Abstract
Histone methylation plays important roles in the regulation of chromatin dynamics and transcription. Steady-state levels of histone lysine methylation are regulated by a balance between enzymes that catalyze either the addition or removal of methyl groups. Using an activity-based biochemical approach, we recently uncovered the JmjC domain as an evolutionarily conserved signature motif for histone demethylases. Furthermore, we demonstrated that Jhd1, a JmjC domain-containing protein in Saccharomyces cerevisiae, is an H3K36-specific demethylase. Here we report further characterization of Jhd1. Similar to its mammalian homolog, Jhd1-catalyzed histone demethylation requires iron and α-ketoglutarate as cofactors. Mutation and deletion studies indicate that the JmjC domain and adjacent sequences are critical for Jhd1 enzymatic activity, while the N-terminal PHD domain is dispensable. Overexpression of JHD1 results in a global reduction of H3K36 methylation in vivo. Finally, chromatin immunoprecipitation-coupled microarray studies reveal subtle changes in the distribution of H3K36me2 upon overexpression or deletion of JHD1. Our studies establish Jhd1 as a histone demethylase in budding yeast and suggest that Jhd1 functions to maintain the fidelity of histone methylation patterns along transcription units.
In eukaryotic cells, DNA is wrapped around histone octamers to form the nucleosome. Various posttranslational modifications of histone tails function as epigenetic marks that control chromatin structure and gene expression (18). Among these modifications, histone methylation plays an important role in many chromatin-based processes, including transcriptional regulation, heterochromatin formation, and X-chromosome inactivation (28, 29). In contrast to acetylation, methylation was formerly considered an “irreversible” mark due to its low turnover rate. However, recent studies have revealed that the methyl groups on histone tails can be removed through active demethylation. Shi and colleagues demonstrated that LSD1 (lysine-specific demethylase), a member of the nuclear amine oxidase family, can catalyze removal of the mono- and dimethyl groups from lysine 4 of histone H3 through a flavin adenine dinucleotide-dependent amine oxidative reaction (40). However, this family of proteins cannot remove methyl groups from a trimethyl-lysine, because formation of an imine intermediate requires a protonated lysine for the amine oxidation. In addition, no apparent homolog of LSD1 exists in Saccharomyces cerevisiae.
Using a novel in vitro histone demethylation assay coupled with protein fractionation, we have recently identified a novel class of histone demethylases (45). This class of histone demethylases is characterized by the presence of a JmjC domain, an evolutionarily conserved motif (8). We have demonstrated that, in the presence of α-ketoglutarate (α-KG) and Fe(II), the JmjC domain-containing histone demethylases can remove the methyl groups from lysine residues to produce succinate and formaldehyde (45). Unlike LSD1, the JmjC domain-containing proteins comprise a large protein family that is conserved from bacteria to humans (19). In addition, the JmjC domain-containing demethylases remove methyl groups from lysine residues through a hydroxylation reaction and, therefore, have the capacity to demethylate trimethyl states (9, 20, 47).
Three lysine residues of histone H3 (K4, K36, and K79) in the budding yeast S. cerevisiae are subjected to methylation (29). Genome-wide location studies indicate that H3K4 trimethylation is mainly enriched at promoter and transcriptional initiation sites of active genes (4, 6, 21, 27, 33, 35, 38), while H3K36 methylation is enriched in the open reading frames (ORFs) of protein-encoding genes (2, 35, 36). Consistent with this distribution, the enzymes responsible for these methylation states, Set1 and Set2, have been linked to the PAF complex and to active RNA polymerase during transcription initiation and elongation, respectively (21, 22, 24, 25, 33, 50). To determine whether histone methylation in yeast is subjected to active demethylation, we screened the five S. cerevisiae JmjC domain-containing proteins for histone demethylation activity. Here we report the identification and characterization of Jhd1, the S. cerevisiae homolog of human JHDM1A. We demonstrate that Jhd1 is an H3K36-specific demethylase. Deletion and mutation studies indicate that both the JmjC domain and its adjacent sequences are required for the demethylase activity. Chromatin immunoprecipitation (ChIP)-coupled microarray analyses reveal that overexpression of JHD1 results in a subtle 3′ shift of the H3K36me2 pattern in transcription units. In contrast, deletion of JHD1 causes a more uniform distribution of H3K36me2 across ORFs. Our work uncovers Jhd1 as an H3K36-specific demethylase and suggests that Jhd1 fine-tunes the distribution of H3K36me2.
MATERIALS AND METHODS
Histone methylation and demethylation assays.
Core histones and nucleosomes were purified from HeLa nuclei as previously described (11). Expression and purification of glutathione S-transferase (GST)-SET7, CBP-Set2-Flag, and GST-hDOT1L have been described elsewhere (43, 45, 46). The in vitro histone methyltransferase assay and in vitro histone demethylase assay were performed as previously described (11, 45).
Strains and culture conditions.
All strains, except those in the telomeric silencing assay, were derived from BY4741. Strains used in telomeric silencing assay were of the YCB647 background (42). Strains harboring individual C-terminal 3× Flag tags of each of the five genes encoding JmjC-domain proteins were generated by PCR (12). The jhd1Δ and JHD1 overexpression strains were constructed using standard protocols. Unless otherwise described, yeast were grown with shaking at 30°C in yeast YPD medium (1% yeast extract, 2% peptone, 2% dextrose) to an optical density at 600 nm of 0.8 to 1.0.
Preparation of yeast whole-cell extracts and immunoprecipitation.
For small-scale experiments, cell pellets were resuspended in lysis buffer and lysed with an equal volume of glass beads using a mini-bead beater. For larger-scale experiments, cell pellets were resuspended in an equal volume using 2× lysis buffer (0.3 M HEPES-KOH, 2 mM EDTA, 40% glycerol, 20 mM β-glycerophosphate, 1 mM NaF, 2 mM dithiothreitol, 100 mM KCl, and 2× protease inhibitor cocktail; pH 7.6). The resulting yeast paste was extruded from a syringe into liquid nitrogen and crushed manually with a mortar and pestle. Broken cells were resuspended in an equal volume of 1× lysis buffer, and the final KCl concentration was adjusted to 200 mM. The subsequent anti-Flag M2 immunoprecipitation was performed as previously described (30), and the immunoprecipitated samples were analyzed using both our in vitro histone demethylation assay and sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis followed by Western blotting using anti-Flag M2 antibodies.
Antibodies and ChIP assays.
Histone H3K36me2 rabbit polyclonal antibodies have been previously described (45), and the following other methyl-lysine-specific histone antibodies were obtained commercially from Abcam: H3K36me3 (Ab9050), H3K36me1(Ab9048), and H3 (Ab1791). Flag M2 antibody and M2 agarose were purchased from Sigma (F3165). ChIP assays were performed as described previously (36).
DNA amplification and labeling.
All samples and references were amplified using a random primed PCR-based method (5). The first amplification round involved the use of primer A (5′-GTTTCCCAGTCACGATCNNNNNNNNN-3′) in conjunction with Sequenase, a modified T7 DNA polymerase. In the second round, primer B (5′-GTTTCCCAGTCACGATC-3′) was used with Taq DNA polymerase in 25 cycles of PCR. In the final round, the fluorescent nucleotide Cy3-dUTP or Cy5-dUTP was then incorporated directly into the reference or sample in an additional 25 cycles of PCR by using primer B and Taq DNA polymerase.
DNA microarray hybridization and scanning.
Labeled ChIP DNA was purified and hybridized to DNA microarrays as previously described (17). DNA microarrays were manufactured using a robotic arrayer to print PCR products on poly-l-lysine-coated glass slides as described elsewhere (17). The microarray consisted of two types of probes. The first type represents the whole genome, based on annotated functional boundaries. These PCR-amplified products represent ORFs, intergenic regions, and other noncoding regions (rDNA, tRNA, transposons, transposon long terminal repeats, telomeres, centromeres, and introns). Generally, each ORF was represented from start codon to stop codon. The intergenic regions consisted of the DNA between annotated ORFs divided such that PCR products were not longer than 1.5 kb, with a few exceptions. The noncoding regions conform to boundaries as annotated by the Saccharomyces Genome Database (SGD; http://www.yeastgenome.org) as of the year 2000. Mitochondrial segments did not necessarily conform to annotated functional boundaries. The second type of probes are higher-resolution PCR-amplified products that span almost all of chromosome III at 200-bp resolution with additional overlapping products covering one-third of the chromosome at 100-bp resolution (coordinates 10,000 to 83,000). Images were acquired using a GenePix 4000B scanner and Genepix software (Molecular Devices).
Data acquisition and normalization.
Acquired images were inspected visually to remove low-quality probes. Raw data were submitted to the University of North Carolina (UNC) Microarray Database (http://genome.unc.edu). We retrieved the value from each probe as the log2-normalized ratio of the median intensity of sample pixels divided by the median intensity of reference pixels, and only the probes with a regression correlation of >0.6 (i.e., those comprised of pixels with consistent ratio values) were downloaded. The log2 ratio of each probe was transformed to a z-score by using the formula zx = (X − μ)/σ, where X is a retrieved probe value, μ is the mean of all retrieved probes from one array, and σ is the standard deviation of all retrieved probes from that same array. Following z-score transformation, any technical replicates from dye-swap experiments were averaged and treated as one biological replicate, followed by averaging all biological replicates. Each ChIP-chip experiment was performed using three biological replicates.
Microarray data accession number.
Raw microarray data are available from the UNC Microarray Database (UMD; https://genome.unc.edu). The data have also been deposited in the National Center for Biotechnology Information's Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) and are accessible through GEO series accession number GSE7627.
RESULTS
Identification of demethylase activity in S. cerevisiae.
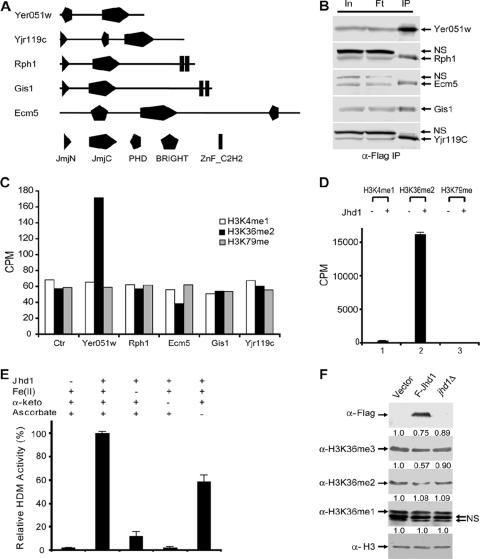
Using an activity-based biochemical approach, we previously identified the evolutionarily conserved JmjC domain as a signature motif for histone demethylases (45, 52). Subsequently, we and others have demonstrated that members of this protein family have the capacity to demethylate a trimethyl-lysine state (9, 20, 47). Given that no apparent LSD1 homolog exists in S. cerevisiae, we suspected that JmjC domain-containing proteins were responsible for histone demethylation in this organism. Five JmjC domain-containing proteins are encoded by the S. cerevisiae genome (19) (Fig. 1A). To determine whether any of the five proteins possesses histone demethylase activity, we generated five yeast strains, each of which expressed C-terminal epitope-tagged alleles of one of the five JmjC domain-containing proteins. After immunoprecipitation of the Flag-tagged proteins from whole-cell extracts (Fig. 1B), the immunoprecipitates were incubated with radioactive histone substrates corresponding to known lysine methylation states in yeast. By monitoring release of radioactive formaldehyde, we found that only the Flag-Yer051w-containing immunoprecipitates exhibited consistent demethylase activity towards H3K36 methylated substrates. However, the same immunoprecipitates did not show any activity toward H3K4 or H3K79 methylated substrates (Fig. 1C), indicating that the Yer051w protein is likely an H3K36-specific demethylase. Consistent with this possibility, sequence analysis indicated that the JmjC domain Yer051w is most similar to the human H3K36 demethylase JHDM1. Consequently, we have named the yeast protein Jhd1 (JmjC domain-containing histone demethylase 1).
FIG. 1.
Jhd1 is an H3K36-specific demethylase. (A) Schematic representation of the five JmjC domain-containing proteins in S. cerevisiae. (B) Western blot analysis of immunoprecipitates from five Flag-tagged JmjC domain-containing proteins using whole-cell extracts. Input (In), flowthrough (Ft), and immunoprecipitates (IP) were fractionated by SDS-polyacrylamide gel electrophoresis and analyzed by Western blotting using an anti-Flag antibody. NS, nonspecific cross-reactive band. (C) In vitro radioactive formaldehyde release assays using immunoprecipitates analyzed in panel B and substrates that are methylated on H3K4me1, H3K36me2, or H3K79me, using GST-SET7, CBP-Set2-Flag, and GST-hDOT1L, respectively. (D) In vitro histone demethylase activities of recombinant GST-Jhd1 toward different radioactive histone substrates. About 2 μg (∼24 pmol) of recombinant GST-Jhd1 protein was incubated with 4 μg (∼37 pmol, based on the molecular weight of histone octamer) of radioactively labeled H3K36me2 substrates (100,000 cpm total) at 37°C for 1 h and analyzed in formaldehyde release assays. Demethylation activities are presented as averages of three independent experiments with standard deviations. (E) Effects of cofactors Fe(II), α-ketoglutarate (α-keto), and ascorbate on the enzymatic activity of Jhd1. Demethylase assays were performed under the same conditions as for panel D. Results presented are averages of three independent experiments with standard deviations. (F) Overexpression of Jhd1 results in a subtle decrease of H3K36me2 and me3 methylation levels, but the jhd1 deletion does not. Equal amount of whole-cell lysates from wild-type, JHD1 overexpression, or jhd1Δ strains were analyzed by Western blotting using the various antibodies indicated. Changes in methylation levels were quantified with NIH ImageJ software and normalized with histone H3. Relative methylation levels are indicated above each blot. Overexpression of JHD1 was confirmed by anti-Flag antibodies (top panel), and deletion of JHD1 was confirmed by PCR (data not shown). NS, two nonspecific cross-reactive bands.
Jhd1 specifically demethylates histone H3K36 in a Fe(II)- and α-KG-dependent manner.
To verify that Jhd1, but not a protein that coimmunoprecipitates with Jhd1, is responsible for the detected activity, we generated a recombinant Jhd1 protein. Consistent with our previous report (45), recombinant Jhd1 exhibited robust demethylase activity toward methylated H3K36me2 substrates. However, it did not exhibit any detectable activity towards H3K4me1 or H3K79me methylated substrates generated by hSET7 or hDOT1L (Fig. 1D). These data confirm that the JmjC domain-containing protein Jhd1 is an H3K36-specific histone demethylase in vitro.
We have previously demonstrated that JmjC domain-containing protein-mediated histone demethylation uses the same oxidative demethylation mechanism as that used by the AlkB family of DNA demethylases (45), which requires Fe(II) and α-KG as cofactors. To confirm the same oxidation mechanism applies to Jhd1, we tested the importance of the cofactors for the Jhd1-mediated demethylation reaction by omitting each cofactor individually from the demethylation reaction. As expected, omitting either Fe(II) or α-KG from the reaction mixture resulted in a significant decrease of the enzymatic activity, whereas omitting ascorbate alone had a mild effect (Fig. 1E). These data indicated that both Fe(II) and α-KG are critical for the enzymatic activity of Jhd1, while ascorbate can stimulate the activity, most likely due to its ability to regenerate Fe(II) from Fe(III). The above results demonstrate that Jhd1, the S. cerevisiae counterpart to human JHDM1, uses the same oxidative mechanism to specifically demethylate H3K36 in vitro.
Jhd1 is capable of modulating H3K36 methylation levels in vivo.
Having demonstrated that Jhd1 is an H3K36-specific histone demethylase in vitro, we then asked whether Jhd1 has the capacity to demethylate H3K36 in vivo. For this purpose, we generated a construct expressing Flag-tagged Jhd1 under control of the ADH1 promoter on a high-copy-number plasmid. JHD1 RNA levels in our overexpression strain were increased 13- to 24-fold relative to the wild type based on DNA microarray measurements of relative transcript levels (data not shown). We then analyzed the H3K36 methylation levels in strains carrying this construct using H3K36 methylation state-specific antibodies. We observed a moderate decrease in H3K36me2 and a slight decrease in H3K36me3 along with unaltered H3K36me1 levels in the JHD1 overexpression strain, whereas all three H3K36 methylation states remained relatively unchanged in the jhd1Δ strain comparing with the wild type (Fig. 1F). These results indicate that Jhd1, at least when overexpressed, can modulate H3K36 methylation levels in vivo.
The JmjC domain and adjacent sequences are required for Jhd1 demethylase activity.
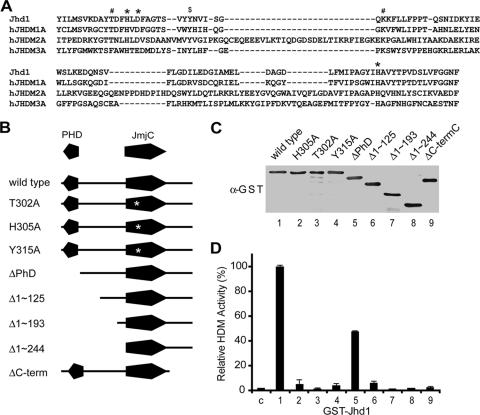
Having established that Jhd1 is an H3K36-specific histone demethylase, we next evaluated the domain requirements for its demethylase activity. The crystal structure of the histone demethylase JMJD2A/JHDM3A revealed that the active center of the JmjC domain consists of a highly conserved H-X-D/E-Xn-H signature motif that is required for Fe(II) ion binding (7). In addition, structural studies and modeling also revealed the location of α-KG binding sites (44). Sequence alignment of Jhd1 with known active JmjC domain-containing histone demethylases indicated that all Fe(II) and α-KG binding sites of Jhd1 are conserved (Fig. 2A). Given the cofactor requirements (Fig. 1E), we hypothesized that amino acid substitutions within the predicted cofactor binding sites would abrogate the demethylase activity. Results shown in Fig. 2B to D (lanes 2 and 3) confirmed that the single amino acid substitutions T302A and H305A greatly impaired Jhd1's enzymatic activity. We conclude that the JmjC domain is critical for its demethylase activity.
FIG. 2.
The JmjC domain and its adjacent sequences are important for Jhd1 enzymatic activity. (A) Sequence alignment of the JmjC domain of Jhd1 with that from several known human histone demethylases. *, conserved Fe(II) binding site; #, α-KG binding site; $, Tyr315. Gaps are indicated by dashes. (B) Schematic representation of different Jhd1 mutants. The PHD domain and JmjC domain are indicated. The point mutation in the JmjC domain is indicated with an asterisk. (C) Purified wild-type and mutant Jhd1 proteins quantified by anti-GST Western blotting. (D) Equal molar amounts (about 12 pmol) of wild-type and mutant Jhd1 proteins were incubated with about 2 μg (∼18 pmol, based on the molecular weight of histone octamer) of Set2-methylated substrates, and their histone demethylase activities were compared. The relative demethylase activities are presented as averages of three independent experiments, and the standard deviations are indicated by error bars. The bar numbers correlate with the lane numbers in panel C. C, control.
In addition to the cofactor binding sites, we also discovered that Tyr315 of Jhd1 (Fig. 2A) is conserved in all known active histone demethylases and in Epe1, the Jhd1 homolog in Schizosaccharomyces pombe. Although Epe1 is not an active demethylase in vitro (45), genetic studies indicated that the epe1-Y307A allele could not complement the epe1-1 mutation (1). This prompted us to test whether Tyr315 is critical for the demethylase activity of Jhd1. Indeed, the Y315A mutation dramatically reduced the enzymatic activity of Jhd1 (Fig. 2B to D, lane 4). Based on the crystal structure of JMJD2A/JHDM3A (7), the corresponding tyrosine does not contact cofactors directly but instead is located in a β-sheet beside Asn198, whose side chain could form a hydrogen bond with α-KG. It is therefore possible that this conserved tyrosine could affect the enzyme-α-KG interaction. Alternatively, it may influence the folding of the JmjC domain.
We next evaluated whether any other sequences outside the JmjC domain are required for Jhd1 demethylase activity. To this end, we generated a series of N-terminal and C-terminal Jhd1 deletion mutants and evaluated their enzymatic activities in the formaldehyde release assay. Deletion of the 82 amino acids at the extreme C terminus of the JmjC domain almost completely abrogated the enzymatic activity of Jhd1 (Fig. 2B to D, lane 9). On the contrary, deletion of the 72 amino acids at the N terminus that includes the PHD domain only resulted in a 50% loss of Jhd1 activity (Fig. 2B to D, lane 5). Further deletion of the sequences N-terminal to the JmjC domain significantly impaired its enzymatic activity (Fig. 2B to D, lane 6 to 8). These results indicate that sequences adjacent to the JmjC domain are required for demethylase activity. It is possible that these sequences are required for the correct folding of the JmjC domain.
Jhd1 does not form a stable protein complex with other proteins.
Having established that Jhd1 is an H3K36-specific demethylase, we attempted to identify its putative in vivo functional protein partners. Previous studies indicated that the human homolog of Jhd1 copurifies with a 20-kDa polypeptide (45). Therefore, we asked whether Jhd1 might associate with other proteins to mediate its biological function. To this end, we fractionated yeast whole-cell lysates from a Flag-tagged Jhd1 strain through six columns (Fig. 3A). Based on its elution profile on a Superdex 200 gel filtration column, although the predicted molecular mass of a Jhd1 monomer is 56.5 kDa, native Jhd1 occurs in a complex with an apparent molecular mass of 250 kDa (Fig. 3B). Several proteins appear to coelute with Jhd1 on this column. However, further purification on a heparin column indicated that these proteins do not coelute with Jhd1 (Fig. 3C). A protein of about 100 kDa cofractionates with Jhd1 on the heparin column (Fig. 3C) but does not coelute with Jhd1 on the gel filtration column (Fig. 3B). Furthermore, recombinant Jhd1 exhibited a similar elution profile as that of the native Jhd1 on the Superdex 200 column (data not shown).
FIG. 3.
Purification of endogenous Jhd1 from yeast whole-cell extract. (A) Schematic representation of steps used to purify Jhd1. The numbers represent the salt concentration (in mM) at which Jhd1 eluted from different columns. (B) Silver staining of a polyacrylamide-SDS gel (top panel) and Western blot analysis (bottom panel) of the fractions derived from gel filtration with a Superdex 200 column. Fraction numbers and elution profiles of the protein markers are indicated on the top of the gel. Jhd1 is indicated by an asterisk. (C) Silver staining of an SDS gel (top panel) and Western blot analysis (bottom panel) of the fractions derived from a heparin column after gel filtration. Fraction numbers are indicated on the top of the gel, and Jhd1 is indicated by an asterisk. The # indicates a protein of about 100 kDa that cofractionated with Jhd1 on the heparin column but did not coelute with Jhd1 on the gel filtration column. (D) Hydrodynamic properties of Jhd1. Recombinant Jhd1 was fractionated and analyzed over a 5 to 20% sucrose gradient together with standard molecular weight markers. The sedimentation coefficient of Jhd1 (∼21.37S) was calculated from this sucrose gradient. The Stoke's radius (∼5.16 nm) of Jhd1 was calculated from size-exclusion chromatography. The molecular mass (453.8 kDa) and frictional ratio (1.02) of Jhd1 were calculated using formulas derived by Siegel and Monty (41).
These data indicate that Jhd1 does not exist in a stable complex with other proteins but instead forms an oligomer. To confirm this possibility, we analyzed recombinant Jhd1 by sucrose gradient sedimentation and calculated the sedimentation coefficient of Jhd1 as 21.37S (Fig. 3D). The Stoke's radius of Jhd1 from the size exclusion column was calculated to be 5.16 nm. Based on Siegel and Monty formulas (41), the molecular mass of recombinant Jhd1 was calculated as 453.8 kDa (Fig. 3D). Given that the theoretical mass of Jhd1 is 56.5 kDa, we conclude that Jhd1 likely functions as an oligomer.
Phenotypic analysis in jhd1Δ and JHD1 overexpression strains.
In an attempt to reveal the biological function of Jhd1, we performed phenotypic analysis in JHD1 knockout and overexpression strains. Given that Jhd1 is an H3K36-specific demethylase, deletion of JHD1 could result in phenotypes opposite to those observed for the H3K36 methyltransferase set2Δ strain. Previous studies indicated that deletion of SET2 confers reduced sensitivity to the IMP dehydrogenase inhibitor 6-azauracil (6-AU), which is thought to inhibit transcription elongation (25, 51). We therefore tested the 6-AU sensitivity of the jhd1Δ strain. As a positive control, we included the 6-AU-sensitive strain harboring a deletion of the transcription elongation factor RTF1 (restores TBP function) (10). This analysis, however, did not reveal any noticeable difference in 6-AU sensitivity between wild-type and jhd1Δ strains, although a difference between the set2Δ or rtf1Δ strains and the wild-type strain was observed (Fig. 4A). Similarly, overexpression of JHD1 also did not confer a significant change in 6-AU sensitivity (Fig. 4B). Further phenotypic analysis under a variety of conditions (15) revealed no obvious defects upon deletion of JHD1 (Fig. 4C). These data suggest that processes affected by Jhd1 might be regulated by another redundant gene, or Jhd1 might function in a specialized process yet to be revealed.
FIG. 4.
Phenotype analysis of jhd1Δ and JHD1 overexpression strains. (A) Cells from BY4741 (wt), jhd1Δ, rtf1Δ(10), and set2Δ strains (51) containing plasmid pRS426 were spotted onto synthetic complete (SC) medium containing 30 μg/ml or 100 μg/ml 6-AU, after normalization at A600. Plates were incubated at 30°C for 2 days. (B) The wild-type strain used in this assay was the same as that used for panel A. JHD1 overexpression strains (JHD1 OE) were generated by transforming a high-copy-number plasmid (2μm ori, URA3) expressing Flag-Jhd1 under the control of the ADH1 promoter. Two independent JHD1 overexpression strains were prepared and assayed as described for panel A. (C) List of phenotypes tested for jhd1Δ. Strains spt4Δ (3), snf2Δ (32), spt7Δ (34), htz1Δ (30), and sir2Δ (42) served as positive controls for the tested phenotypes.
JHD1 overexpression causes a subtle 3′ shift of H3K36me2.
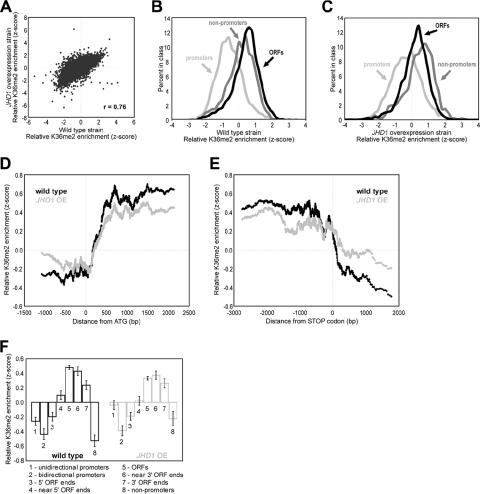
Given that overexpression of JHD1 results in a slight decrease in bulk H3K36me2 and H3K36me3 levels (Fig. 1F), we performed ChIP-chip experiments to examine the genome-wide distribution of H3K36me2 during JHD1 overexpression. To this end, H3K36me2-associated chromatin from wild-type and JHD1 overexpression strains was isolated using an affinity-purified H3K36me2-specific antibody (45). Total genomic DNA was prepared in parallel as a reference. The ChIP and genomic DNA were amplified, fluorescently labeled, and comparatively hybridized to DNA microarrays. Each microarray contained both “low-resolution” probes that spanned the entire genome at ∼800 bp and higher-resolution probes spanning most of chromosome III at ∼200-bp resolution (see Materials and Methods). Data from each microarray were log2 transformed and converted to z-scores by centering the mean and normalizing the variance (see Materials and Methods). A previous ChIP-chip study demonstrated that H3K36me2 was enriched over ORFs, beginning at a stereotypic distance into the transcribed region and continuing 3′ until termination of transcription (36). The H3K36me2 pattern we obtained for this study using the wild-type strain BY4741 matched the pattern described in the prior study, despite using a different yeast strain and antibody.
H3K36me2 ChIP-chip data from the JHD1 overexpression strain correlated highly with data from the wild-type strain (Fig. 5A), revealing ORFs enriched relative to promoters (Fig. 5B). However, H3K36me2 was more enriched in nonpromoter regions (defined as intergenic regions downstream of two convergently transcribed genes) relative to ORFs in the JHD1 overexpression strain (compare Fig. 5C to B). The greater enrichment of nonpromoters raised the possibility that the pattern of H3K36me2 was shifted 3′ along transcriptional units in the JHD1 overexpression strains.
FIG. 5.
JHD1 overexpression promotes a subtle 3′ shift of H3K36me2. (A) Scatter plot of H3K36me2 ChIP enrichment in wild-type yeast versus the JHD1 overexpression strain. (B) Lined histograms of the z-score distribution for SGD-annotated ORFs, promoters, and nonpromoters in the wild-type yeast strain BY4741. (C) Same analysis as for panel B, except this is for the JHD1 overexpression strain. Note the enrichment of nonpromoters. (D) Tiling probes spaced at ∼200 bp along chromosome III (see Materials and Methods) were sorted according to their distance to the closest start codon. A moving average of the data from H3K36me2 ChIPs was then calculated using a window of 100 and step size of 1. Any probe lying within an ORF but calculated to be upstream of its closest start codon or any probe lying within an intergenic region but downstream of a start codon was removed from this analysis. (E) Same analysis as in panel D, except the data were aligned to the translation stop codon of each gene. Any probe lying within an ORF but downstream of its calculated closest stop codon or any probe lying within an intergenic region but upstream of a stop codon was removed from this analysis. (F) Higher-resolution probes that tile chromosome III were assigned to annotations as labeled on the x axis. Annotations were based on the May 2006 version of the SGD. ORF 5′ or 3′ ends indicate probes that contain the start or stop codons, respectively. ORFs near the 5′ or 3′ ends represent the probes adjacent to those containing start or stop codons, respectively. The enrichment level indicates the average for all probes of that type, and the error bars indicate standard errors of the means.
To explore this possibility further, we analyzed our higher-resolution probes, which tile chromosome III at 200-bp resolution (some regions of the chromosome were tiled at 100 bp; see Materials and Methods). To examine the H3K36me2 pattern on the “average” transcription unit, the probes were sorted according to their distance to the nearest translation start codon (Fig. 5D) or translation stop codon (Fig. 5E). The distribution of H3K36me2 upon JHD1 overexpression did not change near translation start codons at the 5′ ends of genes (Fig. 5D). However, when probes were aligned relative to their translation stops (Fig. 5E), the flatter shape of the JHD1 overexpression plot relative to the wild type indicated a more gradual reduction in H3K36me2 after the translation stop codon. This provides evidence for subtle 3′ shift of H3K36me2 upon JHD1 overexpression or, alternatively, a greater abundance of H3K36me2 downstream of coding regions relative to coding regions of genes.
For further evidence of a 3′ shift, H3K36me2 distribution was measured with higher-resolution probes that tile chromosome III. Each probe was assigned to one of eight annotations that reflect their position in the genome with respect to ORFs (Fig. 5F). In the wild type, the peak of H3K36me2 is present in the central region of ORFs, while in the JHD1 overexpression strain, peak levels of H3K36me2 are found in the ORF probes adjacent to the stop codon. Furthermore, higher levels of H3K36me2 were observed in JHD1 overexpression strains at unidirectional promoters (which lie 3′ of one gene) and nonpromoters (3′ of two genes) but not bidirectional promoters (not 3′ of any gene), providing more evidence for a subtle 3′ shift of H3K36me2 upon JHD1 overexpression.
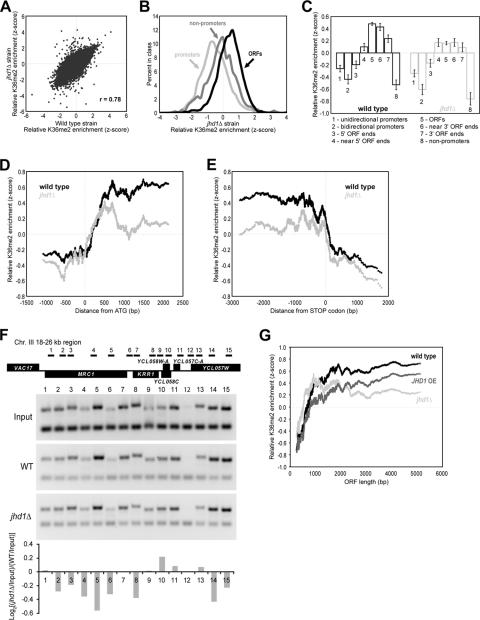
JHD1 knockout changes the stereotypic H3K36me2 distribution, resulting in a more uniform distribution across ORFs.
We also explored the pattern of H3K36me2 distribution in the jhd1Δ strain, despite a lack of detectable change in bulk histone methylation levels (Fig. 1F). We performed H3K36me2 ChIP-chip using extracts derived from the jhd1Δ strain, and we again found the H3K36me2 enrichment was highly correlated to the wild-type data (Fig. 6A). As in the wild type, ORFs were enriched relative to nonpromoters and promoters (Fig. 6B; compare to Fig. 5B). To detect any subtle changes in H3K36me2, we again analyzed our higher-resolution chromosome III probes. When these probes were assigned to one of eight annotations that reflect their position in the genome with respect to ORFs, we found that in contrast to the sharp increase in H3K36me2 observed just within the ORFs in the wild-type strain, in the jhd1Δ strain H3K36me2 was more uniformly distributed between the start and stop codons (Fig. 6C). Furthermore, aligning the higher-resolution probes relative to the translation start or stop site revealed changes in the relative distribution of H3K36me2 within genes (Fig. 6D and E). To validate our microarray observations further, we analyzed an 8-kb region located on chromosome III that exhibited differences in H3K36 methylation profiles after deletion of JHD1. Consistent with the ChIP-chip results presented above, in the jhd1Δ strain the H3K36me2 level was generally decreased within the ORF (Fig. 6F, lanes 1 to 5) and flat or slightly enriched on the 5′ ends of some genes (Fig. 6F, lanes 9 to 13).
FIG. 6.
JHD1 knockout promotes a more uniform distribution of H3K36me2 across ORFs and a subtle 5′ shift. (A) Scatter plot of H3K36me2 ChIP enrichment in wild-type yeast versus the jhd1Δ strain. (B) Same analysis as in Fig. 5B, except that this is for the jhd1Δ strain. Overall, H3K36me2 distributions were very similar to wild type. (C) Same analysis as for Fig. 5F, except that this is for the jhd1Δ strain. Note the more uniform H3K36me2 levels across ORFs. (D) Same analysis as for Fig. 5D, except that this is for the jhd1Δ strain. Note a subtle shift toward the 5′ end in the KO strain and the lower overall difference between enrichment values in ORFs versus upstream regions. (E) Same analysis as for Fig. 5E, except that this is for the jhd1Δ strain. Note the lower relative H3K36me2 enrichment values within the ORF but no evidence for a 3′ spread. (F) Validation of the microarray results by conventional ChIP for H3K36me2 levels across the 18- to 26-kb region on chromosome III. PCR products derived from input DNA and anti- as top bands on the gel pictures. H3K36me2 ChIP DNA from the wild-type and jhd1Δ strains are shown bottom. The amplified band is from a nonspecific intergenic region on chromosome V (ChV) that served as an internal control. The bar graph represents the average of two independent experiments and was generated by analyzing the normalized quantification results from wild-type (WT) and knockout (jhd1Δ) strains by using [log2(jhd1Δ/input)/(WT/input)]. (G) ORFs were sorted by their length, and a moving average (window of 100, step of 1) of enrichment in H3K36me2 ChIPs was plotted.
We sought further confirmation of our interpretation by examining the lower-resolution array probes that cover the entire genome. Because the H3K36me2 mark is deposited at a stereotypic distance from the start of transcription regardless of ORF length and because the mark is deposited from that point to the end of the transcription unit, H3K36me2 enrichment from a wild-type strain appears to increase as a function of ORF length when probes that cover the entire ORF are employed to detect ChIP enrichment (36). We again observed this phenomenon in the wild-type strains using our independent data (Fig. 6G). As was illustrated in Fig. 6 of reference 36, “the prediction of higher ratios for longer ORFs is based entirely on the relative proportion of the arrayed element that is dimethylated, not the absolute length of the arrayed element or genomic feature per se. For example, if the entire ORF were modified, or if the distance at which the modification began from transcriptional initiation were proportional to ORF length, equal ratios would be obtained for long and short ORFs.” We also plotted this relationship in the jhd1Δ strain (Fig. 6G). Apparent H3K36me2 enrichment is observed at smaller ORFs in the jhd1Δ strain and levels out more quickly with increasing ORF length (Fig. 6G). This is consistent with a subtle 5′ shift in the point at which H3K36me2 begins to be found along genes.
Furthermore, a subtle 3′ shift in the point at which H3K36me2 begins and ends along genes is predicted in the JHD1 overexpression strain. By the metric in Fig. 6G, one would expect the relationship between ORF size and apparent H3K36me2 enrichment to begin at longer ORFs and to proceed with a more gradual slope, which is in fact what we observe in the JHD1 overexpression strain (Fig. 6G). These differences between the wild type and strains with an altered JHD1 status provide additional evidence for Jhd1 function in maintaining a precise pattern of H3K36me2 distribution along genes.
DISCUSSION
Recent studies have demonstrated that histone methylation can be dynamically regulated through active demethylation (40, 45). Identification of the evolutionarily conserved JmjC domain as a signature motif for histone demethylases raised the possibility that active histone demethylation also occurs in single-celled eukaryotes. To identify potential histone demethylases in S. cerevisiae, we analyzed all five JmjC domain-containing proteins in S. cerevisiae using an in vitro formaldehyde release assay (45). Here we report the first characterization of the histone demethylase Jhd1 of S. cerevisiae. We demonstrate that, similar to its mammalian homolog, Jhd1 specifically targets H3K36 for demethylation (Fig. 1). Targeted deletions and mutations of Jdh1 indicate that both the JmjC domain and its adjacent sequences are required for the enzymatic activity of the Jhd1 protein (Fig. 2). Although phenotypic screening failed to reveal the nature of Jhd1's functions, chromatin immunoprecipitation coupled with microarray analysis suggests that Jhd1-mediated histone demethylation functions to maintain a precise H3K36me2 pattern along transcription units (Fig. 5 and 6). The lack of an overt phenotype in jhd1 mutants may indicate that Jhd1-mediated H3K36 demethylation is regulated by a redundant pathway. Alternatively, it is possible that Jhd1-mediated H3K36 demethylation only occurs in a particular biological process yet to be identified.
In S. cerevisiae, histone H3K4 methylation and H3K36 methylation are mediated by Set1 and Set2, respectively (6, 37, 43). Previous studies indicate that Set1 and Set2 are recruited to gene coding regions by the PAF complex and play important roles in transcription initiation and elongation (21, 22, 24, 25, 33, 50). During transcriptional initiation, the Set1 complex is first recruited to the promoters where it trimethylates lysine 4 of histone H3. Set2 is then recruited by the PAF complex and travels with RNA polymerase II to methylate H3K36 during the elongation phase of transcription (13, 16). The data we present suggest that H3K36me2 levels are subject to another layer of regulation by Jhd1-mediated demethylation.
Limitations of our methods.
While the in vitro radioactive formaldehyde release assay used in our screen is very sensitive, it has its limitations. For example, the methylation state of the substrates is limited by the methyltransferases used to generate the substrates. Given that histone demethylases identified so far exhibit both site and methylation-state specificity (20, 40, 52), it is possible that some of the JmjC domain-containing proteins that did not exhibit enzymatic activity in this study could be active enzymes when alternative substrates are used. Indeed, we have demonstrated using the radioactive formaldehyde release assay that another yeast JmjC domain-containing protein, Yjr119c/Jhd2, cannot demethylate substrates generated by the H3K4me1 methyltransferase hSET7 (23, 48, 49) but does have activity towards H3K4me3 substrates (26). A second limitation of the assay used in this study is the amount of enzyme obtained from the immunoprecipitation (Fig. 1). Although the immunoprecipitated proteins can be detected by Western blotting, the amounts may not be sufficient to catalyze detectable histone demethylation. Nevertheless, our study reveals Jhd1 as an active S. cerevisiae H3K36 demethylase.
We also stress that the results from our ChIP-chip studies reveal relative redistributions of H3K36me2 in response to changes in Jhd1 status, and these changes are most easily observed when many genes are analyzed in aggregate. Nevertheless, the relative changes we observed seem robust, since they were faithfully reproduced among three biological replicates for each strain. We caution against interpreting the data in terms of absolute H3K36me2 levels. For example, enrichment of H3K36me2 is apparently lower throughout the ORFs in JHD1 overexpression strains (Fig. 5D and E). However, because all of the measurements are relative, greater amounts of H3K36me2 elsewhere in the genome could cause an apparent decrease in ORF H3K36me2, even if the absolute amount of H3K36me2 in ORF chromatin remained constant.
A speculative model for Jhd1 function.
In addition to being methylated by Set2, a recent study has demonstrated that H3K36 is acetylated by Gcn5p (31), raising the intriguing possibility that Jhd1 plays an important role in an H3K36 “switch” from methylation to acetylation. Such a modification “switch” is also supported by the results of two other recent studies. Proteome-wide analysis in yeast has shown that the PHD finger domain of Jhd1 is capable of specific binding to H3K4me3 but shows much less binding to H3K4me2 and no binding at all to H3K4me1, H3K36me1-3, or H3K79me1-3 (39). The H3K4me3 mark is predominantly found at the 5′ end of transcription units where the transition between H3K36ac and H3K36me occurs. It is possible that the PHD domain of Jhd1 might facilitate recruitment of Jhd1 to the 5′ end of genes by binding to the H3K4me3 mark, where it maintains low H3K36me2 levels. Furthermore, Gcn5p, which is responsible for H3K36ac, has been demonstrated to promote trimethylation of H3K4 during transcription induction (14). Collectively, our study and other recent studies suggest an intriguing but speculative model in which the transition from H3K36 acetylation to H3K36 methylation in 5′ genic regions may proceed as follows: (i) Gcn5p as part of the SAGA complex is recruited to promoters; (ii) Gcn5p stimulates H4K4me3 by Set1; (iii) Jhd1 is recruited to H3K4me3 to ensure a demethylated state at H3K36; (iv) Gcn5p acetylates H3K36 in the promoter region; and (v) methylation of H3K36 proceeds by association of Set2 with RNA polymerase II during transcriptional elongation.
This model would be consistent with the more-5′ “early start” of H3K36me2 observed in the JHD1 knockout and with the “delayed start” or more-3′ distribution of H3K36me2 observed in the JHD1 overexpression strains. In the knockout, H3K36 would fail to be demethylated at promoters, resulting in an apparent 5′ shift in H3K36me2. Thus, the apparent 5′ shift is actually caused by failure to remove methyl groups from H3K36 rather than appearance of “new” H3K36me2. In the overexpression strain, we speculate that Jhd1 is overabundant and fails to be restricted precisely within the 5′ end of transcription units, causing demethylation to extend further into the gene. This results in an apparent 3′ shift in H3K36me2 upon Jhd1 overexpression. We speculate that the loss of methylation at 5′ ends of genes is actual and that, due to the relative nature of our measurements, this decrease causes 3′ regions to appear more enriched in comparison. This would explain how an enzyme that we postulate to act only at 5′ ends of genes could affect measured methylation in both 5′ and 3′ regions.
Based on this model, Jhd1 could play a role in transcription initiation by indirectly facilitating hyperacetylation of promoter nucleosomes. Future studies will allow this model to be tested and the function of Jhd1 in transcription to be revealed.
Acknowledgments
We thank Brian Strahl for providing the BY4741, set2Δ, spt4Δ, rtf1Δ, snf2Δ, spt7Δ, htz1Δ, and sir2Δ strains and for his generous help.
This work was supported by NIH grants GM68804 to Y.Z. and GM072518 to J.D.L. Y.Z. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Published ahead of print on 30 April 2007.
REFERENCES
- 1.Ayoub, N., K. Noma, S. Isaac, T. Kahan, S. I. Grewal, and A. Cohen. 2003. A novel jmjC domain protein modulates heterochromatization in fission yeast. Mol. Cell. Biol. 23:4356-4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bannister, A. J., R. Schneider, F. A. Myers, A. W. Thorne, C. Crane-Robinson, and T. Kouzarides. 2005. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J. Biol. Chem. 280:17732-17736. [DOI] [PubMed] [Google Scholar]
- 3.Basrai, M. A., J. Kingsbury, D. Koshland, F. Spencer, and P. Hieter. 1996. Faithful chromosome transmission requires Spt4p, a putative regulator of chromatin structure in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:2838-2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernstein, B. E., E. L. Humphrey, R. L. Erlich, R. Schneider, P. Bouman, J. S. Liu, T. Kouzarides, and S. L. Schreiber. 2002. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl. Acad. Sci. USA 99:8695-8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bohlander, S. K., R. Espinosa III, M. M. Le Beau, J. D. Rowley, and M. O. Diaz. 1992. A method for the rapid sequence-independent amplification of microdissected chromosomal material. Genomics 13:1322-1324. [DOI] [PubMed] [Google Scholar]
- 6.Briggs, S. D., M. Bryk, B. D. Strahl, W. L. Cheung, J. K. Davie, S. Y. Dent, F. Winston, and C. D. Allis. 2001. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 15:3286-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, Z., J. Zang, J. Whetstine, X. Hong, F. Davrazou, T. G. Kutateladze, M. Simpson, Q. Mao, C. H. Pan, S. Dai, J. Hagman, K. Hansen, Y. Shi, and G. Zhang. 2006. Structural insights into histone demethylation by JMJD2 family members. Cell 125:691-702. [DOI] [PubMed] [Google Scholar]
- 8.Clissold, P. M., and C. P. Ponting. 2001. JmjC: cupin metalloenzyme-like domains in jumonji, hairless and phospholipase A2β. Trends Biochem. Sci. 26:7-9. [DOI] [PubMed] [Google Scholar]
- 9.Cloos, P. A., J. Christensen, K. Agger, A. Maiolica, J. Rappsilber, T. Antal, K. H. Hansen, and K. Helin. 2006. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature 442:307-311. [DOI] [PubMed] [Google Scholar]
- 10.Desmoucelles, C., B. Pinson, C. Saint-Marc, and B. Daignan-Fornier. 2002. Screening the yeast “disruptome” for mutants affecting resistance to the immunosuppressive drug, mycophenolic acid. J. Biol. Chem. 277:27036-27044. [DOI] [PubMed] [Google Scholar]
- 11.Fang, J., H. Wang, and Y. Zhang. 2004. Purification of histone methyltransferases from HeLa cells. Methods Enzymol. 377:213-226. [DOI] [PubMed] [Google Scholar]
- 12.Gelbart, M. E., T. Rechsteiner, T. J. Richmond, and T. Tsukiyama. 2001. Interactions of Isw2 chromatin remodeling complex with nucleosomal arrays: analyses using recombinant yeast histones and immobilized templates. Mol. Cell. Biol. 21:2098-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerber, M., and A. Shilatifard. 2003. Transcriptional elongation by RNA polymerase II and histone methylation. J. Biol. Chem. 278:26303-26306. [DOI] [PubMed] [Google Scholar]
- 14.Govind, C. K., F. Zhang, H. Qiu, K. Hofmeyer, and A. G. Hinnebusch. 2007. Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Mol. Cell 25:31-42. [DOI] [PubMed] [Google Scholar]
- 15.Hampsey, M. 1997. A review of phenotypes in Saccharomyces cerevisiae. Yeast 13:1099-1133. [DOI] [PubMed] [Google Scholar]
- 16.Hampsey, M., and D. Reinberg. 2003. Tails of intrigue: phosphorylation of RNA polymerase II mediates histone methylation. Cell 113:429-432. [DOI] [PubMed] [Google Scholar]
- 17.Iyer, V. R., C. E. Horak, C. S. Scafe, D. Botstein, M. Snyder, and P. O. Brown. 2001. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature 409:533-538. [DOI] [PubMed] [Google Scholar]
- 18.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 19.Klose, R. J., E. M. Kallin, and Y. Zhang. 2006. JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 7:715-727. [DOI] [PubMed] [Google Scholar]
- 20.Klose, R. J., K. Yamane, Y. Bae, D. Zhang, H. Erdjument-Bromage, P. Tempst, J. Wong, and Y. Zhang. 2006. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature 442:312-316. [DOI] [PubMed] [Google Scholar]
- 21.Krogan, N. J., J. Dover, A. Wood, J. Schneider, J. Heidt, M. A. Boateng, K. Dean, O. W. Ryan, A. Golshani, M. Johnston, J. F. Greenblatt, and A. Shilatifard. 2003. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell 11:721-729. [DOI] [PubMed] [Google Scholar]
- 22.Krogan, N. J., M. Kim, A. Tong, A. Golshani, G. Cagney, V. Canadien, D. P. Richards, B. K. Beattie, A. Emili, C. Boone, A. Shilatifard, S. Buratowski, and J. Greenblatt. 2003. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 23:4207-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwon, T., J. H. Chang, E. Kwak, C. W. Lee, A. Joachimiak, Y. C. Kim, J. Lee, and Y. Cho. 2003. Mechanism of histone lysine methyl transfer revealed by the structure of SET7/9-AdoMet. EMBO J. 22:292-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, B., L. Howe, S. Anderson, J. R. Yates III, and J. L. Workman. 2003. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 278:8897-8903. [DOI] [PubMed] [Google Scholar]
- 25.Li, J., D. Moazed, and S. P. Gygi. 2002. Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. J. Biol. Chem. 277:49383-49388. [DOI] [PubMed] [Google Scholar]
- 26.Liang, G., R. J. Klose, K. E. Gardner, and Y. Zhang. 2007. Yeast Jhd2p is a histone H3 Lys4 trimethyl demethylase. Nat. Struct. Mol. Biol. 14:243-245. [DOI] [PubMed] [Google Scholar]
- 27.Liu, C. L., T. Kaplan, M. Kim, S. Buratowski, S. L. Schreiber, N. Friedman, and O. J. Rando. 2005. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 3:e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Margueron, R., P. Trojer, and D. Reinberg. 2005. The key to development: interpreting the histone code? Curr. Opin. Genet. Dev. 15:163-176. [DOI] [PubMed] [Google Scholar]
- 29.Martin, C., and Y. Zhang. 2005. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 6:838-849. [DOI] [PubMed] [Google Scholar]
- 30.Mizuguchi, G., X. Shen, J. Landry, W. H. Wu, S. Sen, and C. Wu. 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303:343-348. [DOI] [PubMed] [Google Scholar]
- 31.Morris, S. A., B. Rao, B. A. Garcia, S. B. Hake, R. L. Diaz, J. Shabanowitz, D. F. Hunt, C. D. Allis, J. D. Lieb, and B. D. Strahl. 2006. Identification of histone H3 lysine 36 acetylation as a highly conserved histone modification. J. Biol. Chem. 282:7632-7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neigeborn, L., and M. Carlson. 1984. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics 108:845-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ng, H. H., F. Robert, R. A. Young, and K. Struhl. 2003. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11:709-719. [DOI] [PubMed] [Google Scholar]
- 34.Patton-Vogt, J. L., and S. A. Henry. 1998. GIT1, a gene encoding a novel transporter for glycerophosphoinositol in Saccharomyces cerevisiae. Genetics 149:1707-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pokholok, D. K., C. T. Harbison, S. Levine, M. Cole, N. M. Hannett, T. I. Lee, G. W. Bell, K. Walker, P. A. Rolfe, E. Herbolsheimer, J. Zeitlinger, F. Lewitter, D. K. Gifford, and R. A. Young. 2005. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122:517-527. [DOI] [PubMed] [Google Scholar]
- 36.Rao, B., Y. Shibata, B. D. Strahl, and J. D. Lieb. 2005. Dimethylation of histone H3 at lysine 36 demarcates regulatory and nonregulatory chromatin genome-wide. Mol. Cell. Biol. 25:9447-9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roguev, A., D. Schaft, A. Shevchenko, W. W. Pijnappel, M. Wilm, R. Aasland, and A. F. Stewart. 2001. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 20:7137-7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Santos-Rosa, H., R. Schneider, A. J. Bannister, J. Sherriff, B. E. Bernstein, N. C. Emre, S. L. Schreiber, J. Mellor, and T. Kouzarides. 2002. Active genes are tri-methylated at K4 of histone H3. Nature 419:407-411. [DOI] [PubMed] [Google Scholar]
- 39.Shi, X., I. Kachirskaia, K. L. Walter, J. H. Kuo, A. Lake, F. Davrazou, S. M. Chan, D. G. Martin, I. M. Fingerman, S. D. Briggs, L. Howe, P. J. Utz, T. G. Kutateladze, A. A. Lugovskoy, M. T. Bedford, and O. Gozani. 2006. Proteome-wide analysis in S. cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J. Biol. Chem. 282:2450-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi, Y., F. Lan, C. Matson, P. Mulligan, J. R. Whetstine, P. A. Cole, R. A. Casero, and Y. Shi. 2004. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119:941-953. [DOI] [PubMed] [Google Scholar]
- 41.Siegel, L. M., and K. J. Monty. 1966. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochim. Biophys. Acta 112:346-362. [DOI] [PubMed] [Google Scholar]
- 42.Smith, J. S., C. B. Brachmann, I. Celic, M. A. Kenna, S. Muhammad, V. J. Starai, J. L. Avalos, J. C. Escalante-Semerena, C. Grubmeyer, C. Wolberger, and J. D. Boeke. 2000. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl. Acad. Sci. USA 97:6658-6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strahl, B. D., P. A. Grant, S. D. Briggs, Z. W. Sun, J. R. Bone, J. A. Caldwell, S. Mollah, R. G. Cook, J. Shabanowitz, D. F. Hunt, and C. D. Allis. 2002. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol. Cell. Biol. 22:1298-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trewick, S. C., P. J. McLaughlin, and R. C. Allshire. 2005. Methylation: lost in hydroxylation? EMBO Rep. 6:315-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsukada, Y., J. Fang, H. Erdjument-Bromage, M. E. Warren, C. H. Borchers, P. Tempst, and Y. Zhang. 2006. Histone demethylation by a family of JmjC domain-containing proteins. Nature 439:811-816. [DOI] [PubMed] [Google Scholar]
- 46.Wang, H., R. Cao, L. Xia, H. Erdjument-Bromage, C. Borchers, P. Tempst, and Y. Zhang. 2001. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol. Cell 8:1207-1217. [DOI] [PubMed] [Google Scholar]
- 47.Whetstine, J. R., A. Nottke, F. Lan, M. Huarte, S. Smolikov, Z. Chen, E. Spooner, E. Li, G. Zhang, M. Colaiacovo, and Y. Shi. 2006. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 125:467-481. [DOI] [PubMed] [Google Scholar]
- 48.Wilson, J. R., C. Jing, P. A. Walker, S. R. Martin, S. A. Howell, G. M. Blackburn, S. J. Gamblin, and B. Xiao. 2002. Crystal structure and functional analysis of the histone methyltransferase SET7/9. Cell 111:105-115. [DOI] [PubMed] [Google Scholar]
- 49.Xiao, B., C. Jing, J. R. Wilson, P. A. Walker, N. Vasisht, G. Kelly, S. Howell, I. A. Taylor, G. M. Blackburn, and S. J. Gamblin. 2003. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature 421:652-656. [DOI] [PubMed] [Google Scholar]
- 50.Xiao, T., H. Hall, K. O. Kizer, Y. Shibata, M. C. Hall, C. H. Borchers, and B. D. Strahl. 2003. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev. 17:654-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiao, T., C. F. Kao, N. J. Krogan, Z. W. Sun, J. F. Greenblatt, M. A. Osley, and B. D. Strahl. 2005. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol. Cell. Biol. 25:637-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamane, K., C. Toumazou, Y. Tsukada, H. Erdjument-Bromage, P. Tempst, J. Wong, and Y. Zhang. 2006. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 125:483-495. [DOI] [PubMed] [Google Scholar]