Abstract
The cell cycle regulatory retinoblastoma (Rb) protein is a key regulator of neural precursor proliferation; however, its role has been expanded to include a novel cell-autonomous role in mediating neuronal migration. We sought to determine the Rb-interacting factors that mediate both the cell cycle and migration defects. E2F1 and E2F3 are likely Rb-interacting candidates that we have shown to be deregulated in the absence of Rb. Using mice with compound null mutations of Rb and E2F1 or E2F3, we asked to what extent either E2F1 or E2F3 interacts with Rb in neurogenesis. Here, we report that E2F1 and E2F3 are both functionally relevant targets in neural precursor proliferation, cell cycle exit, and laminar patterning. Each also partially mediates the Rb requirement for neuronal survival. Neuronal migration, however, is specifically mediated through E2F3, beyond its role in cell cycle regulation. This study not only outlines overlapping and distinct functions for E2Fs in neurogenesis but also is the first to establish a physiologically relevant role for the Rb/E2F pathway beyond cell cycle regulation in vivo.
Neurogenesis is a highly regulated process by which neural precursors divide and differentiate, giving rise to the cells that make up the nervous system (reviewed in references 24 and 25). While the role of cell cycle genes in regulating proliferation of neural precursor cells is well appreciated, accumulating data point convincingly to their unique roles in regulating diverse cellular processes, independent of cell cycle regulation (reviewed in reference 63). The retinoblastoma (Rb) tumor suppressor is a key cell cycle regulator that we along with others have shown to play a number of roles in neurodevelopment including proliferation, survival, and, more recently, neuronal migration (7, 8, 10, 18, 19, 33, 40, 52). Differentiating Rb-deficient neural precursor cells exhibit delayed cell cycle exit, while the absence of Rb in the telencephalon leads to ectopic proliferation of neural precursor cells and enhanced brain size at midgestation (7, 19, 52). In a recent study we described a role for Rb in regulating the survival of discrete neuronal subpopulations and a novel cell autonomous role for Rb in regulating neuronal migration (18).
The mechanism by which Rb regulates neurogenesis and the extent to which defects in migration and survival are the result of cell cycle deregulation remain unknown. While Rb is known to interact with numerous proteins (reviewed in reference 67), many of which are expressed in quiescent cells or have cell cycle-independent functions, members of the cell cycle regulatory E2F family are likely targets in neurogenesis. The E2F family of transcription factors is comprised of E2Fs 1 to 8; however, E2F1, E2F2, and E2F3, the so-called activating E2Fs, are key Rb-interacting targets best known for their role in promoting cell cycle progression (9, 14, 17, 48, 49, 54; reviewed in reference78). Both E2F1 and E2F3 are likely candidates involved in Rb-mediated regulation of neurogenesis. Deficiency of either E2F1 or E2F3 was observed to correct the ectopic proliferation observed in the central nervous system (CNS) in germ line Rb deficiency alone, and both E2F1 and E2F3 are grossly deregulated in proliferating neural precursors in the absence of Rb (7, 75, 81, 93).
While E2F1 and E2F3 are key regulatory targets in the Rb signaling pathway, the extent to which each contributes to Rb-mediated neurogenesis is unknown. Whether E2F1 and E2F3 are functionally redundant or are capable of unique function is still subject to debate and likely depends on the context examined. Individually, E2F1 is a tumor suppressor, and its deficiency results in mice that are viable but develop tumors at an advanced age (92). E2F1 expression is cell cycle regulated, with expression peaking at G1/S (reviewed in reference 78). A role for E2F1 in neurogenesis is indicated in the adult, where mice deficient for E2F1 exhibit decreased precursor cell division in the proliferative regions of the lateral ventricle and hippocampus (12). By contrast, E2F3 is not known as a tumor suppressor, but mice lacking E2F3 do exhibit a developmental phenotype (11). E2F3-deficient mice survive postnatally at a frequency of 25% on a mixed 129/Sv × C57BL/6 genetic background, and no E2F3-deficient mice are born on a pure 129/Sv genetic background (11, 30). Additionally, the E2F3 locus expresses two distinct transcripts, full-length E2F3a and N-terminal-truncated E2F3b transcribed from an intronic promoter within the E2F3 locus (27, 41). E2F3a expression is cell cycle regulated and is similar to that of E2F1 (27, 41). E2F3b, however, is expressed equivalently in quiescent and proliferating cells, is a specific partner for Rb in quiescent cells and thus may have an opposing role to E2F3a in cell cycle control (27, 41).
As both E2F1 and E2F3 are expressed in the developing cortex starting from embryonic day 11.5 (E11.5) and are deregulated in the absence of Rb (7, 13), we sought to determine the extent to which each is a target in Rb-mediated neurogenesis. Using mice with compound null mutations of Rb and E2F1 or E2F3, we describe both overlapping and unique functions for each. Here, we report that E2F1 and E2F3 are both functionally relevant targets in neural precursor proliferation, cell cycle exit, and laminar patterning. Each can partially mediate the Rb requirement for neuronal survival. Neuronal migration, however, is specifically mediated through E2F3. This study not only outlines overlapping and distinct functions for E2Fs in neurogenesis but also is the first to establish a physiologically relevant requirement for the Rb/E2F pathway beyond cell cycle regulation in vivo.
MATERIALS AND METHODS
Mice.
Germ line E2F1 null mice were generated previously (20) and obtained from the Jackson Laboratory (Bar Harbor, ME) and maintained on a C57BL/6 genetic background. Germ line E2F3 null mice were generated previously (42) and maintained on a mixed 129/Sv and C57BL/6 genetic background. To generate E2F1- and E2F3-deficient mice, heterozygous (E2F1+/− or E2F3+/−) mice were crossed. Embryos and animals were genotyped according to standard protocols with previously published primers for E2F1 (20) and E2F3 (42). Telencephalon-specific Rb-deficient mice were generated by crossing floxed Rb-F19 (58, 84) and Foxg1-cre mice (28), and mice were genotyped according to standard protocols with previously published primers (18, 19). Telencephalon-specific Rb-deficient/germ line-deficient E2F1 (Rb E2F1 DKO) mice were generated by interbreeding flox Rb/Foxg1-cre mice with flox Rb/germ line E2F1-deficient mice, both maintained on an FVBN background. Telencephalon-specific Rb-deficient/telencephalon-specific E2F3-deficient (Rb E2F3 DKO) mice were generated by crossing flox Rb/Foxg1-cre mice with flox E2F3 mice (91), both maintained on an FVBN genetic background. Mice were genotyped for flox E2F3 according to standard protocols with previously published primers for flox E2F3 (91). For embryonic time points, the time of plug identification was counted as E0.5. All experiments were approved by the University of Ottawa's Animal Care ethics committee adhering to the Guidelines of the Canadian Council on Animal Care.
Tissue fixation and cryoprotection.
Pregnant female mice and adult mice were euthanized with a lethal injection of sodium pentobarbital. Embryos were dissected and fixed overnight in 4% paraformaldehyde (PFA) in 1× phosphate-buffered saline (PBS), pH 7.4; cryoprotected in sequential solutions of 12, 16, and 22% sucrose in 1× PBS, followed by embedding in OCT (TissueTek 4583); and frozen on liquid N2. Adult mice were perfused with 1× PBS followed by cold 4% PFA, and brains were removed. Brains were postfixed overnight in 4% PFA, cryoprotected in 22% sucrose in 1× PBS, and frozen. Sections from either embryos or adults were collected as 14-μm coronal cryosections on Superfrost Plus slides (catalog no. 12-550-15; Fisher Scientific).
BrdU labeling, immunohistochemistry, and in situ hybridization.
To assess neural progenitor proliferation in adult mice (12 weeks old), intraperitoneal injections of bromodeoxyuridine ([BrdU] dissolved in 0.007 N NaOH in 0.9% NaCl; 50 mg/kg of body mass) (B-5002; Sigma) were given every 2 h over a 10-h period. Mice were euthanized 30 min after the last injection (80, 82). BrdU detection was performed with a mouse monoclonal anti-BrdU (1:100 dilution; catalog no. 347580; BD Biosciences) as previously described (19). BrdU-positive cells were counted in the subependyma of the lateral ventricles in every 10th coronal cryosection (14 μm thick) from the most caudal crossing of the corpus callosum to the start of the third ventricle (crossing of the anterior commissure). A two-tailed t test was performed to compare the mean numbers of BrdU-positive cells, and significant differences were assessed at α values of 0.05. To assess neural progenitor proliferation in embryos, pregnant females were injected intraperitoneally with 50 μg of BrdU/g of body mass and processed as above. BrdU-labeled cells were quantified over a 650-μm region of dorsal cortex with a minimum of three matched sections counted per embryo. To assess cells in mitotic M phase, phospho-histone H3 (PH3) labeling was performed with rabbit polyclonal anti-PH3 (dilution of 1:100; catalog no. 06-570; Upstate Biotechnology) as previously described (19). To assess cell death, either terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) (in situ end labeling kit; Roche) or active caspase-3 ([AC-3] 1:500; 559565 rabbit polyclonal; BD Pharmingen) immunohistochemistry combined with Hoescht nuclear staining was performed according to standard protocols (19). To quantify cell death specific to the marginal zone, AC-3-labeled cells in the marginal zone were counted from the cingulate cortex to the dorsal-ventral boundary. Both hemispheres were quantified, and counts are expressed as the mean of the two hemispheres from four matched sections per embryo. To quantify cell death in the ventral telencephalon, AC-3-labeled cells were counted below the dorsal-ventral boundary from four matched sections per embryo. Reelin and calbindin immunolabeling were performed with the mouse monoclonal anti-reelin G10 (1:500; catalog no. 553731; Calbiochem) and rabbit polyclonal anticalbindin (D-28; 1:1,000) (item AB1778; Chemicon) as previously described (18). Reelin-labeled cells were quantified along a 500-μm region of dorsal cortex and temporal cortex from a minimum of four matched sections per embryo. Calbindin-labeled cells were quantified within the marginal zone or within the same area comprising all cells within the migratory route (“total”) of each hemisphere from four matched sections per embryo and expressed as cells per 500-μm length. For all immunohistochemistry, secondary antibodies were obtained from Molecular Probes and used at a concentration of 1:500. Cresyl violet staining was performed according to standard protocols, and cells in the marginal zone were quantified along a 500-μm region of dorsal cortex and expressed as the mean from a minimum of four matched sections per embryo. Nonradioactive in situ hybridization and digoxigenin probe labeling were performed according to previously described protocols (85). Tbr1 antisense riboprobe was used, as previously described (6), and neogenin riboprobes were generous gifts of Helen Cooper, University of Queensland (22), and Elke Stein, Yale University. E2F1 and E2F3 digoxigenin-labeled riboprobes were generated from pBS-IIKS-E2F1 and pBS-IIKS-E2F3 templates, containing 0.65-kb and 0.72-kb cDNA inserts, respectively, which were amplified by PCR with primers E2F1 (Forward, ATCGGAATTCTCTCTTTGACTGTGACT; Reverse, ATTAAAGCTTCGATCGGAAAACTT) and E2F3 (Forward, ATCGAAGCTTAGACTTGGCTTCTAACAACT; Reverse, TGGCAGAATTCCATTCCGTGGTAG) and verified by sequencing.
Microarray analysis.
For microarray analysis, total RNA was extracted from tissue derived from ganglionic eminences at E14.5 from control and conditional Rb mutants using Trizol reagent according to the manufacturer's instructions (Invitrogen, San Diego, CA). Samples from embryos (n = 6) were pooled for each genotype. RNA was sent to the Ottawa Genomics Innovation Centre Microarray Facility, where the Affymetrix Mouse Genome 430 2.0 Array was used for analysis.
EMSA.
Electrophoretic mobility shift assay (EMSAs) were performed on total protein extracts from neural precursor cells as described previously (7), with the following modifications. Total cell protein was extracted in a lysis buffer (buffer A) and assayed by the method of Bradford (Bio-Rad protein assay reagent, catalog no. 500-0006). A 20-μg aliquot of lysate was incubated with an excess of 32P-labeled double-stranded DNA probe (70,000 cpm/0.2 ng of DNA) containing a single E2F-binding site: 5′-GGATTTAAGTTTCGCGCCCTTTCTCAA-3′. The binding reaction (25 μl) was carried out at room temperature for 20 min in binding buffer (20 mM HEPES, pH 7.6, 4% Ficoll, 2.5% MgCl2, 40 mM KCl, 0.1 mM EGTA, 0.5 mg/ml acetylated bovine serum albumin, 0.5 mM dithiothreitol). To control for binding specificity, a 10-fold excess of unlabeled wild-type oligonucleotide was added to the binding reaction mixture and incubated for 20 min before the addition of labeled probe. To identify the composition of the complexes, tissue culture supernatant or purified antibody was added to the reaction mixture. Complexes were resolved on a 5.0% gel run for 4 h, dried, and visualized by autoradiography. The tissue culture supernatant containing the monoclonal pRb antibody 21C9 was a gift from David Cobrinik (77). All other antibodies were purchased from Santa Cruz Biotechnology Inc. (E2F1, sc 193; E2F3, sc878 and sc878x). Immunoprecipitation-EMSA was performed as described previously (31) with the following modifications. For immunoprecipitation, 200 μg of total protein from neural precursors was incubated with either 2 μg of mouse monoclonal anti-human Rb antibody (catalog no. 554136; BD Biosciences) or equivalent mouse serum (Sigma M-5905) conjugated to protein G-Sepharose beads (17-0618-01; Amersham/GE Healthcare) in 1× shift buffer (20 mM HEPES, pH 7.9, 40 mM KCl, 6 mM MgCl2, 1 mM EGTA, 0.4 mM sodium vanadate, 0.4 mM sodium fluoride, 0.1% NP-40, 1 mM dithiothreitol, and protease inhibitors) for 1 h with gentle rotation. Beads were washed three to four times in 1× shift buffer, followed by treatment with 16 μl of 0.8% deoxycholate (DOC) for 10 min on ice to dissociate the E2F complexes. Following neutralization with 4 μl of 6% NP-40, 5μl of the supernatant was used for E2F EMSA as described above.
Microscopy.
Sections treated for immunohistochemistry were examined by a Zeiss Axioskop 2 microscope with standard fluorescence and bright-field or dark-field settings with 5× (numerical aperture, 0.17) or 20× (numerical aperture, 0.17) objectives, respectively. Images were captured using a digital black and white camera with Northern Eclipse software. For confocal microscopy, a Zeiss LSM 510 META on an Axiovert 200 M inverted microscope was used with images captured through the manufacturer's integrated digital imaging software. Figures were compiled using Adobe Photoshop CS2. Manipulations of brightness and intensity were made equally to all treatment groups.
RESULTS
E2F3 is a positive regulator of neural precursor proliferation.
Previous studies have described a role for E2F1 in regulating neural precursor proliferation in vivo (12); however, little is known regarding the role of E2F3. As E2F3 is known as a major regulator of cellular proliferation (30; reviewed in reference 78), we first asked if E2F3 is also capable of regulating neural precursor proliferation using mice lacking E2F3. As cell cycle time of neural progenitors in the adult brain has been estimated to be 12.7 h (68), we administered a series of BrdU injections over 10.5 h (82). Similar to what has been reported for E2F1, E2F3−/− mice exhibited 35% fewer neural progenitors lining the lateral ventricles relative to littermate controls (Fig. 1) (E2F3+/+, 662 ± 25.5 cells; E2F3−/−, 434 ± 14.5 cells; n = 3 per genotype). To determine whether the decrease in the number of BrdU-labeled cells was due to an increase in apoptosis, we performed TUNEL and AC-3 staining on E2F3−/− and littermate controls. No difference was observed in the number of dying cells along the lateral ventricles (data not shown). In the embryo, a similar 30% reduction in the number of proliferating cells and no difference in cell death were observed (data not shown). These results indicate that E2F3 is a positive regulator of neural precursor proliferation and point to functional redundancy among E2Fs in regulating neural precursor proliferation.
FIG. 1.
E2F3 is a positive regulator of neural precursor proliferation. (A and B) BrdU was administered over 10.5 h at 2-h intervals to adult male E2F3−/− and wild-type littermates to label dividing progenitor cells. Sections were labeled with an antibody to BrdU. BrdU-labeled cells lining one lateral ventricle were counted every 10th section between anatomical landmarks, and the total number of BrdU-labeled cells counted was expressed as the mean ± standard error of the mean (C). Approximately one-third fewer BrdU-labeled cells were observed in E2F3−/− compared to wild-type littermates (n = 3). Significance was determined using a two-tailed t test. *, P < 0.05. Bar, 100 μm.
E2F1 and E2F3 are physiologically relevant Rb-interacting partners in vivo.
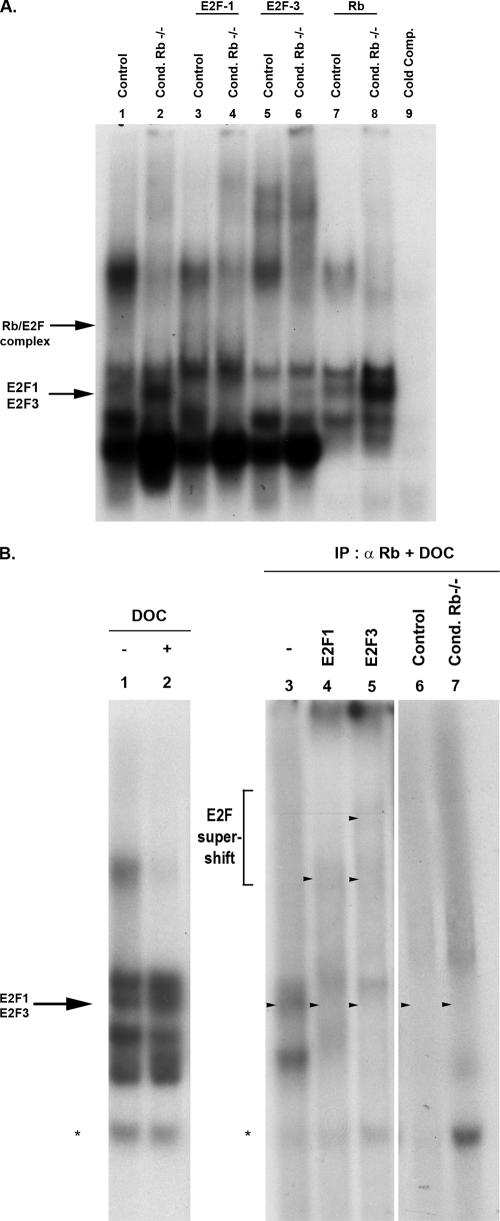
While both E2F1 and E2F3 function independently in regulating neural precursor proliferation, the context-dependent nature of Rb/E2F interaction led us to ask whether E2F1 and E2F3 are physiologically relevant Rb-interacting partners in the developing nervous system. The nature of Rb/E2F complexes was examined in protein extracts of neural precursors derived from embryonic forebrain through EMSA, and complexes were identified using antibodies specific to E2F family members (Fig. 2A). In wild-type tissue, both free E2F1 and E2F3 as well as bound Rb/E2F activity are observed (Fig. 2A, lane 1), indicating that E2F1 and E2F3 are active in the context of neurodevelopment. Here, we also observe that Rb is bound predominantly to E2F3 and E2F1 as antibodies to E2F1 (Fig. 2A, lane 3) and E2F3 (Fig. 2A, lane 5) displace the Rb band; however, E2F3 appears to be the more predominant Rb binding partner. To confirm that Rb is indeed binding to both E2F1 and E2F3 in neural precursor cells, we performed an immunoprecipitation for Rb from neural precursor cells, followed by DOC treatment to release the associated E2F activity, and subjected this extract to EMSA (31, 41) (Fig. 2B). Immunoprecipitation for Rb followed by DOC treatment gave rise to a pattern of free E2F binding activity (Fig. 2B, lane 3) similar to that of DOC treatment on extracts alone (Fig. 2B, lane 2). Supershifts of the immunoprecipitation extract with E2F1 and E2F3 antibodies demonstrate that both E2F1 (Fig. 2B, lane 4) and E2F3 (Fig. 2B, lane 5) bind to Rb in the nervous system. We next asked what consequences disrupting Rb activity would have on E2F1 and E2F3 activity. We hypothesized that if E2F1 and E2F3 are significant Rb binding partners in wild-type tissue, then the absence of Rb should lead to an increase in their free activity. Indeed, extracts of brain tissue from Rb mutants exhibit a gross deregulation of E2F1 and E2F3 activity relative to the control (Fig. 2A, lane 2). This increase appears specific to free E2F1 and E2F3 activity as supershifts with E2F1 and E2F3 antibodies displace the free E2F1 and E2F3 band (Fig. 2A, lanes 4 and 6). Thus, together these data provide biochemical evidence that E2F1 and E2F3 are physiological Rb-interacting factors in the developing nervous system in vivo.
FIG. 2.
E2F1 and E2F3 are physiologically relevant Rb-interacting partners in vivo. (A) For EMSA experiments, total protein was extracted from proliferating neural precursors in conditional Rb mutant and controls. Total protein extracts were incubated alone or in the presence of E2F antibodies prior to incubation with double-stranded 32P-labeled E2F consensus probe. Antibodies used for supershift are indicated above the corresponding lane. In control extracts, Rb is bound to both E2F1 and E2F3 as antibodies to both E2F1 and E2F3 displace the Rb band (lanes 3 and 5). In the absence of Rb, an obvious increase in free E2F1 and E2F3 binding activity is noted compared to control (lane 2; E2F1 and E2F3 supershifts are shown in lanes 4 and 6, respectively). (B) For immunoprecipitation (IP)-EMSA experiments, total protein extracts from control proliferating neural precursors were subjected to immunoprecipitation for Rb followed by DOC treatment to release E2F associated with Rb, as described in Materials and Methods. The released material was subjected to EMSA (lane 3) and assayed for E2F1 (lane 4) or E2F3 (lane 5) binding activity. As controls, IP-EMSA was repeated using control mouse serum in place of Rb-immunoprecipitating antibodies (lane 6) or using Rb-deficient neural precursor protein extracts instead of control extracts (lane 7). Finally, a sample of protein extract was subject to EMSA (lane 1) or directly treated with DOC and then assayed for E2F binding activity (lane 2). α, anti; Cond, conditional.
E2F1 and E2F3 exhibit overlapping patterns of expression in the developing telencephalon in vivo.
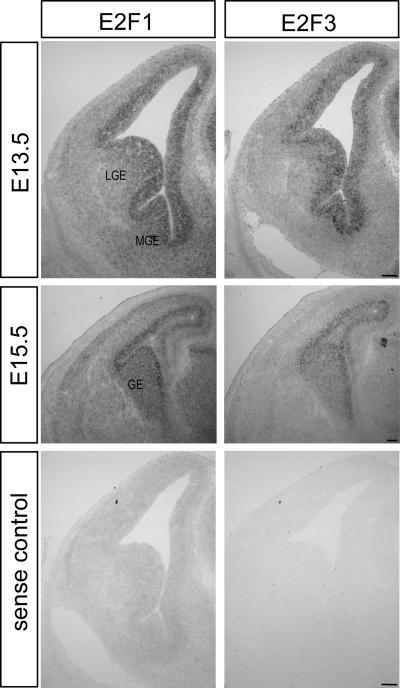
As E2F1 and E2F3 are both functionally relevant interacting partners in neural precursor cells, we next asked if E2F1 and E2F3 are expressed in the same cell populations in the developing telencephalon. Sections from control tissue at E13.5 and E15.5 were subjected to in situ hybridization for E2F1 and E2F3. At each age, both E2F1 and E2F3 are expressed in similar overlapping patterns (Fig. 3). This region of robust expression encompasses both the dorsal and ventral ventricular/subventricular zones where proliferating and newly committed neurons reside (reviewed in reference 25). In addition, at E15.5, the expression of both E2F1 and E2F3 is observed throughout the ganglionic eminences, the region which gives rise to migrating populations of interneurons, interneurons that ultimately exhibit aberrant migration in Rb deficiency (18). Hence, these data support the hypothesis that E2F1 and E2F3 could each be functional targets in both Rb-mediated proliferation and migration in the developing telencephalon.
FIG. 3.
E2F1 and E2F3 exhibit overlapping patterns of expression in the developing telencephalon in vivo. Control E13.5 and E15.5 sections were subjected to in situ hybridization for E2F1 and E2F3 using antisense riboprobes or sense riboprobes as a control. At both time points, both E2F1 and E2F3 are expressed in overlapping patterns in the developing telencephalon. At E13.5 expression of both E2F1 and E2F3 is largely confined to the developing ventricular and subventricular zones comprised of proliferating and postmitotic cells lining the lateral ventricles. At E15.5 expression is highest within the ventricular and subventricular zones, but for both E2F1 and E2F3 expression is also observed similarly throughout the ganglionic eminences ([ge] n = 4 embryos for each E2F). Note the absence of signal in the sense control for each probe. LGE, lateral ganglionic eminence; MGE, medial ganglionic eminence. Bar, 200 μm.
Both E2F1 and E2F3 are functional targets in Rb-mediated neural precursor proliferation.
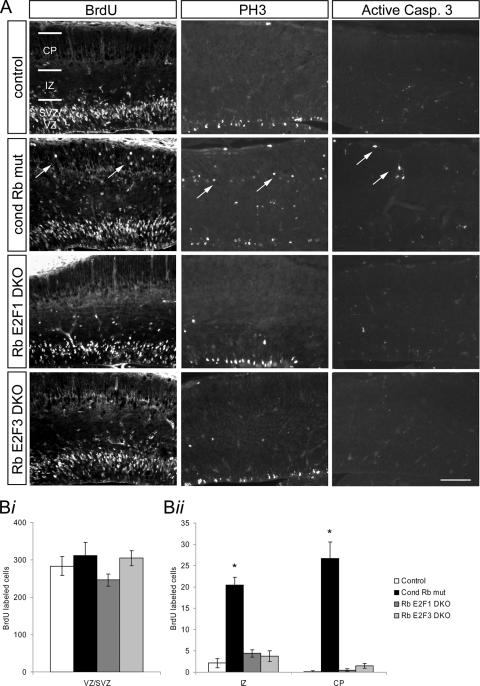
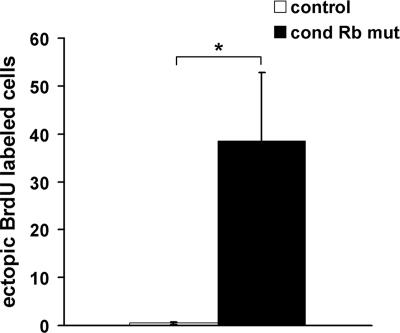
Our previous studies have demonstrated that Rb deficiency in the telencephalon leads to ectopic proliferation of neural precursor cells without the widespread apoptosis observed in germ line Rb deficiency (19). The gross deregulation of E2F1 and E2F3 binding activity in Rb-deficient neural precursors and their overlapping patterns of expression in proliferating neural precursor cells in vivo suggest that these E2Fs could be targets in Rb-mediated neural precursor proliferation. To ask if the proliferation defect observed in the absence of Rb could be attributed to deregulated E2F1 or E2F3 activity, we generated mice with (i) compound null mutations for Rb and E2F3 in the telencephalon (Rb E2F3 DKO) and (ii) an absence of Rb in the telencephalon and whole-embryo E2F1 deficiency (Rb E2F1 DKO). Pregnant females were subjected to a BrdU injection 2 h prior to sacrifice at E15.5, and embryonic sections were subjected to BrdU immunohistochemistry. In contrast to the conditional Rb mutant where proliferating cells are observed in the ventricular zone and postmitotic regions, in both Rb E2F1 DKO and Rb E2F3 DKO sections, BrdU-labeled cells were largely confined to the ventricular and subventricular zones with a minor proportion observed in the intermediate zone (Fig. 4A). Quantification of BrdU-labeled cells confirms that the Rb E2F1 DKO and Rb E2F3 DKO mutations lead to a rescue of ectopically proliferating cells caused by Rb deficiency alone (Fig. 4B graph ii). Similar observations were made in sections labeled with the M phase marker, PH3 (Fig. 4A). Whereas PH3-labeled cells are observed in the cortical plate, intermediate zone, and subventricular zone in the Rb mutant, mitotically active cells are largely observed in the ventricular and subventricular zones of Rb E2F1 DKO and Rb E2F3 DKO cells, indicative of a rescue of the failed cell cycle exit in committed neurons. To determine whether the rescue of cell cycle exit was due to an increase in apoptosis specific to the population of ectopically positioned proliferating cells, we performed a TUNEL assay and AC-3 staining on Rb E2F1 DKO and Rb E2F3 DKO tissues (Fig. 4A). No increase in cell death was observed in Rb E2F1 DKO or Rb E2F3 DKO cells relative to the Rb mutant or control (Fig. 4A; data not shown). Together, these data demonstrate that the absence of either E2F1 or E2F3 in Rb deficiency leads to a rescue of the ectopic proliferation. These findings indicate that E2F1 and E2F3 are each functionally relevant targets in Rb-mediated regulation of neural precursor proliferation and cell cycle exit.
FIG. 4.
Both E2F1 and E2F3 are functional targets in Rb-mediated neural precursor proliferation. (A) Sections from E15.5 conditional Rb mutant, Rb E2F1 DKO, and Rb E2F3 DKO tissues and their corresponding controls were labeled with an antibody to BrdU to label cells in S phase, PH3 to label cells in M phase, or AC-3 to label dying cells. While conditional Rb mutants exhibit BrdU- and PH3-labeled cells in the ventricular zone/subventricular zone (VZ/SVZ), intermediate zone (IZ), and cortical plate (CP), both Rb E2F1 DKO and Rb E2F3 DKO sections exhibit BrdU and PH3 labeling confined to the VZ/SVZ. No difference was observed in AC-3 labeling between conditional Rb mutant, Rb E2F1 DKO, and Rb E2F3 DKO and their corresponding controls. (B) BrdU-labeled cells were quantified along a 650-μm region of dorsal cortex and classified according to zone, from four matched sections per embryo. The number of BrdU-labeled cells counted was expressed as the mean ± standard error of the mean. Whereas no difference in the number of labeled cells was observed between conditional (cond) Rb mutant, Rb E2F1 DKO, and Rb E2F3 DKO and their corresponding controls in the VZ/SVZ (graph i), significantly fewer BrdU-labeled cells were observed in the IZ and CP of Rb E2F1 DKO and Rb E2F3 DKO sections relative to conditional Rb mutants but not different relative to their respective controls (graph ii). Significance was determined using a single-factor analysis of variance with a Tukey posthoc test. *, P < 0.05, n = 4 all genotypes. Bar, 100 μm.
Rb-mediated regulation of radial migration and laminar patterning occurs through interactions with E2F1 and E2F3.
We have recently reported that the loss of Rb leads to defective radial migration and laminar patterning in the developing cortex (18). Specifically, in the conditional Rb mutant we observe the absence of a clear cortical plate-intermediate zone boundary in histological sections and expanded expression of cortical plate markers into the intermediate zone. To determine whether radial migration and laminar patterning are mediated through the Rb/E2F cell cycle regulatory pathway, we therefore asked whether compound Rb E2F1 or Rb E2F3 deficiency is sufficient to correct the laminar patterning defects observed in the Rb mutant. Sections of Rb E2F1 DKO or Rb E2F3 DKO tissues at E15.5 were stained with cresyl violet. In control tissue, a clear boundary exists between the packed cells of the developing cortical plate and the underlying intermediate zone (Fig. 5). Compared to the conditional Rb mutant, where the developing cortical plate appeared to be comprised of loosely packed cells that are intermingled with cells of the intermediate zone, both the Rb E2F1 DKO and Rb E2F3 DKO tissues exhibited a cortical plate with a similar structure of densely packed cells and a clear cortical plate-intermediate zone division as the control (Fig. 5). No defects in laminar patterning were observed in either E2F1 or E2F3 single-deficiency tissue (data not shown). We next asked whether the layer-specific defects we observe in the conditional Rb mutant were also restored. Sections from Rb E2F1 DKO and Rb E2F3 DKO tissues were subjected to in situ hybridization with Tbr1, a layer-specific marker demarcating the preplate and layer 6 from the intermediate zone (6, 29), which exhibited vivid defects in the conditional Rb mutant (18). Consistent with the restoration of gross overall morphological structure, sections from Rb E2F1 DKO and Rb E2F3 DKO tissues exhibited Tbr1 expression that was largely confined to the developing cortical plate and exhibited a clear division between the cortical plate and intermediate zone, similar to the division observed in control sections (Fig. 5). This Tbr1 expression pattern in Rb E2F1 DKO and Rb E2F3 DKO tissues is in contrast to the conditional Rb mutant, where Tbr1 expression is expanded beyond the confines of the cortical plate into the intermediate zone (Fig. 5). These data demonstrate that the compound absence of either Rb E2F1 or Rb E2F3 is sufficient to restore cortical structure and laminar patterning. Furthermore, as both the Rb E2F1 DKO and Rb E2F3 DKO mutants are capable of rescuing both the cell cycle and laminar patterning defects, these data are consistent with the interpretation that Rb-mediated radial migration and cortical structure occur as a result of defects in cell cycle regulation.
FIG. 5.
Rb-mediated radial migration and laminar patterning defects occur through interactions with E2F1 and E2F3. E15.5 sections of conditional Rb mutant, Rb E2F1 DKO, and Rb E2F3 DKO tissues and their corresponding controls were stained with cresyl violet and subjected to in situ hybridization for Tbr1. In cresyl violet-stained sections, conditional Rb mutants exhibit the absence of a clear boundary between cortical plate and intermediate zone compared to control (arrows). This defect appears corrected in both Rb E2F1 DKO and Rb E2F3 DKO cells. Similarly, whereas Tbr1 expression is expanded beyond the cortical plate and into the intermediate zone in the conditional Rb mutant (arrows), both Rb E2F1 DKO and Rb E2F3 DKO tissues exhibit Tbr1 expression which is confined to the cortical plate, similar to that observed in the control (n = 3 per genotype). MZ, marginal zone; CP, cortical plate; IZ, intermediate zone; SVZ, subventricular zone; and VZ, ventricular zone. Bar, 100 μm.
The Rb-mediated requirement for survival of a subset of neurons is partially mediated through the Rb/E2F pathway.
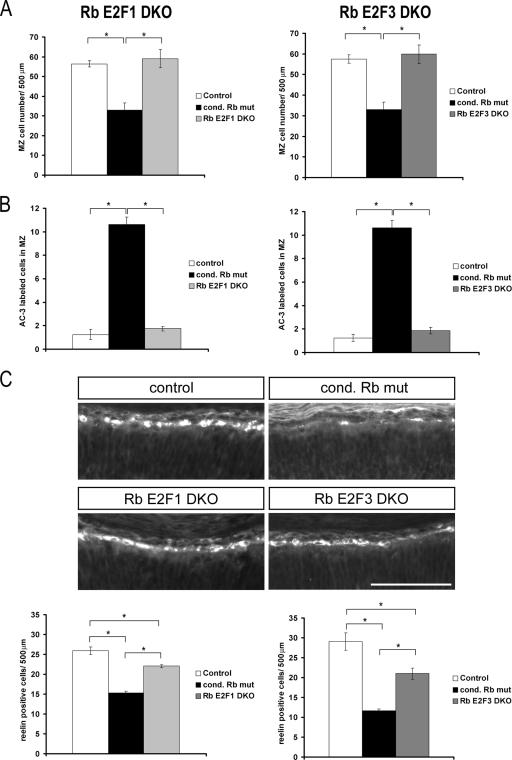
While conditional Rb mutants have shown that Rb is not required for widespread neuronal survival, we have recently demonstrated that Rb is required for survival of neurons within the marginal zone of the developing cortex (18, 19, 52). As E2F1, in particular, is a well-known mediator of survival (reviewed in reference 60), we asked to what extent marginal zone neuronal survival could be mediated through Rb interactions with E2F1 or E2F3. Total cells within the marginal zone were quantified from histological sections stained with cresyl violet of conditional Rb mutant, Rb E2F1 DKO, and Rb E2F3 DKO tissues. Both Rb E2F1 DKO and Rb E2F3 DKO sections exhibited an increased number of marginal zone cells relative to Rb mutants and no difference relative to the control (Fig. 6A). To determine if this increase was the result of increased survival, we examined cell death by quantifying AC-3-labeled cells within the marginal zone. Consistent with the increase in marginal zone cell numbers, both Rb E2F1 DKO and Rb E2F3 DKO tissues exhibited a decrease in the number of AC-3-labeled cells relative to conditional Rb mutants and no difference relative to the control (Fig. 6B). These data suggest that E2F1 and E2F3 are both capable of mediating the Rb requirement for neuron survival within the marginal zone.
FIG. 6.
The Rb-mediated requirement for survival of CR neurons is only partially mediated through the Rb/E2F pathway. (A) Marginal zone cells in cresyl violet-stained sections of E15.5 conditional Rb mutant, Rb E2F1 DKO, and Rb E2F3 DKO tissues and their corresponding controls were quantified along a 500-μm region of dorsal cortex from a minimum of three matched sections per embryo and expressed as mean ± standard error of the mean (n = 4 embryos per genotype). Whereas conditional Rb mutants exhibit decreased numbers of cells in the marginal zone, both Rb E2F1 DKO and Rb E2F3 DKO sections exhibit an increased number of cells in the marginal zone, similar to that observed in their respective controls. (B) AC-3-labeled cells of conditional Rb mutant, Rb E2F1 DKO, and Rb E2F3 DKO tissues and their corresponding controls were quantified within the marginal zone from the cingulate cortex to the dorsal-ventral boundary. Both hemispheres were quantified, and counts are expressed as the mean of the two hemispheres from four matched sections per embryo. Bars represent mean ± standard error of the mean (n = 8 per genotype). Whereas conditional Rb mutants exhibit increased numbers of AC-3-positive cells in the marginal zone, both Rb E2F1 DKO and Rb E2F3 DKO samples exhibit a decreased number of AC-3-positive cells in the marginal zone, similar to that observed in their respective controls. (C) A noticeable increase in reelin-labeled cells was observed in Rb E2F1 DKO and Rb E2F3 DKO tissues relative to the conditional Rb mutant. Reelin-labeled cells were quantified along a 500-μm region of dorsal cortex and temporal cortex and expressed as the mean from a minimum of four matched sections per embryo. Whereas conditional Rb mutant tissues exhibit decreased numbers of reelin-labeled cells in the marginal zone, both Rb E2F1 DKO and Rb E2F3 DKO sections exhibit an increased number of cells in the marginal zone relative to the conditional Rb mutant yet still significantly less than that observed in their respective controls. Bars represent mean ± standard errors of the mean (n = 3 for the control and conditional Rb mutant; n = 4 for Rb E2F1 DKO; and n = 5 for Rb E2F3 DKO). In all cases significance was determined using a single-factor analysis of variance with a Tukey posthoc test. *, P < 0.05. MZ, marginal zone; cond, conditional. Bar, 100 μm.
The marginal zone is a complex layer comprised of a het erogeneous population of cells including Cajal-Retzius (CR) neurons (59, 64). We have previously demonstrated that the conditional Rb mutants exhibit a specific loss of CR neurons by cell death which contributes to the overall reduction of cells within the marginal zone (18). Thus, to determine if Rb-mediated neuronal survival is also mediated through Rb interactions with E2F1 and E2F3, we examined CR neurons within the marginal zone in Rb E2F1 DKO and Rb E2F3 DKO tissues. Sections were subjected to immunohistochemistry for reelin, a CR neuron-specific protein (reviewed in reference 64). Whereas both Rb E2F1 DKO and Rb E2F3 DKO sections exhibited increased numbers of reelin-positive cells relative to conditional Rb mutants, fewer reelin-positive cells were observed relative to the control (Fig. 6C). These data indicate that E2F1 and E2F3 only partially mediate the Rb requirement for CR neuron survival. As CR neurons themselves represent a heterogeneous population of cells (4), these data support the hypothesis that the Rb/E2F pathway mediates survival of a subtype of CR neurons.
E2F3 specifically mediates the aberrant tangential migration of interneurons in Rb mutants.
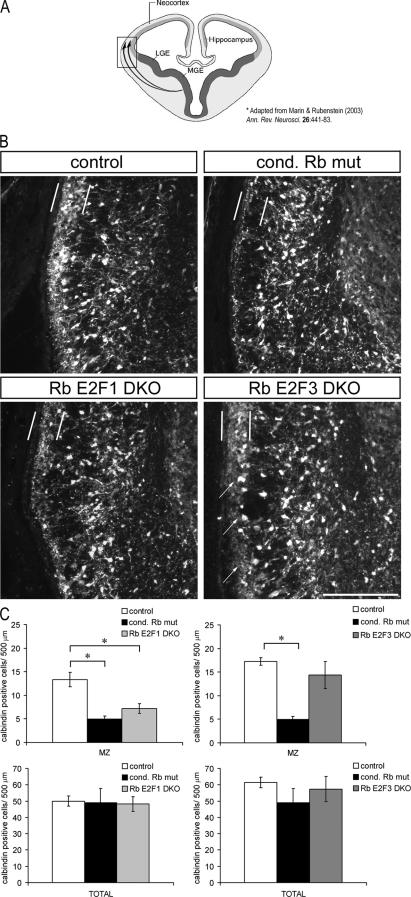
Interneurons are key regulators of neuronal function that act by modulating the activity of major excitatory neural circuits (reviewed in reference 62). Interneuron dysfunction and/or aberrant migration of interneurons during development has been implicated in a wide range of neurological disorders including autism, epilepsy, schizophrenia, and bipolar disorder (3; reviewed in reference 2). In a recent study, we demonstrated that interneurons arising from the ventral telencephalon exhibit aberrant tangential migration to the dorsal cortex in conditional Rb deficiency. Specifically, calbindin-labeled cells, a marker of GABAergic interneurons, are absent in Rb mutants along the marginal zone migratory route that is taken by these cells (18). As many of the neurodevelopmental defects described in conditional Rb mutants appear to be mediated through both E2F1 and E2F3 and as both are expressed in the ventral ganglionic eminences, where interneurons originate, we questioned whether the E2F pathway could also be mediating migration. To assess the degree to which E2F1 and E2F3 could contribute to the tangential migration defect in conditional Rb mutants, we examined the calbindin cell population along its migratory route in Rb E2F1 DKO and Rb E2F3 DKO tissues (Fig. 7A). At E15.5, calbindin-labeled cells are reduced or absent along the marginal zone migratory route in Rb E2F1 DKO sections, similar to that observed in the conditional Rb mutant (Fig. 7B). By contrast, an abundance of calbindin-labeled cells is observed in Rb E2F3 DKO sections along the marginal zone migratory route at the dorsal ventral boundary (Fig. 7B). Quantification of the number of calbindin-positive cells specifically within the marginal zone indicates that Rb E2F1 DKO sections exhibited significantly fewer calbindin-labeled cells within the marginal zone, similar to that observed in the conditional Rb mutant, while Rb E2F3 DKO sections exhibited no difference in number of calbindin-labeled cells relative to the control (Fig. 7C). Further, no difference in the distribution of calbindin-labeled cells was observed in single E2F3 deficiency at E15.5 (data not shown).
FIG. 7.
E2F3 specifically mediates the aberrant tangential migration of interneurons in Rb mutants. (A) Schematic diagram of superficial marginal zone and deeper intermediate zone routes taken by migrating calbindin-labeled interneurons originating from the medial ganglionic eminence (modified from reference 56). Box indicates region of magnification in panel B. Calbindin labeling was examined in conditional Rb mutant, Rb E2F1 DKO, and Rb E2F3 DKO tissues and their respective controls along the marginal and intermediate zone migratory routes at E15.5. Calbindin-positive cells are reduced or absent along the marginal zone migratory route in Rb E2F1 DKO and are present at a level similar to that observed in the conditional Rb mutant. By contrast, an abundance of calbindin-labeled cells is observed in Rb E2F3 DKO tissue along the marginal zone migratory route at the dorsal ventral boundary (arrows). (C) Calbindin-positive cells were quantified within the marginal zone (indicated by vertical bars) or along the same length within the complete migratory route as demarcated by the marginal zone and external capsule as lateral and medial boundaries (boxed area in panel A) for the total region. The total region was comprised of the marginal zone, as well as the intermediate zone, and cortical plate and labeled cells were counted on each hemisphere from four matched sections per embryo and expressed as cells per 500-μm length. Quantification confirms that calbindin cells positioned in the marginal zone are restored in Rb E2F3 DKO but not in Rb E2F1 DKO cells, yet the total number of calbindin-labeled cells is the same across all groups. Bars represent mean ± standard error of the mean (n = 4 embryos per genotype). Significance was determined using a single-factor analysis of variance with a Tukey posthoc test. *, P < 0.05. LGE, lateral ganglionic eminence; MGE, medial ganglionic eminence; cond, conditional. Bar, 100 μm.
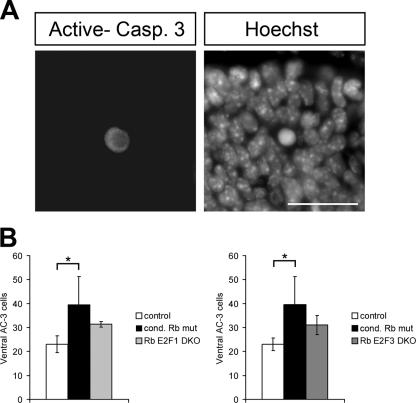
While these results are suggestive of a specific role for Rb acting through E2F3 in mediating neuronal migration, to rule out the possibility that cell death was involved, we examined both the total number of calbindin-positive cells within the migrating region and the level of apoptosis within the ventral telencephalon in Rb E2F1 DKO and Rb E2F3 DKO tissues. As we have previously observed (18), quantification of total calbindin-labeled cells revealed no significant differences between the control and conditional Rb mutant. Additionally, we observed no difference between Rb E2F1 DKO or Rb E2F3 DKO embryo and the control or conditional Rb mutants in the total number of calbindin-labeled cells, suggestive of a population of similar size among all groups. We also quantified cell death as measured by AC-3 labeling in the ventral telencephalon including the ventricular zone, where interneurons originate; in the marginal zone and cortical plate, along the route of migration; and points in between (Fig. 8A). While a low level of cell death was observed overall, consistent with our previous results (19), we observed a small but significant increase in the overall level of cell death in the conditional Rb mutant relative to the control (Fig. 8B). Quantification of cell death within the same regions of the ventral telencephalon in Rb E2F1 DKO and RbE2F3 DKO tissues revealed similar levels of cell death for each genotype, which was observed at a level between that of the control and the conditional Rb mutant (Fig. 8B). Together, these results support our interpretation that the specific rescue of calbindin-labeled cells within the marginal zone of Rb E2F3 DKO tissues represents a rescue of the aberrant migration of calbindin-labeled cells in the conditional Rb mutant and is not the result of altered cell death or other defects in the calbindin cell population. Thus, these results reveal a unique function for E2F3 as the Rb target that mediates migration, a function distinct from the role of E2F1. Furthermore, these findings indicate that neuronal migration is mediated via an E2F-dependent mechanism and, hence, point toward novel targets specific for E2F3-mediated transcription.
FIG. 8.
Rb E2F1 DKO and Rb E2F3 DKO tissues exhibit similar levels of cell death in the ventral telencephalon. E15.5 sections of conditional Rb mutant, Rb E2F1 DKO, and Rb E2F3 DKO and their corresponding controls were stained with AC-3 and Hoescht to examine cell death in the ventral telencephalon. Labeled cells were counted along the ventricular zone, where interneurons originate; the marginal zone and cortical plate along the route of migration; and points in between from four matched sections per embryo. A similar level of cell death is observed between Rb E2F1 DKO and Rb E2F3 DKO cells. Each exhibited a level of cell death between that of control and conditional (cond) Rb mutant. Bars represent the mean of total number of AC-3-labeled cells counted ± standard deviation (n = 4 embryos per genotype). Significance was determined using a single-factor analysis of variance with a Tukey posthoc test. *, P < 0.05. Bar, 50 μm.
Rb/E2F3 mediates neuronal migration in a manner beyond cell cycle regulation.
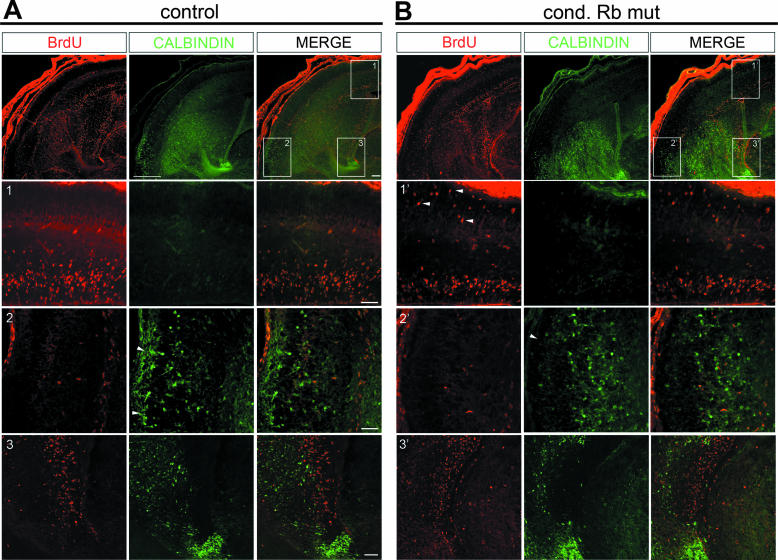
Since Rb/E2F interaction is best characterized for its role in cell cycle regulation, our observation that Rb interaction with E2F3 is capable of mediating interneuron migration led us to question whether Rb-mediated migration could be the result of defects in cell cycle control. To address this issue, we performed a thorough analysis by confocal microscopy of the location of proliferating and calbindin-labeled cells in conditional Rb mutants at two developmental time points: at E15.5 when defective migration is observed and at an earlier developmental time point, E13.5. We hypothesized that if the aberrant migration of calbindin interneurons is the result of defects in cell cycle control, then a population of calbindin-labeled cells should be double labeled with BrdU after a standard 2-h pulse. In E15.5 control embryos at low magnification, proliferating cells are seen largely confined to the dorsal and ventral ventricular zones (Fig. 9A, top). In the conditional Rb mutants, ectopically proliferating cells appear largely confined to the postmitotic region of the dorsal cortex (Fig. 9B, 1′). The dorsal cortex region corresponds to the region where aberrant laminar patterning and radial migration are perturbed, thus further supporting the hypothesis that Rb-mediated regulation of radial migration and cortical structure occurs as a result of defects in cell cycle signaling. By contrast, migrating calbindin-labeled cells are observed in the ventral telencephalon, a distinct neuroanatomical region, beyond the regions of normal and ectopic proliferation (Fig. 9A, rows 2 and 3, and B, rows 2′ and 3′). Using confocal microscopy we examined three distinct regions to see if calbindin-labeled cells were ectopically proliferating: the dorsal cortex (Fig. 9A, row 1, and B, row 1′), where ectopically proliferating cells are observed in conditional Rb mutants; the migratory route within the ventral telencephalon (Fig. 9A, row 2, and B, row 2′), where calbindin labeled cells are aberrantly localized within the conditional Rb mutants; and the ventral ventricular zone within ganglionic eminences (Fig. 9A, row 3, and B, row 3′), where calbindin-labeled cells originate. Close inspection from at least three sections from multiple embryos did not reveal BrdU and calbindin double-labeled cells in either control or conditional Rb mutants in any of the three regions examined.
FIG. 9.
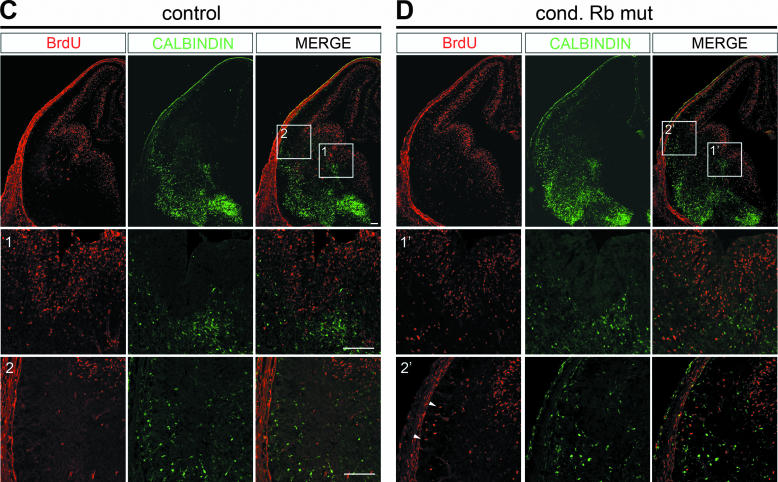
Rb/E2F3-mediated tangential migration is not the result of cell cycle deregulation. Sections from conditional Rb mutant and control were double labeled with BrdU (red) and calbindin (green) at E15.5 (A and B) and E13.5 (C and D). At E15.5 at low magnification, BrdU labeling is largely observed surrounding the ventricle, whereas calbindin-labeled cells are localized largely in the ventral telencephalon. At higher magnification, ectopic proliferation in conditional (cond) Rb mutant is observed largely confined to the dorsal cortex (B1′), while no difference in the low level of BrdU labeling between the conditional Rb mutant and control is observed in the ventral telencephalon (B2′ and B3′ versus A2 and A3, respectively). Instead, in this region where BrdU labeling is not detected, an absence of calbindin-labeled cells is observed in the marginal zone, and the absence of calbindin-labeled cells along the marginal zone migratory route in conditional Rb mutants is observed (B2′) (arrowheads). By confocal microscopy no BrdU calbindin-double-labeled cells are observed in the telencephalon at any of the three regions examined. At E13.5 ectopic proliferation is prevalent in conditional Rb mutants in the dorsal and ventral telencephalon (D, top panel and 2′); however, by confocal microscopy in either control or the conditional Rb mutant, we do not detect BrdU-calbindin double labeling at either the ganglionic eminence, where calbindin cells originate (C1 and D1′) or at the future ventro-lateral migratory route (C2 and D2′). Double-labeled cells, however, were occasionally observed within the blood vessel-rich pial layer outside of the telencephalon and are likely blood cells. Bar, 100 μm.
To determine if calbindin-labeled cells that are migrating at E15.5 were ectopically proliferating at an earlier time point, we performed a similar confocal microscopy analysis of BrdU-calbindin double labeling at E13.5 (Fig. 9C and D). At this time point, we do observe ectopic BrdU-labeled cells in the ventral telencephalon of conditional Rb mutants along the route of migration of calbindin-labeled interneurons (Fig. 9D, row 2′). Quantification of ectopically proliferating cells demonstrates a significant increase in ventral ectopic proliferation at this age (Fig. 10); however, while several distinct neuronal subtypes exist in this region (reviewed in reference 89), none of these ectopically dividing progenitors was colabeled with calbindin. Specifically, we examined distinct regions at E13.5 to see if calbindin-labeled cells were ectopically proliferating, focusing on the ventral regions: the future route of migration of aberrantly migrating cells within the ventral telencephalon (Fig. 9C, row 2, and D, row 2′); and the ventral ventricular zone within medial ganglionic eminence (Fig. 9C, row 1, and D, row 1′), where calbindin-labeled cells originate. Similar to what we observed at E15.5, at E13.5, using confocal microscopy we did not observe BrdU and calbindin double-labeled cells in either control or conditional Rb mutants at either region examined. While we cannot unequivocally rule out that aberrantly migrating cells were once ectopically proliferating, since we did not observe calbindin colocalized with BrdU in any region at either time point examined, these data suggest that calbindin-labeled cells have successfully exited the cell cycle in conditional Rb mutants. Hence, our data support the hypothesis that Rb mediates tangential migration through E2F3 in a manner beyond cell cycle regulation.
FIG. 10.
Quantification of ectopic proliferation within the ventral telencephalon at E13.5, comprising the region of migration in the control and conditional (cond) Rb mutant. BrdU cells within the region were counted in four matched sections per embryo. Bars represent means of the average number of BrdU-labeled cells within a 2,000-μm length along the ventrolateral boundary comprising the region of migration between the outermost edge of the marginal zone and the external capsule as lateral and medial boundaries ± standard deviation (n = 3 embryos per genotype). Significance was determined using a two-tailed t test. *, P < 0.05.
Rb mediates the expression of genes involved in regulating neuronal migration.
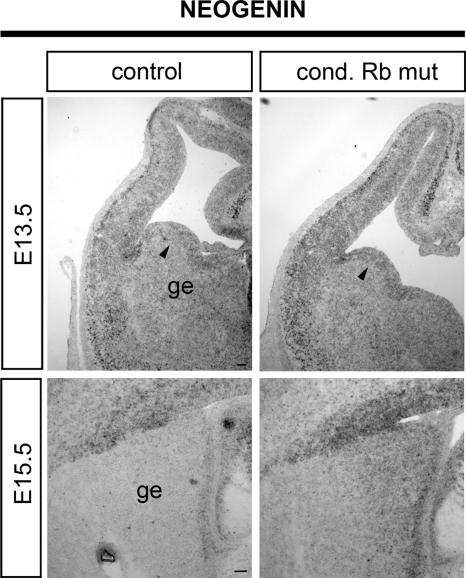
Our data demonstrating that Rb mediates migration through E2F3 in vivo represent physiological evidence in support of the hypothesis that Rb/E2F could regulate the transcription of novel genes unrelated to cell cycle regulation. In an effort to identify candidate genes, we performed microarray analyses on neural precursor cells from control and conditional Rb mutants derived from the medial ganglionic eminences, the region which gives rise to migrating populations of interneurons which ultimately exhibit aberrant migration in Rb deficiency (18). We hypothesized that if Rb/E2F3 mediates migration of interneurons in a cell-autonomous manner as our previous data demonstrated (18), then the absence of Rb would lead to deregulation of genes required to regulate this process. Through our microarray analysis, we have identified several candidate genes that are deregulated in con ditional Rb mutants which have been shown to mediate neu ronal migration, including migration of interneurons, such as members of the neogenin/netrin/repulsive guidance molecule (RGM) signaling pathway, the semaphorin/neuropilin signaling pathway, and the Slit/Robo pathway (Table 1). While all candidates remain plausible targets, we have focused our initial attention on neogenin, a cell surface receptor and member of the immunoglobulin superfamily (83) with well-known (72, 87) and hypothesized roles in regulating neuronal migration, including interneuron migration (21). By microarray, neogenin expression was increased threefold, consistent with the role of E2F3 as a transcriptional activator. We next confirmed this observation through in situ hybridization for neogenin in conditional Rb mutants (Fig. 11). At E13.5, a moderate increase in neogenin expression in the conditional Rb mutant was observed throughout the telencephalon including the ganglionic eminences (Fig. 11). At E15.5 this increase in neogenin expression is more pronounced, particularly within the ganglionic eminences, where interneurons originate. Together, these data confirm that absence of Rb leads to deregulation of known genes required for interneuron migration within the population of migrating cells during the time of migration and, in addition, establishes neogenin as a potential target gene in Rb/E2F-mediated interneuron migration.
TABLE 1.
Candidate molecules identified in microarray from control and conditional Rb mutant ventral precursor cellsa
| Molecule name | Pathway | Relative changeb | Migration function | Reference |
|---|---|---|---|---|
| Neogenin | Netrin/RGM | 3-fold increase | Binds netrin and RGM; repels temporal retinal axons through RGM | 72 |
| 61 | ||||
| Sema3d | Neuropilin/semaphorin | Moderate increase | Guides retinal axon along DV axis in zebrafish; can be repulsive or attractive | 47 |
| 88 | ||||
| VLDLR | Reelin signaling | Moderate increase | Receptors for reelin; VLDLR and ApoER2 KO mice have reeler-like phenotype with inverted cortical lamina structure | 79 |
| ApoE | Reelin signaling | 2-fold increase | Out-competes reelin for binding to ApoER2? | NA |
| CCK | 2.5-fold increase | A marker of GABAergic interneurons; CCK also reduces migration of GnRH neurons | 50 | |
| 23 | ||||
| TWIST1 | bHLH transcription factor | Moderate decrease | Loss-of-function mutant leads to defect in neural crest cell migration | 70 |
| Twist neighbor | bHLH transcription factor | 2-fold increase |
VLDLR, very-low-density lipoprotein receptor; CCK, cholecystokinin; bHLH, basic helix-loop-helix; DV, dorsal-ventral; ApoER2, ApoE receptor 2; NA, not applicable.
Increase/decrease in mutant relative to control.
cTwist neighbor.
FIG. 11.
Neogenin, a microarray-identified gene, exhibits deregulated expression in conditional Rb mutants (cond Rb mut). Control and conditional Rb mutant E13.5 and E15.5 sections were subjected to in situ hybridization for neogenin. At both time points, neogenin expression appears increased in the conditional Rb mutant relative to the control. At E13.5 expression appears increased overall including in the ganglionic eminences (arrowheads). At E15.5 this increase in neogenin expression appears more pronounced, particularly within the ganglionic eminences (ge), where interneurons originate (n = 4 embryos for each genotype). Bar, 100 μm.
DISCUSSION
The mammalian nervous system is comprised of a complex array of different cell types and subtypes (reviewed in references 24 and 25). Neurogenesis, the process by which neural precursor cells divide and differentiate to give rise to all the different cell types, occurs in a highly regulated manner (reviewed in references 24 and 25). The cortex, a model of neurogenesis, is comprised of a series of layers that form in an inside-out manner populated with excitatory projection neurons and inhibitory interneurons (reviewed in reference 25). In both cases, neural precursors divide along the germinal zone lining the ventricles (reviewed in reference 24). Once committed to a neuronal fate, cells exit the cell cycle and leave the germinal zone to migrate toward their final destination with projection neurons migrating radially to form the layers of the cortex and cortical interneurons migrating from the ventral telencephalon along tangential routes into the dorsal cortex (reviewed in references 24, 37, 55, and 56). The fundamental processes of neurogenesis, namely, proliferation differentiation, migration, and maturation, are controlled by the precise coordination of various genetic pathways (reviewed in references 24 and 25). Normal development of the cortex is essential for proper brain function as abnormal development has been hypothesized as an underlying cause in a number of neurological and psychiatric disorders (2, 34-36, 43-45, 71, 76) Numerous studies have demonstrated the pivotal roles that Rb plays in nervous system development, and in addition, much insight has been gained into Rb function itself by studying its role in the nervous system (8, 10, 19, 33, 39, 40, 46, 51-53, 57, 66, 73, 86).
In this study we evaluated the in vivo contributions of E2Fs as regulatory targets for Rb-mediated neurogenesis, and our results herein support a number of conclusions. First, this study establishes that E2Fs are indeed major physiological targets in Rb-mediated neurogenesis, mediating both cell cycle-dependent processes and roles beyond cell cycle regulation. We also demonstrate that functional redundancy exists among E2Fs in regulating cell cycle exit, laminar patterning, and radial migration, as well as cell survival. Finally, our data demonstrating that Rb mediates neuronal migration specifically through E2F3 represent the first physiologically relevant requirement for the Rb/E2F pathway beyond cell cycle regulation in vivo.
E2Fs are physiological targets in Rb-mediated neurogenesis regulating cell cycle-dependent processes and mediating roles beyond cell cycle regulation.
Rb is known to interact with numerous proteins, many of which are expressed in quiescent cells or have cell cycle-independent functions. Thus, the search for Rb-interacting proteins in neurogenesis represents a potentially long list (reviewed in reference 67). We hypothesized that members of the cell cycle regulatory E2F family represent functional targets of Rb in neurogenesis in vivo for a number of reasons. First, E2F1 and E2F3 have both been established as Rb targets in regulating neural precursor proliferation as each is capable of rescuing the ectopic proliferation in the CNS in germ line Rb deficiency (75, 81, 93). While these studies support the hypothesis that E2F1 and E2F3 are targets in Rb-mediated neurogenesis, the use of germ line Rb-deficient mice limited the interpretation due to the widespread defects that were the result of non-cell-autonomous requirements for Rb during development (19, 52, 90). To circumvent this issue, we examined the role of E2F1 and E2F3 as targets in Rb-mediated neurogenesis using telencephalon-specific Rb-deficient mice crossed with E2F1-deficient or telencephalon-specific E2F3-deficient mice. It is possible that some of the differences observed between the Rb E2F1 DKO and Rb E2F3 DKO models are due to systemic loss of E2F1 and effects outside the nervous system. The justification for using a telencephalon-specific model of E2F3 deficiency, however, was out of necessity due to the vital role of E2F3 during development (11, 30). As E2F1 does not have such a vital role (92), no analogous tissue-specific model of E2F1 deficiency was available.
Here, our results demonstrate that E2F1 and E2F3 are physiologically relevant Rb targets in neurogenesis in vivo. First, we observed that both are expressed in overlapping patterns in the developing telencephalon. In vitro, we showed that Rb interacts predominantly with E2F1 and E2F3 in extracts of neural tissue. The significance of this interaction is demonstrated in vivo where we not only show that E2F1 and E2F3 are targets of Rb-mediated neural precursor proliferation but also establish that E2Fs play a major role as Rb targets in laminar patterning, radial migration, cell survival, and tangential migration of interneurons. As both Rb E2F1 DKO and Rb E2F3 DKO cells are capable of rescuing the proliferation defect, along with the laminar patterning and radial migration defects observed in telencephalon-specific Rb deficiency, our data are consistent with the interpretation that the radial migration and laminar patterning defects may occur as the result of cell cycle deregulation. Rb-mediated tangential migration, however, appears to be mediated specifically through E2F3. Thus, with these results, we establish roles for E2F1 and E2F3 as the physiological targets in Rb-mediated neurogenesis.
Functional redundancy exists among E2F1 and E2F3 in vivo.
Our results demonstrate that functional redundancy exists among E2F1 and E2F3 in the context of neural precursor proliferation, cell cycle exit, and survival in vivo. First, we show that E2F3 alone is a positive regulator of neural precursor proliferation, similar to what has been reported for E2F1 (12). Next, as both Rb E2F1 DKO and Rb E2F3 DKO cells are capable of rescuing the proliferation and survival defects observed in Rb deficiency in the telencephalon, our data support the hypothesis that E2F1 and E2F3 are functionally equivalent targets in Rb-mediated cell cycle exit and survival.
The idea that E2Fs are functionally redundant is still debated within the field. In vitro studies have indicated that, individually, E2F1 and E2F3 are capable of regulating the expression of distinct genes, likely as a result of differences within the marked box domain (5, 26). In the context of Rb interaction, however, it remains unresolved as to whether Rb interaction is equivalent with each E2F (15, 74). In vivo, absence of either E2F1 or E2F3 results in individual and distinct phenotypes, yet in the context of Rb interaction, both have been shown to rescue many of the proliferation, apoptosis, and midgestational survival defects associated with germ line Rb deficiency (81, 93). Specificity, however, has been reported to exist in Rb-mediated phenotypes where a unique function for E2F1 has been reported in mediating apoptosis in the Rb-deficient lens and retina (75). Thus, the function of E2F1 and E2F3 as targets in Rb-mediated neurogenesis should be viewed as context dependent, even within the CNS.
Finally, our observation that E2F1 and E2F3 only partially mediate the Rb requirement for subtype-specific neuronal survival raises a number of questions. First, it is unlikely that the partial rescue in CR neurons in Rb E2F1 DKO or Rb E2F3 DKO cells is related to the E2F3-mediated rescue of Rb-mediated tangential migration. While CR neurons have been hypothesized to have roles in regulating tangential migration of interneurons (65), the decrease in CR neurons observed in conditional Rb-deficient embryos is unlikely to influence migration of interneurons as our previous findings demonstrated that the role for regulating Rb is cell autonomous (18). Rather, as CR neurons themselves are a heterogeneous population (4), these data support the hypothesis that the Rb/E2F pathway mediates survival of only a subtype of CR neurons. Two possible explanations are hypothesized. First E2F1 and E2F3 may mediate survival of nonoverlapping populations of CR cells that together mediate survival of the entire population of CR cells that is absent in the conditional Rb mutant. Alternatively, it is also possible that other non-E2F Rb-interacting factors are contributing to survival. Id2 is a non-E2F Rb-interacting factor that has been shown to mediate many of the neurological defects arising in Rb mutants, including apoptosis (38); thus, it is possible that E2F and Id2 are acting along parallel pathways to regulate CR neuron survival. The latter is a particularly provocative hypothesis as there is little discussion in the literature about a possible E2F-independent role for Rb in cell survival.
Unique physiological requirement for Rb/E2F3 beyond cell cycle regulation exists in mediating neuronal migration in vivo.
Here, we demonstrate that Rb-mediated migration of interneurons is indeed mediated through the E2F pathway, specifically, through E2F3. These results support a model for specificity among E2Fs in nervous system development. Further, it is this specificity of E2F function which underlies the hypothesis that E2F3-specific mediated interneuron migration represents a novel physiologically relevant requirement beyond cell cycle regulation for the Rb/E2F pathway in vivo. In support of this hypothesis, we observe that E2F1 and E2F3 are expressed in overlapping patterns within the ganglionic eminences, where interneurons originate, and that both Rb E2F1 DKO and Rb E2F3 DKO cells are capable of rescuing the proliferation defects, yet only Rb E2F3 DKO cells can rescue the migration defect. In addition, aberrantly migrating interneurons do not incorporate BrdU either during (E15.5) or before (E13.5) migration. While we cannot unequivocally rule out that aberrantly migrating cells were once ectopically proliferating, the absence of double-labeled cells suggests that the population of calbindin interneurons has successfully exited the cell cycle in conditional Rb mutants.
Further support for role for E2F3 beyond cell cycle regulation can be inferred from the known function of E2F3 itself. E2F3 is one of the more intriguing E2Fs as the locus expresses two distinct transcripts: full-length E2F3a, whose expression is cell cycle regulated and acts as a transcriptional activator, and E2F3b, which is expressed equivalently in quiescent and proliferating cells and is a specific partner for Rb in quiescent cells (27, 41). While it is possible to hypothesize that Rb/E2F3-mediated neuronal migration could be regulated through E2F3b, complementary to our work, it has been shown that a defect in the differentiation of Rb-deficient cholinergic neurons in the retina is mediated through E2F3a (D. Chen, R. Opaskvy, M. Pacal, N. Tanimoto, P. Wenzel, M. W. Seeliger, G. Legne, and R. Bremner, unpublished data), an effect also shown to be cell cycle independent. Thus, together our data suggest a common mechanism through which Rb exhibits roles beyond cell cycle regulation through E2F3 during nervous system development.
Finally, further support for a novel, in vivo function for the Rb/E2F pathway beyond cell cycle regulation comes from our search for novel E2F-regulated target genes in the context of neuronal migration. E2F-mediated regulation of cell cycle-independent genes is an emerging concept. In vivo, studies examining individual E2F-deficient mice demonstrated a vast array of tissue-specific defects in development and differentiation, suggesting that E2Fs may be regulating non-cell cycle-related genes (reviewed in references 1 and 16). These studies were limited, however, as the individual phenotypes could be the result of E2F acting independently from interactions with Rb. Here, we have performed microarray analysis on neural precursor cells from the medial ganglionic eminences, the region which gives rise to migrating populations of interneurons which ultimately exhibit aberrant migration in Rb deficiency. With this strategy, we identified a number of putative target genes that are deregulated in conditional Rb mutants and that have been shown to mediate neuronal migration, including migration of interneurons. Further, we have validated our microarray results for one candidate gene, neogenin. Neogenin is of particular interest, not only because of its known and hypothesized roles in regulating neuronal migration (21, 72, 87) but also because of its identification through other E2F microarray studies (69). Even more significant, through sensitive subtractive screening assays, neogenin, was recently shown to be a novel, direct target gene of E2F1, with expression induced in a cell cycle-independent manner (32). As E2F1 is capable of inducing E2F3 expression, it is possible that these genes may be also induced by E2F3 (reviewed in reference 78). While previous studies have provided evidence in support of roles for the Rb/E2F pathway in regulating cell cycle-independent functions, evidence for deregulation of such genes in vivo has not been reported. Thus, our data demonstrating that Rb interacts specifically through E2F3 to mediate neuronal migration represent the first physiological demonstration that such an in vivo role for the Rb/E2F pathway beyond cell cycle regulation exists.
In conclusion, our results demonstrate that both functionally redundant and unique roles exist for E2F1 and E2F3 in regulating Rb-mediated neurogenesis. As Rb-mediated migration is mediated specifically through E2F3, our results represent a novel, physiologically relevant requirement for the Rb/E2F pathway beyond cell cycle regulation in vivo, pointing toward novel targets specific for E2F3-mediated transcription in the context of neuronal migration.
Acknowledgments
We thank Laszlo Jakoi, Duke University, and Fred Dick and Laurie Anne Seifried, University of Western Ontario, for advice regarding the immunoprecipitation and immunoprecipitation gel shift assays; Sheng Hou, NRC-Ottawa for helpful discussions; and Helen Cooper, University of Queensland, and Elke Stein, Yale University, for in situ probes for neogenin. We thank Noel Ghanem for a critical review of the manuscript and assistance. We thank J. G. MacLaurin, W. C. McIntosh, and L. Jui for excellent technical assistance.
This work was funded by a CIHR grant to R.S.S., and the microarray was funded by the Stem Cell Network. K.A.M. is a recipient of a CIHR Canada Graduate Doctoral Research Award, V.A.R. is a recipient of OGS and SCN studentships, J.L.V. is a recipient of an HSFC fellowship, and K.L.F. is a recipient of CIHR doctoral and postdoctoral research awards.
Footnotes
Published ahead of print on 23 April 2007.
REFERENCES
- 1.Attwooll, C., E. Lazzerini Denchi, and K. Helin. 2004. The E2F family: specific functions and overlapping interests. EMBO J. 23:4709-4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benes, F. M., and S. Berretta. 2001. GABAergic interneurons: implications for understanding schizophrenia and bipolar disorder. Neuropsychopharmacology 25:1-27. [DOI] [PubMed] [Google Scholar]
- 3.Bentivoglio, M., L. Tassi, E. Pech, C. Costa, P. F. Fabene, and R. Spreafico. 2003. Cortical development and focal cortical dysplasia. Epileptic Disord. 5(Suppl. 2):S27-S34. [PubMed] [Google Scholar]
- 4.Bielle, F., A. Griveau, N. Narboux-Neme, S. Vigneau, M. Sigrist, S. Arber, M. Wassef, and A. Pierani. 2005. Multiple origins of Cajal-Retzius cells at the borders of the developing pallium. Nat. Neurosci. 8:1002-1012. [DOI] [PubMed] [Google Scholar]
- 5.Black, E. P., T. Hallstrom, H. K. Dressman, M. West, and J. R. Nevins. 2005. Distinctions in the specificity of E2F function revealed by gene expression signatures. Proc. Natl. Acad. Sci. USA 102:15948-15953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bulfone, A., S. M. Smiga, K. Shimamura, A. Peterson, L. Puelles, and J. L. Rubenstein. 1995. T-brain-1: a homolog of Brachyury whose expression defines molecularly distinct domains within the cerebral cortex. Neuron 15:63-78. [DOI] [PubMed] [Google Scholar]
- 7.Callaghan, D. A., L. Dong, S. M. Callaghan, Y. X. Hou, L. Dagnino, and R. S. Slack. 1999. Neural precursor cells differentiating in the absence of Rb exhibit delayed terminal mitosis and deregulated E2F 1 and 3 activity. Dev. Biol. 207:257-270. [DOI] [PubMed] [Google Scholar]
- 8.Chen, D., I. Livne-bar, J. L. Vanderluit, R. S. Slack, M. Agochiya, and R. Bremner. 2004. Cell-specific effects of RB or RB/p107 loss on retinal development implicate an intrinsically death-resistant cell-of-origin in retinoblastoma. Cancer Cell 5:539-551. [DOI] [PubMed] [Google Scholar]
- 9.Christensen, J., P. Cloos, U. Toftegaard, D. Klinkenberg, A. P. Bracken, E. Trinh, M. Heeran, L. Di Stefano, and K. Helin. 2005. Characterization of E2F8, a novel E2F-like cell-cycle regulated repressor of E2F-activated transcription. Nucleic Acids Res. 33:5458-5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clarke, A. R., E. R. Maandag, M. van Roon, N. M. van der Lugt, M. van der Valk, M. L. Hooper, A. Berns, and H. te Riele. 1992. Requirement for a functional Rb-1 gene in murine development. Nature 359:328-330. [DOI] [PubMed] [Google Scholar]
- 11.Cloud, J. E., C. Rogers, T. L. Reza, U. Ziebold, J. R. Stone, M. H. Picard, A. M. Caron, R. T. Bronson, and J. A. Lees. 2002. Mutant mouse models reveal the relative roles of E2F1 and E2F3 in vivo. Mol. Cell. Biol. 22:2663-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooper-Kuhn, C. M., M. Vroemen, J. Brown, H. Ye, M. A. Thompson, J. Winkler, and H. G. Kuhn. 2002. Impaired adult neurogenesis in mice lacking the transcription factor E2F1. Mol. Cell Neurosci. 21:312-323. [DOI] [PubMed] [Google Scholar]
- 13.Dagnino, L., C. J. Fry, S. M. Bartley, P. Farnham, B. L. Gallie, and R. A. Phillips. 1997. Expression patterns of the E2F family of transcription factors during mouse nervous system development. Mech. Dev. 66:13-25. [DOI] [PubMed] [Google Scholar]
- 14.de Bruin, A., B. Maiti, L. Jakoi, C. Timmers, R. Buerki, and G. Leone. 2003. Identification and characterization of E2F7, a novel mammalian E2F family member capable of blocking cellular proliferation. J. Biol. Chem. 278:42041-42049. [DOI] [PubMed] [Google Scholar]
- 15.Dick, F. A., and N. Dyson. 2003. pRB contains an E2F1-specific binding domain that allows E2F1-induced apoptosis to be regulated separately from other E2F activities. Mol. Cell 12:639-649. [DOI] [PubMed] [Google Scholar]
- 16.Dimova, D. K., and N. J. Dyson. 2005. The E2F transcriptional network: old acquaintances with new faces. Oncogene 24:2810-2826. [DOI] [PubMed] [Google Scholar]
- 17.Di Stefano, L., M. R. Jensen, and K. Helin. 2003. E2F7, a novel E2F featuring DP-independent repression of a subset of E2F-regulated genes. EMBO J. 22:6289-6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferguson, K. L., K. A. McClellan, J. L. Vanderluit, W. C. McIntosh, C. Schuurmans, F. Polleux, and R. S. Slack. 2005. A cell-autonomous requirement for the cell cycle regulatory protein, Rb, in neuronal migration. EMBO J. 24:4381-4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferguson, K. L., J. L. Vanderluit, J. M. Hebert, W. C. McIntosh, E. Tibbo, J. G. MacLaurin, D. S. Park, V. A. Wallace, M. Vooijs, S. K. McConnell, and R. S. Slack. 2002. Telencephalon-specific Rb knockouts reveal enhanced neurogenesis, survival and abnormal cortical development. EMBO J. 21:3337-3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Field, S. J., F. Y. Tsai, F. Kuo, A. M. Zubiaga, W. G. Kaelin, Jr., D. M. Livingston, S. H. Orkin, and M. E. Greenberg. 1996. E2F-1 functions in mice to promote apoptosis and suppress proliferation. Cell 85:549-561. [DOI] [PubMed] [Google Scholar]
- 21.Fitzgerald, D. P., S. J. Cole, A. Hammond, C. Seaman, and H. M. Cooper. 2006. Characterization of neogenin-expressing neural progenitor populations and migrating neuroblasts in the embryonic mouse forebrain. Neuroscience 142:703-716. [DOI] [PubMed] [Google Scholar]
- 22.Gad, J. M., S. L. Keeling, A. F. Wilks, S. S. Tan, and H. M. Cooper. 1997. The expression patterns of guidance receptors, DCC and Neogenin, are spatially and temporally distinct throughout mouse embryogenesis. Dev. Biol. 192:258-273. [DOI] [PubMed] [Google Scholar]
- 23.Giacobini, P., A. S. Kopin, P. M. Beart, L. D. Mercer, A. Fasolo, and S. Wray. 2004. Cholecystokinin modulates migration of gonadotropin-releasing hormone-1 neurons. J. Neurosci. 24:4737-4748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gotz, M., and W. B. Huttner. 2005. The cell biology of neurogenesis. Nat Rev. Mol. Cell Biol. 6:777-788. [DOI] [PubMed] [Google Scholar]
- 25.Guillemot, F. 2005. Cellular and molecular control of neurogenesis in the mammalian telencephalon. Curr. Opin. Cell Biol. 17:639-647. [DOI] [PubMed] [Google Scholar]
- 26.Hallstrom, T. C., and J. R. Nevins. 2003. Specificity in the activation and control of transcription factor E2F-dependent apoptosis. Proc. Natl. Acad. Sci. USA 100:10848-10853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He, Y., M. K. Armanious, M. J. Thomas, and W. D. Cress. 2000. Identification of E2F-3B, an alternative form of E2F-3 lacking a conserved N-terminal region. Oncogene 19:3422-3433. [DOI] [PubMed] [Google Scholar]
- 28.Hebert, J. M., and S. K. McConnell. 2000. Targeting of cre to the Foxg1 (BF-1) locus mediates loxP recombination in the telencephalon and other developing head structures. Dev. Biol. 222:296-306. [DOI] [PubMed] [Google Scholar]
- 29.Hevner, R. F., L. Shi, N. Justice, Y. Hsueh, M. Sheng, S. Smiga, A. Bulfone, A. M. Goffinet, A. T. Campagnoni, and J. L. Rubenstein. 2001. Tbr1 regulates differentiation of the preplate and layer 6. Neuron 29:353-366. [DOI] [PubMed] [Google Scholar]
- 30.Humbert, P. O., R. Verona, J. M. Trimarchi, C. Rogers, S. Dandapani, and J. A. Lees. 2000. E2f3 is critical for normal cellular proliferation. Genes Dev. 14:690-703. [PMC free article] [PubMed] [Google Scholar]
- 31.Ikeda, M. A., L. Jakoi, and J. R. Nevins. 1996. A unique role for the Rb protein in controlling E2F accumulation during cell growth and differentiation. Proc. Natl. Acad. Sci. USA 93:3215-3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iwanaga, R., H. Komori, S. Ishida, N. Okamura, K. Nakayama, K. I. Nakayama, and K. Ohtani. 2006. Identification of novel E2F1 target genes regulated in cell cycle-dependent and independent manners. Oncogene 25:1786-1798. [DOI] [PubMed] [Google Scholar]
- 33.Jacks, T., A. Fazeli, E. M. Schmitt, R. T. Bronson, M. A. Goodell, and R. A. Weinberg. 1992. Effects of an Rb mutation in the mouse. Nature 359:295-300. [DOI] [PubMed] [Google Scholar]
- 34.Kalanithi, P. S., W. Zheng, Y. Kataoka, M. DiFiglia, H. Grantz, C. B. Saper, M. L. Schwartz, J. F. Leckman, and F. M. Vaccarino. 2005. Altered parvalbumin-positive neuron distribution in basal ganglia of individuals with Tourette syndrome. Proc. Natl. Acad. Sci. USA 102:13307-13312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kato, M., and W. B. Dobyns. 2005. X-linked lissencephaly with abnormal genitalia as a tangential migration disorder causing intractable epilepsy: proposal for a new term, “interneuronopathy.” J. Child Neurol. 20:392-397. [DOI] [PubMed] [Google Scholar]
- 36.Keverne, E. B. 1999. GABA-ergic neurons and the neurobiology of schizophrenia and other psychoses. Brain Res. Bull. 48:467-473. [DOI] [PubMed] [Google Scholar]
- 37.Kriegstein, A. R., and S. C. Noctor. 2004. Patterns of neuronal migration in the embryonic cortex. Trends Neurosci. 27:392-399. [DOI] [PubMed] [Google Scholar]
- 38.Lasorella, A., M. Noseda, M. Beyna, Y. Yokota, and A. Iavarone. 2000. Id2 is a retinoblastoma protein target and mediates signalling by Myc oncoproteins. Nature 407:592-598. [DOI] [PubMed] [Google Scholar]
- 39.Lee, E., N. Hu, S. S. F. Yuan, L. A. Cox, A. Bradley, W. Lee, and K. Herrup. 1994. Dual roles of the retinoblastoma protein in cell cycle regulation and neuron differentiation. Genes Dev. 8:2008-2021. [DOI] [PubMed] [Google Scholar]
- 40.Lee, E. Y., C. Y. Chang, N. Hu, Y. C. Wang, C. C. Lai, K. Herrup, W. H. Lee, and A. Bradley. 1992. Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature 359:288-294. [DOI] [PubMed] [Google Scholar]
- 41.Leone, G., F. Nuckolls, S. Ishida, M. Adams, R. Sears, L. Jakoi, A. Miron, and J. R. Nevins. 2000. Identification of a novel E2F3 product suggests a mechanism for determining specificity of repression by Rb proteins. Mol. Cell. Biol. 20:3626-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leone, G., R. Sears, E. Huang, R. Rempel, F. Nuckolls, C. H. Park, P. Giangrande, L. Wu, H. I. Saavedra, S. J. Field, M. A. Thompson, H. Yang, Y. Fujiwara, M. E. Greenberg, S. Orkin, C. Smith, and J. R. Nevins. 2001. Myc requires distinct E2F activities to induce S phase and apoptosis. Mol. Cell 8:105-113. [DOI] [PubMed] [Google Scholar]
- 43.Levitt, P. 2005. Disruption of interneuron development. Epilepsia 46(Suppl. 7):22-28. [DOI] [PubMed] [Google Scholar]
- 44.Levitt, P., K. L. Eagleson, and E. M. Powell. 2004. Regulation of neocortical interneuron development and the implications for neurodevelopmental disorders. Trends Neurosci. 27:400-406. [DOI] [PubMed] [Google Scholar]
- 45.Lewis, D. A., and P. Levitt. 2002. Schizophrenia as a disorder of neurodevelopment. Annu. Rev. Neurosci. 25:409-432. [DOI] [PubMed] [Google Scholar]
- 46.Lipinski, M. M., K. F. Macleod, B. O. Williams, T. L. Mullaney, D. Crowley, and T. Jacks. 2001. Cell-autonomous and non-cell-autonomous functions of the Rb tumor suppressor in developing central nervous system. EMBO J. 20:3402-3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu, Y., J. Berndt, F. Su, H. Tawarayama, W. Shoji, J. Y. Kuwada, and M. C. Halloran. 2004. Semaphorin3D guides retinal axons along the dorsoventral axis of the tectum. J. Neurosci. 24:310-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Logan, N., L. Delavaine, A. Graham, C. Reilly, J. Wilson, T. R. Brummelkamp, E. M. Hijmans, R. Bernards, and N. B. La Thangue. 2004. E2F-7: a distinctive E2F family member with an unusual organization of DNA-binding domains. Oncogene 23:5138-5150. [DOI] [PubMed] [Google Scholar]
- 49.Logan, N., A. Graham, X. Zhao, R. Fisher, B. Maiti, G. Leone, and N. B. La Thangue. 2005. E2F-8: an E2F family member with a similar organization of DNA-binding domains to E2F-7. Oncogene 24:5000-5004. [DOI] [PubMed] [Google Scholar]
- 50.Lopez-Bendito, G., K. Sturgess, F. Erdelyi, G. Szabo, Z. Molnar, and O. Paulsen. 2004. Preferential origin and layer destination of GAD65-GFP cortical interneurons. Cereb. Cortex 14:1122-1133. [DOI] [PubMed] [Google Scholar]
- 51.Maandag, E. C., M. van der Valk, M. Vlaar, C. Feltkamp, J. O'Brien, M. van Roon, N. van der Lugt, A. Berns, and H. te Riele. 1994. Developmental rescue of an embryonic-lethal mutation in the retinoblastoma gene in chimeric mice. EMBO J. 13:4260-4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.MacPherson, D., J. Sage, D. Crowley, A. Trumpp, R. T. Bronson, and T. Jacks. 2003. Conditional mutation of Rb causes cell cycle defects without apoptosis in the central nervous system. Mol. Cell. Biol. 23:1044-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.MacPherson, D., J. Sage, T. Kim, D. Ho, M. E. McLaughlin, and T. Jacks. 2004. Cell type-specific effects of Rb deletion in the murine retina. Genes Dev. 18:1681-1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maiti, B., J. Li, A. de Bruin, F. Gordon, C. Timmers, R. Opavsky, K. Patil, J. Tuttle, W. Cleghorn, and G. Leone. 2005. Cloning and characterization of mouse E2F8, a novel mammalian E2F family member capable of blocking cellular proliferation. J. Biol. Chem. 280:18211-18220. [DOI] [PubMed] [Google Scholar]
- 55.Marin, O., and J. L. Rubenstein. 2003. Cell migration in the forebrain. Annu. Rev. Neurosci. 26:441-483. [DOI] [PubMed] [Google Scholar]
- 56.Marin, O., and J. L. Rubenstein. 2001. A long, remarkable journey: tangential migration in the telencephalon. Nat. Rev. Neurosci. 2:780-790. [DOI] [PubMed] [Google Scholar]
- 57.Marino, S., D. Hoogervoorst, S. Brandner, and A. Berns. 2003. Rb and p107 are required for normal cerebellar development and granule cell survival but not for Purkinje cell persistence. Development 130:3359-3368. [DOI] [PubMed] [Google Scholar]
- 58.Marino, S., M. Vooijs, H. van Der Gulden, J. Jonkers, and A. Berns. 2000. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev. 14:994-1004. [PMC free article] [PubMed] [Google Scholar]
- 59.Marin-Padilla, M. 1998. Cajal-Retzius cells and the development of the neocortex. Trends Neurosci. 21:64-71. [DOI] [PubMed] [Google Scholar]
- 60.Matsumura, I., H. Tanaka, and Y. Kanakura. 2003. E2F1 and c-Myc in cell growth and death. Cell Cycle 2:333-338. [PubMed] [Google Scholar]
- 61.Matsunaga, E., S. Tauszig-Delamasure, P. P. Monnier, B. K. Mueller, S. M. Strittmatter, P. Mehlen, and A. Chedotal. 2004. RGM and its receptor neogenin regulate neuronal survival. Nat. Cell Biol. 6:749-755. [DOI] [PubMed] [Google Scholar]
- 62.McBain, C. J., and A. Fisahn. 2001. Interneurons unbound. Nat. Rev. Neurosci. 2:11-23. [DOI] [PubMed] [Google Scholar]
- 63.McClellan, K. A., and R. S. Slack. 2006. Novel functions for cell cycle genes in nervous system development. Cell Cycle 5:1506-1513. [DOI] [PubMed] [Google Scholar]
- 64.Meyer, G., A. M. Goffinet, and A. Fairen. 1999. What is a Cajal-Retzius cell? A reassessment of a classical cell type based on recent observations in the developing neocortex. Cereb. Cortex 9:765-775. [DOI] [PubMed] [Google Scholar]
- 65.Morante-Oria, J., A. Carleton, B. Ortino, E. J. Kremer, A. Fairen, and P. M. Lledo. 2003. Subpallial origin of a population of projecting pioneer neurons during corticogenesis. Proc. Natl. Acad. Sci. USA 100:12468-12473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Morgenbesser, S. D., B. O. Williams, T. Jacks, and R. A. DePinho. 1994. p53-dependent apoptosis produced by Rb-deficiency in the developing mouse lens. Nature 371:72-74. [DOI] [PubMed] [Google Scholar]
- 67.Morris, E. J., and N. J. Dyson. 2001. Retinoblastoma protein partners. Adv. Cancer Res. 82:1-54. [DOI] [PubMed] [Google Scholar]
- 68.Morshead, C. M., and D. van der Kooy. 1992. Postmitotic death is the fate of constitutively proliferating cells in the subependymal layer of the adult mouse brain. J. Neurosci. 12:249-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Muller, H., A. P. Bracken, R. Vernell, M. C. Moroni, F. Christians, E. Grassilli, E. Prosperini, E. Vigo, J. D. Oliner, and K. Helin. 2001. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 15:267-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.O'Rourke, M. P., and P. P. Tam. 2002. Twist functions in mouse development. Int. J. Dev. Biol. 46:401-413. [PubMed] [Google Scholar]
- 71.Polleux, F., and J. M. Lauder. 2004. Toward a developmental neurobiology of autism. Ment. Retard. Dev. Disabil. Res. Rev. 10:303-317. [DOI] [PubMed] [Google Scholar]
- 72.Rajagopalan, S., L. Deitinghoff, D. Davis, S. Conrad, T. Skutella, A. Chedotal, B. K. Mueller, and S. M. Strittmatter. 2004. Neogenin mediates the action of repulsive guidance molecule. Nat. Cell Biol. 6:756-762. [DOI] [PubMed] [Google Scholar]
- 73.Robanus-Maandag, E., M. Dekker, M. van der Valk, M. L. Carrozza, J. C. Jeanny, J. H. Dannenberg, A. Berns, and H. te Riele. 1998. p107 is a suppressor of retinoblastoma development in pRb-deficient mice. Genes Dev. 12:1599-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rubin, S. M., A. L. Gall, N. Zheng, and N. P. Pavletich. 2005. Structure of the Rb C-terminal domain bound to E2F1-DP1: a mechanism for phosphorylation-induced E2F release. Cell 123:1093-1106. [DOI] [PubMed] [Google Scholar]
- 75.Saavedra, H. I., L. Wu, A. de Bruin, C. Timmers, T. J. Rosol, M. Weinstein, M. L. Robinson, and G. Leone. 2002. Specificity of E2F1, E2F2, and E2F3 in mediating phenotypes induced by loss of Rb. Cell Growth Differ. 13:215-225. [PubMed] [Google Scholar]
- 76.Sherr, E. H. 2003. The ARX story (epilepsy, mental retardation, autism, and cerebral malformations): one gene leads to many phenotypes. Curr. Opin. Pediatr. 15:567-571. [DOI] [PubMed] [Google Scholar]
- 77.Templeton, D., S. H. Park, L. Lanier, and R. A. Weinberg. 1991. Nonfunctional mutants of the retinoblastoma protein are characterized by defects in phosphorylation, viral oncoprotein association, and nuclear tethering. Proc. Natl. Acad. Sci. USA 88:3033-3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Trimarchi, J. M., and J. A. Lees. 2002. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol. 3:11-20. [DOI] [PubMed] [Google Scholar]
- 79.Trommsdorff, M., M. Gotthardt, T. Hiesberger, J. Shelton, W. Stockinger, J. Nimpf, R. E. Hammer, J. A. Richardson, and J. Herz. 1999. Reeler/Disabled-like disruption of neuronal migration in knockout mice lacking the VLDL receptor and ApoE receptor 2. Cell 97:689-701. [DOI] [PubMed] [Google Scholar]
- 80.Tropepe, V., C. G. Craig, C. M. Morshead, and D. van der Kooy. 1997. Transforming growth factor-alpha null and senescent mice show decreased neural progenitor cell proliferation in the forebrain subependyma. J. Neurosci. 17:7850-7859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tsai, K. Y., Y. Hu, K. F. Macleod, D. Crowley, L. Yamasaki, and T. Jacks. 1998. Mutation of E2f-1 suppresses apoptosis and inappropriate S phase entry and extends survival of Rb-deficient mouse embryos. Mol. Cell 2:293-304. [DOI] [PubMed] [Google Scholar]
- 82.Vanderluit, J. L., K. L. Ferguson, V. Nikoletopoulou, M. Parker, V. Ruzhynsky, T. Alexson, S. M. McNamara, D. S. Park, M. Rudnicki, and R. S. Slack. 2004. p107 regulates neural precursor cells in the mammalian brain. J. Cell Biol. 166:853-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vielmetter, J., J. F. Kayyem, J. M. Roman, and W. J. Dreyer. 1994. Neogenin, an avian cell surface protein expressed during terminal neuronal differentiation, is closely related to the human tumor suppressor molecule deleted in colorectal cancer. J. Cell Biol. 127:2009-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vooijs, M., M. van der Valk, H. te Riele, and A. Berns. 1998. Flp-mediated tissue-specific inactivation of the retinoblastoma tumor suppressor gene in the mouse. Oncogene 17:1-12. [DOI] [PubMed] [Google Scholar]
- 85.Wallace, V. A., and M. C. Raff. 1999. A role for Sonic hedgehog in axon-to-astrocyte signalling in the rodent optic nerve. Development 126:2901-2909. [DOI] [PubMed] [Google Scholar]
- 86.Williams, B. O., E. M. Schmitt, L. Remington, R. T. Bronson, D. M. Albert, R. A. Weinberg, and T. Jacks. 1994. Extensive contribution of Rb-deficient cells to adult chimeric mice with limited histopathological consequences. EMBO J. 13:4251-4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wilson, N. H., and B. Key. 2006. Neogenin interacts with RGMa and netrin-1 to guide axons within the embryonic vertebrate forebrain. Dev. Biol. 296:485-498. [DOI] [PubMed] [Google Scholar]
- 88.Wolman, M. A., Y. Liu, H. Tawarayama, W. Shoji, and M. C. Halloran. 2004. Repulsion and attraction of axons by semaphorin3D are mediated by different neuropilins in vivo. J. Neurosci. 24:8428-8435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wonders, C. P., and S. A. Anderson. 2006. The origin and specification of cortical interneurons. Nat. Rev. Neurosci 7:687-696. [DOI] [PubMed] [Google Scholar]
- 90.Wu, L., A. de Bruin, H. I. Saavedra, M. Starovic, A. Trimboli, Y. Yang, J. Opavska, P. Wilson, J. C. Thompson, M. C. Ostrowski, T. J. Rosol, L. A. Woollett, M. Weinstein, J. C. Cross, M. L. Robinson, and G. Leone. 2003. Extra-embryonic function of Rb is essential for embryonic development and viability. Nature 421:942-947. [DOI] [PubMed] [Google Scholar]
- 91.Wu, L., C. Timmers, B. Maiti, H. I. Saavedra, L. Sang, G. T. Chong, F. Nuckolls, P. Giangrande, F. A. Wright, S. J. Field, M. E. Greenberg, S. Orkin, J. R. Nevins, M. L. Robinson, and G. Leone. 2001. The E2F1-3 transcription factors are essential for cellular proliferation. Nature 414:457-462. [DOI] [PubMed] [Google Scholar]
- 92.Yamasaki, L., T. Jacks, R. Bronson, E. Goillot, E. Harlow, and N. J. Dyson. 1996. Tumor induction and tissue atrophy in mice lacking E2F-1. Cell 85:537-548. [DOI] [PubMed] [Google Scholar]
- 93.Ziebold, U., T. Reza, A. Caron, and J. A. Lees. 2001. E2F3 contributes both to the inappropriate proliferation and to the apoptosis arising in Rb mutant embryos. Genes Dev. 15:386-391. [DOI] [PMC free article] [PubMed] [Google Scholar]