Abstract
The pharmacological effect of morphine as a painkiller is mediated mainly via the mu opioid receptor (MOR) and is dependent on the number of MORs in the cell surface membrane. While several studies have reported that the MOR gene is regulated by various cis- and trans-acting factors, many questions remain unanswered regarding in vivo regulation. The present study shows that epigenetic silencing and activation of the MOR gene are achieved through coordinated regulation at both the histone and DNA levels. In P19 mouse embryonal carcinoma cells, expression of the MOR was greatly increased after neuronal differentiation. MOR expression could also be induced by a demethylating agent (5′-aza-2′-deoxycytidine) or histone deacetylase inhibitors in the P19 cells, suggesting involvement of DNA methylation and histone deacetylation for MOR gene silencing. Analysis of CpG DNA methylation revealed that the proximal promoter region was unmethylated in differentiated cells compared to its hypermethylation in undifferentiated cells. In contrast, the methylation of other regions was not changed in either cell type. Similar methylation patterns were observed in the mouse brain. In vitro methylation of the MOR promoters suppressed promoter activity in the reporter assay. Upon differentiation, the in vivo interaction of MeCP2 was reduced in the MOR promoter region, coincident with histone modifications that are relevant to active transcription. When MeCP2 was disrupted using MeCP2 small interfering RNA, the endogenous MOR gene was increased. These data suggest that DNA methylation is closely linked to the MeCP2-mediated chromatin structure of the MOR gene. Here, we propose that an epigenetic mechanism consisting of DNA methylation and chromatin modification underlies the cell stage-specific mechanism of MOR gene expression.
Opioids exert their pharmacological and physiological effects through binding to their endogenous receptors. Three types of opioid receptors, mu (μ), delta (δ), and kappa (κ), all belonging to the G-protein-coupled receptor superfamily, have been cloned. Upon agonist binding, these receptors couple to G proteins and affect several signal transduction pathways thought to mediate a broad range of functions and pharmacological effects of endogenous and exogenous opioids (51). Previous studies suggested that the μ opioid receptor (MOR) plays a key role in mediating the major clinical effects of analgesics, such as morphine, as well as the development of tolerance and physical dependence upon prolonged administration (39). MOR is mainly expressed in the central nervous system, with densities varying greatly in different regions, which can display different functional roles (55). During mouse embryonic development, the MOR message was specifically observed as early as embryonic day 8.5 (E8.5) using the reverse transcription (RT)-PCR method (44). In contrast, MOR transcripts were detected only beginning at E12 using the radioligand binding method (70) and at E10.5 by in situ hybridization (85). Transcript levels gradually increased throughout embryonic and postnatal stages, reaching a plateau at adulthood (44). To achieve its unique expression pattern spatially and temporally, the expression of MOR must be tightly regulated.
In mammals, DNA methylation and histone modifications represent the major epigenetic mechanisms implicated in the regulation of gene transcription. For instance, DNA methylation is a prominent feature of vertebrate genomes. This methylation occurs predominantly at cytosine residues in cytosine-guanine dinucleotides (CpGs) (25). Cell-type-specific DNA methylation patterning is one of the epigenetic events generating diverse cell types in the body (73). Methylation of DNA is essential for mammalian development (52) and is associated with gene silencing in conjunction with histone core modifications, probably through chromatin remodeling (3, 7, 17, 18, 36, 56). Several findings support the premise that hypomethylation of the DNA surrounding the promoter region is a prerequisite for gene activation, whereas heavy methylation leads to gene silencing (31). There are a number of ways in which DNA methylation can repress transcription. Many of the trans-acting factors known to bind to sequences containing CpG dinucleotides do not bind when the CpG doublets are methylated (82). Alternatively, methyl-CpG-binding proteins (MBPs), such as MBP 2 (MeCP2), bind preferentially to methylated DNA and directly repress transcription, inhibit the binding of other trans factors, structurally modify the DNA, or recruit corepressor complexes (19, 21, 62).
The assembly of higher-order chromatin structure has been linked to the covalent modification of histone tails. The combinatorial nature of histone N-terminal modifications, or the histone code, represents an additional pathway of epigenetic regulation and considerably extends the information potential of the genetic code (35). For instance, hyperacetylation of the lysine residues of H3 and H4 histones is generally associated with the promoters of actively transcribed genes, whereas hypoacetylated histones have been correlated with gene silencing (74). Intriguingly, the lysine residues on histones can be acetylated and methylated. For example, histone H3 lysine 4 methylation has been correlated with active gene expression (83), whereas H3 lysine 9 methylation has been linked to gene silencing and the assembly of heterochromatin (50). It is now appreciated that DNA methylation pathways and the histone code are functionally interactive. Through the binding of MeCP2 to 5-methyl-CpG, dinucleotides can recruit transcriptional corepressors with histone deacetylase (HDAC) activity, providing a link between DNA methylation and histone deacetylation. MeCP2 has also been shown to associate with histone H3 lysine 9 methyltransferase activity, providing a mechanism for targeting repressive histone methylation to DNA-methylated promoters (19).
Embryonal carcinoma (EC) P19 cells are a murine cell line derived from teratocarcinomas induced in C3H/HC mice and are one of the best-characterized pluripotent EC cell lines (57). These cells are capable of differentiating into a variety of tissues when injected into healthy embryos (58). P19 cells treated with retinoic acid (RA) can differentiate into different cell types, including neurons and astroglia, which are normally derived from embryonic neuroectoderm. Therefore, P19 cells are commonly used as an in vitro model system for studying mammalian developmental processes, such as cell differentiation of neurons and muscle cells (59). MORs are not expressed in undifferentiated P19 cells, but expression is greatly increased after neuronal differentiation (10), suggesting that MOR gene expression is associated with cell differentiation or neural development. Thus, the P19 cell system provides an excellent model to study the mechanisms that regulate MOR gene expression temporally, as it resembles the in vivo transcriptional regulation of the MOR gene.
In this study, we present evidence that hypermethylation of CpG dinucleotides, along with histone modifications at the MOR promoter, is mechanistically linked to the lack of MOR mRNA induction in mouse P19 cells. In addition, our findings suggest that differential DNA methylation of undifferentiated and differentiated P19 cells at the proximal promoter (PP) contributes to the marked difference in MOR inducibility in both normal P19 and differentiated P19 cell types. Collectively, this work defines roles for DNA- and chromatin-based mechanisms in the control of mouse MOR gene expression.
MATERIALS AND METHODS
Cell culture.
P19 cells were purchased from ATCC and cultured in α-minimal essential medium containing 7.5% newborn calf serum and 2.5% fetal calf serum at 37°C in a humidified atmosphere of 5% CO2. The procedures to differentiate P19 cells have been described previously (10). Briefly, cells were aggregated in petri dishes in the presence of 0.5 μM RA for 4 days. After aggregation, the cells were trypsinized and plated in tissue culture grade flasks in a chemically defined medium (N2) at a density of 5 × 106 cells/10-cm dish. Cells were harvested at different time points for protein and total-RNA purification. For cytosine arabinoside-treated P19 cultures, cytosine arabinoside was added to the culture medium 24 h after the cells were plated to a final concentration of 10 μM and removed 72 h after plating. For 5′-aza-2′-deoxycytidine (5-aza-dC) (Sigma) treatment experiments, cells were split to low density (105 cells/well in six-well culture plates) 24 h before treatment. The cells were then treated with 5-aza-dC (0.5, 1, 2, or 5 μM) or mock treated with the same volume (2.5 μl) of dimethyl sulfoxide for 72 h, and the medium was changed every 24 h. Cells were harvested on day 4 for RNA and protein determination and analysis of the methylation status. For trichostatin A (TSA) (Sigma) and valproic acid (VPA) (2-propylpentanoic acid; Sigma) treatments, cells were split to a density of 5 × 105 cells/well in six-well culture plates 24 h before treatment. The cells were then treated with TSA (5 to 50 nM) or VPA (1 to 10 mM) or mock treated with the same volume of ethanol for 6 or 24 h and harvested for RNA determinations.
RT-PCR and real-time quantitative RT-PCR (qRT-PCR).
Total RNA was isolated according to the supplier's protocol (TRI Reagent; Molecular Research Center) and analyzed by RT-PCR using MOR gene-specific primers (12). RT-PCR was performed using the QIAGEN OneStep RT-PCR Kit and MOR PCR primers (mMOR-S1 and mMOR-AS1) (Table 1). A similar reaction was carried out using primers for β-actin (9) as an internal control, except the number of cycles was reduced to 20. Primers for δ (DOR) and κ (KOR) opioid receptors were as described previously (67, 80). Primers for N-cadherin, βIII-tubulin, and glial fibrillary acidic protein (GFAP) are described in Table 1. The PCR products were electophoresed in a 2% agarose gel and quantified with ImageQuant version 5.2 (Amersham). The DNA sequences of PCR products were confirmed by sequencing.
TABLE 1.
Primers used in this study
| Name | Primer (5′→3′) | Locationa | Note |
|---|---|---|---|
| mMOR-S1 | CCCTCTATTCTATCGTGTGTGT | +218 | Located at exon 1, P2-Se |
| mMOR-AS1 | AGAAGAGAGGATCCAGTTGCA | +568 | Located at exon 2, P2-ASe |
| MS-630 | GTGGGTAAAGGATAATATTAATAATTTT | −630 | MSPb |
| MAS + 65 | CAACTTACAAAAACTAAAAAATCAAAAC | +65 | MSP |
| MS-1754 | GAGAAGAAGATTGAGGTAAAGTAGTAT | −1754 | MSP |
| MAS-927 | CATACCAAATCTACTCTCCTAAACCTAC | −927 | MSP |
| MS + 19 | GTAGTAAGTATTTAGAATTATGGATAGTAG | +19 | MSP |
| MAS + 352 | CTCTATCACTCATAAAAAAACTTTATC | +352c | MSP |
| S-1318d | AAACTTTCTACAACCAAATGGGAG | −1318 | ChIP primer a |
| AS-1099 | CCTTAAGCAAGATCCTTGAGGAGC | −1099 | ChIP primer a |
| S-1091 | ACACTCAAAAGAGCTGTGGAGTTTC | −1091 | ChIP primer b |
| AS-947 | GGACCTGCTTAGGAAGATTTATC | −947 | ChIP primer b |
| S-731 | CTTTGAACAGGTTTGTGGGGTTG | −731 | ChIP primer c |
| AS-623 | TTACCCACATCCCCATATCTGA | −623 | ChIP primer c |
| S-565 | AAGAGTGTTGAGGTATAAGGGT | −565 | ChIP primer d |
| AS-447 | GAGGGTGGGAAGAGAGACTCTAAG | −447 | ChIP primer d, D1-ASe |
| S-342 | CACAATCCACTCCTTCTCTCTCCTC | −342 | ChIP primer e |
| AS-229 | AGGCGCATCCTTAGCATCCCCA | −229 | ChIP primer e |
| md2-S | GCACATGAAACAGGCTTCTTTCAC | −789 | D1-Se |
| md3-S | CCAGTCCTAATTAATTGCATATGG | −602 | D2-Se |
| mp1-AS | CATCCCCAAAGCGCCACTCTCTGAG | −243 | D2-ASe |
| mp2-S | CTCCGTGTACTTCTAAGGTGGGAG | −230 | P1-Se |
| mtm1-AS | AGAATAGAGGGCCATGATGGTGAT | +228 | P1-ASe |
| H19-s | GCAGAAGGCAGGACACCTATG | 1072 | ChIP primer H19 |
| H19-as | AGCGCAGCAATTTGGTCTTTC | 1201 | gbf AY849916 |
| actb-S | ATATCGCTGCGCTGGTCGTC | +10 | ChIP primer β-actin |
| actb-AS | TCACTTACCTGGTGCCTAGGG | +131 | NC_000071.4 |
| mMeCP2-S | GAGCGGCACTGGGAGACC | +761 | gb NM_010788 |
| mMeCP2-AS | GAGTGATGGTGGTGATGATGG | +1300 | |
| mMBD2-S | GGATGGAAGAAGGAGGAAGTG | +710 | gb NM_010773 |
| mMBD2-AS | CTAAGTCCTTGTAGCCTCTTCTC | +1110 | |
| βIII-tubulin-S | ATGTCTATGAAGGAGGTGGACG | +1005 | gb BC088749 |
| βIII-tubulin-AS | TCTCGGCCTCGGTGAACTC | +1263 | |
| N-cadherin-S | ATGCGGATGATCCAAATGCCC | +1210 | Reference 20 |
| N-cadherin-AS | CATGGCAGTAAACTCTGGAGG | +1487 | |
| GFAP-s | TTCTCCTTGTCTCGAATGAC | +114 | gb NM_010277.2 |
| GFAP-as | GGTTTCATCTTGGAGCTTCT | +482 |
The location (starting at the 5′ end) of each primer is designated relative to +1 (as the ATG start codon).
M as the first capital letter of a primer name indicates a primer specific for methylated (methylation-specific PCR [MSP]) MOR DNA.
The base distance passing exon 1 into intron 1 was calculated in genomic MOR DNA from the start codon at +1.
S and AS in the primer name indicate sense and antisense primers, respectively.
The primers under alternative names were used for DP- and PP-mediated transcripts in Fig. 4.
gb indicates GenBank accession number.
For real-time qRT-PCR, 5 μg of total RNA was treated with DNase I and reverse transcribed using reverse transcriptase (Roche) and primers combined with oligo(dT) and random hexamer. One-fortieth of this reaction mixture was used for real-time qRT-PCR analysis of gene expression, using SYBR Green I dye chemistry. Real-time qRT-PCR was performed in an iCycler (Bio-Rad) using SYBR Green (Quantitect SYBR Green PCR kit; QIAGEN). To calculate relative mRNA gene expression, amplification curves of a test sample and standard samples that contained 101 to 108 molecules of the gene of interest (e.g., the MOR expression plasmid pmMuEG, constructed in our laboratory) were monitored, and the number of target molecules in the test sample was analyzed using qCalculator version 1.0 software (23) (http://www.gene-quantification.de/download.html#qcalculator) based on the mathematical model of Pfaffl (68). The number of target molecules was normalized against that obtained for β-actin, used as an internal control. Primer sequences are shown in Table 1. The specificities of RT-PCR primers were determined using a melting curve after the amplification to show that only a single species of qRT-PCR product resulted from the reaction. Single PCR products were also verified on an agarose gel.
Northern blot analysis.
Total RNA was obtained from mouse P19 cells with the TRI Reagent according to the manufacturer's instructions (Molecular Research Center). The Northern blot analysis was performed as described in the manufacturer's manual (NorthernMax Kit; Ambion). In brief, 20 μg of total RNA per lane was loaded in a 1% formaldehyde-agarose gel and transferred to a Hybond-N+ membrane (Amersham). The membrane was hybridized with 32P-labeled probes produced with the Random Labeling Kit (Amersham) using [α-32P]dCTP. Probe DNA for the neuron-specific gene N-cadherin was generated using the gene-specific PCR primers listed in Table 1 after the products were confirmed by sequence analysis. The membrane was scanned using a PhosphorImager (Storm 840; Molecular Dynamics).
Western blot analysis.
For Western blotting, equal amounts of total proteins were separated on a NuPAGE Novex 3 to 8% Tris-acetate gel (Invitrogen) and transferred to an Immobilon-P (polyvinylidene difluoride; Millipore) membrane. The proteins were detected with anti-MeCP2 (kindly provided by Weidong Wang, NIH), anti-N-cadherin (33-3900; Zymed Lab), anti-βIII-tubulin (PRB-435P; Covance), and anti-β-actin (4967; Cell Signaling) antibodies using ECF substrate (Amersham). The membrane was scanned using a PhosphorImager (Storm 840; Molecular Dynamics).
In vitro methylation of reporter plasmid and reporter gene assays.
In vitro methylation of reporter plasmids was carried out as reported previously (79, 80). Briefly, methylases SssI and HpaII were used to methylate MOR promoter/luciferase reporter constructs following the recommendations of the manufacturer (New England Biolabs). Complete methylation was determined by digesting the DNA constructs with the methylation-sensitive restriction enzyme HpaII (New England Biolabs) and running the products on an agarose gel. Only DNA that was completely methylated was used. The construction of all luciferase fusion plasmids (pL450, pL1.3k, and pLup) used in this study has been described previously (46). P19 cells were plated 24 h prior to transfection at a density of 3 × 105 cells/well in six-well culture plates. Transfection was carried out using the Effectene transfection reagent (QIAGEN) as described by the manufacturer. Cells were washed and lysed with lysis buffer (Promega) 48 h after transfection. To correct differences in transfection efficiency, a one-fifth molar ratio of a pCH110 plasmid (Amersham Biosciences) containing the β-galactosidase gene under the simian virus 40 promoter was included in the transfection to normalize values. The luciferase and galactosidase activities of each lysate were determined as described by the manufacturers (Promega and Tropix, respectively). Each normalized value represents the average of at least three independent determinations, and the error bars represent standard errors of the mean.
Methylation analysis.
Genomic DNA from the P19 cells was isolated using the Wizard Genomic DNA Purification Kit (Promega) and linearized with the restriction enzyme EcoRV. Bisulfite treatment of DNA was carried out according to the manufacturer's recommendations (EZ DNA Methylation-Gold Kit; Zymo Research). The resulting bisulfite-modified DNA was amplified by PCR. The primer sequences for the amplification of MOR (i.e., bisulfite-sequencing primers) are listed in Table 1. The PCR conditions were as follows: 94°C for 2 min, followed by 35 cycles of 94°C for 30 seconds, 60°C for 30 seconds, and 72°C for 1 min, and finally 30 min at 72°C. After PCR amplification, the PCR products were purified using a gel extraction kit (QIAGEN) and cloned into the pCR2.1-TOPO vector (Invitrogen) according to the manufacturer's instructions. Twenty clones containing an insert of the correct size from each set of P19 cells were randomly chosen for DNA sequencing. The sequencing reactions were performed with Applied Biosystems model 377 DNA sequencers by the Advanced Genetic Analysis Center at the University of Minnesota using T7 and SP6 universal primers. For statistical analyses (see Fig. 3B), the data are representative of three independent experiments, with sequencing data for at least 10 clones for each sample, which were used to quantify the percentage of methylation in particular CpG sites.
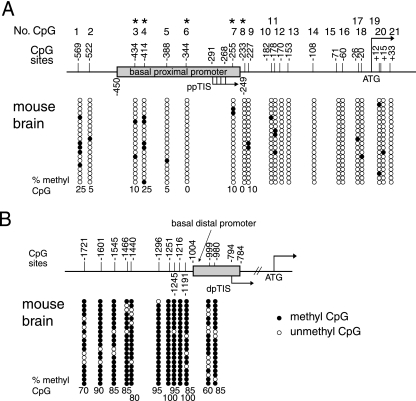
FIG. 3.
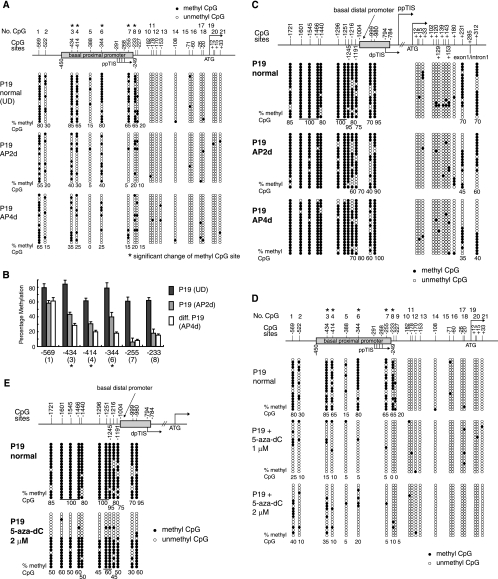
Methylation statuses of the promoter regions of the MOR gene in P19 cells. (A) The 5′-flanking region of the MOR gene contains 21 putative methyl CpG sites from −569 to +33 (with the ATG start codon designated +1). The numbers at the top of the figure (No. CpG) are arbitrary designations to indicate each methyl CpG site. ppTIS indicates the TISs of the major MOR PP containing four sites (61). The methylation statuses of the MOR gene in normal P19 (UD), AP2d (2 days after plating, i.e., intermediately differentiated P19 cells), and AP4d (4 days after plating, i.e., fully differentiated P19 cells) were determined by bisulfite genomic sequencing. Methylation-specific PCR was performed using primers MS-630 and MAS + 65 (Table 1), followed by TA cloning (Invitrogen). Each row of circles represents a single cloned allele, and each circle indicates a single CpG site at a specific location. The methylation statuses of 20 individual clones were analyzed for each cell type. The filled and open circles represent the methylated and unmethylated CpG sites, respectively. The percentages of methyl CpG versus unmethylated CpG are indicated for the first nine CpG sites. (B) Differentiation-dependent methylation changes within the MOR promoter. The percentages of methylation at CpG sites in the MOR promoter from the region of base pairs −233 to −569 (*, P < 0.05 for fully differentiated samples compared with undifferentiated [UD] samples; n ≥ 3). For statistical analysis, the data are representative of three independent experiments with sequencing data of at least 10 clones for each sample, which were used to quantify the percentage of methylation in the above-mentioned CpG sites. (C) Methylation statuses of the DP and coding exon 1 region of the MOR gene in P19 cells. For the DP and its upstream region, methylation-specific PCR was performed using the primers MS-1754 and MAS-927 (Table 1), followed by TA cloning (Invitrogen) as described for panel A. Methylation-specific PCR primers MS + 19 and MAS + 352 (Table 1) were also used for the coding exon 1 and a part of intron 1. dpTIS indicates a TIS of the DP (46). Except as noted above, experiments were performed as described for panel A. The junction site of exon 1 and intron 1 is +285, as indicated. (D and E) Demethylation of the MOR gene promoters in P19 cells after 5-aza-dC treatment. The DNA methylation statuses of the basal proximal (D) and distal (E) MOR promoters in P19 cells after treatment with 5-aza-dC are shown. The results of 20 clones assayed by bisulfite-sequencing analyses are presented. The filled and open circles indicate the methylated and unmethylated CpG sites, respectively.
ChIP assay.
Chromatin immunoprecipitation (ChIP) assays were performed by using a modified protocol from Upstate Biotechnology as previously reported (33, 41). Cells were treated for 10 min with 1% formaldehyde at room temperature. The cells were lysed in sodium dodecyl sulfate (SDS) lysis buffer (50 mM Tris-HCl [pH 8.1], 10 mM EDTA, 1% SDS). The lysates were sonicated under conditions yielding fragments ranging from 200 to 500 bp. Two percent of each lysate was used for input, and the residual lysate was subjected to the following immunoprecipitation. Samples (about 25 μg chromatin after determination of the amount of protein) were subsequently precleared at 4°C with recombinant protein G-agarose beads (Upstate Biotechnology) coated with salmon sperm DNA. Precleared lysates (100 μl) diluted in immunoprecipitation buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl [pH 8.1], 167 mM NaCl) were immunoprecipitated overnight at 4°C with 2 μg of antibodies against each of the following: anti-AcH3 (06-599), anti-AcH4 (06-866), anti-HDAC1 (06-720), anti-H3dmK4 (07-030), and anti-H3dmK9 (07-441) (all from Upstate Biotechnology); anti-Pol II (sc-5943) and anti-mSin3A (sc-5299x) (both from Santa Cruz); and anti-MeCP2 (kindly provided by Weidong Wang, NIH). All ChIP assays were controlled by performing parallel experiments with no antibody, with normal rabbit serum (NRS), or with gal4 antibody (sc-577; Santa Cruz) pull downs. The complexes were collected for 1 h by using recombinant protein G-agarose beads coated in salmon sperm DNA. After thorough washing of the beads and elution, formaldehyde cross-linking was reversed with 6 h of incubation at 65°C. After the reverse cross-linking step, DNA was extracted with phenol-chloroform, precipitated in ethanol, and dissolved in 50 μl of TE buffer (10 mM Tris-HCl and 1 mM EDTA, pH 8.0). PCR mixtures contained 2 μl of each immunoprecipitated chromatin sample with the primers described in Table 1 in a total volume of 50 μl. After 32 cycles of amplification, 15 μl of each PCR product was analyzed on a 2% agarose gel.
For scanning ChIP (SChIP) assays, experiments were carried out as described above, except that real-time qPCR and several primer sets (as described in Table 1; see also Fig. 7 and 9) were used to scan the rough locations of the indicated proteins on the MOR promoter. For real-time qPCR of SChIP, the threshold cycle for each sample was chosen from the linear range and converted to a starting quantity by interpolation from a standard curve run for each set of primers. Calibration (standard) curves were constructed for each primer pair using P19 genomic DNA templates with fivefold dilutions ranging from 16 picograms to 250 nanograms. Calibration curves with linearity R2 values of at least 0.98 were used to determine the factor by which ChIP samples changed relative to each input. Single PCR products were verified both by assessing that the melting temperature of the product had a single value and by viewing the PCR product on an agarose gel.
FIG. 7.
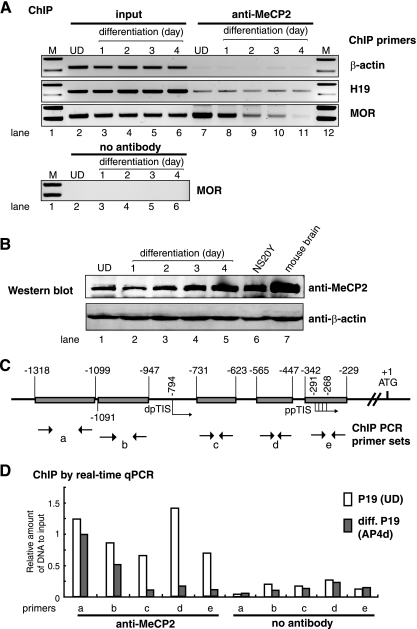
ChIP analysis of the release of MeCP2 from the MOR promoter induced by neuronal differentiation. (A) Primers specific for the MOR gene promoter (especially the PP region overlapped by primer set e), the β-actin gene, and the H19 promoter (16) were used to amplify genomic DNA sequences that were present in each immunoprecipitate with 32 cycles of PCR. Recruitment of the MeCP2 to the MOR gene promoter was reduced in a time-dependent manner during P19 cell differentiation (lanes 7 to 11). Two percent of each lysate was used as an input control. A “no antibody” control for ChIP was included in a separate parallel run. (B) The specificity of MeCP2 antibody was assessed by Western blot analysis in normal and differentiated (including AP1d to AP3d) P19 cells. MeCP2 protein levels in neuroblastoma NS20Y cells and mouse brain are also shown. Anti-β-actin was used as a control. (C) The locations (shaded boxes) of five different PCR primer sets (a to e) (Table 1) indicate their 5′-flanking regions of the MOR gene. Left- and right-direction arrows indicate sense and antisense PCR primers, respectively. (D) SChIP by real-time qPCR for MeCP2 interaction. Association of the MOR promoter with MeCP2 during P19 cell differentiation was reduced specifically in the PP region covered by primer sets c, d, and e relative to normal P19 cells. Amplification of soluble chromatin before precipitation was used as an input control. Amplification of each primer set was normalized against its input after calculating individual amounts of real-time qPCR product based on each standard curve (see details in Materials and Methods). A “no antibody” control for ChIP was performed separately in parallel.
FIG. 9.
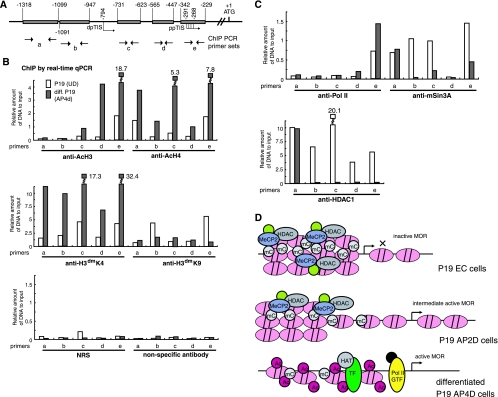
SChIP by real-time qPCR analysis for the statuses of histone modifications and corepressors associated with the MOR gene promoter. (A) Five PCR primer sets were used (as in Fig. 7). (B) The results shown in the graph were from normal P19 cells (UD) and differentiated P19 cells (AP4d) as described in the legend to Fig. 7, but using different antibodies. The chromatin modification status of the MOR promoter region was examined by SChIP assays with anti-AcH3, anti-AcH4, anti-H3dmK4, and anti-H3dmK9 antibodies. The DNAs precipitated by either NRS or the nonspecific antibody anti-gal4 were amplified with the same primers as negative controls. (C) The interaction of corepressors was also analyzed using anti-HDAC1 and mSin3A. RNA polymerase II (using anti-Pol II antibody) was bound more strongly to primer e locations (where the PP-driven TIS is localized) in differentiated P19 cells than in normal P19 cells. (D) A proposed regulation mechanism for the MOR gene. In P19 normal (EC) cells, hypermethylation of CpGs around the PP is coincident with densely interacted MeCP2 recruitment of corepressors. This might lead to compaction of the chromatin structure after histone modifications, followed by silencing of the MOR gene in these cells. In AP2d cells (i.e., intermediately differentiated P19 cells), demethylation of CpGs around the PP begins as MeCP2 and its corepressors start to dissociate, concurrent with histone modifications; this results in intermediate MOR expression. In fully differentiated P19 cells, nearly complete demethylation of the CpGs around the PP region is observed as MeCP2 and its corepressors dissociate. Hyperacetylation of histones also occurs in the promoter, suggesting active transcription of the MOR gene in the cells. All the components for active transcription shown in the figure, e.g., HAT (histone acetyltransferase), TF (transcription factors), and GTF (general transcription factors associated with Pol II), are putative factors for many genes, based on current knowledge from numerous studies.
siRNA “knockdown” experiments.
MeCP2 small interfering RNA (siRNA) was purchased from Ambion (Silencer Predesigned siRNA; identifier, 156283). MBD2 and scrambled (scb) siRNAs (sc-35866 and sc-37007, respectively) were purchased from Santa Cruz. siRNAs were transfected into P19 cells using Lipofectamine 2000 transfection reagent (Invitrogen) according to the supplier's protocol. The concentrations of siRNAs were optimized to 50 pmol in each transfection for P19 cells in six-well plates with a cell density of 105 per well. The siRNA transfection was repeated 24 hours after the first transfection to obtain maximal silencing effects. Forty-eight hours after the first transfection, total RNA and protein were prepared using TRI Reagent (Molecular Research Center). RT-PCR was performed to determine MOR, MeCP2, MBD2, and β-actin expression at the mRNA level using the PCR conditions described above. Western blots were also performed as described previously.
RESULTS
Induction of the μ opioid gene during P19 cell differentiation.
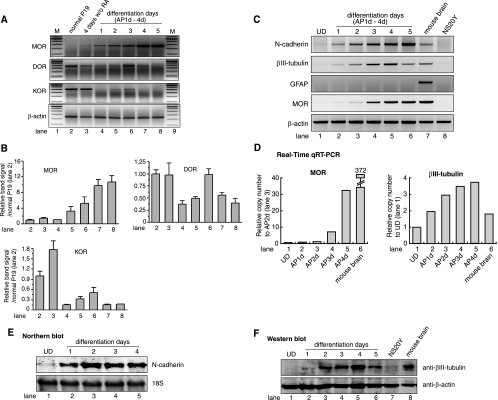
We have previously reported that MOR message increases significantly after RA-induced differentiation of P19 cells, while MOR does not increase in undifferentiated P19 cells (10). In the present study, we have further optimized the differentiation procedures for MOR induction. MOR was undetectable in undifferentiated (normal) P19 cells (Fig. 1A, lane 2). During differentiation, the expression level gradually increased and reached a maximum 4 days after plating (AP4d; Fig. 1A, lane 7). In contrast, both DOR and KOR exhibited their highest levels in undifferentiated P19 cells (Fig. 1A, lane 2) and decreased at the beginning of differentiation (lanes 4 and 5). However, the expressions of both DOR and KOR gradually recovered 2 to 3 days after plating and decreased again after 4 days after plating (Fig. 1A), indicating that MOR gene regulation during P19 cell development is quite different from that of the other opioid receptor genes. Quantitative analysis of triplicate independent experiments is shown in Fig. 1B. Therefore, differentiated P19 cells provide an excellent model system for studying opioid receptors, especially the regulation of the MOR gene at the transcriptional level. The increased expression of MOR mRNA from undifferentiated to differentiated P19 cells was also confirmed by real-time qRT-PCR (Fig. 1D). The levels of MOR mRNA increased in differentiated cells in a time-dependent manner compared to normal P19 cells, as confirmed by the real-time qRT-PCR analysis. Adult mouse brain RNA was included as a positive control.
FIG. 1.
Differential opioid receptor expression during neuronal differentiation of P19 EC cells. To induce neuronal differentiation, P19 EC cells were plated on bacterial petri dishes and allowed to aggregate for 4 days in the presence of 0.5 μM RA and then replated on tissue culture dishes without RA. For each day after plating, the cells were harvested and used for RNA or protein isolations as described in Materials and Methods. (A) The sizes of MORs (mMOR-S1 and mMOR-AS1 [Table 1] spanning exons 1 and 2, respectively) and β-actin (9) PCR fragments are 350 bp and 230 bp, respectively. PCRs for opioid receptors (MOR, DOR, and KOR) and β-actin consisted of 32 and 20 cycles, respectively. RA, all-trans retinoic acid. The DNA sequences of PCR products were confirmed by sequencing. Lanes 1 and 9 (M) are 100-bp size markers from Invitrogen. Samples for lane 3 consisted of control P19 cells cultured identically to the differentiated culture but without RA treatment. Undifferentiated cells (normal) are shown as a control. P19 cells cultured from 1 to 5 days after plating are indicated in lanes 4 to 8. Expression profiles of other opioid receptor genes (δ as DOR and κ as KOR) in this P19 differentiation were also included and analyzed by RT-PCR. (B) Quantitative analyses were performed on the receptor PCR band signal. Data are presented with the receptor signal normalized to the β-actin signal and the relative band signal to the normal P19 signal. The data are shown as means ± standard errors of the mean from three independent experiments. (C) In order to determine if neuronal differentiation of P19 cells occurred, PCR primers (Table 1) for two neuronal markers (N-cadherin and βIII-tubulin) and GFAP as a glial cell marker were used for RT-PCR. The MOR PCR was included to monitor the integrity of the experiment, and β-actin PCR was used as a control. Adult mouse brain and NS20Y cells were used as positive and negative control samples, respectively. UD, undifferentiated P19 cells. (D) Expression of the MOR and βIII-tubulin genes in P19 cells analyzed by real-time qRT-PCR. Levels of MOR mRNA were determined by real-time PCR analysis using normal and differentiated P19 cells. Five micrograms of total RNA was treated with DNase I and reverse transcribed using reverse transcriptase (Roche) and primers combined with oligo(dT) and random hexamer. One-fortieth of this cDNA sample was used for real-time qRT-PCR analysis of gene expression, using the Quantitect SYBR Green PCR kit (QIAGEN) in an iCycler (Bio-Rad). The relative expression of mRNA species was calculated using the comparative threshold cycle method as described in Materials and Methods after normalization against β-actin as an internal control. Primer sequences are shown in Table 1. (E) Northern blot analysis of a neuronal marker, N-cadherin, during neuronal differentiation using a PCR probe generated from the primers described for panel C. N-cadherin levels increase early in neuronal differentiation and steadily throughout differentiation (lanes 2 to 5). (F) Induction of βIII-tubulin, as revealed by Western blot analysis, confirmed the proper neuronal differentiation of P19 EC cells.
In order to ensure that P19 cells were differentiated from neuron-like cells, we used neuron-specific markers in RT-PCR, Northern blot, and Western blot analyses. In RT-PCR studies (including real-time qRT-PCR studies) (Fig. 1D), mRNA transcripts of N-cadherin (a neuron-specific cell adhesion molecule) and βIII-tubulin (Fig. 1C) gradually increased with differentiation of the P19 cells, as observed previously (20, 26, 54). These two neuronal markers were also used for either Northern or Western blots (Fig. 1E and F) to further determine neuronal differentiation of P19 cells in this study. The expression of the βIII-tubulin protein (Fig. 1F) gradually increased as P19 cells differentiated, as reported previously (54). The glial cell marker GFAP was not detected in any P19 cells (Fig. 1C), indicating no glial cell differentiation from P19 cells, as has been observed in other studies (38, 76).
Derepression of the MOR gene in P19 cells by treatment with reagents affecting epigenetic status.
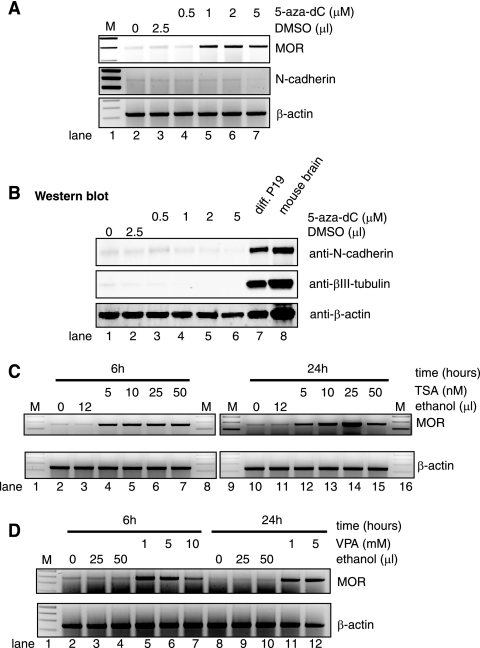
To investigate whether induction of the MOR gene is mediated by epigenetic mechanisms, we treated P19 cells with an inhibitor of DNA methylation, 5-aza-dC, and with inhibitors of histone deacetylation, TSA and VPA. Up-regulation of MOR expression in undifferentiated P19 cells was induced by all three drugs (5-aza-dC, TSA, and VPA) (Fig. 2). Induction of MOR was not significantly different in treatments with 5 to 50 nM TSA for 6 h (Fig. 2C, lanes 4 to 7). However, with treatment for 24 h, TSA caused a dose-dependent increase in expression of the MOR gene in P19 cells. Maximal MOR expression was induced by treatment with 25 nM TSA for 24 h (Fig. 2C, lane 14). Treatment with 5-aza-dC (0.5 to 5 μM) for 2 days had little influence on MOR gene expression in P19 cells (data not shown). Extending the treatment of cells with 5-aza-dC (0.5 to 5 μM) to 3 days induced MOR expression (Fig. 2A). We also used VPA (a known HDAC inhibitor), a drug that has been used for decades in the treatment of epilepsy and as a mood stabilizer. Interestingly, VPA has been reported to trigger replication-independent active demethylation of DNA (15). As shown in Fig. 2D, VPA treatments (both 6- and 24-hour treatments with 1 and 5 mM VPA) induced the endogenous MOR gene. Further studies are required to determine whether VPA action is mediated by histone deacetylation or demethylation. These results suggest that both DNA methylation and histone deacetylation are involved in the repression of the MOR gene.
FIG. 2.
TSA, VPA, and 5-aza-dC treatment induced MOR gene expression in P19 cells. The results of RT-PCR analyses on the MOR mRNA levels in P19 cells treated with different doses of 5-aza-dC (A), TSA (C), or VPA (D) using the procedure described in Materials and Methods are shown. The TSA and VPA samples were treated for 6 h and 24 h, and 5-aza-dC samples were treated for 72 h (3 days) to induce maximal effects on MOR levels in P19 cells. The neuronal markers N-cadherin and βIII-tubulin were included in RT-PCR (N-cadherin in panel A) and Western blotting (N-cadherin and βIII-tubulin in panel B) analyses to demonstrate that P19 cell differentiation was not induced by 5-aza-dC treatment. β-Actin was used as a control.
Since the MOR gene is induced by prolonged treatment (3 days) with 5-aza-dC, we decided to determine whether the drug itself also causes neuronal differentiation in P19 cells. Neither the protein (Fig. 2B) nor mRNA (Fig. 2A) levels of N-cadherin were increased in the 5-aza-dC-treated cells, indicating no neuronal differentiation in P19 cells treated with the drug. Another neuronal cell marker, βIII-tubulin, also showed no increase (Fig. 2B). Adult mouse brain and differentiated P19 cells (AP4d) were used as positive controls in Fig. 2B (lanes 8 and 7, respectively), showing high expression of the proteins in the Western blot, along with the internal control (β-actin).
Methylation status of the MOR gene promoter in P19 cells.
Since the DNA-demethylating agent induced the MOR gene, we attempted to analyze the methylation statuses of the MOR gene promoters in P19 cells. There are 21 CpG sites in the 5′-flanking region (−569 to +31, covering the PP region, with the ATG start codon designated +1) of the MOR gene. To elucidate the epigenetic mechanisms that participate in silencing or activating MOR gene transcription, we first utilized normal (undifferentiated) P19 and fully differentiated P19 (i.e., AP4d)cells (Fig. 1A, lane 7). The DNA methylation statuses of the MOR gene promoter in normal, intermediately differentiated (AP2d), and fully differentiated (AP4d) P19 cells were assessed by utilizing bisulfite treatment and sequencing analyses. Evaluation of 20 individual clones from each cell type revealed that six CpG sites, located at −569, −434, −414, −344, −255, and −233, were highly methylated (over 65% of the clones) in P19 cells while other sites were partially methylated or unmethylated (Fig. 3A). Five of these CpG sites, all except the first CpG site at −569, were gradually unmethylated as P19 differentiation proceeded from normal to 4 days after plating (Fig. 3A). The first CpG site, at −569, remained heavily methylated (over 55% for all types of P19 cells). As described in Materials and Methods, the differential methylated domains were observed in the three CpG sites (−434, −414, and −344) as statistically significant for the three stages of the cells in the percentage of methylation (Fig. 3B). These three sites are located upstream, close to the PP-derived transcription initiation site (TIS).
We further examined whether the change in methylation status was specific to this PP region (known to be a major promoter for the MOR gene) or if it also occurred in other regions. Analysis of the methylation status for the upstream region of the distal promoter (DP) (from −980 to −1721, containing 12 CpG sites) was performed (Fig. 3C). These sites were highly methylated in normal P19 cells (more than 70% methylation; as high as 100%). This hypermethylation status was unchanged in AP2d and AP4d P19 cells (Fig. 3C). Coding exon 1 and the junction region between exon 1 and intron 1 (from +12 to +312, containing 12 CpG sites) were also analyzed for methylation status (Fig. 3C). The analyzed region of the first three sites (+12, +15, and +33) overlapped with the region shown in Fig. 3A. Both experiments showed that the three sites were unmethylated, indicating the integrity of the methylation experiments (Fig. 3A and C). The 10 CpG sites in coding exon 1 (except the +231 site) were unmethylated in all types of P19 cells, while two sites (+231 and +312) in the exon/intron junction region were gradually unmethylated as the cells differentiated. This might indicate that lack of methylation in this junction region could provide access to transcriptional machinery in the region to be transcribed in the differentiated cells, whereas hypermethylation blocks the transcription machinery in normal P19 cells. These findings suggested that the level of DNA methylation might be responsible for silencing or reactivation of MOR gene expression.
Treatment of P19 cells with 5-aza-dC results in demethylation of the MOR promoter.
Repression of MOR expression in undifferentiated P19 cells was alleviated by 5-aza-dC treatment (Fig. 2). Thus, it was important to determine whether the methylation state of the MOR gene promoter was changed in P19 cells treated with 5-aza-dC. Bisulfite-sequencing analyses revealed that the methylated PP was demethylated in the presence of 5-aza-dC (Fig. 3D). Similar levels of demethylation were achieved with either 1 or 2 μM 5-aza-dC. This correlates with the MOR mRNA induction observed by treatment with 5-aza-dC (Fig. 2A). The DP and its upstream regions were also demethylated about 50% by 2 μM 5-aza-dC (Fig. 3E).
Preferential transcription from the PP is due to the unmethylation of the promoter.
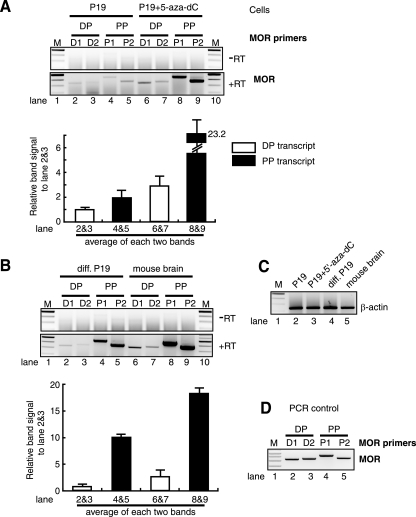
Previous reports have shown that the PP acts as the major promoter compared to the DP for MOR transcription, with a ratio of 20:1 in the mouse brain (44, 46). It is unknown whether a similar ratio exists in the induced MOR expression of differentiated P19 cells. It is also unclear why DP-mediated transcription is so much less active than PP-mediated transcription. To answer these questions, we performed the following differential RT-PCR for DP and PP transcriptions, using gene-specific PCR primers (two primer pairs per transcript: D1, D2, P1, and P2) (Fig. 4 and Table 1). As shown in Fig. 4A, the expression of PP-mediated MOR (lanes 8 and 9) in 5-aza-dC-treated P19 cells increased compared to the PP expression in P19 cells (lanes 4 and 5). DP-mediated MOR expression in the treated cells (lanes 6 and 7) remained low, albeit higher than that of P19 cells (lanes 2 and 3). These results probably reflect the fact that hypermethylated CpGs in the PP region of the P19 cells (Fig. 3A) became unmethylated in the 5-aza-dC-treated P19 cells (Fig. 3D). The methylation percentage (65 to 85% for the five sites) of the PP region in P19 cells was reduced to 5 to 35% methylation in cells treated with 2 μM 5-aza-dC (Fig. 3D). However, in the DP region and its upstream region, hypermethylation of all 12 sites in P19 cells was reduced about 50% by 5-aza-dC treatment (Fig. 3E). It was interesting that 5-aza-dC treatment was more effective in the PP region than in the DP region. We speculate that its demethylation effect is less active in more highly methylated sites (80 to 100% for the DP region) than in 65 to 85% methylated sites for the PP region (see Discussion). As mentioned above, in differentiated P19 cells, high methylation in the DP region was unchanged relative to normal P19 cells (Fig. 3C) while the PP region was unmethylated (Fig. 3A). As shown in Fig. 4B, PP-mediated MOR expression in differentiated cells (lanes 4 and 5) was increased relative to that seen in normal P19 cells (Fig. 4A, lanes 2 and 3), while the expression of DP-mediated transcripts remained low in the differentiated cells (Fig. 4B, lanes 2 and 3).
FIG. 4.
Differential expression from dual promoters of MOR. (A) Total RNAs from P19 and 5-aza-dC-treated P19 cells were used for DP- and PP-mediated transcription by RT-PCR using their specific PCR primers. Two primer sets were used for each transcript: D1 and D2 for DP transcript, P1 and P2 for PP transcript (Table 1). −RT and +RT indicate samples analyzed without or with reverse transcriptase, respectively. Quantitative analysis is shown below the figure as a graph representing the average of each transcript from two sets of primers. The error bars indicate standard errors of the mean. (B) Analysis of differentiated P19 cells and mouse brain tissue by RT-PCR, as described above. (C) Control RT-PCR performed using β-actin primers shows equal amounts of total RNA used. (D) Control PCR performed to show PCRs using DP- and PP-specific primers and MOR cDNA template.
In adult mouse brain tissue, DP-MOR transcription was lower than that of PP-MOR (about sevenfold). The methylation status analysis from the mouse brain revealed that all the CpGs of the PP region, including the region downstream of the PP TIS, were unmethylated (less than 25%) (Fig. 5A). However, the CpGs of the DP and its upstream region were highly methylated (Fig. 5B), similar to the results seen for the same region in differentiated P19 cells (Fig. 3C). Taken together, these data show that DP-mediated MOR transcription in differentiated P19 cells occurs at a low level, similar to that seen in the mouse brain. This low level of DP-mediated expression is possibly due, in part but not entirely, to retention of hypermethylated CpGs in the DP region of the differentiated cells, as well as in the mouse brain. PP-mediated MOR transcription may be induced preferentially in both differentiated P19 cells and mouse brain tissue because the CpGs of the PP region are more efficiently unmethylated, although other mechanisms may be involved. Figure 4C shows similar levels of β-actin expressed in all four tested samples, indicating the use of similar amounts of RNA as RT-PCR templates. Control PCR experiments were also performed, as seen in Fig. 4D, using the corresponding template. The four primer sets (D1, D2, P1, and P2) had similar amplification efficiencies (Fig. 4D); similar efficiencies were also observed by using various amounts of templates.
FIG. 5.
Methylation statuses of the promoter regions of the MOR gene in mouse brain. (A) Methylation analysis of the PP and its downstream regions from adult mouse brain. (B) Similar methylation analysis of the DP and its upstream regions from adult mouse brain. The experiments were performed as described for Fig. 3.
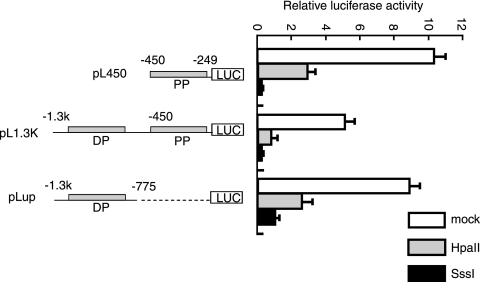
In vitro methylation of the MOR promoter represses its promoter activity.
The relevance of MOR gene expression patterns to its promoter activities in P19 cells were further investigated by reporter gene analyses. To mimic the endogenous methylation status of the MOR gene promoter, MOR promoter plasmids (46) fused with luciferase containing pL1.3k (DP and PP, kbp −1.3 to bp 249), pL450 (PP only, bp −450 to bp 249), or pLup (DP only, kbp −1.3 to bp −775) were methylated by either partial methylase HpaII or full methylase SssI in vitro. The resulting in vitro-methylated MOR promoter/reporter constructs were transfected into P19 cells individually. An unmodified promoter (mock) was also introduced into P19 cells (Fig. 6). Partial methylation of the luciferase constructs induced about 80% reduction in promoter activity compared with the mock-methylated constructs, whereas full methylation of the constructs of pL450 and pL1.3k suppressed the promoter activity completely (Fig. 6). Full methylation of pLup (DP only) repressed the promoter activity slightly less than in the two other constructs (pL450 and pL1.3k), indicating a reduced effect of methylation in the DP region. These in vitro methylation data suggest that methylation of the promoter may contribute to the repression of MOR gene transcription in a methylation density-dependent manner.
FIG. 6.
Repression of MOR promoter-driven transcription by CpG methylation. Three different luciferase (LUC) constructs (pL450, pGL1.3k, and pLup) were mock methylated (mock) or in vitro methylated with HpaII (partial) or SssI (full) methylase and transfected into P19 cells. The results are given as luciferase activity normalized against cotransfected pCH110 β-galactosidase activity. The data shown are the means of three independent experiments with at least two different plasmid preparations. The error bars indicate the range of standard errors.
Interaction of MeCP2 in the MOR promoter region is reduced in differentiated P19 cells.
A major mechanism of methylation-induced transcriptional repression is known to be the direct binding of specific MBPs to methylated DNA (4). Therefore, we examined whether the transcriptional repression conferred by the methylated MOR promoter correlates with its affinity for MBPs. We used a ChIP assay to investigate the association of endogenous MeCP2 with MOR promoters. The results showed that in P19 cells, the PP-mediated MOR promoter (primer set e overlaps the PP region) was associated with MeCP2. Furthermore, this association gradually decreased in a time-dependent manner from undifferentiated P19 cells to differentiated cells (AP4d) (Fig. 7A). This was consistent with the results of methylation status studies (Fig. 3A). This represents a specific binding of MeCP2 with the MOR promoter, because NRS and nonspecific antibody (anti-gal4) failed to immunoprecipitate MOR promoter sequences (see Fig. 9B). The release of MeCP2 was specific to the MOR promoter, because the interaction of MeCP2 with the imprinted gene H19 (16) was not affected by neuronal differentiation of P19 cells (Fig. 7A). As a negative control, we performed ChIP assays with β-actin, which is actively expressed in all P19 cells independent of methylation in its regulatory region. MeCP2 was not associated with the β-actin gene (Fig. 7A). Another negative control using no antibody also showed no MeCP2 interaction with the MOR promoter (Fig. 7A and D). The specificity of the antibodies against MeCP2 and the expression levels was demonstrated by Western blotting (Fig. 7B).
To pinpoint the location of MeCP2 binding reduction in the 5′-flanking region of the MOR gene, we employed a modified ChIP assay, namely, the SChIP, assisted with real-time qPCR. This assay was used to determine the location that exhibited the greatest change in binding for a single target protein in the promoter region using several PCR primer sets. Primer sets a and b (Fig. 7C and Table 1), spanning the DP and its upstream region, showed little change in MeCP2 interactions between normal P19 and differentiated P19 cells (Fig. 7D). The hypermethylation status of this region was not changed between normal and differentiated P19 cells (Fig. 3). However, the in vivo interactions of MeCP2 with regions of the upstream regulatory region and the PP (primer set c, d, and e) were decreased in differentiated P19 cells relative to normal P19 cells (Fig. 7D). This reduction in MeCP2 binding might be caused by hypomethylation in these regions in differentiated P19 cells, as the binding ability of MeCP2 proteins to DNA is known to be dependent on the methylation status of CpG DNA. A control with no added antibody was included in parallel with the ChIP assay to demonstrate the specificity of MeCP2 binding.
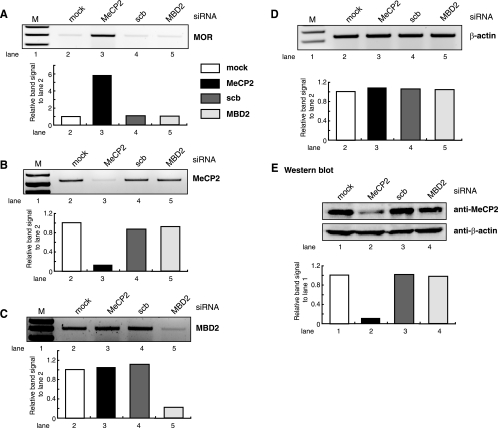
siRNA-mediated knockdown of MeCP2 in P19 cells implicates MeCP2 in hypermethylation-dependent MOR repression.
To test whether MeCP2 was responsible for repression of transcription from hypermethylated MOR promoter alleles, an siRNA-based strategy was employed to silence the endogenous MeCP2 expression in P19 cells. The concentration of siRNAs was optimized to 50 pmol in each transfection for P19 cells. Transfection of MeCP2 siRNA resulted in about 90% silencing of the MeCP2 gene at the transcription and translation levels (Fig. 8B and E). As a control, siRNA transfection with the related MBD family member MBD2 also showed a significant reduction of its transcription (Fig. 8C). Transfection of MBD2 siRNA and scb siRNA as a nonspecific sequence control had no effect on MeCP2 expression compared to the “no siRNA” transfection control (mock) (Fig. 8B and E). Expression of the target MOR was increased with MeCP2 siRNA transfection at the mRNA level (Fig. 8A), compared to mock, scb, and MBD2 siRNA transfection, showing in vivo evidence of the MeCP2 function on endogenous MOR gene regulation. The β-actin gene was not affected by the siRNA transfection (Fig. 8D).
FIG. 8.
Alleviation of repression from hypermethylated MOR promoters after targeted reduction of MeCP2 using siRNA. P19 cells were transfected with siRNA-targeting mRNA encoding MeCP2, scb control, and MBD2. (A) After siRNA treatment, MOR transcription was assessed by RT-PCR as described in Materials and Methods. (B and C) Reductions in the levels of the targeted mRNA (MeCP2 [B] and MBD2 [C]) were monitored by RT-PCR analysis using the corresponding gene-specific primers (Table 1). (D) β-Actin was included as a control. (E) Reduction of MeCP2 protein was monitored by Western blot analysis using MeCP2 antibody and β-actin antibody as a control. Quantitative analyses of the RT-PCR and Western blot experiments measured changes in mRNA and protein levels, shown as a graph below each result. The data were normalized against β-actin levels. The graph was generated by using Kodak molecular imaging software version 4.0 (Kodak) for RNA and ImageQuant TL (Amersham) for protein.
MOR activation correlates with histone modifications and corepressor dissociation in differentiated P19 cells.
The MBP MeCP2 is known to mediate long-term gene silencing, in part by recruitment of histone-modifying enzymes, such as deacetylases and methyltransferases (19, 36, 62). Therefore, we considered the possibility that MeCP2 might regulate MOR promoter activation by altering the chromatin architecture of the MOR gene. SChIP analysis with real-time qPCR revealed that, before neuronal differentiation of P19 cells, MOR promoter-associated histone H3 in the primer b and e regions (overlapping the DP and PP, respectively), was methylated at lysine 9 (H3dmK9) (Fig. 9B), a modification that on the H19 gene is correlated with gene inactivation (49) and facilitated by MeCP2 (19). Furthermore, SChIP analysis of MOR promoter sequences showed reduced methylation of H3 dimethyl K9 and concurrent increases in dimethylation at lysine 4 (H3dmK4). This suggests a more global increase in the promoter (for H3dmK4 as a hallmark of active transcription) (35) in the differentiated cells, consistent with the observed release of MeCP2 from the MOR promoter in these cells. These results suggest that histone code in a repressive or active state is established, not only at the PP, but also in the broad 5′-flanking region of the MOR gene in differentiated P19 cells with repressed or active MOR expression.
To further characterize the histone modifications, we repeated the ChIP experiments using antibodies for acetylated histone H3 and acetylated histone H4. Chromatin from P19 cells immunoprecipitated with serum against acetylated histone H3 displayed little enrichment of any sequences in the 5′-flanking region of the MOR compared to chromatin from differentiated P19 cells (Fig. 9B). The hyperacetylated histone H3 in differentiated P19 cells was more densely localized to the primer set d and e region, which covers the PP and its TIS. In contrast, acetylated histone H4 was localized to all the examined regions relative to undifferentiated P19 cells. This suggests that for MOR activation, histones H3 and H4 must be hyperacetylated, but such hyperacetylations are differentially localized between histones H3 and H4 (Fig. 9B).
As more acetylation of H3 and H4 was observed in differentiated P19 cells, we asked whether this correlates with the dissociation of transcriptional corepressors, such as HDAC1 and mSin3A (Fig. 9C). Interactions of both corepressors were abolished in the regions including primer sets b to e, while the interactions for the region of primer a remained similar in normal and differentiated P19 cells. Primer sets b to e encompass two MOR promoters (distal and proximal) and their TISs, while primer set a is located upstream of the DP (Fig. 9A). These results correlate with the enrichment of acetylated H3 and H4 and dissociation of corepressors at the promoter, as well as unmethylation of CpGs located at the promoter region in differentiated P19 cells. Interaction of RNA polymerase II (Pol II) was increased in the primer e region that covers the PP-driven TIS in the differentiated cells, suggesting more active transcription in the region. Normal rabbit serum and gal4 antibody included in SChIP-real-time qPCR assays as controls showed no PCR signal. In summary, we propose that the release of MeCP2 from the MOR gene in response to unmethylation of the PP region may allow the assembled transcriptional activator complex to remodel local chromatin structure through the acetylation or methylation of specific histone residues, thereby promoting MOR transcription.
DISCUSSION
Several nonneuronal cell lines exist that express the mouse MOR gene endogenously, such as the immune murine macrophage-like RAW264.7 cell line (33). Similarly, there are endogenous MOR-expressing cells from different species, e.g., SY-SH5Y and NMB as human neuroblastoma cell lines and the MOR-positive C6 glioma cells from the rat (5). In addition, regulation of the MOR gene has been studied in various physiological and neuronal states in several species. For example, mRNA levels of MOR are regulated by activation of protein kinase C in a neuronal model with SH-SY5Y cells (22). MORs of human and monkey lymphocytes were upregulated by morphine treatment at the transcriptional and translational levels (75). Interleukin-1β treatment increased the level of MOR mRNA by 55 to 75% in primary astrocyte-enriched cultures derived from rat striatum, cerebellum, and hippocampus, but not in cultures derived from the cortex or hypothalamus (71). These results suggest a capacity for astrocytes to differentially regulate MOR mRNA in response to an immune factor in a brain region-dependent fashion. The expression of MOR in the rat mesentery was induced by treatment with a bacterial endotoxin, lipopolysaccharide (8). This induction may be mediated through the actions of interleukin 1 on MOR (8). The levels of MOR transcripts in discrete brain regions are also regulated by the administration of other nonopioid drugs (e.g., dopaminergic drugs, such as cocaine and haloperidol) (1, 14), indicating that the transcriptional regulation of the MOR gene is closely associated with the dopaminergic system in the brain.
Until now, there has not been a mouse neuronal cell line expressing the MOR gene endogenously to serve as a model for the study of mouse MOR gene regulation at the transcriptional level. The promoter sequences of MORs from mouse, rat, and human are not exactly homologous, except for some regions known to be conserved (12, 33, 34, 41, 45, 47, 48). To study regulation of the mouse MOR gene, we had to find a mouse model cell line in which the MOR gene could be regulated while the endogenous MOR gene was not expressed constitutively. Previously, we reported that the MOR message was significantly increased in P19 cells in which differentiation was triggered by RA treatment (10). In this study, we further optimized the differentiation conditions for MOR induction (along with DOR and KOR genes). The MOR gradually increased in a time-dependent manner and reached maximum expression on the fourth day after plating. As described in several studies (37, 59), this is known to be the time when P19 cells fully differentiate into neuron-like cells. We also confirmed the neuronal differentiation from the P19 cells using neuronal markers (N-cadherin and βIII-tubulin) at the RNA or protein level (Fig. 1). Different expression profiles for DOR and KOR genes were observed during differentiation, relative to the MOR gene. In mouse embryonic development, these three opioid receptors show very different expression patterns both temporally and spatially (85), suggesting unique functional roles. Thus, we chose the P19 cell system in which to study the temporal regulation of the MOR gene as a model for in vivo regulation. The P19 cell model could also be useful for studying the regulation of DOR and KOR genes (32, 53, 67).
CpG DNA methylation in the promoter region affects gene regulation in early embryonic development and stem cells. We therefore examined the possibility that CpG sites might be present in the MOR promoter. While gene promoters in mammals are generally localized upstream of the TIS, the downstream region of the MOR TIS was included in our studies. This region contains several transcriptional regulatory elements, including a region overlapping the ATG start codon containing NRSE (41) and Sp3 (40) functional sites. Among the 21 putative CpG sites in the region spanning −569 to +31 of the MOR gene, we found six CpG sites, located at −569, −434, −414, −344, −255, and −233, that were hypermethylated in normal P19 cells. Other sites were partially methylated or unmethylated (Fig. 3). In intermediately differentiated (AP2d) and fully differentiated (AP4d) P19 cells, five of these sites (except the first site at −569) were gradually unmethylated. In contrast, the upstream region of the DP (from −980 to −1721) was hypermethylated, and the hypermethylation status did not change as the cells differentiated. Coding exon 1 and the junction region between exon 1 and intron 1 (from +12 to +312) were also analyzed for methylation status. The 10 CpG sites in coding exon 1 (from +12 to +180) were unmethylated in all stages of P19 cell development, while two sites (+231 and +312) in the exon/intron junction region were gradually unmethylated as the cells differentiated.
The MBP MeCP2 can bind preferentially to a single symmetrically methylated CpG (43, 63). The BDNF gene is regulated by MeCP2 in response to neuronal depolarization through its methylated promoter (11, 56). The BDNF promoters seem to have scattered CpGs similar to those of the MOR promoter. Mouse BDNF IV promoter contains 11 CpGs in a 191-bp region (11.5% in dinucleotide) (56), and the rat BDNF III promoter contains 9 CpGs in 167 bp (10.7%) (11). The mouse MOR promoter contains 21 CpGs in 557 bp (7.5%). Interestingly, the first CpG (−148 bp) in both BDNF promoters remains hypermethylated after depolarization with KCl treatment (with no impact on promoter activities). Similarly, the first CpG (−569 bp) in the MOR promoter also remains hypermethylated upon differentiation of P19 cells, suggesting similar mechanisms for these genes. Taken together, these results demonstrate a distinct methylation status of the MOR gene promoter in normal P19 versus differentiated P19 cells.
As shown in Fig. 3D and E, treatment with 5-aza-dC resulted in incomplete demethylation (about 50%) of the sites in the DP region. 5-Aza-dC is a known inhibitor of DNA methyltransferase (13). Among the four DNA methyltransferases (i.e., Dnmt1, Dnmt2, Dnmt3a, and Dnmt3b), the effect of 5-aza-2dC is primarily mediated by Dnmt3a and Dnmt3b (65). Oka et al. reported that Dnmt3a-Dnmt3b double-null embryonic stem cells were highly resistant to 5-aza-dC compared to its effects in wild-type, Dnmt3a null, Dnmt3b null, or Dnmt1 null embryonic stem cells (65). This suggests that 5-aza-dC may exert its effects only on a specific Dnmt enzyme. Dnmt3a and Dnmt3b also have distinct substrate preferences for certain genomic loci, including major and minor satellite repeats (66). Therefore, treatment with 5-aza-dC may have differential demethylation effects, depending on the locations of the CpG sites in the genome. This may explain the results observed for the MOR promoter, especially with regard to demethylation of the DP region.
In mammals, there are two general mechanisms by which DNA methylation inhibits gene expression. First, modification of cytosine bases can inhibit the association of some DNA-binding factors with their cognate DNA recognition sequences (81). Second, proteins that recognize methyl-CpG can elicit the repressive potential of methylated DNA (6, 28). The silencing of methylated promoters usually requires MBPs that specifically recognize symmetrically methylated CpG. To date, five such MBPs with homologous DNA-binding domains have been identified (27-29, 42). MBD1, MBD2, and MeCP2 recruit HDACs to repress the methylated promoters. MBD3 is a component of the nuclear remodeling and HDAC complex, Mi-2-NuRD, that is recruited to the methylated promoter by interacting with MBD2 (77, 78, 84). MBD4 codes for a uracil DNA glycosylase that repairs methylated CpG-TpG mismatch pairs (2, 30). One report has suggested that proteins without an MBD can also bind methylated DNA (69). Kaiso, a protein that interacts with β-catenin, is a second methylated DNA-binding component of MeCP2. This protein uses its POZ zinc finger domain to bind methylated DNA. MeCP2 can bind to a single methylated CpG, whereas other MBPs (e.g., MeCP1) (60), generally bind to the DNA containing at least 12 symmetrically methylated CpGs (62). Because only five scattered CpGs (at −434, −414, −344, −255, and −233) out of the 21 sites in the MOR promoter showed changes in their methylation status, we turned our attention to MeCP2 as a possible mediator of transcriptional repression. The interaction of MeCP2 did indeed gradually decrease as the P19 cells differentiated. SChIP assays showed that the reduction of MeCP2 interaction occurred primarily in the area of primer sets c, d, and e (from −229 to −731), which contains the PP and its regulatory regions in differentiated cells. The five demethylated sites were also localized to this MeCP2 reduction region. SChIP analysis in differentiated cells with primer sets a and b (covering from −1318 to −947) showed a slight but insignificant decrease in MeCP2 binding in the DP and its upstream regions. This is consistent with the observation that the hypermethylation status in the DP and its upstream regions was unchanged during differentiation. Several studies (4) have shown that MeCP2 binds to DNA in a methyl-CpG-dependent manner. Phosphorylation of MeCP2 also causes release of MeCP2 from the methyl CpG site of the BDNF promoter (11). It will be interesting to determine whether MeCP2 is phosphorylated at the time of MeCP2 reduction on unmethylated sites of the MOR promoter. It will also be necessary to define which of the five methyl CpG sites has the highest affinity for MeCP2 protein or whether all five sites are required for MeCP2 binding.
MeCP2 directly binds to the corepressor mSin3A, which interacts with HDAC1, and recruits it to methyl-CpG regions. This suppresses transcription through histone deacetylation, resulting in more compaction of nucleosome complexes on the promoter (4, 42, 62). Our results show a decrease in the interactions of both HDAC1 and mSin3A in the proximal and DP regions of differentiated P19 cells relative to normal P19 cells. Hyperacetylation of histone H3 was localized to the region of primer sets d and e (i.e., the PP region), while hyperacetylation of histone H4 was detected in all areas tested. This suggests that histone H3 may have a different regulation system from histone H4. Indeed, differential acetylation of histones H3 and H4 on the U2af1-rs1 gene has been reported (24). Taken together, these studies suggest that MBPs use transcriptional corepressor molecules to silence transcription and to modify surrounding chromatin, providing a link between DNA methylation and chromatin modification, as described in other studies (36, 62, 64, 72, 78, 84).
In conclusion, we propose the following regulatory mechanism for the control of MOR expression (Fig. 9D). In normal P19 cells, hypermethylation of CpG sites in the PP region leads to binding of MeCP2. MeCP2 subsequently recruits repressors, such as HDAC1 and mSin3A, that result in deacetylation of histones. This tightly compacts the nucleosome complex around the promoter region, silencing the MOR gene at the transcriptional level. Demethylation of the CpGs begins as the cells differentiate, leading to dissociation of MeCP2 and its repressors from the promoter and subsequent intermediate MOR expression. In fully differentiated P19 cells, complete demethylation of CpGs in the PP region causes dissociation of MeCP2 from the promoter and leads to histone modifications, such as hyperacetylation or dimethylation. This results in chromatin modification (e.g., loosening or disassembling of nucleosomes) that permits the general transcription machinery to become active. These observations provide insight into the epigenetic regulation of MOR gene expression and the diverse mechanisms of transcriptional regulation using the methylated MOR promoter.
Acknowledgments
This work was supported by NIH grants DA000564, DA001583, DA011806, and K05-DA070554 (H.H.L.) and DA011190 and DA013926 (L.-N.W.) and by the A&F Stark Fund of the Minnesota Medical Foundation.
We thank Weidong Wang (NIH) for kindly providing MeCP2 antibody. We also thank Vida Gavino and Martin Winer for critical review of the manuscript.
Footnotes
Published ahead of print on 23 April 2007.
REFERENCES
- 1.Azaryan, A. V., B. J. Clock, J. G. Rosenberger, and B. M. Cox. 1998. Transient upregulation of mu opioid receptor mRNA levels in nucleus accumbens during chronic cocaine administration. Can. J. Physiol. Pharmacol. 76:278-283. [PubMed] [Google Scholar]
- 2.Bader, S., M. Walker, B. Hendrich, A. Bird, C. Bird, M. Hooper, and A. Wyllie. 1999. Somatic frameshift mutations in the MBD4 gene of sporadic colon cancers with mismatch repair deficiency. Oncogene 18:8044-8047. [DOI] [PubMed] [Google Scholar]
- 3.Berger, S. L. 2001. An embarrassment of niches: the many covalent modifications of histones in transcriptional regulation. Oncogene 20:3007-3013. [DOI] [PubMed] [Google Scholar]
- 4.Bird, A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16:6-21. [DOI] [PubMed] [Google Scholar]
- 5.Bohn, L. M., M. M. Belcheva, and C. J. Coscia. 1998. Evidence for kappa- and mu-opioid receptor expression in C6 glioma cells. J. Neurochem. 70:1819-1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyes, J., and A. Bird. 1991. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell 64:1123-1134. [DOI] [PubMed] [Google Scholar]
- 7.Chan, Y., J. E. Fish, C. D'Abreo, S. Lin, G. B. Robb, A. M. Teichert, F. Karantzoulis-Fegaras, A. Keightley, B. M. Steer, and P. A. Marsden. 2004. The cell-specific expression of endothelial nitric-oxide synthase: a role for DNA methylation. J. Biol. Chem. 279:35087-35100. [DOI] [PubMed] [Google Scholar]
- 8.Chang, S. L., B. Felix, Y. Jiang, and M. Fiala. 2001. Actions of endotoxin and morphine. Adv. Exp. Med. Biol. 493:187-196. [DOI] [PubMed] [Google Scholar]
- 9.Chen, H. C., and H. H. Loh. 2001. μ-Opioid receptor gene expression: the role of NCAM. Neuroscience 108:7-15. [DOI] [PubMed] [Google Scholar]
- 10.Chen, H. C., L. N. Wei, and H. H. Loh. 1999. Expression of mu-, kappa- and delta-opioid receptors in P19 mouse embryonal carcinoma cells. Neuroscience 92:1143-1155. [DOI] [PubMed] [Google Scholar]
- 11.Chen, W. G., Q. Chang, Y. Lin, A. Meissner, A. E. West, E. C. Griffith, R. Jaenisch, and M. E. Greenberg. 2003. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science 302:885-889. [DOI] [PubMed] [Google Scholar]
- 12.Choi, H. S., C. K. Hwang, C. S. Kim, K. Y. Song, P. Y. Law, L. N. Wei, and H. H. Loh. 2005. Transcriptional regulation of mouse mu opioid receptor gene: Sp3 isoforms (M1, M2) function as repressors in neuronal cells to regulate the mu opioid receptor gene. Mol. Pharmacol. 67:1674-1683. [DOI] [PubMed] [Google Scholar]
- 13.Christman, J. K. 2002. 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene 21:5483-5495. [DOI] [PubMed] [Google Scholar]
- 14.Delfs, J. M., L. Yu, G. D. Ellison, T. Reisine, and M. F. Chesselet. 1994. Regulation of mu-opioid receptor mRNA in rat globus pallidus: effects of enkephalin increases induced by short- and long-term haloperidol administration. J. Neurochem. 63:777-780. [DOI] [PubMed] [Google Scholar]
- 15.Detich, N., V. Bovenzi, and M. Szyf. 2003. Valproate induces replication-independent active DNA demethylation. J. Biol. Chem. 278:27586-27592. [DOI] [PubMed] [Google Scholar]
- 16.Drewell, R. A., C. J. Goddard, J. O. Thomas, and M. A. Surani. 2002. Methylation-dependent silencing at the H19 imprinting control region by MeCP2. Nucleic Acids Res. 30:1139-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fish, J. E., C. C. Matouk, A. Rachlis, S. Lin, S. C. Tai, C. D'Abreo, and P. A. Marsden. 2005. The expression of endothelial nitric-oxide synthase is controlled by a cell-specific histone code. J. Biol. Chem. 280:24824-24838. [DOI] [PubMed] [Google Scholar]
- 18.Fujita, N., S. Watanabe, T. Ichimura, S. Tsuruzoe, Y. Shinkai, M. Tachibana, T. Chiba, and M. Nakao. 2003. Methyl-CpG binding domain 1 (MBD1) interacts with the Suv39h1-HP1 heterochromatic complex for DNA methylation-based transcriptional repression. J. Biol. Chem. 278:24132-24138. [DOI] [PubMed] [Google Scholar]
- 19.Fuks, F., P. J. Hurd, D. Wolf, X. Nan, A. P. Bird, and T. Kouzarides. 2003. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J. Biol. Chem. 278:4035-4040. [DOI] [PubMed] [Google Scholar]
- 20.Gao, X., W. Bian, J. Yang, K. Tang, H. Kitani, T. Atsumi, and N. Jing. 2001. A role of N-cadherin in neuronal differentiation of embryonic carcinoma P19 cells. Biochem. Biophys. Res. Commun. 284:1098-1103. [DOI] [PubMed] [Google Scholar]
- 21.Georgel, P. T., R. A. Horowitz-Scherer, N. Adkins, C. L. Woodcock, P. A. Wade, and J. C. Hansen. 2003. Chromatin compaction by human MeCP2. Assembly of novel secondary chromatin structures in the absence of DNA methylation. J. Biol. Chem. 278:32181-32188. [DOI] [PubMed] [Google Scholar]
- 22.Gies, E. K., D. M. Peters, C. R. Gelb, K. M. Knag, and R. A. Peterfreund. 1997. Regulation of mu opioid receptor mRNA levels by activation of protein kinase C in human SH-SY5Y neuroblastoma cells. Anesthesiology 87:1127-1138. [DOI] [PubMed] [Google Scholar]
- 23.Gilsbach, R., M. Kouta, H. Bonisch, and M. Bruss. 2006. Comparison of in vitro and in vivo reference genes for internal standardization of real-time PCR data. BioTechniques 40:173-177. [DOI] [PubMed] [Google Scholar]
- 24.Gregory, R. I., T. E. Randall, C. A. Johnson, S. Khosla, I. Hatada, L. P. O'Neill, B. M. Turner, and R. Feil. 2001. DNA methylation is linked to deacetylation of histone H3, but not H4, on the imprinted genes Snrpn and U2af1-rs1. Mol. Cell. Biol. 21:5426-5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gruenbaum, Y., R. Stein, H. Cedar, and A. Razin. 1981. Methylation of CpG sequences in eukaryotic DNA. FEBS Lett. 124:67-71. [DOI] [PubMed] [Google Scholar]
- 26.Hamada-Kanazawa, M., K. Ishikawa, K. Nomoto, T. Uozumi, Y. Kawai, M. Narahara, and M. Miyake. 2004. Sox6 overexpression causes cellular aggregation and the neuronal differentiation of P19 embryonic carcinoma cells in the absence of retinoic acid. FEBS Lett. 560:192-198. [DOI] [PubMed] [Google Scholar]
- 27.Hendrich, B., C. Abbott, H. McQueen, D. Chambers, S. Cross, and A. Bird. 1999. Genomic structure and chromosomal mapping of the murine and human Mbd1, Mbd2, Mbd3, and Mbd4 genes. Mamm. Genome 10:906-912. [DOI] [PubMed] [Google Scholar]
- 28.Hendrich, B., and A. Bird. 1998. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell. Biol. 18:6538-6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hendrich, B., and A. Bird. 2000. Mammalian methyltransferases and methyl-CpG-binding domains: proteins involved in DNA methylation. Curr. Top. Microbiol. Immunol. 249:55-74. [DOI] [PubMed] [Google Scholar]
- 30.Hendrich, B., U. Hardeland, H. H. Ng, J. Jiricny, and A. Bird. 1999. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature 401:301-304. [DOI] [PubMed] [Google Scholar]
- 31.Hsieh, C. L. 1994. Dependence of transcriptional repression on CpG methylation density. Mol. Cell. Biol. 14:5487-5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu, X., J. Bi, H. H. Loh, and L. N. Wei. 2001. An intronic Ikaros-binding element mediates retinoic acid suppression of the kappa opioid receptor gene, accompanied by histone deacetylation on the promoters. J. Biol. Chem. 276:4597-4603. [DOI] [PubMed] [Google Scholar]
- 33.Hwang, C. K., C. S. Kim, H. S. Choi, S. R. McKercher, and H. H. Loh. 2004. Transcriptional regulation of mouse mu opioid receptor gene by PU 1. J. Biol. Chem. 279:19764-19774. [DOI] [PubMed] [Google Scholar]
- 34.Hwang, C. K., X. Wu, G. Wang, C. S. Kim, and H. H. Loh. 2003. Mouse mu opioid receptor distal promoter transcriptional regulation by SOX proteins. J. Biol. Chem. 278:3742-3750. [DOI] [PubMed] [Google Scholar]
- 35.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 36.Jones, P. L., G. J. Veenstra, P. A. Wade, D. Vermaak, S. U. Kass, N. Landsberger, J. Strouboulis, and A. P. Wolffe. 1998. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 19:187-191. [DOI] [PubMed] [Google Scholar]
- 37.Jones-Villeneuve, E. M., M. W. McBurney, K. A. Rogers, and V. I. Kalnins. 1982. Retinoic acid induces embryonal carcinoma cells to differentiate into neurons and glial cells. J. Cell Biol. 94:253-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kazama, H., T. Kodera, S. Shimizu, H. Mizoguchi, and K. Morishita. 1999. Ecotropic viral integration site-1 is activated during, and is sufficient for, neuroectodermal P19 cell differentiation. Cell Growth Differ. 10:565-573. [PubMed] [Google Scholar]
- 39.Kieffer, B. L., and C. J. Evans. 2002. Opioid tolerance—in search of the holy grail. Cell 108:587-590. [DOI] [PubMed] [Google Scholar]
- 40.Kim, C. S., H. S. Choi, C. K. Hwang, K. Y. Song, B. K. Lee, P. Y. Law, L. N. Wei, and H. H. Loh. 2006. Evidence of the neuron-restrictive silencer factor (NRSF) interaction with Sp3 and its synergic repression to the mu opioid receptor (MOR) gene. Nucleic Acids Res. 34:6392-6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim, C. S., C. K. Hwang, H. S. Choi, K. Y. Song, P. Y. Law, L. N. Wei, and H. H. Loh. 2004. Neuron-restrictive silencer factor (NRSF) functions as a repressor in neuronal cells to regulate the mu opioid receptor gene. J. Biol. Chem. 279:46464-46473. [DOI] [PubMed] [Google Scholar]
- 42.Klose, R. J., and A. P. Bird. 2006. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 31:89-97. [DOI] [PubMed] [Google Scholar]
- 43.Klose, R. J., S. A. Sarraf, L. Schmiedeberg, S. M. McDermott, I. Stancheva, and A. P. Bird. 2005. DNA binding selectivity of MeCP2 due to a requirement for A/T sequences adjacent to methyl-CpG. Mol. Cell 19:667-678. [DOI] [PubMed] [Google Scholar]
- 44.Ko, J. L., H. C. Chen, and H. H. Loh. 2002. Differential promoter usage of mouse mu-opioid receptor gene during development. Brain Res. Mol. Brain Res. 104:184-193. [DOI] [PubMed] [Google Scholar]
- 45.Ko, J. L., H. C. Liu, S. R. Minnerath, and H. H. Loh. 1998. Transcriptional regulation of mouse mu-opioid receptor gene. J. Biol. Chem. 273:27678-27685. [DOI] [PubMed] [Google Scholar]
- 46.Ko, J. L., S. R. Minnerath, and H. H. Loh. 1997. Dual promoters of mouse mu-opioid receptor gene. Biochem. Biophys. Res. Commun. 234:351-357. [DOI] [PubMed] [Google Scholar]
- 47.Kraus, J., C. Borner, E. Giannini, and V. Hollt. 2003. The role of nuclear factor κB in tumor necrosis factor-regulated transcription of the human mu-opioid receptor gene. Mol. Pharmacol. 64:876-884. [DOI] [PubMed] [Google Scholar]
- 48.Kraus, J., G. Horn, A. Zimprich, T. Simon, P. Mayer, and V. Hollt. 1995. Molecular cloning and functional analysis of the rat mu opioid receptor gene promoter. Biochem. Biophys. Res. Commun. 215:591-597. [DOI] [PubMed] [Google Scholar]
- 49.Lachner, M., and T. Jenuwein. 2002. The many faces of histone lysine methylation. Curr. Opin. Cell Biol. 14:286-298. [DOI] [PubMed] [Google Scholar]
- 50.Lachner, M., D. O'Carroll, S. Rea, K. Mechtler, and T. Jenuwein. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116-120. [DOI] [PubMed] [Google Scholar]
- 51.Law, P. Y., Y. H. Wong, and H. H. Loh. 2000. Molecular mechanisms and regulation of opioid receptor signaling. Annu. Rev. Pharmacol. Toxicol. 40:389-430. [DOI] [PubMed] [Google Scholar]
- 52.Li, E., T. H. Bestor, and R. Jaenisch. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69:915-926. [DOI] [PubMed] [Google Scholar]
- 53.Li, J., S. W. Park, H. H. Loh, and L. N. Wei. 2002. Induction of the mouse kappa-opioid receptor gene by retinoic acid in P19 cells. J. Biol. Chem. 277:39967-39972. [DOI] [PubMed] [Google Scholar]
- 54.Lyu, J., F. Costantini, E. H. Jho, and C. K. Joo. 2003. Ectopic expression of Axin blocks neuronal differentiation of embryonic carcinoma P19 cells. J. Biol. Chem. 278:13487-13495. [DOI] [PubMed] [Google Scholar]
- 55.Mansour, A., C. A. Fox, H. Akil, and S. J. Watson. 1995. Opioid-receptor mRNA expression in the rat CNS: anatomical and functional implications. Trends Neurosci. 18:22-29. [DOI] [PubMed] [Google Scholar]
- 56.Martinowich, K., D. Hattori, H. Wu, S. Fouse, F. He, Y. Hu, G. Fan, and Y. E. Sun. 2003. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science 302:890-893. [DOI] [PubMed] [Google Scholar]
- 57.McBurney, M. W. 1993. P19 embryonal carcinoma cells. Int. J. Dev. Biol. 37:135-140. [PubMed] [Google Scholar]
- 58.McBurney, M. W., E. M. Jones-Villeneuve, M. K. Edwards, and P. J. Anderson. 1982. Control of muscle and neuronal differentiation in a cultured embryonal carcinoma cell line. Nature 299:165-167. [DOI] [PubMed] [Google Scholar]
- 59.McBurney, M. W., K. R. Reuhl, A. I. Ally, S. Nasipuri, J. C. Bell, and J. Craig. 1988. Differentiation and maturation of embryonal carcinoma-derived neurons in cell culture. J. Neurosci. 8:1063-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Meehan, R. R., J. D. Lewis, S. McKay, E. L. Kleiner, and A. P. Bird. 1989. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell 58:499-507. [DOI] [PubMed] [Google Scholar]
- 61.Min, B. H., L. B. Augustin, R. F. Felsheim, J. A. Fuchs, and H. H. Loh. 1994. Genomic structure analysis of promoter sequence of a mouse mu opioid receptor gene. Proc. Natl. Acad. Sci. USA 91:9081-9085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nan, X., H. H. Ng, C. A. Johnson, C. D. Laherty, B. M. Turner, R. N. Eisenman, and A. Bird. 1998. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393:386-389. [DOI] [PubMed] [Google Scholar]
- 63.Nan, X., P. Tate, E. Li, and A. Bird. 1996. DNA methylation specifies chromosomal localization of MeCP2. Mol. Cell. Biol. 16:414-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ng, H. H., Y. Zhang, B. Hendrich, C. A. Johnson, B. M. Turner, H. Erdjument-Bromage, P. Tempst, D. Reinberg, and A. Bird. 1999. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat. Genet. 23:58-61. [DOI] [PubMed] [Google Scholar]
- 65.Oka, M., A. M. Meacham, T. Hamazaki, N. Rodic, L. J. Chang, and N. Terada. 2005. De novo DNA methyltransferases Dnmt3a and Dnmt3b primarily mediate the cytotoxic effect of 5-aza-2′-deoxycytidine. Oncogene 24:3091-3099. [DOI] [PubMed] [Google Scholar]
- 66.Oka, M., N. Rodic, J. Graddy, L. J. Chang, and N. Terada. 2006. CpG sites preferentially methylated by Dnmt3a in vivo. J. Biol. Chem. 281:9901-9908. [DOI] [PubMed] [Google Scholar]
- 67.Park, S. W., M. D. Huq, H. H. Loh, and L. N. Wei. 2005. Retinoic acid-induced chromatin remodeling of mouse kappa opioid receptor gene. J. Neurosci. 25:3350-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Prokhortchouk, A., B. Hendrich, H. Jorgensen, A. Ruzov, M. Wilm, G. Georgiev, A. Bird, and E. Prokhortchouk. 2001. The p120 catenin partner Kaiso is a DNA methylation-dependent transcriptional repressor. Genes Dev. 15:1613-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rius, R. A., J. Barg, W. T. Bem, C. J. Coscia, and Y. P. Loh. 1991. The prenatal development profile of expression of opioid peptides and receptors in the mouse brain. Brain Res. Dev. Brain Res. 58:237-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ruzicka, B. B., and H. Akil. 1997. The interleukin-1β-mediated regulation of proenkephalin and opioid receptor messenger RNA in primary astrocyte-enriched cultures. Neuroscience 79:517-524. [DOI] [PubMed] [Google Scholar]
- 72.Sarraf, S. A., and I. Stancheva. 2004. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol. Cell 15:595-605. [DOI] [PubMed] [Google Scholar]
- 73.Shiota, K., Y. Kogo, J. Ohgane, T. Imamura, A. Urano, K. Nishino, S. Tanaka, and N. Hattori. 2002. Epigenetic marks by DNA methylation specific to stem, germ and somatic cells in mice. Genes Cells 7:961-969. [DOI] [PubMed] [Google Scholar]
- 74.Struhl, K. 1998. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 12:599-606. [DOI] [PubMed] [Google Scholar]
- 75.Suzuki, S., T. Miyagi, T. K. Chuang, L. F. Chuang, R. H. Doi, and R. Y. Chuang. 2000. Morphine upregulates mu opioid receptors of human and monkey lymphocytes. Biochem. Biophys. Res. Commun. 279:621-628. [DOI] [PubMed] [Google Scholar]
- 76.Tang, K., J. Yang, X. Gao, C. Wang, L. Liu, H. Kitani, T. Atsumi, and N. Jing. 2002. Wnt-1 promotes neuronal differentiation and inhibits gliogenesis in P19 cells. Biochem. Biophys. Res. Commun. 293:167-173. [DOI] [PubMed] [Google Scholar]
- 77.Wade, P. A. 2001. Methyl CpG-binding proteins and transcriptional repression. Bioessays 23:1131-1137. [DOI] [PubMed] [Google Scholar]
- 78.Wade, P. A., A. Gegonne, P. L. Jones, E. Ballestar, F. Aubry, and A. P. Wolffe. 1999. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat. Genet. 23:62-66. [DOI] [PubMed] [Google Scholar]
- 79.Wang, G., T. Liu, L. N. Wei, P. Y. Law, and H. H. Loh. 2005. DNA methylation-related chromatin modification in the regulation of mouse delta-opioid receptor gene. Mol. Pharmacol. 67:2032-2039. [DOI] [PubMed] [Google Scholar]
- 80.Wang, G., L. N. Wei, and H. H. Loh. 2003. Transcriptional regulation of mouse delta-opioid receptor gene by CpG methylation: involvement of Sp3 and a methyl-CpG-binding protein, MBD2, in transcriptional repression of mouse delta-opioid receptor gene in Neuro2A cells. J. Biol. Chem. 278:40550-40556. [DOI] [PubMed] [Google Scholar]
- 81.Watt, F., and P. L. Molloy. 1988. Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes Dev. 2:1136-1143. [DOI] [PubMed] [Google Scholar]
- 82.Wenger, R. H., I. Kvietikova, A. Rolfs, G. Camenisch, and M. Gassmann. 1998. Oxygen-regulated erythropoietin gene expression is dependent on a CpG methylation-free hypoxia-inducible factor-1 DNA-binding site. Eur. J. Biochem. 253:771-777. [DOI] [PubMed] [Google Scholar]
- 83.Zegerman, P., B. Canas, D. Pappin, and T. Kouzarides. 2002. Histone H3 lysine 4 methylation disrupts binding of nucleosome remodeling and deacetylase (NuRD) repressor complex. J. Biol. Chem. 277:11621-11624. [DOI] [PubMed] [Google Scholar]
- 84.Zhang, Y., H. H. Ng, H. Erdjument-Bromage, P. Tempst, A. Bird, and D. Reinberg. 1999. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 13:1924-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhu, Y., M. S. Hsu, and J. E. Pintar. 1998. Developmental expression of the mu, kappa, and delta opioid receptor mRNAs in mouse. J. Neurosci. 18:2538-2549. [DOI] [PMC free article] [PubMed] [Google Scholar]