Abstract
Bmi-1 and Mel-18 are structural homologues that belong to the Polycomb group of transcriptional regulators and are believed to stably maintain repression of gene expression by altering the state of chromatin at specific promoters. While a number of clinical and experimental observations have implicated Bmi-1 in human tumorigenesis, the role of Mel-18 in cancer cell growth has not been investigated. We report here that short hairpin RNA-mediated knockdown of either Bmi-1 or Mel-18 in human medulloblastoma DAOY cells results in the inhibition of proliferation, loss of clonogenic survival, anchorage-independent growth, and suppression of tumor formation in nude mice. Furthermore, overexpression of both Bmi-1 and Mel-18 significantly increases the clonogenic survival of Rat1 fibroblasts. In contrast, stable downregulation of Bmi-1 or Mel-18 alone does not affect the growth of normal human WI38 fibroblasts. Proteomics-based characterization of Bmi-1 and Mel-18 protein complexes isolated from cancer cells revealed substantial similarities in their respective compositions. Finally, gene expression analysis identified a number of cancer-relevant pathways that may be controlled by Bmi-1 and Mel-18 and also showed that these Polycomb proteins regulate a set of common gene targets. Taken together, these results suggest that Bmi-1 and Mel-18 may have overlapping functions in cancer cell growth.
The Polycomb group (PcG) genes were first identified by their roles in ensuring appropriate temporal and spatial expression of Homeotic (Hox) genes, which act as transcriptional repressors required for correct body patterning during development. Members of the PcG family also play important regulatory roles in adult organisms, including stem cell maintenance, differentiation, and malignant transformation (for a review, see references 15, 23, and 42).
PcG members are generally subdivided into two groups, Polycomb repressive complex 1 (PRC1) and PRC2, and are associated with distinct multiprotein complexes (13, 30). It is believed that a coordinated action between PRC1 and PRC2 complexes introduces heritable histone modifications at certain promoters, followed by the recruitment of additional transcriptional repressors to these sites. The PRC2 complex, which includes Ezh2 and EED, possesses histone H3 lysine 9 and 27 methyltransferase activity, a “silencing” histone mark (8, 10, 38). PRC1 components subsequently bind to methylated H3K27, inhibit SWI/SNF remodeling, and recruit histone deacetylases to silence gene expression (11, 47). Biochemical histone modification by components of the PRC1 complex has also been described: reconstituted PRC1-like complexes containing Bmi1 and RING1B contain an E3 ubiquitin ligase activity capable of ubiquitinating histone H2A (7, 46).
Bmi-1 and Mel-18 are highly homologous constituents of the PRC1 complex. They share 65% amino acid identity and contain a RING finger domain in the N terminus, as well as proline-rich sequences and a helix-turn-helix domain in the C-terminal tail. While they are viable and have certain distinguishable features, Bmi-1−/− and Mel-18−/− mice display similar phenotypes, including severe combined immunodeficiency, growth retardation, and skeletal malformations (2, 43). Furthermore, double Bmi-1/Mel-18 nullizygous mutants die in utero and exhibit strong exacerbation of the single Bmi-1 or Mel-18 mutant phenotype (3). Bmi-1 has a well-established role in supporting self-renewal of hematopoietic and neural stem cells by, at least in part, suppressing the expression of the negative growth regulators p16/INK4A and p19/ARF (34, 37). To a lesser extent, Mel-18−/− hematopoetic progenitor cells are unable to fully reconstitute hematopoiesis in sublethally irradiated recipient mice (20). Mel-18 can also suppress p16 expression (21); however, the precise mechanisms of its contribution to stem cell maintenance have not yet been fully defined.
Although they appear to function similarly in normal cells, it is not known whether Bmi-1 and Mel-18 contribute to cancer cell growth and survival in similar ways. Bmi-1 was originally identified as a proto-oncogene that strongly cooperated with c-myc in mouse lymphomagenesis (17, 22, 44). In human cancers, overexpression of Bmi-1 has been found in medulloblastoma, lung and breast tumors, and cell lines (12, 29, 45). Gene amplification of Bmi-1 has been identified in a subset of human B-cell lymphomas (4). In addition, Bmi-1 expression has been detected at the invading front of breast tumors (25) and was found to be a strong predictor of metastasis and poor survival in a number of cancers (16).
Even though Mel-18 was originally cloned from B16 mouse melanoma cells and its expression was detected in transformed cells, but not in normal cells (41), a later study suggested that it might function as a tumor suppressor (24). To date, no systematic investigation of whether Mel-18 plays a role in the transformation or maintenance of a malignant phenotype has been undertaken. The consequences of long-term Bmi-1 or Mel-18 depletion in cancer cells in vitro and in vivo have also not been elucidated. We set out to investigate the roles of these Polycomb homologues in cancer cell growth.
Using tandem affinity purification (TAP), we isolated the respective Bmi-1 and Mel-18 protein complexes from human cancer cells, and we show that these Polycomb repressors associate with strikingly similar repertoires of proteins. Overexpression of Bmi-1 and Mel-18 in Rat1 fibroblasts results in growth stimulation, while short hairpin RNA (shRNA)-mediated ablation of Bmi-1 and Mel-18 expression in a number of cancer cell lines significantly inhibits their growth and survival in vitro. Gene-profiling experiments in DAOY medulloblastoma cells with defined Bmi-1 or Mel-18 status revealed a substantial proportion of commonly regulated genes, as well as a number of cancer-relevant pathways that might be controlled by these gene products. Most importantly, downregulation of Bmi-1 and Mel-18 in human medulloblastoma xenografts results in tumor growth retardation in vivo. Taken together, our data suggest that Bmi-1 and Mel-18 might have overlapping functions in the establishment or maintenance of tumorigenic phenotypes in medulloblastoma and other human cancers.
MATERIALS AND METHODS
Cloning and retroviral and lentiviral infection.
Open reading frames of Bmi-1 and Mel-18 were cloned into the pNTAP vector (Stratagene). TAP-tagged Bmi-1 and Mel-18 were then subcloned into the pLNCX2 retroviral vector (Clontech). To obtain retroviruses, pLNCX2-TAP-Bmi-1 and pLNCX2-TAP-Mel-18 were cotransfected with pCG10-VSVG into GP2-293 packaging cells using Transit-293 reagent (Mirus). Media were exchanged the following day, and the virus-containing supernatant was harvested 72 h later and used to infect proliferating HeLa cells in the presence of Polybrene (10 μg/ml). Stable lines were generated by G418 selection (1 mg/ml).
Two shRNA constructs targeting either Bmi-1 or Mel-18 were purchased from Sigma (Bmi-1, NM_005180.5-693s1c1; Bmi-1-A, NM_005180.5-1061s1c1; Mel-18, NM_007144.2-840s1c1; Mel-18-A, NM_007144.2-441s1c1). shRNA targeting anaplastic lymphoma kinase, which is only expressed in lymphoid cells (ALK, NM_004304.x-5706s1c1), served as a negative control in all experiments. Lentiviruses were generated by cotransfection of shRNAs with pCG-VSVG and pCMV-Δ8.9 helper plasmids into a 293T packaging cell line using Transit-293 reagent (Mirus) according to the manufacturer's instructions. Stable shRNA-expressing cell lines were generated by infection with lentivirus in the presence of Polybrene (10 μg/ml) and puromycin selection (1 μg/ml).
TAP of protein complexes.
Purification of TAP-Bmi-1, TAP-Mel-18, and control protein complexes was carried out from 5- by 500-cm2 confluent dishes of HeLa stable lines for each condition using the InterPlay system (Stratagene) according to the manufacturer's instructions with minor modifications. HeLa cells stably expressing the green fluorescent protein were used as a negative control. Briefly, cells were lysed by several freeze-thaw cycles in the InterPlay lysis buffer containing 0.4 M NaCl. For incubation with streptavidin resin, the salt concentration was adjusted to 0.15 M NaCl and the protein content was normalized across samples. Protein complexes bound to streptavidin resin were eluted from the beads after several washes using biotin solution, and the resulting fraction was applied to calmodulin resin in the presence of Ca2+. Bound protein complexes were liberated following several washes by elution with the high-salt, EGTA-containing buffer. The eluates were dialyzed against phosphate-buffered saline (PBS) and concentrated using Microcon YM-3 devices (Amicon). Samples were further processed and analyzed by mass spectrometry as described in the supplemental material.
Tissue culture and in vitro phenotypic studies.
All cell lines were purchased from ATCC and maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS) (Invitrogen) and 1% antibiotic-antimycotic solution (Invitrogen). For Rat1 colony formation assays, cells were transfected in 60-mm dishes with pLNCX2-TAP-Bmi-1 and pLNCX2-TAP-Mel-18 using Lipofectamine 2000 reagent (Invitrogen). Twenty-four hours posttransfection, cells were trypsinized and counted. One thousand cells were plated onto 60-mm dishes in triplicate. Following 7 to 10 days of incubation in G418 (1 mg/ml)-containing media, colonies were fixed, stained with crystal violet, and counted using ImagePro software (MediaCybernetics). For Rat1 stable-line generation, plasmids were linearized prior to Lipofectamine 2000-mediated transfection, followed by G418 selection for 7 days.
For colony formation assays and phenotypic evaluation of stable shRNA-mediated knockdown in cancer cells, transduced cells were seeded in six-well dishes in triplicate (750 cells/well) and incubated for 7 days in the presence of puromycin. Colonies were fixed, stained, and counted as described above. Cell proliferation was measured using the MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium] assay as supplied in the CellTiter 96 Aqueous One Solution Cell Proliferation kit (Promega). Briefly, 500 to 1,000 cells were seeded into 96 wells in replicates of six. The media were exchanged 24 h later for either 10% or 0.5% FBS, as indicated. The first MTS assay was performed on the same day (day 1) and then repeated on days 3, 5, and 7. For soft-agar assays, stable cell lines were seeded in 0.4% agar overlaid on top of 1% agar and incubated for 14 to 21 days. Fresh medium containing an appropriate selection antibiotic was added every 4 to 5 days. The resulting colonies were stained with Hoechst 33342 (1 μg/ml) and photographed by fluorescence microscopy (Nikon). Colonies were counted using ImagePro software.
Apoptosis and cell cycle analysis.
shRNA-expressing DAOY cells (50,000) were plated in triplicate onto six-well dishes and incubated for 5 days. The media and the cells were harvested, fixed in 70% ice-cold ethanol, and stained with propidium iodide (50 μg/ml) in the presence of RNase (100 μg/ml). Ten thousand cells were analyzed by fluorescence-activated cell sorting (FACS).
Western blot analysis.
Protein expression was analyzed by Western blotting as previously described (48). Briefly, cells were harvested by scraping them into cold PBS and lysed using RIPA buffer (Teknova) supplemented with protease and phosphatase inhibitor cocktails. Following sodium dodecyl sulfate-polyacrylamide gel electrophoresis, the proteins were transferred onto nitrocellulose membranes and analyzed using the following antibodies: anti-Bmi-1 (229F6; Upstate), anti-Mel-18 (sc10744; Santa Cruz), and anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (Novus Biologicals).
Gene expression analysis.
Total RNA was isolated using affinity resin (QIAGEN RNeasy Mini Kit; QIAGEN AG). RNA integrity and purity were assessed with the RNA 6000 Nano LabChip system on a Bioanalyzer 2100 (Agilent Technologies). Gene array analysis was conducted using Gene Chip Human Genome 133 2.0 Plus Expression Array (Affymetrix Inc.). A detailed description of the gene array experimental procedure and statistical analysis of gene array data are available in the supplemental material.
Quantitative RT-PCR.
Expression of mRNA was measured using TaqMan Gene Expression Assays on an ABI PRISM 7900 HT Sequence Detection System (Applied Biosystems). Total RNA was isolated from samples with RNAeasy columns (QIAGEN) and treated with DNase prior to cDNA amplification. cDNA was synthesized according to the manufacturer's conditions (iScript; Bio-Rad). Reverse transcription (RT)-PCR primers and 6-carboxyfluorescein-labeled target gene probes were purchased from Applied Biosystems. The ABI 7900HT sequence detection system was used under normal amplification conditions for 40 cycles. VIC-MGB B-Action control primers and probe were coamplified as internal controls. All data were collected in triplicate and normalized to β-actin levels. Relative mRNA expression is given by the formula 2−(CT of sample − CT of β-actin), where CT (cycle count) is the threshold cycle value.
Tumor xenografts.
All animals were handled in strict accordance with the internal, local, state, and federal regulations governing the use of laboratory animals in research. DAOY cells (2 × 106) were seeded in 10 T-150 flasks for each condition and transduced with appropriate shRNA lentiviruses. Following selection in 1 μg/ml of puromycin for 4 days and confirmation of knockdown by Western blotting, 107 shRNA-expressing cells were subcutaneously injected into the right flanks of 4- to 6-week-old nude female mice (eight mice per group). Satisfactory implantation was confirmed by the formation of a well-localized skin hillock. Tumors were measured every week with calipers. On day 54 postimplantation, all animals were euthanized, and each resulting tumor was excised and cleaned of external connective tissue. A portion of each tumor was fixed in 4% buffered formalin for histology analysis or snap-frozen in liquid nitrogen for protein extraction and Western blotting.
Histology and immunohistochemistry analysis of tumor xenografts.
Eight animals, two from each of the four knockdown groups, were examined. Animals were selected for examination based on a high degree of knockdown on Western blotting. Formalin-fixed tumors were embedded in paraffin, and 5-μm-thick sections were stained with hematoxylin and eosin or Masson's trichrome stain (Polyscientific Research and Development Inc.). The Chemicon ApopTag Peroxidase In Situ Apoptosis Detection Kit was used for apoptosis evaluation (terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling [TUNEL] staining). Von Willebrand factor staining was performed using rabbit anti-von Willebrand factor from AbCam laboratories. Ki-67 immunohistochemistry was performed using rabbit anti-Ki67 antibody from Vector Laboratories. Image analysis for TUNEL and Ki-67 was performed on the Aperio Image Analysis system. All histology results were evaluated by a board-certified pathologist (R.M.).
Microarray data accession number.
The microarray data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus under accession number GSE 7578.
RESULTS
Bmi-1 and Mel-18 compose nearly identical core protein complexes.
Bmi-1 is believed to exert its biological effects in the context of the multiprotein PRC1. Although Mel-18 has also been detected in Drosophila PRC1, the constituents of the Mel-18 complex in human cells are presently unknown. Moreover, although a number of PRC1 members have been identified by a variety of indirect means (e.g., two-hybrid screening and coimmunoprecipitation studies), comparative evaluation of the compositions and biological roles of such complexes in mammalian cells has not been carried out.
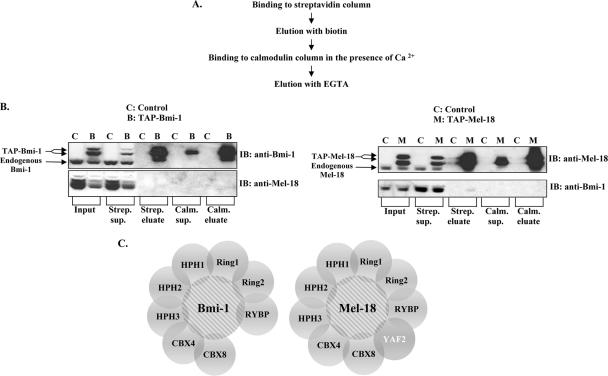
Using the TAP method, we sought to comprehensively compare the compositions of Bmi-1 and Mel-18 multiprotein complexes purified from human cancer cells. Briefly, Bmi-1 and Mel-18 cDNAs were cloned into a vector containing tandem streptavidin- and calmodulin-binding peptide sequences, resulting in N-terminally tagged TAP-Bmi-1 and TAP-Mel-18 fusion proteins. Complexes containing either the TAP-Bmi-1 or TAP-Mel-18 fusion protein were purified from whole-cell lysates of stably expressed HeLa cell lines, according to the scheme depicted in Fig. 1A. Western blot analysis of samples taken at various steps of purification indicated significant (approximately 30-fold) enrichment for both Bmi-1 and Mel-18 in the final fraction (Fig. 1B). Coomassie blue staining revealed a number of proteins that coeluted with Bmi-1 or Mel-18 but were absent from the control sample (data not shown). Mass spectrometry analysis of these eluted samples identified several peptides that were present with high confidence in the Bmi-1 or Mel-18 complexes but not in the negative control sample (see Table S1 in the supplemental material). These results enabled us to construct a putative protein interaction map for Bmi-1 and Mel-18 (Fig. 1C).
FIG. 1.
Bmi-1 and Mel-18 share multiple constituents of their respective protein complexes isolated from human cancer cells. (A) Outline of the TAP procedure used to isolate Bmi-1 and Mel-18 protein complexes from HeLa cells stably expressing TAP-tagged Bmi-1 and Mel-18. (B) Bmi-1 (left) and Mel-18 (right) were followed throughout TAP by Western blotting (IB) using the indicated antibodies. TAP-tagged proteins migrate slightly above their endogenous counterparts. (C) Graphic representation of Bmi-1 and Mel-18 putative protein complexes following mass spectrometry-based identification of their respective constituents. YAF2 is unique to the Mel-18 protein complex.
Our data show that Bmi-1 and Mel-18 appear to be present in nearly identical but distinct protein complexes, consisting of a number of proteins previously identified as direct interactors with Bmi-1 (i.e., Ring1, Ring2, Hph1, Hph2, and Cbx8) or as proteins that bind these interacting proteins (i.e., RYBP, YAF2, and Cbx4), thus potentially indirectly associating with Bmi-1 or Mel-18. Also, since no dedicated effort to identify Mel-18-interacting proteins has been undertaken previously, it is striking to observe that although Bmi-1 and Mel-18 appear to associate with almost indistinguishable repertoires of proteins, they are not found in a complex with each other, suggesting that although these proteins may have overlapping or complementary functions, they exist in independent, distinguishable complexes. In addition, YAF2, a YY1-interacting protein, was found exclusively in the Mel-18 but not the Bmi-1 complex.
Bmi-1 and Mel-18 overexpression enhances proliferation and survival of Rat1 fibroblasts.
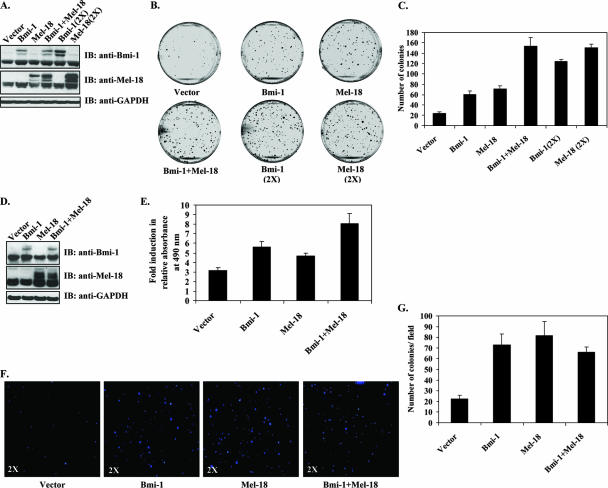
Proteomic analysis revealed that Bmi-1 and Mel-18 assemble largely identical protein complexes comprised of core PRC1 constituents. The identification of these two nearly identical complexes raised the possibility that Bmi-1 and Mel-18 might have significantly overlapping functions in oncogenesis. To test this hypothesis, Bmi-1 and Mel-18 were transiently overexpressed in Rat1 fibroblasts either alone or in combination. Exogenous protein expression was confirmed by Western blotting (Fig. 2A). When plated at low density, Rat1 cells transiently expressing either Bmi-1 or Mel-18 formed more colonies than empty-vector-expressing cells. The number of colonies was dose dependent with respect to the amount of transfected Bmi-1- or Mel-18-expressing plasmid. Furthermore, Bmi-1 and Mel-18 were able to substitute for each other in causing this phenotype, since concomitant expression of both Bmi-1 and Mel-18 resulted in nearly identical numbers of colonies, double the amount of transfected Bmi-1 or Mel-18 alone (Fig. 2B, C).
FIG. 2.
Overexpression of Bmi-1 and Mel-18 enhances proliferation and survival of Rat1 fibroblasts. (A) Western blot (IB) analysis of Rat1 fibroblasts transiently transfected with Bmi-1 or Mel-18 as indicated. Exogenous proteins appear above endogenous Bmi-1 and Mel-18 due to the presence of a TAP tag. (B) Transiently transfected Rat1 cells were plated onto 60-mm dishes and cultured for 10 days in the presence of antibiotic selection (1 mg/ml of G418), and the resulting colonies were stained with crystal violet. Representative images are shown. (C) Colonies were counted using ImagePro software (triplicates; means plus standard deviations). (D) Stable Bmi-1- or Mel-18-expressing Rat1 cell lines were generated by transfection of linearized plasmid DNA, followed by G418 selection until no untransfected cells remained. Expression of exogenous proteins was confirmed by Western blotting. (E) Survival of stable cell lines in 0.1% FBS-containing media was assessed by the MTS assay. The results are shown as induction (n-fold) in relative cell numbers on day 10 over day 1 (replicates of six; means plus standard deviations). (F) Stable Rat1 cell lines were seeded in soft agar and incubated for 21 days. Colonies were visualized with Hoechst 33342 viability dye. Images of representative wells are shown at ×2 magnification. (G) Soft agar colonies were counted in triplicate wells using ImagePro software (means of 18 visual fields per sample plus standard deviations are shown).
We next asked if stable expression of Bmi-1 or Mel-18 could confer resistance to growth factor withdrawal in Rat1 fibroblasts. Rat1 cell lines stably expressing exogenous Bmi-1 or Mel-18 alone or in combination were generated (Fig. 2D) and cultured for 10 days in 0.1% serum. As shown in Fig. 2E, exogenous Bmi-1 or Mel-18 alone only marginally improved cell survival in growth factor-depleted medium. However, simultaneous expression of both Bmi-1 and Mel-18 afforded significant proliferative advantage in Rat1 cells under low-serum conditions.
Finally, to determine if overexpression of Bmi-1 or Mel-18 could allow anchorage-independent growth, a hallmark of transformation, Rat1 fibroblasts stably expressing Bmi-1, Mel-18, or both were seeded in semisolid medium. After a 3-week incubation period, a greater number of small colonies was detectable in Bmi-1- or Mel-18-expressing cells than in vector-expressing controls (Fig. 2F and G). Simultaneous expression of Bmi-1 and Mel-18 did not result in greater numbers of colonies than either protein overexpressed alone.
These data demonstrate that overexpression of either Bmi-1 or Mel-18 results in similar growth-enhancing phenotypes in Rat1 cells, thus further implying functional homology between the two proteins. Furthermore, in contrast to previously reported tumor-suppressive properties, these results strongly suggest an oncogenic function for Mel-18.
Stable shRNA-mediated knockdown of Bmi-1 and Mel-18 inhibits cancer cell growth in vitro but is not universally cytotoxic.
Given extensive commonality of constituents in the Bmi-1 and Mel-18 protein complexes and similarity of phenotypes observed upon their overexpression in Rat1 cells, we hypothesized that Bmi-1 and Mel-18 might also have overlapping roles in the maintenance of the tumorigenic phenotype in cancer cells.
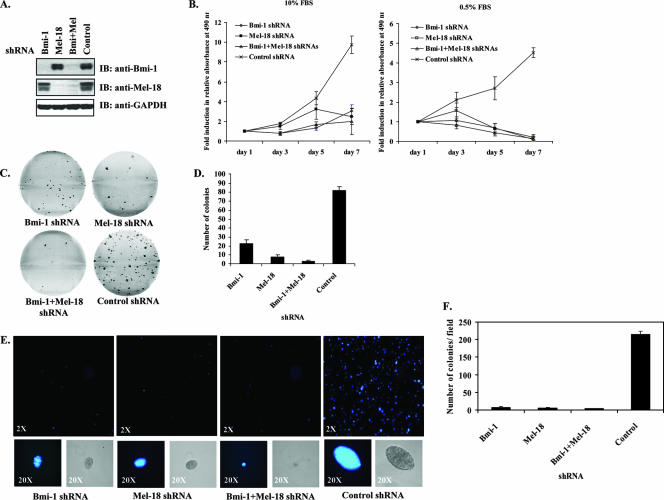
To investigate this, we aimed to downregulate Bmi-1 and Mel-18 expression using shRNA in a number of cancer cell lines. Bmi-1 has been previously implicated in the pathogenesis of medulloblastomas and other cancers. In contrast, little is known about the role of Mel-18 in tumor cell growth. We therefore utilized lentiviral shRNA to stably knock down Bmi-1 and Mel-18 expression in DAOY human medulloblastoma cells either separately or in combination. Levels of Bmi-1 and Mel-18 proteins were significantly reduced in stable shRNA-expressing DAOY cells, whereas an irrelevant control shRNA had no such effect (Fig. 3A). Knockdown of either Bmi-1 or Mel-18 resulted in the inhibition of cell proliferation in 10% FBS-containing media; this effect was further accentuated under reduced-serum conditions (Fig. 3B). In a clonogenic survival assay, Bmi-1 or Mel-18 knockdown resulted in decreased colony formation, with the combination of Bmi-1 and Mel-18 shRNAs having the most pronounced effect (Fig. 3C and D). Furthermore, while negative control shRNA-expressing cells formed numerous large, multicellular clusters in soft agar, Bmi-1 and Mel-18 shRNA-transduced colonies were fewer and, on average, smaller (Fig. 3E and F). To rule out off-target effects, phenotypic analysis was also confirmed in DAOY cells, using additional independent shRNA sequences that effectively abrogated Bmi-1 and Mel-18 expression (see Fig. S1 in the supplemental material).
FIG. 3.
shRNA-mediated knockdown of Bmi-1 and Mel-18 inhibits growth of DAOY medulloblastoma cells. (A) Knockdown of Bmi-1 and Mel-18 was evaluated in DAOY cells stably transduced with the indicated lentiviral shRNA by Western blotting (IB) using the indicated antibodies. (B) shRNA-expressing DAOY cells were cultured in 96-well plates (500 cells/well in replicates of six) in either 10% or 0.5% FBS-containingmedium for 7 days. Cell proliferation was assessed over time by the MTS assay. The graphs depict induction (n-fold) in relative absorbance at 490 nm at each indicated time point relative to the initial reading on day 1 (six replicates; means ± standard deviations). (C) The colony formation ability of shRNA-expressing DAOY cells was examined by plating the cells in triplicate at low density (750 cells/well) onto six-well plates. Following a 7- to 8-day incubation period, the resulting colonies were stained by crystal violet. Representative images are shown. (D) Colonies were counted using ImagePro software (triplicates; means plus standard deviations). (E) The anchorage-independent growth of shRNA-expressing DAOY cells was assessed in the soft-agar assay as described in Materials and Methods. Following a 21- to 25-day incubation period, colonies were stained with Hoechst 33342 viability dye and photographed using a camera attached to a fluorescence microscope. Representative wells are shown at ×2 magnification; examples of individual colonies are shown at ×20 as both phase-contrast (right) and fluorescent (left) images. (F) Soft-agar colonies were counted using ImagePro software (each sample in triplicate; means of 18 visual fields per sample plus standard deviations are shown).
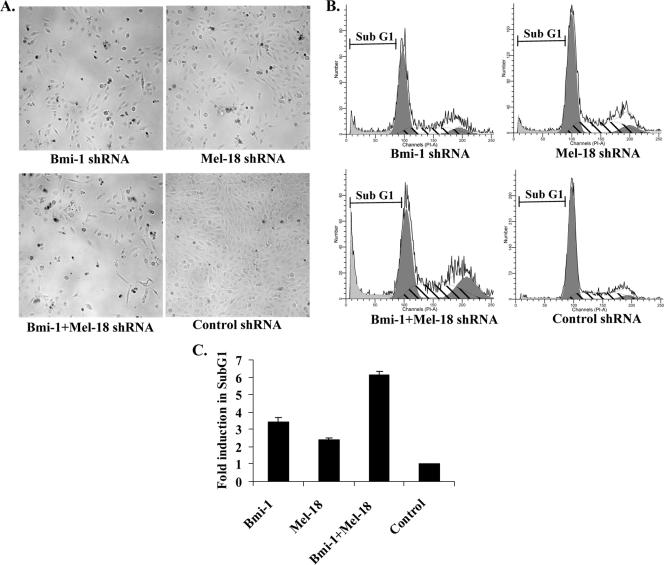
shRNA-mediated knockdown of Bmi-1 or Mel-18 in DAOY cells, alone or in combination, resulted in phenotypes associated with cellular growth retardation and cell death, such as cell rounding and membrane blebbing (Fig. 4A). The effects of Bmi-1 and Mel-18 knockdown in DAOY cells were also analyzed by FACS to assess cell cycle and apoptosis. Individual knockdown of either Bmi-1 or Mel-18 resulted in a two- to threefold increase in the sub-G1 population in DAOY cells, indicative of apoptosis. Simultaneous abrogation of both Bmi-1 and Mel-18 expression caused further increase in the apoptotic fraction, up to sixfold compared with negative controls (Fig. 4B and C).
FIG. 4.
Abrogation of Bmi-1 and Mel-18 expression causes apoptotic cell death of DAOY cells. (A) DAOY cells stably transduced with the indicated shRNAs were plated onto six-well dishes (50,000 cells/well) and incubated for 5 days. Phase-contrast photographs of representative wells are shown. (B) Both the adherent and detached cells were harvested, fixed, and stained with propidium iodide (50 μg/ml in PBS) in the presence of RNase. Cell cycle profiles were obtained by FACS analysis. Cells in the sub-G1 population represent the apoptotic fraction, as shown in representative FACS profiles. (C) Induction in the sub-G1 population was calculated for every sample relative to control shRNA (triplicates; means plus standard deviations).
We also examined the effects of Bmi-1 and Mel-18 knockdown on the growth of additional cancer cell lines, as well as untransformed human lung fibroblasts. Since Bmi-1 and Mel-18 have recently been shown to be coupregulated in primary breast cancers (40), we generated MCF7 human breast carcinoma cell lines stably expressing shRNAs targeting Bmi-1 and Mel-18 alone or in combination. Similar to the effects observed in DAOY cells, knockdown of either Bmi-1 or Mel-18 inhibited proliferation, clonogenic survival, and anchorage-independent growth of MCF7 cells. Simultaneous suppression of Bmi-1 and Mel-18 by shRNA consistently had a more dramatic effect on MCF7 cell growth than individual knockdowns (see Fig. S2 in the supplemental material).
The phenotypic effects of Bmi-1 and Mel-18 knockdown in SK-OV-3 human ovarian carcinoma and U2OS human osteosarcoma cell lines were also investigated. The growth and survival of these cells in a colony formation assay were not significantly affected by loss of either Bmi-1 or Mel-18 expression, whereas simultaneous suppression of both Bmi-1 and Mel-18 resulted in approximately 50% reduction in colony numbers (see Fig. S3A and B in the supplemental material). It must be noted, however, that residual Bmi-1 protein levels were slightly higher in U2OS and SK-OV-3 shRNA-expressing cell lines than in DAOY or MCF7 knockdown cells, which might account for differences in the observed phenotypes. Importantly, the growth of normal lung WI-38 fibroblasts was not significantly affected by nearly complete abrogation of Bmi-1 or Mel-18 expression (see Fig. S3C in the supplemental material).
Taken together, these results indicate that while it had no discernible effect on the growth of normal human fibroblasts, loss of Bmi-1 and Mel-18 expression was deleterious to the growth and survival of multiple cancer cell lines. However, the cancer cell lines examined here varied in their sensitivities to Bmi-1 or Mel-18 depletion, perhaps due to different degrees of cellular dependence on Polycomb proteins and the ability of Bmi-1 and Mel-18 to substitute for each other in certain cellular contexts. Nonetheless, all cancer cell lines tested during the course of this study were sensitive to simultaneous knockdown of both Bmi-1 and Mel-18.
Bmi-1 and Mel-18 knockdown causes global changes in gene expression.
Although the repertoire of genes regulated by PRC proteins in murine and human embryonic stem cells has recently been extended beyond Hox clusters and now includes multiple developmental regulators (5, 6, 28), such comprehensive determination of gene expression patterns mediated by Bmi-1 and Mel-18 in cancer cells has not been undertaken. To better understand the phenotypes associated with Bmi-1 and Mel-18 depletion and to identify biological processes regulated by these Polycomb proteins in medulloblastoma cells, we surveyed gene expression changes in DAOY shRNA-expressing cells using an Affymetrix gene chip (HG-U133_Plus_2) containing approximately 55,000 gene probes, accounting for nearly the entire human genome.
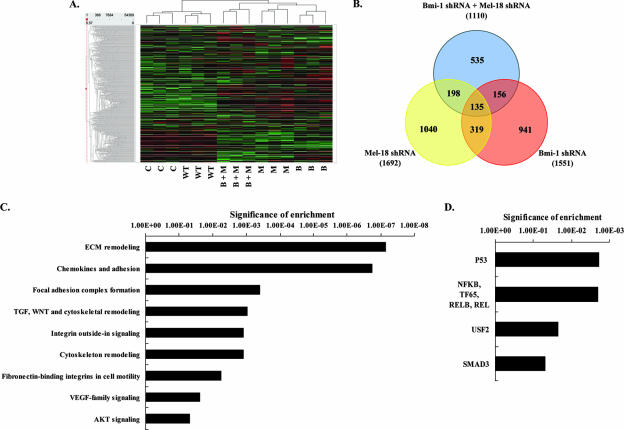
Hierarchical clustering of transcription expression profiles revealed that wild-type (untransduced) and control shRNA-transduced cell lines subclustered relative to shRNA-Bmi-1 and shRNA-Mel-18 cell lines. All technical replicates also subclustered together (Fig. 5A). Further statistical analysis revealed significant changes in expression (either up- or downregulation) of 1,551 genes in the Bmi-1 shRNA sample, 1,692 genes in the Mel-18 shRNA sample, and 1,110 genes in the Bmi-1 plus Mel-18 shRNA sample compared to either control shRNA or wild-type controls. A significant change in expression was defined as a P value of ≤0.05 for the Benjamini Hochberg (BH) false-discovery rate-adjusted t test score (26). Notably, no significant differences in gene expression were detected between wild-type and control shRNA-expressing cells (all P values were determined to be ≥0.2 for the BH adjusted t test score) (data not shown). Strikingly, there was a highly significant (P = 6.7 × 10−167) overlap among the genes regulated by Bmi-1 and Mel-18: 27% of Bmi-1-regulated genes were also found to be regulated by Mel-18, whereas 29% of Mel-18-regulated genes appeared to be coregulated by Bmi-1 (Fig. 5B). These results support the notion of potential functional redundancy between these Polycomb homologues.
FIG. 5.
Transcriptional profiling reveals significant overlap in Bmi-1- and Mel-18-regulated genes in medulloblastoma and identifies novel biological processes under their control. (A) Hierarchical clustering of gene expression profiles of wild-type (WT) DAOY cells or DAOY stable lines expressing control shRNA (C) or shRNAs targeting Bmi-1 (B) and Mel-18 (M) (each group in triplicate). (B) Venn diagram showing the overlap in significantly regulated genes in Bmi-1, Mel-18, and Bmi-1 plus Mel-18 shRNA samples. The numbers in parentheses represent the total number of significantly regulated genes in each category. Overlap was significant for all groups (P ≤ 0.0001 for all comparisons). Genes that were significantly upregulated upon Bmi-1, Mel-18, and Bmi plus Mel shRNA-mediated knockdown in DAOY cells were categorized by their GO biological pathways (C) and transcription factor family (D) terms. Detailed description of categorization procedures and full categorization tables are included in the supplemental material. The significance of enrichment (the y axis) represents log(P [BH]).
Given the well-established role of Polycomb proteins as transcriptional repressors, we further focused on genes whose expression levels were increased upon Bmi-1 and Mel-18 knockdown. In order to identify biological processes that are controlled by Bmi-1 and Mel-18 in DAOY cells, all significantly upregulated genes in Bmi-1, Mel-18, and Bmi-1 plus Mel-18 shRNA samples (significance was defined by a P value [BH] of ≤0.05, regardless of induction) were combined and categorized by their gene ontology (GO) pathways and transcription factor terms. As shown in Fig. 5C (see also Tables S2 and S3 in the supplemental material), Bmi-1 and Mel-18 appear to regulate a significant number of genes associated with extracellular matrix (ECM) remodeling, cell adhesion, and integrin-mediated signaling. In addition, Bmi-1 and Mel-18 knockdown perturbed the expression of constituent members of the TGF, WNT, VEGF, and AKT signaling pathways and revealed an overlap with gene expression-regulatory patterns associated with canonical transcription factors, such as p53, NF-κB, USF2, and SMAD (Fig. 5D).
Among the genes that displayed significant induction upon Bmi-1 or Mel-18 knockdown (twofold or greater; P value [BH] ≤ 0.05) were a number of important developmental regulators and differentiation factors (e.g., TAGLN, NRP2, NAV2, BMP5, TGFB2, NOTCH2, HNT, VANGL1, and DKK1 in Bmi-1 shRNA-expressing cells and HHIP, JAG1, HNT, LTBP4, GAL, WNT5B, NFATC2, ODZ2, DKK3, NRP2, NRP1, NAV2, BMP1, LIF, NOTCH2, FZD8, VANGL1, and TGFB2 in Mel-18 shRNA-expressing cells) (see Table S4 in the supplemental material), further suggesting that Bmi-1 and Mel-18 maintain undifferentiated, “stem-like” cellular states and that abrogation of their activity results in the induction of differentiation programs.
To further confirm the results of gene expression profiling, we used quantitative RT-PCR (qRT-PCR) to assess changes in the expression of a number of upregulated genes, including tissue inhibitor of matrix metalloproteinase 3 (TIMP-3), hedgehog interacting protein (HHIP), and inhibin A (INHBA) genes (see Fig. S4 in the supplemental material). Upregulation of these genes was also confirmed by qRT-PCR in an independent experiment in DAOY cells expressing Bmi-1 or Mel-18 shRNA (data not shown), thus further validating the microarray data. Preliminary chromatin immunoprecipitation experiments suggested that Bmi-1 and Mel-18 may interact directly with some of the genes identified as targets of regulation by gene expression analysis (data not shown).
Downregulation of Bmi-1 and Mel-18 suppresses the growth of human medulloblastoma xenografts in vivo.
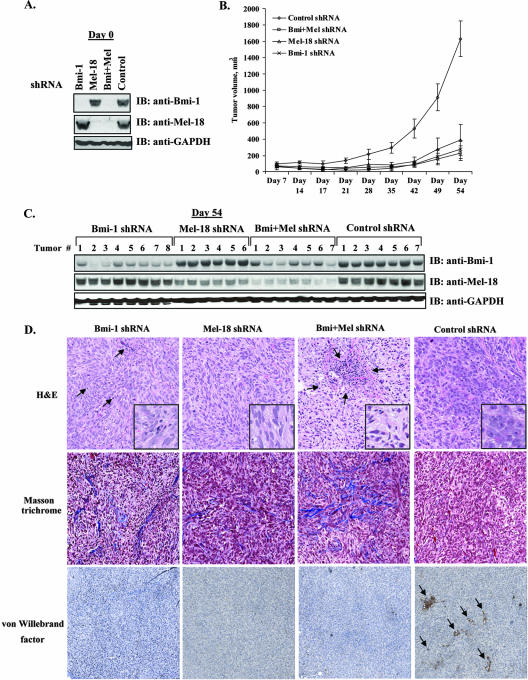
Our results have demonstrated that knockdown of either Bmi-1 or Mel-18 negatively affects the growth of DAOY and other cancer cell lines in vitro. In order to ascertain their roles in medulloblastoma growth in vivo, 2 × 106 DAOY cells were transduced with Bmi-1 or Mel-18 shRNA, followed by rapid puromycin selection for 4 days prior to implantation. This approach was designed to circumvent the need to substantially expand transduced cells in order to have enough cells for implantation. Following confirmation of satisfactory knockdown in the resulting stable lines by Western blotting (Fig. 6A) and qRT-PCR (data not shown), cells were harvested and implanted subcutaneously into nude mice. Tumor growth was monitored by weekly caliper measurement of tumor volume. DAOY xenografts in which the expression of Bmi-1 or Mel-18 was suppressed by shRNA exhibited substantially slower growth than their control counterparts over the 54-day observation period (Fig. 6B). Moreover, Western blot analysis of tumor tissue harvested from animals upon termination of the experiment revealed persistent, albeit reduced, downregulation of Bmi-1 and Mel-18, thus confirming that the observed differences in tumor growth were due to suppression of Polycomb protein expression (Fig. 6C). In addition, since the tumors were not microdissected and both anti-Bmi-1 and anti-Mel-18 antibodies recognize human and mouse proteins, we cannot rule out the possibility that mouse tissue contamination may account for some of the Bmi-1 and Mel-18 protein expression observed.
FIG. 6.
Downregulation of Bmi-1 and Mel-18 by specific shRNA suppresses medulloblastoma xenograft growth in vivo. (A) Western blot (IB) analysis of stable DAOY cell lines expressing control shRNA or shRNAs against Bmi-1, Mel-18, or both prior to implantation into nude mice. (B) Cells (1 × 107) from each knockdown group were implanted subcutaneously in female nude mice (eight animals per group). Tumors failed to grow in two animals in the “Mel-18 shRNA” group and in one animal each in the “Bmi plus Mel shRNA” and “control shRNA” groups. The growth of all other xenografts was monitored every week with calipers. Shown are means ± standard deviations of tumor volumes. (C) Animals were sacrificed on day 54 postimplantation, and total tissue extracts from all resulting tumors were analyzed by Western blotting (IB) for Bmi-1 and Mel-18 knockdown. (D) Xenograft tissues were also sectioned and stained with hematoxylin and eosin (H&E; top row). The arrows in the Bmi-1 shRNA sample indicate examples of inflammatory cells found throughout the tumor. Well-defined areas of inflammatory infiltrate were also found in the Bmi-plus-Mel shRNA xenografts, as indicated by arrows. The inset images are shown at higher magnification to highlight morphological differences between control shRNA and Bmi-1 and Mel-18 shRNA tumor xenografts. Masson's trichrome, which stains collagen fiber blue and cytoplasm, keratin, and muscle red, was used to reveal increased collagen deposition in the Bmi-1 and Bmi-plus-Mel knockdown tumors (middle panels). Anti-von Willebrand factor immunostaining was used to identify intratumoral blood vessels, as indicated by arrows (lower panels).
Representative xenografts were analyzed using established histological and immunohistochemical techniques. Hematoxylin and eosin staining revealed that control shRNA xenografts had plump epithelioid cells with focal spindling. In contrast, xenografts with either individual or combined knockdown of Bmi-1 and Mel-18 showed an increased number of cells with angular, hyperchromatic nuclei and contained areas of focal and diffused mixed chronic inflammatory infiltrate, which could be indicative of past apoptotic events (18) (Fig. 6D, top row). Quantitative analysis of the Ki-67-positive nuclei and TUNEL staining for apoptosis revealed no statistically significant differences in the percentages of positive cells between the control and experimental groups (data not shown). However, it is likely that by the end point of this experiment (54 days postimplantation), most affected cells had undergone apoptosis, consistent with both the growth effects observed in vitro and with the histological features described above.
Gene expression analysis identified ECM remodeling as one of the major biological processes regulated by Bmi-1 and Mel-18 in DAOY cells. We therefore utilized Masson's trichrome stain to evaluate extracellular collagen contents in shRNA-expressing tumors. Consistent with the results of gene expression profiling, Masson's stain revealed a moderate increase in collagen deposition in Bmi-1 and Bmi-1 plus Mel-18 knockdown tumors (Fig. 6D, second row). In addition, we hypothesized that Polycomb knockdown might affect neovascularization of tumor xenografts, since the gene chip analysis identified significant changes in the expression of a number of angiogenesis regulators in DAOY cells. von Willebrand factor staining for intratumoral blood vessels revealed different patterns of vascularization in control shRNA and Bmi-1 and Mel-18 shRNA xenografts. Control shRNA samples showed von Willebrand factor immunoreactivity across the entire section, associated with both presumed vascular spaces; the Bmi-1 and Mel-18 knockdown tumors exhibited an overall reduction in von Willebrand factor staining structures (Fig. 6D, third row).
Collectively, these data show that shRNA-mediated downregulation of Bmi-1 and Mel-18 results in significant inhibition of medulloblastoma xenograft growth in vivo. In addition to quantitative differences in rates of progression, Bmi-1 and Mel-18 shRNA-expressing tumors appear to be morphologically different from their control counterparts, display alterations in collagen deposition, and are less vascularized. These results are consistent with our gene expression profiling observations and suggest that Bmi-1 and Mel-18 contribute to both tumor cell growth and tumor-stromal interactions.
DISCUSSION
This study was designed to characterize the relative contributions of the Polycomb homologue genes Bmi-1 and Mel-18 to cancer cell growth in vitro and in vivo. Bmi-1 has been proposed to play a role in the pathogenesis of several human cancers, including medulloblastoma, breast cancer, and hematologic malignancies. The role of its close structural homologue Mel-18 in tumorigenesis has been unclear. We utilized several human tumor cell lines, including DAOY medulloblastoma cells, to intensively investigate the consequences of Bmi-1 and Mel-18 shRNA-mediated downregulation both in vitro and in vivo. The DAOY cell line was chosen as a representative medulloblastoma cell line because previously described comparative gene expression profiling had demonstrated that the expression profile of DAOY cells closely resembles that of human medulloblastoma tumors (32).
shRNA-mediated depletion of either Bmi-1 or Mel-18 had a significant effect on DAOY cell growth, which made it difficult to detect any additive or synergistic effects upon simultaneous knockdown of both Bmi-1 and Mel-18. However, we did consistently observe a slight but reproducible further decrease in DAOY proliferation and colony formation when both Bmi-1 and Mel-18 were downregulated. A similar response was noted in MCF7 cells upon simultaneous knockdown of Bmi-1 and Mel-18. In contrast, SK-OV-3 and U2OS cells were refractory to either Bmi-1 or Mel-18 knockdown but exhibited growth retardation phenotypes when both genes were downregulated. These data imply that the consequences of Bmi-1 and Mel-18 downregulation might vary depending on the cell lineage or other cellular factors and that Bmi-1 and Mel-18 might (at least in certain cells) perform overlapping functions. Importantly, normal human WI38 fibroblasts were insensitive to either individual or combined downregulation of Bmi-1 and Mel-18. These data are consistent with a recently published description of an acute small interfering RNA-mediated knockdown of Bmi-1, which resulted in apoptosis in cancer cells but not in normal cells (31).
The increase in apoptosis that was observed in DAOY cells upon shRNA-mediated knockdown of Bmi-1 and Mel-18 is similar to previously noted moderate cell death in Bmi-1-null hematopoietic stem cells (20) and Mel-18-null early T-cell progenitors (33). In addition, combined Bmi-1/Mel-18 shRNA-mediated knockdown resulted in substantial apoptosis in DAOY cells, akin to the previously reported cell death phenotype in double-null Bmi-1/Mel-18 embryos (3). Apoptosis in Bmi-1−/− hematopoietic stem cells has been proposed to be independent of p19/ARF induction, since concurrent p19/ARF ablation did not rescue these cells. We have examined the expression status of p16 and p19 in DAOY and other cancer cell lines utilized in this study following Bmi-1 or Mel-18 knockdown and failed to observe any appreciable increase in the levels of these tumor suppressors (data not shown). These observations, as well as previous studies indicating that the p16 promoter is methylated and p19 is altogether deleted in DAOY cells (14, 19), suggest that silencing of this locus in cancer cells may occur via mechanisms that do not require Bmi-1. Furthermore, in previous studies of human mantle cell lymphomas with Bmi-1 gene amplifications and a wild-type INK4A/ARF locus, no correlation was observed between p16 levels and Bmi-1 overexpression (4). It is possible that distinct regulatory mechanisms may govern the activity of the INK4A/ARF locus during development (or in stem cells) and in cancer cells.
We attempted to identify downstream targets responsible for the observed growth-inhibitory phenotypes in Polycomb shRNA-expressing DAOY cells by carrying out a comprehensive gene-profiling experiment. Microarray analysis and GO categorization of significantly upregulated genes revealed that both Bmi-1 and Mel-18 appear to regulate a large number of genes involved in ECM remodeling and cell adhesion. A number of such genes have additional well-documented roles in tumor cell proliferation and survival in vitro and in vivo. One such example is TIMP3, which, while acting primarily as an inhibitor of matrix metalloproteinases, also induces apoptosis in tumor cells through the death receptor pathway (1) and inhibits tumor vascularization in vivo via direct competition with VEGF for its cognate receptor binding (39). Indeed, several other upregulated genes in the Bmi-1 and Mel-18 shRNA-expressing cells are capable of inducing apoptosis in addition to having other functions in the cell (e.g., HHIP and inhibin A). Strikingly, both HHIP and inhibin A have also been shown to serve as bona fide angiogenesis inhibitors in vivo (35, 36, 39). Whether altered expression of these or other genes is directly responsible for the phenotypes observed upon Bmi-1 or Mel-18 knockdown in DAOY cells is currently under investigation.
While the ability of cancer cells to migrate and adhere does play a role in cell survival and growth in semisolid medium in vitro, it is particularly important to tumor maintenance in vivo. Consistent with the observed changes in the expression of genes controlling tumor-stromal interactions, our data showed that loss of Bmi-1 and Mel-18 expression results in an increase in extracellular collagen content and drastically reduced angiogenesis. These findings might also help explain previously reported strong correlation between Bmi-1 overexpression and distant metastases in a variety of human tumors (25).
Several lines of experimental evidence presented here point to functional similarities between Bmi-1 and Mel-18, at least in the context of the cancer cell lines utilized in this study. Bmi-1 and Mel-18 protein complexes isolated by us also showed a striking degree of similarity, suggesting overlapping functions of these proteins. Our data confirm the results of several earlier immunoprecipitation and yeast two-hybrid studies aimed at characterizing the constituents of the mammalian PRC1 complex. In addition, our results are consistent with previously published observations that Bmi-1 and Mel-18 are capable of independently assembling identical protein complexes in vivo from overexpressed PRC1 core constituents (7). We have now shown that this is true for endogenous PRC1 proteins. Since core Polycomb complexes are highly conserved throughout evolution (30), we expect that Bmi-1 and Mel-18 complexes purified by us from HeLa cells are similar to those found in other cell types.
Additional support for the notion of functional redundancy between Bmi-1 and Mel-18 was obtained when the overexpression phenotypes of Bmi-1 and Mel-18 were found to be similar in Rat1 fibroblasts. Consistent with previous reports that both Bmi-1 and Mel-18 can enhance the proliferation of rodent fibroblasts (9, 27), we have now shown that Bmi-1 and Mel-18 can fully substitute for each other in increasing the proliferative potential of Rat1 cells and cooperate in rescuing these cells from death in growth factor-depleted media. Taken together with the results of shRNA-based experiments, these results suggest for the first time an oncogenic function for Mel-18. This is in contrast with previously published observations of increased tumorigenicity of immortal NIH 3T3 fibroblasts upon antisense-mediated Mel-18 downregulation (24). In addition, no epidemiological data currently exist to directly implicate Mel-18 in human tumorigenesis. These observations would justify a more intensive investigation of Mel-18 in primary human tumors.
Supplementary Material
Acknowledgments
We are grateful to Akos Szilvasi, Alan Ho, and Deborah Ahern-Ridlon for expert assistance with flow cytometry and to Helen He, Ron Meyer, and Kim Merriam for assistance with histology analysis. We thank William Sellers and Francesco Hofmann (Novartis Oncology Research) and Peter Howley (Harvard Medical School) for critical review of the manuscript.
D.W. is a Presidential Postdoctoral Fellow at the Novartis Institutes for BioMedical Research.
Footnotes
Published ahead of print on 23 April 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Ahonen, M., M. Poukkula, A. H. Baker, M. Kashiwagi, H. Nagase, J. E. Eriksson, and V. M. Kahari. 2003. Tissue inhibitor of metalloproteinases-3 induces apoptosis in melanoma cells by stabilization of death receptors. Oncogene 22:2121-2134. [DOI] [PubMed] [Google Scholar]
- 2.Akasaka, T., M. Kanno, R. Balling, M. A. Mieza, M. Taniguchi, and H. Koseki. 1996. A role for mel-18, a Polycomb group-related vertebrate gene, during the anteroposterior specification of the axial skeleton. Development 122:1513-1522. [DOI] [PubMed] [Google Scholar]
- 3.Akasaka, T., M. van Lohuizen, N. van der Lugt, Y. Mizutani-Koseki, M. Kanno, M. Taniguchi, M. Vidal, M. Alkema, A. Berns, and H. Koseki. 2001. Mice doubly deficient for the Polycomb Group genes Mel18 and Bmi1 reveal synergy and requirement for maintenance but not initiation of Hox gene expression. Development 128:1587-1597. [DOI] [PubMed] [Google Scholar]
- 4.Bea, S., F. Tort, M. Pinyol, X. Puig, L. Hernandez, S. Hernandez, P. L. Fernandez, M. van Lohuizen, D. Colomer, and E. Campo. 2001. BMI-1 gene amplification and overexpression in hematological malignancies occur mainly in mantle cell lymphomas. Cancer Res. 61:2409-2412. [PubMed] [Google Scholar]
- 5.Boyer, L. A., K. Plath, J. Zeitlinger, T. Brambrink, L. A. Medeiros, T. I. Lee, S. S. Levine, M. Wernig, A. Tajonar, M. K. Ray, G. W. Bell, A. P. Otte, M. Vidal, D. K. Gifford, R. A. Young, and R. Jaenisch. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441:349-353. [DOI] [PubMed] [Google Scholar]
- 6.Bracken, A. P., N. Dietrich, D. Pasini, K. H. Hansen, and K. Helin. 2006. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 20:1123-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao, R., Y. Tsukada, and Y. Zhang. 2005. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol. Cell 20:845-854. [DOI] [PubMed] [Google Scholar]
- 8.Cao, R., L. Wang, H. Wang, L. Xia, H. Erdjument-Bromage, P. Tempst, R. S. Jones, and Y. Zhang. 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298:1039-1043. [DOI] [PubMed] [Google Scholar]
- 9.Cohen, K. J., J. S. Hanna, J. E. Prescott, and C. V. Dang. 1996. Transformation by the Bmi-1 oncoprotein correlates with its subnuclear localization but not its transcriptional suppression activity. Mol. Cell. Biol. 16:5527-5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Czermin, B., R. Melfi, D. McCabe, V. Seitz, A. Imhof, and V. Pirrotta. 2002. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111:185-196. [DOI] [PubMed] [Google Scholar]
- 11.Dellino, G. I., Y. B. Schwartz, G. Farkas, D. McCabe, S. C. Elgin, and V. Pirrotta. 2004. Polycomb silencing blocks transcription initiation. Mol. Cell 13:887-893. [DOI] [PubMed] [Google Scholar]
- 12.Dimri, G. P., J. L. Martinez, J. J. Jacobs, P. Keblusek, K. Itahana, M. Van Lohuizen, J. Campisi, D. E. Wazer, and V. Band. 2002. The Bmi-1 oncogene induces telomerase activity and immortalizes human mammary epithelial cells. Cancer Res. 62:4736-4745. [PubMed] [Google Scholar]
- 13.Francis, N. J., A. J. Saurin, Z. Shao, and R. E. Kingston. 2001. Reconstitution of a functional core polycomb repressive complex. Mol. Cell 8:545-556. [DOI] [PubMed] [Google Scholar]
- 14.Fruhwald, M. C., M. S. O'Dorisio, Z. Dai, S. M. Tanner, D. A. Balster, X. Gao, F. A. Wright, and C. Plass. 2001. Aberrant promoter methylation of previously unidentified target genes is a common abnormality in medulloblastomas—implications for tumor biology and potential clinical utility. Oncogene 20:5033-5042. [DOI] [PubMed] [Google Scholar]
- 15.Gil, J., D. Bernard, and G. Peters. 2005. Role of polycomb group proteins in stem cell self-renewal and cancer. DNA Cell Biol. 24:117-125. [DOI] [PubMed] [Google Scholar]
- 16.Glinsky, G. V., O. Berezovska, and A. B. Glinskii. 2005. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J. Clin. Investig. 115:1503-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haupt, Y., W. S. Alexander, G. Barri, S. P. Klinken, and J. M. Adams. 1991. Novel zinc finger gene implicated as myc collaborator by retrovirally accelerated lymphomagenesis in E mu-myc transgenic mice. Cell 65:753-763. [DOI] [PubMed] [Google Scholar]
- 18.Hoeflich, K. P., D. C. Gray, M. T. Eby, J. Y. Tien, L. Wong, J. Bower, A. Gogineni, J. Zha, M. J. Cole, H. M. Stern, L. J. Murray, D. P. Davis, and S. Seshagiri. 2006. Oncogenic BRAF is required for tumor growth and maintenance in melanoma models. Cancer Res. 66:999-1006. [DOI] [PubMed] [Google Scholar]
- 19.Inda, M. M., J. Munoz, P. Coullin, D. Fauvet, G. Danglot, T. Tunon, A. Bernheim, and J. S. Castresana. 2006. High promoter hypermethylation frequency of p14/ARF in supratentorial PNET but not in medulloblastoma. Histopathology 48:579-587. [DOI] [PubMed] [Google Scholar]
- 20.Iwama, A., H. Oguro, M. Negishi, Y. Kato, Y. Morita, H. Tsukui, H. Ema, T. Kamijo, Y. Katoh-Fukui, H. Koseki, M. van Lohuizen, and H. Nakauchi. 2004. Enhanced self-renewal of hematopoietic stem cells mediated by the polycomb gene product Bmi-1. Immunity 21:843-851. [DOI] [PubMed] [Google Scholar]
- 21.Jacobs, J. J., K. Kieboom, S. Marino, R. A. DePinho, and M. van Lohuizen. 1999. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature 397:164-168. [DOI] [PubMed] [Google Scholar]
- 22.Jacobs, J. J., B. Scheijen, J. W. Voncken, K. Kieboom, A. Berns, and M. van Lohuizen. 1999. Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes Dev. 13:2678-2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacobs, J. J., and M. van Lohuizen. 2002. Polycomb repression: from cellular memory to cellular proliferation and cancer. Biochim. Biophys. Acta 1602:151-161. [DOI] [PubMed] [Google Scholar]
- 24.Kanno, M., M. Hasegawa, A. Ishida, K. Isono, and M. Taniguchi. 1995. mel-18, a Polycomb group-related mammalian gene, encodes a transcriptional negative regulator with tumor suppressive activity. EMBO J. 14:5672-5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, J. H., S. Y. Yoon, S. H. Jeong, S. Y. Kim, S. K. Moon, J. H. Joo, Y. Lee, I. S. Choe, and J. W. Kim. 2004. Overexpression of Bmi-1 oncoprotein correlates with axillary lymph node metastases in invasive ductal breast cancer. Breast 13:383-388. [DOI] [PubMed] [Google Scholar]
- 26.Klipper-Aurbach, Y., M. Wasserman, N. Braunspiegel-Weintrob, D. Borstein, S. Peleg, S. Assa, M. Karp, Y. Benjamini, Y. Hochberg, and Z. Laron. 1995. Mathematical formulae for the prediction of the residual beta cell function during the first two years of disease in children and adolescents with insulin-dependent diabetes mellitus. Med. Hypotheses 45:486-490. [DOI] [PubMed] [Google Scholar]
- 27.Kranc, K. R., S. D. Bamforth, J. Braganca, C. Norbury, M. van Lohuizen, and S. Bhattacharya. 2003. Transcriptional coactivator Cited2 induces Bmi1 and Mel18 and controls fibroblast proliferation via Ink4a/ARF. Mol. Cell. Biol. 23:7658-7666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, T. I., R. G. Jenner, L. A. Boyer, M. G. Guenther, S. S. Levine, R. M. Kumar, B. Chevalier, S. E. Johnstone, M. F. Cole, K. Isono, H. Koseki, T. Fuchikami, K. Abe, H. L. Murray, J. P. Zucker, B. Yuan, G. W. Bell, E. Herbolsheimer, N. M. Hannett, K. Sun, D. T. Odom, A. P. Otte, T. L. Volkert, D. P. Bartel, D. A. Melton, D. K. Gifford, R. Jaenisch, and R. A. Young. 2006. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125:301-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leung, C., M. Lingbeek, O. Shakhova, J. Liu, E. Tanger, P. Saremaslani, M. Van Lohuizen, and S. Marino. 2004. Bmi1 is essential for cerebellar development and is overexpressed in human medulloblastomas. Nature 428:337-341. [DOI] [PubMed] [Google Scholar]
- 30.Levine, S. S., A. Weiss, H. Erdjument-Bromage, Z. Shao, P. Tempst, and R. E. Kingston. 2002. The core of the Polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol. Cell. Biol. 22:6070-6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, L., L. G. Andrews, and T. O. Tollefsbol. 2006. Loss of the human polycomb group protein BMI1 promotes cancer-specific cell death. Oncogene 25:4370-4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacDonald, T. J., K. M. Brown, B. LaFleur, K. Peterson, C. Lawlor, Y. Chen, R. J. Packer, P. Cogen, and D. A. Stephan. 2001. Expression profiling of medulloblastoma: PDGFRA and the RAS/MAPK pathway as therapeutic targets for metastatic disease. Nat. Genet. 29:143-152. [DOI] [PubMed] [Google Scholar]
- 33.Miyazaki, M., H. Kawamoto, Y. Kato, M. Itoi, K. Miyazaki, K. Masuda, S. Tashiro, H. Ishihara, K. Igarashi, T. Amagai, R. Kanno, and M. Kanno. 2005. Polycomb group gene mel-18 regulates early T progenitor expansion by maintaining the expression of Hes-1, a target of the Notch pathway. J. Immunol. 174:2507-2516. [DOI] [PubMed] [Google Scholar]
- 34.Molofsky, A. V., R. Pardal, T. Iwashita, I. K. Park, M. F. Clarke, and S. J. Morrison. 2003. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature 425:962-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olsen, C. L., P. P. Hsu, J. Glienke, G. M. Rubanyi, and A. R. Brooks. 2004. Hedgehog-interacting protein is highly expressed in endothelial cells but down-regulated during angiogenesis and in several human tumors. BMC Cancer 4:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Panopoulou, E., C. Murphy, H. Rasmussen, E. Bagli, E. K. Rofstad, and T. Fotsis. 2005. Activin A suppresses neuroblastoma xenograft tumor growth via antimitotic and antiangiogenic mechanisms. Cancer Res. 65:1877-1886. [DOI] [PubMed] [Google Scholar]
- 37.Park, I. K., D. Qian, M. Kiel, M. W. Becker, M. Pihalja, I. L. Weissman, S. J. Morrison, and M. F. Clarke. 2003. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature 423:302-305. [DOI] [PubMed] [Google Scholar]
- 38.Peters, A. H., J. E. Mermoud, D. O'Carroll, M. Pagani, D. Schweizer, N. Brockdorff, and T. Jenuwein. 2002. Histone H3 lysine 9 methylation is an epigenetic imprint of facultative heterochromatin. Nat. Genet. 30:77-80. [DOI] [PubMed] [Google Scholar]
- 39.Qi, J. H., Q. Ebrahem, N. Moore, G. Murphy, L. Claesson-Welsh, M. Bond, A. Baker, and B. Anand-Apte. 2003. A novel function for tissue inhibitor of metalloproteinases-3 (TIMP3): inhibition of angiogenesis by blockage of VEGF binding to VEGF receptor-2. Nat. Med. 9:407-415. [DOI] [PubMed] [Google Scholar]
- 40.Silva, J., J. M. Garcia, C. Pena, V. Garcia, G. Dominguez, D. Suarez, F. I. Camacho, R. Espinosa, M. Provencio, P. Espana, and F. Bonilla. 2006. Implication of polycomb members Bmi-1, Mel-18, and Hpc-2 in the regulation of p16INK4a, p14ARF, h-TERT, and c-Myc expression in primary breast carcinomas. Clin. Cancer Res. 12:6929-6936. [DOI] [PubMed] [Google Scholar]
- 41.Tagawa, M., T. Sakamoto, K. Shigemoto, H. Matsubara, Y. Tamura, T. Ito, I. Nakamura, A. Okitsu, K. Imai, and M. Taniguchi. 1990. Expression of novel DNA-binding protein with zinc finger structure in various tumor cells. J. Biol. Chem. 265:20021-20026. [PubMed] [Google Scholar]
- 42.Valk-Lingbeek, M. E., S. W. Bruggeman, and M. van Lohuizen. 2004. Stem cells and cancer; the polycomb connection. Cell 118:409-418. [DOI] [PubMed] [Google Scholar]
- 43.van der Lugt, N. M., J. Domen, K. Linders, M. van Roon, E. Robanus-Maandag, H. te Riele, M. van der Valk, J. Deschamps, M. Sofroniew, M. van Lohuizen, et al. 1994. Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev. 8:757-769. [DOI] [PubMed] [Google Scholar]
- 44.van Lohuizen, M., S. Verbeek, B. Scheijen, E. Wientjens, H. van der Gulden, and A. Berns. 1991. Identification of cooperating oncogenes in E mu-myc transgenic mice by provirus tagging. Cell 65:737-752. [DOI] [PubMed] [Google Scholar]
- 45.Vonlanthen, S., J. Heighway, H. J. Altermatt, M. Gugger, A. Kappeler, M. M. Borner, M. van Lohuizen, and D. C. Betticher. 2001. The bmi-1 oncoprotein is differentially expressed in non-small cell lung cancer and correlates with INK4A-ARF locus expression. Br. J. Cancer 84:1372-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, H., L. Wang, H. Erdjument-Bromage, M. Vidal, P. Tempst, R. S. Jones, and Y. Zhang. 2004. Role of histone H2A ubiquitination in Polycomb silencing. Nature 431:873-878. [DOI] [PubMed] [Google Scholar]
- 47.Wang, L., J. L. Brown, R. Cao, Y. Zhang, J. A. Kassis, and R. S. Jones. 2004. Hierarchical recruitment of polycomb group silencing complexes. Mol. Cell 14:637-646. [DOI] [PubMed] [Google Scholar]
- 48.Wiederschain, D., H. Kawai, A. Shilatifard, and Z. M. Yuan. 2005. Multiple mixed lineage leukemia (MLL) fusion proteins suppress p53-mediated response to DNA damage. J. Biol. Chem. 280:24315-24321. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.