn2+ suppressor mutants possess WT RH cleavage activity.
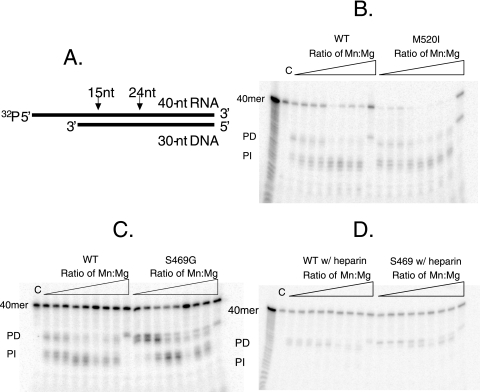
To determine whether the suppressor point mutations affected the RH activity or cleavage specificity of the mutants, the RH activity of GST tag-purified mutants was examined on a randomly selected heteropolymeric RNA-DNA hybrid in the presence of 5 mM Mg2+ and various Mn2+ concentrations. The assay employs a previously described radiolabeled 32P-labeled RNA 40-nucleotide-DNA 30-nucleotide substrate (18). Ty1 RT/RH binds to the DNA 3′ OH of the hybrid template and cleaves the RNA, liberating the radiolabeled 5′ end at a distance thought to reflect the physical distance between the RT and RH active sites. This cleavage product represents “polymerase-dependent” hydrolysis, and further cleavage of this product by RH is characterized as “polymerase independent.” Unlike the case for Ty3 RH, which liberates RNA oligonucleotides of 31 and 23 nucleotides (18), the shorter distance between the Ty1 RT and RH active sites results in products of ∼24 and 15 nucleotides in length (Fig. 5A), corresponding to the “polymerase-dependent” and “polymerase-independent” modes of hydrolysis, respectively (13, 23). In contrast to HIV-1 (7), but like Ty3 (18), polymerase-independent hydrolysis predominates with WT RH, especially at higher Mn2+ concentrations. This predominance, however, can be reversed by the addition of heparin to the reaction mixture (Fig. 5D). Despite the location of the “manganese suppressor” mutations in the RH domain, little or no difference was observed in the absolute levels of RH activity or cleavage specificity of these mutants in these assays (M520I is representative of these mutants [Fig. 5B]), with the notable exceptions of the two suppressor mutants containing the S469G mutation (Fig. 5C). Unlike the WT or the other suppressor mutants, the S469G and S469G S392C suppressor mutants seem to possess only polymerase-dependent hydrolysis, except at high concentrations of Mn2+. This correlates with the fact that the S469G mutant shows the most extreme pmr1 mutation dependence (Table 3). Lener et al. (18) have indicated that the levels of polymerase-dependent and polymerase-independent modes of hydrolysis in Ty3 could be switched in favor of polymerase-dependent hydrolysis under conditions that hinder multiple binding events by the RT/RH, and thus the S469G (and possibly the S392C) mutation might represent a defect in RH turnover in the absence of high Mn2+ levels. Another interesting possibility is that the S469G mutation results in a loss of cooperative divalent metal cation binding by RH. Recent work with HIV-1 RT/RH has revealed that cooperative binding of more than one divalent metal cation is required for generation of the “polymerase-independent” products (14). The S469G mutation presumably lowers the affinity of one of the binding sites for Mn2+, an idea consistent with the location of the S469G mutation immediately adjacent to the metal ion-chelating D468 residue in RH. We, however, favor a model in which S469G is impaired in RH turnover, as WT RT/RH and the S469G suppressor mutant display identical cleavage patterns when heparin is added to block multiple binding events by the RT/RH.
FIG. 5.
RH assay using “generic” RNA-DNA hybrid template (adapted from reference 18). (A) Diagram illustrating the RNA-DNA hybrid template and the expected Ty1 RH cut sites. (B) Comparison of WT and M520I RH cleavages. (C) Comparison of WT and S469G RH cleavages. (D) Comparison of WT and S469G RH cleavages in the presence of heparin. (B to D) RH assays were performed in 10 mM Tris-HCl (pH 7.8), 80 mM NaCl, 5 mM DTT, 5 mM MgCl2, and increasing concentrations of MnCl2 (Mn/Mg ratios of 0, 0.0002, 0.001, 0.002, 0.01, 0.02, 0.1, 0.2, and 1). C, uncut substrate. A ladder was generated by subjecting uncut template to 0.1 M sodium carbonate for 4 min at 100°C. Hydrolysis was initiated by the addition of equal activities (normalized by RT activity) of WT or mutant enzymes at 22°C for 1 h in a final volume of 30 μl.
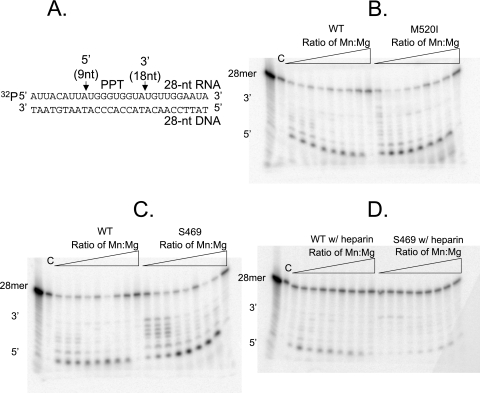
To assess cleavage specificity on a specific substrate, namely, the Ty1 PPT-containing template, a previously described radiolabeled 32P-labeled 28-nucleotide RNA-28-nucleotide DNA duplex (28) was employed. This 28-mer RNA-DNA hybrid consists of the Ty1 PPT, with flanking upstream sequence from the Ty1 3′ untranslated region and downstream sequence from the 3′ U3 segment (Fig. 6A). Cleavage with WT RT/RH yielded the expected 5′ and 3′ cleavage products (Fig. 6B). As observed with the previously used “generic” template, RH cleavage with a representative suppressor mutant, M520I, also yielded a cleavage pattern nearly identical to that of the WT (Fig. 6B). However, once again, differing cleavage specificity was observed with either of the S469G-containing suppressor mutants (Fig. 6C). Interestingly, abundant 5′ cleavage was not observed with either of these suppressor mutants until the Mn2+ concentration was elevated, which therefore may reveal a certain metal ion stoichiometry requirement for 5′ PPT cleavage. Furthermore, although only 3′ cleavage of the PPT is thought to be essential for priming, proper hydrolysis at the 5′ terminus of the PPT helps to prevent aberrant internal cleavage (24). As proper cleavage of the PPT is necessary for plus-strand synthesis, failure of 5′ cleavage at physiological concentrations of Mn2+ likely explains the manganese dependence of the S469G mutants observed in the quantitative transposition assays (Table 3). In addition, the dependence of the S469G mutants on elevated Mn2+ for retrotransposition in yeast may also reveal a requirement for proper 5′ cleavage of the PPT for efficient reverse transcription. When heparin was added to favor single binding events, little to no difference was observed with WT RT/RH or the representative suppressor mutants (Fig. 6D).
FIG. 6.
RH assay using a specific PPT-containing RNA-DNA hybrid template (adapted from reference 28). (A) Diagram illustrating the PPT-containing RNA-DNA hybrid template and the expected Ty1 RH cut sites. (B) Comparison of WT and M520I RH cleavages. (C) Comparison of WT and S469G RH cleavages. (D) Comparison of WT and S469G RH cleavages in the presence of heparin. RH assays were performed in 10 mM Tris-HCl (pH 7.8), 80 mM NaCl, 5 mM DTT, 5 mM MgCl2, and increasing concentrations of MnCl2 (Mn/Mg ratios of 0, 0.0002, 0.001, 0.002, 0.01, 0.02, 0.1, 0.2, and 1). C, uncut substrate. A ladder was generated by subjecting uncut template to 0.1 M sodium carbonate for 4 min at 100°C. Hydrolysis was initiated by the addition of equal activities (normalized by RT activity) of WT or mutant enzymes at 22°C for 1 h in a final volume of 30 μl.
DISCUSSION
Ty1 retrotransposons are genomic parasites that replicate in S. cerevisiae by converting Ty1 transcripts into full-length cDNA copies by the actions of RT. The complete reverse transcription reaction requires primers, templates, and dNTPs, as well as a divalent cation, either Mg2+ or Mn2+. However, Mg2+ is strongly preferred. Previous work suggests that Ty1 transposition is exquisitely sensitive to the levels of intracellular free Mn2+ in the cell and that elevated levels of Mn2+ inhibit Mg2+-catalyzed in vitro Ty1 and HIV-1 RT activity (5). The sensitivity to intracellular free Mn2+, inferred from genetic experiments showing that pmr1 mutations block Mn2+ export, allowed us to screen for Ty1 RT/RH mutants capable of transposition in the pmr1Δ mutant, a yeast mutant that accumulates fivefold-higher levels of total cellular manganese and displays 100-fold less transposition than PMR1 cells.
Mapping of suppressor mutations reveals the surprising finding that suppressor point mutations localize not to the RT but to the RH domain. These results were unexpected, as previous in vitro work had indicated that Mn2+ inhibits RT-polymerizing activity. Quantitative transposition using HIS3 marker mobilization as an indication of transposition frequency revealed that the Mn2+ suppressor mutants increased Ty1 transposition by 2.5- to 23.5-fold over that of WT Ty1 in pmr1Δ cells. Interestingly, the three strongest suppressor mutants (M520I, S469G, and S469G S392C) have mutations that lie within one or two residues of the metal-chelating amino acids. Furthermore, quantitative transposition assays with PMR1 cells indicates that two of the suppressor mutants (S469G and S469G S392C) are strongly Mn2+ dependent for Ty1 transposition. This Mn2+ dependency is likely explained by the altered RH cleavage specificity of these mutants, which is partly restored in vitro in the presence of elevated Mn2+ levels.
We have demonstrated that the Mn2+ suppressor mutants possess in vitro RT polymerase resistance to Mn2+. The ratio of intracellular free Mn2+ to Mg2+ (1 μM/1 mM = 0.001) falls along the steepest part of the curve, and thus any perturbation in the free Mn2+concentration, which was inferred to occur in pmr1 mutants, results in a dramatic drop in RT-polymerizing activity. In the suppressor mutants, much more Mn2+ is needed to achieve the same drop in polymerizing activity as seen in the WT. It has been proposed (5) that Ty1 RT possesses two metal binding sites with different affinities for Mg2+ and Mn2+, with one site possessing a high affinity for Mg2+ or low affinity for Mn2+ and the other site having high affinity for Mn2+ or low affinity for Mg2+. This would be consistent with the metal ion stoichiometry of HIV-1 RT/RH, which possesses a high-affinity binding site for Mg2+ and a high-affinity site for Mn2+ in the RT-polymerizing domain (8), as well as a metal binding site for Mn2+ and a second mutually exclusive site for either Mg2+ or Mn2+ in the RH domain. Under normal reaction conditions, both of these sites are occupied by magnesium due to the ∼1,000-fold excess of free Mg2+ to Mn2+, and Ty1 RT activity is high. However, when one of these sites becomes occupied by Mn2+, such as might occur in the case of a pmr1Δ mutant in which intracellular levels of Mn2+ are increased fivefold, Ty1 RT activity is inhibited. Thus, one possible explanation for the Mn2+ resistance observed in the Mn2+ suppressor mutants is that the suppressor mutations in the RH domain somehow alter the affinity of one or both of these sites in the polymerase domain to increase the affinity for Mg2+ or lower the affinity for Mn2+. This would require some biochemical communication between two distinct domains of the protein.
Unlike the case for RT-polymerizing activity, most suppressor mutants display little or no difference in RH activity or cleavage specificity on either a “generic” or a PPT-containing substrate. The lone exceptions are the S469G and S469G S392C mutants. Both of these mutants display a loss of polymerase-independent hydrolysis on the generic 40-nucleotide RNA-30-nucleotide DNA substrate. Furthermore, both of these mutants show loss of 5′ cleavage at the PPT that can be restored upon addition of Mn2+ back to the reaction mixture, which may reveal a certain metal ion stoichiometry requirement for both 5′ PPT cleavage and efficient reverse transposition. As formation of the PPT primer for plus-strand synthesis is an essential step for retrotransposition, this result likely explains why these two mutants display a large degree of Mn2+ dependence in the quantitative transposition assays.
We have considered three plausible models to explain how Mn2+ suppressor mutations in the RH domain communicate with the RT domain. (i) The first model involves allosteric communication between the RH domain with the RT catalytic domain. (ii) The second model involves a ball-and-chain mechanism by which RH binds too tightly to the primer-template and thereby limits the movement of polymerizing RT in the presence of elevated Mn2+ levels. The HIV-1, and likely the Ty1 as well, RT/RH resembles a right hand with fingers, palm, and thumb domains joined to the RH domain by a connection or tether domain, and the RT/RH makes contacts with its template through both the polymerase thumb domain and the RH primer grip (25). It has been previously demonstrated for HIV-1 that mutation of certain metal-chelating amino acids in the RH domain can cause much tighter binding of the RT to RNA-DNA templates (9), at least in the presence of low Mg2+ levels. If such tighter binding also occurred in the presence of elevated Mn2+ levels, suppressor mutations would thus serve to alleviate such inhibitory binding. (iii) The third model involves metal ion exchange between the RT and RH domains. As most suppressor mutations cluster around the RH metal binding pocket, any specificity obtained at the RH domain could be communicated to the RT domain in this model. This scenario, however, seems unlikely, as the RT/RH nucleic acid binding cleft, which extends from the polymerase active site to the RH active site, can be up to ∼60 Å in length (25).
Although we have been unable to show gross conformational differences between WT and suppressor mutant RT/RH proteins by limited proteolysis with proteinase K (data not shown), we favor the first proposed model for several reasons. Foremost, the Y299C connector domain mutation is not in the RH domain, a seeming prerequisite for the other models. Though this mutation could possibly represent a different class of Mn2+ suppressor mutants, the location of this suppressor mutation in the “connector” domain is highly consistent with an allosteric model. Second, numerous attempts to make in-frame deletions in the connector domain resulted in proteins completely lacking RT activity (data not shown), suggesting that a specific structure of this domain is indeed important for active Ty1 RT/RH. Furthermore, analysis of RH evolution (19) has provided some surprising insights. This study showed that retrotransposon RH domains are fundamentally different from both retroviral and cellular RH domains, lacking a critical histidine. Retroviruses have a cellular-like RH domain, and Malik and Eickbush (19) infer on phylogenetic grounds that this is a late addition to retroviral genomes. Their analyses suggest that the connection domain in retroviral RT/RH proteins is actually a “fossil” ancestral retrotransposon-type RH domain that has likely been maintained due to some required structural/functional roles, such as the conformational changes required in the HIV-1 p66/p51 heterodimer for reverse transcription (1). Presumably, the retrotransposon RH domain has similar structural roles along with its enzymatic activities. Finally, recent work has revealed that various deletions in Ty1 integrase (29) can have profound effects on RT activity, a result consistent with the idea of Ty1 RT being a target of allosteric communication, working in a network of cooperating proteins rather than an isolated enzyme activity.
Acknowledgments
This work was supported in part by NIH grant GM36481 to J.D.B.
We thank Stuart Le Grice for helpful discussions.
Footnotes
Published ahead of print on 30 May 2007.
REFERENCES
- 1.Bahar, I., B. Erman, R. L. Jernigan, A. R. Atilgan, and D. G. Covell. 1999. Collective motions in HIV-1 reverse transcriptase: examination of flexibility and enzyme function. J. Mol. Biol. 285:1023-1037. [DOI] [PubMed] [Google Scholar]
- 2.Barat, C., V. Lullien, O. Schatz, G. Keith, M. T. Nugeyre, F. Gruninger-Leitch, F. Barre-Sinoussi, S. F. LeGrice, and J. L. Darlix. 1989. HIV-1 reverse transcriptase specifically interacts with the anticodon domain of its cognate primer tRNA. EMBO J. 8:3279-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belcourt, M. F., and P. J. Farabaugh. 1990. Ribosomal frameshifting in the yeast retrotransposon Ty: tRNAs induce slippage on a 7 nucleotide minimal site. Cell 62:339-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boeke, J. D., D. J. Garfinkel, C. A. Styles, and G. R. Fink. 1985. Ty elements transpose through an RNA intermediate. Cell 40:491-500. [DOI] [PubMed] [Google Scholar]
- 5.Bolton, E. C., A. S. Mildvan, and J. D. Boeke. 2002. Inhibition of reverse transcription in vivo by elevated manganese ion concentration. Mol. Cell 9:879-889. [DOI] [PubMed] [Google Scholar]
- 6.Chapman, K. B., A. S. Bystrom, and J. D. Boeke. 1992. Initiator methionine tRNA is essential for Ty1 transposition in yeast. Proc. Natl. Acad. Sci. USA 89:3236-3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cirino, N. M., C. E. Cameron, J. S. Smith, J. W. Rausch, M. J. Roth, S. J. Benkovic, and S. F. Le Grice. 1995. Divalent cation modulation of the ribonuclease functions of human immunodeficiency virus reverse transcriptase. Biochemistry 34:9936-9943. [DOI] [PubMed] [Google Scholar]
- 8.Cowan, J. A., T. Ohyama, K. Howard, J. W. Rausch, S. M. Cowan, and S. F. Le Grice. 2000. Metal-ion stoichiometry of the HIV-1 RT ribonuclease H domain: evidence for two mutually exclusive sites leads to new mechanistic insights on metal-mediated hydrolysis in nucleic acid biochemistry. J. Biol. Inorg. Chem. 5:67-74. [DOI] [PubMed] [Google Scholar]
- 9.Cristofaro, J. V., J. W. Rausch, S. F. Le Grice, and J. J. DeStefano. 2002. Mutations in the ribonuclease H active site of HIV-RT reveal a role for this site in stabilizing enzyme-primer-template binding. Biochemistry 41:10968-10975. [DOI] [PubMed] [Google Scholar]
- 10.Curcio, M. J., and D. J. Garfinkel. 1991. Single-step selection for Ty1 element retrotransposition. Proc. Natl. Acad. Sci. USA 88:936-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friant, S., T. Heyman, F. X. Wilhelm, and M. Wilhelm. 1996. Role of RNA primers in initiation of minus-strand and plus-strand DNA synthesis of the yeast retrotransposon Ty1. Biochimie 78:674-680. [DOI] [PubMed] [Google Scholar]
- 12.Garfinkel, D. J., J. D. Boeke, and G. R. Fink. 1985. Ty element transposition: reverse transcriptase and virus-like particles. Cell 42:507-517. [DOI] [PubMed] [Google Scholar]
- 13.Gopalakrishnan, V., J. A. Peliska, and S. J. Benkovic. 1992. Human immunodeficiency virus type 1 reverse transcriptase: spatial and temporal relationship between the polymerase and RNase H activities. Proc. Natl. Acad. Sci. USA 89:10763-10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klumpp, K., J. Q. Hang, S. Rajendran, Y. Yang, A. Derosier, P. Wong Kai In, H. Overton, K. E. Parkes, N. Cammack, and J. A. Martin. 2003. Two-metal ion mechanism of RNA cleavage by HIV RNase H and mechanism-based design of selective HIV RNase H inhibitors. Nucleic Acids Res. 31:6852-6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lapinskas, P. J., K. W. Cunningham, X. F. Liu, G. R. Fink, and V. C. Culotta. 1995. Mutations in PMR1 suppress oxidative damage in yeast cells lacking superoxide dismutase. Mol. Cell. Biol. 15:1382-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lauermann, V., K. Nam, J. Trambley, and J. D. Boeke. 1995. Plus-strand strong-stop DNA synthesis in retrotransposon Ty1. J. Virol. 69:7845-7850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Grice, S. F., C. E. Cameron, and S. J. Benkovic. 1995. Purification and characterization of human immunodeficiency virus type 1 reverse transcriptase. Methods Enzymol. 262:130-144. [DOI] [PubMed] [Google Scholar]
- 18.Lener, D., S. R. Budihas, and S. F. Le Grice. 2002. Mutating conserved residues in the ribonuclease H domain of Ty3 reverse transcriptase affects specialized cleavage events. J. Biol. Chem. 277:26486-26495. [DOI] [PubMed] [Google Scholar]
- 19.Malik, H. S., and T. H. Eickbush. 2001. Phylogenetic analysis of ribonuclease H domains suggests a late, chimeric origin of LTR retrotransposable elements and retroviruses. Genome Res. 11:1187-1197. [DOI] [PubMed] [Google Scholar]
- 20.Mellor, J., M. H. Malim, K. Gull, M. F. Tuite, S. McCready, T. Dibbayawan, S. M. Kingsman, and A. J. Kingsman. 1985. Reverse transcriptase activity and Ty RNA are associated with virus-like particles in yeast. Nature 318:583-586. [DOI] [PubMed] [Google Scholar]
- 21.Muhlrad, D., R. Hunter, and R. Parker. 1992. A rapid method for localized mutagenesis of yeast genes. Yeast 8:79-82. [DOI] [PubMed] [Google Scholar]
- 22.Muller, F., K. H. Bruhl, K. Freidel, K. V. Kowallik, and M. Ciriacy. 1987. Processing of TY1 proteins and formation of Ty1 virus-like particles in Saccharomyces cerevisiae. Mol. Gen. Genet. 207:421-429. [DOI] [PubMed] [Google Scholar]
- 23.Peliska, J. A., and S. J. Benkovic. 1992. Mechanism of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Science 258:1112-1118. [DOI] [PubMed] [Google Scholar]
- 24.Rausch, J. W., and S. F. Le Grice. 2004. ‘Binding, bending and bonding’: polypurine tract-primed initiation of plus-strand DNA synthesis in human immunodeficiency virus. Int. J. Biochem. Cell. Biol. 36:1752-1766. [DOI] [PubMed] [Google Scholar]
- 25.Sarafianos, S. G., K. Das, C. Tantillo, A. D. Clark, Jr., J. Ding, J. M. Whitcomb, P. L. Boyer, S. H. Hughes, and E. Arnold. 2001. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J. 20:1449-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sherman, F., G. R. Fink, and J. Hicks. 1986. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 27.Wilhelm, M., M. Boutabout, and F. X. Wilhelm. 2000. Expression of an active form of recombinant Ty1 reverse transcriptase in Escherichia coli: a fusion protein containing the C-terminal region of the Ty1 integrase linked to the reverse transcriptase-RNase H domain exhibits polymerase and RNase H activities. Biochem. J. 348:337-342. [PMC free article] [PubMed] [Google Scholar]
- 28.Wilhelm, M., O. Uzun, E. H. Mules, A. Gabriel, and F. X. Wilhelm. 2001. Polypurine tract formation by Ty1 RNase H. J. Biol. Chem. 276:47695-47701. [DOI] [PubMed] [Google Scholar]
- 29.Wilhelm, M., and F. X. Wilhelm. 2005. Role of integrase in reverse transcription of the Saccharomyces cerevisiae retrotransposon Ty1. Eukaryot. Cell. 4:1057-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiong, Y., and T. H. Eickbush. 1990. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 9:3353-3362. [DOI] [PMC free article] [PubMed] [Google Scholar]