Abstract
P-TEFb, comprised of CDK9 and a cyclin T subunit, is a global transcriptional elongation factor important for most RNA polymerase II (pol II) transcription. P-TEFb facilitates transcription elongation in part by phosphorylating Ser2 of the heptapeptide repeat of the carboxy-terminal domain (CTD) of the largest subunit of pol II. Previous studies have shown that P-TEFb is subjected to negative regulation by forming an inactive complex with 7SK small RNA and HEXIM1. In an effort to investigate the molecular mechanism by which corepressor N-CoR mediates transcription repression, we identified HEXIM1 as an N-CoR-interacting protein. This finding led us to test whether the P-TEFb complex is regulated by acetylation. We demonstrate that CDK9 is an acetylated protein in cells and can be acetylated by p300 in vitro. Through both in vitro and in vivo assays, we identified lysine 44 of CDK9 as a major acetylation site. We present evidence that CDK9 is regulated by N-CoR and its associated HDAC3 and that acetylation of CDK9 affects its ability to phosphorylate the CTD of pol II. These results suggest that acetylation of CDK9 is an important posttranslational modification that is involved in regulating P-TEFb transcriptional elongation function.
Transcription by RNA polymerase II (pol II) is a multistep process including preinitiation, initiation, promoter clearance, elongation, and termination (48). Phosphorylation of the carboxy-terminal domain (CTD) of the largest subunit of pol II plays a critical role in transition from transcriptional initiation to elongation as well as in coordinating transcription elongation and RNA maturation (7, 30). P-TEFb, a positive transcription elongation factor originally identified based on its ability to stimulate 5,6-dichloro-1-β-d-ribofuranosyl-benzimidazole-sensitive transcription of long transcripts in vitro (36), stimulates transcription elongation by preferentially phosphorylating Ser2 of the heptapeptide repeat of the CTD of the largest subunit of pol II (35). P-TEFb is a heterodimeric complex comprised of cyclin-dependent kinase 9 (CDK9) and a regulatory cyclin subunit of the T family, namely T1, T2, or K1 (13, 43). CDK9 is a cdc2-related, ubiquitously expressed kinase protein (8). P-TEFb also enhances transcriptional elongation by phosphorylating and counteracting the inhibitory factors DSIF and NELF (5, 24, 51, 54).
In addition to its function as a global transcriptional elongation factor important for most pol II transcription (3, 47), activation of CDK9 kinase activity has also been linked to specific events such as human immunodeficiency virus (HIV) replication (20, 65), cardiac hypertrophy (45), and activation of lymphocytes (19). In this regard, P-TEFb is also known as TAK, an HIV Tat-associated kinase (20). Regulation of HIV transcription is primarily mediated by the HIV Tat protein, which recruits TAK/P-TEFb to enhance productive elongation of viral transcripts (11, 12, 64). Activation of P-TEFb induces heart hypertrophy in transgenic mice and induces myocyte enlargement in tissue culture (44, 46).
As a complex important for pol II transcription, regulation of the kinase activity of P-TEFb is a subject of intensive study. Biochemical experiments suggest that cellular P-TEFb exists in two forms, the active heterodimeric CDK9/cyclin T and an inactive form containing the 7SK small nuclear RNA and HEXIM1 protein (37, 41, 56, 57). In vitro studies have shown that HEXIM1 binds to 7SK first, and this interaction allows the subsequent association of 7SK and HEXIM1 with P-TEFb, leading to sequestration of P-TEFb into an inactive complex (10, 33). Contrary to this negative regulation, recent studies indicate that the bromodomain protein Brd4 positively regulates P-TEFb kinase activity (26, 55). Brd4 interacts with P-TEFb through its double bromodomain, and this interaction enhances P-TEFb-dependent phosphorylation of the pol II CTD and transcriptional activation (26, 55).
N-CoR and its related SMRT protein were identified initially as corepressors for nuclear receptors such as thyroid hormone receptors (TR) and retinoic acid receptors (RAR) (4, 21). Subsequent studies have implicated these proteins in repression by many different transcription factors, including Mad/Mxi, BCL6/LAZ3, ETO, and CBF (for a review, see reference 14). Repression mediated by SMRT and N-CoR is sensitive to trichostatin A (TSA), a histone deacetylase (HDAC) inhibitor, suggesting a dependence on HDAC activity for repression (6, 18, 53). Consistent with this observation, SMRT and N-CoR have been reported to associate with a number of HDACs (18, 22, 29, 39). However, biochemical studies have provided compelling evidence that both SMRT and N-CoR exist in stable protein complexes containing mainly HDAC3, GPS2, and TBL1 (transducin beta-like protein 1) and TBLR1 (TBL1-related protein) (17, 32, 52, 58, 62). HDAC3 is required, at least in the case of the thyroid hormone receptor, for transcriptional repression mediated by SMRT and N-CoR (23, 58).
To better understand the biological function of N-CoR, we have used a biochemical approach to purify and identify N-CoR-associated proteins (32, 58, 59). Among the N-CoR-associated proteins, we identified HEXIM1. We found that a subfraction of N-CoR is associated with P-TEFb. This finding has led us to test whether the P-TEFb complex is regulated by acetylation. We demonstrate that CDK9 is acetylated in cells and present evidence that acetylation of CDK9 enhances its ability to phosphorylate the CTD of pol II and promotes transcriptional elongation.
MATERIALS AND METHODS
Plasmids and constructs.
The HIV-1 long terminal repeat (LTR)-based luciferase reporter G5-83-HIV-luc contains the LTR sequences from −83 to +82, with five GAL4 binding sites inserted at position −83 (34), and was kindly provided by L. Lania (University of Naples Federico II, Naples, Italy). The CDK9 mutants (K44R, K48/49R, and K44/48/49R) were generated by site-directed mutagenesis based on pcDNA3-CDK9 and were kindly provided by M. Sano (Baylor College of Medicine). The wild-type and mutant CDK9 were then cloned into pSG5-Flag for transient transfection, pCMV-GAL4-DBD (DNA binding domain) for expression as Gal(DBD) fusion proteins, and pcDNA5/FRT/TO for generating Tet-inducible stable cell lines. In addition, the full-length CDK9 was cloned into pGEX-4T1 for preparation of glutathione S-transferase (GST)-CDK9 from bacteria. GST-CDK9N and its related mutants were generated by cloning the corresponding DNA sequences encoding the first 55 amino acids (aa 1 to 55) of CDK9 into pGEX4T1. The coding region of human HEXIM1 was amplified by reverse transcription-PCR and cloned into pGEX-4T1 or pCMV-Gal4-DBD.
Identification of HEXIM1 as an N-CoR-associated protein.
The purification of the N-CoR complex from HeLa nuclear extracts was previously reported (59). The presence of two peptides derived from HEXIM1 was not recognized at the time and was identified when the data were reexamined later on.
IP, Western blotting, and gel filtration.
Immunoprecipitation (IP) and Western blot analysis (IP-Western) with HeLa nuclear extracts was essentially as described previously (31). For IP-Western analysis using whole-cell extracts, HeLa cells or other cells were washed with phosphate-buffered saline (PBS) two times and lysed with ice-cold EBC buffer (20 mM Tris-HCl, pH 8.0, 125 mM NaCl, 2 mM EDTA, and 0.5% NP-40) with protease inhibitors for 15 min. The lysates were centrifuged at 13,000 rpm for 15 min at 4°C to remove cell debris. The whole-cell extracts were precleared with protein A/G-conjugated Sepharose beads for 2 h at 4°C with gentle agitation. After a high-speed centrifugation to remove beads, the extracts were incubated overnight at 4°C with rotation with a specific antibody as indicated. The protein A/G-conjugated Sepharose beads were then added and incubated at 4°C for another 1 h. The immune complexes were collected and washed extensively with immunoprecipitation washing buffer. The samples were resolved by sodium dodecyl sulfate-7.5% polyacrylamide gel electrophoresis (SDS-7.5% PAGE) or SDS-10% PAGE and transferred to nitrocellulose membranes for subsequent Western blot analysis. For multiple rounds of blotting, the membranes were stripped in stripping buffer (100 mM β-mercaptoethanol, 2% SDS, and 62.5 mM Tris-Cl, pH 6.7) for 30 min at 50°C. The antibodies against N-CoR and HDAC3 were as described previously (59), and antibody against HEXIM1 was raised against GST-HEXIM1. Anti-Flag monoclonal antibody was purchased from Sigma and CDK9, and cyclin T1 antibodies were from Santa Cruz Biotechnology.
Gel filtration analysis of HeLa nuclear extracts using a Superose 6 column was performed as previously described (58). Prior to gel filtration analysis, the HeLa nuclear extracts were treated either with or without RNase A at room temperature for 30 min.
CTD kinase assay.
Immune complex kinase assays for P-TEFb were performed essentially as described previously (44), using monoclonal antibody against FLAG or CDK9. Kinase assays were performed by adding 25 μl of kinase buffer containing 50 mM Tris-HCl (pH 7.4), 10 mM MgCl2, 5 mM dithiothreitol, 2.5 mM MnCl2, 5 μM ATP, 5 μCi [γ-32P]ATP, and 200 ng GST-CTD to the beads and incubation at room temperature with rotation for 60 min. The complexes were resolved by SDS-PAGE, and CTD phosphorylation was visualized by autoradiography.
GST pulldown assay.
GST pulldown assays were done essentially as previously described (61). In brief, both GST and GST-HEXIM1 fusion proteins were expressed in BL21 Escherichia coli cells, and equal amounts of each protein were immobilized onto glutathione-Sepharose 4B beads (Amersham Biosciences). The beads were incubated for 2 h at 4°C with 35S-labeled SMRT, N-CoR, HDAC3, TBL1 and TBLR1, or N-CoR fragments synthesized using a TNT in vitro transcription/translation kit (Promega) according to the manufacturer's instructions. The beads were washed five times with washing buffer (20 mM Tris-HCl, 150 mM NaCl, 1 mM dithiothreitol, 1 mM EDTA, 0.1% NP-40 with protease inhibitors). The samples were then separated by SDS-PAGE and visualized by autoradiography.
Cell culture and siRNA.
HeLa cells and 293T cells were routinely maintained in Dulbecco's modified Eagle medium (Invitrogen Inc.) supplemented with 10% fetal bovine serum and 1% antibiotics at 37°C under 5% CO2. For the small interfering RNA (siRNA) experiments, HeLa cells were seeded the night before transfection at such a density that cells reach about 30 to 40% confluence by the time of transfection. siN-CoR and siHDAC3 were previously described (58). siHEXIM1 (AAGUUCGACGAGAAACAGA) was synthesized by Dharmacon Research. siHEXIM1 (20 nM) was used for transfection using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. siRNA-transfected cells were cultured for 3 days and then transfected with reporter DNA and CDK9 expression constructs as indicated in the figure legends. The efficiency of the siRNA knockdown was determined at the end of each experiment by Western blot analysis using corresponding specific antibodies.
Generation of the CDK9 inducible stable cell line.
Inducible stable 293 cell lines expressing Flag-CDK9 or CDK9 mutants (K44R, K48/49R, and K44/48/49R, respectively) were generated using the Flp-In-T-REx system from Invitrogen. The wild-type CDK9 or CDK mutants were cloned into pcDNA5/FRT/TO expression vector and transfected into Flp-In-293 host cells, and the stable cells with integrated CDK9 or mutant expression constructs were selected according to the manufacturer's instructions (Invitrogen).
RPA.
RNase protection assay (RPA) was performed essentially as described previously, with minor modifications (34). In brief, total RNAs were isolated from transfected cells 24 h posttransfection. Approximately 30 μg of total RNA for each sample was used for the RNase protection assay. To make the HIV LTR probe (120 bp), the 175-bp-long PCR amplification fragment was produced using G5-83-HIV-luc as a template by the primer pair of RPA1 (AACCTTGGCACTATGTTACTGGGTCTCTCTGGTT) and RPA2 (TAATACGACTCACTATGGGTCCTATGGCATAAGTCTTCCATGGTGGCTTTAC) that also contains a T7 promoter. The T7 polymerase was used to produce [α-32P]UTP-labeled RNA probe according to the MAXIscript in vitro transcription kit (Ambion). The RNase protection assay was performed using the RPA III RNase protection assay kit (Ambion). The protected fragments were separated on a 10% polyacrylamide-8 M urea sequencing gel and analyzed using the Molecular Dynamics PhosphorImager System.
In vivo acetylation assay.
For metabolic labeling of acetylated proteins in HeLa cells, HeLa cells (2 × 108) were washed twice with cold PBS, resuspended in Dulbecco's modified Eagle's medium-based labeling medium (1 mCi of [3H]sodium acetate/ml and 2 μM trichostatin A [TSA] in 5 ml of DMEM) and incubated at 37°C for 1 h. Cells were washed twice with cold PBS and lysed with the EBC buffer. After a high-speed centrifugation to remove cell debris, the extracts were prepared and processed for immunoprecipitation with antibody against CDK9, HEX1M1, or cyclin T1. For metabolic labeling of stable 293 inducible Flag-CDK9 or mutant cell lines, cells were first induced to express Flag-CDK9 or mutants with 1 μg/ml doxycycline overnight and then metabolically labeled with [3H]sodium acetate as described above. The cell lysates were prepared either as described above (see Fig. 3B) or with radioimmunoprecipitation assay (RIPA) buffer (see Fig. 4D), centrifuged to remove insoluble debris, and used for IP using M2 agarose beads. The immunopurified Flag-CDK9 or its mutants were resolved on SDS-10% PAGE, fixed with 7% glacial acetic acid and 25% methanol for 30 min, and enhanced with a commercial fluorography enhancing solution (Amplify) (Amersham Pharmacia Biotech) for 30 min. Gels were then dried and subjected to autoradiography at −70°C for 7 to 14 days.
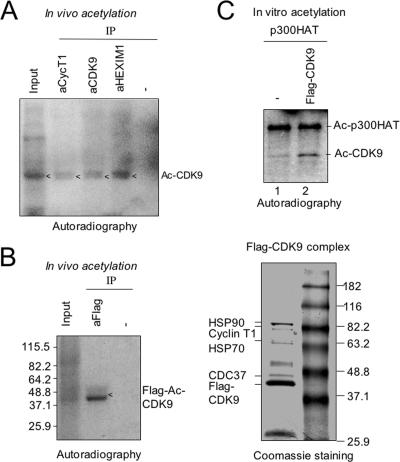
FIG. 3.
In vivo and in vitro acetylation of CDK9. (A) CDK9 is an acetylated (Ac) protein in HeLa cells. The acetylated proteins in HeLa cells were metabolically labeled with [3H]sodium acetate for 1 h. The P-TEFb complex was isolated from the whole-cell extract by IP using anti-cyclin T1, -CDK9, or -HEXIM1 antibody and resolved by SDS-10% PAGE. The acetylation status of P-TEFb was revealed by autoradiography. The size of the acetylated protein matched with CDK9. (B) A stable Flag-CDK9 cell line was metabolically labeled with [3H]sodium acetate, and Flag-CDK9 was isolated from the whole-cell extract using anti-Flag M2 beads. The acetylation of Flag-CDK9 was detected by autoradiography following SDS-PAGE. As a specificity control, the extract was also immunoprecipitated using rabbit anti-mouse immunoglobulin G (−). The left panel shows Flag-CDK9 complex purified from the control unlabeled cells revealed by Coomassie staining. The identities of the proteins were determined by mass spectrometry analysis. (C) Flag-CDK9 was isolated from the whole-cell extract as described above and subjected to in vitro acetylation by p300HAT (HAT domain). The whole-cell extract derived from the parent 293 Flp-In cells (−) was also subjected to purification using anti-Flag M2 beads and used as a negative control. The acetylation of Flag-CDK9 was detected by autoradiography following SDS-PAGE.
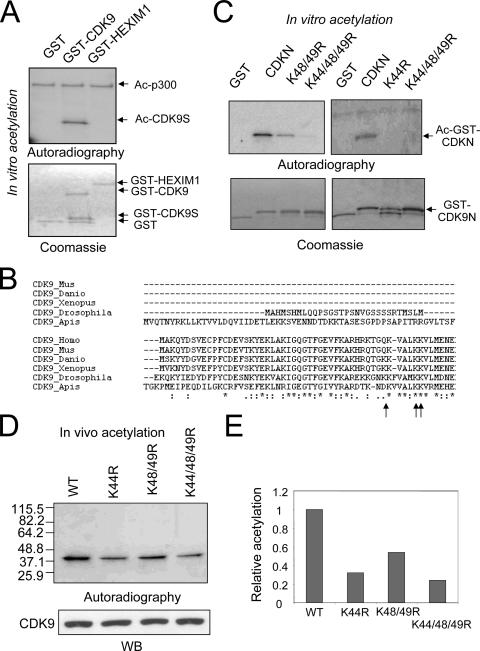
FIG. 4.
CDK9 is mainly acetylated at the site of K44. (A) p300HAT acetylated the CDK9 N-terminal sequence in vitro. The recombinant GST, GST-CDK9, and GST-HEXIM1 were incubated with p300HAT in acetylation reactions, and the resulting acetylated proteins were resolved by SDS-PAGE and visualized by autoradiography. Note that the size of the acetylated protein in the GST-CDK9 lane matched with a short protein (GST-CDK9S) but not with the full-length GST-CDK9. Ac-p300, acetylated p300. (B) The alignment of CDK9 sequences reveals highly conserved K residues in the N-terminal domain. The accession numbers for each of the proteins in the alignment are as follows: NP_001252 (Homo sapiens), NP_570930 (Mus musculus), AAH55634 (Danio rerio), AAH45274 (Xenopus laevis), AAM50669 (Drosophila melanogaster), and XP_392973 (Apis mellifera). The conserved K residues are indicated by arrows. (C) The CDK9 N-terminal sequence (aa 1 to 55) was expressed as a GST fusion (GST-CDK9N). Each of the conserved K residues was mutated to R individually or in combination as indicated. The resulting GST fusion proteins were purified from bacteria and subjected to in vitro acetylation with p300HAT using [3H]acetyl-coenzyme A as substrate. The upper panel shows the results of acetylation detected by autoradiography, whereas the lower panel shows recombinant GST, GST-CDK9N, and its related mutants revealed by Coomassie staining. (D) Stable cell lines expressing wild-type (WT) Flag-CDK9 or Flag-CDK9 bearing K44R, K48/49R, or K44/48/49R mutation were metabolically labeled with [3H]sodium acetate, and the whole-cell extracts were prepared with RIPA buffer. Flag-CDK9 and each mutant CDK9 were isolated from the whole-cell extracts and divided into two aliquots, one for detection of CDK9 acetylation by autoradiography and one for quantitating CDK9 proteins by Western blotting (WB). (E) The results shown in panel D were quantified and are shown as relative levels of acetylation of different CDK9 mutants in comparison to the wild-type CDK9.
In vitro acetylation assay.
In vitro acetylation was performed using either recombinant GST-CDK9, GST-CDK9N (residues 1 to 55), GST-CDK9N mutants, or immunoprecipitated CDK9 as substrate. Bacterially expressed p300 fragment (residues 1195 to 1810) was used as acetyltransferase, and the acetylation reactions were essentially as described previously (31). Reaction products were resolved by SDS-PAGE. Gels were stained with Coomassie brilliant blue, destained, and then treated with Amplify at room temperature for 30 min, dried, and subjected to autoradiography at −70°C for 2 to 15 days.
RESULTS
HEXIM1 as an N-CoR-interacting protein.
Through a combination of conventional chromatography and antibody affinity purification, we had purified the N-CoR complex(es) from HeLa nuclear extracts and identified TBL1/TBLR1 as its associated proteins through mass spectrometry (59). Careful examination of the large number of peptides deriving from the TBL1/TBLR1 band identified two peptides belonging to HEXIM1 protein (Fig. 1A). Because HEXIM1 was known to associate with and regulate P-TFEb (37, 57), a positive transcriptional elongation factor, the association of N-CoR with HEXIM1 raised the possibility that, in addition to histone deacetylation, N-CoR complex may repress transcription through negative regulation of P-TEFb. To confirm its association with N-CoR, we raised an antibody against GST-HEXIM1, and the specificity of this antibody was confirmed by IP-Western analysis (Fig. 1B). When this antibody was used for IP-Western analysis, we found that HEXIM1 was detected in IP of HeLa nuclear extracts with antibodies against N-CoR or HDAC3 but not a control rabbit anti-mouse immunoglobulin G (Fig. 1C). In the same IP, we found that HDAC3 but not HDAC1/2 was coprecipitated with HEXIM1 (data not shown; also see Fig. 2A).
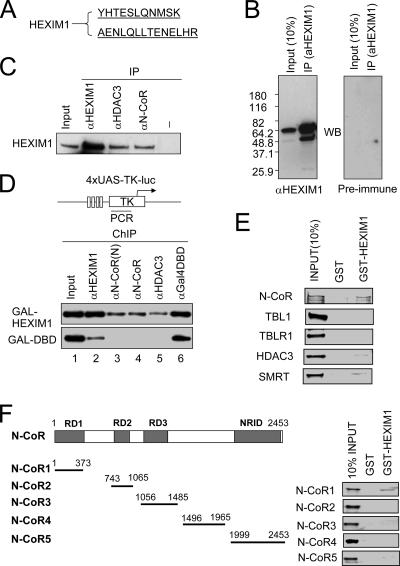
FIG. 1.
N-CoR interacts with HEXIM1. (A) HEXIM1 copurified with N-CoR from HeLa nuclear extracts. The purification of N-CoR complex was reported previously (59). The identity of the protein(s) in each band was determined by mass spectrometry. Two peptides derived from the TBL1/TBLR1 band were found to match with HEXIM1. (B) A rabbit anti-HEXIM1 antibody was raised against GST-HEXIM1. IP-Western analysis was performed using HeLa nuclear extract and anti-HEXIM1 antibody or preimmune serum. WB, Western blot. (C) Reciprocal IP showing the association of HEXIM1 with N-CoR complex. HeLa nuclear extracts were used for IP using antibodies as indicated, and the presence of HEXIM1 was detected by Western blot analysis. Input, 10% extract used for IP. The control (−) lane was IP using rabbit anti-mouse immunoglobulin G. (D) Gal-HEXIM1 recruited N-CoR complex to reporter DNA bearing Gal-DBD binding sites. Gal-HEXIM1 or control Gal-DBD(aa1-147) was transfected with the 4XUAS-TK-luc reporter into HeLa cells, and 24 h after transfection the association of N-CoR complex with the reporter DNA was analyzed by ChIP assay. The weak HEXIM1 signal from the control Gal-DBD sample is likely nonspecific. Note that N-CoR and HDAC3 were found to associate with the reporter DNA only in the presence of Gal-HEXIM1. (E) Analysis of the interaction between HEXIM1 and individual subunits of N-CoR/SMRT complexes using in vitro GST pulldown. Each subunit of the N-CoR complex was in vitro synthesized and labeled with [35S]methionine. The binding to GST-HEXIM1 was detected by autoradiography. (F) Each N-CoR fragment was in vitro synthesized and labeled with [35S]methionine. The binding to GST-HEXIM1 was detected by autoradiography.
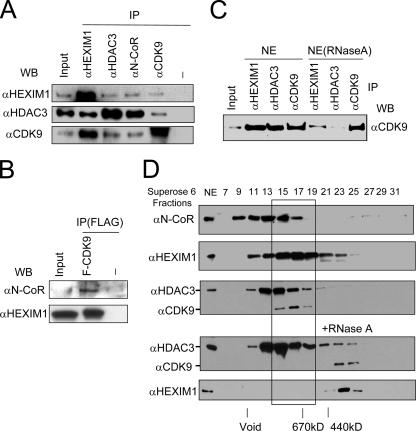
FIG. 2.
Subfraction of P-TEFb associates with the N-CoR complex. (A) CDK9 coimmunoprecipitated with N-CoR complex. HeLa nuclear extracts were used for IP using antibodies as indicated, and Western blotting (WB) was performed with anti-HEXIM1, -HDAC3, and -CDK9, respectively. (B) N-CoR and HEXIM1 coimmunoprecipitated with Flag-tagged CDK9. Whole-cell extracts were prepared from HeLa cells transfected with or without Flag-CDK9, and IP was performed using anti-Flag M2 agarose beads. The co-IP of N-CoR and HEXIM1 was then detected by Western blotting. (C) 7SK RNA is likely required for the association between N-CoR and P-TEFb. HeLa nuclear extracts (NE) were treated with or without RNase A for 30 min and then used for IP-Western analysis. (D) N-CoR complex partially cofractionated with CDK9 and P-TEFb in gel filtration experiments. HeLa nuclear extracts were treated with or without RNase A and fractionated through a fast-protein liquid chromatography Superose 6 sizing column. Different fractions were analyzed by Western blotting using antibodies as indicated. The peak fractions for molecular size markers are indicated.
To further substantiate the interaction between HEXIM1 and N-CoR complex, we next tested if tethering HEXIM1 to DNA through a Gal4 DNA binding domain was able to recruit N-CoR complex to DNA. We transfected a luciferase reporter (4XUAS-TK-luc) with Gal4-HEXIM1 or control Gal4-DBD into HeLa cells and carried out chromatin immunoprecipitation (ChIP) assays as described previously (61). The results in Fig. 1D demonstrate that, while both Gal4-DBD and Gal4-HEXIM1 bound to the reporter DNA (lane 6), N-CoR and HDAC3 were only detected in the presence of Gal4-HEXIM1. The weak signal from HEXIM1 antibody for Gal-DBD is likely the nonspecific background, since it was not detected when less HEXIM1 antibody was used (data not shown). Thus, tethering HEXIM1 to DNA is sufficient to recruit N-CoR complex, demonstrating further an interaction between HEXIM1 and N-CoR complex.
We next analyzed which subunit of the N-CoR complex interacts with HEXIM1 using an in vitro GST pulldown assay. We found that in vitro-translated N-CoR bound to GST-HEXIM1, whereas no binding of TBL1 and TBLR1 was detected (Fig. 1E). A weak binding was also detected for HDAC3. In addition, the N-CoR-related SMRT protein also exhibited a weak binding in this assay. Thus, the in vitro pulldown assay indicates that HEXIM1 likely associates with N-CoR complex through a direct interaction with N-CoR. Consistent with this idea, we have further mapped the HEXIM1 interaction domain of N-CoR to the N-CoR RD1 region (Fig. 1F).
Association of N-CoR complex with CDK9.
Given that HEXIM1 associates with and negatively regulates P-TEFb, we next tested if N-CoR complex also interacts with P-TEFb. We performed IP-Western analysis using HeLa nuclear extracts and antibodies against HEXIM1, N-CoR, HDAC3, and CDK9. We found that, like HEXIM1, a low level of CDK9 was coprecipitated with N-CoR and HDAC3 (Fig. 2A). Furthermore, HDAC3 was also detected in IP with anti-CDK9 antibody. Consistent with previous reports (37, 57, 58), CDK9 was abundantly coprecipitated with HEXIM1, and HDAC3 was readily coprecipitated with N-CoR. These results suggest that a subfraction of N-CoR/HDAC3 complex was associated with HEXIM1/P-TEFb. In support of association with P-TEFb complex, cyclin T1 could be detected in IP with antibody against N-CoR or HDAC3 (data not shown). The association of N-CoR with P-TEFb was further demonstrated by the result that both N-CoR and HEXIM1 were specifically coprecipitated from the HeLa cell extracts overexpressing a Flag-tagged CDK9 (Fig. 2B). The association between N-CoR and P-TEFb is at least in part dependent on 7SK small nuclear RNA, because addition of RNase A to HeLa nuclear extracts substantially reduced the amount of CDK9 coprecipitated with HEXMI1 and CDK9 but had no effect on IP of CDK9 itself (Fig. 2C). Finally, when HeLa nuclear extracts were fractionated using a Superose 6 sizing column, we found that there was an overlap between fractions containing N-CoR/HDAC3 complex and HEXIM1/P-TEFb (Fig. 2D). RNase A treatment of HeLa nuclear extract resulted in significant change in the fractionation pattern of CDK9 and HEXIM1 (Fig. 2D, lower panel), suggesting that the large CDK9/HEXIM1 complex(es) contains an RNA component, most likely 7SK. Taken together, these results indicate that a subfraction of cellular HEXIM1/P-TEFb is associated with N-CoR/HDAC3 complex.
CDK9 can be acetylated both in vivo and in vitro.
N-CoR/HDAC3 is well documented as a corepressor complex that represses transcription through its ability to deacetylate histones in chromatin (17, 32, 52, 58, 62). Given the interaction between N-CoR/HDAC3 and P-TEFb complex, we hypothesized that P-TEFb complex may be regulated by acetylation and deacetylation. To test this possibility, we metabolically labeled the acetylated proteins in HeLa cells with [3H]sodium acetate for 1 h. We then immunoprecipitated P-TEFb complex from the cellular extracts and determined if any component of the cellular P-TEFb complex was acetylated by autoradiography. As shown in Fig. 3A, we found that antibodies against both cyclin T1 and CDK9 precipitated an acetylated protein with an expected size of CDK9. Consistent with an interaction between HEXIM1 and P-TEFb, the same protein was also precipitated with anti-HEXIM1 antibody (Fig. 3A). To ensure that this acetylated protein is CDK9, we established an inducible Flag-CDK9 stable cell line using the 293 Flp-In system from Invitrogen. This cell line was subjected to overnight induction of Flag-CDK9 with doxycycline (1 μg/ml), followed by metabolic labeling with [3H]sodium acetate. Immunoprecipitation with anti-Flag antibody followed by autoradiography clearly detected an acetylated protein with the expected size of Flag-CDK9 (Fig. 3B, left panel). Under similar conditions, purification of Flag-CDK9 from control unlabeled cells yielded a Flag-CDK9 complex containing cyclin T1, HSP90, HSP70, and CDC37 and other known CDK9-associated proteins (Fig. 3B, right panel). Together these results indicate that in the P-TEFb complex only CDK9 is acetylated in vivo.
Having established that CDK9 within the P-TEFb complex is acetylated in vivo, we next tested if CDK9 could be acetylated in vitro. We isolated Flag-CDK9 from our Flag-CDK9 inducible cell line and subjected it to acetylation by recombinant p300HAT. The results in Fig. 3C show that an acetylated protein was detected from samples derived from the Flag-CDK9 stable cell line (lane 2) but not from the control parental cell line (lane 1). The size of these acetylated proteins again matched perfectly with Flag-CDK9. As previously reported, p300HAT underwent autoacetylation in these reactions (50). Taken together, these results show that CDK9 is an acetylated protein in vivo and can be acetylated by p300 in vitro.
Identification of CDK9 acetylation sites.
To identify potential acetylation sites in CDK9, we decided to first map the CDK9 acetylation sites catalyzed by p300HAT in vitro. For this purpose, we expressed and purified GST-CDK9 fusion protein from bacteria and tested if it could be acetylated by p300HAT. The autoradiography results in Fig. 4A revealed a strongly acetylated band in the reaction with GST-CDK9. However, careful comparison with protein bands in the Coomassie staining gel indicated this acetylated band did not match with the full-length GST-CDK9 but with a smaller-sized protein band we termed GST-CDK9S. By Western blot analysis using anti-GST antibody, we verified that this protein contained GST (data not shown). Based on its size and the presence of GST, we surmised this acetylated protein was a spontaneously degraded GST-CDK9 containing a portion of CDK9 N-terminal sequence. As a specificity control, we found that both GST and GST-HEXIM1 were not acetylated by p300HAT under the same experimental condition. As an internal control for the acetylation reaction, p300HAT domain was autoacetylated to the same extent in all three reactions. Since the data in Fig. 3C show that Flag-CDK9 was acetylated by p300HAT in vitro, the lack of acetylation for the full-length GST-CDK9 by p300HAT in this experiment was likely a consequence of steric hindrance caused by the presence of GST. The acetylation site(s) in the N terminus of CDK9 was likely exposed when a large portion of CDK9 was spontaneously deleted from GST-CDK9. Nevertheless, this fortuitous data suggested to us that CDK9 is likely acetylated at its N terminus.
To search for potential acetylation sites in the N terminus of CDK9, we first aligned CDK9 sequences from different organisms (Fig. 4B). This analysis revealed three evolutionally conserved lysine (K) residues at positions 44, 48, and 49 of human CDK9. To test if these sites could be acetylated by p300 in vitro, we generated GST-CDK9N(aa1-55) fusion protein as well as the fusion proteins containing specific K-to-arginine (R) mutations at the sites indicated. We chose K-to-R mutation because this change eliminates acetylation but likely maintains the protein conformation. When these GST fusion proteins were subjected to acetylation by p300HAT, we found that the K48/49R mutant was less efficiently acetylated and the K44/48/49R mutant was poorly acetylated (Fig. 4C). Like the K44/48/49R mutant, the K44R mutant was also poorly acetylated, suggesting that K44 is the major site acetylated by p300HAT in vitro. Thus, our in vitro studies identified within CDK9N(aa1-55) K48/49 as potential sites and K44 as the major site for acetylation by p300HAT.
K44 is the major acetylation site of CDK9 in vivo.
To test if the acetylation occurs in cells at the sites identified in vitro, we generated stable cell lines expressing Flag-tagged K44R, K48/49R, or K44/48/49R CDK9 mutant, respectively. These cell lines were subjected to in vivo metabolic labeling as described above. To ensure that the observed acetylated protein was indeed Flag-tagged CDK9 or mutants, the cell extracts were prepared with stringent RIPA buffer and used for IP with anti-Flag M2 beads. The results depicted in Fig. 4D show that K48/49R mutation resulted in ∼50% reduction of acetylation. The K44R mutant displayed a more significant reduction of acetylation (∼75%), whereas the level of acetylation of K44/48/49R mutant was similar to that of K44R. As a control, Western blot analysis using anti-Flag antibody showed that the difference in the levels of acetylation was not due to the difference in the levels of proteins. Thus, the in vivo labeling experiment demonstrated the critical role for K44 in CDK9 acetylation, in full agreement with our in vitro data in Fig. 4C. Since the K44/48/49R mutant still retained ∼20 to 30% of acetylation compared to Flag-CDK9 (Fig. 4D), CDK9 was also acetylated at another unidentified site(s).
Acetylation on K44 of CDK9 affects P-TEFb kinase activity.
The above results identified K44 as the major acetylation site of CDK9. To examine how K44 acetylation affects CDK9 function, we isolated the P-TEFb complex from the inducible stable cell lines expressing either the wild type or K44R mutant using anti-Flag M2 beads and assayed their kinase activity using GST-CTD substrate as described previously (44). The results in Fig. 5A show that the kinase activity of the K44R mutant was significantly crippled compared to that of the wild-type CDK9. Since the kinase activity of CDK9 is dependent on its association with the cyclin T subunit (42, 43), we performed IP-Western analysis to test if K44R mutation affected the association between CDK9 and cyclin T1. The results shown in Fig. 5B indicate that K44R mutation did not affect the interaction between CDK9 and cyclin T1. In addition, the K44R mutant exhibited normal association with HEXIM1, thus excluding aberrant association with HEXIM/7SK as an explanation for the reduced kinase activity (Fig. 5B). Given that K44 is acetylated in vivo and K-to-R mutation is likely to maintain CDK9 conformation but eliminate acetylation, we suggest that acetylation on K44 of CDK9 positively regulates P-TFEb kinase activity.
FIG. 5.
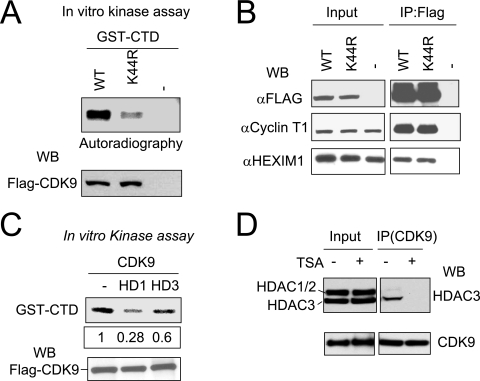
CDK9 acetylation positively regulates its kinase activity. (A) The K44R mutation in CDK9 significantly impaired its kinase activity. The P-TEFb complex was isolated from the whole-cell extracts expressing Flag-CDK9 or Flag-CDK9(K44R) mutant and divided into two aliquots, one for a kinase activity assay using GST-CTD as substrate and [γ-32P]ATP and the other for Western blot (WB) analysis. (B) IP-Western analysis showing that K44R mutation did not affect the association of CDK9 with cyclin T1 and HEXIM1. (C) Expression of HDAC1 or HDAC3 reduced CDK9 kinase activity. Flag-CDK9 was cotransfected without or with an HDAC1 or HDAC3 expression plasmid into HeLa cells, and the Flag-CDK9-containing P-TFEb complex was recovered from the whole-cell extracts using anti-Flag M2 beads and assayed for phosphorylation of GST-CTD as described above. The levels of phosphorylation were quantified using a densitometer. The Western blot analysis of Flag-CDK9 showed that a comparable amount of Flag-CDK9 was recovered from each sample. (D) TSA treatment abolished the association between CDK9 and HDAC3. HeLa cells were treated with or without TSA (330 nM) for 12 h, and the association between CDK9 and HDAC3 was analyzed by IP-Western analysis. WT, wild type.
To further substantiate that acetylation regulates P-TEFb kinase activity, we next tested if the P-TEFb kinase activity is modulated by HDACs. For this purpose, Flag-CDK9 was coexpressed in HeLa cells with or without HDAC1 or HDAC3 by transfection. Flag-CDK9 was then isolated from the transfected cells, and the kinase activity was assayed using GST-CTD as substrate as described above. A representative result in Fig. 5C shows that Flag-CDK9 isolated from cells expressing HDAC1 or HDAC3 exhibited reduced kinase activity toward GST-CTD. Control Western blot analysis demonstrated that an equal amount of CDK9 was isolated and used in the kinase reactions (Fig. 5C, lower panel). Interestingly, in this experiment HDAC1 inhibited CDK9 kinase activity more than HDAC3, although endogenous HDAC1 was not found to associate with CDK9 (see Fig. 5D). This discrepancy could be explained if overexpression of HDAC1 allowed HDAC1 to deacetylate CDK9 and by the possibility that overexpression of HDAC3 may not significantly increase HDAC3 activity, since the HDAC3 activity is dependent on its incorporation into the SMRT/N-CoR complex (16, 52, 62). As additional evidence that CDK9 is regulated by HDAC3, we found that TSA treatment of HeLa cells led to dissociation of endogenous CDK9 from HDAC3 (Fig. 5D). This result suggests that HDAC3 is likely directly involved in the interaction with P-TEFb complex. Together these results support that the kinase activity of P-TEFb is regulated by acetylation of the CDK9 subunit.
Acetylation on K44 of CDK9 affects P-TEFb transcriptional elongation activity.
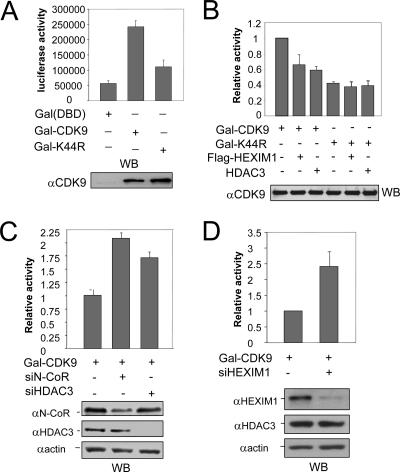
Previous studies indicated that transcription from the HIV LTR gives rise to primarily short, abortive transcripts 55 to 70 nucleotides in length in the absence of Tat activator (27, 34). Tat is known to recruit P-TEFb, which in turn promotes transcriptional elongation and production of full-length polyadenylated transcripts. Importantly, direct targeting of CDK9 or CycT1 as a Gal4(DBD) fusion protein to a reporter driven by the HIV LTR promoter activates transcription primarily by promoting elongation, and the kinase activity of CDK9 is strictly required for this activity (1, 34, 49). We thus used this system to assay if K44R mutation impaired CDK9 transcriptional elongation activity. Consistent with previous results (34), we found that expression of Gal-CDK9 enhanced the transcription ∼4.6-fold from an HIV LTR-driven (G5-83-HIV-luc) reporter bearing five Gal4 binding sites (Fig. 6A). Under the same conditions, the K44R mutant had only ∼30% of the wild-type CDK9 transcriptional activity (Fig. 6A). Western blot analysis revealed a similar level of Gal-CDK9 and Gal-K44R mutant, indicating that K44R mutation significantly reduced its ability to enhance transcriptional elongation.
FIG. 6.
CDK9 transcriptional activity is negatively regulated by N-CoR/HDAC3. (A) The CDK9 bearing K44R mutation exhibited reduced transcriptional activity when tethered to reporter DNA as a Gal-DBD fusion protein. 293T cells were transfected with expression vector for control Gal-DBD, Gal-CDK9, or Gal-K44R mutant and G5-83-HIV-luc reporter as indicated. The luciferase activities were determined, and results are presented as the means ± standard errors of three independent experiments. The expression of Gal-CDK9 fusions was detected by Western blotting (WB) using anti-CDK9 antibody. (B) Expression of HEXIM1 or HDAC3 repressed the transcriptional activity of the wild-type CDK9 but not the K44R mutant. The results are presented as relative activities, with the value of Gal-CDK9 set as 1. Also shown is the level of Gal-CDK9 revealed by Western blotting using anti-CDK9 antibody. (C) Knockdown of N-CoR or HDAC3 enhanced the transcriptional activity of CDK9. HeLa cells were treated with or without siN-CoR or siHDAC3 as indicated, followed by transfection with Gal4-CDK9 and G5-83-HIV-luc reporter. The knockdown of N-CoR or HDAC3 was verified by Western blot analysis. (D) Knockdown of HEXIM1 enhanced the transcriptional activity of CDK9. HeLa cells were treated with or without siHEXIM1 for two days before transfection with Gal4-CDK9 and G5-83-HIV-luc reporter. The knockdown of HEXIM1 was verified by Western blot analysis.
To gain further evidence that the reduced transcriptional activity of K44R mutant is linked to its defect in K44 acetylation, we compared the effect of expression of HEXIM1 or HDAC3 on the transcriptional activity of Gal-CDK9 and Gal-K44R mutant. As shown in Fig. 6B, expression of either HEXIM1 or HDAC3 reduced the transcriptional activity of Gal-CDK9. However, expression of HEXIM1 or HDAC3 had a minimal effect on the transcriptional activity of the Gal-K44R mutant. Western blot analysis detected a comparable level of Gal-CDK9 or Gal-K44R mutant from each sample. This result suggests that K44 of CDK9 is required for HDAC3 to inhibit CDK9 transcriptional activity, implying that HDAC3 inhibits CDK9 transcriptional activity mainly through deacetylation of K44.
To test if endogenous N-CoR/HDAC3 repressed CDK9 transcriptional activity, we made use of siRNA specific for N-CoR or HDAC3. We found that knockdown of either N-CoR or HDAC3 resulted in increased transcriptional activity for Gal-CDK9 (Fig. 6C). The knockdown of N-CoR or HDAC3 by its corresponding siRNA was confirmed by Western blot analysis (Fig. 6C, lower panel). Consistent with HEXIM1 as a negative regulator of P-TEFb (57), we found that knockdown of HEXIM1 also led to increased transcriptional activity for Gal-CDK9 (Fig. 6D).
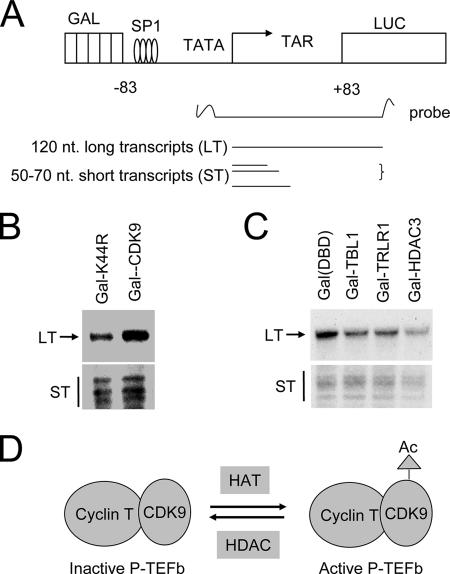
To directly examine that K44R mutation impaired the transcriptional elongation function of CDK9, we performed the RNase protection assays depicted in Fig. 7A to measure both long transcripts and abortive short transcripts as described previously (34). A representative result in Fig. 7B shows that there was no significant difference between two samples in the levels of short transcripts. However, the level of long transcripts was significantly lower for the sample derived from Gal-K44R mutant-transfected cells. This result is consistent with the previous data that CDK9 enhances transcription from the HIV LTR reporter by promoting transcriptional elongation.
FIG. 7.
Evidence that K44R mutation in CDK9 impairs the transcriptional elongation and that tethering components of N-CoR complex to DNA suppresses transcriptional elongation. (A) Schematic representation of G5-83-HIV-luc reporter and RNase protection assay for transcriptional elongation. CDK9 kinase activity is required for overcoming transcriptional pausing at the proximal region. nt, nucleotide. (B) HeLa cells were transfected with Gal-CDK9 or Gal-CDK9(K44R) mutant together with G5-83-HIV-luc reporter. After overnight incubation, RNase protection assays were performed to detect both the short transcripts resulting from RNA pol II pausing and long transcripts as a consequence of processive transcriptional elongation. (C) HeLa cells were transfected with Gal-TBL1, Gal-TBLR1, or Gal-HDAC3, and RNase protection assays were performed as described for panel B. (D) A working model illustrates that P-TEFb transcriptional activity is regulated by dynamic acetylation (Ac) of CDK9.
Targeting components of N-CoR complex to the HIV LTR reporter suppresses transcriptional elongation.
So far we have shown that N-CoR complex interacts with P-TEFb and potentially regulates CDK9 transcriptional elongation activity through deacetylation of CDK9. To test if N-CoR complex could repress P-TEFb-dependent transcriptional elongation, we tested if tethering components of N-CoR complex to the G5-83-HIV-luc reporter inhibited the production of long transcripts by RNase protection. The results in Fig. 7C show that Gal-TBL1, Gal-TBLR1, and Gal-HDAC3 all significantly inhibited the production of long forms of transcripts but had much less effect on the short transcripts. These results show a role for N-CoR complex in repression of transcriptional elongation, presumably through its inhibition of P-TEFb activity.
DISCUSSION
While histone acetylation clearly plays a critical role in transcriptional regulation, growing evidence indicates that acetylation of non-histone proteins also plays a role in transcriptional regulation. More and more proteins have been found to be acetylated in vitro and/or in vivo, including general transcriptional factors, transcription factors, chaperones, and many others (for reviews, see references 15 and 63). In this study, we found that a subfraction of N-CoR/HDAC3 complex is associated with P-TEFb. We further showed that CDK9 is an acetylated protein in vivo and can be acetylated by p300 in vitro. Through both in vitro and in vivo experiments, we identified the highly conserved K44 of CDK9 as a major acetylation site and demonstrated that a single K44R mutation crippled CDK9 kinase activity. We also showed that the K44R mutation significantly reduced the ability for CDK9 to enhance transcriptional elongation from an HIV LTR promoter-driven reporter (Fig. 6A and 7B). Previous studies have shown that the transcriptional activity of the HIV LTR promoter is primarily regulated at the step of transcriptional elongation and that targeting CDK9 to an HIV LTR promoter-driven reporter activates transcription by promoting elongation in a CDK9 kinase-dependent manner (1, 27, 34, 49). Together our data support a role for acetylation of K44 of CDK9 in control of P-TEFb kinase and transcriptional elongation activity.
In this study we have also provided multiple lines of evidence that N-CoR complex negatively regulates P-TEFb activity. First, we found that a subfraction of N-CoR complex is associated with P-TEFb. Second, we showed that coexpression of HDAC3 with CDK9 reduced the CDK9 kinase activity (Fig. 5C). Third, using transcriptional activation of the HIV LTR promoter by Gal-CDK9 as a functional readout for P-TEFb kinase and transcriptional elongation activity, we found that overexpression of HDAC3 repressed and knockdown of HDAC3 or N-CoR enhanced the P-TEFb activity (Fig. 6B and C). It is noteworthy that overexpression of HDAC3 did not repress the transcriptional activity of the CDK9 K44R mutant (Fig. 6B). Since K44 is the major acetylation site of CDK9 and CDK9 is the only acetylated protein in the P-TEFb complex, this result suggests that N-CoR/HDAC3 is likely to repress P-TEFb activity through deacetylation of K44 of CDK9. Finally, we showed that targeting subunits of N-CoR complex to the HIV LTR-driven reporter also preferentially inhibited the production of long transcripts (Fig. 7C). Since transcriptional elongation from this reporter is dependent on P-TEFb, we took these results as inhibition of P-TEFb activity by the N-CoR complex. Given that P-TEFb is a global transcriptional elongation factor important for most pol II transcription (2, 47), it is tempting to propose that, besides its ability to repress transcription through histone deacetylation, N-CoR/HDAC3 can also repress transcription by suppressing P-TEFb activity and consequently transcriptional elongation. However, due to the lack of a specific antibody recognizing acetylated K44 of CDK9, we have not been able to directly demonstrate that acetylation on K44 is regulated by N-CoR/HDAC3. Attempts to generate such antibodies have so far failed. For unknown reasons, the commercial anti-acetylated K antibodies (Abcam, Upstate, and Cell Signaling) do not appear to recognize acetylated CDK9 (data not shown).
As a kinase important for positive transcriptional elongation, multiple mechanisms have been shown to regulate CDK9 kinase activity. First, the CDK9 kinase activity is dependent on its association with a cyclin T subunit (42, 43). Second, the CDK9 kinase activity is regulated by phosphorylation. Like many other CDKs, phosphorylation on the T-loop by CDK9 itself or another kinase(s) activates CDK9 kinase activity (40). Third, the CDK9 kinase activity is inhibited by association with 7SK small nuclear RNA and HEXIM1 (37, 41, 56, 57). Previous studies indicate that about half of cellular P-TEFb is found to be associated with 7SK and HEXIM1 (38). Interestingly, we found that almost all CDK9 and HEXIM1 in HeLa nuclear extract behaved as an ∼700-kDa complex, and RNase A treatment converted all CDK9 and HEXIM1 to a smaller complex (Fig. 2D). Since half of P-TEFb was shown to associate with 7SK RNA, this result raises the possibility that P-TEFb may associate with additional unidentified RNA. Fourth, two recent studies indicated that P-TEFb is positively regulated by Brd4, a double bromodomain-containing protein (26, 55). Brd4 was found to associate only with the 7SK/HEXIM1-free P-TEFb, and this interaction requires the double bromodomain of Brd4. The bromodomain is a signature motif for binding of acetylated lysine (25, 60), and Brd4 has been shown to bind acetylated histones through double bromodomain (9, 28). In this study, we found that CDK9 is subjected to acetylation and deacetylation. We present evidence that acetylation promotes, whereas deacetylation by N-CoR/HDAC3 inhibits, the P-TEFb transcriptional elongation function. Given Brd4 is a double bromodomain associated with the active form of P-TEFb, it is tempting to suggest that, like acetylated histones, Brd4 may bind preferentially to the acetylated form of CDK9 and maintains its kinase activity in transcriptional elongation (Fig. 7D). On the contrary, deacetylation of CDK9 by N-CoR/HDAC3 or other HDACs may reduce the association of CDK9 with Brd4 and thus promotes the interaction of P-TEFb with 7SK and HEXIM1 to form an inactive complex. Thus, acetylation of CDK9 could play a regulatory role in partitioning P-TEFb into an active or inactive complex. In support of this idea, we show that N-CoR/HDAC3 mainly associates with the 7SK/HEXIM1-containing P-TEFb complex (Fig. 2C). In addition, TSA treatment dissociates HDAC3 from P-TEFb (Fig. 5D). Alternatively, CDK9 acetylation may affect T-loop phosphorylation and thus P-TEFb kinase activity.
In sum, we find in this study that a fraction of the cellular N-CoR complex is associated with and regulates P-TEFb. Our study suggests that the P-TEFb activity is likely regulated by reversible acetylation of CDK9. To our knowledge, our work identifies CDK9 as the first CDK whose activity is regulated by acetylation. In addition, our study suggests that, besides its ability to repress transcription through histone deacetylation, N-CoR/HDAC3 complex may also repress transcriptional elongation through its ability to regulate P-TEFb.
Acknowledgments
We thank Motoaki Sano at Baylor College of Medicine for the CDK9 construct and Luigi Lania for the G5-83-HIV-luc reporter. We also thank Bert O'Malley, Ming-jer Tsai, Sophia Tsai, and members of J. Wong's laboratory for valuable discussions.
This work was supported by DK58679 to J.W.
Footnotes
Published ahead of print on 23 April 2007.
REFERENCES
- 1.Bieniasz, P. D., T. A. Grdina, H. P. Bogerd, and B. R. Cullen. 1999. Recruitment of cyclin T1/P-TEFb to an HIV type 1 long terminal repeat promoter proximal RNA target is both necessary and sufficient for full activation of transcription. Proc. Natl. Acad. Sci. USA 96:7791-7796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chao, S. H., K. Fujinaga, J. E. Marion, R. Taube, E. A. Sausville, A. M. Senderowicz, B. M. Peterlin, and D. H. Price. 2000. Flavopiridol inhibits P-TEFb and blocks HIV-1 replication. J. Biol. Chem. 275:28345-28348. [DOI] [PubMed] [Google Scholar]
- 3.Chao, S. H., and D. H. Price. 2001. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J. Biol. Chem. 276:31793-31799. [DOI] [PubMed] [Google Scholar]
- 4.Chen, J. D., and R. M. Evans. 1995. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature 377:454-457. [DOI] [PubMed] [Google Scholar]
- 5.Chodosh, L. A., A. Fire, M. Samuels, and P. A. Sharp. 1989. 5,6-Dichloro-1-beta-D-ribofuranosylbenzimidazole inhibits transcription elongation by RNA polymerase II in vitro. J. Biol. Chem. 264:2250-2257. [PubMed] [Google Scholar]
- 6.Ciana, P., G. G. Braliou, F. G. Demay, M. von Lindern, D. Barettino, H. Beug, and H. G. Stunnenberg. 1998. Leukemic transformation by the v-ErbA oncoprotein entails constitutive binding to and repression of an erythroid enhancer in vivo. EMBO J. 17:7382-7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dahmus, M. E. 1995. Phosphorylation of the C-terminal domain of RNA polymerase II. Biochim. Biophys. Acta 1261:171-182. [DOI] [PubMed] [Google Scholar]
- 8.de Falco, G., and A. Giordano. 1998. CDK9 (PITALRE): a multifunctional cdc2-related kinase. J. Cell. Physiol. 177:501-506. [DOI] [PubMed] [Google Scholar]
- 9.Dey, A., F. Chitsaz, A. Abbasi, T. Misteli, and K. Ozato. 2003. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc. Natl. Acad. Sci. USA 100:8758-8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Egloff, S., E. Van Herreweghe, and T. Kiss. 2006. Regulation of polymerase II transcription by 7SK snRNA: two distinct RNA elements direct P-TEFb and HEXIM1 binding. Mol. Cell. Biol. 26:630-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flores, O., G. Lee, J. Kessler, M. Miller, W. Schlief, J. Tomassini, and D. Hazuda. 1999. Host-cell positive transcription elongation factor b kinase activity is essential and limiting for HIV type 1 replication. Proc. Natl. Acad. Sci. USA 96:7208-7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujinaga, K., T. P. Cujec, J. Peng, J. Garriga, D. H. Price, X. Grana, and B. M. Peterlin. 1998. The ability of positive transcription elongation factor B to transactivate human immunodeficiency virus transcription depends on a functional kinase domain, cyclin T1, and Tat. J. Virol. 72:7154-7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garriga, J., and X. Grana. 2004. Cellular control of gene expression by T-type cyclin/CDK9 complexes. Gene 337:15-23. [DOI] [PubMed] [Google Scholar]
- 14.Glass, C. K., and M. G. Rosenfeld. 2000. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14:121-141. [PubMed] [Google Scholar]
- 15.Glozak, M. A., N. Sengupta, X. Zhang, and E. Seto. 2005. Acetylation and deacetylation of non-histone proteins. Gene 363:15-23. [DOI] [PubMed] [Google Scholar]
- 16.Guenther, M. G., O. Barak, and M. A. Lazar. 2001. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol. Cell. Biol. 21:6091-6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guenther, M. G., W. S. Lane, W. Fischle, E. Verdin, M. A. Lazar, and R. Shiekhattar. 2000. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 14:1048-1057. [PMC free article] [PubMed] [Google Scholar]
- 18.Heinzel, T., R. M. Lavinsky, T. M. Mullen, M. Soderstrom, C. D. Laherty, J. Torchia, W. M. Yang, G. Brard, S. D. Ngo, J. R. Davie, E. Seto, R. N. Eisenman, D. W. Rose, C. K. Glass, and M. G. Rosenfeld. 1997. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature 387:43-48. [DOI] [PubMed] [Google Scholar]
- 19.Herrmann, C. H., R. G. Carroll, P. Wei, K. A. Jones, and A. P. Rice. 1998. Tat-associated kinase, TAK, activity is regulated by distinct mechanisms in peripheral blood lymphocytes and promonocytic cell lines. J. Virol. 72:9881-9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herrmann, C. H., and A. P. Rice. 1995. Lentivirus Tat proteins specifically associate with a cellular protein kinase, TAK, that hyperphosphorylates the carboxyl-terminal domain of the large subunit of RNA polymerase II: candidate for a Tat cofactor. J. Virol. 69:1612-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horlein, A. J., A. M. Naar, T. Heinzel, J. Torchia, B. Gloss, R. Kurokawa, A. Ryan, Y. Kamei, M. Soderstrom, C. K. Glass, et al. 1995. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature 377:397-404. [DOI] [PubMed] [Google Scholar]
- 22.Huang, E. Y., J. Zhang, E. A. Miska, M. G. Guenther, T. Kouzarides, and M. A. Lazar. 2000. Nuclear receptor corepressors partner with class II histone deacetylases in a Sin3-independent repression pathway. Genes Dev. 14:45-54. [PMC free article] [PubMed] [Google Scholar]
- 23.Ishizuka, T., and M. A. Lazar. 2003. The N-CoR/histone deacetylase 3 complex is required for repression by thyroid hormone receptor. Mol. Cell. Biol. 23:5122-5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ivanov, D., Y. T. Kwak, J. Guo, and R. B. Gaynor. 2000. Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Mol. Cell. Biol. 20:2970-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobson, R. H., A. G. Ladurner, D. S. King, and R. Tjian. 2000. Structure and function of a human TAFII250 double bromodomain module. Science 288:1422-1425. [DOI] [PubMed] [Google Scholar]
- 26.Jang, M. K., K. Mochizuki, M. Zhou, H. S. Jeong, J. N. Brady, and K. Ozato. 2005. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell 19:523-534. [DOI] [PubMed] [Google Scholar]
- 27.Jones, K. A., and B. M. Peterlin. 1994. Control of RNA initiation and elongation at the HIV-1 promoter. Annu. Rev. Biochem. 63:717-743. [DOI] [PubMed] [Google Scholar]
- 28.Kanno, T., Y. Kanno, R. M. Siegel, M. K. Jang, M. J. Lenardo, and K. Ozato. 2004. Selective recognition of acetylated histones by bromodomain proteins visualized in living cells. Mol. Cell 13:33-43. [DOI] [PubMed] [Google Scholar]
- 29.Kao, H. Y., M. Downes, P. Ordentlich, and R. M. Evans. 2000. Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes Dev. 14:55-66. [PMC free article] [PubMed] [Google Scholar]
- 30.Kobor, M. S., and J. Greenblatt. 2002. Regulation of transcription elongation by phosphorylation. Biochim. Biophys. Acta 1577:261-275. [DOI] [PubMed] [Google Scholar]
- 31.Li, J., B. W. O'Malley, and J. Wong. 2000. p300 requires its histone acetyltransferase activity and SRC-1 interaction domain to facilitate thyroid hormone receptor activation in chromatin. Mol. Cell. Biol. 20:2031-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, J., J. Wang, Z. Nawaz, J. M. Liu, J. Qin, and J. Wong. 2000. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 19:4342-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, Q., J. P. Price, S. A. Byers, D. Cheng, J. Peng, and D. H. Price. 2005. Analysis of the large inactive P-TEFb complex indicates that it contains one 7SK molecule, a dimer of HEXIM1 or HEXIM2, and two P-TEFb molecules containing Cdk9 phosphorylated at threonine 186. J. Biol. Chem. 280:28819-28826. [DOI] [PubMed] [Google Scholar]
- 34.Majello, B., G. Napolitano, A. Giordano, and L. Lania. 1999. Transcriptional regulation by targeted recruitment of cyclin-dependent CDK9 kinase in vivo. Oncogene 18:4598-4605. [DOI] [PubMed] [Google Scholar]
- 35.Marshall, N. F., J. Peng, Z. Xie, and D. H. Price. 1996. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J. Biol. Chem. 271:27176-27183. [DOI] [PubMed] [Google Scholar]
- 36.Marshall, N. F., and D. H. Price. 1995. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J. Biol. Chem. 270:12335-12338. [DOI] [PubMed] [Google Scholar]
- 37.Michels, A. A., A. Fraldi, Q. Li, T. E. Adamson, F. Bonnet, V. T. Nguyen, S. C. Sedore, J. P. Price, D. H. Price, L. Lania, and O. Bensaude. 2004. Binding of the 7SK snRNA turns the HEXIM1 protein into a P-TEFb (CDK9/cyclin T) inhibitor. EMBO J. 23:2608-2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michels, A. A., V. T. Nguyen, A. Fraldi, V. Labas, M. Edwards, F. Bonnet, L. Lania, and O. Bensaude. 2003. MAQ1 and 7SK RNA interact with CDK9/cyclin T complexes in a transcription-dependent manner. Mol. Cell. Biol. 23:4859-4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagy, L., H. Y. Kao, D. Chakravarti, R. J. Lin, C. A. Hassig, D. E. Ayer, S. L. Schreiber, and R. M. Evans. 1997. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell 89:373-380. [DOI] [PubMed] [Google Scholar]
- 40.Napolitano, G., B. Majello, and L. Lania. 2003. Catalytic activity of Cdk9 is required for nuclear co-localization of the Cdk9/cyclin T1 (P-TEFb) complex. J. Cell. Physiol. 197:1-7. [DOI] [PubMed] [Google Scholar]
- 41.Nguyen, V. T., T. Kiss, A. A. Michels, and O. Bensaude. 2001. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature 414:322-325. [DOI] [PubMed] [Google Scholar]
- 42.Peng, J., N. F. Marshall, and D. H. Price. 1998. Identification of a cyclin subunit required for the function of Drosophila P-TEFb. J. Biol. Chem. 273:13855-13860. [DOI] [PubMed] [Google Scholar]
- 43.Peng, J., Y. Zhu, J. T. Milton, and D. H. Price. 1998. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev. 12:755-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sano, M., M. Abdellatif, H. Oh, M. Xie, L. Bagella, A. Giordano, L. H. Michael, F. J. DeMayo, and M. D. Schneider. 2002. Activation and function of cyclin T-Cdk9 (positive transcription elongation factor-b) in cardiac muscle-cell hypertrophy. Nat. Med. 8:1310-1317. [DOI] [PubMed] [Google Scholar]
- 45.Sano, M., and M. D. Schneider. 2004. Cyclin-dependent kinase-9: an RNAPII kinase at the nexus of cardiac growth and death cascades. Circ. Res. 95:867-876. [DOI] [PubMed] [Google Scholar]
- 46.Sano, M., S. C. Wang, M. Shirai, F. Scaglia, M. Xie, S. Sakai, T. Tanaka, P. A. Kulkarni, P. M. Barger, K. A. Youker, G. E. Taffet, Y. Hamamori, L. H. Michael, W. J. Craigen, and M. D. Schneider. 2004. Activation of cardiac Cdk9 represses PGC-1 and confers a predisposition to heart failure. EMBO J. 23:3559-3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shim, E. Y., A. K. Walker, Y. Shi, and T. K. Blackwell. 2002. CDK-9/cyclin T (P-TEFb) is required in two postinitiation pathways for transcription in the C. elegans embryo. Genes Dev. 16:2135-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sims, R. J., III, R. Belotserkovskaya, and D. Reinberg. 2004. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 18:2437-2468. [DOI] [PubMed] [Google Scholar]
- 49.Taube, R., X. Lin, D. Irwin, K. Fujinaga, and B. M. Peterlin. 2002. Interaction between P-TEFb and the C-terminal domain of RNA polymerase II activates transcriptional elongation from sites upstream or downstream of target genes. Mol. Cell. Biol. 22:321-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thompson, P. R., D. Wang, L. Wang, M. Fulco, N. Pediconi, D. Zhang, W. An, Q. Ge, R. G. Roeder, J. Wong, M. Levrero, V. Sartorelli, R. J. Cotter, and P. A. Cole. 2004. Regulation of the p300 HAT domain via a novel activation loop. Nat. Struct. Mol. Biol. 11:308-315. [DOI] [PubMed] [Google Scholar]
- 51.Wada, T., T. Takagi, Y. Yamaguchi, D. Watanabe, and H. Handa. 1998. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 17:7395-7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wen, Y. D., V. Perissi, L. M. Staszewski, W. M. Yang, A. Krones, C. K. Glass, M. G. Rosenfeld, and E. Seto. 2000. The histone deacetylase-3 complex contains nuclear receptor corepressors. Proc. Natl. Acad. Sci. USA 97:7202-7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wong, J., D. Patterton, A. Imhof, D. Guschin, Y. B. Shi, and A. P. Wolffe. 1998. Distinct requirements for chromatin assembly in transcriptional repression by thyroid hormone receptor and histone deacetylase. EMBO J. 17:520-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yamaguchi, Y., T. Takagi, T. Wada, K. Yano, A. Furuya, S. Sugimoto, J. Hasegawa, and H. Handa. 1999. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97:41-51. [DOI] [PubMed] [Google Scholar]
- 55.Yang, Z., J. H. Yik, R. Chen, N. He, M. K. Jang, K. Ozato, and Q. Zhou. 2005. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell 19:535-545. [DOI] [PubMed] [Google Scholar]
- 56.Yang, Z., Q. Zhu, K. Luo, and Q. Zhou. 2001. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature 414:317-322. [DOI] [PubMed] [Google Scholar]
- 57.Yik, J. H., R. Chen, R. Nishimura, J. L. Jennings, A. J. Link, and Q. Zhou. 2003. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol. Cell 12:971-982. [DOI] [PubMed] [Google Scholar]
- 58.Yoon, H. G., D. W. Chan, Z. Q. Huang, J. Li, J. D. Fondell, J. Qin, and J. Wong. 2003. Purification and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1 and TBLR1. EMBO J. 22:1336-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yoon, H. G., D. W. Chan, A. B. Reynolds, J. Qin, and J. Wong. 2003. N-CoR mediates DNA methylation-dependent repression through a methyl CpG binding protein Kaiso. Mol. Cell 12:723-734. [DOI] [PubMed] [Google Scholar]
- 60.Zeng, L., and M. M. Zhou. 2002. Bromodomain: an acetyl-lysine binding domain. FEBS Lett. 513:124-128. [DOI] [PubMed] [Google Scholar]
- 61.Zhang, D., H. G. Yoon, and J. Wong. 2005. JMJD2A is a novel N-CoR-interacting protein and is involved in repression of the human transcription factor achaete scute-like homologue 2 (ASCL2/Hash2). Mol. Cell. Biol. 25:6404-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang, J., M. Kalkum, B. T. Chait, and R. G. Roeder. 2002. The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol. Cell 9:611-623. [DOI] [PubMed] [Google Scholar]
- 63.Zhang, K., and S. Y. Dent. 2005. Histone modifying enzymes and cancer: going beyond histones. J. Cell. Biochem. 96:1137-1148. [DOI] [PubMed] [Google Scholar]
- 64.Zhou, Q., D. Chen, E. Pierstorff, and K. Luo. 1998. Transcription elongation factor P-TEFb mediates Tat activation of HIV-1 transcription at multiple stages. EMBO J. 17:3681-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu, Y., T. Pe'ery, J. Peng, Y. Ramanathan, N. Marshall, T. Marshall, B. Amendt, M. B. Mathews, and D. H. Price. 1997. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 11:2622-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]