Abstract
Faithful mitotic partitioning of the Golgi apparatus and the centrosome is critical for proper cell division. Although these two cytoplasmic organelles are probably coordinated during cell division, supporting evidence of this coordination is still largely lacking. Here, we show that the RAD50-interacting protein, RINT-1, is localized at the Golgi apparatus and the centrosome in addition to the endoplasmic reticulum. To examine the biological roles of RINT-1, we found that the homozygous deletion of Rint-1 caused early embryonic lethality at embryonic day 5 (E5) to E6 and the failure of blastocyst outgrowth ex vivo. About 81% of the Rint-1 heterozygotes succumbed to multiple tumor formation with haploinsufficiency during their average life span of 24 months. To pinpoint the cellular function of RINT-1, we found that RINT-1 depletion by RNA interference led to the loss of the pericentriolar positioning and dispersal of the Golgi apparatus and concurrent centrosome amplification during the interphase. Upon mitotic entry, RINT-1-deficient cells exhibited multiple abnormalities, including aberrant Golgi dynamics during early mitosis and defective reassembly at telophase, increased formation of multiple spindle poles, and frequent chromosome missegregation. Mitotic cells often underwent cell death in part due to the overwhelming cellular defects. Taken together, these findings suggest that RINT-1 serves as a novel tumor suppressor essential for maintaining the dynamic integrity of the Golgi apparatus and the centrosome, a prerequisite to their proper coordination during cell division.
The endoplasmic reticulum (ER) and Golgi apparatus are both fundamental organelles for cellular activities. The ER performs specialized functions including protein synthesis, folding and modification, the insertion of proteins into the membrane, lipid biosynthesis, vesicular trafficking, and calcium sequestration (16, 19). The ER is in a dynamic equilibrium with the Golgi apparatus, which is involved in several major functions essential for growth, homeostasis, and the division of eukaryotic cells (3). The Golgi apparatus operates as a site for processing and modifying proteins and lipids in the secretory pathway, sorting and transporting proteins, and conveying vesicles from the ER to the appropriate subcellular destinations (17). It also serves as a membranous scaffold for diverse signaling, sorting, and cytoskeleton proteins (2, 14, 23).
In addition to the close dynamic relationship between the ER and Golgi apparatus, a growing interest has been focused on the connection between the Golgi apparatus and the centrosome (46), a complex and dynamic organelle that functions as the major microtubular organization center (18). In mammalian cells, both the Golgi apparatus and the centrosome play important roles in mitosis. The centrosome plays a key role in chromosome segregation and cell cycle checkpoint control (30), while the fragmentation of the Golgi apparatus is required for the mitotic phase entry (9, 50). An intriguing question is how the Golgi apparatus collaborates with the centrosome during cell division. It is likely that the pericentriolar Golgi organization in mammalian cells may serve as a means to connect the Golgi apparatus and the centrosome for the purpose of division. A class of proteins, including Golgi spectrin, ankyrins, GMAP210, and dynein, tether the Golgi ribbon to the microtubule network. Then microtubules and the associated motor and nonmotor proteins anchor the Golgi apparatus in proximity to the centrosome. This pericentriolar localization is thought to be a consequence of the gathering of Golgi vesicles toward the centrosome by minus end-directed motors along microtubules. When microtubules are depolymerized or the associated proteins are inactivated, the Golgi apparatus becomes fragmented and dispersed from the pericentriolar position (8, 21, 25, 34). However, it is speculated that additional proteins may function in linking the Golgi apparatus more directly with the centrosome and regulating cross talk between the two organelles. Molecules commonly localized to the Golgi apparatus and centrosome may be implicated in Golgi organization and positioning. The Golgi apparatus resident proteins golgin 97 and trans-Golgi network protein 38 are associated with the centrosome throughout the cell cycle and even after the Golgi dispersion, suggesting that these two proteins may be involved in the pericentriolar nucleation of the Golgi apparatus by interacting with the centrosome (52). However, the biological significance of these proteins remains unclear.
RINT-1 was originally identified as a RAD50-interacting protein of 792 amino acids (56). The overexpression of an N-terminally truncated RINT-1 protein leads to a defect in the radiation-induced G2/M checkpoint, suggesting a role for RINT-1 in cell cycle progression (56). Yeast two-hybrid screening showed that RINT-1 also interacts with an Rb family protein, p130, via a region spanning amino acids 358 to 440. The p130 protein interacts with RAD50 in a RINT-1-dependent manner, indicating that RINT-1 serves as a bridge between p130 and RAD50. A loss of either p130 or RINT-1 in human foreskin fibroblasts results in telomere elongation without affecting telomerase activity, suggesting that p130 and RINT-1 regulate the telomerase-independent telomere length (29). In addition, the N-terminal region of RINT-1 interacts with a dynamitin-interacting protein, Zeste white 10 (ZW10), which is located at the ER and the Golgi apparatus, throughout the cell cycle (4, 24, 55). ZW10 and RINT-1 form a complex with syntaxin 18, an ER-localized t-SNARE (target membrane-associated SNAP receptor), and mediate membrane trafficking between the ER and the Golgi apparatus (4). In cells depleted of either RINT-1 or ZW10, the Golgi structure becomes dispersed during interphase, suggesting that both proteins are involved in the formation of Golgi architecture (4). These results suggest that RINT-1 has diverse functions involving the cell cycle checkpoint, telomere length, and membrane trafficking through discrete interactions with RAD50, p130, and ZW10. However, the fundamental biological functions of RINT-1 remain to be elucidated.
In this study, we found that RINT-1 localized not only at the ER and the Golgi apparatus but also at the centrosome, implicating a complex yet coordinate role for RINT-1 in association with these organelles. To investigate its biological function, we depleted cells of RINT-1 by RNA interference (RNAi) and observed the dispersal and loss of the pericentriolar positioning of the Golgi apparatus and increased centrosome amplification during the interphase. Severe mitotic cell death was concomitantly elicited, probably as a result of overwhelming mitotic defects, including impaired Golgi apparatus disassembly and reassembly, the formation of multiple spindle poles, and frequent chromosome missegregation. Importantly, the homozygous deletion of Rint-1 alleles in mice caused early embryonic lethality at embryonic day 5.5 (E5.5). Interestingly, 81% of Rint-1+/− mice developed multiple tumors with haploinsufficiency, indicating a potential role for RINT-1 as a tumor suppressor. These findings suggest that RINT-1 serves as a tumor suppressor and maintains the dynamic integrity of the Golgi apparatus and the centrosome critical for cell survival.
MATERIALS AND METHODS
Cell culture and cell synchronization.
Human cervical cancer HeLa and osteosarcoma U2OS cell lines were cultured in high-glucose-content Dulbecco modified Eagle medium containing 10% fetal bovine serum. Human B-cell lymphoma LEM cells were cultured in RPMI 1640 medium containing 10% fetal bovine serum. Cell synchronization at M phase was achieved by treatment with 0.1 μg of nocodazole/ml for 8 h. Mitotic cells were then collected and replated at time 0. Cell synchronization at the G1/S boundary was achieved by 19 h of treatment with 2 mM thymidine followed by 12 h of drug release and 24 h of retreatment with 2 mM thymidine. For the centrosome duplication study, cells were arrested at the G1/S boundary by treatment with 2 mM hydroxyurea (HU) for the indicated time periods (see Fig. 5).
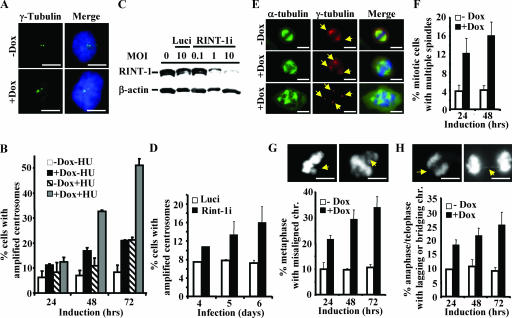
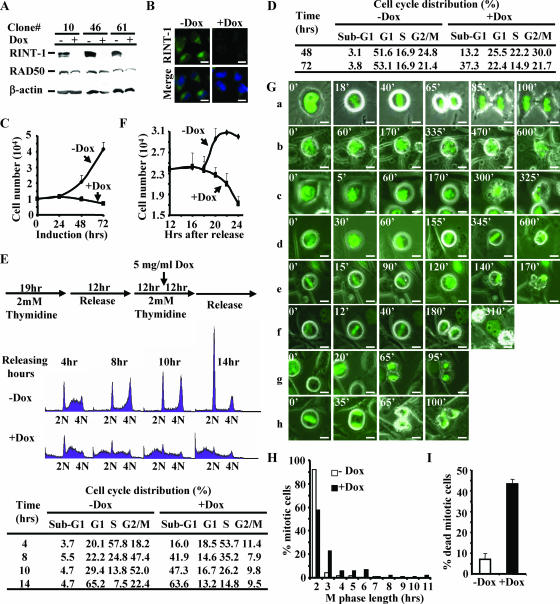
FIG. 5.
Depletion of RINT-1 results in centrosome amplification and consequential chromosome instability. (A) Amplified centrosomes in HeLa-RINT-1i cells induced with 5 μg of doxycycline/ml were detected by immunostaining using anti-γ-tubulin (green) and DAPI for the nucleus (blue). +Dox, with doxycycline; −Dox, without doxycycline. Bars, 10 μm. (B) The percentages of cells with centrosome amplification were determined at the indicated times after induction. HeLa-RINT-1i cells were induced with 5 μg of doxycycline/ml and/or 2 mM HU for the indicated time periods. Three hundred cells were counted at each time point. Error bars, standard deviations. (C) The expression of RINT-1 protein in U2OS cells was abolished by infection with rAd-RINT-1i but not with rAd-Luci. Cells were infected with adenoviruses at the indicated multiplicities of infection (MOI) for 72 h, and the cell lysates were analyzed by Western blotting. β-actin served as an internal control. (D) The percentages of U2OS cells infected with rAd-RINT-1i with centrosome amplification were increased compared to those of cells infected with rAd-Luci. Two hundred to 300 cells were counted at each time point. Error bars, standard deviations. (E) Multiple spindles in HeLa-RINT-1i cells were detected upon induction with 5 μg of doxycycline/ml. Cells were coimmunostained with anti-α-tubulin for the mitotic asters and anti-γ-tubulin for the centrosomes (arrows). Bars, 10 μm. (F) The percentages of HeLa-RINT-1i cells with multiple spindles upon doxycycline induction were increased compared to those of control cells. Two hundred cells were counted at each time point. Error bars, standard deviations. (G and H) The percentages of RINT-1-depleted cells with aberrant chromosome segregation, including a misaligned chromosome(s) at metaphase (G) and a lagging or bridging chromosome(s) at telophase (H), were increased compared to those of control cells. The arrows indicate the misaligned, lagging, or bridging chromosomes. Cells at metaphase (n > 200) and anaphase/telophase (n > 200) were examined at each time point. Representative images are shown at the tops of two histograms. All the data are representative of results from two independent experiments. Bars, 10 μm. Error bars, standard deviations.
RINT-1 siRNA construction and adenovirus production.
A RINT-1 small interfering RNA (siRNA) vector was constructed by inserting a DNA fragment derived from two annealed synthetic oligonucleotides (5′-GAT CCC GAG ATC GAA CGT CAT CTT GTT CAA GAG ACA AGA TGA CGT TCG ATC TCT TTT TGG AAA-3′ and 5′-AGC TTT TCC AAA AAG AGA TCG AAC GTC ATC TTG TCT CTT GAA CAA GAT GAC GTT CGA TCT CGG-3′) into the pTER+ vector (54) to generate pTER-(RINT-1i). Four tandem copies of the RINT-1 siRNA expression cassette under an H1 promoter [(RINT-1i)4] were then subcloned into the pTER+ vector to generate pTER-(RINT-1i)4. An EcoRI/NotI fragment containing (RINT-1i)4 from pTER-(RINT-1i)4 was subcloned into the pAdTrack vector to generate pAd-(RINT-1i)4. The adenovirus was then produced in 293 cells by following the protocol described previously (22). The viral titer was determined by plaque formation on 293 cells and expressed as the number of PFU per milliliter. An adenovirus sample with the titer of 109 to 1010 PFU/ml was collected.
Generation of inducible stable clones.
pPUR-(RINT-1i)4 was generated by inserting the (RINT-1i)4 fragment from pTER-(RINT-1i)4 into the pPUR vector. Stable clones were established by the cotransfection of HeLa cells with the pCDNA6TR plasmid, which expresses the Tet repressor, and the pPUR-(RINT-1i)4 plasmid by using Lipofectin transfection reagent (Invitrogen, Carlsbad, CA). Stable cell lines were established by selection with 5.0 μg of blasticidin/ml and 3.3 μg of puromycin/ml. The stable clones, named HeLa-RINT-1i, were treated with 5 μg of doxycycline/ml for different periods of time to induce siRNA expression, and the downregulation of RINT-1 was verified by Western blot analysis. Unless indicated otherwise, HeLa-RINT-1i clone 46 was used throughout the study.
Cell proliferation assay.
HeLa-RINT-1i cells were seeded either as a random or a synchronized population into 12-well plates in duplicate and induced with 5 μg of doxycycline/ml. The viable cells were counted by a trypan blue exclusion assay at designated time points.
Western blot analysis.
Inducible stable siRNA clones or cells infected with adenoviral RNAi were harvested at different time points for Western blot analysis. Cells were resuspended in lysis 250 buffer (50 mM Tris [pH 7.4], 250 mM NaCl, 5 mM EDTA, 0.1% NP-40, 50 mM NaF, and protease inhibitors), subjected to three cycles of freezing and thawing, and centrifuged at 14,000 × g for 2 min at room temperature. An equal amount of the whole-cell lysate was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by immunoblotting. Antibodies used for Western blot analysis were as follows: RINT-1 rabbit antibody PCIN2, RAD50 monoclonal antibody 13B3, and β-actin monoclonal antibody (Genetex, San Antonio, TX).
Immunostaining, immunofluorescence microscopy, and time-lapse imaging.
Cells cultured on coverslips were washed with phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde in PBS containing 0.5% Triton X-100 for 30 min or with chilled methanol for 3 or 5 min. Fixed cells were permeabilized with 0.05% saponin at room temperature for 30 min. After blocking with 10% goat serum in PBS for 1 h, cells were incubated with primary antibodies at room temperature for 2 h or at 4°C overnight. Subsequently, cells were incubated for 1 h with goat anti-mouse or anti-rabbit immunoglobulin G secondary antibodies conjugated with Alexa Fluor 488 or Alexa Fluor 595 dye (Molecular Probes, Eugene, OR) and then stained with 1 μg of DAPI (4′,6′-diamidino-2-phenylindole)/ml for 5 min. Cells were washed, mounted on glass slides with prolonged antifade reagent (Molecular Probes, Eugene, OR), and observed under an Axiovert 200 M inverted microscope (Carl Zeiss Inc., Thornwood, NY). Primary antibodies used were as follows: RINT-1 rabbit antibody PCIN2 for the ER and centrosome staining and mouse antibody PIN no. 3 for the Golgi apparatus staining, rabbit antibody to histone H3 phosphorylated at serine 10 (Upstate, Lake Placid, NY), GM130 mouse antibody (BD Biosciences, San Jose, CA), giantin rabbit antibody (Genetex, San Antonio, TX), calnexin rabbit antibody (Genetex, San Antonio, TX), γ-tubulin mouse or rabbit antibody (Sigma, St. Louis, MO), and α-tubulin mouse antibody (Sigma, St. Louis, MO).
HeLa-RINT-1i cells were infected with retrovirus which expresses histone H2B-green fluorescent protein (GFP) to mark the histone for visualization (32). Chromosome behaviors in living cells were monitored under an Axiovert 200 M inverted microscope using a 20× objective and Axiovision LE 4.3 software (Carl Zeiss Inc. Thornwood, NY). For time-lapse imaging, cells were cultured in CO2-independent medium (GIBCO-BRL, Grand Island, NY) and photographed manually on a heated stage.
Cell cycle analysis by flow cytometry.
Exponentially growing or synchronized cells were treated with 5 μg of doxycycline/ml for various periods of time. Cells were trypsinized, fixed with chilled 70% ethanol, and stained for 30 min with propidium iodide solution (50 μg of propidium iodide/ml, 0.1% sodium citrate, 50 μg of RNase A/ml, 0.03% NP-40 in PBS) at room temperature. Flow cytometric analysis was performed using a FACSCalibur flow cytometer, and the cell cycle distribution was analyzed using CellQuest software (BD Biosciences, San Jose, CA). For each sample, 10,000 events were analyzed.
Isolation and analysis of the centrosome.
The isolation of human centrosomes from human leukemia LEM cells was carried out by following previously described protocols (7). Centrosomes were further purified by using a discontinuous sucrose gradient (65, 50, and 40% sucrose solution), and collected fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by immunoblotting with anti-RINT-1 or anti-γ-tubulin antibody. For immunofluorescence analysis of centrosomes, the centrosome-containing fractions were spun onto coverslips, fixed with chilled methanol for 6 min, and probed with anti-RINT-1 and anti-γ-tubulin antibodies by using protocols described previously (39).
Constructing targeting vectors and establishing the knockout mouse.
The Rint-1 gene was isolated by screening a λDASH mouse genomic library derived from the 129/Sv mouse strain (kindly provided by Tom Doetschman, University of Cincinnati) by using a 2.4-kb fragment of human RINT-1 cDNA as the probe. A 9-kb BamHI/ScaI fragment of the mouse Rint-1 gene containing exons 5 and 6 was subcloned into the pBluescript SK vector, and the region spanning exons 5 and 6 was replaced with a Neo gene. This construct was used for the targeted disruption of the Rint-1 gene in mice according to procedures described previously (31).
Embryo collection, PCR genotyping, and histology of embryos from Rint-1+/− intercross.
F1 Rint-1 heterozygous mice were intercrossed for timed pregnancy. For F2 progenies at the age of 21 days, toes were cut and animals were genotyped by PCR. For embryos at E12.5, the visceral yolk sac was collected for PCR genotyping. For embryos between E5.5 and E7.5, entire uteri were fixed in 4% paraformaldehyde at 4°C overnight. Uterine horns were excised and dehydrated through a graded ethanol series, cleared in chloroform, and then infiltrated and embedded in Paraplast X-tra (Polysciences). Sections (4 μm) were collected and stained with Mayer's hematoxylin and eosin. Embryonic tissues were microdissected, lysed, and subjected to PCR genotyping. The E3.5 blastocysts were flushed from maternal uteri for PCR genotyping. To detect the wild-type Rint-1 allele, a 162-bp fragment was PCR amplified using a sense oligonucleotide (5′-CAC TGT CTT AGC CTG TGT TCT ATT GC-3′) and an antisense oligonucleotide (5′-CCA GTA CCA CAT CGG CCT GTA TGC-3′) within intron 6 of Rint-1. To detect the disrupted Rint-1 allele, a 236-bp fragment was PCR amplified using a sense oligonucleotide (5′-TGA TAT TGC TGA AGA GCT TGG CGG C-3′) and an antisense oligonucleotide (5′-TGG GAG TGG CAC CTT CCA GGG TCA A-3′) within the pgkneopA cassette.
In vitro blastocyst culture.
Blastocysts were cultured as described previously (35). Blastocysts were flushed from the uteri of Rint-1+/− females at 3.5 days postcoitum and cultured for 7 days in Dulbecco modified Eagle medium plus 20% fetal bovine serum, supplemented with 4 mg of bovine serum albumin/ml, glutamine, and 0.1 mM 2-mercaptoethanol. Blastocyst outgrowths were photographed to monitor their development. They were lysed and genotyped by PCR as described above.
RESULTS
Homozygous Rint-1 deletion is lethal in early embryonic development.
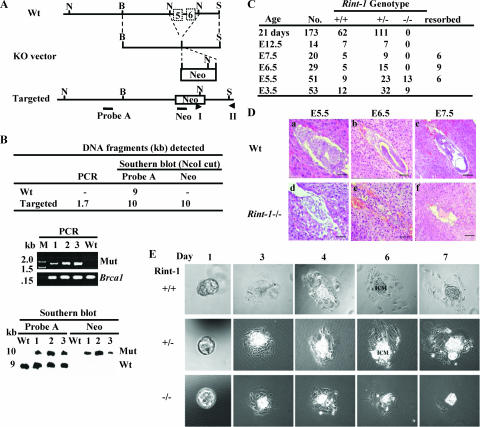
To address the biological functions of RINT-1 during mouse development, an inactivated Rint-1 allele in embryonic stem (ES) cells was generated by the replacement of exons 5 and 6 of the Rint-1 gene, encoding amino acids 172 to 280, with the Neo gene (Fig. 1A). Recombinant ES cell clones were identified by genomic PCR and Southern blotting (Fig. 1B), and then the correct clone was used to generate chimeric mice and, subsequently, heterozygous mice. When we intercrossed Rint-1 heterozygous mice, no Rint-1−/− mice were observed among 173 progenies analyzed, indicating that the homozygotes succumbed to death during embryogenesis (Fig. 1C). To determine the exact stage at which the Rint-1 null embryos died, Rint-1+/− females conceived by intercrosses were sacrificed and the fetuses were examined at different gestation times between E5.5 and E12.5. At E5.5, Rint-1−/− embryos were poorly developed, while the wild-type embryos showed normal growth and the elongation of the egg cylinder (Fig. 1C and D). From E6.5 to E12.5, only empty deciduas and the remains of resorbed embryos were found, and no sizable Rint-1−/− embryo was detected (Fig. 1C and D). These results suggest that Rint-1−/− embryos fail to develop at very early stages following implantation.
FIG. 1.
Targeted disruption of the mouse Rint-1 allele leads to embryonic lethality. (A) Gene-targeting strategy. The region spanning exons 5 and 6 of the Rint-1 gene was replaced with the Neo gene after targeted disruption. Wt, wild type; KO, knockout; B, BamHI; N, NcoI; S, ScaI. Probe A and Neo were probes used for Southern blot analysis. I and II were primers for PCR. (B) Identification of the recombinant ES clones by digesting the genomic DNA with NcoI and probing with probe A or Neo. (Upper panel) Sizes of DNA fragments detected by PCR and Southern blot analyses; middle panel, PCR analysis to confirm the positive clones (the Brca1 gene served as a control); (lower panel) Southern blot analysis to indicate the correct recombinant ES cell clones. M, marker; Mut, mutant; −, none. (C) Genotypic distribution of embryos and mice from Rint-1 heterozygous intercrosses. (D) Histological sections of wild-type and Rint-1−/− embryos grown in the uterus. The embryos in the uterus were fixed, sectioned, and stained with hematoxylin and eosin. Rint-1 null embryos manifested abnormal development at E5.5 (d). The resorbed embryos (e and f) were too small to be genotyped and were presumed to be Rint-1−/−. Bars: panels a and d, 50 μm; panels b, e, and f, 100 μm; and panel c, 200 μm. (E) Blastocysts at E3.5 isolated from Rint-1+/− intercrosses and cultured for 7 days. Wild-type (Rint-1+/+), Rint-1+/−, and Rint-1−/− blastocysts appeared to be morphologically comparable at day 1. However, while the ICM cells of Rint-1+/+ and Rint-1+/− embryos continued to expand through the 7-day culture period, Rint-1−/− ICM cells stopped expanding by day 4 and died later, along with the trophoblastic giant cells.
To test whether Rint-1 is essential for the proliferation of blastocysts, we collected and cultured blastocysts from intercrossed Rint-1 heterozygous females at E3.5. As shown in Fig. 1E, adherent sheets of trophoblastic giant cells and the outgrowth of the inner cell mass (ICM) in 44 cultured Rint-1+/+ and Rint-1+/− blastocysts were observed, while the ICM cells in 9 cultured Rint-1−/− blastocysts failed to expand by day 4 and died later, along with the trophoblastic giant cells. These results suggest that Rint-1 is essential for the proliferation and viability of blastocysts during early embryogenesis.
Rint-1 heterozygotes are prone to multiple tumor formation.
To investigate whether Rint-1 deficiency exerts a physiological impact, a cohort of 31 Rint-1+/− mice was longitudinally monitored. Of these mice, 81% succumbed to multiple tumors, including infrequent mammary gland tumors and hepatocellular carcinomas, during their average life span of 24 months (Fig. 2A). The tumor incidence in Rint-1+/− mice was even higher than that in Brca1+/− mice, which have a tumor incidence of 70% during their life span with a similar genetic background (26), suggesting that Rint-1 is a bona fide tumor susceptibility gene.
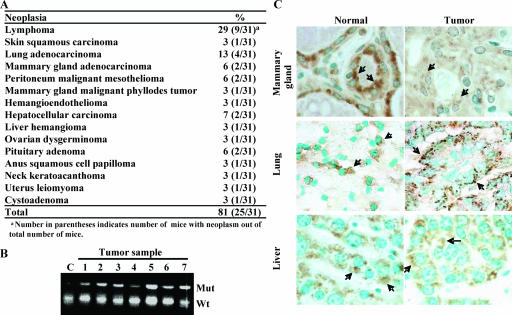
FIG. 2.
Rint-1 heterozygous mice develop multiple tumors. (A) Tumor incidence in Rint-1+/− mice. (B) PCR analysis of microdissected tumor samples showed no loss of heterozygosity in tumors developed in Rint-1+/− mice. C, wild-type mouse embryonic fibroblasts; Mut, mutant; Wt, wild type. (C) Immunostaining with anti-Rint-1 antibody detected the expression of Rint-1 protein (arrows) in mammary, lung, and liver tumors. Specimens were counterstained with methylene green (magnification, ×400).
To test whether the tumorigenesis in Rint-1-deficient mice follows an established recessive mechanism (45), tumor cells were microdissected and analyzed for the Rint-1 genotype by PCR. All the seven tumors analyzed harbored both the wild-type and mutant alleles of Rint-1 (Fig. 2B). Similarly, all the 15 tumors, in addition to 10 normal tissues, were positively immunostained with anti-Rint-1 antibodies, indicating that the remaining wild-type allele was still expressed (Fig. 2C). Thus, the tumor formation in these mice is attributed to the haploinsufficiency of Rint-1.
RINT-1 localizes at the ER, the Golgi apparatus, and the centrosome.
To explore the mechanisms of how RINT-1 plays an essential role in early embryonic growth and tumor suppression, the subcellular localization of human RINT-1 protein was first examined. As shown in Fig. 3A, RINT-1 exhibited staining patterns in the ER and the Golgi apparatus indicated by colocalization with an ER marker, calnexin (44), and a Golgi marker, giantin, respectively (33), in U2OS cells. These results are consistent with recent findings that RINT-1 localizes at the ER and comigrates with the membrane fractions corresponding to the ER and Golgi vesicles (4, 24).
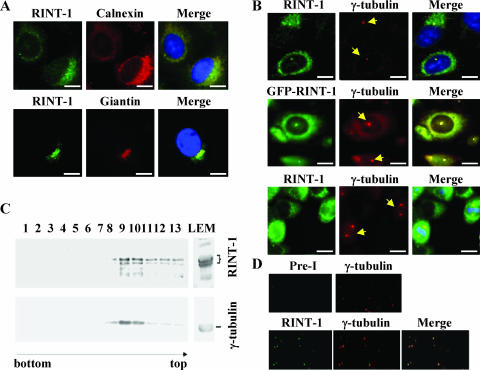
FIG. 3.
RINT-1 is localized to the ER, Golgi apparatus, and centrosome. (A) RINT-1 localized at the ER and Golgi apparatus in U2OS cells. Cells were immunostained with antibodies against RINT-1, calnexin (an ER marker), or Giantin (a Golgi apparatus marker). Bars, 10 μm. (B) RINT-1 localized at the centrosome. (Upper and lower panels) HeLa cells were costained with antibodies against RINT-1 and γ-tubulin, a centrosome marker; (middle panel) GFP-RINT-1 expressed in U2OS cells colocalized with γ-tubulin staining. The arrows indicate the γ-tubulin staining. Bars, 10 μm. (C) Endogenous RINT-1 and γ-tubulin were cofractionated by sucrose density gradient centrifugation. Fractions were immunoblotted with antibodies against either RINT-1 or γ-tubulin. RINT-1 proteins were associated with γ-tubulin through fractions 8 to 13, with a peak in fractions 9 and 10. (D) RINT-1 and γ-tubulin colocalized at the isolated centrosomes. Centrosomes in fractions 9 and 10 were spun onto coverslips and immunostained with preimmune and anti-γ-tubulin antibodies or anti-RINT-1 and anti-γ-tubulin antibodies.
In addition to membrane-localized RINT-1, RINT-1 in speckles costained with a centrosome marker, γ-tubulin, was found in both the interphase and mitotic phase of HeLa cells. (Fig. 3B), suggesting that RINT-1 localizes at the centrosome as well. To further substantiate this possibility, a GFP-tagged RINT-1 protein was stably expressed in U2OS cells (data not shown), and the GFP signal was also detected in the centrosome by colocalization with γ-tubulin (Fig. 3B). Consistent with this result, GFP-RINT-1 ectopically expressed in U2OS cells was also detected at the spindle poles during mitosis (data not shown). To further demonstrate that RINT-1 is a component of the centrosome, cellular fractions containing centrosomes were prepared and analyzed by Western blotting and immunostaining. As shown in Fig. 3C and D, both RINT-1 and γ-tubulin comigrated in the centrosomal fractions and were costained at the purified centrosomes. Preimmune antiserum was used as a control to demonstrate that the centrosomal staining by anti-RINT-1 antibody was specific for RINT-1. These results indicate that RINT-1 is an intrinsic component of the centrosome.
Depletion of RINT-1 by siRNA prolongs M phase and promotes mitotic cell death.
Based on our animal results, RINT-1 is an essential protein for early embryo development and blastocyst proliferation. It is important to understand how an ER-Golgi apparatus-centrosome protein plays such a critical role. To explore the potential mechanism by which Rint-1 inactivation leads to a profoundly lethal phenotype, we established stable HeLa cell lines (HeLa-RINT-1i) with tetracycline-inducible siRNA to modulate RINT-1 expression. As shown in Fig. 4A, upon induction with doxycycline for 48 h, the RINT-1 expression in three independently isolated clones was diminished. Similarly, immunostaining with anti-RINT-1 antibody failed to detect RINT-1 protein in HeLa-RINT-1i cells induced with doxycycline but not in control cells (Fig. 4B).
FIG. 4.
Depletion of RINT-1 leads to a prolonged M phase and severe mitotic cell death. (A) Characterization of three HeLa cell clones with inducible expression of RINT-1 RNAi. Cells were treated with (+) or without (−) 5 μg of doxycycline/ml for 48 h. The cell lysates were analyzed by Western blotting and probed with antibodies against RINT-1, RAD50, and β-actin. The expression of RINT-1 protein was diminished when RINT-1 RNAi was induced with doxycycline. RAD50 and β-actin served as controls. (B) Confirmation of the depletion of RINT-1 protein in HeLa-RINT-1i cells upon addition of 5 μg of doxycycline/ml for 48 h by immunostaining with anti-RINT-1 antibody. −Dox, without doxycycline; +Dox, with doxycycline. Bars, 10 μm. (C) The depletion of RINT-1 inhibited cell growth of the HeLa-RINT-1i clone. Cells were induced with or without 5 μg of doxycycline/ml, and the viable cells were counted by the trypan blue exclusion assay at the indicated times. Each data point represents the average of results for duplicate samples. The data are representative of results from three independent experiments. Error bars, standard deviations. (D) The depletion of RINT-1 caused cell death in a randomly growing population. HeLa-RINT-1i cells were treated with or without 5 μg of doxycycline/ml for the indicated times, and the cell cycle distribution was analyzed by flow cytometry. A significant increase in sub-G1 cells among RINT-1-depleted cells was observed. The data are representative of results from three independent experiments. (E) RINT-1-depleted cells underwent cell death at the G2/M phase. (Top) Diagram showing the experimental scheme for cell cycle synchronization by double thymidine blocking at the G1/S boundary. (Middle) Profiles from fluorescence-activated cell sorter analysis. Cells were collected and analyzed by flow cytometry. 2N, diploid cells; 4N, tetraploid cells. (Bottom) The percentages of cells at each cell cycle phase are indicated. An increase in the sub-G1 population and a decrease in the G2/M population among RINT-1-depleted cells were simultaneously observed. The data are representative of results from three independent experiments. (F) Proliferation curve of one cell cycle division. HeLa-RINT-1i cells were synchronized at mitotic phase by treatment with 0.1 μg of nocodazole/ml for 8 h. The mitotic cells were collected and replated for 4 h, followed by treatment with or without 5 μg of doxycycline/ml. The viable cells were counted by the trypan blue exclusion assay at the indicated times. Each data point represents the average of results for duplicate samples. The data are representative of results from two independent experiments. Error bars, standard deviations. (G) Living HeLa-RINT-1i cells harboring histone H2B-GFP were monitored under a fluorescence microscope at the indicated times after induction with or without 5 μg of doxycycline/ml for 12 h. Chromosome behavior during M phase was recorded. The mitotic progression of control cells is shown in panel a. The mitotic progression of RINT-1-depleted cells is shown in panels b to h. The numbers indicate the minutes passed during the recording. Bars, 10 μm. (H) The mitotic phase of HeLa-RINT-1i cells expressing histone H2B-GFP and treated with doxycycline was prolonged compared to that of control cells. The M phase length was determined from the intervals between chromosome condensation and the completion of cytokinesis. (I) The percentage of HeLa-RINT-1i/H2B-GFP cells treated with doxycycline that died in the mitotic phase was increased compared to that of control cells. Error bars, standard deviations.
Next, we compared the proliferation rates of HeLa-RINT-1i cells with and without doxycycline induction. As shown in Fig. 4C, the proliferation of RINT-1-depleted cells was significantly inhibited compared to that of control cells. Cell cycle analyses of HeLa-RINT-1i cells at days 2 and 3 after induction with doxycycline revealed that approximately 13 and 37% of RINT-1-depleted cells, respectively, were in the sub-G1 fraction, indicative of dead cells. In contrast, less than 4% of control cells were in sub-G1 at both time points. Moreover, the G1-phase population among RINT-1-depleted cells was significantly decreased compared to that among control cells, while the S- and G2/M-phase populations remained relatively constant (Fig. 4D). Similar phenotypes were observed in two other HeLa-RINT-1i clones (data not shown). These results indicate that the depletion of RINT-1 causes cell death at either G1 phase or G2/M phase. To pinpoint at which cell cycle stage RINT-1-depleted cells die, we synchronized HeLa-RINT-1i cells at the G1/S boundary by double thymidine blocking, followed by induction with doxycycline as diagramed in Fig. 4E. Fluorescence-activated cell sorter analysis showed that RINT-1-depleted cells failed to pass through G2/M phase and underwent cell death (Fig. 4E). Notably, a minor delay of S-phase entry and a moderate accumulation of S-phase cells among RINT-1-depleted cells were also observed (Fig. 4E), though the significance of these observations remains unclear. To further confirm that RINT-1-depleted cells die at G2/M phase, we synchronized HeLa-RINT-1i cells at M phase by using nocodazole and counted the viable cells during a single cell cycle division. As shown in Fig. 4F, after release from M phase arrest, the number of control cells increased by up to 34%. In contrast, the number of RINT-1-depleted cells was significantly decreased, suggesting that cells die at G2/M phase but not at G1 or S phase.
Since RINT-1-depleted cells succumb to death at G2/M phase, we examined the mode of mitosis in HeLa-RINT-1i cells marked with histone H2B-GFP by time-lapse imaging. Of all the control cells (n = 135), 92% completed mitosis within 2 h and divided into two normal viable daughter cells (Fig. 4G, panels a, and H). However, of all the RINT-1-depleted cells (n = 93), 43% completed mitosis in a period longer than 2 h, and the average mitotic time was about 3.3 h (Fig. 4G, panels b to f, and H). Furthermore, 94% of all the control cells were viable, whereas 42% of RINT-1-depleted cells were dead (Fig. 4G and I). As shown in Fig. 4G, panels b to h, RINT-1-depleted cells appeared as dense debris at M phase, indicating dying cells at prophase (Fig. 4G, panels b and c), metaphase (Fig. 4G, panels d and e), or postcytokinesis (Fig. 4G, panels f and h). These results strongly suggest that the depletion of RINT-1 leads to a prolonged M phase and mitotic cell death, which is consistent with the earlier observation that Rint-1−/− blastocysts fail to proliferate.
Depletion of RINT-1 leads to centrosome amplification and chromosome missegregation.
To elucidate the relevance of RINT-1 to cell viability during M phase, we first studied a potential role of RINT-1 at the centrosome by examining the centrosome integrity in RINT-1-depleted cells, because severe centrosome abnormality causes aneuploidy and cell death (18, 48, 53). The number of centrosomes was determined by anti-γ-tubulin antibody immunostaining of HeLa-RINT-1i cells induced with or without doxycycline. As shown in Fig. 5A and B, the number of RINT-1-depleted cells having more than two centrosomes was increased by about three- to fourfold compared to that of control cells. Similar patterns of centrosome amplification in different HeLa-RINT-1i clones were also observed (data not shown). In addition, adenovirus carrying RINT-1 RNAi (rAd-RINT-1i) was generated and used to infect U2OS cells. The expression of RINT-1 protein was efficiently abolished in a rAd-RINT-1i dose-dependent manner, while control adenovirus carrying luciferase RNAi (rAd-Luci) did not affect the expression (Fig. 5C). Up to 18.5% of rAd-RINT-1i-infected cells had amplified centrosomes, while the population of cells infected with rAd-Luci had a background level of centrosome amplification of 7% (Fig. 5D). A similar increase in amplified centrosomes in HeLa cells infected with rAd-RINT-1i was also observed (data not shown). These findings demonstrate that the depletion of RINT-1 by siRNA, either in inducible stable cell clones or in cells infected with adenoviruses, results in centrosome amplification, suggesting that RINT-1 is essential for the centrosome integrity. To substantiate the role of RINT-1 involved in centrosome duplication, we treated RINT-1-depleted cells with HU. It has been reported previously that the centrosomes undergo multiple rounds of duplication when cells are arrested at the G1/S boundary by HU (5). As shown in Fig. 5B, the depletion of RINT-1 resulted in significant centrosome amplification at 48 and 72 h post-HU treatment, compared to that in control cells, strongly suggesting that RINT-1 is involved in centrosome duplication.
Since centrosome amplification often results in abnormal spindle formation and consequential chromosome instability (18, 49), we then investigated whether this is the case in RINT-1-depleted cells. As shown in Fig. 5E and F, an increase in the occurrence of multiple spindle formation of about three- to fourfold in HeLa-RINT-1i cells induced with doxycycline compared to control cells was observed. Chromosome missegregation was also frequently detected in RINT-1-depleted cells, as indicated by misaligned chromosomes at metaphase, lagging chromosomes at anaphase, and thin chromatin bridges at telophase (Fig. 5G and H). A modest increase in micronuclei and multinuclei in RINT-1-depleted cells was observed as well (data not shown). These results suggest that the depletion of RINT-1 leads to spindle abnormality and consequential chromosome instability, at least in part contributing to the cell death at M phase.
Depletion of RINT-1 results in premature Golgi structure dispersion at interphase and the failure of Golgi apparatus reassembly at mitosis.
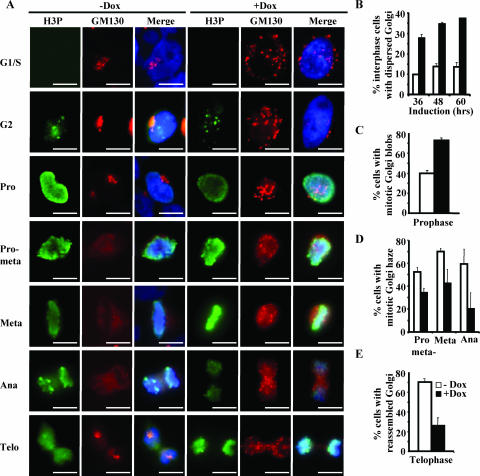
In addition to maintaining the centrosome integrity, RINT-1 may play a critical role in Golgi dynamics because it localizes at the centrosome as well as the Golgi apparatus. The Golgi apparatus is known to undergo a dramatic two-step process of disassembly between late prophase and early anaphase and reassembly at telophase, and the first step of disassembly into mitotic Golgi blobs is essential for cell division to proceed (9, 50). To explore the role of RINT-1 in Golgi dynamics, we examined the organization of the Golgi apparatus at different cell cycle stages by coimmunostaining RINT-1-depleted cells with an antibody against GM130, a Golgi marker (41), and an antibody against phosphorylated histone 3, a marker of mitotic cells, to distinguish the interphase and different mitotic stages (1). As shown in Fig. 6A and B, more RINT-1-depleted cells than control cells had dispersed Golgi structures at interphase, consistent with a recent finding (4). At prophase, a higher percentage of RINT-1-depleted cells than of control cells contained fragmented mitotic Golgi blobs, consisting of tubules and large vesicles (Fig. 6A and C). At prometaphase, metaphase, and early anaphase, the Golgi blobs were further dispersed and diffused completely into the cytosol to form mitotic Golgi haze in control cells, while fewer RINT-1-depleted cells had mitotic Golgi haze formation (Fig. 6A and D). At telophase, 70% of control cells reassembled the Golgi apparatus, while only 26% of RINT-1-depleted cells reassembled it successfully (Fig. 6A and E). These results demonstrate that the depletion of RINT-1 causes the premature dispersion of the Golgi apparatus at interphase and the premature formation of mitotic Golgi blobs at prophase. Moreover, the subsequent formation of mitotic Golgi haze and the reassembly of the Golgi apparatus at the later stages of mitosis are inhibited. Such an aberrant phenotype of RINT-1-deficient cells suggests that RINT-1 mediates the Golgi dynamics during cell cycle progression. The disassociation of Golgi dynamics from cell cycle progression due to RINT-1 deficiency may contribute to the observed prolonged M phase, the mitotic cell death, and the early embryonic lethality.
FIG. 6.
Depletion of RINT-1 leads to abnormal Golgi dynamics during cell cycle progression. (A) Micrographs of HeLa-RINT-1i cells coimmunostained with antibodies against GM130, a Golgi apparatus marker, and phosphorylated histone 3 (H3P), which is a marker of different cell cycle stages. Cells were synchronized at the G1/S boundary by double thymidine blocking and induced with (+Dox) or without (−Dox) 5 μg of doxycycline/ml as depicted in Fig. 4E. Pro, prophase; pro-meta, prometaphase; meta, metaphase; ana, anaphase; telo, telophase. Bars, 10 μm. (B to E) The percentages of HeLa-RINT-1i cells with the dispersed Golgi apparatus at interphase (B); mitotic Golgi blob formation at prophase (C); Golgi haze formation at prometaphase, metaphase, and anaphase (D); and the reassembled Golgi structure at telophase (E) after induction with or without 5 μg of doxycycline/ml were determined. Cells at interphase (n > 300); prophase, prometaphase, or metaphase (n > 200); anaphase (n > 50); and telophase (n > 100) were examined. All the data are representative of results from two independent experiments. Error bars, standard deviations.
Depletion of RINT-1 dislocates the Golgi apparatus from the pericentriolar region.
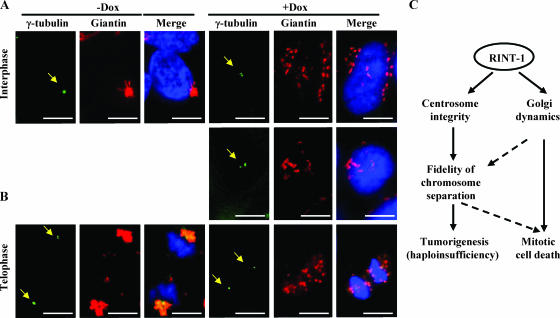
The pericentriolar localization of the Golgi apparatus in mammalian cells is thought to be crucial. It was reported previously that the Golgi complex is dispersed and loses its pericentriolar position when microtubules are depolymerized or microtubule-associated motor or nonmotor proteins are inactivated (8, 21, 25). To test whether RINT-1 is required for the Golgi apparatus positioning at the pericentriolar region, we costained RINT-1-depleted cells with antibodies against γ-tubulin and giantin to mark the centrosome and the Golgi apparatus, respectively. At interphase, the Golgi apparatus was dispersed and lost its compact, perinuclear location and became dislocated from the centrosome, which was frequently amplified in RINT-1-depleted cells (Fig. 7A). Furthermore, at telophase, the Golgi membranes did not reassemble around the centrosome (Fig. 7B). These findings suggest that RINT-1 plays an important role in the pericentriolar Golgi apparatus localization, which may coordinate the Golgi apparatus and the centrosome for the proper division in mammalian cells.
FIG. 7.
Depletion of RINT-1 disrupts the pericentriolar localization of the Golgi apparatus. (A and B) Micrographs of HeLa-RINT-1i cells coimmunostained with antibodies against giantin and γ-tubulin (arrows) upon treating with (+Dox) or without (−Dox) 5 μg of doxycycline/ml. Dissociation of RINT-1 from the pericentriolar region with the dispersed Golgi structure at interphase (A) and with the nonreassembled Golgi structure at telophase (B) was observed in RINT-1-depleted cells (+Dox). Bars, 10 μm. (C) Diagram of proposed RINT-1 functions involved in maintaining centrosome integrity as well as Golgi dynamics. The depletion of RINT-1 leads to mitotic cell death as well as tumorigenesis with haploinsufficiency.
DISCUSSION
In this communication, we report that RINT-1 is located at the ER and the Golgi apparatus as well as the centrosome, implying a potential functional link between the Golgi apparatus-ER and the centrosome for cellular activities. We found that cells depleted of RINT-1 lost the pericentriolar positioning of the Golgi apparatus and displayed increased centrosome amplification during the interphase. In addition, RINT-1 deficiency elicited multiple mitotic aberrancies, including abnormal Golgi dynamics, multipolar spindle structures, and chromosome missegregation. The compound effect of these mitotic defects is likely detrimental, as cells often underwent apoptosis. Consistently, homozygous deletion of Rint-1 alleles in mice resulted in early embryonic lethality at E5.5, while about 81% of Rint-1+/− heterozygous mice developed multiple tumors with haploinsufficiency, implying that RINT-1 is a tumor suppressor. Our results suggest that RINT-1 plays an essential role in cell survival and tumor suppression in part by maintaining the functional integrity of the centrosome and regulating the proper dynamics of the Golgi apparatus and the ER as depicted in Fig. 7C.
RINT-1 mediates Golgi dynamics throughout cell cycle progression.
The ER and Golgi apparatus are two essential organelles critical for coordinating protein synthesis in the ER with the subsequent posttranslational modifications and sorting in the Golgi apparatus. During mitosis, the integrity of the ER network is maintained (16). In contrast, the Golgi apparatus undergoes a series of dramatic transformations (6, 9). A possible dynamic model includes the dispersal of Golgi structures and the formation of blobs at late prophase, the further fragmentation into diffusive cytoplasmic Golgi haze before early anaphase, and the subsequent segregation of Golgi haze and vesicles into daughter cells upon mitotic exit, followed by the reassembly of Golgi ribbons at telophase. (27, 28, 42, 43, 53). Nonetheless, an understanding of the molecular basis of Golgi apparatus disassembly and reassembly is still under vigorous pursuit. The kinases MEK1 and Plk1 and Myt1 and Cdc2 are involved in the sequential steps of disassembly, whereas the two ATPases N-ethylmaleimide-sensitive factor and p97 are required for reassembly (10, 36, 38, 51). As demonstrated in this study, the Golgi apparatus disassembly and reassembly process is severely disturbed in RINT-1-deficient cells. First, the Golgi stacks were prematurely dispersed during the interphase. Second, the formation of the Golgi haze during the mid-mitosis phase was incomplete. Third, the reassembly of Golgi stacks at telophase was significantly impaired. The premature interphase fragmentation would inevitably impair the normal function of the Golgi apparatus in directional protein sorting and transport. The aberrant Golgi dynamics may inhibit normal mitotic progression and is likely detrimental, similar to the Myt1 depletion of Drosophila sp. strain S2 cells, which is associated with incomplete Golgi haze formation during mitosis (11).
What is the role of RINT-1 at the Golgi apparatus? One possibility is that RINT-1 is involved in the anterograde transport from the ER to the Golgi apparatus in part by mediating the dynein motor activity via direct interaction with ZW10 or the dynactin subunit p150Glued (4; our unpublished data). Therefore, in RINT-1-depleted cells the transport from the ER to the Golgi apparatus may be inhibited, and subsequently, retrograde transport from the Golgi apparatus would become trapped in the ER, leading to abnormal Golgi apparatus dispersion. However, this protein trafficking role of RINT-1 cannot adequately explain the defects in the Golgi apparatus reassembly at telophase. It is likely that RINT-1 may alternatively or additionally serve as a scaffold component critical for maintaining the Golgi structural integrity. Moreover, as the Golgi apparatus disassembly and reassembly is an enzymatic reaction, RINT-1 may serve as a key effector of the upstream signaling to control certain steps of structural conversion. Nevertheless, the knowledge of precisely how RINT-1 participates in Golgi apparatus disassembly and reassembly awaits further investigation.
RINT-1 plays a role in maintaining centrosome integrity.
We additionally observed abnormal centrosome amplification in RINT-1-deficient cells. Centrosome amplification in cancer samples is frequently observed and has been linked to deficiencies of numerous proteins involved in DNA repair, such as p53, BRCA1, BRCA2, Mre11, Rad51, XRCC2, and XRCC3 (12, 13, 20, 57, 58). Interestingly, RINT-1 interacts with BRCA1 in a yeast two-hybrid assay (our unpublished data). Therefore, the centrosome amplification observed in this study may be relevant to BRCA1 centrosomal function and/or additionally subject to modulation by ZW10 and the dynein-dynactin motor.
RINT-1 may coordinate the centrosome and Golgi functions.
The interrelationship among the Golgi apparatus, the ER, and the centrosome is of particular interest, given that RINT-1 is concomitantly present in these organelles and required for their functional integrity. The importance of the pericentriolar positioning of the Golgi apparatus in mammalian cells is poorly understood. As this spatial arrangement is not required for the normal Golgi functions, i.e., protein sorting and transport, it is likely designed for other purposes, such as providing a sensor mechanism to ensure the divisibility of cytoplasmic components (9). Clearly, RINT-1 is required for the pericentriolar positioning of the Golgi apparatus (Fig. 7A). It is reasonable to speculate that RINT-1 may coordinate centrosome separation and the Golgi dynamic transformation during the G2/M phase. Interestingly, since RINT-1 participates in the G2/M checkpoint control in the DNA damage response, an additional dimension of cell cycle control may be associated with RINT-1. In this regard, RINT-1 cellular activity is strikingly similar to that of Plk1, whose role in the G2/M checkpoint control during the DNA damage response, the regulation of centrosome separation, and Golgi apparatus fragmentation during the G2/M transition has been well established (51). However, whether RINT-1 may serve as a Plk1 downstream effector or substrate is still an open question, although RINT-1 harbors several potential Plk1 phosphorylation sites.
RINT-1 is an essential gene for cell survival and mouse development.
The depletion of RINT-1 leads to multiple mitotic defects in cells, which are likely overwhelmingly detrimental for cell survival, as supported by our data (Fig. 4). However, we cannot exclude the possibility that other functions of RINT-1, such as the regulation of the G2/M checkpoint in association with RAD50, may also contribute to this phenotype. A defect in the G2/M checkpoint frequently results in embryonic lethality and cell death (37). Similarly, centrosome amplification results in abnormal spindle configuration, chromosome missegregation, and at times cell death (18, 47, 48, 49, 53). Hence, the cell death and embryonic lethality caused by RINT-1 deficiency may be attributed to compound defects in various aspects.
RINT-1 serves as a tumor suppressor.
The increased tumor formation in Rint-1+/− mice associated with haploid insufficiency strongly supports a tumor suppressor role of RINT-1. As the loss of heterozygosity was not detected, the tumors still expressed one Rint-1 allele. How the reduced RinT-1 level may promote tumorigenesis is an intriguing question. We speculate that the reduced Rint-1 gene dosage is sufficient to maintain the cell viability but otherwise inadequate for accurate and efficient mitosis, a frequent source of chromosomal instability. In a stressful event such as DNA damage, RINT-1 is additionally required for the G2/M checkpoint control. Thus, the concomitant requirements for RINT-1 in the DNA damage response, centrosome separation, and Golgi apparatus-ER dynamic regulation may surpass the capacity of Rint-1+/− cells. Consequentially, cancer-promoting chromosomal instability may arise and accumulate in sensitive tissues. We are presently in the process of examining the impact of Rint-1 heterozygosity on the centrosome and Golgi apparatus integrity. Although centrosome and G2/M checkpoint control are seemingly the major effectors of Rint-1 insufficiency in the context of aneuploidy, we do not exclude the contribution of Golgi apparatus defects to tumorigenesis in this unique case. Of special note, the Golgi apparatus has been implicated in regulating the microtubule motor function critical for mitotic processes, the perturbation of which results in unaligned chromosomes and distorted spindle organization (15). Nonetheless, the precise role of RINT-1 in the multiple cellular processes and the respective contribution to genomic stability need to be further dissected in future study. Interestingly, RINT-1 deficiency may be linked to certain human cancers, including acute myeloid leukemia, myelodysplastic syndrome, and breast and ovarian adenocarcinomas, in which the genomic locus 7q22.1 close to the RINT-1 gene (7q22.2) is frequently deleted (40).
Acknowledgments
We thank Qing Zhong, Andrea Fleskin-Nikitin, and Phang-Lang Chen for their contributions at the initial phase of this investigation and Christine Sütterlin, Saori Furuta, and Yung-Ming Jeng for their critical comments.
This work was supported by an NIH grant (R01CA107568) to W.-H.L and a Susan Komen Breast Cancer Foundation grant to G.W.
Footnotes
Published ahead of print on 30 April 2007.
REFERENCES
- 1.Ajiro, K., K. Yoda, K. Utsumi, and Y. Nishikawa. 1996. Alteration of cell cycle-dependent histone phosphorylations by okadaic acid. Induction of mitosis-specific H3 phosphorylation and chromatin condensation in mammalian interphase cells. J. Biol. Chem. 271:13197-13201. [DOI] [PubMed] [Google Scholar]
- 2.Altan-Bonnet, N., R. D. Phair, R. S. Polishchuk, R. Weigert, and J. Lippincott-Schwartz. 2003. A role for Arf1 in mitotic Golgi disassembly, chromosome segregation, and cytokinesis. Proc. Natl. Acad. Sci. USA 100:13314-13319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altan-Bonnet, N., R. Sougrat, and J. Lippincott-Schwartz. 2004. Molecular basis for Golgi maintenance and biogenesis. Curr. Opin. Cell Biol. 16:364-372. [DOI] [PubMed] [Google Scholar]
- 4.Arasaki, K., M. Taniguchi, K. Tani, and M. Tagaya. 2006. RINT-1 regulates the localization and entry of ZW10 to the syntaxin 18 complex. Mol. Biol. Cell 17:2780-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balczon, R., L. Bao, W. E. Zimmer, K. Brown, R. P. Zinkowski, and B. R. Brinkley. 1995. Dissociation of centrosome replication events from cycles of DNA synthesis and mitotic division in hydroxyurea-arrested Chinese hamster ovary cells. J. Cell Biol. 130:105-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barr, F. A. 2004. Golgi inheritance: shaken but not stirred. J. Cell Biol. 164:955-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bornens, M., and M. Moudjou. 1999. Studying the composition and function of centrosomes in vertebrates. Methods Cell Biol. 61:13-34. [DOI] [PubMed] [Google Scholar]
- 8.Burkhardt, J. K., C. J. Echeverri, T. Nilsson, and R. B. Vallee. 1997. Overexpression of the dynamitin (p50) subunit of the dynactin complex disrupts dynein-dependent maintenance of membrane organelle distribution. J. Cell Biol. 139:469-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colanzi, A., C. Suetterlin, and V. Malhotra. 2003. Cell-cycle-specific Golgi fragmentation: how and why? Curr. Opin. Cell Biol. 15:462-467. [DOI] [PubMed] [Google Scholar]
- 10.Colanzi, A., C. Sutterlin, and V. Malhotra. 2003. RAF1-activated MEK1 is found on the Golgi apparatus in late prophase and is required for Golgi complex fragmentation in mitosis. J. Cell Biol. 161:27-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cornwell, W. D., P. J. Kaminski, and J. R. Jackson. 2002. Identification of Drosophila Myt1 kinase and its role in Golgi during mitosis. Cell. Signal. 14:467-476. [DOI] [PubMed] [Google Scholar]
- 12.Deng, C. X. 2006. BRCA1: cell cycle checkpoint, genetic instability, DNA damage response and cancer evolution. Nucleic Acids Res. 34:1416-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dodson, H., E. Bourke, L. J. Jeffers, P. Vagnarelli, E. Sonoda, S. Takeda, W. C. Earnshaw, A. Merdes, and C. Morrison. 2004. Centrosome amplification induced by DNA damage occurs during a prolonged G2 phase and involves ATM. EMBO J. 23:3864-3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donaldson, J. G., and J. Lippincott-Schwartz. 2000. Sorting and signaling at the Golgi complex. Cell 101:693-696. [DOI] [PubMed] [Google Scholar]
- 15.Echeverri, C. J., B. M. Paschal, K. T. Vaughan, and R. B. Vallee. 1996. Molecular characterization of the 50-kD subunit of dynactin reveals function for the complex in chromosome alignment and spindle organization during mitosis. J. Cell Biol. 132:617-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Estrada de Martin, P., P. Novick, and S. Ferro-Novick. 2005. The organization, structure, and inheritance of the ER in higher and lower eukaryotes. Biochem. Cell Biol. 83:752-761. [DOI] [PubMed] [Google Scholar]
- 17.Farquhar, M. G., and G. E. Palade. 1998. The Golgi apparatus: 100 years of progress and controversy. Trends Cell Biol. 8:2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fukasawa, K. 2005. Centrosome amplification, chromosome instability and cancer development. Cancer Lett. 230:6-19. [DOI] [PubMed] [Google Scholar]
- 19.Goetz, J. G., and I. R. Nabi. 2006. Interaction of the smooth endoplasmic reticulum and mitochondria. Biochem. Soc. Trans. 34:370-373. [DOI] [PubMed] [Google Scholar]
- 20.Griffin, C. S., P. J. Simpson, C. R. Wilson, and J. Thacker. 2000. Mammalian recombination-repair genes XRCC2 and XRCC3 promote correct chromosome segregation. Nat. Cell Biol. 2:757-761. [DOI] [PubMed] [Google Scholar]
- 21.Harada, A., Y. Takei, Y. Kanai, Y. Tanaka, S. Nonaka, and N. Hirokawa. 1998. Golgi vesiculation and lysosome dispersion in cells lacking cytoplasmic dynein. J. Cell Biol. 141:51-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He, T. C., S. Zhou, L. T. da Costa, J. Yu, K. W. Kinzler, and B. Vogelstein. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA 95:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hicks, S. W., and C. E. Machamer. 2005. Golgi structure in stress sensing and apoptosis. Biochim. Biophys. Acta 1744:406-414. [DOI] [PubMed] [Google Scholar]
- 24.Hirose, H., K. Arasaki, N. Dohmae, K. Takio, K. Hatsuzawa, M. Nagahama, K. Tani, A. Yamamoto, M. Tohyama, and M. Tagaya. 2004. Implication of ZW10 in membrane trafficking between the endoplasmic reticulum and Golgi. EMBO J. 23:1267-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Infante, C., F. Ramos-Morales, C. Fedriani, M. Bornens, and R. M. Rios. 1999. GMAP-210, a cis-Golgi network-associated protein, is a minus end microtubule-binding protein. J. Cell Biol. 145:83-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeng, Y. M., S. Cai-Ng, A. Li, S. Furuta, H. Chew, P. L. Chen, E. Y. Lee, and W. H. Lee. 9 April 2007. Brca1 heterozygous mice have shortened life span and are prone to ovarian tumorigenesis with haploinsufficiency upon ionizing irradiation. Oncogene. doi: 10.1038/sj.onc.1210451. [DOI] [PubMed]
- 27.Jesch, S. A., A. J. Mehta, M. Velliste, R. F. Murphy, and A. D. Linstedt. 2001. Mitotic Golgi is in a dynamic equilibrium between clustered and free vesicles independent of the ER. Traffic 2:873-884. [DOI] [PubMed] [Google Scholar]
- 28.Jokitalo, E., N. Cabrera-Poch, G. Warren, and D. T. Shima. 2001. Golgi clusters and vesicles mediate mitotic inheritance independently of the endoplasmic reticulum. J. Cell Biol. 154:317-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kong, L. J., A. R. Meloni, and J. R. Nevins. 2006. The Rb-related p130 protein controls telomere lengthening through an interaction with a Rad50-interacting protein, RINT-1. Mol. Cell 22:63-71. [DOI] [PubMed] [Google Scholar]
- 30.Lange, B. M. 2002. Integration of the centrosome in cell cycle control, stress response and signal transduction pathways. Curr. Opin. Cell Biol. 14:35-43. [DOI] [PubMed] [Google Scholar]
- 31.Lee, E. Y., C. Y. Chang, N. Hu, Y. C. Wang, C. C. Lai, K. Herrup, W. H. Lee, and A. Bradley. 1992. Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature 359:288-294. [DOI] [PubMed] [Google Scholar]
- 32.Lin, Y. T., Y. Chen, G. Wu, and W. H. Lee. 2006. Hec1 sequentially recruits Zwint-1 and ZW10 to kinetochores for faithful chromosome segregation and spindle checkpoint control. Oncogene 25:6901-6914. [DOI] [PubMed] [Google Scholar]
- 33.Linstedt, A. D., and H. P. Hauri. 1993. Giantin, a novel conserved Golgi membrane protein containing a cytoplasmic domain of at least 350 kDa. Mol. Biol. Cell 4:679-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lippincott-Schwartz, J., L. C. Yuan, J. S. Bonifacino, and R. D. Klausner. 1989. Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell 56:801-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu, C. Y., A. Flesken-Nikitin, S. Li, Y. Zeng, and W. H. Lee. 1996. Inactivation of the mouse Brca1 gene leads to failure in the morphogenesis of the egg cylinder in early postimplantation development. Genes Dev. 10:1835-1843. [DOI] [PubMed] [Google Scholar]
- 36.Lowe, M., C. Rabouille, N. Nakamura, R. Watson, M. Jackman, E. Jamsa, D. Rahman, D. J. Pappin, and G. Warren. 1998. Cdc2 kinase directly phosphorylates the cis-Golgi matrix protein GM130 and is required for Golgi fragmentation in mitosis. Cell 94:783-793. [DOI] [PubMed] [Google Scholar]
- 37.Luo, G., M. S. Yao, C. F. Bender, M. Mills, A. R. Bladl, A. Bradley, and J. H. Petrini. 1999. Disruption of mRad50 causes embryonic stem cell lethality, abnormal embryonic development, and sensitivity to ionizing radiation. Proc. Natl. Acad. Sci. USA 96:7376-7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meyer, H. H. 2005. Golgi reassembly after mitosis: the AAA family meets the ubiquitin family. Biochim. Biophys. Acta 1744:108-119. [DOI] [PubMed] [Google Scholar]
- 39.Mitchison, T., and M. Kirschner. 1984. Microtubule assembly nucleated by isolated centrosomes. Nature 312:232-237. [DOI] [PubMed] [Google Scholar]
- 40.Mitelman, F., F. Mertens, and B. Johansson. 1997. A breakpoint map of recurrent chromosomal rearrangements in human neoplasia. Nat. Genet. 15(Spec. No.):417-474. [DOI] [PubMed] [Google Scholar]
- 41.Nakamura, N., C. Rabouille, R. Watson, T. Nilsson, N. Hui, P. Slusarewicz, T. E. Kreis, and G. Warren. 1995. Characterization of a cis-Golgi matrix protein, GM130. J. Cell Biol. 131:1715-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nelson, W. J. 2000. W(h)ither the Golgi during mitosis? J. Cell Biol. 149:243-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rabouille, C., and E. Jokitalo. 2003. Golgi apparatus partitioning during cell division. Mol. Membr. Biol. 20:117-127. [DOI] [PubMed] [Google Scholar]
- 44.Rajagopalan, S., Y. Xu, and M. B. Brenner. 1994. Retention of unassembled components of integral membrane proteins by calnexin. Science 263:387-390. [DOI] [PubMed] [Google Scholar]
- 45.Riley, D. J., E. Y. Lee, and W. H. Lee. 1994. The retinoblastoma protein: more than a tumor suppressor. Annu. Rev. Cell Biol. 10:1-29. [DOI] [PubMed] [Google Scholar]
- 46.Rios, R. M., and M. Bornens. 2003. The Golgi apparatus at the cell centre. Curr. Opin. Cell Biol. 15:60-66. [DOI] [PubMed] [Google Scholar]
- 47.Sankaran, S., and J. D. Parvin. 2006. Centrosome function in normal and tumor cells. J. Cell. Biochem. 99:1240-1250. [DOI] [PubMed] [Google Scholar]
- 48.Sato, N., K. Mizumoto, M. Nakamura, H. Ueno, Y. A. Minamishima, J. L. Farber, and M. Tanaka. 2000. A possible role for centrosome overduplication in radiation-induced cell death. Oncogene 19:5281-5290. [DOI] [PubMed] [Google Scholar]
- 49.Sluder, G., and J. J. Nordberg. 2004. The good, the bad and the ugly: the practical consequences of centrosome amplification. Curr. Opin. Cell Biol. 16:49-54. [DOI] [PubMed] [Google Scholar]
- 50.Sutterlin, C., P. Hsu, A. Mallabiabarrena, and V. Malhotra. 2002. Fragmentation and dispersal of the pericentriolar Golgi complex is required for entry into mitosis in mammalian cells. Cell 109:359-369. [DOI] [PubMed] [Google Scholar]
- 51.Sutterlin, C., C. Y. Lin, Y. Feng, D. K. Ferris, R. L. Erikson, and V. Malhotra. 2001. Polo-like kinase is required for the fragmentation of pericentriolar Golgi stacks during mitosis. Proc. Natl. Acad. Sci. USA 98:9128-9132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takatsuki, A., M. Nakamura, and Y. Kono. 2002. Possible implication of Golgi-nucleating function for the centrosome. Biochem. Biophys. Res. Commun. 291:494-500. [DOI] [PubMed] [Google Scholar]
- 53.Tsuchihara, K., V. Lapin, C. Bakal, H. Okada, L. Brown, M. Hirota-Tsuchihara, K. Zaugg, A. Ho, A. Itie-Youten, M. Harris-Brandts, R. Rottapel, C. D. Richardson, S. Benchimol, and T. W. Mak. 2005. Ckap2 regulates aneuploidy, cell cycling, and cell death in a p53-dependent manner. Cancer Res. 65:6685-6691. [DOI] [PubMed] [Google Scholar]
- 54.van de Wetering, M., I. Oving, V. Muncan, M. T. Pon Fong, H. Brantjes, D. van Leenen, F. C. Holstege, T. R. Brummelkamp, R. Agami, and H. Clevers. 2003. Specific inhibition of gene expression using a stably integrated, inducible small-interfering-RNA vector. EMBO Rep. 4:609-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Varma, D., D. L. Dujardin, S. A. Stehman, and R. B. Vallee. 2006. Role of the kinetochore/cell cycle checkpoint protein ZW10 in interphase cytoplasmic dynein function. J. Cell Biol. 172:655-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xiao, J., C. C. Liu, P. L. Chen, and W. H. Lee. 2001. RINT-1, a novel Rad50-interacting protein, participates in radiation-induced G(2)/M checkpoint control. J. Biol. Chem. 276:6105-6111. [DOI] [PubMed] [Google Scholar]
- 57.Xu, X., Z. Weaver, S. P. Linke, C. Li, J. Gotay, X. W. Wang, C. C. Harris, T. Ried, and C. X. Deng. 1999. Centrosome amplification and a defective G2-M cell cycle checkpoint induce genetic instability in BRCA1 exon 11 isoform-deficient cells. Mol. Cell 3:389-395. [DOI] [PubMed] [Google Scholar]
- 58.Yamaguchi-Iwai, Y., E. Sonoda, M. S. Sasaki, C. Morrison, T. Haraguchi, Y. Hiraoka, Y. M. Yamashita, T. Yagi, M. Takata, C. Price, N. Kakazu, and S. Takeda. 1999. Mre11 is essential for the maintenance of chromosomal DNA in vertebrate cells. EMBO J. 18:6619-6629. [DOI] [PMC free article] [PubMed] [Google Scholar]