Abstract
Astrocytes play important roles in brain development and injury response. Transcription factors STAT3 and Smad1, activated by leukemia inhibitory factor (LIF) and bone morphogenetic protein 2 (BMP2), respectively, form a complex with the coactivator p300 to synergistically induce astrocytes from neuroepithelial cells (NECs) (K. Nakashima, M. Yanagisawa, H. Arakawa, N. Kimura, T. Hisatsune, M. Kawabata, K. Miyazono, and T. Taga, Science 284:479-482, 1999). However, the mechanisms that govern astrogliogenesis during the determination of the fate of neural stem cells remain elusive. Here we found that LIF induces expression of BMP2 via STAT3 activation and leads to the consequent activation of Smad1 to efficiently promote astrogliogenic differentiation of NECs. The BMP antagonist Noggin abrogated LIF-induced Smad1 activation and astrogliogenesis by inhibiting BMPs produced by NECs. NECs deficient in suppressor of cytokine signaling 3 (SOCS3), a negative regulator of STAT3, readily differentiated into astrocytes upon activation by LIF not only due to sustained activation of STAT3 but also because of the consequent activation of Smad1. Our study suggests a novel LIF-triggered positive regulatory loop that enhances astrogliogenesis.
Astrocytes are not only a structural component of the central nervous system (CNS) (7) but also important participants in the generation and regeneration of the CNS, for instance, in the regulation of synaptic formation, neurotransmitter transport, metabolic functions, and the response to CNS injury (7). A recent study showed that glial fibrillary acidic protein (GFAP)-positive reactive astrocytes contribute to tissue repair and motor function restoration during the subacute phase of CNS injury by forming glial scars (25). Astrocytes arise from neural stem cells that also produce neurons and oligodendrocytes. Cytokines and growth factors play critical roles in the cell fate specification of neural stem cells (31). Neuroepithelial cells (NECs) isolated from mouse embryonic telencephalons are rich in neural stem cells. Differentiation of NECs into GFAP-positive mature astrocytes is induced by interleukin 6 (IL-6) family cytokines (e.g., leukemia inhibitory factor [LIF] and ciliary neurotrophic factor) and bone morphogenetic protein (BMP) family cytokines (e.g., BMP2 and BMP4) (2, 8), which activate distinct downstream transcription factors, STAT3 and Smad1 (or Smad5 or Smad8), respectively (17, 36). A molecular basis for the cooperative action of these two cytokines is suggested to be the formation of a STAT3-Smad1 complex with a coactivator, p300, that initiates astrocyte-specific gene expression (21, 22). Significant reductions in the levels of GFAP-positive astrocytes were observed in mice deficient in gp130, a STAT3-upstream signal-transducing receptor component for IL-6 family cytokines (20), and in NECs overexpressing Noggin, a BMP antagonist that inhibits Smad1 signaling (33). These results indicate the importance of cross talk between STAT3 and Smad1 signaling pathways in astrogliogenesis, but the molecular mechanisms that govern astrogliogenesis are still largely unknown.
In an attempt to identify molecules crucial for the regulation of cytokine-mediated astrogliogenesis, we prepared NECs from embryonic day 14.5 (E14.5) mice and used DNA microarray analysis to compare the gene expression profiles under culture conditions with and without LIF and BMP2, an astrogliogenic combination of cytokines. Genes whose expression was upregulated in response to LIF plus BMP2 included glial cell lineage-related genes (STAT3, CD44, and AMOG [9, 15, 26]) and previously reported cytokine response genes in neural precursor cells (Id1, Id3, c-fos, C/EBPδ, suppressor of cytokine signaling 2 [SOCS2], and SOCS3 [4, 19, 39, 41]). Since SOCS3 is known as a negative regulator of the LIF-downstream transcription factor STAT3, we focused on SOCS3.
Members of the SOCS family of proteins are known as cytokine-inducible factors that negatively regulate the JAK-STAT pathway (14). Representatives of the SOCS family SOCS1, SOCS2, and SOCS3 have a centrally located SH2 domain and a C-terminal SOCS box as their common feature. Cytokine-induced SOCS proteins interact with their upstream receptors and inhibit the activation of relevant members of the STAT family of transcription factors, which attenuate signal transduction, thereby establishing negative feedback loops in the cytokine response. In the present study, experiments on the function of SOCS3 in NECs provided us with a clue to identify a previously unsuspected signal-modifying mechanism governing astrogliogenesis: LIF induces expression of its cooperative partner cytokine in astrocyte differentiation, i.e., BMP2, and leads to consequent Smad1 activation to efficiently form the astrogliogenic transcription factor complex.
MATERIALS AND METHODS
Animals, cell culture, and cytokines.
The Socs3 gene was conditionally deleted by Cre recombinase expressed under the control of the nestin gene promoter (18). For experiments comparing the properties of wild-type cells with those of SOCS3-deficient cells, littermates were first obtained by crossing Nes-creSocs3flox/flox mice (with a conditional knockout regulated by nestin expression) and Socs3flox/flox mice (wild type) and then genotyped according to a previous report (18). Mice were treated according to the guidelines of the Center for Animal Resources and Development, Kumamoto University. NECs were isolated from the telencephala of E14.5 mice and cultured for 4 days in N2-supplemented Dulbecco's modified Eagle medium-F-12 containing FGF2 (10 ng/ml) as described previously (21). SOCS3-deficient mouse embryonic fibroblasts (MEFs) were maintained as described previously (16). Cytokines were obtained from the following companies: FGF2 from Peprotech, LIF (ESGRO) from Invitrogen, and BMP4 and Noggin/Fc chimera from R&D Systems. BMP2 was kindly provided by Astellas Pharma Inc.
DNA microarray analysis.
The Affymetrix GeneChip system was used to identify cytokine response genes. NECs, maintained as described above, were detached and replated on 10-cm-diameter dishes (1.5 × 107 cells/dish). On the following day, cells were treated with both LIF and BMP2 (80 ng/ml each) for 6 h. NECs cultured in the absence of the cytokines were used as a control. Probes were prepared using 3.5 μg of purified poly(A)+ RNA and were hybridized with murine genome U74Av2 arrays according to the recommended protocol. After the hybridization, DNA chips were processed by the fluidics station and scanner. Affymetrix Microarray Suite 5.0 was used to compare the gene expression profiles of cytokine-treated NECs with those of untreated cells. DNA microarray data are available upon request.
Expression plasmids and RT-PCR analysis.
The cDNA for mouse SOCS3 was obtained by reverse transcription-PCR (RT-PCR) using the oligonucleotides listed below as described previously (6) and was cloned into the pcDNA3.1 expression vector (Invitrogen). Other expression plasmids are described in our previous reports (6, 21). Semiquantitative PCR was performed using KOD-plus (Toyobo) or Phusion (Finzymes) DNA polymerase. Primer sequences used for RT-PCR are as follows: SOCS3-F, 5′-CCGGAATTCATGGTCACCCACAGCAAGTT-3′; SOCS3-R, 5′-TGCTCTAGATTAAAGTGGAGCATCATACT-3′; BMP2-F, 5′-AGAGATGAGTGGGAAAACGG-3′; BMP2-R, 5′-GAAGTCCACATACAAAGGGT-3′; GAPDH-F, 5′-ACCACAGTCCATGCCATCAC-3′; GAPDH-R, 5′-TCCACCACCCTGTTGCTGTA-3′.
Luciferase assay.
NECs replated on 12-well plates were transfected using TransIT-LT1 (Mirus) as described previously (21). MEFs were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. On the day following the transfection, cells were treated with BMP2, BMP4, or LIF (50 ng/ml) for 8 h. The GFAP promoter-luciferase reporter plasmid, GF1L-pGL3, is described in our previous report (21). 4xB2wt-pGL3 and 4xB2mut-pGL3 were made by subcloning two oligonucleotide repeats (wild-type, 5′-TCGACATCCTTCAGAGAATTCTGATCCTTCAGAGAATTCTGGGTAC-3′; mutant, 5′-TCGACATCCCCAAGAGAATTCTGATCCCCAAGAGAATTCTGGGTAC-3′; mutations are underlined) in front of the junB minimal promoter with the luciferase gene as described previously (37). These oligonucleotides contain two repeats of a potential STAT3 binding site identified in the 5′ region of the mouse Bmp2 gene (boldfaced) and additional SalI and KpnI sites at each end. As an internal control, pEF-Rluc (21) or phRL-TK (Promega) was used.
Antibodies and Western blot analysis.
The following antibodies were used in this study: anti-SOCS3 (1:50; catalog no. 18391; Immuno-Biological Laboratories), anti-GFP (1:200 [catalog no. G6539; Sigma] and 1:500 [catalog no. 598; MBL]), anti-GFAP (1:500 [catalog no. G3893; Sigma] and 1:1,000 [catalog no. 031223; Advanced ImmunoChemical]), nestin (1:1,000; catalog no. 556309; BD Pharmingen), anti-phospho-STAT3 Tyr705 (1:500; catalog no. 9131; Cell Signaling), anti-STAT3 (1:1,000; catalog no. sc482; Santa Cruz), anti-phospho-ERK1/2 Thr202/Tyr204 (1:500; catalog no. 9106; Cell Signaling), anti-ERK1 (1:1,000; catalog no. sc93; Santa Cruz), anti-ERK2 (1:1,000; catalog no. sc154; Santa Cruz), anti-phospho-Smad1 Ser463/465 (1:200 for immunocytochemistry and 1:500 for Western blot analysis; catalog no. 06-702; Upstate), and anti-Smad1 (1:1,000; catalog no. sc7153; Santa Cruz). Western blot analysis was performed as described previously (6). For the repeated reprobing in Western blotting, WB stripping solution (Nacalai Tesque) was used.
Immunostaining.
For immunocytochemistry, NECs expanded in the presence of FGF2 were counted and replated on the appropriate culture dishes or chamber slides. Six hours later, cytokines (LIF, 80 ng/ml; Noggin, 200 ng/ml) were added as indicated. After the incubation, cells were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) and stained as described previously (6). For immunohistochemistry, postnatal day 1.5 (P1.5) mice were transcardially perfused with a 4% paraformaldehyde PBS solution. Isolated brains were postfixed with the paraformaldehyde solution for 3 h and submerged in PBS overnight. Tissues were then serially submerged in PBS containing 10% and 20% sucrose for 24 h and embedded in OCT compound. Frozen tissue was cut into 15-μm-thick sections using a cryostat. Nuclei were stained with Hoechst 33258 (Nacalai Tesque).
RESULTS
SOCS3-deficient NECs readily differentiate into astrocytes.
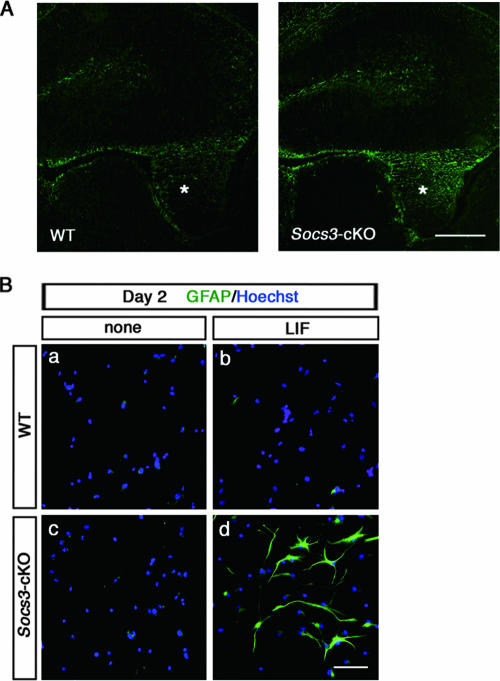
RT-PCR analysis confirmed that SOCS3 mRNA was induced in NECs by LIF but not by BMP2 (data not shown). We analyzed the properties of NECs lacking the Socs3 gene in astrogliogenesis. The complete loss of the Socs3 gene results in embryonic lethality due to a placental defect (30). Therefore, further analyses of the SOCS3 function in astrogliogenesis were done with NECs prepared from E14.5 Socs3-conditional knockout (Socs3-cKO) mice (18), in which Cre recombinase was expressed under the control of the nestin promoter to delete the loxP-flanked Socs3 locus specifically in neural precursor cells. In normal mouse development, GFAP-positive astrocytes appear in the brain and spinal cord around the time of birth. For Socs3-cKO mice on P1.5, we found that GFAP immunoreactivity was significantly denser than that for normal littermates (Fig. 1A).
FIG. 1.
SOCS3-deficient NECs readily differentiate into astrocytes upon induction by LIF. (A) Enhanced astrogliogenesis in the SOCS3-deficient neonatal mouse brain. Immunohistochemical analysis of GFAP expression was performed using cryosection of wild-type and Socs3-cKO mice at P1.5. Asterisks indicate fimbriae. Bar, 500 μm. (B) NECs derived from wild-type and Socs3-cKO mice were cultured in the absence (a and c) or presence (b and d) of LIF (80 ng/ml) for 2 days. Cells were fixed and stained with an anti-GFAP antibody (green). Nuclei were stained with Hoechst 33258 (blue). Bar, 50 μm.
NECs from E14.5 wild-type and Socs3-cKO mice were cultured for 2 days in the absence or presence of LIF and stained for GFAP. No immunoreactivity to GFAP was observed in cultured NECs without LIF, regardless of the genotype (Fig. 1Ba and c). In the presence of LIF, GFAP immunoreactivity was obvious in SOCS3-deficient cells but not in wild-type cells (Fig. 1Bb and d). Considering the negative regulatory role of SOCS3, this result seemed reasonable at first glance, but in fact its interpretation was not as simple as was first thought: in 2-day cultures of wild-type NECs, GFAP-positive astrocytic differentiation has never been observed, even with a sufficiently high concentration of LIF, when BMP2 (or a BMP ligand) was not added (21). The result shown in Fig. 1Bd resembled the data obtained with the 2-day culture of NECs treated with LIF plus BMP2 (21). It was thus presumed that BMP signaling is unexpectedly activated in the LIF-stimulated, SOCS3-deficient NECs.
SOCS3 deficiency leads to unexpected Smad1 activation following LIF stimulation.
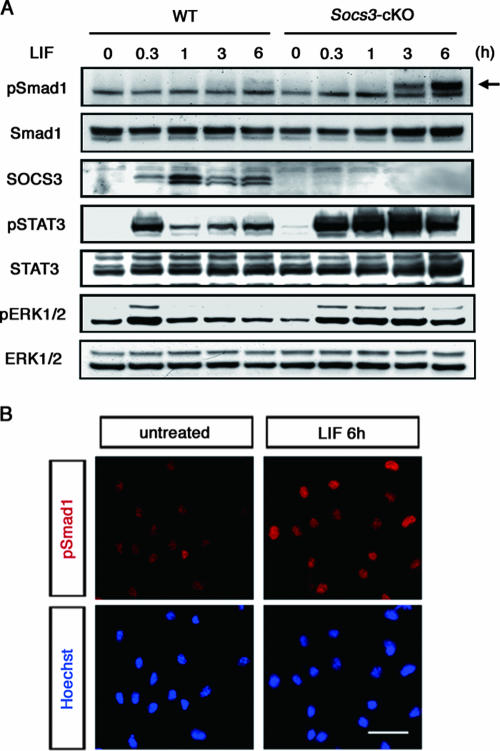
In LIF-stimulated SOCS3-deficient NECs, we observed Smad1 activation, as detected by its serine phosphorylation, with a certain lag time compared to the expected sustained activation of STAT3, Erk1, and Erk2 (Fig. 2A). Immunocytochemistry confirmed that phosphorylated Smad1 had accumulated in the nuclei of LIF-treated SOCS3-deficient cells (Fig. 2B). Direct involvement of SOCS3 in the Smad signaling pathway was excluded, because wild-type and SOCS3-deficient cells exhibited comparable responses to BMP2 and BMP4 (see Fig. S1A in the supplemental material). In addition, overexpression of SOCS3 in NECs completely suppressed GFAP promoter activation by LIF, whereas SOCS3 did not have such a suppressive effect on BMP2-induced activation (see Fig. S1B in the supplemental material). When wild-type and SOCS3-deficient NECs were treated with BMP2, phosphorylation of Smad1 was detected within 30 min at a comparable level (see Fig. S1C in the supplemental material). Taken together, these findings implied that SOCS3 deficiency did not directly change the sensitivity of the cells to BMP2. Western blot analysis showed that in SOCS3-deficient NECs, phosphorylation of Smad1 began to be detected at around 3 h, peaked at around 6 h, and lasted until at least 12 h after LIF treatment (Fig. 2A and data not shown). Since the LIF-induced phosphorylation of Smad1 occurred much more slowly than that of STAT3, Erk1, and Erk2, it was suggested that LIF has a potential to activate (presumably in an indirect fashion) the Smad signaling pathway, which in normal cells is suppressed by SOCS3. A possibility is that LIF induces the production of BMP2 (and related BMP ligands, if any), which is under negative feedback regulation involving SOCS3.
FIG. 2.
SOCS3 deficiency leads to unexpected Smad1 activation following LIF stimulation. (A) Phosphorylation of STAT3, ERK1/2, and Smad1 in response to LIF. Wild-type and SOCS3-deficient cells were treated with LIF (50 ng/ml) for the indicated periods and solubilized. Cell lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and subjected to Western blot analysis using the indicated antibodies by repeated reprobing. Arrow indicates the phosphorylated form of Smad1. (B) Nuclear localization of Smad1 after LIF treatment in SOCS3-deficient cells. Cells were either left untreated or exposed to LIF (50 ng/ml) and cultured for 6 h. The phosphorylated form of Smad1 and nuclei were detected by the anti-phospho-Smad1 antibody (red) and Hoechst 33258 staining (blue), respectively. Background levels of signals are shown in the upper left panel. Bar, 20 μm.
JAK-STAT signaling upregulates BMP2 production in NECs.
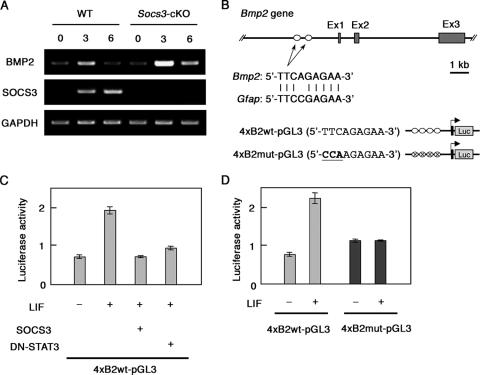
We therefore assessed mRNAs for BMP2 in NECs upon LIF stimulation. As shown in Fig. 3A, BMP2 mRNA was upregulated at 3 h after LIF stimulation, and this upregulation was dramatically enhanced in the absence of SOCS3. Two identical potential STAT3 binding sites (5′-TTCAGAGAA-3′) were found in the 5′ flanking region of the mouse Bmp2 gene (Fig. 3B). LIF treatment actually upregulated reporter expression downstream of the potential STAT3 binding sites, and overexpression of SOCS3 or a dominant-negative form of STAT3 (DN-STAT3) inhibited this LIF-induced reporter activation (Fig. 3C). Introduction of the mutation into this site (TTCAGAGAA→CCAAGAGAA) abrogated responsiveness to LIF (Fig. 4D). These results suggest the presence of a LIF-triggered positive pararegulatory loop involving STAT3-mediated induction of BMP2 expression and consequent activation of Smad1, which leads to efficient formation of an astrogliogenic nuclear complex containing STAT3, p300, and Smad1 (21).
FIG. 3.
LIF induces transcriptional activation of the Bmp2 gene. (A) The induction of BMP2 transcripts in response to LIF was analyzed by RT-PCR. NECs were treated with LIF (50 ng/ml) for the indicated periods, and total RNAs were isolated. After the RT reaction, cDNA for BMP2 was amplified by PCR. cDNA for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was analyzed as a loading control. (B) Structures of the mouse Bmp2 gene and reporter constructs used in this study. There are two identical potential STAT3 binding sites (5′-TTCAGAGAA-3′) in the 5′ region of the mouse Bmp2 gene. Details about the reporter plasmids are given in Materials and Methods. (C) SOCS3-deficient MEFs were cotransfected with 4xB2wt-pGL3 and phRL-TK together with or without the expression plasmid of SOCS3 or DN-STAT3. Data are means ± standard deviations (error bars) derived from each assay performed in triplicate. (D) SOCS3-deficient MEFs were cotransfected with 4xB2wt-pGL3 or 4xB2mut-pGL3, together with phRL-TK. A luciferase assay was performed as for panel C.
FIG. 4.
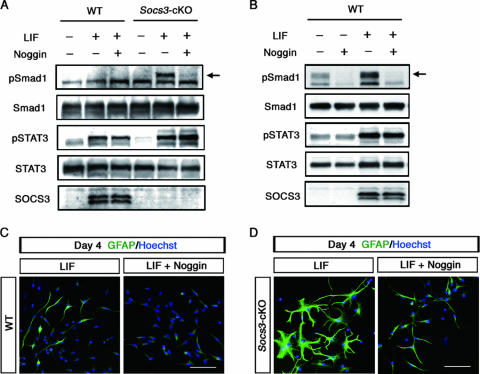
LIF induces Smad1 activation through the expression of BMP2. (A) Effect of Noggin on LIF-induced phosphorylation of Smad1. Wild-type and SOCS3-deficient cells were treated with LIF (50 ng/ml) together with or without Noggin (200 ng/ml). Cell lysates were prepared after 6 h of incubation. Western blot analysis was performed as for Fig. 2A. Arrow indicates the phosphorylated form of Smad1. (B) Phosphorylation of Smad1 by LIF after the 3-day culture. A total of 2 × 105 wild-type cells were plated on 3.5-cm-diameter dishes and treated with LIF (80 ng/ml) together with or without Noggin (200 ng/ml). (C) Noggin suppressed the LIF-induced differentiation of GFAP-positive astrocytes from wild-type NECs. NECs derived from wild-type mice were cultured with LIF (80 ng/ml) in the absence (left) or in the presence (right) of Noggin (200 ng/ml). Cells were fixed and stained with an anti-GFAP antibody (green). Nuclei were stained with Hoechst 33258 (blue). Bar, 50 μm. (D) Effects of Noggin on the morphological changes of SOCS3-deficient NECs after the 4-day culture with LIF. Noggin treatment and immunostaining were performed as for panel C.
To confirm this hypothesis, NECs were stimulated for 6 h by LIF in the presence of recombinant Noggin, which would block the effect of endogenously produced BMP2. As shown in Fig. 4A, LIF-induced phosphorylation of Smad1 in SOCS3-deficient cells was canceled by the addition of Noggin. When wild-type NECs were cultured for a much longer period (3 days) in the absence of LIF, a phosphorylated form of Smad1 was weakly detected (Fig. 4B), possibly due to endogenously produced and accumulated BMP ligands. Importantly, the amount of phosphorylated Smad1 was significantly enhanced by the addition of LIF, which was inhibited by Noggin (Fig. 4B).
We previously reported that, in 4-day NEC cultures, LIF alone or BMP2 alone could induce differentiation of GFAP-positive astrocytes, while medium alone had no such effect (6, 22, 38). Our interpretation was that this was due to accumulation of BMP ligands that had been produced spontaneously by NECs during the 4-day culture and that cooperate with exogenously added LIF (22). Actually, RT-PCR analysis showed that NECs express LIF and BMP2, though at relatively low levels, and GFAP-positive astrocytic differentiation was observed in a 16-day culture of NECs without exogenously added LIF or BMP2 (22). As shown in Fig. 4C, Noggin suppressed the differentiation of GFAP-positive astrocytes from wild-type NECs in the 4-day LIF-containing culture. Our current results provide a revised interpretation for astrocytic differentiation of NECs by LIF alone in the 4-day cultures: LIF upregulated the expression of BMP2 and cooperated with the accumulated BMP protein to induce astrocytes. SOCS3-deficient NECs after 4-day culture with LIF displayed more-intense GFAP immunoreactivity than wild-type cells, and this immunoreactivity was reduced by Noggin (Fig. 4D). Notably, the GFAP-expressing cells of these two different genotypes represented morphologically distinct cell types: wild-type cells showed an elongated bipolar morphology in most cases (Fig. 4C, left panel), while, in marked contrast, SOCS3-deficient cells displayed a widespread and highly branched stellate morphology (Fig. 4D, left panel). This morphological observation could not simply be explained by the enhanced LIF-STAT3 signaling due to SOCS3 deficiency; it reminded us of a recent report showing that BMP4 treatment of neural progenitors leads to the appearance of GFAP-expressing cells with a widespread stellate morphology having an increased number of processes, whereas LIF treatment induces a bipolar or tripolar morphology (1).
DISCUSSION
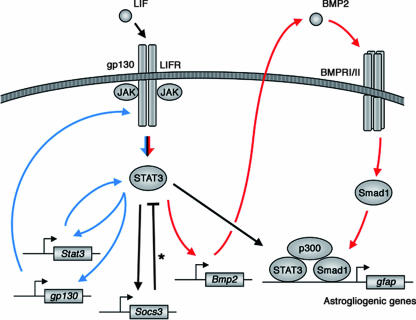
Our present study revealed a positive pararegulatory loop involving STAT3-mediated induction of BMP2 expression and subsequent Smad1 activation (Fig. 5). This newly found loop appears to function as a signal booster for astrogliogenesis triggered by IL-6 family cytokines, in the sense that STAT3 induces Smad1 activation to form the astrogliogenic STAT3-p300-Smad complex. Noggin treatment, which blocked the pararegulatory loop and abrogated astrogliogenic differentiation (Fig. 4C), indicated that this signal booster mechanism is crucial for astrogliogenesis. In addition to this loop, a recent study proposed the presence of a positive autoregulatory loop of JAK-STAT signaling that involves STAT3-mediated upregulated expression of STAT1, STAT3, and gp130 (9) (Fig. 5). Both of these positive loops triggered by IL-6 family cytokines are under the negative feedback regulation of SOCS3 (Fig. 5).
FIG. 5.
Model of LIF-triggered activation of JAK-STAT and Smad signaling pathways in astrogliogenesis. Red arrows indicate the positive pararegulatory loop shown in this study. Asterisk indicates negative feedback. Blue arrows indicate the previously reported positive autoregulatory loop of JAK-STAT signaling (9).
Although LIF and BMP2 cooperate in that they synergistically induce GFAP-positive astrocytes, they affect astrocyte morphology differently (1). The morphological difference in BMP-treated and LIF-treated neural progenitors has also been reported previously (22, 29). LIF-stimulated SOCS3-deficient cells in our current study (Fig. 4D) displayed a morphology similar to that observed for cells under BMP stimulation (1). A notable change in morphology was observed in the Noggin-containing culture; LIF-stimulated SOCS3-deficient NECs exhibited a bipolar or tripolar shape (Fig. 4D, right panel) compared to the widespread and well-branched shape without Noggin (Fig. 4D, left panel). Our present study indicated that the BMP-Smad pathway was activated, in addition to the LIF-STAT3 pathway, in astrogliogenesis. Mechanisms for such morphological changes may involve cytokine-inducible cytoskeletal rearrangements, such as BMP-induced activation of LIM kinase 1, a key regulator of actin dynamics (5).
LIF has been reported to promote astrogliogenesis and, paradoxically, to support neural stem cell renewal as well (11, 13, 27). Further molecular characterization of the pararegulatory loop clarified in this study may explain the differential effect of LIF on neural precursor cells. In nonneural cell systems, it has recently been shown that STAT3 upregulates transforming growth factor β1 expression in hepatocytes and T cells, which is downregulated by SOCS3 (12, 24). Two other groups have shown a negative effect of STAT1 and STAT3 on Smad signaling through the transcriptional upregulation of Smad7 (10, 40). In this sense, JAK-STAT signaling is supposed to have two distinct roles as a modulator of transforming growth factor β superfamily signaling pathways, depending on the cellular context.
Proteins of the SOCS family show broad spatial patterns of expression in the developing and adult mouse CNS (28). Emery et al. in 2006 reported properties of SOCS3-deficient neural precursor cells derived from E10.5 embryos by mating Socs3 heterozygous mutant mice (4). In their report, Northern blot analysis indicated that LIF-induced GFAP mRNA expression was potentiated in SOCS3-deficient cells, consistent with our present observation. However, SOCS3-deficient cells cultured in the presence of LIF grew as small aggregates (Fig. 6 in reference 4) rather than displaying the widespread and highly branched stellate morphology shown in our study (Fig. 4D). The reason for this discrepancy is not known at the moment, but different culture conditions and cell preparation procedures may have influenced the expression of LIF-downstream genes besides GFAP. It is of interest whether AMOG, which was identified as a LIF response gene in our microarray study, regulates cell-cell and cell-substrate interactions in NECs as in human glioma cells (32). In contrast to SOCS3 (3), SOCS2 had no inhibitory effect on LIF-induced astrocytic differentiation (see Fig. S2 in the supplemental material). Considering the previously reported role of SOCS2 in the neuronal cell lineage (39), SOCS3 and SOCS2 appear to have different roles in the brain depending on cell types and upstream cytokines.
In addition to the developing brain, SOCS3-upstream cytokines function in tissues and organs under inflammatory conditions. Injury in the adult spinal cord results in rapid expression of IL-6 family cytokines (35), followed by the induction of SOCS3 (23). Okada et al. recently observed escalated accumulation of reactive astrocytes in the injured spinal cords of Socs3-cKO mice compared to wild-type mice (25). It is possible that SOCS3 prevents the development and/or accumulation of reactive astrocytes at the injured site by inhibiting both the autoregulatory and the pararegulatory loop (see Fig. 5). Upregulation of BMP2 and BMP7 mRNAs was observed after spinal cord injury (33, 34); the kinetics of this upregulation were slow and biphasic (34), which may be attributed, in part, to the IL-6 family cytokine-induced pararegulatory positive loop involving BMP production found in the present study. Further investigations to elucidate the orchestration of the cytokine network would provide valuable clues not only for fuller understanding of the regulation of neural stem cells but also for the development of a therapeutic target for cytokine-induced nerve degeneration and regeneration.
Supplementary Material
Acknowledgments
We thank H. Okano (Keio University) and S. Noguchi (Meiji Milk Products) for Nestin-Cre transgenic mice, T. Kitamura (University of Tokyo) for the retroviral system, and Astellas Pharma Inc. for BMP2. We also thank I. Nobuhisa for valuable discussions; M. Teramoto, N. Oga, and C. Okamura for secretarial assistance; and Y. Saiki, K. Fujimoto, S. Iwaki, and S. Usuki for technical help.
This work was supported by 21st Century COE grant “Cell Fate Regulation Research and Education Unit,” Grant-in-Aid for Scientific Research (B), and by Grant-in-Aid for Scientific Research on Priority Areas “Molecular Brain Science” and “Self-Renewal and Pluripotency of Stem Cells” from MEXT Japan. This work was also supported in part by CREST, JST.
Footnotes
Published ahead of print on 23 April 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bonaguidi, M. A., T. McGuire, M. Hu, L. Kan, J. Samanta, and J. A. Kessler. 2005. LIF and BMP signaling generate separate and discrete types of GFAP-expressing cells. Development 132:5503-5514. [DOI] [PubMed] [Google Scholar]
- 2.Bonni, A., Y. Sun, M. Nadal-Vicens, A. Bhatt, D. A. Frank, I. Rozovsky, N. Stahl, G. D. Yancopoulos, and M. E. Greenberg. 1997. Regulation of gliogenesis in the central nervous system by the JAK-STAT signaling pathway. Science 278:477-483. [DOI] [PubMed] [Google Scholar]
- 3.Cao, F., R. Hata, P. Zhu, Y. J. Ma, J. Tanaka, Y. Hanakawa, K. Hashimoto, M. Niinobe, K. Yoshikawa, and M. Sakanaka. 2006. Overexpression of SOCS3 inhibits astrogliogenesis and promotes maintenance of neural stem cells. J. Neurochem. 98:459-470. [DOI] [PubMed] [Google Scholar]
- 4.Emery, B., T. D. Merson, C. Snell, K. M. Young, M. Ernst, and T. J. Kilpatrick. 2006. SOCS3 negatively regulates LIF signaling in neural precursor cells. Mol. Cell. Neurosci. 31:739-747. [DOI] [PubMed] [Google Scholar]
- 5.Foletta, V. C., M. A. Lim, J. Soosairajah, A. P. Kelly, E. G. Stanley, M. Shannon, W. He, S. Das, J. Massague, and O. Bernard. 2003. Direct signaling by the BMP type II receptor via the cytoskeletal regulator LIMK1. J. Cell Biol. 162:1089-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fukuda, S., T. Kondo, H. Takebayashi, and T. Taga. 2004. Negative regulatory effect of an oligodendrocytic bHLH factor OLIG2 on the astrocytic differentiation pathway. Cell Death Differ. 11:196-202. [DOI] [PubMed] [Google Scholar]
- 7.Jessen, K. R., and W. D. Richardson. 2001. Glial cell development: basic principles and clinical relevance, 2nd ed. Oxford University Press, New York, NY.
- 8.Gross, R. E., M. F. Mehler, P. C. Mabie, Z. Zang, L. Santschi, and J. A. Kessler. 1996. Bone morphogenetic proteins promote astroglial lineage commitment by mammalian subventricular zone progenitor cells. Neuron 17:595-606. [DOI] [PubMed] [Google Scholar]
- 9.He, F., W. Ge, K. Martinowich, S. Becker-Catania, V. Coskun, W. Zhu, H. Wu, D. Castro, F. Guillemot, G. Fan, J. de Vellis, and Y. E. Sun. 2005. A positive autoregulatory loop of Jak-STAT signaling controls the onset of astrogliogenesis. Nat. Neurosci. 8:616-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jenkins, B. J., D. Grail, T. Nheu, M. Najdovska, B. Wang, P. Waring, M. Inglese, R. M. McLoughlin, S. A. Jones, N. Topley, H. Baumann, L. M. Judd, A. S. Giraud, A. Boussioutas, H. J. Zhu, and M. Ernst. 2005. Hyperactivation of Stat3 in gp130 mutant mice promotes gastric hyperproliferation and desensitizes TGF-β signaling. Nat. Med. 11:845-852. [DOI] [PubMed] [Google Scholar]
- 11.Kamakura, S., K. Oishi, T. Yoshimatsu, M. Nakafuku, N. Masuyama, and Y. Gotoh. 2004. Hes binding to STAT3 mediates crosstalk between Notch and JAK-STAT signalling. Nat. Cell Biol. 6:547-554. [DOI] [PubMed] [Google Scholar]
- 12.Kinjyo, I., H. Inoue, S. Hamano, S. Fukuyama, T. Yoshimura, K. Koga, H. Takaki, K. Himeno, G. Takaesu, T. Kobayashi, and A. Yoshimura. 2006. Loss of SOCS3 in T helper cells resulted in reduced immune responses and hyperproduction of interleukin 10 and transforming growth factor-β1. J. Exp. Med. 203:1021-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koblar, S. A., A. M. Turnley, B. J. Classon, K. L. Reid, C. B. Ware, S. S. Cheema, M. Murphy, and P. F. Bartlett. 1998. Neural precursor differentiation into astrocytes requires signaling through the leukemia inhibitory factor receptor. Proc. Natl. Acad. Sci. USA 95:3178-3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kubo, M., T. Hanada, and A. Yoshimura. 2003. Suppressors of cytokine signaling and immunity. Nat. Immunol. 4:1169-1176. [DOI] [PubMed] [Google Scholar]
- 15.Liu, Y., S. S. Han, Y. Wu, T. M. Tuohy, H. Xue, J. Cai, S. A. Back, L. S. Sherman, I. Fischer, and M. S. Rao. 2004. CD44 expression identifies astrocyte-restricted precursor cells. Dev. Biol. 276:31-46. [DOI] [PubMed] [Google Scholar]
- 16.Lu, Y., S. Fukuyama, R. Yoshida, T. Kobayashi, K. Saeki, H. Shiraishi, A. Yoshimura, and G. Takaesu. 2006. Loss of SOCS3 gene expression converts STAT3 function from anti-apoptotic to pro-apoptotic. J. Biol. Chem. 281:36683-36690. [DOI] [PubMed] [Google Scholar]
- 17.Massague, J., and D. Wotton. 2000. Transcriptional control by the TGF-β/Smad signaling system. EMBO J. 19:1745-1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mori, H., R. Hanada, T. Hanada, D. Aki, R. Mashima, H. Nishinakamura, T. Torisu, K. R. Chien, H. Yasukawa, and A. Yoshimura. 2004. Socs3 deficiency in the brain elevates leptin sensitivity and confers resistance to diet-induced obesity. Nat. Med. 10:739-743. [DOI] [PubMed] [Google Scholar]
- 19.Nakashima, K., T. Takizawa, W. Ochiai, M. Yanagisawa, T. Hisatsune, M. Nakafuku, K. Miyazono, T. Kishimoto, R. Kageyama, and T. Taga. 2001. BMP2-mediated alteration in the developmental pathway of fetal mouse brain cells from neurogenesis to astrocytogenesis. Proc. Natl. Acad. Sci. USA 98:5868-5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakashima, K., S. Wiese, M. Yanagisawa, H. Arakawa, N. Kimura, T. Hisatsune, K. Yoshida, T. Kishimoto, M. Sendtner, and T. Taga. 1999. Developmental requirement of gp130 signaling in neuronal survival and astrocyte differentiation. J. Neurosci. 19:5429-5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakashima, K., M. Yanagisawa, H. Arakawa, N. Kimura, T. Hisatsune, M. Kawabata, K. Miyazono, and T. Taga. 1999. Synergistic signaling in fetal brain by STAT3-Smad1 complex bridged by p300. Science 284:479-482. [DOI] [PubMed] [Google Scholar]
- 22.Nakashima, K., M. Yanagisawa, H. Arakawa, and T. Taga. 1999. Astrocyte differentiation mediated by LIF in cooperation with BMP2. FEBS Lett. 457:43-46. [DOI] [PubMed] [Google Scholar]
- 23.Nesic, O., N. M. Svrakic, G. Y. Xu, D. McAdoo, K. N. Westlund, C. E. Hulsebosch, Z. Ye, A. Galante, P. Soteropoulos, P. Tolias, W. Young, R. P. Hart, and J. R. Perez-Polo. 2002. DNA microarray analysis of the contused spinal cord: effect of NMDA receptor inhibition. J. Neurosci. Res. 68:406-423. [DOI] [PubMed] [Google Scholar]
- 24.Ogata, H., T. Chinen, T. Yoshida, I. Kinjyo, G. Takaesu, H. Shiraishi, M. Iida, T. Kobayashi, and A. Yoshimura. 2006. Loss of SOCS3 in the liver promotes fibrosis by enhancing STAT3-mediated TGF-β1 production. Oncogene 25:2520-2530. [DOI] [PubMed] [Google Scholar]
- 25.Okada, S., M. Nakamura, H. Katoh, T. Miyao, T. Shimazaki, K. Ishii, J. Yamane, A. Yoshimura, Y. Iwamoto, Y. Toyama, and H. Okano. 2006. Conditional ablation of Stat3 or Socs3 discloses a dual role for reactive astrocytes after spinal cord injury. Nat. Med. 12:829-834. [DOI] [PubMed] [Google Scholar]
- 26.Pagliusi, S. R., M. Schachner, P. H. Seeburg, and B. D. Shivers. 1990. The adhesion molecule on glia (AMOG) is widely expressed by astrocytes in developing and adult mouse brain. Eur. J. Neurosci. 2:471-480. [DOI] [PubMed] [Google Scholar]
- 27.Pitman, M., B. Emery, M. Binder, S. Wang, H. Butzkueven, and T. J. Kilpatrick. 2004. LIF receptor signaling modulates neural stem cell renewal. Mol. Cell. Neurosci. 27:255-266. [DOI] [PubMed] [Google Scholar]
- 28.Polizzotto, M. N., P. F. Bartlett, and A. M. Turnley. 2000. Expression of “suppressor of cytokine signalling” (SOCS) genes in the developing and adult mouse nervous system. J. Comp. Neurol. 423:348-358. [PubMed] [Google Scholar]
- 29.Rajan, P., D. M. Panchision, L. F. Newell, and R. D. McKay. 2003. BMPs signal alternately through a SMAD or FRAP-STAT pathway to regulate fate choice in CNS stem cells. J. Cell Biol. 161:911-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roberts, A. W., L. Robb, S. Rakar, L. Hartley, L. Cluse, N. A. Nicola, D. Metcalf, D. J. Hilton, and W. S. Alexander. 2001. Placental defects and embryonic lethality in mice lacking suppressor of cytokine signaling 3. Proc. Natl. Acad. Sci. USA 98:9324-9329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sauvageot, C. M., and C. D. Stiles. 2002. Molecular mechanisms controlling cortical gliogenesis. Curr. Opin. Neurobiol. 12:244-249. [DOI] [PubMed] [Google Scholar]
- 32.Scheidenhelm, D. K., J. Cresswell, C. A. Haipek, T. P. Fleming, R. W. Mercer, and D. H. Gutmann. 2005. Akt-dependent cell size regulation by the adhesion molecule on glia occurs independently of phosphatidylinositol 3-kinase and Rheb signaling. Mol. Cell. Biol. 25:3151-3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Setoguchi, T., K. Nakashima, T. Takizawa, M. Yanagisawa, W. Ochiai, M. Okabe, K. Yone, S. Komiya, and T. Taga. 2004. Treatment of spinal cord injury by transplantation of fetal neural precursor cells engineered to express BMP inhibitor. Exp. Neurol. 189:33-44. [DOI] [PubMed] [Google Scholar]
- 34.Setoguchi, T., K. Yone, E. Matsuoka, H. Takenouchi, K. Nakashima, T. Sakou, S. Komiya, and S. Izumo. 2001. Traumatic injury-induced BMP7 expression in the adult rat spinal cord. Brain Res. 921:219-225. [DOI] [PubMed] [Google Scholar]
- 35.Streit, W. J., S. L. Semple-Rowland, S. D. Hurley, R. C. Miller, P. G. Popovich, and B. T. Stokes. 1998. Cytokine mRNA profiles in contused spinal cord and axotomized facial nucleus suggest a beneficial role for inflammation and gliosis. Exp. Neurol. 152:74-87. [DOI] [PubMed] [Google Scholar]
- 36.Taga, T., and T. Kishimoto. 1997. gp130 and the interleukin-6 family of cytokines. Annu. Rev. Immunol. 15:797-819. [DOI] [PubMed] [Google Scholar]
- 37.Takeda, T., K. Nakajima, H. Kojima, and T. Hirano. 1994. E1A repression of IL-6-induced gene activation by blocking the assembly of IL-6 response element binding complexes. J. Immunol. 153:4573-4582. [PubMed] [Google Scholar]
- 38.Takizawa, T., K. Nakashima, M. Namihira, W. Ochiai, A. Uemura, M. Yanagisawa, N. Fujita, M. Nakao, and T. Taga. 2001. DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev. Cell 1:749-758. [DOI] [PubMed] [Google Scholar]
- 39.Turnley, A. M., C. H. Faux, R. L. Rietze, J. R. Coonan, and P. F. Bartlett. 2002. Suppressor of cytokine signaling 2 regulates neuronal differentiation by inhibiting growth hormone signaling. Nat. Neurosci. 5:1155-1162. [DOI] [PubMed] [Google Scholar]
- 40.Ulloa, L., J. Doody, and J. Massague. 1999. Inhibition of transforming growth factor-beta/SMAD signalling by the interferon-gamma/STAT pathway. Nature 397:710-713. [DOI] [PubMed] [Google Scholar]
- 41.Wright, L. S., J. Li, M. A. Caldwell, K. Wallace, J. A. Johnson, and C. N. Svendsen. 2003. Gene expression in human neural stem cells: effects of leukemia inhibitory factor. J. Neurochem. 86:179-195. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.