An estimated 3% of the world population is infected with hepatitis C virus (HCV) (5, 232). In most infected individuals, this remarkable RNA virus evades the immune system and establishes a chronic infection that can lead to cirrhosis, liver cancer, and death. While advances have been made in treating HCV, the current therapy, a combination of pegylated alpha interferon and ribavirin, is poorly tolerated and is effective in only 50% of genotype 1-infected patients (reviewed in reference 53). The quest for new HCV therapies has driven research in academia and industry, which has led to significant progress in understanding basic virus replication mechanisms and in developing specific candidate antiviral compounds. In this minireview, we highlight recent advances in cell culture model systems used to study HCV and describe new insights into the virus life cycle that have been gleaned from these and other recent studies. This review is limited in scope to recent developments in HCV biology and is by no means comprehensive. Throughout the article, we have attempted to direct the reader to a number of high-quality review articles covering various aspects of HCV biology in greater detail.
EVOLUTION OF MODEL SYSTEMS FOR STUDYING RNA REPLICATION AND INFECTION
After the landmark report of the first HCV cDNA clones in 1989 (30), many expected HCV molecular virology to quickly advance to a state rivaling that of other positive-strand RNA viruses. More than 15 years later, this predication is only beginning to be realized. Significant advances in understanding the key steps of the HCV life cycle have been made in recent years, although many steps remain enigmatic (Fig. 1). Early attempts to coax replication in cell culture provided glimmers of hope, but none of this work provided tractable systems that became widely adopted (see reference 130 for a review). It was not until 1997, after the discovery that the original HCV cDNA clones lacked a highly conserved 3′-terminal genome fragment (109), that the first full-length functional cDNA clones were reported (108). In the absence of permissive cell culture systems, the infectivity of RNA transcripts from these clones was assessed by intrahepatic inoculation of chimpanzees (108). Attempts to demonstrate replication of these RNAs in cell culture failed. Through the use of bioinformatics, chimpanzee infections, surrogate expression systems, and biochemical analysis, the structure of the HCV genome, the polyprotein processing mechanisms, the protein topology, and some protein functions were defined, all without the ability to monitor RNA replication in a cell culture environment (Fig. 2). The advent of the subgenomic genotype 1b (isolate Con1) replicon system, first reported by Lohmann et al. in 1999 (134), established persistent HCV RNA replication in a human hepatoma cell line (Huh-7) (Fig. 3A). The inefficiency with which RNA replication was initiated in this system limited its utility, but this breakthrough provided a basis for further optimization. Blight et al. isolated subclones of replicon-transduced Huh-7 cells cured by alpha interferon treatment that showed enhanced permissiveness for HCV RNA replication (19). The most famous of these subclones, Huh-7.5, appears to harbor a defect in the retinoic-acid inducible gene I (RIG-I) intrinsic cellular antiviral response pathway (204). In addition, sequencing of HCV RNAs in replicon-containing cell clones identified a spectrum of adaptive mutations in nonstructural (NS) proteins that could dramatically enhance RNA replication (17; reviewed in reference 9). Adaptive mutations that rendered other genotype 1 RNAs (such as 1a, strain H77) replication competent in cell culture were soon identified (18). Unfortunately, full-length HCV replicons incorporating these changes, despite robust RNA replication, failed to yield infectious virus (18, 84, 178).
FIG. 1.
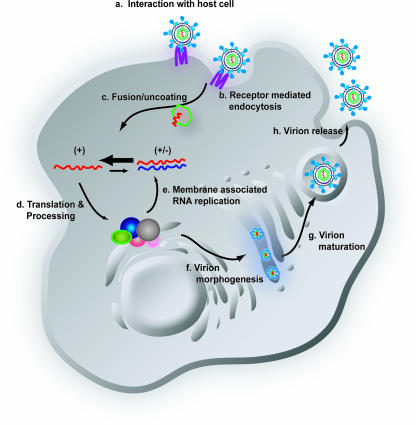
Schematic diagram of the HCV life cycle. The life cycle of HCV is similar to that of other members of the Flaviviridae family. Extracellular HCV virions interact with receptor molecules at the cell surface (a) and undergo receptor-mediated endocytosis (b) into a low-pH vesicle. Following HCV glycoprotein-mediated membrane fusion, the viral RNA is released into the cytoplasm (c). The genomic RNA is translated to generate a single large polyprotein that is processed into the 10 mature HCV proteins in association with a virus-derived ER-like membrane structure termed the membranous web (d). The mature HCV proteins replicate the RNA genome via a minus-strand replicative intermediate to produce progeny RNA. A portion of this newly synthesized RNA is packaged into nucleocapsids and associated with the HCV glycoproteins, leading to budding into the ER (f). Virions follow the cellular secretory pathway (g) and, during this transit, maturation of particles occurs (g). Mature virions are released from the cell, completing the life cycle (h). +, positive-sense genomic RNA; +/−, minus-strand replicative intermediate associated with positive-strand genomic RNA.
FIG. 2.
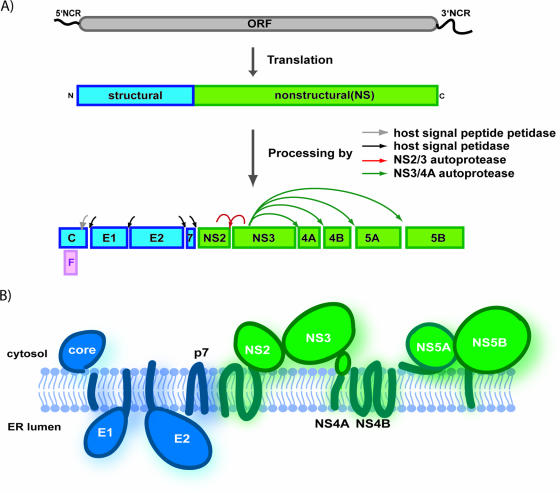
HCV genome organization, polyprotein processing, and protein topology. (A) The HCV genome is a single-stranded RNA encoding a single large open reading frame (ORF) of roughly 3,000 amino acids, flanked by structured 5′ and 3′ NCRs. The translation of the open reading frame, via the activity of an IRES element in the 5′ NCR, generates a large polyprotein that is organized with structural proteins in the amino-terminal third of the polyprotein, followed by the NS replication proteins. The polyprotein undergoes a complex co- and posttranslational series of cleavage events, catalyzed by both host and viral proteases, to produce the 10 individual HCV proteins. (B) The topology of the HCV proteins relative to the ER membrane.
FIG. 3.
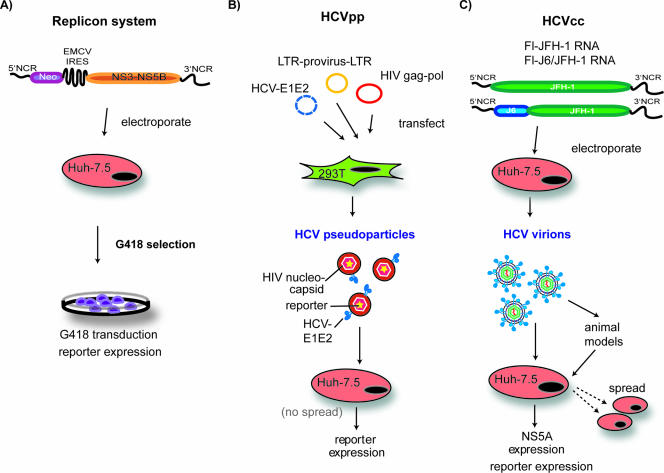
Systems for the study of HCV replication, entry, and infectivity. (A) HCV replicon systems, shown here in one of their simplest iterations, allow for productive viral RNA replication in cell culture. Bicistronic replicon RNAs, encoding a selectable marker (Neor) under control of the HCV IRES in the first cistron and the HCV replicase proteins (NS3-NS5B) under control of a heterologous IRES from encephalomyocarditis virus in the second cistron, are delivered to Huh-7-based cell lines by electroporation. Replication of these RNAs leads to production of the selectable marker and allows for selection of colonies containing active RNA replication. Transduction of resistance to the drug G418 is shown in this figure, but replicons expressing a number of reporter genes have been developed, as have methods to efficiently measure HCV proteins and RNA from these systems. (B) The HCV pseudoparticle system (HCVpp) provides a method to investigate glycoprotein-mediated events in the HCV life cycle. In this system, recombinant retroviruses that contain HCV functional glycoproteins on their surface are generated in 293T cells. These particles can be used to infect permissive cell lines, such as Huh-7.5. The retrovirus genomes have been engineered to express a reporter gene, such as luciferase, allowing for a quantitative measure of cell entry. (C) The HCVcc infectious virus system uses either JFH-1 HCV genomic RNA or chimeras of this genome with heterologous sequences (such as J6). These RNAs are electroporated into permissive cell lines and yield infectious HCV virions that can be used to infect naïve cells or animal models. Productive infection can be monitored by detection of the expression of NS5A, by a number of reporter genes, or by direct measure of viral RNA.
The inability to reproduce the complete HCV life cycle in cell culture led to the development of model systems that have yielded many advances. C-terminally truncated secreted forms of E2 (sE2) proved valuable for probing virus-cell interactions and identifying putative receptors (56, 152, 179, 190). Other systems useful for studying envelope protein functions included cell surface-expressed E2 (58), E1-E2 liposomes (32), virus-like particles generated in insect cells (213, 235), and vesicular stomatitis virus pseudotyped with chimeric glycoproteins consisting of the ectodomains of HCV E1 and E2 fused to the transmembrane domain of the vesicular stomatitis virus G glycoprotein (26, 116, 144). A major advance in studying HCV cell entry was the generation of retroviral pseudotypes bearing unmodified HCV glycoproteins (HCVpp) (12, 79). HCVpp are produced by cotransfection of 293T cells with expression vectors encoding (i) HCV E1E2, (ii) the Gag-Pol proteins of either murine leukemia virus or human immunodeficiency virus, and (iii) a retroviral genome encoding a reporter to detect subsequent productive entry (Fig. 3B). Cell entry of HCVpp is HCV glycoprotein mediated and thought to closely resemble the cell entry properties of genuine HCV virions.
Despite the advances in understanding HCV infection by use of these surrogate systems, the inability to generate authentic infectious HCV in cell culture remained a major roadblock. Probing the mode of action of replicon-adaptive mutations provided possible clues as to why virion production might be compromised. Adaptive mutations alter the degree of NS5A phosphorylation, with highly adapted replicons favoring a hypophosphorylated over a hyperphosphorylated NS5A (6, 17, 49). Remarkably, chemical inhibition or genetic silencing of the cellular kinase responsible for NS5A hyperphosphorylation, casein kinase Iα, allows replication of HCV genomes lacking adaptive mutations (162, 183, 184). Additionally, the mutation of sites of hyperphosphorylation in NS5A can enhance the RNA replication of subgenomic replicons (6). It is now clear that NS5A hypophosphorylation is essential for HCV RNA replication and that hyperphosphorylation is a negative modulator of this process, yet the exact mechanisms of these observations are not yet known (see reference 82 for a recent review of NS5A phosphorylation). One theory of the role of phosphorylation in the HCV life cycle involves the maintenance of replicase stability. The cellular target membrane-associated soluble N-ethylmaleimide-sensitive factor attachment protein receptor, human vesicle-associated membrane protein-associated protein A, interacts with NS5A and NS5B and appears to function in the assembly and localization of the HCV RNA replication complex (65, 217), and the interaction between hVAP-A and NS5A is abrogated by NS5A hyperphosphorylation (49). Thus, adaptive mutations that prevent NS5A hyperphosphorylation encourage replicase assembly and maintenance, possibly at the expense of late viral life cycle events, such as assembly and packaging. In support of this idea, most genomes with adaptive mutations do not yield infectious particles in cell culture despite efficient replication (18, 84, 178) and are impaired or noninfectious in chimpanzees (25). This model further predicts that HCV isolates capable of replication in cell culture without adaptation might yield infectious virus.
Recently, Date et al. (36) and Kato et al. (95, 96) identified an HCV isolate capable of efficient RNA replication in multiple cell types without adaptation. This genotype 2a strain, called JFH-1, came from a Japanese patient with a rare case of acute fulminant hepatitis. Surprisingly, when full-length JFH-1 RNA was transfected into Huh-7 cells, viral particles, termed HCVcc (for HCV cell culture), that were filterable and capable of infecting naïve cells were released (Fig. 3C) (225). JFH-1 produced low HCVcc titers, but subsequently it was found that higher titers could be obtained using Huh-7.5 cells and derived sublines (128, 254). The kinetics and yield of HCVcc were enhanced by creating a chimera using the C-NS2 region of different genotype 2a isolate, J6 (128). Even higher titers (about 1 log) were obtained by altering the fusion junction from NS2/NS3 to breakpoint within NS2 after the first predicted transmembrane segment (177). This finding could be generalized and was used to produce viable JFH-1 chimeras of different HCV genotypes (1a, 1b, and 3a), albeit at lower titers than the best 2a chimera (Jc1). Cell culture-grown HCVcc is infectious in chimpanzees and uPA-SCID mice transplanted with human hepatocytes (129, 225), and the virus recovered from these animals is still infectious in cell culture. This establishes the first system to study the entry and neutralization properties of HCV produced in vivo and indicates that it should be possible to find other isolates like JFH-1 by screening isolates for their ability to infect and replicate in Huh-7.5 cells. Intriguingly, virus recovered from HCVcc-infected animals displayed altered biophysical properties and increased specific infectivity. This may be of particular importance given the association of HCV with serum lipoproteins, which may enhance or modulate infectivity (4, 78, 211, 212).
Since the development of the JFH-1-based infectious cell culture systems, a number of additional infectious systems have been developed. These include cDNA-launched genotype 1b (CG), 1a (H77), and 2a (JFH-1) systems in which authentic HCV RNA is produced via the action of ribozymes following cellular transcription from HCV-encoding episomal plasmid DNA or from chromosomally integrated cDNA (27, 97). In addition, HCVcc has been produced after transfection with genotype 1a RNA, H77S (249). H77S HCVcc titers are very low (between 1 and 2 logs lower than JFH-1), and the H77S genome harbors five cell culture-adaptive mutations (247). This finding conflicts with the previous idea that adaptive mutations impair virus release. However, it is possible that differences will be observed for different HCV genotypes and genetic backgrounds and that high-level RNA replication and protein expression (as seen for JFH-1 and H77S) may also be important prerequisites for particle assembly and release in cell culture.
The current panel of cell culture systems have been very useful tools in understanding the HCV life cycle. Significant progress has been made in the areas of receptor identification, virus entry, cis-acting RNA elements in replication, replication proteins, host cell requirements for replication, and virus assembly, and we will highlight these areas by following the steps as they occur in HCV infection.
VIRIONS AND CELLULAR RECEPTORS
With the development of HCVpp, it was quickly noted that only a few cell lines, all of which were derived from human liver cells, could be infected (12, 79, 252). What precisely defines this narrow tropism is as yet unclear; however, numerous cellular factors have been put forward as potential HCV receptors or coreceptors. Although evidence exists for interactions between HCV and the low-density lipoprotein (LDL) receptor (3, 156, 237), asialoglycoprotein receptor (189), and heparan sulfate (11, 112), none of these has been conclusively proven to be essential for HCV entry.
The strongest evidence exists for CD81, a member of the tetraspanin superfamily, which was initially identified as a candidate HCV receptor based on its ability to bind sE2 (41, 42, 56, 77, 179). Several lines of evidence leave little doubt that CD81 is indeed an essential component of the HCV receptor complex. (i) The human hepatoma cell line HepG2 does not express CD81 and cannot be infected with HCVpp or HCVcc but becomes infectible upon transduction with CD81 (12, 128, 252). (ii) Knockdown of CD81 expression using small interfering RNA abrogates the susceptibility of liver cells to HCVpp and HCVcc infection (112, 252). (iii) Antibodies against CD81, as well as soluble forms of CD81, block HCVpp and HCVcc infection in a dose-dependent manner (12, 14, 128, 225, 254). (iv) CD81 is a limiting factor for HCVcc infection in some Huh-7 sublines (112, 255). The requirement for CD81 is conserved for all tested HCV genotypes (119, 147). However, CD81 is not the only cellular factor required for HCV entry, as CD81 alone is not sufficient for HCVpp entry and its wide tissue distribution does not explain the liver tropism of HCV.
Like CD81, scavenger receptor BI (SR-BI) was first identified as a candidate HCV receptor based on its ability to bind sE2 (190). SR-BI, expressed at high levels in the liver, steroidogenic tissues, placenta, small intestine, and monocytes/macrophages, mediates selective uptake of cholesteryl esters from high-density lipoprotein (HDL) into the cellular membrane (1, 188) and, possibly, endocytosis of entire HDL particles (195). Experiments to define the role of SR-BI in HCV cell entry have been challenging. SR-BI-negative cell lines are rare, and one that becomes susceptible to HCVpp upon SR-BI transfection has not been identified. Recently, Grove et al. demonstrated that the level of SR-BI expression in Huh-7.5 cells modulates the level of productive HCVcc infection (74). Antibodies directed against SR-BI and small interfering RNAs targeting SR-BI inhibit HCVpp infection (12, 119), but both effects vary between HCV genotypes and are less striking than the results obtained for CD81 (119). Adding to the complexity, two natural ligands of SR-BI modulate HCV infectivity in different ways: HDLs moderately enhance HCVpp infectivity (13, 150, 222), and this effect depends on SR-BI being present and functional on the target cell (13, 164, 222). In contrast, oxidized low-density lipoproteins are potent inhibitors of both HCVpp and HCVcc entry (224). However, the coexpression of CD81 and SR-BI is not sufficient to confer HCVpp and HCVcc susceptibility (12, 79), and Kapadia et al. have recently shown that a high level of membrane cholesterol is also a requirement (92).
Very recently, Evans et al. identified claudin-1 (CLND1), a component of cellular tight junctions expressed at high levels in liver cells, as an additional factor required for HCV entry (50). Overexpression of CLDN1 in some, but not all, nonhepatic cells that express CD81 (but not endogenous CLDN1) is capable of rendering these cells infectible by HCVpp. CLDN1 may contribute to the liver tropism of HCV, as it is highly expressed in this organ, and even moderate inhibition of CLDN1 expression renders otherwise-permissive cells uninfectible by HCVcc and HCVpp. However, some human cell lines remain HCV resistant in the presence of CD81, SR-BI, and CLDN1, suggesting that either additional cellular entry factors remain to be discovered or that these cell types express a transdominant inhibitory factor(s).
ENTRY AND MEMBRANE FUSION
By analogy to related viruses for which structures have been obtained, such as dengue virus (113), tick-borne encephalitis virus (54), and West Nile virus (159), HCV particles are thought to consist of an icosahedral lattice of E1-E2 glycoprotein heterodimers anchored in an envelope surrounding the viral nucleocapsid. E1 and E2 are type I transmembrane proteins with extensive N-terminal ectodomains decorated with N-linked glycans (40, 43, 167). Several findings directly implicate E2 in HCV entry: first, sE2 binds both CD81 (179) and SR-BI (190), suggesting that it mediates key interactions between the incoming virus and functional (co)receptors. Glycosylation at specific sites of E2 is essential for HCVpp infectivity (68). Finally, antibody binding to a number of distinct E2 epitopes can neutralize HCVpp (12, 47, 79, 98, 167, 192).
The role of E1 is less well defined, mostly due to a lack of tools to study its function. Based on sequence analysis and computational modeling using the known structure of tick-borne encephalitis virus E protein, some have proposed that HCV E1 may be a truncated class II fusion protein (57, 66, 185). Others, however, have argued that the fusion activity may reside in E2 (124, 240). There is also limited evidence for the existence of neutralizing determinants in E1 (98).
Recent synchronous infection/blocking studies have begun to define the early steps of HCV entry. Glycosaminoglycans appear to be involved in initial HCV binding (112), with CD81 (14, 50, 112) and CLDN1 (50) playing postbinding roles in entry. Antibody-blocking experiments indicate that CLDN1 may function downstream of HCV-CD81 interaction, but prior to membrane fusion (50). The sensitivity of HCVpp and HCVcc entry to inhibitors of endosomal acidification suggests entry via the endosomal route followed by acid-triggered fusion (12, 79, 216). In keeping with this hypothesis, HCV glycoprotein-mediated fusion occurs most efficiently at pH 5 to 5.5 (107). However, HCVcc infectivity prior to cell binding is insensitive to low pH, suggesting that postbinding interactions are required to activate the virion for fusion (112, 149, 216). As might be expected, clathrin is important for HCVcc and HCVpp entry in hepatoma cells (16, 33, 149). Thus far, the mechanism of HCV glycoprotein-mediated membrane fusion is unknown.
The HCVcc systems have also been exploited to examine the phenomenon of superinfection exclusion (SE). Based on previous work with the related bovine viral diarrhea virus, it was assumed that HCV would display SE at the levels of entry and replication (122). Previous studies have shown that competition exists between HCV replicons in cells, suggesting that SE might exist at the level of RNA replication (48, 133). Studies using either HCVcc to infect cells harboring subgenomic replicons or distinguishable HCVcc constructs have revealed strong SE for HCV, but the effect is largely downstream of entry and primary translation of incoming genome RNA (191, 215). Studies have also reported the downregulation of CD81 expression in HCVcc-infected cell populations (255), providing a potential mechanism for SE. However, this does not occur during acute infection and appears to be due to HCVcc-induced cytotoxicity and a selection for nonpermissive cells with low CD81 expression (215). Interestingly, and possibly quite relevant for HCV treatment, the SE is transient and disappears when HCV replication is effectively inhibited. This allows for reinfection of previously resistant cells.
cis RNA ELEMENTS
As for all positive-strand RNA viruses, the HCV genome RNA is the centerpiece of replication, serving as a substrate for translation, replication, and packaging. These processes are controlled by “cis-acting” or “cis replication” RNA elements (CREs) located within the 5′ and 3′ noncoding regions (NCRs) and the polyprotein coding region (Fig. 4). Potential CREs were first identified by phylogenic comparisons and thermodynamic RNA structure modeling (174, 196, 219, 226), followed by functional studies. The best understood of the CREs is the HCV internal ribosome entry site (IRES) (Fig. 4A). The IRES is composed of RNA elements within the 5′ NCR and the core protein coding sequence. These elements have been termed 5′ SLII, SLIII, and SLIV (198). SLIII has been further divided into subdomains, SLIIIa, -b, -c, -d, -e, and -f, which form a four-way junction and a pseudoknot (120, 229, 230). The structures of five of these subdomains and the SLIIIabc junction have been determined by nuclear magnetic resonance (NMR) or X-ray crystallography (34, 99, 106, 139, 140, 186). A high-resolution structure for the entire HCV IRES still eludes us. In the current model of HCV translation initiation, the 40S ribosome subunit interacts with the HCV IRES pseudoknot, positioning the polyprotein start codon in the 40S P site (99, 100, 140, 141, 168, 169, 180). Subsequently, eIF3 interacts with the IRES-40S complex, followed by recruitment of the eIF2/GTP/Met-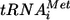 complex, forming a 48S complex (169). Following GTP hydrolysis, the initiation factors are released and the 60S subunit joins to form the IRES-80S complex (169). The HCV IRES can assemble 80S complexes and initiate translation without initiation factors under increased cation concentrations, and this pathway may be used in cells if initiation factors become limiting or modified by the antiviral responses (118). Exciting work using cryoelectron microscopy (cryo-EM) has elucidated the structures of the HCV IRES complexed with the 40S ribosomal subunit, the 40S subunit-complexed eIF3 and the HCV IRES, and the IRES complexed with the 80S ribosome (21, 197, 201). These structures reveal contacts between the IRES and ribosome and the conformational changes that occur during assembly of these complexes. The details of these structures have been discussed recently in an excellent review (60). Determining an atomic-resolution structure of the complete IRES associated with the entire translation complex is the ultimate goal but is a daunting task. The immediate future will likely see these cryo-EM structures pushed to a higher resolution, potentially allowing fitting of the various IRES subdomain structures into the cryo-EM electron density map with great positional certainty. This line of investigation is revealing fundamental similarities and differences between translation of cellular mRNAs and the HCV genome that can be exploited for developing new antivirals that target this early step in intracellular replication.
complex, forming a 48S complex (169). Following GTP hydrolysis, the initiation factors are released and the 60S subunit joins to form the IRES-80S complex (169). The HCV IRES can assemble 80S complexes and initiate translation without initiation factors under increased cation concentrations, and this pathway may be used in cells if initiation factors become limiting or modified by the antiviral responses (118). Exciting work using cryoelectron microscopy (cryo-EM) has elucidated the structures of the HCV IRES complexed with the 40S ribosomal subunit, the 40S subunit-complexed eIF3 and the HCV IRES, and the IRES complexed with the 80S ribosome (21, 197, 201). These structures reveal contacts between the IRES and ribosome and the conformational changes that occur during assembly of these complexes. The details of these structures have been discussed recently in an excellent review (60). Determining an atomic-resolution structure of the complete IRES associated with the entire translation complex is the ultimate goal but is a daunting task. The immediate future will likely see these cryo-EM structures pushed to a higher resolution, potentially allowing fitting of the various IRES subdomain structures into the cryo-EM electron density map with great positional certainty. This line of investigation is revealing fundamental similarities and differences between translation of cellular mRNAs and the HCV genome that can be exploited for developing new antivirals that target this early step in intracellular replication.
FIG. 4.
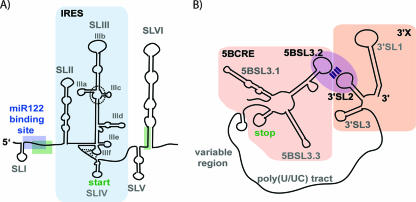
Schematic diagram of the 5′ and 3′ HCV NCRs. (A) The 5′ NCR contains six stem loop structures (SLI to SLVI). The position of the miR122 binding site within the 5′ spacer region is indicated by a blue box. The interaction site of this spacer region with the base of SLVI is also shown (green boxes). The bulk of the 5′ NCR comprises the HCV IRES element (light blue shading). The position of the four-way junction, which connects components of the SLIII loop and is involved in EIF3 binding, is indicated by a dashed circle. The pseudoknot structure within IIIf is shown by dashed lines. The position of the 5′ end of the HCV genome is shown, as is the location of the start site of polyprotein translation. (B) The 3′ NCR has a tripartite structure, containing a variable region, a poly(U/UC) tract, and the 3′ X region (further divided into 3′ stem loop structures, 3′ SL1, 3′ SL2, and 3′ SL3). The 3′ SL2 stem loop interacts via a kissing loop interaction with the 5BSL3.2 stem loop in the cis-acting RNA element in the NS5B coding sequence (5BCRE).
The 5′ NCR contains additional CREs required for replication (Fig. 4A). Nucleotides (nt) 1 to 43, containing the 5′ SLI (nt 5 to 20) and an unstructured spacer (nt 21 to 43), are dispensable for translation but required for HCV RNA replication (63, 103, 187). A recent study uncovered an unexpected requirement for liver-specific microRNA 122 (miR122) in HCV replication (90). miR122 basepairs with the 5′-proximal end of the unstructured spacer. This interaction is required for efficient HCV RNA replication but, surprisingly, does not affect translation (90). A region (nt 22 to 40) that overlaps the miR122 binding site participates in a long-range RNA-RNA interaction with the base of SLVI in the core coding region and has been reported to inhibit IRES function (104). SLII, although part of the IRES, is also involved in RNA replication (63). How these interactions regulate the viral life cycle remains to be determined, but it is tempting to speculate that they may help to orchestrate the switch between translation and replication. The region of the 5′ NCR involved in progeny plus-strand genome synthesis is likely found at the 3′ end of the complementary minus strand. Given the dearth of experimental systems to study plus- and minus-strand initiation, little is known about this region of the RNA, except that predicted and experimentally probed RNA folds are quite different from those in the complementary plus-strand 5′ region (193, 199). New assays are needed to dissect the functional requirements of this region in RNA amplification.
The HCV 3′ NCR has a tripartite structure, containing a region that varies among genotypes, an internal poly(U/UC) tract, and the highly conserved 3′ X region (20, 109, 206, 207) (Fig. 4B). Deletion of the variable region impairs RNA replication, and this region, in conjunction with the poly(U/UC) tract and a portion of the 3′ X region, has been reported to enhance translation from the HCV IRES (86, 87, 145, 153). This finding is controversial, as others have reported that this region has no effect on HCV translation (52, 61, 85, 111). Recent studies suggest that the enhancement of translation by the 3′ NCR seems to be human liver cell specific (200), occurs after initiation, and may increase the efficiency of termination and ribosome recycling (22). Much of these discrepancies in the role of the 3′ NCR in translation likely reside in the choice of experimental system used in each individual study. This idea is supported by recent findings suggesting that the 3′ NCR increases IRES translation only in the context of monocistronic reporter RNAs or HCV genomes with a precise, authentic 3′ terminus (200). The poly(U/UC) tract is variable in length and composition (109), but a minimal length of 26 to 50 nucleotides is required for efficient replication in cell culture (61, 246). Whether this region acts as a spacer to position other elements or interacts directly with replicase machinery is not known. Both NS5A and NS3, which bind RNA, are candidates for binding the poly(U/UC) tract (7, 81, 91). The 3′ X region is highly conserved and consists of two metastable stem loops (3′ SL2 and 3′ SL3) and a highly stable 3′-terminal stem loop (3′ SL1) that is reminiscent of that found for classical flaviviruses (131). Despite an early report to the contrary (76), the 3′ X region is absolutely required for replication in cell culture (61, 246, 248) and for infectivity in chimpanzees (242). The 3′ X region, as well as other 3′ NCR components, likely functions to direct de novo initiation of minus-strand synthesis, probably via interaction with one or more viral and host proteins to form the minus-strand replicase. The mechanistic details of this process remain to be defined.
A number of CREs have also been found within the polyprotein coding sequence (218, 219). One of these CREs in the NS5B coding sequence, termed 5BSL3.2, is required for RNA replication (62, 121, 250) (Fig. 4B). The loop region of 5BSL3.2 is a key determinant of its function (250) via a long-range RNA-RNA or “kissing” interaction with a complementary sequence in the 3′ X region located in 3′ SL2 (62). The poly(U/UC) tract lies between 5BSL3.2 and the 3′ X region and could act as a spacer to allow formation of this interaction. The binding of viral or host replicase components to the poly U/UC tract may modulate this interaction. 5BSL3.2 can also function when inserted in the variable region of the 3′ NCR (62, 121, 250).
The core protein coding region has been an area of intense recent interest, given the existence of a conserved alternative open reading frame that could encode additional HCV proteins that have been collectively termed alternative reading frame proteins (ARFP) (8, 23, 29, 220, 221, 227, 238, 239). The ARFP/C coding sequence also includes several predicted RNA stem-loop structures (SLV and SLVI) (23, 218, 219, 227). Although serologic and T-cell responses against ARFP have been detected in HCV patients (214), a comparison of ARFP codon usage and sequences among HCV genotypes suggests a lack of evolutionary constraints (35), and its function, if any, remains unclear. Recently, the ARF/C SLVI RNA element, rather than ARFP expression, was shown to be important for HCV replication in cell culture and a chimpanzee infection (148).
RNA REPLICATION MACHINERY
Much research has been directed toward defining the roles of the viral gene products in the replication process. One of the most interesting developments in our thinking about HCV replication has come from biochemical studies using crude replicase preparations (155, 182), which indicate that only a tiny fraction of the HCV replicase proteins are directly involved in genome amplification. Another emerging theme is the existence of oligomeric forms of the replicase proteins. Such oligomers may be important in regulating the assembly of a functional RNA replication machine or for modulating host cell functions to set the stage before RNA replication is initiated. Another interesting observation is the interaction of HCV replicase proteins with a variety of cellular chaperones and peptidyl-prolyl-cis-trans-isomerase (PPI)-like proteins. This might indicate that a majority of HCV proteins represent misfolded dead-end products, and their interaction with these host cell foldases reflects limiting factors needed for proper HCV protein maturation. However, given the relatively low levels of HCV proteins and the economy of RNA viruses, this explanation seems unlikely. A more intriguing possibility is that excess HCV NS proteins are involved in modulating host cell functions, such as antagonizing intrinsic antiviral pathways. The following paragraphs detail recent progress on HCV replicase components and highlight several such examples.
NS2 harbors one of two distinct viral proteinase activities required for HCV polyprotein processing. NS2 is an integral membrane protein that teams up with the N-terminal third of NS3 to catalyze a single autocatalytic cleavage that separates NS2 from NS3 (70). This cleavage is essential for HCV replication but probably only to liberate the N terminus of NS3, since subgenomic replicons require only NS3-NS5B to replicate efficiently (134). Lorenz et al. have recently described the postcleavage structure of the protease domain of NS2 (called NS2pro), revealing a novel cysteine protease (137). NS2pro is a dimer, with residues comprising the active-site catalytic triad provided by both monomers. The chimeric active site, requiring protein dimerization for protease activity, provides a potential mechanism to regulate formation of an active replicase. Another interesting feature of the NS2 structure is a cis-proline near the active site. A cellular PPI may be needed to generate this rare conformation, raising questions about the kinetics of polyprotein translation, NS2 folding, oligomerization, and proteolytic cleavage. In this regard, it is worth noting that NS5B associates with cyclophilin B, a PPI, and NS5A associates with FKBP8, a protein with some similarity to cellular PPI enzymes, and both these interactions modulate RNA replication (165, 234). What impact these interactions may have, if any, on NS2 is not known. The recent data indicating that the N-terminal portion of NS2 can affect the efficiency of infectious virus production (discussed below) (177) represent another exciting development.
NS3 is a multifunctional protein composed of two domains; a chymotrypsin-like serine protease resides in its N-terminal third (independent of the NS2-3 proteinase activity), and an NTPase/helicase occupies the C-terminal portion. The NS3 serine protease activity is modulated by NS4A (51, 171), which contributes a β strand to the NS3 serine protease (reviewed in reference 38) and whose N-terminal hydrophobic helix provides a membrane anchor for the NS3-4A complex (236). NS3-4A is perhaps the best characterized of the HCV proteins, with numerous crystal structures available (28, 101, 102, 138, 241, 243, 244). NS3-4A catalyzes cleavages at the NS3/NS4A, NS4A/NS4B, NS4B/NS5A, and NS5A/NS5B polyprotein junctions (71, 72). The NS3-4A protease also antagonizes activation of the RIG-I and the Toll-like receptor 3 (TLR3) intrinsic cellular antiviral pathways. In both cases, the NS3-4A cleaves key adapter proteins (reviewed in references 64 and 89). In the case of RIG-I, NS3-4A cleaves Cardif (also known as MAVS, IPS-1, and VISA) (151), a mitochondrion-associated membrane protein required for activation of IRF-3 and NF-KB (24, 59, 94, 204). NS3-4A also cleaves the Toll-interleukin-1 receptor domain-containing adapter inducing beta interferon to inactivate the TLR3 pathway (55). Additional cellular substrates for NS3-4A have yet to be reported, but it remains possible that HCV manipulates other cellular cascades in this manner. The multifaceted roles of NS3-4A in cleaving polyproteins and antagonizing cellular antiviral responses suggest that NS3-4A protease inhibitors in development for treatment may function at multiple levels to control or eliminate HCV infection (123).
The NS3 NTPase/helicase domain is a member of helicase superfamily 2 (28, 101, 243, 244) that binds to a single-stranded tail and unwinds double-stranded nucleic acids in a 3′-to-5′ direction (205). Recent biophysical studies demonstrate that this helicase activity can unpair long (as much as 18-bp) double-stranded RNA segments in a step-wise fashion (44, 125, 126, 194). A recent cocrystal structure of the NS3 helicase complexed with a large nucleic acid substrate suggests the protein may function as an oligomer, providing yet another example of an oligomeric HCV replicase protein (143). Despite these recent advances in understanding mechanistic details, it is still unclear what the helicase is doing in HCV replication. Does it unwind double-stranded RNA replication intermediates, melt local areas of RNA secondary structure, or function as a single-strand RNA translocase that displaces bound proteins? Regardless, the helicase activity of NS3 is essential for HCV RNA replication in replicons (117) and productive infection in chimpanzees (110).
NS4B is an integral membrane protein that may contain multiple transmembrane segments (83). NS4B can be palmitoylated, and this modification may facilitate its oligomerization (251). Palmitoylation of NS4B is essential for HCV RNA replication (251). NS4B likely functions as a scaffold for assembly of viral RNA replicase complex. The expression of NS4B is sufficient to induce the formation of convoluted membrane structures that mimic the membranous web (45, 83) in replicon cells where RNA replication complexes localize (69). A nucleotide-binding motif is present in a cytoplasmic loop of NS4B, and in vitro GTPase activity has been demonstrated (46). While it is intriguing to speculate that this activity plays a role in membrane reorganization, this seems unlikely given the identification of an adaptive mutation that disrupts this motif (133). NS4B-induced membrane reorganization might also involve an interaction with Rab5, a regulator of membrane fusion, as well as other components of early endosomal compartments (202). Interestingly, NS4B may also modulate the endoplasmic reticulum (ER) unfolded protein response, thereby avoiding cell death triggered by HCV-induced ER overload or membrane alterations (253).
NS5A is a hydrophilic phosphoprotein of unknown function (208). At least one kinase likely responsible for NS5A phosphorylation is casein kinase Iα (183, 184) and, as discussed above, NS5A phosphorylation isoforms and RNA replication appear to be intimately linked. NS5A has been modeled as a three-domain protein (209). Recent structural characterization of NS5A includes an NMR structure for the N-terminal membrane anchor (an amphipathic α helix) and the crystal structure of the bulk of domain I (175, 210). Domain I coordinates a zinc atom that is required for RNA replication, possibly by playing a structural role in the NS5A fold (209). NS5A domain I is present as a dimer in the crystal structure, with a large basic groove generated at the dimer interface forming a possible RNA binding cleft. NS5A does indeed interact with RNA with a preference for RNAs rich in U and G (81). NS5A domains II and III are more variable among HCV genotypes and poorly characterized. A failed attempt at determining an NMR structure for domain II may indicate significant conformational flexibility (127). Domain III appears even more plastic, as this region can tolerate large insertions and deletions without disrupting RNA replication (6, 132, 146, 158). Replicons selected for partial resistance to ribavirin possess mutations in domain III (176). NS5A is perhaps best known for its putative interactions with a large constellation of host proteins. These include, most famously, PKR, and a number of other proteins involved in signaling, apoptosis, and lipid trafficking. The reader is directed to an excellent review for a full discussion of this topic (142). Here, we highlight two recently described NS5A interaction partners, FBL-2 and KFBP8.
A hallmark of acute HCV infection is the induction of changes in cellular lipid metabolism (see reference 163 for a review). Drugs that modify lipid levels can affect HCV replication (reviewed in references 15, 88, 136, and 203) and infectious virus production (80). Inhibitors of geranylgeranyl transferase I are potent HCV replication inhibitors, yet no HCV protein has an acceptor motif for this lipid modification (93, 203, 245). Recently, an interaction of NS5A with the geranylated host F box protein FBL-2 has been shown to be required for HCV RNA replication (228). FBL-2 is believed to target certain proteins for degradation, although its specific substrates remain to be identified. It is interesting to speculate that FBL-2, in conjunction with NS5A, might be responsible for targeting key components of the host antiviral response machinery or other pathways for degradation. This finding, in conjunction with the large number of previously described activities of NS5A in manipulating cellular pathways, suggests that this protein plays a significant role in immune escape and persistence.
NS5A also interacts with FKBP8, an immunophilin that shares similarity with the cyclophilin family of PPIs (although FKBP8 lacks PPI activity) (165). Interestingly, FKBP8 binds to NS5A as a trimeric complex with the chaperone HSP90 to modulate HCV RNA replication (165). FKBP8 is the second PPI or PPI-like protein identified as an interaction partner of an HCV protein (cyclophilin B has been shown to bind NS5B [234], as discussed below). The significance of these interactions for HCV protein folding or posttranslational modification has yet to be explored.
NS5B, the viral RNA-dependent RNA polymerase is, of course, essential for HCV replication in vivo and in cell culture (17, 110, 134). This fact, combined with extensive structural (see references 39 and 75 for reviews) and biochemical (see references 37, 75, and 135 for reviews) characterizations have made NS5B a highly attractive target for development of specific antivirals. NS5B is a member of the tail-anchored class of membrane proteins, and the C-terminal anchor sequence of NS5B is required for membrane localization and replication but not required for enzymatic activity (157). NS5B forms oligomers and exhibits cooperativity in enzyme assays (231). Perhaps the most exciting observation in recent years is the interaction of NS5B with cyclophilins, a class of PPIs. This interaction was uncovered when cyclosporine (CsA) was found to suppress HCV RNA replication in a dose-dependent manner (160, 233). CsA apparently disrupts the interaction of NS5B with cyclophilin B (234), which is required for efficient recruitment and replication of HCV RNA. Knockdown of the expression of cyclophilin B, as well as that of other cyclophilins, has been shown to significantly suppress HCV replication with induction of the cellular unfolded protein response, possibly contributing to suppression of HCV replication (161). Notably, these studies and a more recent one (170) with a nonimmunosuppressive CsA derivative, DEBI0-025, demonstrated that CsA immunosuppressive activity can be separated from its anti-HCV effects, possibly yielding a new class of HCV inhibitors with therapeutic potential.
VIRUS ASSEMBLY AND EXIT
The final stages in the HCV life cycle—genome packaging, virion assembly, and secretion of infectious particles—have been difficult to study. The development of HCVcc systems has made this aspect of the life cycle tractable, and this area has become one of the most exciting areas in HCV research. While the area is still in its infancy, a number of intriguing observations have been made regarding determinants of virion production and the biophysical properties of infectious particles.
One determinant that affects virus production is the cellular environment, with higher titers of JFH-1 produced when virus is grown in Huh-7.5 cells and derived subpopulations than in Huh-7 cells (128, 254). On the virus side, a surprising observation was the fact that a chimeric genotype 2a full-length genome, expressing the core through the NS2 region of the HCJ6 isolate coupled to the JFH-1 replicase (J6/JFH) outperformed the full-length “native” JFH-1 genome (128). The kinetics of JFH-1 virus release in culture, with a rise in infectious particle titers following prolonged culturing of infected cells, suggested the emergence of an adapted virus (10, 254). This result could reflect an adaptation to cell culture conditions or the correction of a replication-impairing defect present in the original JFH-1 consensus cDNA. Recently, a sequence analysis of these passaged viruses has identified a mutation, G451R in E2, that leads to increased infectious virus production while not affecting RNA replication (255). Further investigation of how this mutation functions should provide insights into HCV infectivity.
Another clue to determinants that control infectious HCV production comes from studies to optimize the polyprotein breakpoint for chimeric intergenotype HCVcc. Higher titers were obtained with a fusion junction after the first predicted transmembrane segment of NS2, suggesting important protein-protein interactions between the N-terminal portion of NS2 and the structural proteins and the remaining portion of NS2 and the replicase region that influence, most likely, virus assembly (177). The nature of these interactions is unknown, but these data loosely parallel those seen for the classical flaviviruses and pestiviruses, where NS2A and NS3 or uncleaved NS2-3, respectively, are important determinants of virion production (2, 114).
Recent work has provided yet another interesting clue in infectious HCV biogenesis. Gastaminza et al. have shown that HCV virions form within the cell and that production of intracellular infectious virions precedes secretion of infectious virus (67). Secreted particles differ in buoyant density from intracellular virions (1.03 to 1.16 g/ml for secreted particles versus 1.15 to 1.20 g/ml for intracellular particles), suggesting that virions go through a maturation process that alters particle density. Interestingly, the passage of HCVcc in animals produces virus of lower density than HCVcc produced in cell culture (129). The specific infectivity of HCVcc samples produced in vivo can be significantly higher than that obtained for cell culture-produced HCVcc and, when combined with the lighter density observed for animal-derived HCVcc, suggests that a low-density component associates with virions and is important for infectivity (129). It is intriguing to speculate that maturation and export of highly infectious virus requires association with lipids or lipoproteins. This is consistent with recent data where inhibition of pathways required for very-low-density lipoprotein production (apolipoprotein B and microsomal triglyceride transfer protein) decreased infectious HCV production (80). A complete compositional analysis of infectious particles from cell culture and animal systems is eagerly awaited.
The availability of HCVcc systems will catalyze a detailed analysis of additional requirements in virion assembly in the coming years. Little is known about the process of nucleocapsid assembly, the mechanism of budding, the maturation process of virions, or the cellular pathways used for virus egress. In vitro nucleocapsid assembly systems have been reported, and it will be interesting to compare the properties of these nucleocapsids with those produced using the HCVcc system (105, 115). Another area of intense interest will likely be that of E1-E2 oligomerization and how it relates to virus budding (166, 167, 223). Considerable information has been generated regarding the processing, oligomerization, and maturation of the HCV structural proteins from surrogate systems, and these subjects are ripe for reinvestigation with HCVcc. p7 is thought to function as an ion channel in either the infection or maturation of HCV virions, and direct tests of these possibilities are now possible (31, 73, 154, 172, 173, 181).
WHAT'S NEXT?
Despite these recent advances in our understanding of HCV biology, many questions remain. Identification of the complete set of cellular factors required for HCV entry is a priority for understanding viral tropism and developing permissive small-animal models. While much is known about the viral and cellular factors involved in RNA replication, our understanding of how these components function as a multiprotein RNA recognition complex is still in its infancy. How does HCV genome RNA in polysomes transition to being a substrate for minus-strand synthesis? What determines the fate of newly synthesized genome RNA? What are the steps in infectious virus assembly, where do they occur, and how tight is their association with liver cell lipid metabolism? Beyond the nuts and bolts of RNA replication and virus production, what other virus-host interactions are important for the ability of this virus to establish and maintain persistent infections? The ability of HCV to antagonize the TLR3 and RIG-I pathways is remarkable and revealing, but this is probably only the tip of the iceberg in terms of interactions that this clever RNA virus uses to outwit its supposedly sophisticated host.
Footnotes
Published ahead of print on 23 May 2007.
REFERENCES
- 1.Acton, S., A. Rigotti, K. T. Landschulz, S. Xu, H. H. Hobbs, and M. Krieger. 1996. Identification of scavenger receptor SR-BI as a high density lipoprotein receptor. Science 271:518-520. [DOI] [PubMed] [Google Scholar]
- 2.Agapov, E. V., C. L. Murray, I. Frolov, L. Qu, T. M. Myers, and C. M. Rice. 2004. Uncleaved NS2-3 is required for production of infectious bovine viral diarrhea virus. J. Virol. 78:2414-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agnello, V., G. Abel, M. Elfahal, G. B. Knight, and Q.-X. Zhang. 1999. Hepatitis C virus and other Flaviviridae viruses enter cells via low density lipoprotein receptor. Proc. Natl. Acad. Sci. USA 96:12766-12771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andre, P., K.-P. F. Deforges, S. Perret, M. Berland, J. L. Sodoyer, M. Pol, S. Brechot, C. Paranhos-Baccala, and G. Lotteau. 2002. Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J. Virol. 76:6919-6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anonymous. 2002. National Institutes of Health Consensus Development Conference statement: management of hepatitis C 2002 (June 10-12, 2002). Gastroenterology 123:2082-2099. [DOI] [PubMed] [Google Scholar]
- 6.Appel, N., T. Pietschmann, and R. Bartenschlager. 2005. Mutational analysis of hepatitis C virus nonstructural protein 5A: potential role of differential phosphorylation in RNA replication and identification of a genetically flexible domain. J. Virol. 79:3187-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banerjee, R., and A. Dasgupta. 2001. Specific interaction of hepatitis C virus protease/helicase NS3 with the 3′-terminal sequences of viral positive- and negative-strand RNA. J. Virol. 75:1708-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baril, M., and L. Brakier-Gingras. 2005. Translation of the F protein of hepatitis C virus is initiated at a non-AUG codon in a +1 reading frame relative to the polyprotein. Nucleic Acids Res. 33:1474-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartenschlager, R., M. Frese, and T. Pietschmann. 2004. Novel insights into hepatitis C virus replication and persistence. Adv. Virus Res. 63:71-180. [DOI] [PubMed] [Google Scholar]
- 10.Bartenschlager, R., and T. Pietschmann. 2005. Efficient hepatitis C virus cell culture system: what a difference the host cell makes. Proc. Natl. Acad. Sci. USA 102:9739-9740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barth, H., C. Schafer, M. I. Adah, F. Zhang, R. J. Linhardt, H. Toyoda, A. Kinoshita-Toyoda, T. Toida, T. H. van Kuppevelt, E. Depla, F. von Weizsacker, H. E. Blum, and T. F. Baumert. 2003. Cellular binding of hepatitis C virus envelope glycoprotein E2 requires cell surface heparan sulfate. J. Biol. Chem. 278:41003-41012. [DOI] [PubMed] [Google Scholar]
- 12.Bartosch, B., J. Dubuisson, and F. L. Cosset. 2003. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J. Exp. Med. 197:633-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartosch, B., G. Verney, M. Dreux, P. Donot, Y. Morice, F. Penin, J. M. Pawlotsky, D. Lavillette, and F. L. Cosset. 2005. An interplay between hypervariable region 1 of the hepatitis C virus E2 glycoprotein, the scavenger receptor BI, and high-density lipoprotein promotes both enhancement of infection and protection against neutralizing antibodies. J. Virol. 79:8217-8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bertaux, C., and T. Dragic. 2006. Different domains of CD81 mediate distinct stages of hepatitis C virus pseudoparticle entry. J. Virol. 80:4940-4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bigger, C. B., B. Guerra, K. M. Brasky, G. Hubbard, M. R. Beard, B. A. Luxon, S. M. Lemon, and R. E. Lanford. 2004. Intrahepatic gene expression during chronic hepatitis C virus infection in chimpanzees. J. Virol. 78:13779-13792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blanchard, E., S. Belouzard, L. Goueslain, T. Wakita, J. Dubuisson, C. Wychowski, and Y. Rouille. 2006. Hepatitis C virus entry depends on clathrin-mediated endocytosis. J. Virol. 80:6964-6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 18.Blight, K. J., J. A. McKeating, J. Marcotrigiano, and C. M. Rice. 2003. Efficient RNA replication of hepatitis C virus genotype 1a in cell culture. J. Virol. 77:3181-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blight, K. J., J. A. McKeating, and C. M. Rice. 2002. Highly permissive cell lines for hepatitis C virus genomic and subgenomic RNA replication. J. Virol. 76:13001-13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blight, K. J., and C. M. Rice. 1997. Secondary structure determination of the conserved 98-base sequence at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 71:7345-7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boehringer, D., R. Thermann, A. Ostareck-Lederer, J. D. Lewis, and H. Stark. 2005. Structure of the hepatitis C Virus IRES bound to the human 80S ribosome: remodeling of the HCV IRES. Structure 13:1695-1706. [DOI] [PubMed] [Google Scholar]
- 22.Bradrick, S. S., R. W. Walters, and M. Gromeier. 2006. The hepatitis C virus 3′-untranslated region or a poly(A) tract promote efficient translation subsequent to the initiation phase. Nucleic Acids Res. 34:1293-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Branch, A. D., D. D. Stump, J. A. Gutierrez, F. Eng, and J. L. Walewski. 2005. The hepatitis C virus alternate reading frame (ARF) and its family of novel products: the alternate reading frame protein/F-protein, the double-frameshift protein, and others. Semin. Liver Dis. 25:105-117. [DOI] [PubMed] [Google Scholar]
- 24.Breiman, A., N. Grandvaux, R. Lin, C. Ottone, S. Akira, M. Yoneyama, T. Fujita, J. Hiscott, and E. F. Meurs. 2005. Inhibition of RIG-I-dependent signaling to the interferon pathway during hepatitis C virus expression and restoration of signaling by IKKɛ. J. Virol. 79:3969-3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bukh, J., T. Pietschmann, V. Lohmann, N. Krieger, K. Faulk, R. E. Engle, S. Govindarajan, M. Shapiro, M. St. Claire, and R. Bartenschlager. 2002. Mutations that permit efficient replication of hepatitis C virus RNA in Huh-7 cells prevent productive replication in chimpanzees. Proc. Natl. Acad. Sci. USA 99:14416-14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buonocore, L., K. J. Blight, C. M. Rice, and J. K. Rose. 2002. Characterization of vesicular stomatitis virus recombinants that express and incorporate high levels of hepatitis C virus glycoproteins. J. Virol. 76:6865-6872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cai, Z., C. Zhang, K. S. Chang, J. Jiang, B. C. Ahn, T. Wakita, T. J. Liang, and G. Luo. 2005. Robust production of infectious hepatitis C virus (HCV) from stably HCV cDNA-transfected human hepatoma cells. J. Virol. 79:13963-13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cho, H. S., N. C. Ha, L. W. Kang, K. M. Chung, S. H. Back, S. K. Jang, and B. H. Oh. 1998. Crystal structure of RNA helicase from genotype 1b hepatitis C virus. A feasible mechanism of unwinding duplex RNA. J. Biol. Chem. 273:15045-15052. [DOI] [PubMed] [Google Scholar]
- 29.Choi, J., Z. Xu, and J. H. Ou. 2003. Triple decoding of hepatitis C virus RNA by programmed translational frameshifting. Mol. Cell. Biol. 23:1489-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choo, Q.-L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 31.Clarke, D., S. Griffin, L. Beales, C. S. Gelais, S. Burgess, M. Harris, and D. Rowlands. 2006. Evidence for the formation of a heptameric ion channel complex by the hepatitis C virus p7 protein in vitro. J. Biol. Chem. 281:37057-37068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cocquerel, L., A. Op de Beeck, M. Lambot, J. Roussel, D. Delgrange, A. Pillez, C. Wychowski, F. Penin, and J. Dubuisson. 2002. Topological changes in the transmembrane domains of hepatitis C virus envelope glycoproteins. EMBO J. 21:2893-2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Codran, A., C. Royer, D. Jaeck, M. Bastien-Valle, T. F. Baumert, M. P. Kieny, C. A. Pereira, and J. P. Martin. 2006. Entry of hepatitis C virus pseudotypes into primary human hepatocytes by clathrin-dependent endocytosis. J. Gen. Virol. 87:2583-2593. [DOI] [PubMed] [Google Scholar]
- 34.Collier, A. J., J. Gallego, R. Klinck, P. T. Cole, S. J. Harris, G. P. Harrison, F. Aboul-Ela, G. Varani, and S. Walker. 2002. A conserved RNA structure within the HCV IRES eIF3-binding site. Nat. Struct. Biol. 9:375-380. [DOI] [PubMed] [Google Scholar]
- 35.Cristina, J., F. Lopez, G. Moratorio, L. Lopez, S. Vasquez, L. Garcia-Aguirre, and A. Chunga. 2005. Hepatitis C virus F protein sequence reveals a lack of functional constraints and a variable pattern of amino acid substitution. J. Gen. Virol. 86:115-120. [DOI] [PubMed] [Google Scholar]
- 36.Date, T., T. Kato, M. Miyamoto, Z. Zhao, K. Yasui, M. Mizokami, and T. Wakita. 2004. Genotype 2a hepatitis C virus subgenomic replicon can replicate in HepG2 and IMY-N9 cells. J. Biol. Chem. 279:22371-22376. [DOI] [PubMed] [Google Scholar]
- 37.De Francesco, R., S. E. Behrens, L. Tomei, S. Altamura, and J. Jiricny. 1996. RNA-dependent RNA polymerase of hepatitis C virus. Methods Enzymol. 275:58-67. [DOI] [PubMed] [Google Scholar]
- 38.De Francesco, R., and C. Steinkuhler. 2000. Structure and function of the hepatitis C virus NS3-NS4A serine proteinase. Curr. Top. Microbiol. Immunol. 242:149-169. [DOI] [PubMed] [Google Scholar]
- 39.De Francesco, R., T. L. Altamura, S. Summa, and V. Migliaccio. 2003. Approaching a new era for hepatitis C virus therapy: inhibitors of the NS3-4A serine protease and the NS5B RNA-dependent RNA polymerase. Antiviral Res. 58:1-16. [DOI] [PubMed] [Google Scholar]
- 40.Deleersnyder, V., A. Pillez, C. Wychowski, K. Blight, J. Xu, Y. S. Hahn, C. M. Rice, and J. Dubuisson. 1997. Formation of native hepatitis C virus glycoprotein complexes. J. Virol. 71:697-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drummer, H. E., K. A. Wilson, and P. Poumbourios. 2005. Determinants of CD81 dimerization and interaction with hepatitis C virus glycoprotein E2. Biochem. Biophys. Res. Commun. 328:251-257. [DOI] [PubMed] [Google Scholar]
- 42.Drummer, H. E., K. A. Wilson, and P. Poumbourios. 2002. Identification of the hepatitis C virus E2 glycoprotein binding site on the large extracellular loop of CD81. J. Virol. 76:11143-11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dubuisson, J., H. H. Hsu, R. C. Cheung, H. Greenberg, D. R. Russell, and C. M. Rice. 1994. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J. Virol. 68:6147-6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dumont, S., W. Cheng, V. Serebrov, R. K. Beran, I. Tinoco, Jr., A. M. Pyle, and C. Bustamante. 2006. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature 439:105-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Egger, D., B. Wolk, R. Gosert, L. Bianchi, H. E. Blum, D. Moradpour, and K. Bienz. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76:5974-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Einav, S., M. Elazar, T. Danieli, and J. S. Glenn. 2004. A nucleotide binding motif in hepatitis C virus (HCV) NS4B mediates HCV RNA replication. J. Virol. 78:11288-11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eren, R., D. Landstein, D. Terkieltaub, O. Nussbaum, A. Zauberman, J. Ben-Porath, J. Gopher, R. Buchnick, R. Kovjazin, Z. Rosenthal-Galili, S. Aviel, E. Ilan, Y. Shoshany, L. Neville, T. Waisman, O. Ben-Moshe, A. Kischitsky, S. K. Foung, Z. Y. Keck, O. Pappo, A. Eid, O. Jurim, G. Zamir, E. Galun, and S. Dagan. 2006. Preclinical evaluation of two neutralizing human monoclonal antibodies against hepatitis C virus (HCV): a potential treatment to prevent HCV reinfection in liver transplant patients. J. Virol. 80:2654-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Evans, M. J., C. M. Rice, and S. P. Goff. 2004. Genetic interactions between hepatitis C virus replicons. J. Virol. 78:12085-12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Evans, M. J., C. M. Rice, and S. P. Goff. 2004. Phosphorylation of hepatitis C virus nonstructural protein 5A modulates its protein interactions and viral RNA replication. Proc. Natl. Acad. Sci. USA 101:13038-13043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Evans, M. J., T. von Hahn, D. M. Tscherne, A. J. Syder, M. Panis, B. Wolk, T. Hatziioannou, J. A. McKeating, P. D. Bieniasz, and C. M. Rice. 2007. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature 446:801-805. [DOI] [PubMed] [Google Scholar]
- 51.Failla, C., L. Tomei, and R. De Francesco. 1994. Both NS3 and NS4A are required for proteolytic processing of hepatitis C virus nonstructural proteins. J. Virol. 68:3753-3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fang, J. W., and R. W. Moyer. 2000. The effects of the conserved extreme 3′ end sequence of hepatitis C virus (HCV) RNA on the in vitro stabilization and translation of the HCV RNA genome. J. Hepatol. 33:632-639. [DOI] [PubMed] [Google Scholar]
- 53.Feld, J. J., and J. H. Hoofnagle. 2005. Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature 436:967-972. [DOI] [PubMed] [Google Scholar]
- 54.Ferlenghi, I., M. Clarke, T. Ruttan, S. L. Allison, J. Schalich, F. X. Heinz, S. C. Harrison, F. A. Rey, and S. D. Fuller. 2001. Molecular organization of a recombinant subviral particle from tick-borne encephalitis virus. Mol. Cell 7:593-602. [DOI] [PubMed] [Google Scholar]
- 55.Ferreon, J. C., A. C. Ferreon, K. Li, and S. M. Lemon. 2005. Molecular determinants of TRIF proteolysis mediated by the hepatitis C virus NS3/4A protease. J. Biol. Chem. 280:20483-20492. [DOI] [PubMed] [Google Scholar]
- 56.Flint, M., C. Maidens, L. D. Loomis-Price, C. Shotton, J. Dubuisson, P. Monk, A. Higginbottom, S. Levy, and J. A. McKeating. 1999. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J. Virol. 73:6235-6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Flint, M., and J. A. McKeating. 1999. The C-terminal region of the hepatitis C virus E1 glycoprotein confers localization within the endoplasmic reticulum. J. Gen. Virol. 80:1943-1947. [DOI] [PubMed] [Google Scholar]
- 58.Flint, M., J. M. Thomas, C. M. Maidens, C. Shotton, S. Levy, W. S. Barclay, and J. A. McKeating. 1999. Functional analysis of cell surface-expressed hepatitis C virus E2 glycoprotein. J. Virol. 73:6782-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Foy, E., K. Li, R. Sumpter, Jr., Y. M. Loo, C. L. Johnson, C. Wang, P. M. Fish, M. Yoneyama, T. Fujita, S. M. Lemon, and M. Gale, Jr. 2005. Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proc. Natl. Acad. Sci. USA 102:2986-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fraser, C. S., and J. A. Doudna. 2007. Structural and mechanistic insights into hepatitis C viral translation initiation. Nat. Rev. Microbiol. 5:29-38. [DOI] [PubMed] [Google Scholar]
- 61.Friebe, P., and R. Bartenschlager. 2002. Genetic analysis of sequences in the 3′ nontranslated region of hepatitis C virus that are important for RNA replication. J. Virol. 76:5326-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Friebe, P., J. Boudet, J. P. Simorre, and R. Bartenschlager. 2005. Kissing-loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J. Virol. 79:380-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Friebe, P., V. Lohmann, N. Krieger, and R. Bartenschlager. 2001. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 75:12047-12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gale, M., Jr., and E. M. Foy. 2005. Evasion of intracellular host defence by hepatitis C virus. Nature 436:939-945. [DOI] [PubMed] [Google Scholar]
- 65.Gao, L., H. Aizaki, J. W. He, and M. M. Lai. 2004. Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J. Virol. 78:3480-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Garry, R. F., and S. Dash. 2003. Proteomics computational analyses suggest that hepatitis C virus E1 and pestivirus E2 envelope glycoproteins are truncated class II fusion proteins. Virology 307:255-265. [DOI] [PubMed] [Google Scholar]
- 67.Gastaminza, P., S. B. Kapadia, and F. V. Chisari. 2006. Differential biophysical properties of infectious intracellular and secreted hepatitis C virus particles. J. Virol. 80:11074-11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goffard, A., N. Callens, B. Bartosch, C. Wychowski, F. L. Cosset, C. Montpellier, and J. Dubuisson. 2005. Role of N-linked glycans in the functions of hepatitis C virus envelope glycoproteins. J. Virol. 79:8400-8409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gosert, R., D. Egger, V. Lohmann, R. Bartenschlager, H. E. Blum, K. Bienz, and D. Moradpour. 2003. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 77:5487-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Grakoui, A., D. W. McCourt, C. Wychowski, S. M. Feinstone, and C. M. Rice. 1993. A second hepatitis C virus-encoded proteinase. Proc. Natl. Acad. Sci. USA 90:10583-10587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Grakoui, A., D. W. McCourt, C. Wychowski, S. M. Feinstone, and C. M. Rice. 1993. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J. Virol. 67:2832-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grakoui, A., C. Wychowski, C. Lin, S. M. Feinstone, and C. M. Rice. 1993. Expression and identification of hepatitis C virus polyprotein cleavage products. J. Virol. 67:1385-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Griffin, S. D., L. P. Beales, D. S. Clarke, O. Worsfold, S. D. Evans, J. Jaeger, M. P. Harris, and D. J. Rowlands. 2003. The p7 protein of hepatitis C virus forms an ion channel that is blocked by the antiviral drug, amantadine. FEBS Lett. 535:34-38. [DOI] [PubMed] [Google Scholar]
- 74.Grove, J., T. Huby, Z. Stamataki, T. Vanwolleghem, P. Meuleman, M. Farquhar, A. Schwarz, M. Moreau, J. S. Owen, G. Leroux-Roels, P. Balfe, and J. A. McKeating. 2007. Scavenger receptor BI and BII expression levels modulate hepatitis C virus infectivity. J. Virol. 81:3162-3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hagedorn, C. H., E. H. van Beers, and C. De Staercke. 1999. Hepatitis C virus RNA-dependent RNA polymerase (NS5B polymerase), p. 225-260. In C. Hagedorn and C. M. Rice (ed.), Hepatitis C virus. Current topics in microbiology and immunology, vol. 242. Springer-Verlag, Berlin, Germany. [DOI] [PubMed] [Google Scholar]
- 76.Han, J. H., V. Shyamala, K. H. Richman, M. J. Brauer, B. Irvine, M. S. Urdea, P. Tekamp-Olson, G. Kuo, Q.-L. Choo, and M. Houghton. 1991. Characterization of the terminal regions of hepatitis C viral RNA: identification of conserved sequences in the 5′ untranslated region and poly(A) tails at the 3′ end. Proc. Natl. Acad. Sci. USA 88:1711-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Higginbottom, A., E. R. Quinn, C. C. Kuo, M. Flint, L. H. Wilson, E. Bianchi, A. Nicosia, P. N. Monk, J. A. McKeating, and S. Levy. 2000. Identification of amino acid residues in CD81 critical for interaction with hepatitis C virus envelope glycoprotein E2. J. Virol. 74:3642-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hijikata, M., Y. K. Shimizu, H. Kato, A. Iwamoto, J. W. Shih, H. J. Alter, R. H. Purcell, and H. Yoshikura. 1993. Equilibrium centrifugation studies of hepatitis C virus: evidence for circulating immune complexes. J. Virol. 67:1953-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hsu, M., J. Zhang, M. Flint, C. Logvinoff, C. Cheng-Mayer, C. M. Rice, and J. A. McKeating. 2003. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc. Natl. Acad. Sci. USA 100:7271-7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huang, H., F. Sun, D. M. Owen, W. Li, Y. Chen, M. Gale, Jr., and J. Ye. 2007. Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc. Natl. Acad. Sci. USA 104:5848-5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huang, L., J. Hwang, S. D. Sharma, M. R. Hargittai, Y. Chen, J. J. Arnold, K. D. Raney, and C. E. Cameron. 2005. Hepatitis C virus nonstructural protein 5A (NS5A) is an RNA-binding protein. J. Biol. Chem. 280:36417-36428. [DOI] [PubMed] [Google Scholar]
- 82.Huang, Y., K. Staschke, R. De Francesco, and S. L. Tan. 2007. Phosphorylation of hepatitis C virus NS5A nonstructural protein: a new paradigm for phosphorylation-dependent viral RNA replication? Virology 364:1-9. [DOI] [PubMed] [Google Scholar]
- 83.Hügle, T., F. Fehrmann, E. Bieck, M. Kohara, H.-G. Krausslich, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. The hepatitis C virus nonstructural protein 4B is an integral endoplasmic reticulum membrane protein. Virology 284:70-81. [DOI] [PubMed] [Google Scholar]
- 84.Ikeda, M., M. Yi, K. Li, and S. M. Lemon. 2002. Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells. J. Virol. 76:2997-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Imbert, I., M. Dimitrova, F. Kien, M. P. Kieny, and C. Schuster. 2003. Hepatitis C virus IRES efficiency is unaffected by the genomic RNA 3′NTR even in the presence of viral structural or non-structural proteins. J. Gen. Virol. 84:1549-1557. [DOI] [PubMed] [Google Scholar]
- 86.Ito, T., and M. M. Lai. 1999. An internal polypyrimidine-tract-binding protein-binding site in the hepatitis C virus RNA attenuates translation, which is relieved by the 3′-untranslated sequence. Virology 254:288-296. [DOI] [PubMed] [Google Scholar]
- 87.Ito, T., S. M. Tahara, and M. M. C. Lai. 1998. The 3′-untranslated region of hepatitis C virus RNA enhances translation from an internal ribosomal entry site. J. Virol. 72:8789-8796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jacobs, J. M., D. L. Diamond, E. Y. Chan, M. A. Gritsenko, W. Qian, M. Stastna, T. Baas, D. G. Camp II, R. L. Carithers, Jr., R. D. Smith, and M. G. Katze. 2005. Proteome analysis of liver cells expressing a full-length hepatitis C virus (HCV) replicon and biopsy specimens of posttransplantation liver from HCV-infected patients. J. Virol. 79:7558-7569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Johnson, C. L., and M. Gale, Jr. 2006. CARD games between virus and host get a new player. Trends Immunol. 27:1-4. [DOI] [PubMed] [Google Scholar]
- 90.Jopling, C. L., M. Yi, A. M. Lancaster, S. M. Lemon, and P. Sarnow. 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science 309:1577-1581. [DOI] [PubMed] [Google Scholar]
- 91.Kanai, A., K. Tanabe, and M. Kohara. 1995. Poly(U) binding activity of hepatitis C virus NS3 protein, a putative RNA helicase. FEBS Lett. 376:221-224. [DOI] [PubMed] [Google Scholar]
- 92.Kapadia, S. B., H. Barth, T. Baumert, J. A. McKeating, and F. V. Chisari. 2007. Initiation of hepatitis C virus infection is dependent on cholesterol and cooperativity between CD81 and scavenger receptor B type I. J. Virol. 81:374-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kapadia, S. B., and F. V. Chisari. 2005. Hepatitis C virus RNA replication is regulated by host geranylgeranylation and fatty acids. Proc. Natl. Acad. Sci. USA 102:2561-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Karayiannis, P. 2005. The hepatitis C virus NS3/4A protease complex interferes with pathways of the innate immune response. J. Hepatol. 43:743-745. [DOI] [PubMed] [Google Scholar]
- 95.Kato, T., T. Date, M. Miyamoto, A. Furusaka, K. Tokushige, M. Mizokami, and T. Wakita. 2003. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 125:1808-1817. [DOI] [PubMed] [Google Scholar]
- 96.Kato, T., T. Date, M. Miyamoto, Z. Zhao, M. Mizokami, and T. Wakita. 2005. Nonhepatic cell lines HeLa and 293 support efficient replication of the hepatitis C virus genotype 2a subgenomic replicon. J. Virol. 79:592-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kato, T., T. Matsumura, T. Heller, S. Saito, R. K. Sapp, K. Murthy, T. Wakita, and T. J. Liang. 2007. Production of infectious hepatitis C virus of various genotypes in cell cultures. J. Virol. 81:4405-4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Keck, Z. Y., T. K. Li, J. Xia, B. Bartosch, F. L. Cosset, J. Dubuisson, and S. K. Foung. 2005. Analysis of a highly flexible conformational immunogenic domain A in hepatitis C virus E2. J. Virol. 79:13199-13208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kieft, J. S., K. Zhou, A. Grech, R. Jubin, and J. A. Doudna. 2002. Crystal structure of an RNA tertiary domain essential to HCV IRES-mediated translation initiation. Nat. Struct. Biol. 9:370-374. [DOI] [PubMed] [Google Scholar]
- 100.Kieft, J. S., K. Zhou, R. Jubin, and J. A. Doudna. 2001. Mechansim of ribosome recruitment by hepatitis C IRES RNA. RNA 7:194-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kim, J. L., K. A. Morgenstern, J. P. Griffith, M. D. Dwyer, J. A. Thomson, M. A. Murcko, C. Lin, and P. R. Caron. 1998. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure 6:89-100. [DOI] [PubMed] [Google Scholar]
- 102.Kim, J. L., K. A. Morgenstern, C. Lin, T. Fox, M. D. Dwyer, J. A. Landro, S. P. Chambers, W. Markland, C. A. Lepre, E. T. O'Malley, S. L. Harbeson, C. M. Rice, M. A. Murcko, P. R. Caron, and J. A. Thomson. 1996. Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide. Cell 87:343-355. [DOI] [PubMed] [Google Scholar]
- 103.Kim, Y. K., C. S. Kim, S. H. Lee, and S. K. Jang. 2002. Domains I and II in the 5′ nontranslated region of the HCV genome are required for RNA replication. Biochem. Biophys. Res. Commun. 290:105-112. [DOI] [PubMed] [Google Scholar]
- 104.Kim, Y. K., S. H. Lee, C. S. Kim, S. K. Seol, and S. K. Jang. 2003. Long-range RNA-RNA interaction between the 5′ nontranslated region and the core-coding sequences of hepatitis C virus modulates the IRES-dependent translation. RNA 9:599-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Klein, K. C., S. R. Dellos, and J. R. Lingappa. 2005. Identification of residues in the hepatitis C virus core protein that are critical for capsid assembly in a cell-free system. J. Virol. 79:6814-6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Klinck, R., E. Westhof, S. Walker, M. Afshar, A. Collier, and F. Aboul-Ela. 2000. A potential RNA drug target in the hepatitis C virus internal ribosomal entry site. RNA 6:1423-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kobayashi, M., M. C. Bennett, T. Bercot, and I. R. Singh. 2006. Functional analysis of hepatitis C virus envelope proteins, using a cell-cell fusion assay. J. Virol. 80:1817-1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kolykhalov, A. A., E. V. Agapov, K. J. Blight, K. Mihalik, S. M. Feinstone, and C. M. Rice. 1997. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science 277:570-574. [DOI] [PubMed] [Google Scholar]
- 109.Kolykhalov, A. A., S. M. Feinstone, and C. M. Rice. 1996. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 70:3363-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kolykhalov, A. A., K. Mihalik, S. M. Feinstone, and C. M. Rice. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 74:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kong, L. K., and P. Sarnow. 2002. Cytoplasmic expression of mRNAs containing the internal ribosome entry site and 3′ noncoding region of hepatitis C virus: effects of the 3′ leader on mRNA translation and mRNA stability. J. Virol. 76:12457-12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Koutsoudakis, G., A. Kaul, E. Steinmann, S. Kallis, V. Lohmann, T. Pietschmann, and R. Bartenschlager. 2006. Characterization of the early steps of hepatitis C virus infection by using luciferase reporter viruses. J. Virol. 80:5308-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kuhn, R. J., W. Zhang, M. G. Rossmann, S. V. Pletnev, J. Corver, E. Lenches, C. T. Jones, S. Mukhopadhyay, P. R. Chipman, E. G. Strauss, T. S. Baker, and J. H. Strauss. 2002. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108:717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kümmerer, B. M., and C. M. Rice. 2002. Mutations in the yellow fever virus nonstructural protein NS2A selectively block production of infectious particles. J. Virol. 76:4773-4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kunkel, M., M. Lorinczi, R. Rijnbrand, S. M. Lemon, and S. J. Watowich. 2001. Self-assembly of nucleocapsid-like particles from recombinant hepatitis C virus core protein. J. Virol. 75:2119-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lagging, L. M., K. Meyer, R. J. Owens, and R. Ray. 1998. Functional role of hepatitis C virus chimeric glycoproteins in the infectivity of pseudotyped virus. J. Virol. 72:3539-3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lam, A. M., and D. N. Frick. 2006. Hepatitis C virus subgenomic replicon requires an active NS3 RNA helicase. J. Virol. 80:404-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lancaster, A. M., E. Jan, and P. Sarnow. 2006. Initiation factor-independent translation mediated by the hepatitis C virus internal ribosome entry site. RNA 12:894-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lavillette, D., Y. Morice, G. Germanidis, P. Donot, A. Soulier, E. Pagkalos, G. Sakellariou, L. Intrator, B. Bartosch, J. M. Pawlotsky, and F. L. Cosset. 2005. Human serum facilitates hepatitis C virus infection, and neutralizing responses inversely correlate with viral replication kinetics at the acute phase of hepatitis C virus infection. J. Virol. 79:6023-6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Le, S. Y., N. Sonenberg, and J. V. Maizel, Jr. 1995. Unusual folding regions and ribosome landing pad within hepatitis C virus and pestivirus RNAs. Gene 154:137-143. [DOI] [PubMed] [Google Scholar]
- 121.Lee, H., H. Shin, E. Wimmer, and A. V. Paul. 2004. cis-acting RNA signals in the NS5B C-terminal coding sequence of the hepatitis C virus genome. J. Virol. 78:10865-10877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lee, Y. M., D. M. Tscherne, S. I. Yun, I. Frolov, and C. M. Rice. 2005. Dual mechanisms of pestiviral superinfection exclusion at entry and RNA replication. J. Virol. 79:3231-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lemon, S. M., M. Yi, and K. Li. 2005. “Strong reasons make strong actions”—the antiviral efficacy of NS3/4A protease inhibitors. Hepatology 41:671-674. [DOI] [PubMed] [Google Scholar]
- 124.Lescar, J., A. Roussel, M. W. Wien, J. Navaza, S. D. Fuller, G. Wengler, G. Wengler, and F. A. Rey. 2001. The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell 105:137-148. [DOI] [PubMed] [Google Scholar]
- 125.Levin, M. K., M. Gurjar, and S. S. Patel. 2005. A Brownian motor mechanism of translocation and strand separation by hepatitis C virus helicase. Nat. Struct. Mol. Biol. 12:429-435. [DOI] [PubMed] [Google Scholar]
- 126.Levin, M. K., Y. H. Wang, and S. S. Patel. 2004. The functional interaction of the hepatitis C virus helicase molecules is responsible for unwinding processivity. J. Biol. Chem. 279:26005-26012. [DOI] [PubMed] [Google Scholar]
- 127.Liang, Y., C. B. Kang, and H. S. Yoon. 2006. Molecular and structural characterization of the domain 2 of hepatitis C virus non-structural protein 5A. Mol. Cells 22:13-20. [PubMed] [Google Scholar]
- 128.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 129.Lindenbach, B. D., P. Meuleman, A. Ploss, T. Vanwolleghem, A. J. Syder, J. A. McKeating, R. E. Lanford, S. M. Feinstone, M. E. Major, G. Leroux-Roels, and C. M. Rice. 2006. Cell culture-grown hepatitis C virus is infectious in vivo and can be recultured in vitro. Proc. Natl. Acad. Sci. USA 103:3805-3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1041. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 1. Lippincott-Raven Publishers, Philadelphia, PA. [Google Scholar]
- 131.Lindenbach, B. D., H. J. Thiel, and C. M. Rice. 2007. Flaviviridae: the viruses and their replication, p. 1101-1152. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed., vol. 1. Lippincott-Raven Publishers, Philadelphia, PA. [Google Scholar]
- 132.Liu, S., I. H. Ansari, S. C. Das, and A. K. Pattnaik. 2006. Insertion and deletion analyses identify regions of non-structural protein 5A of hepatitis C virus that are dispensable for viral genome replication. J. Gen. Virol. 87:323-327. [DOI] [PubMed] [Google Scholar]
- 133.Lohmann, V., S. Hoffmann, U. Herian, F. Penin, and R. Bartenschlager. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 77:3007-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lohmann, V., F. Korner, J. O. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 135.Lohmann, V., A. Roos, F. Korner, J. O. Koch, and R. Bartenschlager. 2000. Biochemical and structural analysis of the NS5B RNA-dependent RNA polymerase of the hepatitis C virus. J. Viral Hepat. 7:167-174. [DOI] [PubMed] [Google Scholar]
- 136.Lonardo, A., L. E. Adinolfi, P. Loria, N. Carulli, G. Ruggiero, and C. P. Day. 2004. Steatosis and hepatitis C virus: mechanisms and significance for hepatic and extrahepatic disease. Gastroenterology 126:586-597. [DOI] [PubMed] [Google Scholar]
- 137.Lorenz, I. C., J. Marcotrigiano, T. G. Dentzer, and C. M. Rice. 2006. Structure of the catalytic domain of the hepatitis C virus NS2-3 protease. Nature 442:831-835. [DOI] [PubMed] [Google Scholar]
- 138.Love, R. A., H. E. Parge, J. A. Wickersham, Z. Hostomsky, N. Habuka, E. W. Moomaw, T. Adachi, and Z. Hostomska. 1996. The crystal structure of hepatitis C virus NS3 proteinase reveals a trypsin-like fold and a structural zinc binding site. Cell 87:331-342. [DOI] [PubMed] [Google Scholar]
- 139.Lukavsky, P. J., I. Kim, G. A. Otto, and J. D. Puglisi. 2003. Structure of HCV IRES domain II determined by NMR. Nat. Struct. Biol. 10:1033-1038. [DOI] [PubMed] [Google Scholar]
- 140.Lukavsky, P. J., G. A. Otto, A. M. Lancaster, P. Sarnow, and J. D. Puglisi. 2000. Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Nat. Struct. Biol. 7:1105-1110. [DOI] [PubMed] [Google Scholar]
- 141.Lytle, J. R., L. Wu, and H. D. Robertson. 2002. Domains on the hepatitis C virus internal ribosome entry site for 40s subunit binding. RNA 8:1045-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Macdonald, A., and M. Harris. 2004. Hepatitis C virus NS5A: tales of a promiscuous protein. J. Gen. Virol. 85:2485-2502. [DOI] [PubMed] [Google Scholar]
- 143.Mackintosh, S. G., J. Z. Lu, J. B. Jordan, M. K. Harrison, B. Sikora, S. D. Sharma, C. E. Cameron, K. D. Raney, and J. Sakon. 2006. Structural and biological identification of residues on the surface of NS3 helicase required for optimal replication of the hepatitis C virus. J. Biol. Chem. 281:3528-3535. [DOI] [PubMed] [Google Scholar]
- 144.Matsuura, Y., H. Tani, K. Suzuki, T. Kimura-Someya, R. Suzuki, H. Aizaki, K. Ishii, K. Moriishi, C. S. Robison, M. A. Whitt, and T. Miyamura. 2001. Characterization of pseudotype VSV possessing HCV envelope proteins. Virology 286:263-275. [DOI] [PubMed] [Google Scholar]
- 145.McCaffrey, A. P., K. Ohashi, L. Meuse, S. Shen, A. M. Lancaster, P. J. Lukavsky, P. Sarnow, and M. A. Kay. 2002. Determinants of hepatitis C translational initiation in vitro, in cultured cells and mice. Mol. Ther. 5:676-684. [DOI] [PubMed] [Google Scholar]
- 146.McCormick, C. J., S. Maucourant, S. Griffin, D. J. Rowlands, and M. Harris. 2006. Tagging of NS5A expressed from a functional hepatitis C virus replicon. J. Gen. Virol. 87:635-640. [DOI] [PubMed] [Google Scholar]
- 147.McKeating, J. A., L. Q. Zhang, C. Logvinoff, M. Flint, J. Zhang, J. Yu, D. Butera, D. D. Ho, L. B. Dustin, C. M. Rice, and P. Balfe. 2004. Diverse hepatitis C virus glycoproteins mediate viral infection in a CD81-dependent manner. J. Virol. 78:8496-8505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.McMullan, L. K., A. Grakoui, M. J. Evans, K. Mihalik, M. Puig, A. D. Branch, S. M. Feinstone, and C. M. Rice. 2007. Evidence for a functional RNA element in the hepatitis C virus core gene. Proc. Natl. Acad. Sci. USA 104:2879-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Meertens, L., C. Bertaux, and T. Dragic. 2006. Hepatitis C virus entry requires a critical postinternalization step and delivery to early endosomes via clathrin-coated vesicles. J. Virol. 80:11571-11578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Meunier, J. C., R. E. Engle, K. Faulk, M. Zhao, B. Bartosch, H. Alter, S. U. Emerson, F. L. Cosset, R. H. Purcell, and J. Bukh. 2005. Evidence for cross-genotype neutralization of hepatitis C virus pseudo-particles and enhancement of infectivity by apolipoprotein C1. Proc. Natl. Acad. Sci. USA 102:4560-4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Meylan, E., J. Curran, K. Hofmann, D. Moradpour, M. Binder, R. Bartenschlager, and J. Tschopp. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167-1172. [DOI] [PubMed] [Google Scholar]
- 152.Michalak, J. P., C. Wychowski, A. Choukhi, J. C. Meunier, S. Ung, C. M. Rice, and J. Dubuisson. 1997. Characterization of truncated forms of the hepatitis C virus glycoproteins. J. Gen. Virol. 78:2299-2306. [DOI] [PubMed] [Google Scholar]
- 153.Michel, Y. M., A. M. Borman, S. Paulous, and K. M. Kean. 2001. Eukaryotic initiation factor 4G-poly(A) binding protein interaction is required for poly(A) tail-mediated stimulation of picornavirus internal ribosome entry segment-driven translation but not for X-mediated stimulation of hepatitis C virus translation. Mol. Cell. Biol. 21:4097-4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Mihm, U., N. Grigorian, C. Welsch, E. Herrmann, B. Kronenberger, G. Teuber, M. von Wagner, W. P. Hofmann, M. Albrecht, T. Lengauer, S. Zeuzem, and C. Sarrazin. 2006. Amino acid variations in hepatitis C virus p7 and sensitivity to antiviral combination therapy with amantadine in chronic hepatitis C. Antivir. Ther. 11:507-519. [PubMed] [Google Scholar]
- 155.Miyanari, Y., M. Hijikata, M. Yamaji, M. Hosaka, H. Takahashi, and K. Shimotohno. 2003. Hepatitis C virus non-structural proteins in the probable membranous compartment function in viral genome replication. J. Biol. Chem. 278:50301-50308. [DOI] [PubMed] [Google Scholar]
- 156.Monazahian, M., I. Bohme, S. Bonk, A. Koch, C. Scholz, S. Grethe, and R. Thomssen. 1999. Low density lipoprotein receptor as a candidate receptor for hepatitis C virus. J. Med. Virol. 57:223-229. [DOI] [PubMed] [Google Scholar]
- 157.Moradpour, D., V. Brass, E. Bieck, P. Friebe, R. Gosert, H. E. Blum, R. Bartenschlager, F. Penin, and V. Lohmann. 2004. Membrane association of the RNA-dependent RNA polymerase is essential for hepatitis C virus RNA replication. J. Virol. 78:13278-13284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Moradpour, D., M. J. Evans, R. Gosert, Z. Yuan, H. E. Blum, S. P. Goff, B. D. Lindenbach, and C. M. Rice. 2004. Insertion of green fluorescent protein into nonstructural protein 5A allows direct visualization of functional hepatitis C virus replication complexes. J. Virol. 78:7400-7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Mukhopadhyay, S., B. S. Kim, P. R. Chipman, M. G. Rossmann, and R. J. Kuhn. 2003. Structure of West Nile virus. Science 302:248. [DOI] [PubMed] [Google Scholar]
- 160.Nakagawa, M., N. Sakamoto, N. Enomoto, Y. Tanabe, N. Kanazawa, T. Koyama, M. Kurosaki, S. Maekawa, T. Yamashiro, C. H. Chen, Y. Itsui, S. Kakinuma, and M. Watanabe. 2004. Specific inhibition of hepatitis C virus replication by cyclosporin A. Biochem. Biophys. Res. Commun. 313:42-47. [DOI] [PubMed] [Google Scholar]
- 161.Nakagawa, M., N. Sakamoto, Y. Tanabe, T. Koyama, Y. Itsui, Y. Takeda, C. H. Chen, S. Kakinuma, S. Oooka, S. Maekawa, N. Enomoto, and M. Watanabe. 2005. Suppression of hepatitis C virus replication by cyclosporin A is mediated by blockade of cyclophilins. Gastroenterology 129:1031-1041. [DOI] [PubMed] [Google Scholar]
- 162.Neddermann, P., M. Quintavalle, C. Di Pietro, A. Clementi, M. Cerretani, S. Altamura, L. Bartholomew, and R. De Francesco. 2004. Reduction of hepatitis C virus NS5A hyperphosphorylation by selective inhibition of cellular kinases activates viral RNA replication in cell culture. J. Virol. 78:13306-13314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Negro, F. 2006. Mechanisms and significance of liver steatosis in hepatitis C virus infection. World J. Gastroenterol. 12:6756-6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Nieland, T. J., M. Penman, L. Dori, M. Krieger, and T. Kirchhausen. 2002. Discovery of chemical inhibitors of the selective transfer of lipids mediated by the HDL receptor SR-BI. Proc. Natl. Acad. Sci. USA 99:15422-15427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Okamoto, T., Y. Nishimura, T. Ichimura, K. Suzuki, T. Miyamura, T. Suzuki, K. Moriishi, and Y. Matsuura. 2006. Hepatitis C virus RNA replication is regulated by FKBP8 and Hsp90. EMBO J. 25:5015-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Op De Beeck, A., and J. Dubuisson. 2003. Topology of hepatitis C virus envelope glycoproteins. Rev. Med. Virol. 13:233-241. [DOI] [PubMed] [Google Scholar]
- 167.Op De Beeck, A., C. Voisset, B. Bartosch, Y. Ciczora, L. Cocquerel, Z. Keck, S. Foung, F. L. Cosset, and J. Dubuisson. 2004. Characterization of functional hepatitis C virus envelope glycoproteins. J. Virol. 78:2994-3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Otto, G. A., P. J. Lukavsky, A. M. Lancaster, P. Sarnow, and J. D. Puglisi. 2002. Ribosomal proteins mediate the hepatitis C virus IRES-HeLa 40S interaction. RNA 8:913-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Otto, G. A., and J. D. Puglisi. 2004. The pathway of HCV IRES-mediated translation initiation. Cell 119:369-380. [DOI] [PubMed] [Google Scholar]
- 170.Paeshuyse, J., A. Kaul, E. De Clercq, B. Rosenwirth, J. M. Dumont, P. Scalfaro, R. Bartenschlager, and J. Neyts. 2006. The non-immunosuppressive cyclosporin DEBIO-025 is a potent inhibitor of hepatitis C virus replication in vitro. Hepatology 43:761-770. [DOI] [PubMed] [Google Scholar]
- 171.Pang, P. S., E. Jankowsky, P. J. Planet, and A. M. Pyle. 2002. The hepatitis C viral NS3 protein is a processive DNA helicase with cofactor enhanced RNA unwinding. EMBO J. 21:1168-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Patargias, G., N. Zitzmann, R. Dwek, and W. B. Fischer. 2006. Protein-protein interactions: modeling the hepatitis C virus ion channel p7. J. Med. Chem. 49:648-655. [DOI] [PubMed] [Google Scholar]
- 173.Pavlovic, D., D. C. Neville, O. Argaud, B. Blumberg, R. A. Dwek, W. B. Fischer, and N. Zitzmann. 2003. The hepatitis C virus p7 protein forms an ion channel that is inhibited by long-alkyl-chain iminosugar derivatives. Proc. Natl. Acad. Sci. USA 100:6104-6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Pedersen, J. S., I. M. Meyer, R. Forsberg, P. Simmonds, and J. Hein. 2004. A comparative method for finding and folding RNA secondary structures within protein-coding regions. Nucleic Acids Res. 32:4925-4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Penin, F., V. Brass, N. Appel, S. Ramboarina, R. Montserret, D. Ficheux, H. E. Blum, R. Bartenschlager, and D. Moradpour. 2004. Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 279:40835-40843. [DOI] [PubMed] [Google Scholar]
- 176.Pfeiffer, J. K., and K. Kirkegaard. 2005. Ribavirin resistance in hepatitis C virus replicon-containing cell lines conferred by changes in the cell line or mutations in the replicon RNA. J. Virol. 79:2346-2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Pietschmann, T., A. Kaul, G. Koutsoudakis, A. Shavinskaya, S. Kallis, E. Steinmann, K. Abid, F. Negro, M. Dreux, F. L. Cosset, and R. Bartenschlager. 2006. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc. Natl. Acad. Sci. USA 103:7408-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Pietschmann, T., V. Lohmann, A. Kaul, N. Krieger, G. Rinck, G. Rutter, D. Strand, and R. Bartenschlager. 2002. Persistent and transient replication of full-length hepatitis C virus genomes in cell culture. J. Virol. 76:4008-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Pileri, P., Y. Uematsu, S. Compagnoli, G. Galli, F. Falugi, R. Petracca, A. J. Weiner, M. Houghton, D. Rosa, G. Grandi, and S. Abrignani. 1998. Binding of hepatitis C virus to CD81. Science 282:938-941. [DOI] [PubMed] [Google Scholar]
- 180.Pisarev, A. V., N. E. Shirokikh, and C. U. Hellen. 2005. Translation initiation by factor-independent binding of eukaryotic ribosomes to internal ribosomal entry sites. C. R. Biol. 328:589-605. [DOI] [PubMed] [Google Scholar]
- 181.Premkumar, A., L. Wilson, G. D. Ewart, and P. W. Gage. 2004. Cation-selective ion channels formed by p7 of hepatitis C virus are blocked by hexamethylene amiloride. FEBS Lett. 557:99-103. [DOI] [PubMed] [Google Scholar]
- 182.Quinkert, D., R. Bartenschlager, and V. Lohmann. 2005. Quantitative analysis of the hepatitis C virus replication complex. J. Virol. 79:13594-13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Quintavalle, M., S. Sambucini, C. Di Pietro, R. De Francesco, and P. Neddermann. 2006. The alpha isoform of protein kinase CKI is responsible for hepatitis C virus NS5A hyperphosphorylation. J. Virol. 80:11305-11312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Quintavalle, M., S. Sambucini, V. Summa, L. Orsatti, F. Talamo, R. De Francesco, and P. Neddermann. 2007. Hepatitis C virus NS5A is a direct substrate of casein kinase I-alpha, a cellular kinase identified by inhibitor affinity chromatography using specific NS5A hyperphosphorylation inhibitors. J. Biol. Chem. 282:5536-5544. [DOI] [PubMed] [Google Scholar]
- 185.Rey, F. A., F. X. Heinz, C. Mandl, C. Kunz, and S. C. Harrison. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature 375:291-298. [DOI] [PubMed] [Google Scholar]
- 186.Rijnbrand, R., V. Thiviyanathan, K. Kaluarachchi, S. M. Lemon, and D. G. Gorenstein. 2004. Mutational and structural analysis of stem-loop IIIC of the hepatitis C virus and GB virus B internal ribosome entry sites. J. Mol. Biol. 343:805-817. [DOI] [PubMed] [Google Scholar]
- 187.Rijnbrand, R., Y. Yang, L. Beales, F. Bodola, K. Goettge, L. Cohen, R. E. Lanford, S. M. Lemon, and A. Martin. 2005. A chimeric GB virus B with 5′ nontranslated RNA sequence from hepatitis C virus causes hepatitis in tamarins. Hepatology 41:986-994. [DOI] [PubMed] [Google Scholar]
- 188.Rodrigueza, W. V., S. T. Thuahnai, R. E. Temel, S. Lund-Katz, M. C. Phillips, and D. L. Williams. 1999. Mechanism of scavenger receptor class B type I-mediated selective uptake of cholesteryl esters from high density lipoprotein to adrenal cells. J. Biol. Chem. 274:20344-20350. [DOI] [PubMed] [Google Scholar]
- 189.Saunier, B., M. Triyatni, L. Ulianich, P. Maruvada, P. Yen, and L. D. Kohn. 2003. Role of the asialoglycoprotein receptor in binding and entry of hepatitis C virus structural proteins in cultured human hepatocytes. J. Virol. 77:546-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Scarselli, E., H. Ansuini, R. Cerino, R. M. Roccasecca, S. Acali, G. Filocamo, C. Traboni, A. Nicosia, R. Cortese, and A. Vitelli. 2002. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 21:5017-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Schaller, T., N. Appel, G. Koutsoudakis, S. Kallis, V. Lohmann, T. Pietschmann, and R. Bartenschlager. 2007. Analysis of hepatitis C virus superinfection exclusion by using novel fluorochrome gene-tagged viral genomes. J. Virol. 81:4591-4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Schofield, D. J., B. Bartosch, Y. K. Shimizu, T. Allander, H. J. Alter, S. U. Emerson, F. L. Cosset, and R. H. Purcell. 2005. Human monoclonal antibodies that react with the E2 glycoprotein of hepatitis C virus and possess neutralizing activity. Hepatology 42:1055-1062. [DOI] [PubMed] [Google Scholar]
- 193.Schuster, C., C. Isel, I. Imbert, C. Ehresmann, R. Marquet, and M. P. Kieny. 2002. Secondary structure of the 3′ terminus of hepatitis C virus minus-strand RNA. J. Virol. 76:8058-8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Serebrov, V., and A. M. Pyle. 2004. Periodic cycles of RNA unwinding and pausing by hepatitis C virus NS3 helicase. Nature 430:476-480. [DOI] [PubMed] [Google Scholar]
- 195.Silver, D. L., N. Wang, X. Xiao, and A. R. Tall. 2001. High density lipoprotein (HDL) particle uptake mediated by scavenger receptor class B type 1 results in selective sorting of HDL cholesterol from protein and polarized cholesterol secretion. J. Biol. Chem. 276:25287-25293. [DOI] [PubMed] [Google Scholar]
- 196.Simmonds, P. 2004. Genetic diversity and evolution of hepatitis C virus—15 years on. J. Gen. Virol. 85:3173-3188. [DOI] [PubMed] [Google Scholar]
- 197.Siridechadilok, B., C. S. Fraser, R. J. Hall, J. A. Doudna, and E. Nogales. 2005. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science 310:1513-1515. [DOI] [PubMed] [Google Scholar]
- 198.Smith, D. B., and P. Simmonds. 1997. Characteristics of nucleotide substitution in the hepatitis C virus genome: constraints on sequence change in coding regions at both ends of the genome. J. Mol. Evol. 45:238-246. [DOI] [PubMed] [Google Scholar]
- 199.Smith, R. M., C. M. Walton, C. H. Wu, and G. Y. Wu. 2002. Secondary structure and hybridization accessibility of hepatitis C virus 3′-terminal sequences. J. Virol. 76:9563-9574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Song, Y., P. Friebe, E. Tzima, C. Junemann, R. Bartenschlager, and M. Niepmann. 2006. The hepatitis C virus RNA 3′-untranslated region strongly enhances translation directed by the internal ribosome entry site. J. Virol. 80:11579-11588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Spahn, C. M., J. S. Kieft, R. A. Grassucci, P. A. Penczek, K. Zhou, J. A. Doudna, and J. Frank. 2001. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science 291:1959-1962. [DOI] [PubMed] [Google Scholar]
- 202.Stone, M., S. Jia, W. Do Heo, T. Meyer, and K. V. Konan. 2007. Participation of Rab5, an early endosome protein, in hepatitis C virus RNA replication machinery. J. Virol. 81:4551-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Su, A. I., J. P. Pezacki, L. Wodicka, A. D. Brideau, L. Supekova, R. Thimme, S. Wieland, J. Bukh, R. H. Purcell, P. G. Schultz, and F. V. Chisari. 2002. Genomic analysis of the host response to hepatitis C virus infection. Proc. Natl. Acad. Sci. USA 99:15669-15674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Sumpter, R., Jr., Y. M. Loo, E. Foy, K. Li, M. Yoneyama, T. Fujita, S. M. Lemon, and M. Gale, Jr. 2005. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J. Virol. 79:2689-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Tai, C.-L., W.-K. Chi, D.-S. Chen, and L.-H. Hwang. 1996. The helicase activity associated with hepatitis C virus nonstructural protein 3 (NS3). J. Virol. 70:8477-8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Tanaka, T., N. Kato, M.-J. Cho, and K. Shimotohno. 1995. A novel sequence found at the 3′ terminus of hepatitis C virus genome. Biochem. Biophys. Res. Commun. 215:744-749. [DOI] [PubMed] [Google Scholar]
- 207.Tanaka, T., N. Kato, M.-J. Cho, K. Sugiyama, and K. Shimotohno. 1996. Structure of the 3′ terminus of the hepatitis C virus genome. J. Virol. 70:3307-3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Tanji, Y., T. Kaneko, S. Satoh, and K. Shimotohno. 1995. Phosphorylation of hepatitis C virus-encoded nonstructural protein NS5A. J. Virol. 69:3980-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Tellinghuisen, T. L., J. Marcotrigiano, A. E. Gorbalenya, and C. M. Rice. 2004. The NS5A protein of hepatitis C virus is a zinc metalloprotein. J. Biol. Chem. 279:48576-48587. [DOI] [PubMed] [Google Scholar]
- 210.Tellinghuisen, T. L., J. Marcotrigiano, and C. M. Rice. 2005. Structure of the zinc-binding domain of an essential component of the hepatitis C virus replicase. Nature 435:374-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Thomssen, R., S. Bonk, C. Propfe, K. H. Heermann, H. G. Kochel, and A. Uy. 1992. Association of hepatitis C virus in human sera with beta-lipoprotein. Med. Microbiol. Immunol. 181:293-300. [DOI] [PubMed] [Google Scholar]
- 212.Thomssen, R., S. Bonk, and A. Thiele. 1993. Density heterogeneities of hepatitis C virus in human sera due to the binding of beta-lipoproteins and immunoglobulins. Med. Microbiol. Immunol. 182:329-334. [DOI] [PubMed] [Google Scholar]
- 213.Triyatni, M., B. Saunier, P. Maruvada, A. R. Davis, L. Ulianich, T. Heller, A. Patel, L. D. Kohn, and T. J. Liang. 2002. Interaction of hepatitis C virus-like particles and cells: a model system for studying viral binding and entry. J. Virol. 76:9335-9344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Troesch, M., E. Jalbert, S. Canobio, M. R. Boulassel, J. P. Routy, N. F. Bernard, J. Bruneau, N. Lapointe, M. Boucher, and H. Soudeyns. 2005. Characterization of humoral and cell-mediated immune responses directed against hepatitis C virus F protein in subjects co-infected with hepatitis C virus and HIV-1. AIDS 19:775-784. [DOI] [PubMed] [Google Scholar]
- 215.Tscherne, D. M., M. J. Evans, T. von Hahn, C. T. Jones, Z. Stamataki, J. A. McKeating, B. D. Lindenbach, and C. M. Rice. 2007. Superinfection exclusion in cells infected with hepatitis C virus. J. Virol. 81:3693-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Tscherne, D. M., C. T. Jones, M. J. Evans, B. D. Lindenbach, J. A. McKeating, and C. M. Rice. 2006. Time- and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J. Virol. 80:1734-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Tu, H., L. Gao, S. T. Shi, D. R. Taylor, T. Yang, A. K. Mircheff, Y. Wen, A. E. Gorbalenya, S. B. Hwang, and M. M. C. Lai. 1999. Hepatitis C virus RNA polymerase and NS5A complex with a SNARE-like protein. Virology 263:30-41. [DOI] [PubMed] [Google Scholar]
- 218.Tuplin, A., D. J. Evans, and P. Simmonds. 2004. Detailed mapping of RNA secondary structures in core and NS5B-encoding region sequences of hepatitis C virus by RNase cleavage and novel bioinformatic prediction methods. J. Gen. Virol. 85:3037-3047. [DOI] [PubMed] [Google Scholar]
- 219.Tuplin, A., J. Wood, D. J. Evans, A. H. Patel, and P. Simmonds. 2002. Thermodynamic and phylogenetic prediction of RNA secondary structures in the coding region of hepatitis C virus. RNA 8:824-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Varaklioti, A., N. Vassilaki, U. Georgopoulou, and P. Mavromara. 2002. Alternate translation occurs within the core coding region of the hepatitis C viral genome. J. Biol. Chem. 277:17713-17721. [DOI] [PubMed] [Google Scholar]
- 221.Vassilaki, N., and P. Mavromara. 2003. Two alternative translation mechanisms are responsible for the expression of the HCV ARFP/F/core+1 coding open reading frame. J. Biol. Chem. 278:40503-40513. [DOI] [PubMed] [Google Scholar]
- 222.Voisset, C., N. Callens, E. Blanchard, A. Op De Beeck, J. Dubuisson, and N. Vu-Dac. 2005. High density lipoproteins facilitate hepatitis C virus entry through the scavenger receptor class B type I. J. Biol. Chem. 280:7793-7799. [DOI] [PubMed] [Google Scholar]
- 223.Voisset, C., and J. Dubuisson. 2004. Functional hepatitis C virus envelope glycoproteins. Biol. Cell 96:413-420. [DOI] [PubMed] [Google Scholar]
- 224.von Hahn, T., B. D. Lindenbach, A. Boullier, O. Quehenberger, M. Paulson, C. M. Rice, and J. A. McKeating. 2006. Oxidized low-density lipoprotein inhibits hepatitis C virus cell entry in human hepatoma cells. Hepatology 43:932-942. [DOI] [PubMed] [Google Scholar]
- 225.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Walewski, J. L., J. A. Gutierrez, W. Branch-Elliman, D. D. Stump, T. R. Keller, A. Rodriguez, G. Benson, and A. D. Branch. 2002. Mutation master: profiles of substitutions in hepatitis C virus RNA of the core, alternate reading frame, and NS2 coding regions. RNA 8:557-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Walewski, J. L., T. R. Keller, D. D. Stump, and A. D. Branch. 2001. Evidence for a new hepatitis C virus antigen encoded in an overlapping reading frame. RNA 7:710-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Wang, C., M. Gale, Jr., B. C. Keller, H. Huang, M. S. Brown, J. L. Goldstein, and J. Ye. 2005. Identification of FBL2 as a geranylgeranylated cellular protein required for hepatitis C virus RNA replication. Mol. Cell 18:425-434. [DOI] [PubMed] [Google Scholar]
- 229.Wang, C., S. Y. Le, N. Ali, and A. Siddiqui. 1995. An RNA pseudoknot is an essential structural element of the internal ribosome entry site located within the hepatitis C virus 5′ noncoding region. RNA 1:526-537. [PMC free article] [PubMed] [Google Scholar]
- 230.Wang, C., and A. Siddiqui. 1995. Structure and function of the hepatitis C virus internal ribosome entry site. Curr. Top. Microbiol. Immunol. 203:99-115. [DOI] [PubMed] [Google Scholar]
- 231.Wang, Q. M., M. A. Hockman, K. Staschke, R. B. Johnson, K. A. Case, J. Lu, S. Parsons, F. Zhang, R. Rathnachalam, K. Kirkegaard, and J. M. Colacino. 2002. Oligomerization and cooperative RNA synthesis activity of hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 76:3865-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Wasley, A., and M. J. Alter. 2000. Epidemiology of hepatitis C: geographic differences and temporal trends. Semin. Liver Dis. 20:1-16. [DOI] [PubMed] [Google Scholar]
- 233.Watashi, K., M. Hijikata, M. Hosaka, M. Yamaji, and K. Shimotohno. 2003. Cyclosporin A suppresses replication of hepatitis C virus genome in cultured hepatocytes. Hepatology 38:1282-1288. [DOI] [PubMed] [Google Scholar]
- 234.Watashi, K., N. Ishii, M. Hijikata, D. Inoue, T. Murata, Y. Miyanari, and K. Shimotohno. 2005. Cyclophilin B is a functional regulator of hepatitis C virus RNA polymerase. Mol. Cell 19:111-122. [DOI] [PubMed] [Google Scholar]
- 235.Wellnitz, S., B. Klumpp, H. Barth, S. Ito, E. Depla, J. Dubuisson, H. E. Blum, and T. F. Baumert. 2002. Binding of hepatitis C virus-like particles derived from infectious clone H77C to defined human cell lines. J. Virol. 76:1181-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Wölk, B., D. Sansonno, H. G. Kräusslich, F. Dammacco, C. M. Rice, H. E. Blum, and D. Moradpour. 2000. Subcellular localization, stability, and trans-cleavage competence of the hepatitis C virus NS3-NS4A complex expressed in tetracycline-regulated cell lines. J. Virol. 74:2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Wunschmann, S., J. D. Medh, D. Klinzmann, W. N. Schmidt, and J. T. Stapleton. 2000. Characterization of hepatitis C virus (HCV) and HCV E2 interactions with CD81 and the low-density lipoprotein receptor. J. Virol. 74:10055-10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Xu, Z., J. Choi, W. Lu, and J. H. Ou. 2003. Hepatitis C virus f protein is a short-lived protein associated with the endoplasmic reticulum. J. Virol. 77:1578-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Xu, Z., J. Choi, T. S. Yen, W. Lu, A. Strohecker, S. Govindarajan, D. Chien, M. J. Selby, and J. Ou. 2001. Synthesis of a novel hepatitis C virus protein by ribosomal frameshift. EMBO J. 20:3840-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Yagnik, A. T., A. Lahm, A. Meola, R. M. Roccasecca, B. B. Ercole, A. Nicosia, and A. Tramontano. 2000. A model for the hepatitis C virus envelope glycoprotein E2. Proteins 40:355-366. [DOI] [PubMed] [Google Scholar]
- 241.Yan, Y., Y. Li, S. Munshi, V. Sardana, J. L. Cole, M. Sardana, C. Steinkuehler, L. Tomei, R. De Francesco, L. C. Kuo, and Z. Chen. 1998. Complex of NS3 protease and NS4A peptide of BK strain hepatitis C virus: a 2.2 Å resolution structure in a hexagonal crystal form. Protein Sci. 7:837-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Yanagi, M., M. St. Claire, S. U. Emerson, R. H. Purcell, and J. Bukh. 1999. In vivo analysis of the 3′ untranslated region of the hepatitis C virus after in vitro mutagenesis of an infectious cDNA clone. Proc. Natl. Acad. Sci. USA 96:2291-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Yao, N., T. Hesson, M. Cable, Z. Hong, A. D. Kwong, H. V. Le, and P. C. Weber. 1997. Structure of the hepatitis C virus RNA helicase domain. Nat. Struct. Biol 4:463-467. [DOI] [PubMed] [Google Scholar]
- 244.Yao, N., P. Reichert, S. S. Taremi, W. W. Prosise, and P. C. Weber. 1999. Molecular views of viral polyprotein processing revealed by the crystal structure of the hepatitis C virus bifunctional protease-helicase. Structure 7:1353-1363. [DOI] [PubMed] [Google Scholar]
- 245.Ye, J., C. Wang, R. Sumpter, Jr., M. S. Brown, J. L. Goldstein, and M. Gale, Jr. 2003. Disruption of hepatitis C virus RNA replication through inhibition of host protein geranylgeranylation. Proc. Natl. Acad. Sci. USA 100:15865-15870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Yi, M., and S. M. Lemon. 2003. 3′ nontranslated RNA signals required for replication of hepatitis C virus RNA. J. Virol. 77:3557-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Yi, M., and S. M. Lemon. 2004. Adaptive mutations producing efficient replication of genotype 1a hepatitis C virus RNA in normal Huh7 cells. J. Virol. 78:7904-7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Yi, M., and S. M. Lemon. 2003. Structure-function analysis of the 3′ stem-loop of hepatitis C virus genomic RNA and its role in viral RNA replication. RNA 9:331-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Yi, M., R. A. Villanueva, D. L. Thomas, T. Wakita, and S. M. Lemon. 2006. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc. Natl. Acad. Sci. USA 103:2310-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.You, S., D. D. Stump, A. D. Branch, and C. M. Rice. 2004. A cis-acting replication element in the sequence encoding the NS5B RNA-dependent RNA polymerase is required for hepatitis C virus RNA replication. J. Virol. 78:1352-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Yu, G. Y., K. J. Lee, L. Gao, and M. M. Lai. 2006. Palmitoylation and polymerization of hepatitis C virus NS4B protein. J. Virol. 80:6013-6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Zhang, J., G. Randall, A. Higginbottom, P. Monk, C. M. Rice, and J. A. McKeating. 2004. CD81 is required for hepatitis C virus glycoprotein-mediated viral infection. J. Virol. 78:1448-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Zheng, Y., B. Gao, L. Ye, L. Kong, W. Jing, X. Yang, Z. Wu, and L. Ye. 2005. Hepatitis C virus non-structural protein NS4B can modulate an unfolded protein response. J. Microbiol. 43:529-536. [PubMed] [Google Scholar]
- 254.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 102:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Zhong, J., P. Gastaminza, J. Chung, Z. Stamataki, M. Isogawa, G. Cheng, J. A. McKeating, and F. V. Chisari. 2006. Persistent hepatitis C virus infection in vitro: coevolution of virus and host. J. Virol. 80:11082-11093. [DOI] [PMC free article] [PubMed] [Google Scholar]