Abstract
Cervical carcinoma is associated with certain types of human papillomaviruses expressing the E6 and E7 oncogenes, which are involved in carcinogenesis through their interactions with the p53 and pRB pathways, respectively. A critical event on the path to malignant transformation is often manifested by the loss of expression of the viral E2 transcription factor due to the integration into the host genome of the viral DNA. Using microarrays, we have previously shown that reintroduction of a functional E2 in the HeLa cervical carcinoma cell line activates a cluster of p53 target genes while at the same time severely repressing a group of E2F target genes. In the present study, using new high-density microarrays containing more than 22,000 human cDNA sequences, we identified a novel p63 pathway among E2-activated genes and 38 new mitotic genes repressed by E2. We then sought to determine the pathways through which these genes were modulated and used an approach that relies on small interfering RNA to demonstrate that the p63 target genes were activated through silencing of the E6/E6AP pathway while the mitotic genes were mainly repressed through E7 silencing. Importantly, a subset of the mitotic genes was shown to be significantly induced in biopsies of stage IV cervical cancers, which points to a prominent E7 pathway in cervical carcinoma.
High-risk human papillomaviruses (HPV), such as HPV18, are associated with more than 99% of cervical carcinomas (44). Integration of the HPV18 DNA into the host genome plays a crucial role in carcinogenic progression. This integration usually results in the specific disruption of the E1 and/or E2 viral genes and is a common feature of HPV-associated carcinomas. The E2 protein negatively regulates transcription of the viral E6 and E7 oncogenes through its specific binding to DNA recognition sites located within the promoter sequences (42, 43). Therefore, loss of E2 expression results in overexpression of the viral oncogenes in cervical cancer cells. Conversely, reintroduction of the E2 protein in HPV-associated cervical carcinoma cell lines has been shown to down-regulate E6/E7 transcription and suppress cellular growth, due to cell cycle arrest in G1, senescence, and apoptosis (6, 8-10, 15, 16).
The E6 and E7 oncoproteins act mainly through protein-protein interactions to alter major pathways regulating cell cycle progression and cell proliferation. Through its binding to the E6AP ubiquitin ligase, E6 targets p53 for degradation by the proteasome, which consequently abrogates the p53 transcriptional pathway (36, 37). E6 interaction with E6AP also activates telomerase by inducing human telomerase reverse transcriptase transcription through proteasomal degradation of its transcriptional repressor (14). On the other hand, the E7 protein binds to and inactivates the retinoblastoma (Rb) pocket proteins leading to release of active E2F and activation of E2F target genes (11). E2F target genes have long been known as S-phase genes activated at the G1/S transition necessary for cell proliferation. However, a series of microarrays and chromatin immunoprecipitation (ChIP) analyses have pointed to a new role of E2F in mitosis by activating a cluster of genes of the G2/M transition (18, 25, 26, 30-32). Alterations of the p53 and pRb pathways by the E6 and E7 proteins account for the transforming capacity of high-risk papillomaviruses although the precise mechanisms of action remain elusive. To study the transcriptional impact of E6 and E7 expression in cervical carcinoma, the E2 protein was expressed in an HPV18-associated HeLa cervical carcinoma cell line, and the cellular transcriptome was studied by high-density microarrays containing 13,000 human cDNA sequences. Two series of genes have been found to be regulated. The first one includes p53 target genes, presumably modulated through E6 transcriptional repression, while the second series contains E2F target genes including a large cluster of mitotic genes (41). We assumed that the mitotic genes repressed by E2 in HeLa cells were modulated through repression of E7 transcription, consequently inducing repression of the cellular E2F target genes. However, recent data indicated that transcriptional regulation of mitotic genes might involve other transcription factors such as NF-YB, B-Myb or FoxM1 (4, 22, 24, 38). In addition, it was recently shown that p53 as well as p63 could repress transcription of mitotic genes through direct interaction with the essential NF-Y transcription factor, thus implicating E6 as a potential modulator of the mitotic genes in cervical carcinoma cells (17, 40).
The p63 gene has recently been shown to play a major role in epidermal differentiation, as revealed by the dramatic phenotype of p63-deficient mice which display gross developmental abnormalities including the complete lack of stratified epithelia (27, 46). In addition, heterozygous mutations of the p63 gene are causative for several developmental syndromes of ectodermal dysplasia and orofacial and limb malformations in human (reviewed in reference 33). These results indicated that, despite structural homology between p53 and p63, the functions of the p53 and p63 genes appear to differ substantially. The analysis of p63 function is complicated by the nature of the p63 gene itself, which encodes at least six different isoforms differently expressed in tissues. Recently, global analysis involving microarrays and ChIP-on-chip (ChIP-on-microarray chip) experiments have pointed out a very large number of cellular genes modulated by p63. Although there is an overlap between target genes modulated by p53 and p63, a clear specificity in p63 transcriptional regulation of epithelial cellular adhesion and survival could be found (5). As for the role of p63 in cervical carcinoma, it is not well understood yet, and we therefore decided to investigate whether p63-specific target genes were modulated in HeLa cells expressing E2.
Transcriptional modulation of the cellular genes in HeLa cells, after reintroduction of the E2 repressor gene, was analyzed using new high-density microarrays containing 22,000 human cDNA sequences. We identified 55 new E2-activated genes, among which is a new cluster of p63 target genes involved in cell adhesion. In addition, 77 genes were repressed by E2, including 38 new mitotic genes. A subset of the mitotic genes was significantly activated in cervical biopsies compared to adjacent normal tissue, which contrasts with the lack of consistently modulated p53 target genes. Methods using small interfering RNA (siRNA) established that the p53 and p63 target genes were modulated through the E6/E6AP pathway while mitotic genes were mainly controlled by the E7 pathway.
MATERIALS AND METHODS
Infection of HeLa cells with recombinant adenoviruses.
Recombinant adenoviruses expressing green fluorescent protein (GFP)-E2 and GFP were previously described (7). Cells were infected with purified adenoviruses at a multiplicity of infection of 200 PFU/cell, as described previously (41).
Western blotting.
Infected or transfected cells were collected 24 h postinfection or posttransfection and were denatured in Laemmli denaturing sample buffer, boiled, and separated on 4 to 12% sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis gels. After transfer, the membranes were incubated with the p53 antibody (DO-1) and then with the GFP antibody (TP401; Torrey Pines Biolabs) or beta-actin (A2066; Sigma), as indicated in Fig. 1B, and either mouse or rabbit secondary antibodies coupled to peroxidase. Membranes were revealed by an Amersham ECL plus kit.
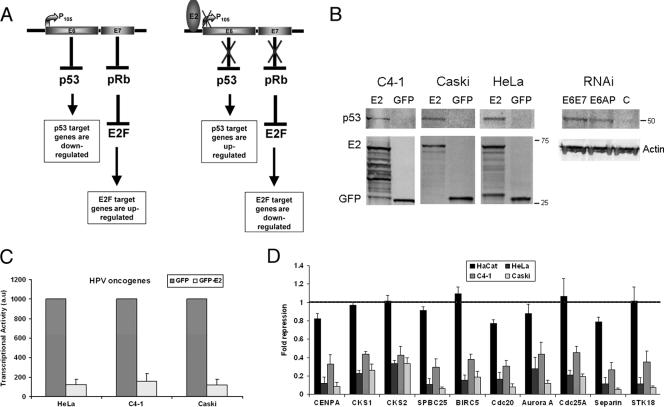
FIG. 1.
P53 stabilization and RT-PCR analysis of a selected group of mitotic genes in cervical carcinoma cell lines and HaCaT cells. (A) Schematic representation of the modulation of cellular genes through the p53 and pRb pathways in cervical carcinoma in absence or presence of the E2 transcriptional repressor. (B) Western blot analyses of the stabilization of p53 in the three cervical carcinoma cell lines expressing E2 as well as in HeLa cells transfected by the E6/E7 and the E6AP siRNA, as indicated. Expression of GFP and GFP-E2 as well as of the beta-actin is shown. (C) RT-PCR of the E6/E7 oncogenes performed in cervical carcinoma cell lines infected by adeno-GFP and adeno-GFP-E2 in three independent experiments. (D) RT-PCR of a series of cellular genes as indicated, in four keratinocyte cell lines: HaCaT not associated with HPV, HeLa and C4-1 associated with HPV18, and Caski associated with HPV16. Values given (au, arbitrary units) are levels of gene expression in adeno-GFP-E2-infected cells compared with adeno-GFP-infected cells in three independent infection experiments.
Construction of microarrays, hybridization, and analysis.
The high-density microarray slides used in this study consist of 20,988 cDNA fragments (expressed sequence tags) and 384 controls (calibration and negative control spots). The clone set was derived from Incyte Genomics (Palo Alto, CA) and the Resource Center and Primary Database (Berlin, Germany) collections, together with 143 additional clones from the MicroArray Facility at the Vlaams Interuniversitair Instituut voor Biotechnologie, Belgium (www.microarray.be). Slides were similarly coated as described previously (29, 41), though clones were arrayed with a different printer (Lucidea Array Spotter; Amersham Biosciences, Buckinghamshire, United Kingdom). The slide blocking process using 3.5% SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.2% SDS, and 1% bovine serum albumin for 10 min at 60°C was replaced with 2× SSPE (1× SSPE is 0.18 M NaCl, 10mM NaH2PO4, and 1 mM EDTA [ph 7.7]) and 0.2% SDS for 30 min at 25°C. A complete description of the array content and the printing procedures is available at ArrayExpress (www.ebi.ac.uk/arrayexpress) under accession number A-MEXP-146. Five micrograms of total RNA of each experiment was amplified and used for hybridization on the microarray. All protocols are available at ArrayExpress under accession numbers P-MEXP-578, P-MEXP-579, P-MEXP-581, and P-MEXP-582 for Cy3 labeling, Cy5 labeling, hybridization, and scanning, respectively.
RNA isolation and quantitative RT-PCR.
Total RNA was extracted with Trizol (Invitrogen) according to the manufacturer's recommendations. A total of 2.5 μg of RNA was reverse transcribed into cDNA and Superscript II (Invitrogen) as recommended by the manufacturer. Of the resulting synthesized single-stranded cDNA, 1/100 was used for each PCR in the presence of a 0.1 μM concentration of specific primers and Syber Green PCR master mix (Applied Biosystems). Quantitative PCR was performed on an MX 3005 P sequence detection system (Stratagene), with cycling conditions of 2 min at 50°C, 10 min at 95°C, and 40 cycles of 15 s at 95°C and 1 min at 60°C. After the last cycle, the temperature was progressively raised in order to provide dissociation curves allowing an assessment of the purity of the amplified product. Each cDNA was normalized with histone deacetylase 1 (HDAC1), GADPH (glyceraldehyde-3-phosphate dehydrogenase (), or 18S ribosomal RNAs. Each PCR was performed in duplicate and fit to a standard curve, providing a mean cycle threshold value, which was translated into an arbitrary DNA concentration. Primers used for the reverse transcription-PCR (RT-PCR) are available upon request.
Construction of E6/E7 pSUPER siRNA and cell transfections.
To induce the silencing of HPV18 E6/E7 mRNAs, pSUPER vector was constructed with a pair of 64-nucleotide (nt) oligonucleotides, each containing a unique 19-nt sequence derived from HPV18 E6/E7 (nt 609 to 627). The 19-nt target appears in both sense and antisense orientation, separated by a 9-nt spacer sequence (5′-GATGGAGTTAATCATCAACttcaagagaGTTGATGATTAACTCCATC-3′; spacer sequence is in lowercase letters). To generate control vector pSUPER-HPV16 E6 siRNA, the double-stranded synthetic oligonucleotide 5′-GAGCTGCAAACAACTATACttcaagagaGTATAGTTGTTTGCAGCTC-3′ containing 19 nt derived from the HPV16 E6 gene (nt 73 to 90 of the mRNA), separated by a 9-nt linker (in lowercase letters) from its reverse complement, was cloned into pSUPER.
HeLa cells were cotransfected with pCDNA-GFP to estimate the efficiency of transfection and with the purified pSUPER control siRNA, pSUPER HPV18 E6/E7 siRNA, or E2 expression plasmid. Total RNA was extracted 40 h after transfection and reverse transcribed into cDNA to perform quantitative PCR.
Alternatively, HeLa cells were transfected with 100 pM of the E6/E7 siRNA (GAUGGAGUUAAUCAUCAACdTdT, where dT is deoxyribosylthymine) or the E6AP siRNA (CAACUCCUGCUCUGAGAUAdTdT) (21) or controlled HPV16 E6/E7 siRNA (GAGCUGCAAACAACUAUACdTdT) from Proligo Sigma, using DharmaFECT1 (Dharmacon); cells were then processed as described for RNA extraction and quantitative PCR.
RESULTS
E2 expression in HeLa cells activates a p63 pathway.
We compared the genes found modulated in the high-density microarrays containing 22,000 human cDNAs used here with the genes that were found modulated in our previous study using microarrays containing 13,000 human cDNAs (41). The 10 known p53 target genes previously found activated by E2 through repression of E6 transcription as schematized in Fig. 1A were all present in the new arrays and were also found activated. They included the p21/CDKN1A, GDF15, Sestrin, CYFIP2, RRM2B, GADD45A, DDB2, MVP, GSN, and TNFRSF10B genes. Three additional p53 target genes were present in the new arrays: Bax, Serpin, and FXYD2. We used stringent criteria to select the cellular genes modulated only in conditions where the E6/E7 transcription is repressed. Cells infected by recombinant adenoviruses expressing the full-length and amino-terminally deleted E2 (transcriptional repression) were compared to cells infected by the recombinant adenoviruses expressing GFP and the TAD domain alone (no transcriptional repression), as previously described (41). Using this protocol, we could establish a list of 55 new cellular genes, not including the p53 target genes, activated by E2 more than twofold in HeLa cells (data not shown).
In light of the recent genomic data deciphering the p63 target genes in epithelial cells (1, 5), we examined whether the E2-activated genes contain p63 target genes. We found 13 of these newly described p63 target genes belonging to the cell adhesion cluster among the 55 positively modulated genes in our arrays (Table 1). Several genes activated in our microarrays could be additional candidates for the p63 pathway as they take part in the cell adhesion program, such as the epidermal growth factor EPS8, the fibroblast activation protein FAP, a fibronectin gene (FLRT2), and the Plexin gene (PLXNB1) (data not shown). In addition, two genes were found modulated in the first microarrays, which are related to the p63 pathway, the laminin LamB2 gene and the collagen Col8 gene. Activation by E2 of several of these genes was confirmed by RT-PCR (Table 1). In all, our data indicated that the p63 pathway is modulated as well as the p53 pathway in cervical carcinoma cell lines.
TABLE 1.
Modulation of p63 target genesa
| Name | Description | Accession no. | Relative activation (n-fold)
|
|
|---|---|---|---|---|
| Microarray | RT-PCR | |||
| COL1A1 | Collagen, type I, alpha 1 | AW577407 | 2.2 | ND |
| COL4A6 | Collagen type IV, alpha 6 | D21337 | 2.5 | ND |
| COL5A1 | Collagen, type V, alpha 1 | BC008760 | 2.2 | 3 |
| COL7A1 | Collagen, type VII, alpha 1 | W68588 | 2.8 | ND |
| COL8A1 | Collagen, type VIII, alpha 1 | NM_001850 | 3 | ND |
| COL9A3 | Collagen, type IX, alpha 3 | BE731782 | 2.3 | ND |
| COL14A1 | Collagen, type XIV, alpha 1 | AA011417 | 2.6 | ND |
| LAMA1 | Laminin alpha 1 | N62886 | 2.2 | ND |
| LAMB2 | Laminin B2 | NM_002292 | 3 | 3.2 |
| FAT | FAT tumor suppressor homolog 1 | W57946 | 2.4 | 2.4 |
| FN1 (×2)b | Fibronectin | AW385690 | 2.5 | 1.8 |
| FBN1 (×2)b | Fibrillin 1 | NM_000138 | 2 | 2.3 |
| FBN2 | Fibrillin 2 | AA452111 | 2 | ND |
| SERPINE1 | Serine proteinase inhibitor E1 | BE812315 | 2 | ND |
| SERPINI1 | Serine proteinase inhibitor I1 | NM_005025 | 2.8 | ND |
According to reference 5. The activation of p63 target genes in E2-expressing cells compared to GFP-expressing cells is given as well as the values of RT-PCR experiments done in cells expressing or not expressing E2. Genes in boldface were found modulated in previous microarray experiments (41).
Genes present twice in the microarray.
E2 expression in HeLa cells represses mitotic genes.
The overlap among genes repressed by E2 that are expected to be modulated through repression of E7 (Fig. 1A), based on previous microarrays and the present one, was extremely good. A total of 190 genes were found repressed by E2 more than twofold (a threshold of 0.5 was used between E2- and GFP-expressing cells) in the present experiment, among which 114 genes were repressed in the previous microarrays. Conversely, the vast majority of the repressed genes of the first microarrays were also repressed in the second experiments (90.6% overlap). An important group of 77 new genes belonging to cell cycle, DNA replication and DNA repair, proliferation, transformation, and motility were found modulated in the new microarrays, which contain 32 known E2F target genes (Table 2). Altogether, results of the previous and new experiments indicate a strong mitotic transcriptional signature in cervical carcinoma cells.
TABLE 2.
Genes of cell cycle, cell proliferation, and motility repressed by E2 in the microarraysa
| E2 target | Name or description | UniGene Hs cluster | Function | Repression (n-fold) |
|---|---|---|---|---|
| Mitosis | ||||
| ANLN | Anillin | 62180 | Actin binding protein involved in cytokinesis | 0.22 |
| BIRC5 | Survivin | 514527 | Inhibitor of apoptosis; senses kinetochore tension | 0.2 |
| BUB1 | Homolog of mitotic checkpoint Saccharomyces erevisiae BUB1 | 469649 | Spindle assembly checkpoint; linked to genetic instability | 0.34 |
| CCNB1 | Cyclin B1 | 23960 | Complexes with p34 cdc2 to form the mitosis promoting factor | 0.31 |
| Cdc25C | Cell cycle division | 656 | M phase inducer phosphatase; activates cdc2 by dephosphorylation | 0.36 |
| CDCA1 | Cell division cycle associated 1 | 234545 | Homolog of yeast Nuf2; component of the NDC80 kinetochore | 0.27 |
| CDCA8 | Cell division cycle associated 8 | 524571 | Component of the chromosomal passenger complex | 0.34 |
| CDKN2C | Cyclin-dependent kinase Inhibitor | 525324 | P18/INK4 inhibits cyclin/cyclin-dependent kinase complexes | 0.46 |
| CENPA | Centromeric protein | 1594 | Component of modified nucleosome; interacts with Aurora A | 0.26 |
| CENPF | Centromeric protein | 497741 | Kinetochore | 0.23 |
| CHK1 | Checkpoint homolog | 24529 | Inhibits cdc25C; prevents activation of cyclinB/cdc2; DNA damage checkpoint | 0.44 |
| CKS1 | Cdc28 protein kinase 1B | 374378 | Activates cdc20 transcription; substrate targeting subunit of the SCF ubiquitin ligase | 0.45 |
| CKS2 | Cdc28 protein kinase | 83758 | Cdc2-associated protein (meiosis) | 0.41 |
| DEPDC1B | DEP domain-containing protein 1B | 482233 | DEP-containing protein | 0.25 |
| DIAPH3 | Diaphanous protein homolog 3 | 283127 | Binds to Rho and to profilin; involved in cytokinesis | 0.36 |
| ECT-2 | Epithelial cell transforming 2 | 518299 | Oncogen; binds to Rhoa protein to activate mitosis (ortholog of Drosophila Pebble) | 0.33 |
| FBXO Emi1 | F box only | 520506 | Interacts with cdc20 and inhibits the APC mitotic ubiquitin ligase | 0.37 |
| HisH4F | Histone H4F | 247816 | Forms centromers with CENPA | 0.2 |
| HURP (DLG7) | Hepatoma up-regulated | 77695 | Cell cycle regulator; up-regulated in G2/M | 0.22 |
| KIF11 | Kinesin family member 11 | 8878 | Spindle assembly | 0.25 |
| KIF22 | Kinesin member 22 | 119324 | Metaphase chromosome alignment | 0.42 |
| KIF4A | Kinesin family member 4 | 279766 | Motor protein that translocates PRC1 | 0.33 |
| LBR | Lamin B receptor | 435166 | Role in chromosome assembly | 0.39 |
| MPHOSDH1 | M phase phosphoprotein | 240 | Kinesin-related protein; M-phase promoting factor | 0.25 |
| NUP188 | Nucleoprotein | 308340 | 0.34 | |
| NUP107 | Nucleoporin | 524574 | Nucleo-cytoplasmic trafficking; part of the nuclear pore and associated to the kinetochores | 0.44 |
| NUSAP1 | Nucleolar and spindle associated protein 1 | 511093 | Involved in mitotic spindle organization | 0.29 |
| PRC1 | Protein regulating cytokinesis | 567385 | Associated with mitotic spindles; associates with KIF4; organizes spindle midzone formation | 0.23 |
| RACGAP1 | Rac GTPase activated protein | 505469 | Master regulator of initiation of cytokinesis | 0.32 |
| SGOL1 | Shugoshin-like 1 | 105153 | Binds and stabilizes microtubules; localized to kinetochores | 0.35 |
| SPBC25 | Kinetochore | 421956 | Component of the NDC80 kinetochore complex | 0.26 |
| SPC24 | Spindle pole body 24 | 381225 | Component of the NDC80 kinetochore complex | 0.33 |
| STK6 | Ser/Thr kinase 6 | 250822 | Aurora A is targeted to the spindle apparatus by TPX2 and regulates its activity | 0.34 |
| STMN1 | Stathmin | 209983 | Sequesters tubulin in a ternary complex; role in depolymerization of mitotic microtubules | 0.28 |
| TTK | TTK protein kinase | 169840 | MPS1L1, PYT, and ESK; required for normal centrosome duplication | 0.44 |
| TMPO | Thymopoietin lamina-associated polypeptide | 11355 | Structural organization of the nucleus; postmitotic nuclear assembly | 0.35 |
| TOPK | Mitotic protein kinase | 104741 | Mitotic Ser/Thr kinase related to mitogen-activated protein kinase kinase | 0.27 |
| TPX2 | Targeting protein for Xklp2 | 244580 | Proliferating associated protein P100; associated with spindle pole and spindle | 0.30 |
| Replication and DNA repair | ||||
| BARD1 | BRCA1-associated ring domain 1 | 54089 | Implicated in BRCA1 tumor suppression and DNA repair | 0.36 |
| CCNE2 | Cyclin E | 567387 | G1/S transition | 0.1 |
| DCC1 | RFC alternative complex | 315167 | Primed loading of PCNA on gapped DNA; defective sister chromatin cohesion | 0.34 |
| DTYMK | Thymidylate kinase | 471873 | DNA synthesis pathway catalyzes dMTP to dTDP | 0.45 |
| HNRPA1 | Heterogenous nuclear ribonucleopreotein | 546261 | Interacts with Fen1 during Okazaki fragment maturation | 0.46 |
| MCM4 | Minichromosome maintenance | 460184 | Replication licensing complex, loaded at the origins before initiation and essential for elongation | 0.47 |
| MELK | Maternal embryonic leucine zipper kinase | 184339 | Implicated in stem cell renewal and cell cycle progression | 0.31 |
| NBS1 | Nibrin | 492208 | Part of the DNA repair complex MRE11/RAD50 | 0.42 |
| Pfs2 | GINS2 | 433180 | DNA replication complex GINS | 0.29 |
| PRIM2A | Primase polypeptide A | 485640 | Polymerase that synthesizes small RNA primers (Okazaki) | 0.45 |
| RFC3 | Replication factor C | 115474 | DNA clamp loading complex; necessary for elongation of primed DNA | 0.33 |
| TREX2 | 3′ Repair exonuclease 2 | 170835 | DNA replication and DNA repair | 0.47 |
| UNG | Uracyl-DNA glycosylase | 191334 | DNA replication and DNA repair | 0.3 |
| USP1 | Ubiquitin specific protease | 35086 | Involved in DNA repair; Fanconi anemia pathway | 0.4 |
| Cell proliferation | ||||
| DHFR | Dihydrofolate reductase | 83765 | Required for synthesis of purines, thymidylic acid, and certain amino acids | 0.32 |
| DEK | DEK oncogene | 110713 | Acute myeloma; oncogene | 0.46 |
| DLEU2 | Deleted in lymphocytic leukemia | 508041 | Candidate tumor suppressor gene | 0.4 |
| HMMR | Hyaluronan mediated motility receptor | 72550 | Involved in cell motility; may also be involved in cellular transformation and metastasis | 0.22 |
| IGSF1 | Immunoglobin superfamily | 22111 | Role in cell adhesion | 0.4 |
| ITGB3BP | Human beta 3 endonexin | 1741 | Coactivator of hormone receptors | 0.44 |
| MLF1 | Myeloid leukemia factor 1 | 85195 | Oncoprotein; negative regulator of cell cycle | 0.47 |
| NET1 | Neuroepithelila cell transforming gene | 25155 | Rho exchange factor | 0.44 |
| RHOG | RAS homolog gene family G | 501728 | GTP-binding protein; activates RAC | 0.48 |
| SHCBP1 | Shc SH2 domain protein | 123253 | Linking activated cell surface receptors to the Ras pathway | 0.32 |
| SHMT1 | Serine hydroxy methyltransferase 1 | 513987 | Plays an essential role in nucleic acids biosynthesis | 0.24 |
| SHMT2 | Serine hydroxy methyltransferase 2 | 75069 | Cytosolic; mitochondrial | 0.45 |
| SRC | v-src sarcoma | 195659 | Protoncogene | 0.24 |
| SYTL2 | Synaptotagmin-like protein | 369520 | Binds to Rab27; involved in vesicle trafficking | 0.42 |
| THBS2 | Thrombospondin | 371147 | Inhibitor of tumor growth and angiogenesis | 0.32 |
| Miscellaneous | ||||
| ASF1B | Anti-silencing function | 26516 | Assembles nucleosome during nucleotide excision repair | 0.33 |
| BCAT1 | Branched-chain amino transferase | 438993 | Direct target of c-Myc regulation | 0.42 |
| CPSF1 | Cleavage polyadenylation specific factor | 493202 | Addition of poly(A) tail to mRNA | 0.43 |
| DDX39 | DEAD box protein 39 | 311609 | ATP-dependent RNA helicase | 0.44 |
| JUNB | JunB proto-oncogene | 25292 | Member of the AP1 transcription factor | 0.41 |
| MNS1 | Meiosis-specific nuclear structural protein | 444483 | Meiosis | 0.42 |
| NF-YB | Nuclear transcription factor Y | 84928 | Binding to the CCAAT motif | 0.41 |
| RBL1 | Retinoblastoma-like; P107 | 207745 | Partner of E2F4/5 | 0.47 |
| SMYD3 | Set and Mynd domain containing 3 | 567571 | Histone methyl transferase | 0.46 |
| SNRPB | Splicing factor | 83753 | Knockout generates mitotic spindle defect; part of snRNP | 0.44 |
Known E2F target genes are in bold. Numbers given in the last column are levels of transcription in E2-expressing cells compared to GFP-expressing cells; underlining indicates that the gene is present more than once in the array.
E2 stabilizes p53 and represses mitotic genes in other HPV-associated cervical carcinoma cell lines.
In order to check whether E2 modulates the E6/E7 pathway in other cervical carcinoma cell lines, we infected HeLa, C4-1, and Caski as well as HaCaT cells as a control, by recombinant adenoviruses expressing either GFP-E2 or GFP alone. The HaCaT cells are human keratinocytes spontaneously transformed and containing no associated HPV, while HeLa and C4-1 are cervical carcinoma cells transformed by HPV18, and Caski cells are transformed by HPV16. We first showed stabilization of the p53 protein in the three cervical carcinoma cell lines expressing E2, as expected from transcriptional repression of E6 (Fig. 1B). We then performed real-time PCR on the viral oncogenes showing strong repression (more than fivefold) of their transcription by E2 in the three cervical carcinoma cell lines (Fig. 1C). A subset of the cellular genes found repressed in the microarrays was then analyzed by RT-PCR, containing known E2F targets such as the CENPA, CKS1, CKS2, and BIRC5 (Survivin) genes, and unknown targets such as the SPBC25 gene (Table 2), as well as four mitotic genes [Cdc20, Aurora A, Separin, and STK18 (Polo) genes] and one G1/S gene (CDC25A), also found modulated in the first arrays (41). These genes were repressed to similar extents (three- to fivefold) by E2 in three cervical carcinoma cell lines (HeLa, C4-1, and Caski) but not in HaCaT cells (Fig. 1D). These experiments show that mitotic genes are highly modulated by E2, not only in HeLa cells but also in other cervical carcinoma cell lines associated with either HPV18 or HPV16.
E6/E7 and E6AP siRNAs differ in repressing the mitotic genes while they similarly activate the p53 and p63 target genes.
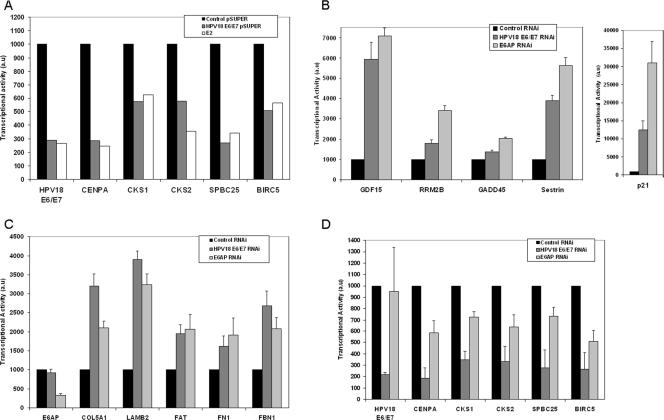
We then compared the efficiency of E2 and siRNA silencing of E6 and E7 transcription in modulating the panel of five mitotic genes that we described earlier. These genes appeared repressed to similar extents (1.6- to 4-fold) in real-time PCR experiments by the E2 expression plasmid and by the E6/E7 siRNA in transfected HeLa cells (Fig. 2A). These results clearly identify the major role of E2 in these experiments to be, indeed, transcriptional repression of E6 and E7.
FIG. 2.
Gene modulation by RT-PCR in E2-transfected HeLa cells compared to E6/E7 and E6AP siRNA-transfected cells. (A) Modulation of a panel of mitotic genes. HeLa cells were transfected with the E6/E7 siRNA cloned in a pSUPER expression vector or with a vector expressing HPV18 GFP-E2. RT-PCR values are given in arbitrary units (au) with the control siRNA set up to an arbitrary value of 1,000. (B) Modulation of p53 target genes. HeLa cells were transfected with siRNA against E6/E7 or E6AP, and the RT-PCR values of gene expression were compared to cells transfected by a control siRNA. Values given are calculated as in panel A. (C) Modulation of a panel of p63 target genes as in panel B. (D) Modulation of a panel of mitotic genes as in panel B.
It is likely that E7 is the major driver in the activation of the mitotic genes through the E2F pathway. However, recent data implicate p53 and p63 in the transcriptional repression of mitotic genes, therefore involving also E6 in the induction of mitotic genes (17, 40). In an attempt to differentiate between the effects of E6 and E7 in the regulation of the mitotic genes, we designed E6-specific siRNAs according to recent reports (3, 39). However, this approach was not satisfactory since transfection of that siRNA in HeLa cells induced concomitant partial silencing of E7 (data not shown). We therefore chose to use an E6AP siRNA instead (21). As expected for the silencing of the E6AP ubiquitin ligase and subsequent stabilization of p53, we found that the transcription of the p53 target genes was activated by transfection of the E6AP siRNA similarly to transfection of the E6/E7 siRNA in HeLa cells (Fig. 2B) and by transfected E2 (data not shown). These results clearly indicated that p53 was stabilized equally well by E2 and by the two siRNAs, as shown in Fig. 1B, and that the E6-mediated p53 degradation pathway was the only pathway involved in modulation of this set of genes as previously described (21). Interestingly, the p63 target genes were similarly activated after transfection of the E6AP siRNA, indicating that these genes were also controlled by the E6/E6AP pathway (Fig. 2C). In contrast, silencing of E6AP did not lead to strong modulation of the mitotic genes compared to silencing of both E6 and E7 with an E6/E7 siRNA (Fig. 2D). The mitotic gene that appeared the most sensitive to silencing of E6AP (50% loss of activity) was the BIRC5 (Survivin) gene, which has been shown to be activated by E2F (19) and repressed by p53 (28). Survivin is not only a member of the inhibitor of apoptosis family but is also required to sense kinetochore tension in metaphase (23, 35). These results thus point to a crucial role of E7 in the regulation of the mitotic genes and of E6 in the modulation of p53- and p63-responsive genes through a common E6AP pathway.
The E7 pathway is predominantly activated in cervical carcinoma biopsies.
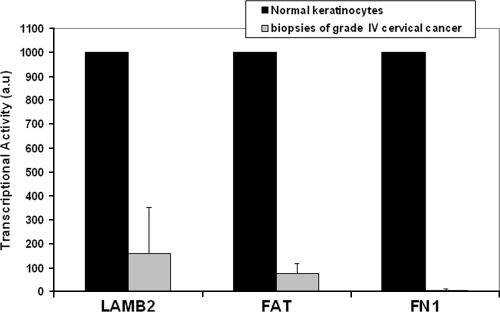
To confirm the data obtained in vitro in microarrays of E2-expressing HeLa cells, we decided to investigate whether some of the cellular genes modulated in HeLa cells were also modulated in biopsies of cervical cancers. Biopsies were obtained from grade IV cervical cancers together with adjacent normal tissues. Seven samples were studied that were associated with various high-risk HPV types including types 16, 18, 45, 33, and 31, as indicated in Table 3. RNA extraction and real-time PCR analyses were done with a subset of the cellular genes described earlier in the microarray analysis as well as with two invariant genes, the GADPH and HDAC1 genes, for each sample. First, we chose to investigate the modulation of the p53 targets, namely, the p21, Sestrin, RRM2B, GADD45, GDF15, and Bax genes, which should be repressed in cancer biopsies compared to normal tissue. Interestingly, however, variations of these target genes did not show significant and reproducible repression in cervical biopsies but, rather, random variations from one sample to the other (data not shown). In contrast, when we examined p63 target genes, we found three of these genes consistently repressed in four cancer biopsies compared to normal keratinocytes (Fig. 3).
TABLE 3.
Modulation of cell cycle genes in cervical cancers biopsies
| Biopsy lesion no. | HPV typea | Level of transcription in cancer biopsies relative to normal adjacent tissue (n-fold)b
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cdc20 | STK12 (Aurora A) | ESPL1 (Separin) | STK18 | Cdc25A | BRRN1 | CKS1 | CKS2 | CENPA | SPBC25 | ||
| I | 16 | 2.5 | 1.6 | 0.7 | 2.2 | 1.2 | 2 | ||||
| II | 16 | 2 | 3 | 0.5 | 4.8 | 1.2 | 3 | ||||
| III | 16 | 5 | 2.7 | 1 | 3 | 2.8 | 3.5 | ||||
| IV | 31 | 4 | 12 | 6 | 3 | 8 | 1.8 | ||||
| V | 45 | 3.5 | 1.2 | 0.6 | 1.2 | 1.2 | 3 | ||||
| VI | 18 | 3.2 | 3.4 | 1 | 5.5 | 0.8 | 6 | 2.5 | 3 | 3.9 | 1.7 |
| VII | 33 | 5.4 | 4.7 | 0.8 | 3.3 | 1.5 | 3.5 | 2.9 | 6.7 | 9 | 3.6 |
| Meanc | 3.6 | 4.1 | 1.5 | 3.3 | 2.4 | 3.3 | |||||
HPV type found associated with the biopsies as determined by RT-PCR.
Values are mean levels of transcription in cancer biopsies compared to normal adjacent tissue.
Mean values were calculated from the six independent samples. The associated P values were 0.008 (Cdc20), 0.008 (STK12), 0.3 (ESPL1), 0.008 (STK18), 0.015 (Cdc25A), and 0.008 (BRRN1) as calculated with GraphPad Prism, version 4, software.
FIG. 3.
RT-PCR analyses of three p63 target genes in four biopsies of grade IV cervical cancer. Levels of gene expression were compared to normal human keratinocytes.
We then investigated the behavior of a panel of genes, found repressed by E2 in microarrays and confirmed by RT-PCR in three cervical carcinoma cell lines (Fig. 1D), that should be activated by the presence of the viral oncogenes in cancer biopsies as depicted in Fig. 1A. In contrast to the p53 target genes, consistent activation of five out of six of these genes, including four mitotic genes and a gene of the G1/S transition (the CDC25A gene), could be observed in cancer biopsies (Table 3). Four additional mitotic genes were studied in two biopsies only and showed very clear activation compared to the adjacent normal tissue (Table 3).
Altogether, these data showed unambiguously that the mitotic genes found modulated in cervical carcinoma cells by E2 were also highly modulated in cervical cancers. Since we have shown that these genes are mainly controlled by the E7 pathway, we could deduce that this pathway is prominent in cervical carcinoma.
DISCUSSION
In the present work we have deciphered a new p63 pathway involving genes of cell-cell and cell-matrix adhesion in cervical carcinoma. It is not unexpected that genes modulating cell adhesion in epithelial cells were down-modulated in cervical carcinoma cells, thus participating in cell transformation. However, the mechanism by which this transcriptional modulation occurs is not yet known, and we can only speculate about potential mechanisms. Unlike p53, p63 status in cervical carcinoma as well as in other cancers is not well established, and it remains unclear whether p63 is a tumor suppressor gene or an oncogene. Point mutations of p63 are rare in cancers, and contradictory reports are present in the literature regarding its specific regulation in transformed cells, although it has been shown to be highly expressed in several epithelial cancers (reviewed in reference 13). The complexity of the studies is due to the existence of multiple isoforms with opposite functions, with TAp63 as a likely tumor suppressor and ΔNp63 as an oncogene. In any case, p63 has not been shown previously to be degraded through the E6/E6AP proteasome pathway, and our results do not necessarily imply a direct involvement of this pathway in p63 modulation, although more experiments are needed to confirm that point. Another option is that modulation of the p63 target genes reflects modulation of the p63 transcription by p53, degradation of p53 leading to transcriptional down-regulation of p63, or, in the presence of E2, p53 stabilization leading to p63 activation.
The microarrays described here have confirmed and extended our previous observation that an important cluster of mitotic genes is modulated in cervical carcinoma. Furthermore, we could deduce that this specific modulation was essentially due to the expression of the E7 viral oncogene and the subsequent modulation of the pRb family of repressors of the E2F transcription factors. A recent study indicated that, indeed, repression of the E7/pRb pathway initiates induction of senescence in HeLa cells expressing the BPV1 E2 protein (20). However, when mitotic genes were examined, no consensus E2F binding sequences could be found in their regulatory regions, contrasting with the genes involved in the G1/S transition (12, 24). This means that other transcription factors are involved in transcriptional activation of the mitotic genes, which themselves are direct E2F target genes. Such transcription factors were found repressed by E2 in our experiments, including NF-YB in the present experiments (Table 2) and B-Myb and FoxM1 in the previous microarray experiments (41). The histone-like NF-Y transcription factor is the paradigm of a constitutive ubiquitous factor that prepares the promoter architecture for other factors to get access to it. It binds to the CCAAT box, which has been found in many cell cycle-regulated promoters (24). In ChIP assays, NF-Y is found sequentially recruited to promoters of cell cycle genes together with E2F1 and E2F4 (4). The B-Myb transcription factor is an E2F-regulated factor induced at the G1/S transition, which plays a major role in activating G2/M genes (47). The FoxM1 transcription factor has been shown to activate a cluster of mitotic target genes, of which six are found modulated here: CENPA, cyclin B1, Nek2, Polo-like kinase, and the ubiquitin-conjugating enzyme 2C (22). In addition, Aurora B, Survivin, CENPA, and cdc25B have been shown in ChIP assays to be direct targets of FoxM1 (45). Modulation of mitotic genes in cervical carcinoma might therefore be essentially owing to modulation of the E2F target genes coding for the NF-Y, Myb, and FoxM1 transcription factors. In contrast, we showed here that the E6 pathway, through modulation of the p53 and p63 transcription factors, has only a minor influence on the mitotic gene regulation.
A recently published study identifies a proliferation gene cluster in cervical carcinoma associated with HPV16 or HPV18. The authors have reported 55 genes activated in cervical carcinoma that are related to the cell cycle, many of which are also E2F target genes. Interestingly, all these genes were also found modulated in our microarrays, indicating a strong overlap between the two studies (34). Another very interesting convergence between the two studies is that, in both cases, the p53 target genes were not found modulated in biopsies of lesions, despite continuous expression of E6. This result may reflect the fact that p53 is not activated in normal cells in the absence of stress, thus inducing no marked difference whether E6 is expressed or not. Additionally, biopsies of cancer and normal tissues are heterogeneous, and modulation of the p53 target genes in E6-expressing cells may have been blurred by signals from contaminated cells, such as cells of the immune system or dermis fibroblasts. In any case, we deduce from our experiments that p53 target genes would be poor markers of carcinogenic progression of HPV-associated lesions of the cervix. In contrast, the large cluster of mitotic genes that were found modulated in established cell lines as well as in biopsies could be regarded as useful biomarkers of the evolution of lesions in cervical carcinoma.
Indeed, comparative analyses with published data indicated that, although E2F target genes of the G2/M cell cycle transition are often found modulated in cancers, they do not form specifically large clusters as in cervical cancer. For example, a global study of the oncogenic pathway signatures in human cancers was recently published by the Bild et al., including an E2F pathway modulated through E2F3 expression in quiescent cells (2). In this work the authors have deciphered five oncogenic pathways including E2F, RAS, MYC, SRC, and beta-cathenin. Strikingly, only 13 genes modulated in our arrays could be found among the genes specific to the E2F3 signature. We can deduce from these data and several comparative studies of other cancers that the HPV E7 signature is very specific and is biased in favor of mitotic gene activation.
Acknowledgments
We thank Jean-Pierre Abastado for critical reading of the manuscript. We thank P. Van Hummelen, Stefan Weckx, and Tom Bogaert from the MicroArray Facility for their assistance with processing of microarrays data and A. Michaux (SCK-CEN) for technical assistance.
This work was supported by Association pour la Recherche contre le Cancer (ARC) and by INCa (project PL052). S.T. was the recipient of an ARC fellowship, and Y.B.K. was a recipient of a fellowship from La Ligue Contre le Cancer. M.M. was an Assistant Scientific Collaborator (AWM) at the SCK-CEN.
Footnotes
Published ahead of print on 20 June 2007.
REFERENCES
- 1.Barbieri, C. E., L. J. Tang, K. A. Brown, and J. A. Pietenpol. 2006. Loss of p63 leads to increased cell migration and up-regulation of genes involved in invasion and metastasis. Cancer Res. 66:7589-7597. [DOI] [PubMed] [Google Scholar]
- 2.Bild, A. H., G. Yao, J. T. Chang, Q. Wang, A. Potti, D. Chasse, M. B. Joshi, D. Harpole, J. M. Lancaster, A. Berchuck, J. A. Olson, Jr., J. R. Marks, H. K. Dressman, M. West, and J. R. Nevins. 2006. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature 439:353-357. [DOI] [PubMed] [Google Scholar]
- 3.Butz, K., T. Ristriani, A. Hengstermann, C. Denk, M. Scheffner, and F. Hoppe-Seyler. 2003. siRNA targeting of the viral E6 oncogene efficiently kills human papillomavirus-positive cancer cells. Oncogene 22:5938-5945. [DOI] [PubMed] [Google Scholar]
- 4.Caretti, G., V. Salsi, C. Vecchi, C. Imbriano, and R. Mantovani. 2003. Dynamic recruitment of NF-Y and histone acetyltransferases on cell-cycle promoters. J. Biol. Chem. 278:30435-30440. [DOI] [PubMed] [Google Scholar]
- 5.Carroll, D. K., J. S. Carroll, C. O. Leong, F. Cheng, M. Brown, A. A. Mills, J. S. Brugge, and L. W. Ellisen. 2006. p63 regulates an adhesion programme and cell survival in epithelial cells. Nat. Cell Biol. 8:551-561. [DOI] [PubMed] [Google Scholar]
- 6.DeFilippis, R. A., E. C. Goodwin, L. Wu, and D. DiMaio. 2003. Endogenous human papillomavirus E6 and E7 proteins differentially regulate proliferation, senescence, and apoptosis in HeLa cervical carcinoma cells. J. Virol. 77:1551-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demeret, C., A. Garcia-Carranca, and F. Thierry. 2003. Transcription-independent triggering of the extrinsic pathway of apoptosis by human papillomavirus (HPV) 18 E2 protein. Oncogene 22:168-175. [DOI] [PubMed] [Google Scholar]
- 8.Desaintes, C., C. Demeret, S. Goyat, M. Yaniv, and F. Thierry. 1997. Expression of the papillomavirus E2 protein in HeLa cells leads to apoptosis. EMBO J. 16:504-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desaintes, C., S. Goyat, S. Garbay, M. Yaniv, and F. Thierry. 1999. Papillomavirus E2 induces p53-independent apoptosis in HeLa cells. Oncogene 18:4538-4546. [DOI] [PubMed] [Google Scholar]
- 10.Dowhanick, J. J., A. A. McBride, and P. M. Howley. 1995. Suppression of cellular proliferation by the papillomavirus E2 protein. J. Virol. 69:7791-7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dyson, N., P. M. Howley, K. Münger, and E. Harlow. 1989. The human papillomavirus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science 243:934-937. [DOI] [PubMed] [Google Scholar]
- 12.Elkon, R., C. Linhart, R. Sharan, R. Shamir, and Y. Shiloh. 2003. Genome-wide in silico identification of transcriptional regulators controlling the cell cycle in human cells. Genome Res. 13:773-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flores, E. R. 2007. The roles of p63 in cancer. Cell Cycle 6:300-304. [DOI] [PubMed] [Google Scholar]
- 14.Gewin, L., H. Myers, T. Kiyono, and D. A. Galloway. 2004. Identification of a novel telomerase repressor that interacts with the human papillomavirus type-16 E6/E6-AP complex. Genes Dev. 18:2269-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodwin, E. C., E. Yang, C. J. Lee, H. W. Lee, D. DiMaio, and E. S. Hwang. 2000. Rapid induction of senescence in human cervical carcinoma cells. Proc. Natl. Acad. Sci. USA 97:10978-10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hwang, E.-S., L. K. Naeger, and D. DiMaio. 1996. Activation of the endogenous p53 growth inhibitory pathway in HeLa cervical carcinoma cells by expression of the bovine papillomavirus E2 gene. Oncogene 12:795-803. [PubMed] [Google Scholar]
- 17.Imbriano, C., A. Gurtner, F. Cocchiarella, S. Di Agostino, V. Basile, M. Gostissa, M. Dobbelstein, G. Del Sal, G. Piaggio, and R. Mantovani. 2005. Direct p53 transcriptional repression: in vivo analysis of CCAAT-containing G2/M promoters. Mol. Cell. Biol. 25:3737-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishida, S., E. Huang, H. Zuzan, R. Spang, G. Leone, M. West, and J. R. Nevins. 2001. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol. Cell. Biol. 21:4684-4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang, Y., H. I. Saavedra, M. P. Holloway, G. Leone, and R. A. Altura. 2004. Aberrant regulation of survivin by the RB/E2F family of proteins. J. Biol. Chem. 279:40511-40520. [DOI] [PubMed] [Google Scholar]
- 20.Johung, K., E. C. Goodwin, and D. Dimaio. 2007. Human papillomavirus E7 repression in cervical carcinoma cells initiates a transcriptional cascade driven by the retinoblastoma family, resulting in senescence. J. Virol. 81:2102-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelley, M. L., K. E. Keiger, C. J. Lee, and J. M. Huibregtse. 2005. The global transcriptional effects of the human papillomavirus E6 protein in cervical carcinoma cell lines are mediated by the E6AP ubiquitin ligase. J. Virol. 79:3737-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laoukili, J., M. R. Kooistra, A. Bras, J. Kauw, R. M. Kerkhoven, A. Morrison, H. Clevers, and R. H. Medema. 2005. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat. Cell Biol. 7:126-136. [DOI] [PubMed] [Google Scholar]
- 23.Lens, S. M., R. M. Wolthuis, R. Klompmaker, J. Kauw, R. Agami, T. Brummelkamp, G. Kops, and R. H. Medema. 2003. Survivin is required for a sustained spindle checkpoint arrest in response to lack of tension. EMBO J. 22:2934-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Linhart, C., R. Elkon, Y. Shiloh, and R. Shamir. 2005. Deciphering transcriptional regulatory elements that encode specific cell cycle phasing by comparative genomics analysis. Cell Cycle 4:1788-1797. [DOI] [PubMed] [Google Scholar]
- 25.Ma, Y., R. Croxton, R. L. Moorer, Jr., and W. D. Cress. 2002. Identification of novel E2F1-regulated genes by microarray. Arch. Biochem. Biophys. 399:212-224. [DOI] [PubMed] [Google Scholar]
- 26.Markey, M. P., S. P. Angus, M. W. Strobeck, S. L. Williams, R. W. Gunawardena, B. J. Aronow, and E. S. Knudsen. 2002. Unbiased analysis of RB-mediated transcriptional repression identifies novel targets and distinctions from E2F action. Cancer Res. 62:6587-6597. [PubMed] [Google Scholar]
- 27.Mills, A. A., B. Zheng, X. J. Wang, H. Vogel, D. R. Roop, and A. Bradley. 1999. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 398:708-713. [DOI] [PubMed] [Google Scholar]
- 28.Mirza, A., M. McGuirk, T. N. Hockenberry, Q. Wu, H. Ashar, S. Black, S. F. Wen, L. Wang, P. Kirschmeier, W. R. Bishop, L. L. Nielsen, C. B. Pickett, and S. Liu. 2002. Human survivin is negatively regulated by wild-type p53 and participates in p53-dependent apoptotic pathway. Oncogene 21:2613-2622. [DOI] [PubMed] [Google Scholar]
- 29.Mori, M., M. A. Benotmane, I. Tirone, E. L. Hooghe-Peters, and C. Desaintes. 2005. Transcriptional response to ionizing radiation in lymphocyte subsets. Cell Mol. Life Sci. 62:1489-1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muller, H., A. P. Bracken, R. Vernell, M. C. Moroni, F. Christians, E. Grassilli, E. Prosperini, E. Vigo, J. D. Oliner, and K. Helin. 2001. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 15:267-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Polager, S., Y. Kalma, E. Berkovich, and D. Ginsberg. 2002. E2Fs up-regulate expression of genes involved in DNA replication, DNA repair and mitosis. Oncogene 21:437-446. [DOI] [PubMed] [Google Scholar]
- 32.Ren, B., H. Cam, Y. Takahashi, T. Volkert, J. Terragni, R. A. Young, and B. D. Dynlacht. 2002. E2F integrates cell cycle progression with DNA repair, replication, and G2/M checkpoints. Genes Dev. 16:245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rinne, T., H. G. Brunner, and H. van Bokhoven. 2007. p63-associated disorders. Cell Cycle 6:262-268. [DOI] [PubMed] [Google Scholar]
- 34.Rosty, C., M. Sheffer, D. Tsafrir, N. Stransky, I. Tsafrir, M. Peter, P. de Cremoux, A. de La Rochefordiere, R. Salmon, T. Dorval, J. P. Thiery, J. Couturier, F. Radvanyi, E. Domany, and X. Sastre-Garau. 2005. Identification of a proliferation gene cluster associated with HPV E6/E7 expression level and viral DNA load in invasive cervical carcinoma. Oncogene 24:7094-7104. [DOI] [PubMed] [Google Scholar]
- 35.Sandall, S., F. Severin, I. X. McLeod, J. R. Yates III, K. Oegema, A. Hyman, and A. Desai. 2006. A Bir1-Sli15 complex connects centromeres to microtubules and is required to sense kinetochore tension. Cell 127:1179-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scheffner, M., J. M. Huibregtse, R. D. Vierstra, and P. M. Howley. 1993. The HPV16 E6 and E6-AP complex functions as ubiquitin-protein ligase in the ubiquitination of p53. Cell 75:495-505. [DOI] [PubMed] [Google Scholar]
- 37.Scheffner, M., B. A. Werness, J. M. Huibregtse, A. J. Levine, and P. M. Howley. 1990. The E6 oncoprotein encoded by human papillomaviruses types 16 and 18 promotes the degradation of p53. Cell 63:1129-1136. [DOI] [PubMed] [Google Scholar]
- 38.Shepard, J. L., J. F. Amatruda, H. M. Stern, A. Subramanian, D. Finkelstein, J. Ziai, K. R. Finley, K. L. Pfaff, C. Hersey, Y. Zhou, B. Barut, M. Freedman, C. Lee, J. Spitsbergen, D. Neuberg, G. Weber, T. R. Golub, J. N. Glickman, J. L. Kutok, J. C. Aster, and L. I. Zon. 2005. A zebrafish bmyb mutation causes genome instability and increased cancer susceptibility. Proc. Natl. Acad. Sci. USA 102:13194-13199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang, S., M. Tao, J. P. McCoy, Jr., and Z. M. Zheng. 2006. The E7 oncoprotein is translated from spliced E6*I transcripts in high-risk human papillomavirus type 16- or type 18-positive cervical cancer cell lines via translation reinitiation. J. Virol. 80:4249-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Testoni, B., and R. Mantovani. 2006. Mechanisms of transcriptional repression of cell-cycle G2/M promoters by p63. Nucleic Acids Res. 34:928-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thierry, F., M. A. Benotmane, C. Demeret, M. Mori, S. Teissier, and C. Desaintes. 2004. A genomic approach reveals a novel mitotic pathway in papillomavirus carcinogenesis. Cancer Res. 64:895-903. [DOI] [PubMed] [Google Scholar]
- 42.Thierry, F., and P. M. Howley. 1991. Functional analysis of E2-mediated repression of the HPV18 P105 promoter. New Biol. 3:90-100. [PubMed] [Google Scholar]
- 43.Thierry, F., and M. Yaniv. 1987. The BPV1-E2 trans-acting protein can be either an activator or a repressor of the HPV18 regulatory region. EMBO J. 6:3391-3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walboomers, J. M., M. V. Jacobs, M. M. Manos, F. X. Bosch, J. A. Kummer, K. V. Shah, P. J. Snijders, J. Peto, C. J. Meijer, and N. Munoz. 1999. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 189:12-19. [DOI] [PubMed] [Google Scholar]
- 45.Wang, I. C., Y. J. Chen, D. Hughes, V. Petrovic, M. L. Major, H. J. Park, Y. Tan, T. Ackerson, and R. H. Costa. 2005. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol. Cell. Biol. 25:10875-10894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang, A., R. Schweitzer, D. Sun, M. Kaghad, N. Walker, R. T. Bronson, C. Tabin, A. Sharpe, D. Caput, C. Crum, and F. McKeon. 1999. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 398:714-718. [DOI] [PubMed] [Google Scholar]
- 47.Zhu, W., P. H. Giangrande, and J. R. Nevins. 2004. E2Fs link the control of G1/S and G2/M transcription. EMBO J. 23:4615-4626. [DOI] [PMC free article] [PubMed] [Google Scholar]