Abstract
Within the human genome there are hundreds of copies of the rRNA gene, but only a fraction of these genes are active. Silencing through epigenetics has been extensively studied; however, it is essential to understand how active rRNA genes are maintained. Here, we propose a role for the methyl-CpG binding domain protein MBD3 in epigenetically maintaining active rRNA promoters. We show that MBD3 is localized to the nucleolus, colocalizes with upstream binding factor, and binds to unmethylated rRNA promoters. Knockdown of MBD3 by small interfering RNA results in increased methylation of the rRNA promoter coupled with a decrease in RNA polymerase I binding and pre-rRNA transcription. Conversely, overexpression of MBD3 results in decreased methylation of the rRNA promoter. Additionally, overexpression of MBD3 induces demethylation of nonreplicating plasmids containing the rRNA promoter. We demonstrate that this demethylation occurs following the overexpression of MBD3 and its increased interaction with the methylated rRNA promoter. This is the first demonstration that MBD3 is involved in inducing and maintaining the demethylated state of a specific promoter.
The cell's ability to properly regulate the levels of rRNA transcription is key for normal cell function. The cell maintains tight control over the transcriptional machinery at many different levels to ensure that the levels of rRNA transcription match the cell's needs for protein production. Within the human genome there are over 400 copies of the rRNA gene, and a fraction of these genes are epigenetically silenced. Therefore, an increase in rRNA transcription may occur either through increased transcription from already active copies of the rRNA gene or through epigenetic activation of an increased number of rRNA genes. The transcriptional activity of rRNA genes varies with cell growth, and in exponentially growing cells the need for ribosome production is high. Accordingly, it is likely that for sustained increases in transcriptional, and downstream translational, capacity the cell responds by increasing the number of active copies of the rRNA gene, as is seen in cancer cells.
Several studies have established a mechanism through which rRNA transcription is epigenetically silenced. The transcription termination factor 1 (Ttf-1) recruits the NoRC complex, a transcriptional repressor complex consisting of Swi/Snf proteins, histone deacetylases, and Ttf-1-interacting protein 5. The binding of NoRC induces deacetylation of histone 4 and dimethylation of histone 3 lysine 9 (43). This then leads to DNA methylation of the promoter, which inhibits the binding of the transcription factor upstream binding factor (UBF) (44). Studies regarding epigenetic activation showed by using the histone deacetylase inhibitor trichostatin A that acetylation of histone 4 results in activation of rRNA genes (25), but the DNA methylation pattern was not examined. The mechanism by which rRNA promoters become demethylated has yet to be elucidated.
Hypomethylation of repetitive sequences is a hallmark of cancerous cells (11); therefore, it is likely that the rRNA repetitive sequences become demethylated in cancer. Indeed, although the transcribed region of the rRNA gene becomes hypermethylated in breast cancer (50), the rRNA promoter was shown to be hypomethylated in tumors (17), consistent with an increase in rRNA transcriptional activity characteristic of cancer cells. Cancer cells typically overexpress the methyltransferase DNMT1 (13), indicating there is no lack of methyltransferase activity in these cells, which raises the question of how demethylated promoters may coexist with hypermethylated transcribed regions? An interesting possibility is the involvement of specific proteins that protect the promoters from methylation or target them for demethylation. Identification of these proteins is critical for understanding how the epigenetic state of rRNA genes is programmed in normal and cancer cells.
Methyl-CpG binding domain (MBD) proteins target methylated DNA. This family of proteins, characterized by the MBD, includes MBD1 to -4 and MeCp2. MBD2 and MeCp2 bind to methylated DNA and recruit transcriptional repressors and histone-modifying enzymes, such as histone deacetylases (HDACs) and histone methyltransferases, resulting in silencing of genes. However, MBD2 has also been characterized as a DNA demethylase by our lab (1, 9, 10), although this finding has been contested by other groups (3, 53). More recently, MBD2 was demonstrated to be involved in activation of specific unmethylated and methylated promoters (12, 15, 18).
MBD3, another member of this family of proteins, has been shown to associate with HDAC1 and to be a component of the transcriptional repressor complex NuRD (41). However, MBD3 does not exhibit selective binding to methylated DNA in vitro, and this difference is probably due to two amino acid sequence differences within the MBD (35, 48). MBD3 was shown to bind to the rRNA promoter (17), and we hypothesized that MBD3 binding is important in the regulation of rRNA genes through DNA methylation. We used a combination of chromatin immunoprecipitation (ChIP) and bisulfite mapping to examine the methylation status of each individual CG within the rRNA promoter bound to MBD3 in living cells. Surprisingly, we found that the population of rRNA promoters bound to MBD3 was less methylated than the unbound fraction. Additionally, we tested whether MBD3 plays a causal role in determining the DNA methylation pattern of both the endogenous rRNA promoter as well as an exogenous transiently transfected rRNA promoter through small interfering RNA (siRNA) knockdown and overexpression. Our data are consistent with the hypothesis that MBD3 plays an important role in maintaining the rRNA promoters in an unmethylated state.
MATERIALS AND METHODS
Cell culture and transfections.
HeLa and HEK 293 cells were maintained as a monolayer in Dulbecco's modified Eagle's medium (Life Technologies, Inc.) containing 10% fetal calf serum (Colorado Serum Co.). HeLa cells were plated at a density of 4 × 105/10-cm tissue culture dish and transiently transfected with 20 nM siRNA (SMARTpool; Dharmacon) using Lipofectamine 2000 (Invitrogen), following the manufacturer's protocol. The cells were replated 24 h later to the same density, and the procedure was repeated three more times over a period of 7 days. HEK 293 cells were plated at a density of 5 × 105/10-cm tissue culture dish and transiently transfected with 5 μg of plasmid DNA using the calcium phosphate precipitation method as described previously (40).
Immunocytochemistry.
HeLa cells were plated at a density of 3 × 104/24-well tissue culture dish on glass coverslips. Cells were fixed for 30 min in 4% paraformaldehyde and blocked for 1 h in 10% serum plus phosphate-buffered saline (PBS)-Tween. Primary antibodies were diluted in 5% serum plus PBS (anti-MBD3, 1:50, Santa Cruz; anti-UBF, 1:50, Santa Cruz; anti-Mi-2, 1:500 [47]; anti-HDAC1, 1:50, Santa Cruz; anti-HDAC2, 1:100, Santa Cruz; anti-Sin3A, 1:400, Santa Cruz; anti-Aurora A, 1:100, BD Biosciences) and incubated for 2 h. Secondary antibodies were diluted (1:400) in 5% serum plus PBS (Molecular Probes) and incubated for 1 h. Visualization was performed using a Zeiss Axioplan 2 imaging fluorescence microscope equipped with a high-resolution color digital camera and connected to a computer with the Zeiss Axiovision 4.1 software (Zeiss Canada).
Bisulfite mapping.
Bisulfite mapping was performed as described previously with minor modifications (6). Five micrograms of sodium bisulfite-treated DNA samples was subjected to PCR amplification using the primers listed in Table 1. The PCR products were either sequenced directly (33, 34) or were subsequently cloned using the Invitrogen TA cloning kit (following the manufacturer's protocol), and the clones were sequenced using the T7 sequencing kit (Amersham Pharmacia Biotech, following the manufacturer's protocol C). The primer sequences are listed in Table 1. The primers for bisulfite mapping do not contain any CG dinucleotide sequences, thereby ensuring that methylated and unmethylated sequences are amplified with equal efficiency. Additionally, to ensure the bisulfite reaction was complete, we only included clones in which 95% of the cytosine residues in non-CpG dinucleotide sequences had been converted to thymidine. Any clones which contained less than 95% bisulfite reaction efficiency were not included in the analysis.
TABLE 1.
Primer sequences
| Primer group and gene | Primer direction | Primer sequence (5′-3′) | Annealing temp (°C) |
|---|---|---|---|
| Bisulfite mapping primers | |||
| rRNA promoter | S | GTTTTTGGGTTGATTAGA | 51 |
| AS | AAAACCCAACCTCTCC | ||
| Luciferase | S | GGAGAGTAATTGTATAAGGTT | 56 |
| AS | AATATTCATACTATTAAACAATTC | ||
| ChIP primers | |||
| rRNA promoter | S | GAGAGAACAGCAGGCCCG | 60 |
| AS | CCTTCGGTCCCTCGTCTC | ||
| rRNA promoter (quantitative PCR) | S | TCCTTGGGTTGACCAGAGGGAC | 60 |
| AS | AGAGGACAGCGTGTCAGCATAAAC | ||
| Luciferase | S | AGAGATACGCCCTGGTTCC | 54 |
| AS | CCAACACCGGCATAAAGAA | ||
| RT-PCR primers | |||
| MBD3 | S | ACGGGCAAGATGCTGATGAGC | 60 |
| AS | CAGCAATGTCGAAGGCGTTCA | ||
| MBD2 | S | CTGGCAAGAGCGATGTC | 53 |
| AS | AGTCTGGTTTACCCTTATTTTG | ||
| β-Actin | S | GTTGCTAGCCAGGCTGTGCT | 60 |
| AS | CGGATGTCCACGTCACACTT | ||
| Luciferase | S | AGAGATACGCCCTGGTTCC | 54 |
| AS | CCAACACCGGCATAAAGAA | ||
| Pre-rRNA | S | TCAGATCGCTAGAGAAGG | 55 |
| AS | AGTGAGACGAGACGAGAC | ||
| GAPDH | S | TGCACCACCAACTGCTTA | 60 |
| AS | GGATGCAGGGATGATGTTC |
aS, sense; AS, antisense.
Western blot analysis.
Total cell extracts were prepared using standard protocols and resolved on a 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. After transferring to nitrocellulose membrane and blocking the nonspecific binding with 0.5% bovine serum albumin, MBD3 protein was detected using an antibody specific for MBD3 (Santa Cruz) followed by anti-goat immunoglobulin G (IgG; Santa Cruz) and an enhanced chemiluminescence detection kit (Amersham Pharmacia Biotech). Actin was detected using an antibody specific for β-actin (Sigma) followed by anti-mouse IgG and use of the enhanced chemiluminescence detection kit.
Chromatin immunoprecipitation.
ChIP assays (7) were performed as described previously (5), using an anti-MBD3 antibody (Santa Cruz), anti-RPA-116 antibody (45), anti-UBF antibody (Santa Cruz), or normal rabbit IgG antibody (Santa Cruz). DNA was purified from both the immunoprecipitated and preimmune (input) samples and was subjected to PCR amplification. In all experiments, 1/10 of the input sample was used for PCR amplification. Samples were taken every 2 cycles from 22 to 32 cycles to ensure the amplification was in the linear range. For amplification of the rRNA promoter, quantitative real-time PCR mixtures (20 μl) containing the immunoprecipitated DNA, FailSafe GREEN real-time PCR capillary enzyme, buffer C8, and 0.4 μM primer were loaded into LightCycler capillaries (Roche Molecular Biochemicals). For amplification of the luciferase sequence, quantitative real-time PCR mixtures (25 μl) containing the reverse-transcribed cDNA, SuperArray enzyme mix, and 0.4 μM primer were loaded into LightCycler capillaries (Roche Molecular Biochemicals). Samples were run in the LightCycler 3.5 (Roche Molecular Biochemicals). To determine the CT ratio, a five-point calibration curve of increasing amounts of DNA (0.5, 1, 2.5, 5, and 7.5 μl) as well as a no-template negative control were performed by using separate tubes for each reaction. The primer sequences are listed in Table 1. The double ChIP experiments were performed as above; however, between the two immunoprecipitations the beads were incubated two times with 20 mM dithiothreitol for 30 min at 37°C in order to release the sample from the beads. The elutions were combined and diluted to 1.5 ml with ChIP dilution buffer before adding the second antibody (10 μg) for immunoprecipitation overnight at 4°C.
RT-PCR.
Reverse transcription (RT) was performed as described previously (31), with one modification. Following extraction, the RNA was subjected to DNase I (Roche) treatment to ensure complete elimination of the DNA from the extract. PCR amplification from RNA in the absence of RT was performed to verify complete elimination of the DNA from the RNA sample. Samples were taken every 2 cycles from 16 to 25 cycles to ensure the amplification was in the linear range. Quantitative real-time RT-PCR mixtures (25 μl) containing the reverse-transcribed cDNA, SuperArray enzyme mix, and 0.4 μM primer were loaded into LightCycler capillaries (Roche Molecular Biochemicals) and run in the LightCycler 3.5 (Roche Molecular Biochemicals). To determine the CT ratio, a five-point calibration curve of increasing amounts of DNA (0.5, 1, 2.5, 5, and 7.5 μl) as well as a no-template negative control were performed by using separate tubes for each reaction. The primer sequences are listed in Table 1.
[3H]leucine incorporation assay.
Cells were plated in a six-well plate (8 × 104/well). For the final 6 h of incubation, 1 μCi/ml [3H]leucine (Perkin-Elmer Life Sciences) was added to the medium. After five washes with ice-cold PBS, the cells were incubated in 10% trichloroacetic acid for 2 h at 4°C, washed for 30 min twice with cold 5% trichloroacetic acid, and then lysed with 0.1 N NaOH and 0.1% sodium dodecyl sulfate. [3H]leucine incorporation was measured using a liquid scintillation counter (LKB Wallac).
In vitro methylation of substrates.
rRNA-pGL3 plasmid was methylated in vitro by incubating 10 μg of plasmid DNA with 20 U of SssI CpG DNA methyltransferase (New England BioLabs, Inc.) in a buffer recommended by the manufacturer containing 160 μM S-adenosylmethionine, at 37°C for 3 h. After repeating this procedure twice, full protection from HpaII digestion was observed.
Actinomycin D treatment.
At 24 h after transfection with 5 μg rRNA-pGL3 or CMV-pGL3 plasmids (as described above), HEK 293 cells were treated with 5 ng/ml of actinomycin D and cells were harvested at 0, 2, 4, or 8 h.
Southern blot analysis.
DNA was first digested with 50 U of EcoRI followed by digestion with 20 U of either HpaII or MspI restriction enzyme (see Fig. S2A in the supplemental material) or DNA was digested with 50 U of DpnI restriction enzyme (see Fig. 7C, below). Southern blot analysis was performed exactly as described previously (8).
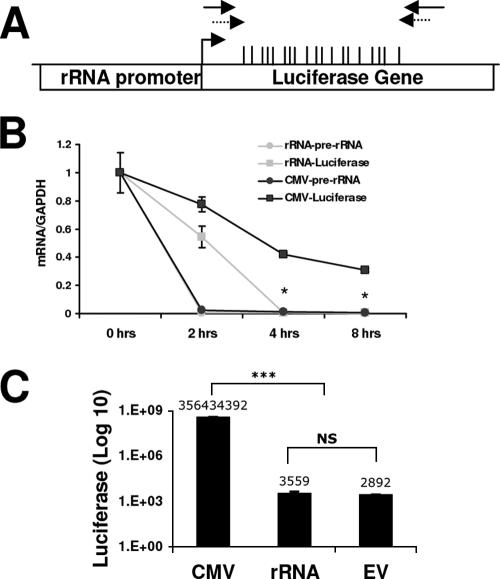
FIG. 7.
rRNA-pGL3 transcription is driven by Pol I. (A) Physical map of the rRNA-pGL3 plasmid containing the rRNA promoter and the luciferase gene. The positions of the CGs within the area examined are indicated by the vertical bars. The primers used for ChIP assay and bisulfite mapping are indicated with solid and dashed arrows, respectively. (B) HEK 293 cells were transfected with CMV-pGL3 or rRNA-pGL3 for 32 h. Prior to harvesting, the cells were treated with 5 μg/ml actinomycin D for 0, 2, 4, or 8 h. RNA was extracted and treated with DNase. Results shown are from quantitative RT-PCR amplification of luciferase (squares) and pre-rRNA (circles) sequences normalized to GAPDH mRNA levels. Student's t test was used to analyze rRNA-pGL3 versus CMV-pGL3 luciferase mRNA amplification (*, P < 0.05). (C) HEK 293 cells were transfected with CMV-pGL3, rRNA-pGL3, or pGL3 (promoterless, empty vector [EV]) for 48 h. Values represent luciferase activities relative to total protein levels and are representative of three independent experiments (mean ± standard error of the mean). Student's t test was used for analysis of CMV-pGL3 versus rRNA-pGL3 or pGL3 (***, P < 0.001). NS, nonsignificant change between the rRNA-pGL3 and pGL3 luciferase values.
Luciferase assay.
HEK 293 cells were plated at a density of 6 × 104 in six-well dishes and transiently transfected with 0.5 μg rRNA-pGL3 or CMV-pGL3 or pGL3 plasmids using the calcium phosphate method as described previously (40). The cells were harvested 72 h after transfection and lysed, and luciferase activity was assayed using the luciferase assay system (Promega) according to the manufacturer's protocol.
RESULTS
The active rRNA promoter is unmethylated in HeLa cells.
Previous studies have shown that methylation of the CpG at −133 of the mouse rRNA promoter prevents the binding of the transcription factor UBF (44). However, the rRNA promoter is highly divergent between species, with very little sequence similarity between the mice and humans. Therefore, we first tested whether DNA methylation correlates with the state of activity of the rRNA promoter in human cells. The diagram in Fig. 1A represents the human rRNA promoter and the area amplified for bisulfite mapping and ChIP analysis. The rRNA promoter consists of two key elements: the upstream control element (UCE) and the core promoter (CP) (21) to which the transcription factor UBF binds (29). The areas amplified for bisulfite mapping and ChIP analysis are shown (Fig. 1). The primers used for PCR amplification were designed using the human rRNA repeat (accession number U13369) and detected the repeat sequence located on chromosomes 13, 14, 15, 21, and 22 in the human genome (22).
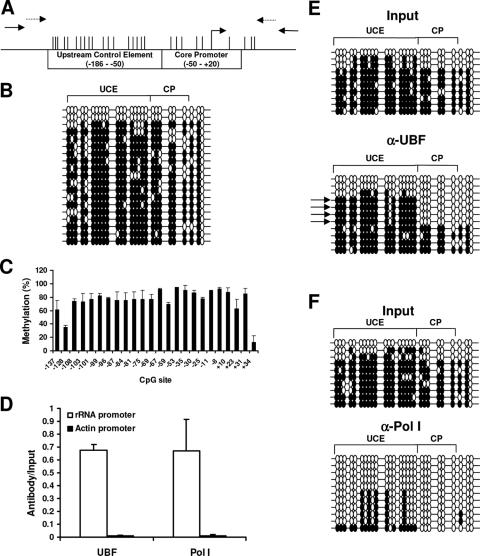
FIG. 1.
The rRNA promoter in HeLa cells is highly methylated; however, the active rRNA promoters bound to Pol I and UBF are unmethylated. (A) Physical map of the rRNA promoter, with positions of the CGs indicated by vertical bars. The primers used for the ChIP assay and bisulfite mapping are indicated with solid and dashed arrows, respectively. (B) Bisulfite analysis of the rRNA promoter from DNA extracted from HeLa cells. Each line represents an independent clone. A filled circle represents a methylated CG dinucleotide, and an empty circle represents an unmethylated CG dinucleotide. (C) Semiquantitative analysis of the C:T peaks of CG dinucleotides as determined through sequencing of the sodium bisulfite PCR product. (D) Chromatin immunoprecipitation analysis of the association between UBF or Pol I binding to the rRNA promoter in HeLa cells. To control for specificity of the antibodies used, the β-ACTIN promoter was amplified for all samples. The graph represents results from quantitative real-time PCR analysis of the rRNA promoter and the β-ACTIN promoter from anti-UBF or anti-Pol I immunoprecipitates normalized to the input samples from three independent experiments. One-tenth of the input sample was used for PCR amplification. (E and F) Bisulfite analysis of rRNA promoters from input samples or from samples immunoprecipitated with anti-UBF (α-UBF) antibody (E) or anti-Pol I antibody (F). The arrows point out the predominant DNA methylation pattern bound to UBF. The results are representative of three independent experiments.
Bisulfite mapping of the rRNA promoter from HeLa cells showed that the majority of the clones were highly methylated and only a fraction of the clones were unmethylated (Fig. 1B), in accordance with a previous report that found that the rRNA promoter is hypermethylated in tissue culture cells compared to primary culture cells (28). This was quite a surprising report, nonetheless, as we expected this promoter, due to its high activity, to be unmethylated. Since the rRNA promoter is present at a high copy number within the cell, it is possible that the 20 clones selected were not representative of the entire population. To try to determine an overall level of DNA methylation of the rRNA promoter, rather than sequencing individual clones we automatically sequenced the PCR product directly and quantified the C versus T peaks at the CG dinucleotides in the entire population (33, 34). These results showed that on average the CG sites in the rRNA promoters are 70 to 80% methylated (Fig. 1C). These data are consistent with the data obtained through bisulfite mapping of individual clones, where 74% of the total number of CGs were methylated, indicating that the rRNA promoter in HeLa cells is highly methylated and that only a fraction of the molecules are unmethylated at a given time.
Since only a fraction of the rRNA genes are active in the cell, we tested the hypothesis that the active copies of the rRNA promoter are unmethylated. To isolate the active promoters, we used ChIP assays with antibodies against the transcription factor UBF or RNA polymerase I (Pol I), the polymerase dedicated to rRNA transcription (Fig. 1D), and the immunoprecipitated DNA was subsequently treated with sodium bisulfite. Additionally, primers specific for the ubiquitously expressed β-ACTIN promoter, which was not predicted to be bound by either UBF or Pol I, were used as negative controls to test the specificity of the antibodies. No binding was found to the β-ACTIN promoter by either anti-UBF or anti-RPA-116, supporting the specificity of the immunoprecipitation (Fig. 1D). The promoters bound to UBF had a distinct pattern of methylation in the UCE, whereas the CP and downstream of the transcription start site were mostly unmethylated (Fig. 1E). The promoters that bound to Pol I, which we presume to be the active copies of the rRNA gene, were mostly unmethylated in both the UCE and the CP (Fig. 1F). Therefore, DNA methylation provides a distinguishing covalent modification of the DNA molecule which differentiates between the active and inactive rRNA promoters. Thus, the DNA methylation of rRNA promoters could be used to define the fraction of active rRNA genes in a given cell. Interestingly, although the CP of rRNA promoters bound to UBF was unmethylated, similar to the promoters bound to Pol I, the UCE was heavily methylated, unlike the promoters bound to Pol I, indicating that UBF may not bind exclusively to fully active promoters but to a distinct intermediary population of rRNA promoters which is less methylated than inactive promoters but more methylated than active promoters. It has been shown that UBF binds throughout the cell cycle, including during mitosis when rRNA transcription is shut down (16), as well as when transcription has been inhibited through actinomycin D treatment or starvation (52), suggesting that UBF binds to both the active and the inactive promoters. The fact that the cell maintains a portion of the rRNA promoters unmethylated while the rest of the copies are methylated raises the question of how these two populations are established and maintained.
MBD3 binds to unmethylated rRNA promoters.
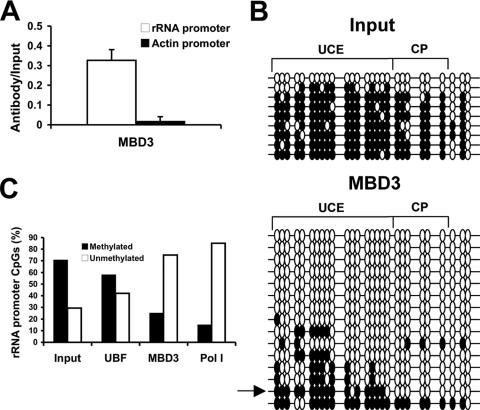
In preliminary immunocytochemistry experiments, we found that MBD3 was the only known methylated DNA binding protein preferentially localized to the nucleolus, indicating that MBD3 may possess a specific nucleolar function. This is consistent with a previous report which showed nucleolar localization of MBD3 (17). We therefore examined whether MBD3 plays a role in regulating the state of activity of the rRNA promoter. ChIP assay results confirmed the association of MBD3 with the rRNA promoter (Fig. 2A). To control for the specificity of the ChIP assays with MBD3, we determined whether the ubiquitously active β-ACTIN promoter was amplified from the MBD3 ChIP. No signal was obtained for the β-ACTIN promoter, supporting the specificity of the immunoprecipitation with the anti-MBD3 antibody (Fig. 2A). MBD3 has been shown to be a part of a transcriptional repressor complex, NuRD, and therefore we hypothesized that it may be binding to methylated rRNA promoters to repress transcription and protect from demethylation. Although reports have shown that MBD3 does not bind to methylated DNA in vitro (41), it has been postulated that it is directed to methylated DNA in living cells through MBD2, which associates with the NuRD complex (53). To determine the methylation status of the rRNA promoters bound to MBD3, the DNA immunoprecipitated with the anti-MBD3 antibody was treated with sodium bisulfite. Surprisingly, we found that MBD3 binds preferentially to hypomethylated rRNA promoters (Fig. 2B). Whereas greater than 70% of CGs in the unbound fraction are methylated, 75% of the MBD3-bound fraction is unmethylated (Fig. 2C). A previous report, however, indicated that MBD3 is bound to methylated copies of the rRNA promoter (17). The discrepancy between the two results may be attributed to the use of a less sensitive technique in the previous report involving HpaII digestion analysis, which only examines CG sites found in CCGG sequences (17), whereas bisulfite mapping examines the methylation status of each individual CG within the sequence amplified by PCR. There are two possible explanations for the preferential interaction of MBD3 with hypomethylated rRNA promoters. First, MBD3 may be acting as a transcriptional repressor, binding to unmethylated promoters to prevent their spurious activation. Alternatively, MBD3 may be binding to the active, hypomethylated rRNA promoters in order to maintain them in an unmethylated state, protecting them from aberrant methylation.
FIG. 2.
MBD3 binds to unmethylated rRNA promoters. (A) Chromatin immunoprecipitation analysis of the association between MBD3 binding to the rRNA promoter sequence in HeLa cells. To control for specificity of the antibody used, the β-ACTIN promoter was amplified for all samples. The graph represents results from quantitative real-time PCR amplification of the rRNA promoter or the β-ACTIN promoter amplified from the anti-MBD3 immunoprecipitates normalized to input samples from three independent experiments. One-tenth of the input sample was used for PCR amplification. (B) Bisulfite analysis of rRNA promoters from input samples or samples immunoprecipitated with the anti-MBD3 antibody. The arrow points out the promoter bound to MBD3 with a similar DNA methylation pattern as was found bound to UBF in Fig. 1E. The results are representative of three independent experiments. (C) Quantification of the individual methylated versus unmethylated CGs in the rRNA promoter from the nonimmunoprecipitated samples (input), combined from all three input samples, and the samples immunoprecipitated with antibodies directed against UBF, Pol I, or MBD3.
MBD3 colocalizes with UBF in the nucleolus.
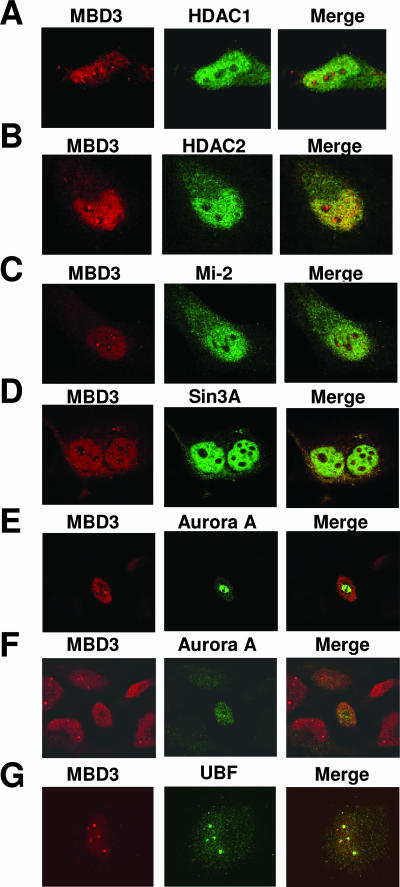
In order to elucidate the function of MBD3 in the nucleolus, we used immunocytochemistry to colocalize MBD3 with other transcriptional repressors or activators in the nucleolus. Since MBD3 has been characterized as a member of the NuRD complex, comprising nucleosomal remodelling enzymes and histone deacetylases, including HDAC1 (53), immunocytochemistry was used to determine whether MBD3 and HDAC1 colocalize in the nucleolus and work together to repress rRNA transcription. Surprisingly, although MBD3 and HDAC1 do colocalize in some areas of the nucleus as previously reported, there is no colocalization in the nucleolus (Fig. 3A).
FIG. 3.
MBD3 colocalizes with UBF and Pol I in the nucleolus. HeLa cells were grown on coverslips and stained with anti-MBD3 antibody (left panel, red). The same cells were also stained with anti-HDAC1 (A), anti-HDAC2 (B), anti-Mi-2 (C), anti-Sin3A (D), anti-Aurora A (E and F, the same field with different focus levels), or anti-UBF (G) antibody (middle panel, green). Right panel, confocal merge of MBD3 with the other proteins examined.
We further confirmed that MBD3 is not associated with the silencing complex NuRD in the nucleolus by using antibodies for other components of the NuRD complex, HDAC2 (49) and Mi-2 (20). Similar to the results with HDAC1, we found that although MBD3 colocalizes with HDAC2 and Mi-2 in different positions in the nucleus, supporting previous reports in which it was found that MBD3, HDAC1/2, and Mi-2 form a complex (30, 41, 53), there was no colocalization detected in the nucleolus (Fig. 3B and C, respectively). It has been previously reported through chromatin immunoprecipitation assays, however, that HDAC1 does bind to the rRNA promoter (54); therefore, a complete lack of these proteins in the nucleolus is unlikely. Additionally, we examined the possibility that MBD3 may be localized with the transcriptional repressor Sin3A, but again we found there was no colocalization within the nucleolus between MBD3 and Sin3A (Fig. 3D). It is possible that transcriptional repressors reside on the periphery of the nucleolus or that the concentration of these proteins within the nucleolus is very low and thus undetectable using immunocytochemistry.
A previous report had indicated that MBD3 localizes with the kinase Aurora A at the centrioles during mitosis (42). In order to examine the possibility that the foci of MBD3 observed in the immunocytochemistry experiment represents centrioles rather than the nucleolus, we performed immunocytochemistry with an antibody against Aurora A, together with an anti-MBD3 antibody. Similar to earlier reports which used exogenously expressed proteins (42), we found colocalization between Aurora A and MBD3 in mitotic cells (Fig. 3E). However, examination of nonmitotic cells within the same field revealed that although Aurora A was not visualized in these interphase cells, MBD3 still localized to the nucleolus (Fig. 3F), suggesting that most of the MBD3 foci detected in our immunochemistry experiment were not colocalized with Aurora in the centrioles.
To obtain further confirmation that MBD3 is localized to the nucleolus, we used antibodies against MBD3 and the nucleolar protein UBF and found that MBD3 colocalizes with this transcriptional activator in the nucleolus (Fig. 3G). This localization to the nucleolus is specific, as control experiments using a blocking peptide against the MBD3 antibody, or using no primary antibody, demonstrated a lack of nuclear and nucleolar signal (see Fig. S1A and B, respectively, in the supplemental material). The fact that MBD3 binds to unmethylated rRNA promoters and that it is not colocalized with the known members of the NuRD repressor complex, HDAC1/2 and Mi-2, in the nucleolus is inconsistent with the hypothesis that MBD3 acts as a transcriptional repressor of rRNA genes. Rather, they suggest that MBD3 may be involved in activating rRNA genes, binding to the unmethylated rRNA promoters to protect them from methylation to allow for Pol I binding and transcription.
MBD3 and UBF bind to the same rRNA promoters.
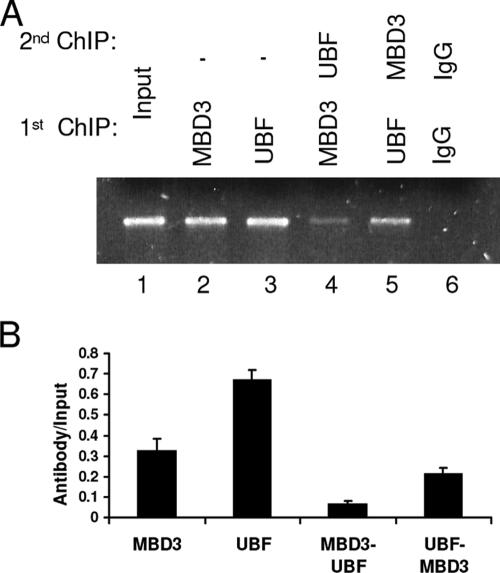
In order to further elucidate the role of MBD3 in activating transcription at the rRNA promoter, we used a double ChIP assay to determine whether MBD3 and UBF are binding to the same rRNA promoter molecules (Fig. 4). We first immunoprecipitated the chromatin with an anti-UBF antibody, followed by immunoprecipitation with an anti-MBD3 antibody (lane 5). These results were confirmed through quantitative real-time PCR (Fig. 4B), using the signal from the first immunoprecipitation as the input for the second immunoprecipitation, and demonstrated that a majority of the rRNA promoter molecules which are bound by UBF are also bound by MBD3. However, when the order of antibodies was reversed, using an anti-MBD3 antibody for the first immunoprecipitation followed by an anti-UBF antibody for the second (lane 4), the binding in the double ChIP was decreased relative to the single ChIPs with either MBD3 or UBF, suggesting that a fraction of the rRNA promoters occupied by MBD3 also binds UBF at the same time. These results are in accordance with the bisulfite mapping data from the immunoprecipitation studies of both UBF (Fig. 1E) and MBD3 (Fig. 2B). While both MBD3 and UBF bind to fully unmethylated rRNA promoters, the predominant methylation pattern of the rRNA promoter bound to UBF, with a highly methylated UCE and highly unmethylated CP, was also found within the population of rRNA promoters bound to MBD3 (Fig. 1E and 2B). Therefore, the bisulfite mapping analysis strongly supports the double ChIP analysis where MBD3 and UBF bind to the same rRNA promoter molecules.
FIG. 4.
MBD3 and UBF bind to the same rRNA promoters. Shown are the results of a double chromatin immunoprecipitation assay of the association with MBD3 and UBF to the same rRNA promoter molecule. The single ChIP samples were the input for the double ChIP samples. One-tenth of the input sample was used for PCR amplification. (A) Immunoprecipitated HEK 293 extracts. Lanes: 1, nonimmunoprecipitated input; 2, single immunoprecipitation with anti-MBD3; 3, single immunoprecipitation with anti-UBF; 4, double immunoprecipitation with anti-MBD3 followed by anti-UBF; 5, double immunoprecipitation with anti-UBF followed by anti-MBD3; 6, double immunoprecipitation with nonimmune IgG. The results are representative of triplicate PCRs from two independent experiments. (B) Quantitative real-time PCR amplification of the rRNA sequence amplified from immunoprecipitated samples normalized to the input samples.
siRNA knockdown of MBD3 results in rRNA promoter hypermethylation, decreased Pol I binding to the rRNA promoter, and decreased pre-rRNA transcription.
The biological role of MBD3 at the rRNA promoter may be either as a transcriptional repressor as previously suggested or as a transcriptional activator as implied by its colocalization with UBF (Fig. 3G) and its binding to the same rRNA promoter molecules as UBF (Fig. 4). In order to test this, we knocked down MBD3 in HeLa cells using a pool of four siRNAs directed against MBD3 (siMBD3). The knockdown was confirmed through both quantitative RT-PCR and Western blot analyses (Fig. 5A and B, respectively). Quantitative RT-PCR of MBD3 (Fig. 5A, left panel) showed a time-dependent decrease in the levels of MBD3 mRNA, while the mRNA levels of the highly similar MBD2 remained unaffected, demonstrating the specificity of the knockdown (Fig. 5A, right panel). Quantitative RT-PCR of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was used as a loading control. A Western blot assay (Fig. 5B, top panel) confirmed the knockdown in mRNA levels corresponded to a knockdown in MBD3 protein levels. The membrane was stripped and reblotted with an antibody against β-actin (bottom panel) to demonstrate equal loading. As an additional control, mock transfections with the vehicle were also performed and showed no change in MBD3 mRNA or protein levels (see Fig. S2A and B in the supplemental material).
FIG. 5.
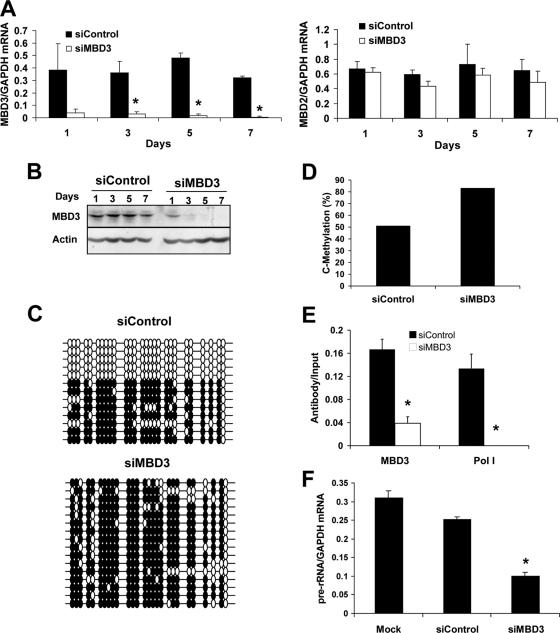
siRNA knockdown of MBD3 over 7 days results in increased methylation of the rRNA promoter, decreased Pol I binding to the rRNA promoter, and decreased pre-rRNA transcription. (A and B) HeLa cells were transfected with 20 nM siControl or siMBD3 for 1, 3, 5, or 7 days. (A) Quantitative RT-PCR of MBD3 (left panel) and MBD2 (right panel) normalized to GAPDH mRNA levels. Student's t test was used to analyze the siMBD3 versus siControl response (*, P < 0.05). (B) Western blot analysis of MBD3 (upper panel) and β-actin (lower panel), a control for total protein loading. (C to G) HeLa cells were transfected with 20 nM siControl or siMBD3 for 7 days. (C) Bisulfite analysis of the rRNA promoter from siControl or siMBD3 transfectants. Each line represents an independent clone. Filled circles, methylated CG dinucleotides; empty circles, unmethylated CG dinucleotides. (D) Quantification of individual methylated CGs in the rRNA promoter from siControl or siMBD3 transfectants. (E) Chromatin immunoprecipitation assays of the association between MBD3 or Pol I binding to the rRNA promoter. Results shown are from quantitative real-time PCR of the rRNA sequence amplified from MBD3- or RPA-116-immunoprecipitated HeLa extracts, normalized to the nonimmunoprecipitated extracts (input), from three independent experiments. One-tenth of the input sample was used for PCR amplification. Student's t test was used for analysis of siControl versus siMBD3 (*, P < 0.05). (F) Quantitative RT-PCR of pre-rRNA sequences normalized to GAPDH mRNA levels from three independent experiments. Student's t test was used for analysis of siMBD3 versus mock or siControl (*, P < 0.05).
We first examined the effects of MBD3 knockdown on the methylation levels of the rRNA promoter following 7 days of treatment using bisulfite sequencing (Fig. 5C). Interestingly, the levels of methylation increased over the siControl-transfected group from 51% to 83% (Fig. 5D). More interestingly, none of the clones examined in the siMBD3-transfected group was completely unmethylated, indicating an overall increase in methylation of the rRNA promoter. These results are consistent with the hypothesis that MBD3 is involved in maintaining the rRNA promoter in an unmethylated state, as there is a rebound hypermethylation in the absence of MBD3.
Next, ChIP assays were used to determine whether MBD3 knockdown had an effect on Pol I occupancy of the rRNA promoter. If MBD3 were acting as a transcriptional repressor, then a knockdown of MBD3 would result in increased Pol I binding to the rRNA promoter, as the unmethylated promoters previously bound to MBD3 would now be accessible for Pol I to bind. Alternatively, if MBD3 were participating in the transcriptional activation of the rRNA promoter, then a decrease in MBD3 binding would also result in decreased Pol I binding. ChIP assays show that a knockdown of MBD3 results in a significant decrease in Pol I occupancy on the rRNA promoter (P < 0.05) (Fig. 5E). Despite almost a complete knock-down in the mRNA and protein levels of MBD3 (Fig. 5A and B), we were still able to detect a small level of MBD3 binding to the rRNA promoter following 7 days of siMBD3 transfection. Additionally, when a ChIP analysis was performed 48 h after siMBD3 transfection, although there was a significant decrease in the protein levels, there was no change in the binding of MBD3 to the rRNA promoter (see Fig. S2C and D in the supplemental material). Therefore, we believe that the turnover of MBD3 bound to the rRNA promoter occurs at a very slow rate.
We next tested whether decreased Pol I binding to the rRNA promoter also results in decreased transcription. Using RNA that was treated with DNase, we measured the levels of pre-rRNA transcription with quantitative RT-PCR, as described in Materials and Methods (Fig. 5F). There was a significant reduction in pre-rRNA levels in siMBD3-transfected cells compared to mock- or siControl-transfected cells (P < 0.05). Together, the increase in DNA methylation of the rRNA promoter and the decrease in both Pol I binding to the rRNA promoter and pre-rRNA transcription are consistent with the hypothesis that MBD3 is important for protecting the rRNA promoter from methylation, thereby allowing Pol I to bind.
HeLa cells deficient in MBD3 have decreased protein production and altered cell morphology.
To define the biological consequences of MBD3 depletion and reductions in pre-rRNA transcription, we examined total protein production by measuring [3H]leucine incorporation in HeLa cells transfected with siMBD3 over 7 days. There was a significant decrease in protein production in the cells deficient in MBD3 compared to the mock- or siControl-transfected cells (see Fig. S3A in the supplemental material), in accordance with the decreased pre-rRNA transcription. Additionally, using 4′,6′-diamidino-2-phenylindole staining, we observed a significant increase in the number of multinucleated cells (cells containing two or more nuclei) when MBD3 was knocked down (see Fig. S3B and C in the supplemental material). Flow cytometry experiments were done to confirm these results. Although there was not a significant change between siMBD3- and siControl-transfected cells in the percentage of cells in the G0/G1 and S phases of the cell cycle (G0/G1, 41% versus 42%; S, 15% versus 16%), there was a reduction in cells found in G2 (24% versus 16%), which could be partially explained by an increase in the multinucleated cell count in the siMBD3 transfectants (4% versus 7%) (see Fig. S3D in the supplemental material). Moreover, we observed striking morphological changes in the cells transfected with siMBD3 compared to siControl transfectants (see Fig. S3E in the supplemental material). The HeLa cells lacking MBD3 became less rounded and developed long processes.
Overexpression of MBD3 resulted in demethylation of the rRNA promoter and increased Pol I binding.
To confirm the results from the MBD3 knockdown, we overexpressed MBD3 in HEK 293 cells to determine whether it would induce effects opposite to the MBD3 depletion. HEK 293 cells were used for the overexpression experiment, since these cells are transfected at very high efficiency. We verified the overexpression of MBD3 using Western blot analysis. Two specific bands were seen when the membrane was blotted with an antibody specific to MBD3 (Fig. 6A, upper panel); the lower band represents the endogenous MBD3, while the upper band represents the exogenous X-press-tagged MBD3. The membrane was stripped and reblotted with anti-β-actin to demonstrate equal loading (Fig. 6A, lower panel).
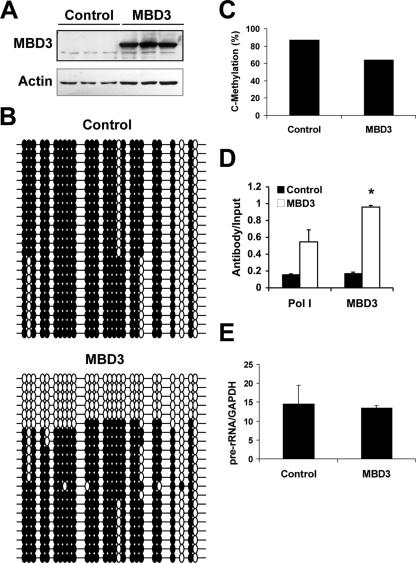
FIG. 6.
MBD3 overexpression in HEK 293 cells results in decreased methylation of the rRNA promoter. HEK 293 cells were transfected with empty vector pEF6 (control) or MBD3-pEF6 for 72 h. (A) Western blot analysis of the endogenous MBD3 (34 kDa) and exogenous MBD3 with an X-press epitope (40 kDa) visualized with an anti-MBD3 antibody (upper panel). The membrane was reprobed for β-actin as a control for loading (lower panel). (B) Bisulfite analysis of the rRNA promoter from control or MBD3 transfectants. Each line represents an independent clone. Filled circles, methylated CG dinucleotides; empty circles, unmethylated CG dinucleotides. (C) Quantification of the individual methylated CGs in the rRNA promoter from control or MBD3 transfectants. (D) Chromatin immunoprecipitation assays of the association between MBD3 or Pol I binding to the rRNA promoter. Results are from quantitative real-time PCR of the rRNA promoter amplified from MBD3- or RPA-116-immunoprecipitated HEK 293 extracts, normalized to the nonimmunoprecipitated extracts (input), from three independent experiments. One-tenth of the input sample was used for PCR amplification. Student's t test was used for analysis of MBD3 versus control (*, P < 0.05). (E) Quantitative RT-PCR of pre-rRNA sequences normalized to GAPDH mRNA levels from three independent experiments.
Bisulfite mapping was used to determine the levels of methylation of the rRNA promoter in response to MBD3 overexpression. In contrast to the results obtained with the MBD3 knockdown in HeLa cells (Fig. 5C), overexpression of MBD3 in HEK 293 cells induced hypomethylation of the rRNA promoter compared to the control transfectants (Fig. 6B and C) with an increase in fully demethylated promoters. The methylation pattern of the rRNA promoter in HEK 293 cells, however, is markedly different than that observed in HeLa cells. Although the promoter is heavily methylated, there are two specific sites that are fully unmethylated in all copies. It is possible that this demethylation is sufficient to allow for Pol I binding, as seen in Fig. 6D. Ectopic expression of MBD3 markedly increased the fraction of rRNA promoters which are fully unmethylated and decreased the average methylation of CGs in the rRNA promoter region from 87% to 63% (Fig. 6C). These data support a role for MBD3 in maintaining the unmethylated state of the rRNA promoters. ChIP assays showed an increase in Pol I binding to the promoter when there are increased levels of MBD3 (Fig. 6D). However, there was no increase in pre-rRNA transcription, as assayed with quantitative RT-PCR (Fig. 6E). These results suggest that although ectopic MBD3 increases the fraction of unmethylated rRNA promoters, other regulatory mechanisms exist to limit rRNA transcription.
Overexpression of MBD3 induces demethylation and expression of an exogenous, in vitro-methylated gene driven by the rRNA promoter.
In order to determine whether ectopic MBD3 expression can induce demethylation of a methylated gene driven by the rRNA promoter, we transfected a plasmid containing the rRNA promoter and the luciferase reporter gene into HEK 293 cells. A decrease in methylation could occur through either a passive loss of methylation during cell division in the absence of DNA methyltransferase or by active removal of the methyl group by an enzymatic demethylation process. The plasmid used here does not replicate during transient transfection; therefore, this model system has been used in our lab to examine replication-independent demethylation in human cells (5). We first verified that expression of the luciferase reporter gene was indeed driven by Pol I and not Pol II. Following transfection of HEK 293 cells with a plasmid containing the luciferase gene driven by either the rRNA promoter (rRNA-pGL3) or the cytomegalovirus promoter (CMV-pGL3), the cells were treated with actinomycin D. Actinomycin D is a potent inhibitor of Pol I transcription, with lesser effects on Pol II-driven transcription (36). Quantitative RT-PCR was used to monitor the rate of disappearance of the luciferase transcript following actinomycin D treatment, and we found that the transcript driven by the rRNA promoter was reduced at a faster rate (t1/2, 2.5 h) compared to the same transcript driven by the CMV promoter (t1/2, 4 h). Since the RNA sequence is identical for both promoters, the difference in mRNA depletion should reflect decreased transcription and not RNA stability (Fig. 7B). There was very little endogenous pre-rRNA transcription following 2 h of actinomycin D treatment in either group (Fig. 7B), indicating that the actinomycin D treatment was effective in both transfectant groups.
Transcripts driven by Pol I are not capped and thus cannot enter the ribosome for translation; therefore, no luciferase protein production is expected. To verify that there is no spurious Pol II-driven transcription from the rRNA-pGL3 construct that would lead to luciferase protein production, a luciferase assay was used (Fig. 7C). As expected, the luciferase activity detected from the rRNA-pGL3 transfectants was similar to the background levels found with the empty vector transfectants. Together, these experiments support that transcription from the rRNA promoter on the luciferase construct is driven by Pol I.
Next, the rRNA-pGL3 plasmid was methylated in vitro using mSssI methyltransferase and transfected with or without MBD3 into HEK 293 cells to determine whether ectopic MBD3 could induce expression of the gene. Using quantitative RT-PCR we found that the levels of luciferase mRNA were significantly increased when MBD3 was overexpressed (Fig. 8A). We next examined the methylation of the luciferase gene driven by the rRNA promoter using Southern blot analysis, a regional methylation assay (data not shown), and bisulfite mapping (Fig. 8B). To differentiate between the exogenous rRNA promoter and the endogenous rRNA promoter, we chose to examine the luciferase gene sequence immediately downstream to the rRNA promoter as readout of demethylation triggered by MBD3 in the rRNA promoter region. Since the demethylation associated with MBD3 is not site specific but spans the entire CG region of the rRNA promoter, we reasoned that this demethylation would extend into the luciferase CG-rich region immediately downstream. Figure 7A depicts the region examined for both ChIP and sodium bisulfite analysis. Increased demethylation of the rRNA-pGL3 plasmid was observed with overexpression of MBD3 (Fig. 8B and C).
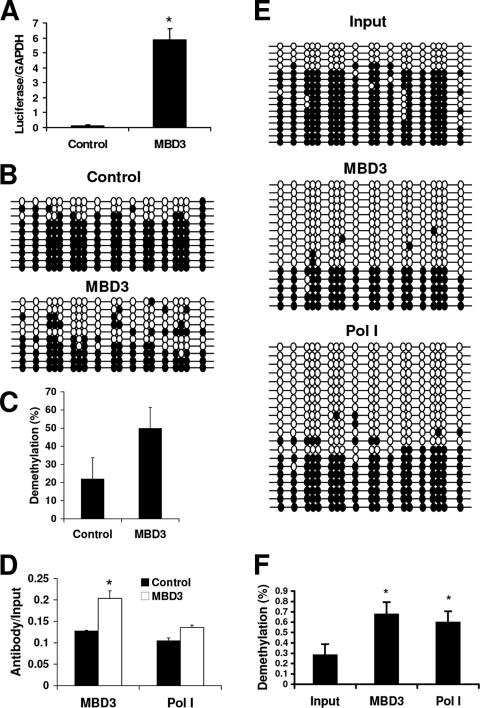
FIG. 8.
MBD3 overexpression can induce expression and demethylation of an in vitro-methylated exogenous gene driven by the rRNA promoter. HEK 293 cells were transfected with in vitro-methylated rRNA-pGL3 and pEF6 (control) or MBD3-pEF6 (MBD3) for 72 h. (A) Quantitative RT-PCR of luciferase mRNA relative to GAPDH mRNA from three independent experiments. Student's t test was used for analysis of MBD3 versus control (*, P < 0.001). (B) Bisulfite analysis of the luciferase gene. Each line represents an independent clone. Filled circles, methylated CG dinucleotides; empty circles, unmethylated CG dinucleotides. (C) Quantification of the individual methylated CGs in the rRNA promoter from the control or MBD3 transfectants. (D) Chromatin immunoprecipitation assays of the association between MBD3 or Pol I binding to the rRNA promoter. Results are from quantitative real-time PCR of the luciferase gene amplified from MBD3- or RPA-116-immunoprecipitated HEK 293 extracts, normalized to the nonimmunoprecipitated extracts (input), from three independent experiments. One-tenth of the input sample was used for PCR amplification. Student's t test was used for analysis of MBD3 versus control (*, P < 0.05). (E) Bisulfite analysis of the luciferase gene immunoprecipitated with anti-MBD3 antibody (MBD3), anti-RPA-116 antibody (Pol I), or nonimmunoprecipitated DNA (input). (F) Percentage of demethylated cytosines within the sequence amplified from the luciferase gene from input, MBD3-, or Pol I-immunoprecipitated samples. Student's t test was used for analysis of input versus MBD3 or Pol I (*, P < 0.05).
MBD3 binds to the exogenous gene driven by the rRNA promoter and is involved in its demethylation.
ChIP assays were used to verify that MBD3 is binding to the exogenous rRNA-luciferase reporter construct. Indeed, both MBD3 and Pol I bind to the in vitro-methylated rRNA-pGL3, and overexpression of MBD3 increased the binding of both MBD3 and Pol I (Fig. 8D). Additionally, sodium bisulfite analysis of the immunoprecipitated DNA revealed that the DNA bound to both MBD3 and Pol I was demethylated when MBD3 was overexpressed (Fig. 8E and F). These data support the hypothesis that MBD3 interacts with a methylated gene driven by the rRNA promoter, induces its demethylation, and activates its expression.
In order to determine whether the actions of MBD3 are specific to the rRNA promoter or are part of a more global mechanism, we in vitro methylated the CMV-pGL3 construct described earlier and transfected it with or without MBD3 into HEK 293 cells. Methylation effectively suppressed luciferase expression by 75-fold (relative luciferase expression of unmethylated CMV-pGL3, 33,964,178 ± 585,291.4 [mean ± standard error of the mean] versus methylated CMV-pGL3, 455,228.4 ± 74,971.43; Student's t test comparing unmethylated versus methylated, P < 0.0001). While we did observe a significant increase in the levels of luciferase production with the overexpression of MBD3 (P < 0.05) (see Fig. S4A in the supplemental material), there was no change in the levels of demethylation of the luciferase gene over that observed in the control sample (see Fig. S4B and C). Therefore, the ability of MBD3 to induce demethylation of the luciferase gene appears to be promoter specific.
MBD3 induces active demethylation of the in vitro-methylated rRNA-luciferase construct.
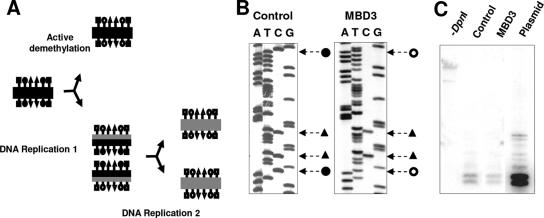
DNA demethylation may occur through either a passive or an active mechanism, as outlined in Fig. 9A. Passive demethylation is dependent on DNA replication. The newly replicated strand is void of methylation until methylated by the maintenance methyltransferase. If the methyltransferases are blocked, the newly replicated strand of DNA will remain unmethylated, resulting in a passive loss of the methylation pattern. Active demethylation on the other hand does not depend on replication of the DNA and involves the removal of the methyl group by an enzyme from the cytosine nucleotide found in the CG dinucleotide sequence.
FIG. 9.
MBD3 induces active demethylation. (A) Active versus passive demethylation. Filled shapes, methylated sites; shapes with white, unmethylated sites; black lines, parent DNA strands; gray lines, daughter strands resultant from DNA replication; squares, DpnI sites; triangles, EcoRII sites; circles, CG dinucleotide sites. (B) Representative sequencing gel with methylated EcoRII sites. (C) Southern blot of DNA extracted from HEK 293 cells transfected with in vitro-methylated rRNA-pGL3 and pEF6 (control) or MBD3-pEF6 (MBD3), digested with DpnI. Untransfected rRNA-pGL3 digested with DpnI was used as a control (plasmid).
To determine whether the demethylation observed with MBD3 overexpression was dependent on DNA replication, we took advantage of the rRNA-pGL3 plasmid, which does not contain a mammalian origin of replication and should not replicate once transfected into mammalian cells. Therefore, any demethylation of the transiently transfected rRNA-pGL3 should be due to an active process. Since the plasmid is raised in bacteria, it acquires the bacterial DNA methylation pattern. Mammalian cells do not contain the DNA methyltransferases which methylate these sites; therefore, they would be lost if the plasmid replicated in mammalian cells. We examined EcoRII sites (CCA/TGG), in which the second C is methylated in bacteria, and DpnI sites (GATC), in which the A is methylated in bacteria. Within the sequence analyzed with bisulfite mapping, there were two EcoRII sites present, and these sequences remained methylated while the adjacent CG sites became demethylated with MBD3 overexpression (Fig. 9B). Additionally, following transient transfection, the plasmid was digested with DpnI enzyme, which cuts the plasmid only if the GATC sites remain methylated. Southern blot analysis following DpnI digestion revealed that rRNA-pGL3 maintained its DpnI sensitivity and did not replicate following transient transfection (Fig. 9C). In summary, the demethylation observed with the overexpression of MBD3 was due to a replication-independent mechanism.
DISCUSSION
The regulation of rRNA transcription is tightly controlled within the cell at many levels, and the fraction of rRNA genes that are either active or silenced is determined through epigenetics. Several studies have addressed the mechanisms responsible for epigenetic inactivation of rRNA genes; however, questions remain as to how the active fraction is protected from silencing and how the fraction silenced through DNA methylation may become reactivated. In this paper we identify the protein MBD3 as a candidate for maintaining active, unmethylated rRNA genes. Previously, MBD3 was shown to be a component of the NuRD complex, which is involved in transcriptional repression (53), and was also proposed to be involved in silencing of rRNA genes (17). However, here we present evidence supporting a role for MBD3 in both the maintenance of unmethylated active rRNA genes and demethylation of rRNA genes.
MBD3 interacts with hypomethylated copies of the rRNA promoter (Fig. 2B), and this binding is likely directed through the activity of UBF (Fig. 4). Depletion of MBD3 resulted in a rebound methylation of the rRNA promoters (Fig. 5C), indicating an essential role for MBD3 in maintaining these promoters in an unmethylated state. Overexpression of MBD3 led to increased demethylation of both the endogenous rRNA promoter (Fig. 6B) and an exogenous gene driven by the rRNA promoter (Fig. 8E), suggesting that MBD3 not only protects unmethylated promoters from remethylation but may also be instrumental in the demethylation of the rRNA promoters. Additionally, the demethylation appears to be occurring in a promoter-specific fashion, as similar experiments with the luciferase gene driven by the CMV promoter did not result in demethylation (see Fig. S4 in the supplemental material). Finally, the observed demethylation of the exogenous gene occurs in an active manner, in the absence of DNA replication (Fig. 9B and C). Together these data strongly support an essential role for MBD3 in the activation of rRNA genes through the induction of hypomethylation of the rRNA promoter.
One possible mechanism that must be considered is that MBD3 acts indirectly on rRNA promoters, through silencing of another gene, which in turn acts as a repressor of rRNA activity. However, we have provided evidence from ChIP assays that MBD3 interacts with the rRNA promoter (Fig. 2A). Moreover, using double ChIP assays, we showed that MBD3 and the transcriptional activator UBF occupy the same rRNA promoters (Fig. 4). Thus, our evidence points to involvement of MBD3 with transcriptional activity and hypomethylation at the rRNA promoter. The question remains as to whether MBD3 is binding directly to the rRNA promoter or whether this interaction is occurring indirectly through the chromatin. MBD3 has been shown to bind nonspecifically to DNA, and therefore it is possible that it may bind directly to the promoter (14, 23). However, while indirect binding of MBD3 to DNA has been shown (19, 46), it is well-documented that wild-type MBD3 has an MBD that does not possess methyl-DNA binding activity, neither as a recombinant protein nor in the NuRD complex purified from mammalian cells (30, 41). From ChIP experiments it cannot be determined whether MBD3 is binding directly to the rRNA promoter or whether it is associated through the chromatin. Our data point to the suggestion that the interaction of MBD3 with the unmethylated rRNA promoter may be occurring through the transcription factor UBF (Fig. 4).
Traditionally, it was thought that unmethylated DNA is passively maintained due to the maintenance methyltransferase being highly inefficient in de novo methylation (38). Demethylation of specific genes has previously been reported, including the immunoglobulin κ gene during B-cell differentiation (27) and α-actin during muscle cell differentiation (51), but it was believed that once a gene is demethylated during differentiation or early development its demethylated state is passively maintained. To our knowledge this is the first report illustrating that a specific protein must protect an unmethylated promoter and that in its absence remethylation may occur, even in somatic cells.
Two theories exist regarding the formation of the preinitiation complex at the rRNA promoter (32). The first involves a stepwise assembly of the numerous factors involved, with binding of UBF regarded as the initial step in the assembly of the preinitiation complex and the binding of Pol I as the final step required for rRNA transcription. The second theory is that the complex binds as a holoenzyme; however, UBF is thought to remain bound to the rRNA promoters regardless of activation. In either case, the binding of UBF precedes the binding of Pol I and transcription initiation. Therefore, the observation that the promoters bound to UBF have higher levels of methylation than the promoters bound to Pol I, particularly in the UCE (Fig. 1E and F, respectively), indicates that there must be a demethylation event occurring after UBF binding which precedes, or occurs concurrently with, Pol I binding.
The overexpression of MBD3 led to increased demethylation of the rRNA promoter and an increase in Pol I binding; however, it did not lead to an increase in pre-rRNA transcription (Fig. 6E). Similarly, it was shown that although the binding of β-actin to Pol I is essential for rRNA transcription, overexpression of β-actin did not lead to increased rRNA transcription (37). The failure of the overexpression of both β-actin and MBD3 to increase rRNA transcription may be attributed either to a saturation of the system by the endogenous proteins or to an additional level of control over the rates of rRNA transcription in the cell. There exists a large excess of Pol I in the cell (2), which may account for the increase in Pol I binding to the newly available unmethylated promoters; however, an increase in pre-rRNA transcription most likely was inhibited through additional regulatory mechanisms.
It is known that MBD3 plays an essential role in development, as MBD3 knockouts in mice and Xenopus laevis are embryo-lethal (24, 26). Therefore, an essential role for MBD3 in the regulation of rRNA transcription is in accordance with the embryo lethality of these knockout models. If MBD3 were required for maintaining unmethylated copies of the rRNA promoter, then a lack of MBD3 would result in increased methylation of the promoter to which Pol I cannot bind, similar to the siRNA knock-down of MBD3 in HeLa cells (Fig. 5C and E). However, since the mouse embryos develop to embryonic day 8.5, it implies that MBD3 is not required for rRNA expression early in embryogenesis (24).
Whether MBD3 causes demethylation itself or if it recruits another enzyme(s) remains unknown. Our data suggest that MBD3 directly interacts with the rRNA promoter and induces their replication-independent active demethylation. MBD3 has been shown to form heterodimers with MBD2 (46), which we have proposed to be a DNA demethylase (1, 4, 39), and has previously been shown to bind to the rRNA promoter (17). Therefore, it is possible that MBD3 is required to recruit MBD2 to the rRNA promoter for demethylation. However, the demethylase activity of MBD2 has been contested by several groups (3, 53), although we have since demonstrated demethylase activity from recombinant MBD2 (9). Alternatively, MBD3 shares over 71% homology with MBD2 (23); therefore, it is possible that MBD3 may also share the as-yet-undetermined catalytic domain for demethylase activity. Experiments are currently under way to determine the in vitro demethylase activity and specificity of MBD3. Whereas the biochemical activity responsible for demethylation of the rRNA promoter remains unknown, our data demonstrate that MBD3 plays a critical role in maintaining the unmethylated rRNA promoters in eukaryotic cells and that in its absence these promoters would be susceptible to silencing through DNA methylation.
Supplementary Material
Acknowledgments
This study was funded by a grant awarded to M.S. by the NCIC. S.E.B. is supported by a fellowship from the CIHR.
We thank I. Grummt for her contribution of the anti-RPA-116 antibody and P. Wade for his contribution of the anti-Mi-2 antibody. As well, we thank Jacynthe Laliberté for her help with the confocal microscopy.
Footnotes
Published ahead of print on 23 April 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bhattacharya, S. K., S. Ramchandani, N. Cervoni, and M. Szyf. 1999. A mammalian protein with specific demethylase activity for mCpG DNA. Nature 397:579-583. [DOI] [PubMed] [Google Scholar]
- 2.Bier, M., S. Fath, and H. Tschochner. 2004. The composition of the RNA polymerase I transcription machinery switches from initiation to elongation mode. FEBS Lett. 564:41-46. [DOI] [PubMed] [Google Scholar]
- 3.Boeke, J., O. Ammerpohl, S. Kegel, U. Moehren, and R. Renkawitz. 2000. The minimal repression domain of MBD2b overlaps with the methyl-CpG-binding domain and binds directly to Sin3A. J. Biol. Chem. 275:34963-34967. [DOI] [PubMed] [Google Scholar]
- 4.Cervoni, N., S. Bhattacharya, and M. Szyf. 1999. DNA demethylase is a processive enzyme. J. Biol. Chem. 274:8363-8366. [DOI] [PubMed] [Google Scholar]
- 5.Cervoni, N., and M. Szyf. 2001. Demethylase activity is directed by histone acetylation. J. Biol. Chem. 276:40778-40787. [DOI] [PubMed] [Google Scholar]
- 6.Clark, S. J., J. Harrison, C. L. Paul, and M. Frommer. 1994. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 22:2990-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crane-Robinson, C., F. A. Myers, T. R. Hebbes, A. L. Clayton, and A. W. Thorne. 1999. Chromatin immunoprecipitation assays in acetylation mapping of higher eukaryotes. Methods Enzymol. 304:533-547. [DOI] [PubMed] [Google Scholar]
- 8.Detich, N., V. Bovenzi, and M. Szyf. 2003. Valproate induces replication-independent active DNA demethylation. J. Biol. Chem. 278:27586-27592. [DOI] [PubMed] [Google Scholar]
- 9.Detich, N., S. Hamm, G. Just, J. D. Knox, and M. Szyf. 2003. The methyl donor S-adenosylmethionine inhibits active demethylation of DNA: a candidate novel mechanism for the pharmacological effects of S-adenosylmethionine. J. Biol. Chem. 278:20812-20820. [DOI] [PubMed] [Google Scholar]
- 10.Detich, N., J. Theberge, and M. Szyf. 2002. Promoter-specific activation and demethylation by MBD2/demethylase. J. Biol. Chem. 277:35791-35794. [DOI] [PubMed] [Google Scholar]
- 11.Dunn, B. K. 2003. Hypomethylation: one side of a larger picture. Ann. N. Y. Acad. Sci. 983:28-42. [DOI] [PubMed] [Google Scholar]
- 12.Ego, T., Y. Tanaka, and K. Shimotohno. 2005. Interaction of HTLV-1 Tax and methyl-CpG-binding domain 2 positively regulates the gene expression from the hypermethylated LTR. Oncogene 24:1914-1923. [DOI] [PubMed] [Google Scholar]
- 13.Etoh, T., Y. Kanai, S. Ushijima, T. Nakagawa, Y. Nakanishi, M. Sasako, S. Kitano, and S. Hirohashi. 2004. Increased DNA methyltransferase 1 (DNMT1) protein expression correlates significantly with poorer tumor differentiation and frequent DNA hypermethylation of multiple CpG islands in gastric cancers. Am. J. Pathol. 164:689-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fraga, M. F., E. Ballestar, G. Montoya, P. Taysavang, P. A. Wade, and M. Esteller. 2003. The affinity of different MBD proteins for a specific methylated locus depends on their intrinsic binding properties. Nucleic Acids Res. 31:1765-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujita, H., R. Fujii, S. Aratani, T. Amano, A. Fukamizu, and T. Nakajima. 2003. Antithetic effects of MBD2a on gene regulation. Mol. Cell. Biol. 23:2645-2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gebrane-Younes, J., N. Fomproix, and D. Hernandez-Verdun. 1997. When rDNA transcription is arrested during mitosis, UBF is still associated with non-condensed rDNA. J. Cell Sci. 110:2429-2440. [DOI] [PubMed] [Google Scholar]
- 17.Ghoshal, K., S. Majumder, J. Datta, T. Motiwala, S. Bai, S. M. Sharma, W. Frankel, and S. T. Jacob. 2004. Role of human ribosomal RNA (rRNA) promoter methylation and of methyl-CpG-binding protein MBD2 in the suppression of rRNA gene expression. J. Biol. Chem. 279:6783-6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goel, A., S. P. Mathupala, and P. L. Pedersen. 2003. Glucose metabolism in cancer. Evidence that demethylation events play a role in activating type II hexokinase gene expression. J. Biol. Chem. 278:15333-15340. [DOI] [PubMed] [Google Scholar]
- 19.Gu, P., D. Le Menuet, A. C. Chung, and A. J. Cooney. 2006. Differential recruitment of methylated CpG binding domains by the orphan receptor GCNF initiates the repression and silencing of Oct4 expression. Mol. Cell. Biol. 26:9471-9483. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Guschin, D., P. A. Wade, N. Kikyo, and A. P. Wolffe. 2000. ATP-dependent histone octamer mobilization and histone deacetylation mediated by the Mi-2 chromatin remodeling complex. Biochemistry 39:5238-5245. [DOI] [PubMed] [Google Scholar]
- 21.Haltiner, M. M., S. T. Smale, and R. Tjian. 1986. Two distinct promoter elements in the human rRNA gene identified by linker scanning mutagenesis. Mol. Cell. Biol. 6:227-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henderson, A. S., D. Warburton, and K. C. Atwood. 1972. Location of ribosomal DNA in the human chromosome complement. Proc. Natl. Acad. Sci. USA 69:3394-3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hendrich, B., and A. Bird. 1998. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell. Biol. 18:6538-6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hendrich, B., J. Guy, B. Ramsahoye, V. A. Wilson, and A. Bird. 2001. Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development. Genes Dev. 15:710-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirschler-Laszkiewicz, I., A. Cavanaugh, Q. Hu, J. Catania, M. L. Avantaggiati, and L. I. Rothblum. 2001. The role of acetylation in rDNA transcription. Nucleic Acids Res. 29:4114-4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iwano, H., M. Nakamura, and S. Tajima. 2004. Xenopus MBD3 plays a crucial role in an early stage of development. Dev. Biol. 268:416-428. [DOI] [PubMed] [Google Scholar]
- 27.Kirillov, A., B. Kistler, R. Mostoslavsky, H. Cedar, T. Wirth, and Y. Bergman. 1996. A role for nuclear NF-κB in B-cell-specific demethylation of the Igκ locus. Nat. Genet. 13:435-441. [DOI] [PubMed] [Google Scholar]
- 28.Kochanek, S., K. Hosokawa, G. Schiedner, D. Renz, and W. Doerfler. 1996. DNA methylation in the promoter of ribosomal RNA genes in human cells as determined by genomic sequencing. FEBS Lett. 388:192-194. [DOI] [PubMed] [Google Scholar]
- 29.Learned, R. M., T. K. Learned, M. M. Haltiner, and R. T. Tjian. 1986. Human rRNA transcription is modulated by the coordinate binding of two factors to an upstream control element. Cell 45:847-857. [DOI] [PubMed] [Google Scholar]
- 30.Le Guezennec, X., M. Vermeulen, A. B. Brinkman, W. A. Hoeijmakers, A. Cohen, E. Lasonder, and H. G. Stunnenberg. 2006. MBD2/NuRD and MBD3/NuRD, two distinct complexes with different biochemical and functional properties. Mol. Cell. Biol. 26:843-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Milutinovic, S., S. E. Brown, Q. Zhuang, and M. Szyf. 2004. DNA methyltransferase 1 knock down induces gene expression by a mechanism independent of DNA methylation and histone deacetylation. J. Biol. Chem. 279:27915-27927. [DOI] [PubMed] [Google Scholar]
- 32.Moss, T., and V. Y. Stefanovsky. 2002. At the center of eukaryotic life. Cell 109:545-548. [DOI] [PubMed] [Google Scholar]
- 33.Murphy, B. C., R. L. O'Reilly, and S. M. Singh. 2005. Site-specific cytosine methylation in S-COMT promoter in 31 brain regions with implications for studies involving schizophrenia. Am. J. Med. Genet. B 133:37-42. [DOI] [PubMed] [Google Scholar]
- 34.Niculescu, M. D., C. N. Craciunescu, and S. H. Zeisel. 2006. Dietary choline deficiency alters global and gene-specific DNA methylation in the developing hippocampus of mouse fetal brains. FASEB J. 20:43-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohki, I., N. Shimotake, N. Fujita, M. Nakao, and M. Shirakawa. 1999. Solution structure of the methyl-CpG-binding domain of the methylation-dependent transcriptional repressor MBD1. EMBO J. 18:6653-6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perry, R. P., and D. E. Kelley. 1970. Inhibition of RNA synthesis by actinomycin D: characteristic dose-response of different RNA species. J. Cell Physiol. 76:127-139. [DOI] [PubMed] [Google Scholar]
- 37.Philimonenko, V. V., J. Zhao, S. Iben, H. Dingova, K. Kysela, M. Kahle, H. Zentgraf, W. A. Hofmann, P. de Lanerolle, P. Hozak, and I. Grummt. 2004. Nuclear actin and myosin I are required for RNA polymerase I transcription. Nat. Cell Biol. 6:1165-1172. [DOI] [PubMed] [Google Scholar]
- 38.Pollack, Y., R. Stein, A. Razin, and H. Cedar. 1980. Methylation of foreign DNA sequences in eukaryotic cells. Proc. Natl. Acad. Sci. USA 77:6463-6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramchandani, S., S. K. Bhattacharya, N. Cervoni, and M. Szyf. 1999. DNA methylation is a reversible biological signal. Proc. Natl. Acad. Sci. USA 96:6107-6112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rouleau, J., G. Tanigawa, and M. Szyf. 1992. The mouse DNA methyltransferase 5′-region. A unique housekeeping gene promoter. J. Biol. Chem. 267:7368-7377. [PubMed] [Google Scholar]
- 41.Saito, M., and F. Ishikawa. 2002. The mCpG-binding domain of human MBD3 does not bind to mCpG but interacts with NuRD/Mi2 components HDAC1 and MTA2. J. Biol. Chem. 277:35434-35439. [DOI] [PubMed] [Google Scholar]
- 42.Sakai, H., T. Urano, K. Ookata, M. H. Kim, Y. Hirai, M. Saito, Y. Nojima, and F. Ishikawa. 2002. MBD3 and HDAC1, two components of the NuRD complex, are localized at Aurora-A-positive centrosomes in M phase. J. Biol. Chem. 277:48714-48723. [DOI] [PubMed] [Google Scholar]
- 43.Santoro, R., and I. Grummt. 2005. Epigenetic mechanism of rRNA gene silencing: temporal order of NoRC-mediated histone modification, chromatin remodeling, and DNA methylation. Mol. Cell. Biol. 25:2539-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Santoro, R., and I. Grummt. 2001. Molecular mechanisms mediating methylation-dependent silencing of ribosomal gene transcription. Mol. Cell 8:719-725. [DOI] [PubMed] [Google Scholar]
- 45.Seither, P., and I. Grummt. 1996. Molecular cloning of RPA2, the gene encoding the second largest subunit of mouse RNA polymerase I. Genomics 37:135-139. [DOI] [PubMed] [Google Scholar]
- 46.Tatematsu, K. I., T. Yamazaki, and F. Ishikawa. 2000. MBD2-MBD3 complex binds to hemi-methylated DNA and forms a complex containing DNMT1 at the replication foci in late S phase. Genes Cells 5:677-688. [DOI] [PubMed] [Google Scholar]
- 47.Wade, P. A., A. Gegonne, P. L. Jones, E. Ballestar, F. Aubry, and A. P. Wolffe. 1999. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat. Genet. 23:62-66. [DOI] [PubMed] [Google Scholar]
- 48.Wakefield, R. I., B. O. Smith, X. Nan, A. Free, A. Soteriou, D. Uhrin, A. P. Bird, and P. N. Barlow. 1999. The solution structure of the domain from MeCP2 that binds to methylated DNA. J. Mol. Biol. 291:1055-1065. [DOI] [PubMed] [Google Scholar]
- 49.Xue, Y., J. Wong, G. T. Moreno, M. K. Young, J. Cote, and W. Wang. 1998. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell 2:851-861. [DOI] [PubMed] [Google Scholar]
- 50.Yan, P. S., F. J. Rodriguez, D. E. Laux, M. R. Perry, S. B. Standiford, and T. H. Huang. 2000. Hypermethylation of ribosomal DNA in human breast carcinoma. Br. J. Cancer 82:514-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yisraeli, J., R. S. Adelstein, D. Melloul, U. Nudel, D. Yaffe, and H. Cedar. 1986. Muscle-specific activation of a methylated chimeric actin gene. Cell 46:409-416. [DOI] [PubMed] [Google Scholar]
- 52.Zatsepina, O. V., R. Voit, I. Grummt, H. Spring, M. V. Semenov, and M. F. Trendelenburg. 1993. The RNA polymerase I-specific transcription initiation factor UBF is associated with transcriptionally active and inactive ribosomal genes. Chromosoma 102:599-611. [DOI] [PubMed] [Google Scholar]
- 53.Zhang, Y., H. H. Ng, H. Erdjument-Bromage, P. Tempst, A. Bird, and D. Reinberg. 1999. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 13:1924-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou, Y., R. Santoro, and I. Grummt. 2002. The chromatin remodeling complex NoRC targets HDAC1 to the ribosomal gene promoter and represses RNA polymerase I transcription. EMBO J. 21:4632-4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.