Abstract
A common feature of animal circadian clocks is the progressive phosphorylation of PERIOD (PER) proteins from hypo- to hyperphosphorylated species, events that are highly dependent on casein kinase 1ɛ (termed DOUBLETIME [DBT] in Drosophila melanogaster) and necessary for normal clock progression. Drosophila PER (dPER) functions in the negative limb of the clockworks by presumably binding to the transcription factor CLOCK (CLK) and inhibiting its transactivation activity. Here, we identify a small region on dPER that is conserved with mammalian PERs and contains the major in vivo DBT binding domain, termed dPDBD (for dPER DBT binding domain). This domain is required for the manifestation of molecular and behavioral rhythms in vivo. In the absence of the dPDBD, the dPER protein is present at constant high levels throughout a daily cycle, undergoes little phosphorylation, and is severely impaired in its ability to function as a transcriptional repressor. Our findings indicate that the binding of dPER to CLK is not sufficient for transcriptional inhibition, implicating a more indirect mode of action whereby dPER acts as a molecular bridge to “deliver” DBT and/or other factors that directly repress CLK-dependent gene expression.
Circadian (≅24-h) rhythms are a pervasive feature of the temporal organization exhibited by life-forms ranging from bacteria to plants and humans (30). These rhythms are driven by intracellular clocks or pacemakers that share a similar mechanism. The central organizing principle is based on interconnected transcriptional-translational feedback loops that produce coordinated rhythms in “clock” RNAs and proteins that are required for the daily progression of clocks, synchronization to local time, and transducing temporal signals to downstream effector pathways (17, 72). Work using Drosophila melanogaster has been instrumental in our understanding of clock mechanisms in general and especially in animals, since analogous clock genes operate in both systems (18, 29).
The intracellular clock mechanism in Drosophila is largely depicted as two interconnected transcriptional feedback loops with overlaying posttranslational regulatory circuits (4, 28). Prominent players in the first or “major” loop are PERIOD (PER; herein referred to as Drosophila PER [dPER]), TIMELESS (TIM), CLOCK (dCLK) and CYCLE (CYC). dCLK and CYC are transcription factors of the basic helix-loop-helix/PAS (Per-Arnt-Sim) superfamily that heterodimerize to stimulate the daily transcription of dper and tim, in addition to other clock and downstream genes (3, 15, 56). A key aspect of the temporal regulation in dCLK-CYC activity involves the phase-specific accumulation of dPER and TIM in the nucleus, whereby dPER (and less likely TIM) interacts with the dCLK-CYC heterodimer, inhibiting transactivation (15, 40, 41, 55). After several hours in the nucleus, the levels of TIM and dPER undergo sharp decreases, relieving autoinhibition and initiating another round of dCLK-CYC-dependent transcription. In the so-called “second” loop, cycles in dClk expression are driven by the alternating actions of two bZip transcription factors, VRILLE and PDP1ɛ (13, 25). A very similar circuitry operates in mammalian circadian clocks, which includes the participation of mammalian CLK (mCLK), BMAL1 (homolog of CYC), and several PER homologs (mammalian PER protein 1 [mPER1], mPER2, and mPER3) (37).
Temporal changes in dPER phosphorylation are thought to play a central role in controlling dPER function such that it operates in a phase-specific manner to inhibit dCLK-CYC-dependent transcription only during certain times in a daily cycle (i.e., from approximately early/mid-night to midday) (4). Newly synthesized dPER is initially present as a hypophosphorylated variant(s) in the late day/early night, progressively increasing in the extent of phosphorylation such that by the late night/early day only hyperphosphorylated species are detected (19). Phosphorylation has been linked to regulation of dPER stability (26, 35, 38, 53), nucleocytoplasmic distribution (2, 7, 14, 44, 45, 54), and possibly transcriptional repressor potency (51).
The DOUBLETIME (DBT) kinase (homolog of mammalian casein kinase 1ɛ [CK1ɛ]) is a key kinase controlling the temporal program underlying dPER phosphorylation and stability (35, 53). In a presumptive dbt-null mutant (dbtP), dPER is hypophosphorylated and attains high constant levels (53). DBT associates with dPER through most or all of its daily life cycle (36) and is thought to modulate the phosphorylation and stability of dPER in both the cytoplasm and the nucleus (53). Newly synthesized dPER in the cytoplasm is rapidly degraded in a DBT-dependent manner. As the levels of TIM increase during the day, it binds dPER (but see reference 48) and somehow protects it against DBT-mediated degradation. The interaction of dPER with TIM also stimulates but is not obligatory for nuclear localization of both proteins (49, 57, 60, 68). Nuclear entry/accumulation of dPER and TIM are also controlled by at least three kinases, DBT, CK2, and SHAGGY/glycogen synthase kinase 3β, with the former two likely directly targeting dPER and the last likely targeting TIM (2, 7, 14, 44, 45, 47). In the nucleus, DBT and TIM continue to modulate the metabolism of dPER, whereby the earlier drop in the abundance of TIM somehow accelerates the progressive multiphosphorylation of dPER (36, 55). Hyperphosphorylated isoforms of dPER are targeted to the 26S proteasome by the F-box protein SLIMB (26, 38). Rapidly declining levels of dPER severely diminish or terminate its repressor function on dCLK-CYC activity. In addition to kinases, protein phosphatase 2A and possibly other phosphatases regulate the phosphorylated state of dPER and modulate the clockworks (58).
mPERs undergo similar cycles in phosphorylation, abundance, and subcellular localization, with prominent roles for CK1ɛ and its very close variant CK1δ (20, 42). Indeed, mutations that affect the phosphorylation of human PER2 (65) or the activity of CK1δ (71) are causally linked to familial sleep disorders. Moreover, it appears that the cyclical phosphorylation of one or more clock proteins is the core biochemical foundation in generating an oscillator with circadian properties (50). Here we identify the major or sole region on dPER that mediates stable interactions with DBT. In the absence of this region (herein termed dPER DBT binding domain [dPDBD]), the altered dPER protein [dPER(Δ)] does not support molecular or behavioral rhythms and is constitutively expressed at elevated levels in a hypophosphorylated manner. Surprisingly, although dPER(Δ) retains its ability to bind TIM and dCLK it is a very poor repressor, indicating that the stable association of dPER with dCLK is not sufficient to block transcription. Our findings offer new perspectives on the role of dPER in the negative limb of the Drosophila clockworks.
MATERIALS AND METHODS
Plasmids.
The pAct-dper, pMT-dClk-V5, and pMT-dbt-V5 plasmids were described previously (9, 34, 38). To construct either full-length or truncated versions of dper-V5, the desired coding regions of dper were amplified by PCR in the presence of pAct-dper and the relevant fragments subcloned into pAc5.1/V5-His (Invitrogen). The pAct-dper(Δ755-809)-V5 construct was generated by site-directed mutagenesis using a QuikChange site-directed mutagenesis kit (Stratagene) and the internally deleted dper fragment subcloned into the pAc5.1/V5-His vector. A similar strategy was used to generate the pAct-dper(S/T9A) plasmid, which encodes a nontagged dPER variant that has nine conserved Ser/Thr residues in the region between amino acids (aa) 755 and 809 changed to Ala residues. All the dper constructs were verified by DNA sequencing. To generate pAct-tim-3HA, the entire tim open reading frame was amplified by PCR in the presence of pAct-tim (9) and exchanged for the slimb fragment in the pAct-slimb-3HA vector (38).
Transgenic flies.
To generate transgenic flies that produce the dPER(Δ) protein, we used a previously described CaSpeR-4-based transformation vector containing a 13.2-kb genomic dper insert that was modified with sequences encoding the hemagglutinin (HA) epitope tag and a stretch of histidine residues just upstream of the dper translation stop signal, termed 13.2(per+-HA10His) (herein referred to as per+-HAHis) (40). Deletion of sequences encoding aa 755 to 809 from dPER was performed using a QuikChange site-directed mutagenesis kit (Stratagene, CA) with an appropriate dper genomic subfragment, confirmed by DNA sequencing, and reconstructed into the above-mentioned transformation vector to yield 13.2(per(Δ)-HA10His) [herein referred to more simply as per(Δ)-HAHis]. Transgenic flies were generated by Genetics Services Inc. (Sudbury, MA) using standard P element-mediated transformation techniques and w1118 embryos as hosts. Three independent germ line transformants bearing the per(Δ)-HAHis transgene in a per+ background (w1118per+; herein referred to as wper+) were obtained and then crossed into a wper01 genetic background to yield wper01; per(Δ)-HAHis. Similar procedures were also performed with the original version of the per+-HAHis transgene (40) to generate appropriate control groups expressing a wild-type version of the recombinant dPER protein in the same w1118 and wper01 genetic backgrounds. For the wild-type control transgenic flies, we show behavioral and molecular results obtained with one of the lines (M16), which are representative of other independent lines bearing the per+-HAHis transgene (data not shown).
Locomotor activity.
The locomotor activities of individual flies were measured as previously described using the monitoring system from Trikinetics (Waltham, MA) (27). Young adult male flies were used for the analysis and kept in incubators at 25°C, exposed to at least 4 days of 12 h of light followed by 12 h of dark (12:12LD, where zeitgeber time zero [ZT0] is defined as the time when the light phase begins) and subsequently kept in constant-darkness conditions (DD) for 5 to 8 days. The locomotor activity data for each individual fly were analyzed using the FaasX software (Fly Activity Analysis Suite for MacOSX), which was generously provided by F. Rouyer. Periods were calculated for each individual fly using chi-square periodogram analysis and pooled to obtain a group average for each independent transgenic line or genotype. Power is a quantification of the relative strength of the rhythm during DD. Individual flies with a power of ≥10 and a “width” value (denoting the number of peaks in 30-min increments above the periodogram 95% confidence line) of 2 or more were considered rhythmic.
Methods using cultured S2 cells.
S2 cells were obtained from Invitrogen and transfected using Cellfectin reagent (Invitrogen) as described previously (38). Exogenous DBT was induced in cells transfected with pMT-dbt by adding CuSO4 to a final concentration of 500 μM in the media. RNA-mediated interference (RNAi) directed against endogenously expressed slimb was performed as described previously (38, 70). Briefly, after the addition of double-stranded RNA against slimb (38), cells were allowed to recover for 2 days, transfected with the desired plasmids, and incubated for a further 1.5 days before being harvested. dCLK-dependent transactivation using a luciferase (luc) reporter assay was performed as described previously, with slight modifications (15). Briefly, S2 cells were placed in 12-well plates and transfected with pAct-dper(1-1224) and/or pAct-dper(Δ) plasmids at the indicated amounts, along with 10 ng of perEluc, 30 ng of pAct-β-gal-V5/His, and 2 ng of pMT-dClk-V5. One day after transfection, dClk expression was induced with 500 μM CuSO4 (final in the media), and after another day cells were washed in phosphate-buffered saline, followed by lysis in 300 μl of reporter lysis buffer (Promega). Aliquots of cell extracts were assayed for β-galactosidase (β-Gal) and luciferase activities using the luciferase assay system and protocols supplied by the manufacturer (Promega).
Immunoblotting.
To prepare cell extracts for immunoblotting of proteins in cultured S2 cells, the cells were washed with phosphate-buffered saline and treated as previously described (34, 38), except that additional buffers were used depending on the protein sought (extracts prepared for immunoprecipitation are described below). When dClk plasmids were not included in the transfection, cells were homogenized in our standard EB1 solution (10 mM HEPES [pH 7.5], 5 mM Tris-HCl [pH 7.5], 50 mM KCl, 10% glycerol, 2.5 mM EDTA, 5 mM dithiothreitol [DTT], 0.2% Triton X-100, and 2.5 mM NaF), with the addition of a protease inhibitor cocktail at the manufacturer's recommended concentration (Roche) (34, 38). When we wanted to detect recombinant dCLK, extracts were prepared in harsher conditions with radioimmunoprecipitation assay (RIPA) buffer (25 mM Tris-HCl [pH 7.5], 50 mM NaCl, 0.5% sodium deoxycholate, 0.5% NP-40, 0.1% sodium dodecyl sulfate [SDS]), with the addition of a protease inhibitor cocktail (Roche). Prior work showed that harsher conditions are more efficient for extracting dCLK (references 34 and 73 and data not shown). In the case of fly material, flies were collected by freezing at the indicated times in LD and total fly head extracts prepared using either mild (EB1) or harsh (RIPA) conditions, as indicated. Gels with differing polyacrylamide concentrations were used to resolve different proteins (6% for dPER, TIM, and dCLK, and 12% for DBT). In the case of dCLK, 5% Criterion gels (Bio-Rad) were also used. Primary antibodies were used at the following dilutions: for anti-V5 (Invitrogen), 1:5,000; for anti-PER (GP73), 1:3,000 (62); for anti-HA (12CA5; Roche), 1:2,000; for anti-HA (3F10; Roche), 1:1,000; for anti-TIM (TR3), 1:2,000 (62); for anti-dCLK (GP208 [34] [see below] or GP-47 [32]), 1:2000; and for anti-DBT (GP292), 1:2,000 (see below).
To generate antibodies to DBT, we used PCR to amplify two dbt cDNA sequences that encode either aa 1 to 177 (dbt-N) or aa 253 to 440 (dbt-C) and cloned the relevant fragments into pET-21b (Novagen). The oligonucleotide primers used in the PCR were as follows (dbt sequences are underlined): 5′-ATCAATTCGATGGAGCTGCGCGTGGG-3′ and 5′-ATAAGCTTGGCAGTGCCCGTGAGGTTC-3′ for dbt-N, and 5′-ATGAATTCGAACTTCTGTCGCCAGATG-3′ and 5′-ATAAGCTTTTTGGCGTTCCCCACGCC-3′ for dbt-C. Purified fusion proteins were used to produce antibodies in guinea pigs (Cocalico Biologicals, Reamstown, PA). The most specific antibodies to DBT were obtained when using DBT-N as the immunogen, and one antiserum (GP292) was used in this study. We also raised novel anti-dCLK antiserum in guinea pigs (Cocalico Biologicals, Reamstown, PA) by use of the same immunogen previously described (40), and in this study we used GP208, which showed the highest dCLK staining intensity with low background (data not shown). Relevant bands on films were quantified using a charge-coupled-device camera and Alphaimager 2200 V5.5 software from Alpha Inotech Corporation.
Immunoprecipitation.
For immunoprecipitations, cell extracts, either from S2 cells or fly heads, were prepared using modified RIPA buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate) with the addition of a protease inhibitor cocktail (Roche). To the extracts, 3 μl of anti-HA (12CA5), anti-V5, or anti-PER (GP73) antibody was added, depending on the target protein sought, and incubated with gentle rotation for 3 to 5 h at 4°C followed by the addition of 5 μl of Dynabeads Protein A (Invitrogen) with a further incubation of 1 to 2 h. Beads were collected using DynalMPC. Immune complexes were mixed with 30 μl of 1× SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer and incubated for 10 min at 65°C, and the resulting supernatants were revolved by immunoblotting as described above. To detect interactions between DBT and dPER in flies (see Fig. 6A), 20 μl of either HA-agarose (Sigma) or the nonspecific V5-agarose (Sigma) was added to total fly head extracts. Immune complexes were mixed with 0.1 M glycine-HCl (pH 2.7) buffer and incubated for 10 min at 25°C to release bound proteins. Beads were collected by light centrifugation and the resulting supernatants mixed with 4× SDS sample buffer and resolved on 12% polyacrylamide gels, followed by immunoblotting in the presence of anti-DBT antibodies. Phosphatase treatment of immune complexes was performed as described previously (40) except that Dynabeads Protein A was used instead of Gammabind Plus (Pharmacia).
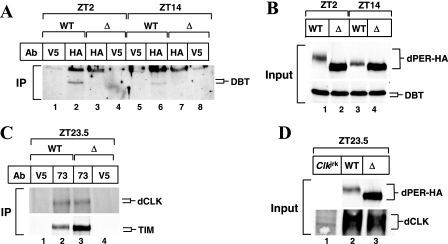
FIG. 6.
dPER(Δ) stably interacts with TIM and dCLK but not with DBT in flies. wper01; per+-HAHis (WT; line M16) and wper01; per(Δ)-HAHis flies (Δ; line F21) or Clkjrk flies were collected at the indicated times in LD. (A, C) Head extracts were prepared and subjected to immunoprecipitation. For panel A, anti-HA-agarose was used to purify HA-tagged versions of dPER, whereas anti-V5-agarose was used as a nonspecific control, and for panel C, anti-dPER (GP73) antibodies were used to immunoprecipitate dPER, whereas anti-V5 antibodies were used as a nonspecific control. (B and D) Head extracts were prepared and directly analyzed by immunoblotting in the presence of anti-HA (3F10, to detect dPER-HA), anti-DBT (GP292), or anti-dCLK (GP208) antibodies. Abbreviations: IP, immunoprecipitation; Ab, antibody.
In vitro phosphorylation.
For the in vitro kinase assay, pAct-dper-V5 or pAct-dper(Δ)-V5 was transfected into S2 cells and incubated for 48 h (see Fig. 2C). Extracts were prepared in EB1 buffer (see above), and dPER or dPER(Δ) was immunoprecipitated using anti-dPER antibodies (GP73) and Gammabind Plus Sepharose beads (Pharmacia), as described above. Subsequently, the immune complexes were equilibrated in CK1 reaction buffer (50 mM Tris-HCl [pH 7.5], 10 mM MgCl2, 5 mM dithiothreitol, and 200 μM ATP) and subjected to an in vitro kinase reaction with purified recombinant CK1δ (New England Biolabs, Beverly, MA) according to the supplier's protocols. Briefly, 500 units of recombinant CK1δ and 10 μCi of [γ-32P]ATP were added to beads containing either dPER or dPER(Δ) in 50 μl of CK1 reaction buffer on ice, followed by incubation for 30 min at 30°C. Reactions were terminated by the addition of 2× SDS-PAGE sample buffer and resolved on 6% polyacrylamide gels. Subsequently, gels were dried and radioactive bands visualized using a Typhoon 9400 Imager, and the intensity was quantified using Imagequant software (Molecular Dynamics).
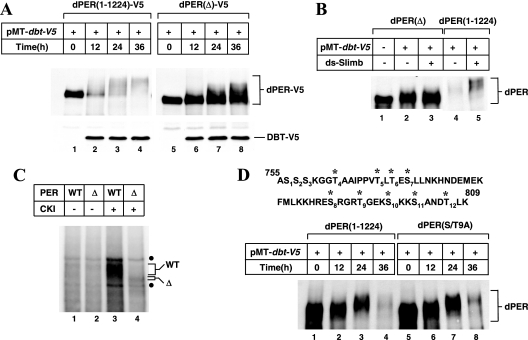
FIG. 2.
Deletion of dPER aa 755 to 809 strongly attenuates DBT-dependent hyperphosphorylation and degradation of dPER in S2 cells. (A to D) S2 cells were transfected with 600 ng of V5-tagged (A) or nontagged (B to D) versions of dper-containing plasmids (as indicated on top of panels). (A, B, and D) The presence (+) or absence (−) of 200 ng of pMT-dbt-V5 is indicated. Exogenous DBT was induced 36 h after transfection by adding to the media 500 μM CuSO4 (final). Cells were harvested at the indicated times, and extracts were analyzed by immunoblotting in the presence of either anti-V5 antibodies (A) or anti-dPER (GP73) antibodies (B and D) to visualize DBT or dPER. (B) Cells were either mock treated (−) or incubated (+) with RNAi directed against endogenous slimb (ds-Slimb). (C) Immunoprecipitated dPER(1-1224) (indicated as WT) or dPER(Δ) (indicated as Δ) were incubated with (+) or without (−) 500 units of CKIδ in the presence of [γ-32P]ATP, and radiolabeled bands were visualized by PAGE and autoradiography, as described in Materials and Methods. •, nonspecific bands. (D) At the top of the panel are shown dPER amino acid sequences from 755 to 809. *, Ser and Thr residues that were mutated to Ala to generate the dPER(S/T9A) version of dPER.
RT-PCR.
The relative mRNA levels of dper and dClk were measured by semiquantitative reverse transcription-PCR (RT-PCR) essentially as described previously (46). Briefly, 2 μg of total RNA was used to synthesize cDNA, and a 2-μl aliquot of the reaction was further processed by PCR in a final volume of 50 μl using either dper- or dClk-specific primers. For dper amplification, primers were P7197 (5′-TCTACATTATCCTCGGCTTGC-3′) and P6869 (5′-TAGTAGCCACACCCGCAGT-3′), as previously described (46). For dClk, we used the following set of primers: dClk6856F (5′-TCCTGAAGTCCACGATAGCC-3′) and dClk7105R (5′-TTCCGAGGCGTAAAAGATGG-3′). To control for sample-to-sample differences in total RNA, we also included primers for the noncycling mRNA coding for CBP20 (cap binding protein 20) by use of the primers CBP495R (5′-CAACAGTTTGCCATAACCCC-3′) and CBP362F (5′-GTCTGATTCGTGTGGACTGG-3′), as previously described (46). PCR products were separated and visualized by electrophoresis on 2% agarose gels containing Gelstar (Cambrex Co.), and the bands were quantified using a Typhoon 9400 Imager.
ChIP assays.
Chromatin immunoprecipitation (ChIP) assays were performed as described previously (73) with the following modifications. Adult flies were frozen at the indicated times in LD, and isolated heads (<900 μl) were gently homogenized for 10 min at 25°C in <5 volumes of XIP homogenization buffer (50 mM HEPES at pH 8.0, 140 mM NaCl, 0.4% Igpel CA-630, 0.2% Triton X-100, 1% HCHO, 1 mM EDTA, 0.5 mM EGTA, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 mM Na3VO4, and 1 mM NaF). Glycine was then added to a final concentration at 0.125 M to stop the cross-linking reaction. Homogenates were filtered with 100-μm nylon meshes to remove cuticle. The cross-linked nuclei were harvested by centrifugation at 800 × g for 5 min, washed three times with XN wash buffer (20 mM Tris at pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1 mM DTT, 0.5 mM PMSF, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 2 μg/ml pepstatin A, 1 mM Na3VO4, and 1 mM NaF), and stored at −80°C for future use. Extracts used for ChIP were prepared by sonicating the cross-linked nuclei in sonication buffer (20 mM Tris at pH 7.5, 150 mM NaCl, 10% glycerol, 1% Triton X-100, 0.4% sodium deoxycholate, 0.1% SDS, 1 mM EDTA, 0.5 mM EGTA, 1 mM DTT, 0.5 mM PMSF, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 2 μg/ml pepstatin A, 1 mM Na3VO4, and 1 mM NaF) 15 times for 10 s each (150 s in total) by use of a Misonix XL2000 sonicator set at 3. The average size of sheared DNA fragments was <500 bp on agarose gels (data not shown). After centrifugation at 25,000 × g for 10 min, the supernatants were quantified by Bradford assay (7a). Fifty micrograms of extract was set aside as input, and 500 μg of extract was precleared and subjected to immunoprecipitation using GP-47 (dCLK antisera made in guinea pig [32]) or normal guinea pig serum (GPS). The preclearance, immunoprecipitation, and DNA extraction were performed as described previously (73). DNA from immunoprecipitates and inputs was suspended in 50 μl of 1× TE. This suspension was subjected to “hot” PCR after being diluted 1:20 for immunoprecipitated samples and 1:300 for input DNA (thus equaling 0.33% of input). For each 20 μl of PCR mixture, 1 μl of diluted DNA was used as the template. Semiquantitative “hot” PCR was performed as described previously (73). Bands from PCR products were resolved on polyacrylamide gels and quantified by densitometry. For each time point and genotype, the background signal from material collected in the presence of GPS antiserum was subtracted from the values obtained using anti-dCLK antibodies. Finally, dCLK-specific values were converted to percentages of input by use of the following formula: % input = (dCLK signal/10 × 20)/(input signal × 300) × 100%.
RESULTS
A region between aa 755 and 809 is essential for DBT-dependent hyperphosphorylation and SLIMB-mediated degradation of dPER in S2 cells.
Prior work using dPER fragments beginning at the amino terminus and fused to β-Gal implicated a stretch of 231 residues between aa 638 and 876 in dPER as necessary for its hyperphosphorylation and temporal instability in flies (16) (Fig. 1A). We previously showed that the DBT-dependent progressive phosphorylation and subsequent SLIMB-mediated degradation of dPER can be recapitulated in a simplified cell culture system by expressing recombinant proteins in Drosophila Schneider (S2) cells (38). In this system, dPER is expressed from a constitutive promoter (pAct), whereas exogenous DBT is induced using the copper-inducible metallothionein promoter (pMT). To further delineate the region on the 231-aa stretch of dPER presumably mediating DBT-dependent hyperphosphorylation and enhanced degradation, we generated additional versions of dPER (fused to the V5 epitope to facilitate detection) with truncations between aa 638 and 876 and evaluated their responses following DBT induction in cultured S2 cells (Fig. 1B).
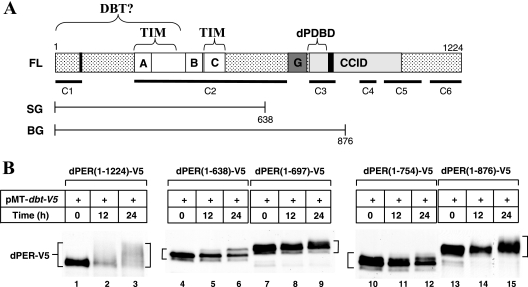
FIG. 1.
A region between aa 638 and 876 on dPER is essential for its DBT-dependent hyperphosphorylation in S2 cells. (A) Schematic representation of the full-length (FL) dPER open reading frame protein used in this study (1 to 1224; accession no. P07663) and the dPER regions in the dPER(1-638)-β-Gal (SG) and dPER(1-876)-β-Gal (BG) fusions previously generated (e.g., reference 16). Shown for the full-length version are the following domains: (i) putative NLS (aa 73 to 77 and 813 to 840; vertical black lines; it is not clear if aa 73 to 77 function as an NLS [see reference 10]), (ii) PAS domain (aa 238 to 512; white box) showing the PAS-A (letter “A”) and -B (letter “B”) repeats (33), (iii) cytoplasmic localization domain located in the PAS domain (aa 452 to 512; letter “C”) (57), (iv) Thr-Gly repeats (dark gray box with letter “G”); and (v) the dCLK-CYC inhibitory domain (aa 764 to 1034; CCID, light gray box) (10). Brackets above dPER indicate putative binding sites for TIM (24, 57) and DBT (35); work presented here indicates that the sole or major DBT binding region is the dPDBD (see text). Black horizontal lines below dPER represent regions that are highly conserved in different Drosophila species (12). (B) S2 cells were transfected with 600 ng of different versions of pAct-dper-V5 (as indicated at the tops of the panels) in combination with 200 ng of pMT-dbt-V5. Recombinant DBT was induced 36 h after transfection by adding to the media a final concentration of 500 μM CuSO4. Cells were harvested at the indicated times (h) after induction and extracts were analyzed by immunoblotting in the presence of anti-V5 antibodies to visualize the different versions of dPER-V5.
As previously shown using this experimental paradigm, the induction of DBT evokes dramatic increases in the phosphorylation of full-length dPER [dPER(1-1224)] that are readily observed as temporal decreases in electrophoretic mobility (38) (Fig. 1B, lanes 1 to 3, and 2A, lanes 1 to 4). Under the conditions used, full-length dPER exhibits a highly smeared pattern after 24 h of inducing dbt, which upon further incubation progresses to exclusively hyperphosphorylated species that are less stable (e.g., Fig. 2A, compare lanes 1 and 4). Notably different from this pattern, dPER(1-638), dPER(1-697), and dPER(1-754) fragments show only narrow mobility shifts that are characterized by the appearance of one or two sharp bands following induction of dbt, most likely due to limited phosphorylation (Fig. 1B, lanes 4 to 12). Although the mobility shifts do not appear as extensive for dPER(1-876) as for dPER(1-1224), this fragment displays a more smeared mobility pattern indicative of multiphosphorylation (lanes 13 to 15), consistent with prior work with transgenic flies expressing the dPER(1-876)-β-Gal fusion (Fig. 1A) (16). We also analyzed an extensive series of other dPER fragments and observed that strong DBT-dependent hyperphosphorylation requires aa 638 to 876 (reference 10 and data not shown). Together, our results suggest that although there are likely to be multiple areas on dPER that are phosphorylated or contribute to phosphorylation, the region between aa 755 and 876 plays a major role in the DBT-dependent hyperphosphorylation of dPER (Fig. 1B).
Based on several phosphorylation site prediction programs, there are multiple plausible CK1ɛ sites clustered between aa 755 and 809 of dPER (data not shown). We thus generated a dPER variant internally deleted from aa 755 to 809 [referred to as dPER(Δ)] and probed its response to DBT induction. Without induction of exogenous dbt, both the wild-type dPER(1-1224) and dPER(Δ) versions are exclusively hypophosphorylated and produced at comparable levels (Fig. 2A, compare lanes 1 and 5). Following dbt induction, the progressive accumulation of highly phosphorylated dPER was markedly delayed or attenuated for dPER(Δ) compared to that for full-length dPER (Fig. 2A). Importantly, the overall levels of dPER(Δ) did not undergo significant changes even after 36 h following induction of DBT, in sharp contrast to the wild-type control protein (Fig. 2A, compare lane 4 to lane 1 and lane 8 to lane 5). These results indicate that the region stimulating DBT-dependent hyperphosphorylation is also necessary for enhanced degradation, consistent with the observation that hyperphosphorylation of dPER is accompanied by rapid decreases in levels (26, 35, 38, 53). Similar results using a slightly smaller internal deletion (Δ768-792, according to the numbering used in this study) were recently obtained by others (51a). Moreover, while the degradation of dPER is significantly attenuated by RNAi targeting endogenous slimb expression as previously shown (38) (Fig. 2B, compare lanes 4 and 5), there was no effect on the levels or phosphorylated status of dPER(Δ) under the same conditions (Fig. 2B, compare lanes 2 and 3). This indicates that our inability to detect highly phosphorylated isoforms of dPER(Δ) is not due to their rapid degradation but most likely reflects inefficient phosphorylation (Fig. 2B). Indeed, unlike full-length dPER purified from S2 cells, which is extensively phosphorylated in vitro by recombinant mammalian CK1ɛ/δ (purified DBT does not exhibit in vitro kinase activity for unknown reasons, as previously noted [e.g., reference 63]), dPER(Δ) is a very poor substrate (Fig. 2C, compare lanes 3 and 4).
We considered the possibility that the region between aa 755 and 809 contains multiple phosphorylation sites that underlie the DBT-mediated hyperphosphorylation of dPER. There are 12 Ser/Thr residues in this region, of which 9 are highly conserved in dPER from other drosophilids (data not shown). We therefore altered all the conserved nine Ser/Thr residues to Ala, generating dPER(S/T9A) (Fig. 2D). Surprisingly, dPER(S/T9A) behaves similar to wild-type dPER in terms of phosphorylation and temporal changes in abundance when coexpressed with DBT. These findings suggest an alternative explanation whereby aa 755 to 809 are important for DBT binding and that this association subsequently facilitates DBT-mediated hyperphosphorylation of dPER at other sites. Indeed, the appearance of a minor fraction of hyperphosphorylated isoforms of dPER(Δ) in some experiments (e.g., that shown in Fig. 2A) further suggests that the region between aa 755 and 809 is not a major site directly modified by multiple phosphorylation events [although we cannot rule out the possibility that the DBT-induced multiphosphorylated species of dPER(Δ) arise from aberrant phosphorylation events].
The region encompassing dPER aa 755 to 809 is a major binding domain for DBT.
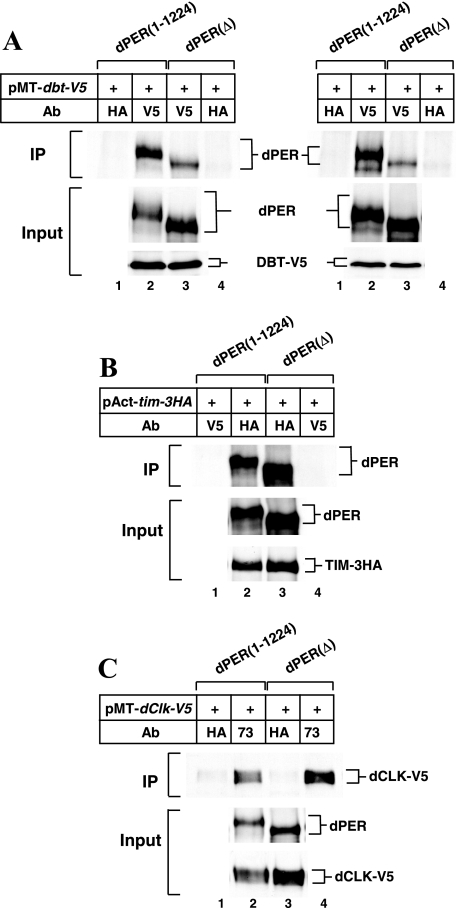
To test if dPER aa 755 to 809 play a role in the association of DBT with dPER, we performed immunoprecipitation assays between recombinant dPER(1-1224) or dPER(Δ) and DBT proteins expressed in S2 cells. Under the experimental conditions used, about two- to threefold less dPER(Δ) copurifies with DBT, even though there is more dPER(Δ) present in the extract than there is dPER(1-1224) (Fig. 3A, lanes 2 and 3; results from two independent experiments are shown). We varied the relative and absolute concentrations of the dbt and dper plasmids during transfection and still obtained very similar results, whereby less dPER(Δ) than dPER(1-1224) copurifies with DBT (data not shown). The observation that the interaction between dPER(Δ) and DBT is severely decreased but not abolished is consistent with the low levels of hyperphosphorylation (Fig. 2A, lanes 5 to 8). Importantly, dPER(Δ) stably interacts with TIM or dCLK with a relative efficiency the same as or better than that of the full-length dPER control (Fig. 3B, lanes 2 and 3, and C, lanes 2 and 4). These results show that deletion of aa 755 to 809 in dPER specifically affects its ability to stably interact with DBT. Similar results were obtained when we used GST-DBT bound to resins to examine the interaction with dPER and dPER(Δ) (data not shown). The fact that dPER(Δ) strongly binds TIM and dCLK indicates that the inability to associate with DBT is not due to nonspecific issues such as gross misfolding of the internally deleted dPER variant. Based on these and other results shown below we refer to the region between aa 755 and 809 as the dPDBD (for dPER DBT binding domain).
FIG. 3.
Strong binding of dPER(Δ) to TIM and dCLK but not DBT in S2 cells. S2 cells were transfected with 600 ng (A) or 400 ng (B, C) of either pAct-dper(1-1224) or pAct-dper(Δ) (as indicated at top of panels) in combination with 200 ng of pMT-dbt-V5 (A), 400 ng of pAct-tim-3HA (B), or 400 ng of pMT-dClk-V5 (C). (A, C) At 36 h after transfection, cells were incubated in media containing 500 μM CuSO4 (final) to induce ectopic expression of target proteins and harvested 12 h (A) or 24 h (C) later. (B) Cells were harvested 48 h after transfection without the addition of CuSO4. Extracts were prepared and either subjected to immunoprecipitation (IP) or analyzed directly (Input). Immune complexes were recovered with anti-V5 (added as the specific antibody [panel A] or as the nonspecific antibody [panel B]), anti-HA (added as the specific antibody [panel B] or as the nonspecific antibody [panels A and C]), or anti-dPER (added as the specific antibody [panel C]) antibodies. Immunoblots were probed with antibodies against dPER (GP73), V5 (to detect DBT or dCLK), or HA (to detect TIM), as indicated. In panel A, two independent experiments are shown side by side. Ab, antibody.
dPER(Δ) is highly impaired in its ability to inhibit dCLK-dependent transcriptional activation.
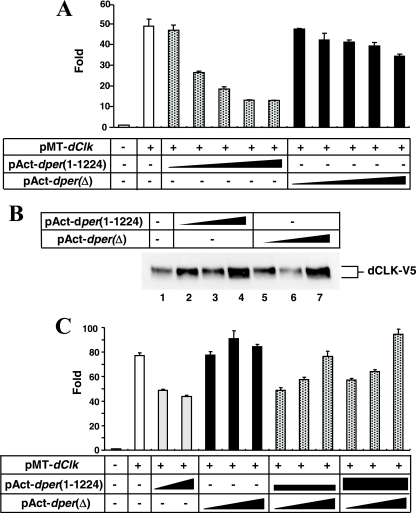
We next sought to evaluate the ability of dPER(Δ) to inhibit dCLK-CYC-mediated transactivation of gene expression using the well-established assay developed with S2 cells (15). In this system, dCLK is exogenously expressed (the levels of endogenously produced CYC are sufficient) and stimulates expression of a luc reporter fused to an E-box containing circadian promoter (usually dper or tim). Recent work has shown that dPER alone (i.e., without coexpression of TIM) acts as a potent repressor of dCLK-dependent transactivation in the cultured S2 cell system (10, 51, 69), consistent with findings with flies (14, 55). Unlike full-length dPER, dPER(Δ) has little repressor effect, exhibiting at most a 20% reduction in reporter gene expression at the highest dose tested (Fig. 4A). Interestingly, the dPDBD falls within a much larger region of dPER (aa 764 to 1034), previously identified as the minimal region giving over 50% inhibition of dCLK-dependent transcription in S2 cells (termed CCID [for CLK-CYC-inhibitory domain]) (10) (Fig. 1A). Variations in the total levels of dPER(1-1224) and dPER(Δ) cannot account for the differences in repressor activity (e.g., Fig. 2A, compare lanes 1 and 5). Furthermore, in our system we did not observe significant differences in the subcellular distributions of dPER(Δ) and to dPER(1-1224) (data not shown and Discussion). The levels of recombinant dCLK in cells coexpressing either dPER(1-1224) or dPER(Δ) (Fig. 4B) are comparable, indicating that the severely attenuated repressor function of dPER(Δ) is not due to secondary effects on dCLK metabolism (for reasons that are still unclear, the levels of dCLK slightly increase with high amounts of dPER [e.g., Fig. 4B, compare lane 1 to 4 and 7]). Finally, although we cannot rule out subtle effects on dCLK phosphorylation, dPER(1-1224) inhibits dCLK activity without any gross increases in dCLK phosphorylation. These results suggest that in our cell culture system, the hyperphosphorylation of dCLK is not absolutely required for its repression by dPER, consistent with our earlier in vitro results (41).
FIG. 4.
dPER(Δ) is a very weak repressor of dCLK-dependent transactivation in S2 cells. (A) Shown are the average values from three independent experiments for relative Luc activity in the absence (−) or presence (+) of pMT-dClk-V5. In addition, some cells were also cotransfected with various amounts of pAct-dper(1-1224) or pAct-dper(Δ), as indicated. Luc activity in the absence of transfecting pMT-dClk-V5 was set to 1, and all other values were normalized. The following different amounts (ng) of pAct-dper(1-1224) or pAct-dper(Δ) were used: 1, 5, 10, 50, and 100. (B) S2 cells were transfected either singly with 50 ng of pMT-dClk-V5 (−) or in combination with 50, 100, or 500 ng of pAct-dper(1-1224) or pAct-dper(Δ). Cells were incubated with 500 μM CuSO4 (final in the media) to induce ectopic expression of dCLK at 36 h after transfection and harvested 24 h later. Immunoblots were probed with antibodies against V5 to visualize dCLK-V5. Note that dCLK-V5 levels are very similar in cells expressing either dPER(1-1224) or dPER(Δ). (C) Shown are the average values from three independent experiments for relative Luc activity. Luc activity in the absence of pMT-dClk-V5 (−) was set to 1, and all other values were normalized. Two nanograms of pMT-Clk-V5 was used (+). pAct-dper(1-1224) and pAct-dper(Δ) were transfected singly or together in the presence (+) of pMT-Clk-V5. The following amounts of dper-containing plasmids were used in the transfections, as represented by the thicknesses of the black horizontal bars or triangles: 5 or 10 ng of pAct-dper(1-1224) and 5, 10, or 50 ng of pAct-dper(Δ).
Thus, the dPDBD is critical for the ability of dPER to act as an efficient repressor of dCLK-CYC (similar results were recently obtained by Nawathean and colleagues [51a]). We note that dPER(Δ) exhibits highly impaired repressor activity even in the absence of dbt induction, where only hypophosphorylated isoforms of dPER(Δ) and wild-type dPER are detected (e.g., Fig. 2A, compare lanes 1 and 5), suggesting that the lack of extensive phosphorylation or hyperphosphorylation does not explain the inability of dPER(Δ) to block dCLK-dependent transcription (see Discussion). Moreover, because dPER(Δ) interacts with dCLK (Fig. 3; also see Fig. 6), it is clear that the binding of dPER to dCLK is not sufficient for transcriptional inhibition. Nonetheless, the ability of dPER(Δ) to bind dCLK might explain why coexpression of dPER(Δ) interferes with the repressor activity of wild-type dPER (Fig. 4C and Table 1; also see below), suggesting that dPER(Δ) acts in a dominant-negative manner with regards to transcriptional autoinhibition.
TABLE 1.
Locomotor activity rhythms of per(Δ) and control per+ transgenic fliesa
| Genotype (transgenic line)b | Period (± SEM) (h)c | Powerd | Rhythmicity (%)e | Total fliesf |
|---|---|---|---|---|
| w1118 | 23.9 ± 0.2 | 49.2 | 91.7 | 12 |
| wper01; per+-HAHis (M16) | 23.4 ± 0.1 | 87.9 | 93.8 | 16 |
| wper01; per(Δ)-HAHis (F6) | AR | 1.4 | 0 | 17 |
| wper01; per(Δ)-HAHis (F11) | AR | 8.7 | 15.4 | 13 |
| wper01; per(Δ)-HAHis (F21) | AR | 4.2 | 4.8 | 21 |
| w+; per(Δ)-HAHis (F6) | 27.3 ± 0.6 | 15.5 | 37.5 | 16 |
| w+; per(Δ)-HAHis (F11) | 26.7 ± 0.3 | 39.7 | 75.0 | 16 |
| w+; per(Δ)-HAHis (F21) | 28.8 ± 0.3 | 11.2 | 26.7 | 15 |
Flies were kept at 25°C and exposed to 4 days of 12:12LD followed by 7 days of DD.
F6, F11, F21, and M16 denote independent transgenic lines.
AR, arhythmic.
Measure of the strength or amplitude of the rhythm.
Percentage of flies with activity rhythms having a power value of ≥10 and a width value of ≥2.
Total number of flies that survived until the end of the testing period.
Arrhythmic behavior in dper(Δ)-expressing flies.
To evaluate the physiological significance of the dPDBD in clock function, we generated transgenic flies harboring a dper transgene internally deleted for sequences encoding aa 755 to 809 of dper. The internal deletion was engineered into a previously characterized vector that contains a 13.2-kb dper genomic fragment [13.2(per+)] and also includes a HA epitope tag and a stretch of His moieties (10His) at the carboxy terminus of the dper open reading frame (per+-HAHis) protein, facilitating purification and surveillance of the recombinant protein (40). Several independent lines of transgenic flies bearing the dper(Δ) transgene [13.2(per(Δ)-HA10His)] were obtained and evaluated for circadian functionality in the per-null wper01 (39) genetic background [wper01; 13.2(per(Δ)-HA10His), herein more simply referred to as wper01; per(Δ)-HAHis]. Prior work showed that transgenic per01 flies harboring 13.2(per+)-based transgenes manifest robust locomotor activity rhythms with periods similar to those of per+ wild-type controls (Table 1).
For behavioral analysis, flies were initially entrained under standard conditions of 12:12LD, where ZT0 is defined as the time when the light phase begins, at 25°C, followed by at least 1 week in DD. In contrast to the wper01; per+-HAHis control transgenic flies, all the independent dper(Δ)-containing transformants were arrhythmic (Table 1 and data not shown). When dper(Δ) was introduced into a per+ background [w; per(Δ)-HAHis)], the proportion of arrhythmic flies significantly increased, and for those displaying rhythmic activity the amplitudes were strongly reduced and the periods lengthened by 3 to 4 h (Table 1). The semidominant effects of dPER(Δ) on behavioral rhythms might arise from an obstruction of the ability of wild-type dPER to inhibit dCLK-dependent transcription, as observed for S2 cells (Fig. 4C).
High constant levels of exclusively hypophosphorylated dPER(Δ) in flies.
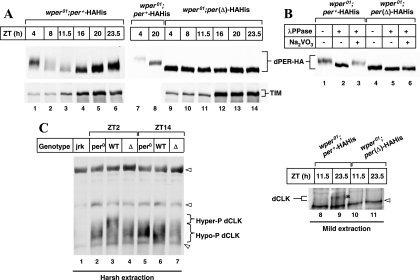
We next investigated the temporal regulation of dPER phosphorylation and abundance by analyzing head extracts prepared from flies collected at different times of day. The levels and time-of-day-specific phosphorylation of the control dPER-HAHis protein in wper01; per+-HAHis transgenic flies undergo daily cycles that are indistinguishable from those for endogenously expressed dPER (Fig. 5A, top panel, lanes 1 to 6, and data not shown). Moreover, TIM also exhibits normal cycles in abundance (Fig. 5A, bottom panel, lanes 1 to 6). Consistent with the behavior results, dPER(Δ)-HAHis does not undergo noticeable changes in mobility or abundance throughout a daily cycle (Fig. 5A, top panel, compare lanes 9 to 14 to lanes 7 and 8). Also, the overall abundance of dPER(Δ)-HAHis is approximately two- to threefold higher than the daily peak values for the wild-type control dPER protein (Fig. 5A, top right panel, compare lanes 8 and 9; also see Fig. 6B and D). In agreement with prior work (19), λ phosphatase treatment revealed that the electrophoretic mobility variants of dPER-HAHis are due to differential phosphorylation (Fig. 5B, lanes 1 to 3). In sharp contrast, dPER(Δ)-HAHis manifests only a slight increase in mobility when treated with λ phosphatase, indicating that the overwhelming majority is hypophosphorylated with some limited phosphorylation (Fig. 5B, compare lanes 4 and 5). Unlike the situation for S2 cells, for which we occasionally detected low levels of highly phosphorylated dPER(Δ) following dbt induction (Fig. 2A), we never observed extensively phosphorylated dPER(Δ) produced in flies, even when films were exposed for long periods (data not shown). It is possible that low-efficiency hyperphosphorylation of dPER(Δ) is possible in S2 cells where the relevant target proteins are highly expressed. TIM in wper01; per(Δ)-HAHis flies displayed low but constant levels during the day and higher constant amounts during the night (Fig. 5A, bottom panel, lanes 9 to 14). This profile is similar to that observed for per0 flies, where circadian cycling of TIM is abolished but daily changes arise from the direct photosensitivity of TIM (49, 74).
FIG. 5.
Elevated and constant levels of exclusively hypophosphorylated dPER(Δ) throughout a daily cycle in flies. Adult flies (genotypes indicated above panels) were collected at the indicated ZT. Head extracts were prepared and either directly analyzed by immunoblotting (A and C) or first subjected to immunoprecipitation using anti-PER (GP73) antibodies (B). (A and C) The following antibodies were used for immunoblotting; anti-HA (3F10) (A, top panel), anti-TIM (TR3) (A, bottom panel), anti-dCLK (C; GP-47, lanes 1 to 7; GP208, lanes 8 to 11). (B) Immune complexes were incubated in the absence (−) or presence (+) of λ phosphatase or Na2VO3 followed by immunoblotting using anti-HA (3F10) antibody to visualize dPER. (C) Extracts were prepared from the indicated genotypes {per0, w1118per01; jrk, Clkjrk; WT, wper01; per+-HAHis [line M16]; Δ, wper01; per(Δ)-HAHis [line F21]} using either harsh (RIPA buffer) or mild (EB1 buffer) extraction conditions as indicated. Clkjrk flies do not produce a full-length dCLK protein and serve as negative controls. *, dPER-dependent late-night-specific hyperphosphorylated isoform of dCLK (panel C, lane 9). arrowheads, nonspecific bands. Similar results, whereby hyperphosphorylated isoforms of dCLK were not observed for wper01; per(Δ)-HAHis, were obtained in at least three independent experiments using two different anti-dCLK antibodies (GP-47 and GP208). To simplify, we designated all the nonhyperphosphorylated dCLK as hypophosphorylated.
We recently showed that when head extracts are prepared using harsh conditions, immunoblotting analysis of dCLK reveals that its levels are relatively constant throughout a daily cycle (Fig. 5C, lanes 1 to 7) (34, 73). Moreover, there are several different dCLK phosphoisoforms (non- or hypophosphorylated to intermediately phosphorylated and hyperphosphorylated). The hyperphosphorylated isoform(s) is detected only in the late night/early day, is absent from per0 flies, and likely requires DBT activity (34, 73). This profile differs from that found in our earlier work, in which we used mild extraction conditions and showed a biochemical cycle in dCLK abundance with trough amounts reached during the late day and peak values attained in the late night/early day (e.g., Fig. 5C, compare lanes 8 and 9) (40). In contrast to harsh conditions, mild treatments do not efficiently release dCLK from chromatin, and it is extracted mainly during times of dPER-mediated repression (mid-night to midday) when dCLK is hyperphosphorylated and displaced from chromatin (73).
To examine the status of dCLK in dPER(Δ)-expressing flies, we collected flies at times in a daily cycle that represent relatively active (e.g., ZT11.5 and 14) and inactive (e.g., ZT23.5 and 2) phases of dCLK-CYC-driven circadian transcription, respectively. When we used harsh extraction conditions, the overall levels of dCLK were comparable in both wper01; per+-HAHis and wper01; per(Δ)-HAHis flies (Fig. 5C, lanes 3, 4, 6, and 7). A notable difference, however, is that hyperphosphorylated dCLK does not accumulate in wper01; per(Δ)-HAHis flies (Fig. 5C, compare lanes 3 and 4; to simplify, we designated all the nonhyperphosphorylated dCLK as hypophosphorylated). These results are consistent with the suggestion that dPER acts as a bridge to enhance dCLK phosphorylation by DBT (73), a function impaired in the dPER(Δ) mutant (Fig. 6A and Discussion). As previously shown (5, 40), with mild extraction conditions dCLK is readily observed in extracts prepared from flies collected at ZT23.5 but not in those from ZT11.5 (Fig. 5C, compare lanes 8 and 9). However, irrespective of daily time, dCLK is not observed in wper01; per(Δ)-HAHis head extracts prepared using mild extraction procedures (Fig. 5C, lanes 10 and 11), reinforcing the contention that dCLK is constitutively bound to chromatin in the presence of dPER(Δ) (Fig. 7).
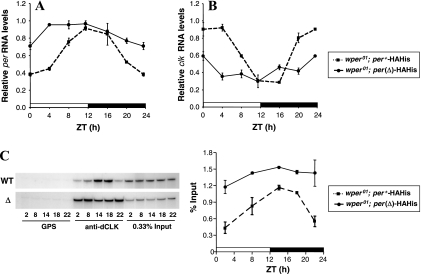
FIG. 7.
dPER(Δ) is highly impaired in its ability to repress dCLK-CYC-mediated expression in flies. wper01; per+-HAHis (M16) or wper01; per(Δ)-HAHis (F21) flies were collected at the indicated times in LD. (A and B) RT-PCR assays were used to measure the relative levels of total dper or dClk RNAs. (C) ChIP assays targeting the dper circadian regulatory sequence were performed in the presence of either nonspecific antibody (GPS) or anti-dCLK antibodies (GP-47). Shown is a representative gel of amplified bands following immunoprecipitation with GPS or anti-dCLK antibodies; also shown is 0.33% of the input DNA material (left panel). Densitometry quantification of bands from hot PCR analysis was calculated as a percentage of the total input DNA, and the results from two independent experiments were averaged (right panel).
dPER(Δ) interacts with dCLK and TIM but not DBT in flies.
To examine the in vivo interaction of dPER(Δ) with DBT, we collected wper01; per(Δ)-HAHis and wper01; per+-HAHis flies at ZT2 and 14, times in a daily cycle when wild-type dPER is present mainly as either hyper- or hypophosphorylated isoforms, respectively (Fig. 6B, top panel, compare lanes 1 and 3). Wild-type dPER stably interacts with DBT throughout a daily cycle (Fig. 6A, lanes 2 and 6) as previously reported (36) (the slightly stronger DBT staining at ZT2 is likely due to the higher overall levels of dPER at this time [Fig. 6B, top panel, compare lanes 1 and 3]). No DBT signal was observed when an irrelevant antibody was used (e.g., Fig. 6A, compare lanes 1 and 2), demonstrating specificity. In sharp contrast, dPER(Δ) does not copurify with DBT (Fig. 6A, compare lane 3 to lane 2 and lane 6 to lane 7), despite the much higher overall levels of dPER(Δ) than of wild-type dPER (Fig. 6B, top panel). Gross differences in DBT levels cannot explain the lack of interaction between dPER(Δ) and DBT (Fig. 6B, bottom panel). As with the difference between dPER(Δ) in flies and that in S2 cells, with more-dramatic effects on hyperphosphorylation seen for the former (compare Fig. 2A and 5A), the interaction between dPER(Δ) and DBT appears completely abolished in flies, unlike the situation for the cultured cells. As noted above, this could simply be a function of the higher concentrations attained in S2 cells or of other in vivo constraints that are not present in S2 cells. For example, the in vivo interaction between dPER and DBT might be stimulated by other factors that directly or indirectly interact with the dPDBD. In any event, our results strongly suggest that the stable interaction of dPER with DBT is required for dPER hyperphosphorylation and that the dPDBD is the major or sole region mediating this association in vivo.
Consistent with the S2 cell results, both dPER and dPER(Δ) bind dCLK and TIM (Fig. 6C, lanes 2 and 3). The higher abundance of TIM copurifying with dPER(Δ) compared to that copurifying with dPER (Fig. 6C, bottom panel, lanes 2 and 3) is likely a result of the increased levels of dPER(Δ) in the extract (Fig. 6B, top panel, compare lanes 1 and 2). In the case of dCLK, equal amounts are copurified with dPER and dPER(Δ) (Fig. 6C, top panel), most likely because dCLK levels are limiting (5). Furthermore, because at ZT23.5 dPER is highly phosphorylated compared to dPER(Δ) (e.g., Fig. 6D, top panel, compare lanes 2 and 3), our results suggest that the phosphorylated state of dPER has little if any effect on its ability to bind dCLK.
dPER(Δ) is inefficient in the negative limb of the transcriptional feedback circuitry.
In wper01; per+-HAHis transgenic flies, dper RNA attains peak abundance at ∼ZT12 and trough levels by ∼ZT0 (Fig. 7A), similar to the endogenous dper temporal profile. Consistent with the luc-based results obtained with S2 cells (Fig. 4), the average daily levels of dper RNA in wper01; per(Δ)-HAHis flies are higher than in the control wper01; per+-HAHis situation (Fig. 7A). Both genotypes show similar peak values for dper transcript levels, suggesting that in the presence of dPER(Δ) the dper negative feedback loop is essentially locked in the “derepressed” mode. Interestingly, during the night phase dper(Δ) transcript levels exhibit a slight but reproducible decline (Fig. 7A). Although the reason for this night-specific decline is not clear, evidence indicates that at least under some circumstances, TIM and/or CRY can function as repressors of dCLK-CYC activity (11, 15, 41). Presumably, the light-induced degradation of TIM and/or CRY would reduce its capacity to inhibit dper expression during the day.
We also examined dClk RNA, which cycles essentially antiphase to that of dper and is expressed at low constant levels in per-null mutants (6). Consistent with the impaired function of dPER(Δ), dClk transcript levels in wper01; per(Δ)-HAHis flies are relatively flat and pegged close to the trough values observed in the wild-type per+ transgenic flies (Fig. 7B). The essentially mirror image response of dClk expression compared to the dper RNA profile further indicates that PER(Δ) has a severely impaired ability to inhibit dCLK-CYC-dependent transcription.
Further evidence that dPER(Δ) is highly impaired in its ability to repress dCLK-driven transcriptional activation was obtained from ChIP assays. It was recently reported that the dCLK-CYC complex rhythmically binds to E-box containing circadian regulatory sequences that drive daily cycles in dper or tim expression, with the highest binding occurring at midday and lowest association during the late night/early morning (73). As expected, the temporal profile for dCLK interaction with the dper circadian relevant E-box element in wper01; per+-HAHis transgenic flies is very similar to that previously obtained with wild-type flies (Fig. 7C). However, in wper01; per(Δ)-HAHis transgenics, the relative binding of dCLK to the same circadian DNA element is high at all times in a daily cycle compared to what is seen for wild-type dPER-expressing transgenic flies. Together, our findings indicate that despite the stable association of dPER(Δ) with dCLK, the dCLK-CYC complex remains bound to circadian relevant promoters and is actively engaged in stimulation of transcription.
DISCUSSION
The first circadian clock protein to be characterized biochemically is dPER from Drosophila melanogaster (19). It was shown to undergo progressive changes in phosphorylation over a significant portion of the day, from hypo- to hyperphosphorylated species. Daily changes in abundance are temporally linked to the phosphorylated state of dPER, whereby the appearance of hyperphosphorylated species precedes rapid decreases in overall levels. More-recent work established that DBT is a major kinase controlling the phosphorylation and stability of dPER (35, 53) and that hyperphosphorylated variants of dPER are targeted to the 26S proteasome/ubiquitin pathway by the F-box protein SLIMB (26, 38). A highly shared mechanism operates in the mammalian system for the mPERs, whereby the mammalian homologs of DBT (CK1ɛ and the δ variant) and SLIMB (β-TrCP1 and possibly β-TrCP2) play major roles in the temporal regulation of mPERs' phosphorylation and stability (37). Herein, we identify the major or sole DBT interaction domain on dPER (termed dPDBD) that is required in vivo for dPER hyperphosphorylation, temporal instability, and repressor function. Similar results were also obtained by others using a slightly smaller internal deletion (51a). Moreover, our findings suggest that the dPDBD plays distinct roles in dPER degradation and repressor function and that dPER has a mode of action in transcriptional inhibition that is more indirect than previously thought (Fig. 8).
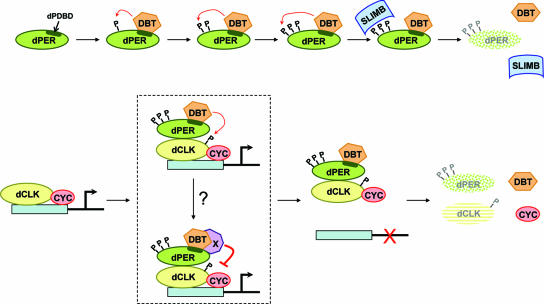
FIG. 8.
Model for the multiple roles of DBT in regulating the levels and activities of dPER and dCLK. (A) Role for DBT in the hyperphosphorylation and degradation of dPER. DBT stably associates with dPER via the dPDBD (dark green stripe within the dPER oval), which promotes the progressive phosphorylation (denoted by the letter “P”) of dPER by DBT and/or other kinases (for simplicity, TIM and other possible factors are not shown). SLIMB preferentially interacts with hyperphosphorylated isoforms of dPER (represented by three P's), targeting them to the 26S proteasome for rapid degradation. (B) In the absence of nucleus-localized dPER, the dCLK-CYC transcription factor binds E-box containing regulatory elements and drives expression of clock and output genes. Various phosphoisoforms of dCLK are observed throughout a daily cycle (hypophosphorylated, intermediately phosphorylated, and hyperphosphorylated), but for simplicity the hypophosphorylated and intermediately phosphorylated variants are represented by the same object (yellow oval with no phosphates), whereas only one phosphate is used to represent the DBT-dependent hyperphosphorylated species of dCLK. Eventually, a heteromeric complex containing DBT bound to dPER enters the nucleus, whereby dPER interacts with dCLK. The interaction of dPER with dCLK enhances the ability of DBT to phosphorylate dCLK, which stimulates its rapid degradation and might directly lead to its dissociation from DNA. It is also possible that additional steps, indicated by the binding of factor “X,” are required to block dCLK-CYC transactivation activity and release from DNA. Factor “X” might directly bind to the dPDBD or interact with DBT (as shown). DBT-dependent hyperphosphorylated dCLK is subject to enhanced degradation, although the putative F-box mediating this targeting is not known. It is also not clear if dissociation from DNA is required for the rapid degradation of hyperphosphorylated dCLK.
Earlier reports suggested that the amino-terminal half of dPER and possibly just the first 385 aa are sufficient to stably bind DBT (35, 36, 63). However, we show that in flies the absence of the dPDBD abolishes any detectable interaction between dPER and DBT (Fig. 6). Based on the effects of DBT binding to dPER(Δ) in S2 cells, which are less dramatic than those seen for flies (compare Fig. 3 and 6), it is possible that the use of high-level expression systems (35, 63) or a highly stable dPER-β-Gal fusion (36) in the earlier studies exaggerated weak interactions. Thus, while it is likely that there are other DBT binding regions on dPER (especially those that might be involved only in transient interactions), our findings indicate that in vivo the DBT-dependent hyperphosphorylation and degradation of dPER have an absolute requirement for the dPDBD, highlighting the tight association between hyperphosphorylation and enhanced degradation. These data are well accommodated by prior work showing that hyperphosphorylated isoforms of dPER are preferentially targeted by SLIMB (26, 38). Presumably, the ability of DBT to promote the hyperphosphorylation of dPER requires a stable and perhaps long-term association with the substrate, consistent with the observation that DBT is bound to dPER during a significant portion of the daily cycle (36). The tight interaction between dPER and DBT is also highlighted by the redistribution of predominately nucleus-localized DBT to the cytoplasm in pacemaker cells only during times in a daily cycle when dPER is present in the cytoplasm (36). Although the ability of CK1 to phosphorylate dPER in an in vitro reconstituted assay (Fig. 2C and data not shown) suggests that DBT and dPER directly interact, our data do not rule out the possibility that the stable interaction between DBT and dPER observed in vivo is stimulated or dependent on other factors that either directly or indirectly interact with the dPDBD.
Likewise, in the mammalian clockworks a region beginning around the middle of mPER1 and mPER2 and extending about 200 aa mediates their stable association with CK1ɛ (65, 67). Here too, removal of major CK1ɛ binding domains (CKBD) on mPER1 or -2 attenuates their CK1ɛ-driven instability and hyperphosphorylation (1, 22), although the physiological relevance of these regions has not been tested in animals. In contrast to mPER1 and -2, mPER3 displays no or weak interactions with CK1ɛ (1, 43). Rather, it is thought that the association of mPER3 with mPER1 enables mPER1 to act as a bridge and bring CK1ɛ into a favorable proximity to phosphorylate mPER3 (43). There is only limited knowledge of the structural features underlying CK1ɛ docking regions, with arguably the best characterized being the F-X-X-X-F motif present in the mammalian NFAT1 transcription factor (52). mPER1 and -2 but not -3 have this signature motif in their CKBD regions, and alteration of this motif greatly attenuates binding to CK1ɛ (52, 61). Intriguingly, when we did a BLAST analysis of our 57-aa region encompassing the dPDBD against the mPER1 and mPER2 protein sequences, it aligned within a subset of their CKBD that included the F-X-X-X-F motifs, but the alignment with mPER3 was noticeably dispersed (Fig. 9). While this finding is highly suggestive, the F-X-X-X-F motif is not found in the dPDBD, and future work will be required to more precisely define the key structural features.
FIG. 9.
Alignment of aa 755 to 809 from dPER against mPER1, mPER2, and mPER3. Pairwise alignment of dPER aa 755 to 809 against mPER1 and mPER2 (A) or mPER3 (B) was done using ClustalW software. Asterisks indicate amino acid identity, colons indicate that conserved substitutions have been observed, and periods indicate that semiconserved substitutions have been observed. Note that the alignment with mPER3 is more dispersed. In panel A, the F-X-X-X-F motif in mPER1 and -2 is denoted by a line on top.
Although the dPDBD promotes DBT-dependent hyperphosphorylation of dPER, it is unlikely to be a major site directly modified by phosphorylation. When the majority of Ser and Thr residues in this region (those conserved with dPER from other Drosophila species) are converted to Ala residues, the modified dPER undergoes progressive phosphorylation and degradation kinetics indistinguishable from those of the wild-type control version (Fig. 2D). This also seems to be the case for mammalian PERs. For example, a more detailed analysis showed that deletion of either of two small regions on mPER2 (aa 582 to 606 or aa 731 to 756) greatly reduced the ability of CK1ɛ to stably interact and promote degradation of mPER2 (22). However, recent work mapping phosphorylation sites on mPER2 did not identify either aa 582 to 606 or aa 731 to 756 as areas modified by phosphorylation (66).
Thus, although there might be structural features that differ between the DBT/CK1ɛ docking sites on mammalian and fly PERs, there are remarkable similarities. Most notable is the presence of one or a few small regions that are not major targets for phosphorylation but serve to stably bind DBT/CK1ɛ. Once stably bound, DBT/CK1ɛ either directly or indirectly (e.g., via other kinases) stimulates progressive phosphorylation at other sites until a threshold of multiphosphorylation is attained that enhances degradation by SLIMB/β-TrCP-mediated pathways (Fig. 8). Obviously, time-of-day-dependent variations in phosphorylation that regulate the levels of key transcriptional repressors constitute one mechanism that can control the timing and relative potency of repressor function, giving rise to cyclical gene expression.
In addition to regulating stability, DBT has been implicated in modulating the subcellular localization of dPER (e.g., references 7, 14, and 54), a function also ascribed to CK1ɛ in the case of mPERs (e.g., reference 67). We did not observe differences in the subcellular distributions between dPER(Δ) and wild-type dPER expressed in S2 cells, and whole-mount staining of adult Drosophila heads showed strong staining of dPER(Δ) in nuclei of dPER-expressing cells (data not shown), consistent with the exclusively nuclear staining of dPER in key pacemaker cells of mutants with abolished or severely reduced dbt activity (14). These results are in agreement with our biochemical studies showing that similar amounts of dCLK, which is the limiting component (5) and exclusively localized in the nucleus of both S2 cells (34) and fly clock cells (32), copurifies with dPER(Δ) and wild-type dPER (Fig. 3 and 6). Nonetheless, recent work by Nawathean and colleagues using an internal deletion smaller than ours suggests that the DBT binding domain enhances the nuclear localization of dPER (51a). The reason for this apparent discrepancy is not clear, but we note that by far the majority of recombinant dPER is found in the cytoplasm when expressed in S2 cells (e.g., references 10, 34, and 51). It is possible that in our S2 cell system the overall proportion of cytoplasmic dPER is higher, making it difficult to detect subtle differences in nuclear staining. Nonetheless, similar to the results obtained by Nawathean et al., when we placed a nuclear localization signal (NLS) on dPER and dPER(Δ), both proteins were highly localized in the nucleus with increased repressor function; however, dPER(Δ) was still approximately twofold less efficient in blocking dCLK-dependent transcription (data not shown). Although we cannot exclude possible effects on subcellular distribution, our results indicate that the strongly attenuated repressor function of dPER(Δ) is not explained simply by a lack of interaction with dCLK.
Why is the binding of dPER(Δ) to dCLK not sufficient for robust inhibition of transactivation activity? A possible answer is based on prior work suggesting that hyperphosphorylated dPER might be a more potent repressor of dCLK-CYC transcriptional activity (51), rendering the mainly hypophosphorylated dPER(Δ) deficient in this capacity. Another non-mutually exclusive possibility is based on our recent findings indicating that dPER is required for the DBT-dependent hyperphosphorylation of dCLK during the night/early day, events that promote the rapid degradation of dCLK and possibly reduce its transactivation activity (34, 73). Presumably, dPER can act as a bridge to enhance the phosphorylation of dCLK by DBT (73) (Fig. 8). Indeed, in per0 flies DBT is located largely in the nucleus of pacemaker neurons (36), yet hyperphosphorylation of dCLK is not observed (34, 73) (Fig. 5C). Our findings greatly strengthen this proposal by showing that dPER(Δ), which can bind dCLK but is impaired in its association with DBT, does not support the hyperphosphorylation of dCLK (Fig. 5C). A scenario where PER acts as a molecular bridge by which DBT can phosphorylate other clock proteins is similar to that proposed for mPER1 in stimulating CK1ɛ phosphorylation of mPER3 (43) and mCRYs (21), suggesting that PERs might have a general role in targeting DBT/CK1ɛ to numerous clock components. Even more impressive, a similar mechanism also occurs in the Neurospora clock, whereby the binding of CK1a (a CK1 homolog) to FREQUENCY is critical for inhibiting the transcriptional activity of the WHITE COLLAR COMPLEX (31). Thus, there is remarkable conservation of posttranslational regulatory pathways operating in widely divergent clocks.
Despite the reasonable possibilities that the dPDBD modulates dPER repressor function by regulating dPER phosphorylation and/or serving as a conduit to facilitate DBT-dependent phosphorylation of dCLK, there is circumstantial evidence from S2 cells and flies suggesting that neither scenario is obligatory for dPER to exhibit repressor capabilities. For example, in our S2 cell culture system, dPER(Δ) displays little repressor function compared to wild-type dPER even in the absence of exogenous DBT (Fig. 4), conditions under which only the hypophosphorylated isoforms of dPER(Δ) and wild-type dPER are observed (Fig. 2A) and in which more dCLK stably interacts with dPER(Δ) (Fig. 3C). Furthermore, in mutant flies with highly impaired DBT activities/levels, evidence suggests that dPER can still inhibit dCLK-CYC-driven transcription (14, 69), though the relative strength of this repression is not known. These results raise the possibility that the dPDBD acts as a docking site for other factors besides DBT that play a more direct role in inhibiting dCLK-CYC-dependent transactivation, e.g., activities involved in chromatin remodeling (8, 23) (Fig. 8). Indeed, the DBT-mediated phosphorylation of dCLK might be involved only in regulating dCLK stability whereas repression of its activity is carried out by other factors. However, it is important to note that in the aforementioned examples based on results obtained with cultured S2 cells or dbt-impaired mutants, the elevated levels of dPER might override stimulatory effects of phosphorylation and/or DBT on the ability of dPER to function as a transcriptional inhibitor. This reasoning might also explain why recombinant dPER was able to block dCLK-CYC binding to E-box containing DNA elements in an in vitro assay (41). Taken together, we speculate that dPER “alone” has low intrinsic repressor capabilities that are enhanced by the dPDBD-dependent assembly of DBT and/or other factors that more directly modulate the ability of dCLK-CYC to stimulate transcription.
Although future work will be required to determine if phosphorylation regulates the transactivation potential of dCLK, our findings highlight a clear qualitative difference in the roles of the dPDBD with regards to dPER instability and repressor function. Namely, whereas differences in the SLIMB-mediated degradation of wild-type dPER and dPER(Δ) are observed only under conditions of DBT-dependent hyperphosphorylation (Fig. 2), this is not the case for transcriptional inhibition (Fig. 4).
A more secondary mode of action for dPER as a transcriptional repressor is reminiscent of how mPERs are thought to function in the negative limb of the mammalian clockworks, where the mCRYs are the major inhibitors. Although mPERs exhibit some repressor capacity in cultured cell assays and they are found in complexes with mammalian CLK-BMAL1, it is likely that their main function in transcriptional regulation is to control the timing of mCRYs nuclear localization (42). This theme of roles as indirect repressors of circadian gene expression is also observed for FREQUENCY in Neurospora (59) and KaiC in cyanobacteria (64). A strikingly common feature of these indirect repressors is that they undergo complex daily changes in phosphorylation that are central to clock progression. Indeed, a daily rhythm in the cyanobacterial clock protein KaiC can be recapitulated in vitro using ATP and recombinant versions of the three critical clock proteins (KaiA, KaiB, and KaiC) from this system (50). While it is likely that the “minimal” circadian phosphorylation programs in eukaryotes are more complex, the bacterial work elegantly suggests that the most basic and ancient building block of circadian clocks is a biochemical oscillator based on time-of-day-specific phosphorylation of one or more key clock proteins. This central biochemical oscillator can be viewed as a semiautonomous subsystem from which temporal coordinates are largely transduced by phase-specific interactions with a variety of regulatory factors that subsequently control the activities of major transcription factors, leading to rhythmic gene expression and ultimately to daily changes in physiology and behavior.
Acknowledgments
We thank D. Chang and S. Reppert for providing a series of dper-containing plasmids that we used in our initial screen for regions that modulate dPER phosphorylation and stability in S2 cells. We thank Pipat Nawathean and Michael Rosbash for sharing unpublished results.
This work was supported by NIH grants awarded to I.E. (NS34958) and P.E.H. (NS052854).
Footnotes
Published ahead of print on 23 April 2007.
REFERENCES
- 1.Akashi, M., Y. Tsuchiya, T. Yoshino, and E. Nishida. 2002. Control of intracellular dynamics of mammalian period proteins by casein kinase I ɛ (CKIɛ) and CKIδ in cultured cells. Mol. Cell. Biol. 22:1693-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akten, B., E. Jauch, G. K. Genova, E. Y. Kim, I. Edery, T. Raabe, and F. R. Jackson. 2003. A role for CK2 in the Drosophila circadian oscillator. Nat. Neurosci. 6:251-257. [DOI] [PubMed] [Google Scholar]
- 3.Allada, R., N. E. White, W. V. So, J. C. Hall, and M. Rosbash. 1998. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell 93:791-804. [DOI] [PubMed] [Google Scholar]
- 4.Bae, K., and I. Edery. 2006. Regulating a circadian clock's period, phase and amplitude by phosphorylation: insights from Drosophila. J. Biochem. (Tokyo) 140:609-617. [DOI] [PubMed] [Google Scholar]
- 5.Bae, K., C. Lee, P. E. Hardin, and I. Edery. 2000. dCLOCK is present in limiting amounts and likely mediates daily interactions between the dCLOCK-CYC transcription factor and the PER-TIM complex. J. Neurosci. 20:1746-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bae, K., C. Lee, D. Sidote, K. Y. Chuang, and I. Edery. 1998. Circadian regulation of a Drosophila homolog of the mammalian Clock gene: PER and TIM function as positive regulators. Mol. Cell. Biol. 18:6142-6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bao, S., J. Rihel, E. Bjes, J. Y. Fan, and J. L. Price. 2001. The Drosophila double-timeS mutation delays the nuclear accumulation of period protein and affects the feedback regulation of period mRNA. J. Neurosci. 21:7117-7126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 8.Brown, S. A., J. Ripperger, S. Kadener, F. Fleury-Olela, F. Vilbois, M. Rosbash, and U. Schibler. 2005. PERIOD1-associated proteins modulate the negative limb of the mammalian circadian oscillator. Science 308:693-696. [DOI] [PubMed] [Google Scholar]
- 9.Ceriani, M. F., J. B. Hogenesch, M. Yanovsky, S. Panda, M. Straume, and S. A. Kay. 2002. Genome-wide expression analysis in Drosophila reveals genes controlling circadian behavior. J. Neurosci. 22:9305-9319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang, D. C., and S. M. Reppert. 2003. A novel C-terminal domain of Drosophila PERIOD inhibits dCLOCK:CYCLE-mediated transcription. Curr. Biol. 13:758-762. [DOI] [PubMed] [Google Scholar]
- 11.Collins, B., E. O. Mazzoni, R. Stanewsky, and J. Blau. 2006. Drosophila CRYPTOCHROME is a circadian transcriptional repressor. Curr. Biol. 16:441-449. [DOI] [PubMed] [Google Scholar]
- 12.Colot, H. V., J. C. Hall, and M. Rosbash. 1988. Interspecific comparison of the period gene of Drosophila reveals large blocks of non-conserved coding DNA. EMBO J. 7:3929-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cyran, S. A., A. M. Buchsbaum, K. L. Reddy, M. C. Lin, N. R. Glossop, P. E. Hardin, M. W. Young, R. V. Storti, and J. Blau. 2003. vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell 112:329-341. [DOI] [PubMed] [Google Scholar]
- 14.Cyran, S. A., G. Yiannoulos, A. M. Buchsbaum, L. Saez, M. W. Young, and J. Blau. 2005. The double-time protein kinase regulates the subcellular localization of the Drosophila clock protein period. J. Neurosci. 25:5430-5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Darlington, T. K., K. Wager-Smith, M. F. Ceriani, D. Staknis, N. Gekakis, T. D. L. Steeves, C. J. Weitz, J. S. Takahashi, and S. A. Kay. 1998. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science 280:1599-1603. [DOI] [PubMed] [Google Scholar]
- 16.Dembinska, M. E., R. Stanewsky, J. C. Hall, and M. Rosbash. 1997. Circadian cycling of a PERIOD-beta-galactosidase fusion protein in Drosophila: evidence for cyclical degradation. J. Biol. Rhythms 12:157-172. [DOI] [PubMed] [Google Scholar]
- 17.Dunlap, J. C. 1999. Molecular bases for circadian clocks. Cell 96:271-290. [DOI] [PubMed] [Google Scholar]
- 18.Edery, I. 2000. Circadian rhythms in a nutshell. Physiol. Genomics 3:59-74.11015601 [Google Scholar]
- 19.Edery, I., L. J. Zwiebel, M. E. Dembinska, and M. Rosbash. 1994. Temporal phosphorylation of the Drosophila period protein. Proc. Natl. Acad. Sci. USA 91:2260-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eide, E. J., H. Kang, S. Crapo, M. Gallego, and D. M. Virshup. 2005. Casein kinase I in the mammalian circadian clock. Methods Enzymol. 393:408-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eide, E. J., E. L. Vielhaber, W. A. Hinz, and D. M. Virshup. 2002. The circadian regulatory proteins BMAL1 and cryptochromes are substrates of casein kinase Iepsilon. J. Biol. Chem. 277:17248-17254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eide, E. J., M. F. Woolf, H. Kang, P. Woolf, W. Hurst, F. Camacho, E. L. Vielhaber, A. Giovanni, and D. M. Virshup. 2005. Control of mammalian circadian rhythm by CKIɛ-regulated proteasome-mediated PER2 degradation. Mol. Cell. Biol. 25:2795-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Etchegaray, J. P., C. Lee, P. A. Wade, and S. M. Reppert. 2003. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature 421:177-182. [DOI] [PubMed] [Google Scholar]
- 24.Gekakis, N., L. Saez, A. M. Delahaye-Brown, M. P. Myers, A. Sehgal, M. W. Young, and C. J. Weitz. 1995. Isolation of timeless by PER protein interaction: defective interaction between timeless protein and long-period mutant PERL. Science 270:811-815. [DOI] [PubMed] [Google Scholar]
- 25.Glossop, N. R., J. H. Houl, H. Zheng, F. S. Ng, S. M. Dudek, and P. E. Hardin. 2003. VRILLE feeds back to control circadian transcription of Clock in the Drosophila circadian oscillator. Neuron 37:249-261. [DOI] [PubMed] [Google Scholar]
- 26.Grima, B., A. Lamouroux, E. Chelot, C. Papin, B. Limbourg-Bouchon, and F. Rouyer. 2002. The F-box protein slimb controls the levels of clock proteins period and timeless. Nature 420:178-182. [DOI] [PubMed] [Google Scholar]
- 27.Hamblen-Coyle, M. J., D. A. Wheeler, J. E. Rutila, M. Rosbash, and J. C. Hall. 1992. Behavior of period-altered rhythm mutants of Drosophila in light:dark cycles. J. Insect. Behav. 5:417-446. [Google Scholar]
- 28.Hardin, P. E. 2005. The circadian timekeeping system of Drosophila. Curr. Biol. 15:R714-R722. [DOI] [PubMed] [Google Scholar]
- 29.Hardin, P. E. 2004. Transcription regulation within the circadian clock: the E-box and beyond. J. Biol. Rhythms 19:348-360. [DOI] [PubMed] [Google Scholar]
- 30.Hastings, J. W., B. Rusak, and Z. Boulos. 1991. Circadian rhythms: the physiology of biological timing, p. 435-546. In C. L. Prosser (ed.), Neural and integrative animal physiology. Wiley-Liss Inc., New York, NY.
- 31.He, Q., J. Cha, Q. He, H. C. Lee, Y. Yang, and Y. Liu. 2006. CKI and CKII mediate the FREQUENCY-dependent phosphorylation of the WHITE COLLAR complex to close the Neurospora circadian negative feedback loop. Genes Dev. 20:2552-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Houl, J. H., W. Yu, S. M. Dudek, and P. E. Hardin. 2006. Drosophila CLOCK is constitutively expressed in circadian oscillator and non-oscillator cells. J. Biol. Rhythms 21:93-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang, Z. J., I. Edery, and M. Rosbash. 1993. PAS is a dimerization domain common to Drosophila period and several transcription factors. Nature 364:259-262. [DOI] [PubMed] [Google Scholar]
- 34.Kim, E. Y., and I. Edery. 2006. Balance between DBT/CKIepsilon kinase and protein phosphatase activities regulate phosphorylation and stability of Drosophila CLOCK protein. Proc. Natl. Acad. Sci. USA 103:6178-6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kloss, B., J. L. Price, L. Saez, J. Blau, A. Rothenfluh, C. S. Wesley, and M. W. Young. 1998. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iepsilon. Cell 94:97-107. [DOI] [PubMed] [Google Scholar]
- 36.Kloss, B., A. Rothenfluh, M. W. Young, and L. Saez. 2001. Phosphorylation of period is influenced by cycling physical associations of double-time, period, and timeless in the Drosophila clock. Neuron 30:699-706. [DOI] [PubMed] [Google Scholar]
- 37.Ko, C. H., and J. S. Takahashi. 2006. Molecular components of the mammalian circadian clock. Hum. Mol. Genet. 15:R271-R277. [DOI] [PubMed] [Google Scholar]
- 38.Ko, H. W., J. Jiang, and I. Edery. 2002. Role for Slimb in the degradation of Drosophila Period protein phosphorylated by Doubletime. Nature 420:673-678. [DOI] [PubMed] [Google Scholar]
- 39.Konopka, R. J., and S. Benzer. 1971. Clock mutants of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 68:2112-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee, C., K. Bae, and I. Edery. 1998. The Drosophila CLOCK protein undergoes daily rhythms in abundance, phosphorylation, and interactions with the PER-TIM complex. Neuron 21:857-867. [DOI] [PubMed] [Google Scholar]
- 41.Lee, C., K. Bae, and I. Edery. 1999. PER and TIM inhibit the DNA binding activity of a Drosophila CLOCK-CYC/dBMAL1 heterodimer without disrupting formation of the heterodimer: a basis for circadian transcription. Mol. Cell. Biol. 19:5316-5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee, C., J. P. Etchegaray, F. R. Cagampang, A. S. Loudon, and S. M. Reppert. 2001. Posttranslational mechanisms regulate the mammalian circadian clock. Cell 107:855-867. [DOI] [PubMed] [Google Scholar]
- 43.Lee, C., D. R. Weaver, and S. M. Reppert. 2004. Direct association between mouse PERIOD and CKIɛ is critical for a functioning circadian clock. Mol. Cell. Biol. 24:584-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin, J. M., V. L. Kilman, K. Keegan, B. Paddock, M. Emery-Le, M. Rosbash, and R. Allada. 2002. A role for casein kinase 2alpha in the Drosophila circadian clock. Nature 420:816-820. [DOI] [PubMed] [Google Scholar]
- 45.Lin, J. M., A. Schroeder, and R. Allada. 2005. In vivo circadian function of casein kinase 2 phosphorylation sites in Drosophila PERIOD. J. Neurosci. 25:11175-11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Majercak, J., W. F. Chen, and I. Edery. 2004. Splicing of the period gene 3′-terminal intron is regulated by light, circadian clock factors, and phospholipase C. Mol. Cell. Biol. 24:3359-3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martinek, S., S. Inonog, A. S. Manoukian, and M. W. Young. 2001. A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell 105:769-779. [DOI] [PubMed] [Google Scholar]
- 48.Meyer, P., L. Saez, and M. W. Young. 2006. PER-TIM interactions in living Drosophila cells: an interval timer for the circadian clock. Science 311:226-229. [DOI] [PubMed] [Google Scholar]
- 49.Myers, M. P., K. Wager-Smith, A. Rothenfluh-Hilfiker, and M. W. Young. 1996. Light-induced degradation of TIMELESS and entrainment of the Drosophila circadian clock. Science 271:1736-1740. [DOI] [PubMed] [Google Scholar]
- 50.Nakajima, M., K. Imai, H. Ito, T. Nishiwaki, Y. Murayama, H. Iwasaki, T. Oyama, and T. Kondo. 2005. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science 308:414-415. [DOI] [PubMed] [Google Scholar]
- 51.Nawathean, P., and M. Rosbash. 2004. The doubletime and CKII kinases collaborate to potentiate Drosophila PER transcriptional repressor activity. Mol. Cell 13:213-223. [DOI] [PubMed] [Google Scholar]
- 51a.Nawathean, P., D. Stoleru, and M. Rosbash. 2007. A small conserved domain of Drosophila PERIOD is important for circadian phosphorylation, nuclear localization, and transcriptional repressor activity. Mol. Cell. Biol. 27:5002-5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Okamura, H., C. Garcia-Rodriguez, H. Martinson, J. Qin, D. M. Virshup, and A. Rao. 2004. A conserved docking motif for CK1 binding controls the nuclear localization of NFAT1. Mol. Cell. Biol. 24:4184-4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Price, J. L., J. Blau, A. Rothenfluh, M. Abodeely, B. Kloss, and M. W. Young. 1998. double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell 94:83-95. [DOI] [PubMed] [Google Scholar]
- 54.Rothenfluh, A., M. Abodeely, and M. W. Young. 2000. Short-period mutations of per affect a double-time-dependent step in the Drosophila circadian clock. Curr. Biol. 10:1399-1402. [DOI] [PubMed] [Google Scholar]
- 55.Rothenfluh, A., M. W. Young, and L. Saez. 2000. A TIMELESS-independent function for PERIOD proteins in the Drosophila clock. Neuron 26:505-514. [DOI] [PubMed] [Google Scholar]
- 56.Rutila, J. E., V. Suri, M. Le, W. V. So, M. Rosbash, and J. C. Hall. 1998. CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell 93:805-814. [DOI] [PubMed] [Google Scholar]
- 57.Saez, L., and M. W. Young. 1996. Regulation of nuclear entry of the Drosophila clock proteins period and timeless. Neuron 17:911-920. [DOI] [PubMed] [Google Scholar]
- 58.Sathyanarayanan, S., X. Zheng, R. Xiao, and A. Sehgal. 2004. Posttranslational regulation of Drosophila PERIOD protein by protein phosphatase 2A. Cell 116:603-615. [DOI] [PubMed] [Google Scholar]
- 59.Schafmeier, T., A. Haase, K. Kaldi, J. Scholz, M. Fuchs, and M. Brunner. 2005. Transcriptional feedback of Neurospora circadian clock gene by phosphorylation-dependent inactivation of its transcription factor. Cell 122:235-246. [DOI] [PubMed] [Google Scholar]
- 60.Shafer, O. T., M. Rosbash, and J. W. Truman. 2002. Sequential nuclear accumulation of the clock proteins period and timeless in the pacemaker neurons of Drosophila melanogaster. J. Neurosci. 22:5946-5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shirogane, T., J. Jin, X. L. Ang, and J. W. Harper. 2005. SCFbeta-TRCP controls clock-dependent transcription via casein kinase 1-dependent degradation of the mammalian period-1 (Per1) protein. J. Biol. Chem. 280:26863-26872. [DOI] [PubMed] [Google Scholar]
- 62.Sidote, D., J. Majercak, V. Parikh, and I. Edery. 1998. Differential effects of light and heat on the Drosophila circadian clock proteins PER and TIM. Mol. Cell. Biol. 18:2004-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Suri, V., J. C. Hall, and M. Rosbash. 2000. Two novel doubletime mutants alter circadian properties and eliminate the delay between RNA and protein in Drosophila. J. Neurosci. 20:7547-7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Takai, N., M. Nakajima, T. Oyama, R. Kito, C. Sugita, M. Sugita, T. Kondo, and H. Iwasaki. 2006. A KaiC-associating SasA-RpaA two-component regulatory system as a major circadian timing mediator in cyanobacteria. Proc. Natl. Acad. Sci. USA 103:12109-12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Toh, K. L., C. R. Jones, Y. He, E. J. Eide, W. A. Hinz, D. M. Virshup, L. J. Ptacek, and Y. H. Fu. 2001. An hPer2 phosphorylation site mutation in familial advanced sleep-phase syndrome. Science 291:1040-1043. [DOI] [PubMed] [Google Scholar]
- 66.Vanselow, K., J. T. Vanselow, P. O. Westermark, S. Reischl, B. Maier, T. Korte, A. Herrmann, H. Herzel, A. Schlosser, and A. Kramer. 2006. Differential effects of PER2 phosphorylation: molecular basis for the human familial advanced sleep phase syndrome (FASPS). Genes Dev. 20:2660-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vielhaber, E., E. Eide, A. Rivers, Z. H. Gao, and D. M. Virshup. 2000. Nuclear entry of the circadian regulator mPER1 is controlled by mammalian casein kinase I epsilon. Mol. Cell. Biol. 20:4888-4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vosshall, L. B., J. L. Price, A. Sehgal, L. Saez, and M. W. Young. 1994. Block in nuclear localization of period protein by a second clock mutation, timeless. Science 263:1606-1609. [DOI] [PubMed] [Google Scholar]
- 69.Weber, F., and S. A. Kay. 2003. A PERIOD inhibitor buffer introduces a delay mechanism for CLK/CYC-activated transcription. FEBS Lett. 555:341-345. [DOI] [PubMed] [Google Scholar]
- 70.Worby, C. A., N. Simonson-Leff, and J. E. Dixon. 2001. RNA interference of gene expression (RNAi) in cultured Drosophila cells. Sci. STKE 2001(95):PL1. [DOI] [PubMed] [Google Scholar]
- 71.Xu, Y., Q. S. Padiath, R. E. Shapiro, C. R. Jones, S. C. Wu, N. Saigoh, K. Saigoh, L. J. Ptacek, and Y. H. Fu. 2005. Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature 434:640-644. [DOI] [PubMed] [Google Scholar]
- 72.Young, M. W., and S. A. Kay. 2001. Time zones: a comparative genetics of circadian clocks. Nat. Rev. Genet. 2:702-715. [DOI] [PubMed] [Google Scholar]
- 73.Yu, W., H. Zheng, J. H. Houl, B. Dauwalder, and P. E. Hardin. 2006. PER-dependent rhythms in CLK phosphorylation and E-box binding regulate circadian transcription. Genes Dev. 20:723-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zeng, H., Z. Qian, M. P. Myers, and M. Rosbash. 1996. A light-entrainment mechanism for the Drosophila circadian clock. Nature 380:129-135. [DOI] [PubMed] [Google Scholar]