Abstract
The N-terminal and C-terminal domains of mitochondrially synthesized cytochrome c oxidase subunit II, Cox2, are translocated through the inner membrane to the intermembrane space (IMS). We investigated the distinct mechanisms of N-tail and C-tail export by analysis of epitope-tagged Cox2 variants encoded in Saccharomyces cerevisiae mitochondrial DNA. Both the N and C termini of a truncated protein lacking the Cox2 C-terminal domain were translocated to the IMS via a pathway dependent upon the conserved translocase Oxa1. The topology of this Cox2 variant, accumulated at steady state, was largely but not completely unaffected in mutants lacking proteins required for export of the C-tail domain, Cox18 and Mss2. C-tail export was blocked by truncation of the last 40 residues from the C-tail domain, indicating that sequence and/or structural features of this domain are required for its translocation. Mss2, a peripheral protein bound to the inner surface of the inner membrane, coimmunoprecipitated with full-length newly synthesized Cox2, whose leader peptide had already been cleaved in the IMS. Our data suggest that the C-tail domain is recognized posttranslationally by a specialized translocation apparatus after the N-tail has been translocated by Oxa1.
Cytochrome c oxidase, the terminal component of the mitochondrial respiratory chain, is a highly complex multisubunit enzyme. In animals, fungi, and many other phyla, the three core subunits are mitochondrially encoded and translated in the matrix, embedded into the inner membrane, elaborated with metal cofactors, and assembled with nuclearly encoded subunits that are imported into mitochondria. Assembly of this complex enzyme in Saccharomyces cerevisiae requires over 20 genes with specific roles in cytochrome c oxidase biogenesis, including RNA specific mitochondrial translation, proteolytic processing, heme a biosynthesis, copper delivery, and insertion of proteins into the inner mitochondrial membrane (reviewed in references 7, 13, and 26). Even the expression of a single mitochondrial gene, COX2, involves several specific factors encoded in the nucleus. Translation of the COX2 mRNA requires the membrane-bound translational activator Pet111, which interacts with the 54-nucleotide 5′-untranslated leader of the COX2 mRNA (18, 33). Pet111 both limits expression of COX2 (18) and localizes translation at the inner membrane surface (34, 44).
Insertion of Cox2 into the inner membrane has been thought to occur cotranslationally (26) and results in the translocation of hydrophilic N- and C-terminal domains to the intermembrane space (IMS) anchored by two transmembrane domains. Nuclearly encoded Oxa1, the founding member of the conserved Oxa1/YidC/Alb3 family of membrane proteins (59), is required for translocation of both N- and C-terminal domains of Cox2 (21, 23). Following translocation of the precursor of yeast Cox2 to the IMS, an N-terminal leader peptide (15 residues) is cleaved by the Imp1/Imp2/Som1 protease complex (27, 38) in a reaction dependent upon the specific chaperone Cox20 (25). Two nuclear genes, COX18 and MSS2, appear to be specifically required for Cox2 C-tail export (6, 46). Cox18 is an integral membrane protein related to Oxa1 (41, 46, 48) that is also functionally conserved among animals and fungi (14). Mss2 is a peripheral membrane protein facing the matrix (6). A third integral inner membrane protein, Pnt1, appears to facilitate Cox2 C-tail domain export but is not essential in S. cerevisiae (22). Cox18, Mss2, and Pnt1 interact genetically and coimmunoprecipitate (46), suggesting that they function together.
The translocated C-tail domain of Cox2 is 144 residues in length and negatively charged. The machinery that recognizes and exports this domain from the matrix can also translocate a soluble passenger protein, Arg8m, when Arg8m is translationally fused to the Cox2 C terminus (21). However, when Arg8m was fused just downstream of the second transmembrane domain, replacing the C-tail domain, it remained in the matrix, suggesting that the Cox2 C-tail domain might contain signals necessary for its own translocation (21).
In this study, we have further investigated translocation of the N- and C-terminal domains of Cox2 through analysis of variants that lack the C-tail domain or portions of it. We constructed mutant cox2 genes encoding epitope-tagged variant proteins, inserted them into yeast mitochondrial DNA (mtDNA), and examined the behavior of their products. These studies revealed that the C-tail domain is not required for insertion of the transmembrane domains of Cox2 but does contain sequences that direct its own translocation across the mitochondrial membrane. Furthermore, we found that Mss2 interacts with newly synthesized full-length Cox2 but not a truncated Cox2 variant entirely lacking this domain. Our data suggest that the C-tail domain is recognized posttranslationally by a specialized translocation apparatus.
MATERIALS AND METHODS
Genetic methods.
Standard yeast nuclear genetic manipulations and complete medium (yeast extract-peptone) containing dextrose, ethanol plus glycerol, or raffinose were as previously described (1, 19). Synthetic complete medium was purchased from Bio101 Systems. Mitochondrial genetic manipulations were performed as described elsewhere (4).
Yeast strains.
S. cerevisiae strains used in this study are listed in Table 1. All strains are congenic to D273-10B (ATCC 25657) except CM1, HLF5-22, SCS32, and NAB33, which are congenic to DBY947 (36). Genes encoding truncated, epitope-tagged variants of Cox2 were constructed in the vector pTZ18u (Bio-Rad), transformed by microprojectile bombardment into mitochondria of [rho°] strain NAB33, and inserted into [rho+] mtDNA by mating the transformants with the cox2 mutant NB40-3C followed by selection of recombinant cytoductants HLF3-60 {[rho+] cox2(1-109)::3xHA} and HLF4-13 {[rho+] cox2(1-211)::3xHA}. YME1 and OXA1 were deleted using deletion constructs pPT45 (52) and pNB60 (3), respectively. COX18 was disrupted using construct pG34/ST12 (48). MSS2 was disrupted with an mss2::LEU2 disruption cassette amplified by PCR from strain SB12 (6). An imp1Δ::kanMX4 mutation was generated as described previously (55, 56). A MYC tag was added to the C terminus of MSS2 and COX18 using plasmid pMPY-3x-MYC (47). Strain HLF5-22 was created by homologous mitochondrial recombination after mating the synthetic [rho−] strain CM1 (cox2[C225H H229C]) (49) with NB41 (5). All strains were verified by functional complementation tests when appropriate and/or by PCR analysis and sequencing.
TABLE 1.
Strains used in this study
| Strain | Nuclear genotype | Mitochondrial genotype | Reference or source |
|---|---|---|---|
| CM1 | MATaade2 ura3-52 | [rho−] cox2(C225H, H229C) | 49 |
| HLF3-60 | MATaura3-52 leu2-3,112 lys2 his3-ΔHindIII arg8-Δ::hisG | [rho+] cox2(1-109)::3xHA | This study |
| HLF4-13 | MATaura3-52 leu2-3,112 lys2 his3ΔHindIII arg8-Δ::hisG | [rho+] cox2(1-211)::3xHA | This study |
| HLF4-17 | MATaura3-52 leu2-3,112 lys2 his3-ΔHindIII arg8-Δ::hisG oxa1Δ::LEU2 | [rho+] cox2(1-109)::3xHA | This study |
| HLF4-30 | MATaura3-52 leu2-3,112 lys2 his3ΔHindIII arg8-Δ::hisG cox18::URA3 | [rho+] cox2(1-109)::3xHA | This study |
| HLF4-63 | MATaura3-52 leu2-3,112 lys2 his3-ΔHindIII arg8-Δ::hisG mss2::LEU2 | [rho+] cox2(1-109)::3xHA | This study |
| HLF4-73 | MATaura3-52 leu2-3,112 lys2 his3ΔHindIII arg8-Δ::hisG imp1Δ::kanMX | [rho+] cox2(1-109)::3xHA | This study |
| HLF4-87 | MATaura3-52 leu2-3,112 lys2 his3-ΔHindIII arg8-Δ::hisG yme1Δ::URA3 | [rho+] cox2(1-109)::3xHA | This study |
| HLF5-08 | MATα ura3-Δ leu2-3,112 his4-519 MSS2::3xMYC | [rho+] | This study |
| HLF5-13 | MATα ura3-Δ leu2-3,112 his4-519 | [rho+] COX2::3xHA | This study |
| HLF5-14 | MATα ura3-Δ leu2-3,112 his4-519 MSS2::3xMYC | [rho+] COX2::3xHA | This study |
| HLF5-17 | MATα ura3-Δ leu2-3,112 his4-519 MSS2::3xMYC | [rho+] cox2(1-109)::3xHA | This study |
| HLF5-22 | MATaade2 ura3-52 | [rho+] cox2 (C225H, H229C) | This study |
| HLF5-32 | MATα ura3-Δ leu2-3,112 his4-519 MSS2::3xMYC | [rho+] cox2(1-211)::3xHA | This study |
| HLF5-65 | MATaura3-52 leu2-3,112 lys2 his3-ΔHindIII arg8-Δ::hisG COX18::3xMYC | [rho+] COX2::3xHA | This study |
| HLF5-81 | MATα ade2 ura3 leu2-3,112 lys2 Δ arg8-Δ::hisG imp1Δ::kanMX | [rho+] cox2(1-211)::3xHA | This study |
| NAB33 | MATα ade2-101 ura3-52 arg8-Δ::hisG kar1-1 | [rho°] | This study |
| NB40-3C | MATaura3-52 leu2,3-112 lys his3ΔHindIII arg8-Δ::hisG | [rho+] cox2-62 | 5 |
| NB41 | MATα ade2-10 ura3-52 leu2-Δ arg8-Δ::URA3 kar1-1 | [rho+] cox2-62 | 5 |
| SAS1A | MATaura3-52 leu2-3,112 lys2 his3-ΔHindIII arg8-Δ::hisG | [rho+] | This study |
| SB12 | MATaade2-101 ura3-52 leu2-Δ mss2::LEU2 | [rho+] | 6 |
| SB151 | MATα ura3-52 leu2-3,112 lys2 his3ΔHindIII arg8-Δ::hisG mss2::LEU2 | [rho+] COX2::3xHA | This study |
| SB19A | MATα ura3-Δ leu2-3,112 his4-519 MSS2::3xHA | [rho+] | 6 |
| SCS101 | MATaura3-52 leu2-3,112 lys2 his3-ΔHindIII arg8-Δ::hisG | [rho+] COX2::3xHA | 45 |
| SCS32 | MATα ade2-101 ura3-52 leu2-Δ arg8-Δ::hisG cox18::URA3 | [rho+] COX2(1-251)::ARG8m | 46 |
| TF215 | MATα ura3-Δ leu2-3,112 his4-519 | [rho+] | 6 |
Mitochondrial purification and protein analysis.
Isolation and purification of mitochondria and protease protection experiments were carried out as previously described (16, 21). Mitochondrial protein concentrations were determined by the Lowry method using the DC protein assay kit (Bio-Rad). For Western blot analysis, proteins were transferred to Immobilon-P polyvinylidene difluoride membrane (Millipore) and incubated with antihemagglutinin (anti-HA) monoclonal antibody 3F10 (Roche), polyclonal anti-cytochrome b2, polyclonal anti-citrate synthase, or monoclonal anti-Cox2 CCO6. Secondary anti-mouse or anti-rabbit antibodies were detected using the ECL+ kit (Amersham Pharmacia).
Translation in isolated mitochondria with [35S]methionine (ICN Biochemicals) was performed essentially as described elsewhere (58). Following translation for 10 min at 30°C, the mitochondria were washed with 0.6 M sorbitol, 20 mM HEPES pH 7.4 and resuspended to 10 mg/ml before further processing. For immunoprecipitations of mitochondrial translation products, the labeled mitochondria were chased with 25 mM unlabeled methionine for 5 min, washed, resuspended in lysis buffer containing 1% digitonin, 100 mM NaCl, and 20 mM Tris pH 7.4 for 10 min, and spun at 40,000 × g for 45 min. The clarified supernatant was incubated with anti-HA (Roche, Inc.) or anti-MYC (Santa Cruz Biotechnology, Inc.) antibody-conjugated beads.
In vivo pulse-labeling of cycloheximide-treated cells with [35S]methionine was performed as previously described (2). As indicated, labeled strains were chased with the addition of unlabeled methionine to 10 mM for 10 min. After labeling, cells were chilled on ice in the presence of 10 mM unlabeled methionine, and mitochondria were prepared using glass bead disruption (11). Radiolabeled mitochondrial proteins were electrophoretically separated on sodium dodecyl sulfate (SDS)-polyacrylamide gels as indicated, dried, and exposed to film.
RESULTS
Removal of the C-terminal domain of Cox2 affects protein stability but not synthesis.
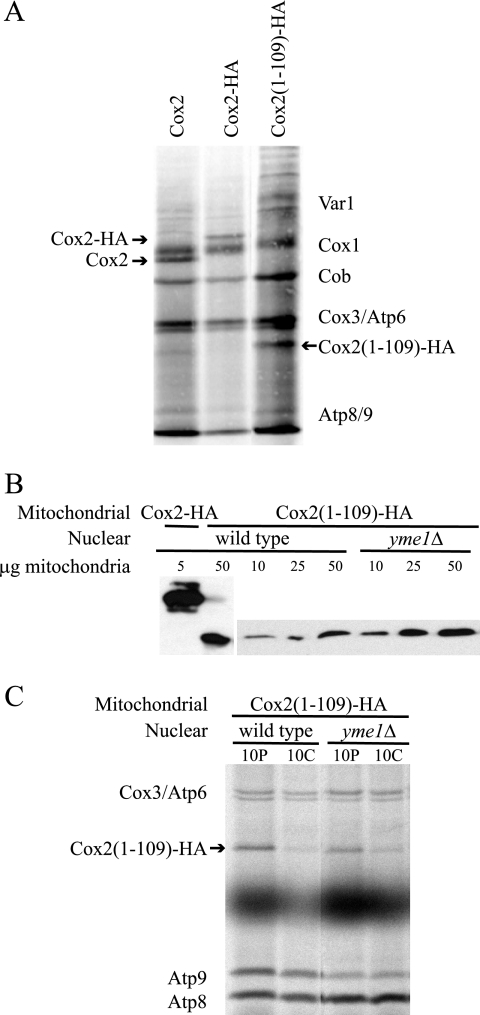
To investigate assembly of Cox2, we have examined full-length Cox2 carrying a C-terminal epitope tag (Cox2-HA) (46) and a cox2 truncation mutation at position 109 that removes the C-tail domain [Cox2(1-109)-HA]. Because elements within the coding sequence of the COX2 mRNA are known to be essential for translation, we examined translation in these cox2 mutants. Purified mitochondria from strains harboring each mitochondrial construct were incubated with [35S]methionine under conditions promoting translation in isolated organelles, and the mitochondrial translation products were resolved by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by autoradiography (Fig. 1A). Considering that the Cox2(1-109)-HA peptide contains half the number of methionines as wild-type Cox2 or Cox2-HA (three versus six methionines for processed mature Cox2 translation products), there was little or no difference in translation rate of the truncated form relative to the full-length proteins. Thus, translational regulation was apparently unaffected by removal of the Cox2 C-tail domain or by the addition of the HA epitope.
FIG. 1.
Synthesis of truncated Cox2 (1-109)-HA is normal, but protein stability is reduced. (A) Translation of Cox2, Cox2-HA, and Cox2(1-109)-HA in isolated mitochondria. Purified mitochondria harboring COX2, COX2::3xHA, and cox2(1-109)::3xHA constructs from strains SAS1A, SCS101, and HLF3-60, respectively, were incubated with [35S]methionine under conditions promoting translation in isolated mitochondria (see Materials and Methods). Translation products were resolved using 12% SDS-PAGE and visualized by autoradiography. The bands corresponding to Cox2, Cox2-HA, and Cox2(1-109)-HA are indicated. (B) Steady-state levels of Cox2-HA and Cox2(1-109)-HA. Purified mitochondria containing Cox2-HA (strain SCS101) or Cox2(1-109)-HA from strains with YME1 (strain HLF3-60) or yme1Δ (strain HLF4-87) nuclear backgrounds were analyzed by Western blot analysis following 12% SDS-PAGE electrophoresis to compare steady-state levels of HA-tagged products. Mitochondrial protein concentrations were normalized by probing the Western blots with antiporin and anti-citrate synthase (not shown). (C) Stability of newly synthesized Cox2(1-109)-HA. Strains carrying the mitochondrial cox2(1-109)::3xHA construct with wild-type (strain HLF3-60) or yme1Δ (strain HLF4-87) nuclear backgrounds were grown in yeast extract-peptone-raffinose and treated with cycloheximide for 5 min, followed by addition of [35S]methionine. Radiolabeling was quenched after 10 min by addition of unlabeled methionine, and mixtures were either placed immediately on ice to terminate translation (10P) or incubated for a 10-min chase period (10C) before terminating translation. Crude mitochondria were prepared using a glass bead cell disruption and centrifugation method, and labeled mitochondrial products were separated by 14 to 20% gradient SDS-PAGE and visualized by autoradiography.
Despite its normal synthesis rate, Cox2(1-109)-HA accumulated to significantly lower steady-state levels than full-length Cox2-HA, reduced more than 10-fold (Fig. 1B). A comparison of Cox2 and Cox2-HA steady-state levels using a C-tail-specific Cox2 monoclonal antibody showed no decrease in accumulation of Cox2 due to the epitope (not shown). The stability of newly synthesized Cox2(1-109)-HA was evaluated by [35S]methionine pulse-labeling of mitochondrial translation products in cycloheximide-treated cells followed by an unlabeled chase. Crude mitochondria were then isolated and separated by SDS-PAGE, and the mitochondrial translation products were visualized by autoradiography. Cox2(1-109)-HA was significantly degraded during a 10-min chase (Fig. 1C), in contrast to wild-type Cox2, which is stable during a 10-min chase (6, 40) (see Fig. 5A, below).
FIG. 5.
Newly synthesized Cox2(1-211)-HA is unstable but N-terminally processed. (A) Stability of newly synthesized Cox2(1-211)-HA. Cells producing either the full-length Cox2-HA (SCS101) or truncated Cox2(1-211)-HA (HLF4-13) proteins were pulse-labeled (10P), chased (10C), and analyzed as described in the legend for Fig. 1C. The labeled proteins are indicated by arrows. A novel unidentified translation product is indicated by the asterisk (see text). (B) Cox2(1-211)-HA is processed by Imp1. Mitochondrially synthesized proteins were radiolabeled either in cycloheximide-treated cells (CHX) or in isolated mitochondria (M), as indicated, and analyzed as described above. The mitochondrial genomes encoded either Cox2-HA or Cox2(1-211)-HA, while the relevant nuclear genotype was either wild type (wt) or imp1Δ, as indicated. Mature Cox2(1-211)-HA is indicated by the filled arrowhead, and its unprocessed precursor is indicated by the open arrowhead. Strains were as follows: lane 1, SCS101; lanes 2 and 4, HLF5-32; lane 3, HLF5-81.
Yme1 is a mitochondrial inner membrane i-AAA protease that is known to target unassembled Cox2 for degradation (31, 35, 39, 57). We looked to see if Cox2(1-109)-HA was stabilized in a yme1 mutant. A yme1 deletion offered no protection to newly synthesized Cox2(1-109)-HA in cycloheximide-treated cells (Fig. 1C) but did cause an approximately fourfold increase in Cox2(1-109)-HA steady-state levels (Fig. 1B). Thus, Yme1 appears to be active in the long-term degradation of this truncated protein. The active site of Yme1 lies in the inner membrane space of the mitochondria (31), but it is able to extract unassembled peptides from the membrane (30).
Translocation of newly synthesized Cox2(1-109)-HA N-tail across the inner membrane is dependent on OXA1 but not COX18 or MSS2.
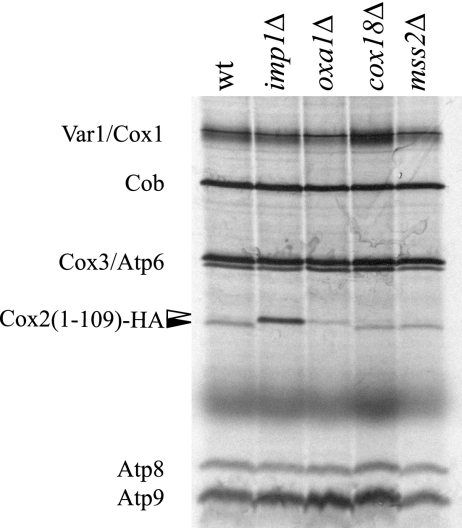
To determine whether the N-tail of the truncated Cox2(1-109)-HA is translocated following synthesis, processing of the peptide in the IMS by the Imp1 protease was evaluated following pulse-labeling of mitochondrial translation products in vivo. Cycloheximide-treated log-phase cells were incubated with [35S]methionine, followed by analysis of labeled mitochondrial proteins. Unprocessed pre-Cox2(1-109)-HA peptide synthesized in cells lacking Imp1 was slightly longer than the processed peptide in the wild type (Fig. 2). As expected, the leader peptide was not processed by Imp1 in an oxa1Δ mutant, showing that N-tail export of the truncated protein required Oxa1. Neither mss2Δ nor cox18Δ mutations affected Imp1 processing of pre-Cox2(1-109)-HA, confirming that Mss2 and Cox18 are not required for N-tail export. Unprocessed pre-Cox2(1-109)-HA contains 40% more labeled methionines (five versus three), explaining the increased labeling of pre-Cox2(1-109)-HA in the imp1Δ background. Reduced signal intensity in the oxa1Δ background is most likely due to decreased stability of Cox2(1-109)-HA (data not shown).
FIG. 2.
The N terminus of Cox2(1-109)-HA is translocated across the inner membrane immediately after synthesis. Log-phase cells grown in synthetic complete medium containing raffinose and lacking methionine were treated with cycloheximide for 5 min prior to incubation with [35S]methionine for 10 min. Crude mitochondria were isolated, electrophoresed on a 14 to 20% gradient SDS gel, and visualized by autoradiography. All strains harbored the cox2(1-109)-HA mitochondrial construct. The relevant nuclear genotypes of strains HLF3-60, HLF4-73, HLF4-17, HLF4-30, and HLF4-63 are indicated. N-terminally processed (filled arrow) and unprocessed (unfilled arrow) Cox2(1-109)-HA peptides are indicated.
N-tail translocation of Cox2(1-109)-HA is inhibited during synthesis in isolated mitochondria.
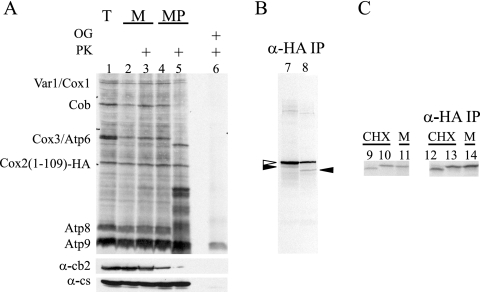
To investigate the fate of translocation of the C terminus of Cox2(1-109)-HA immediately following synthesis, we attempted protease protection experiments following protein synthesis in isolated mitochondria (Fig. 3A). Surprisingly, the band corresponding to Cox2(1-109)-HA remained unaltered following protease treatment of mitoplasts, indicating that neither the N-tail nor the C-tail of this truncated protein was exported under these conditions. To ensure that we were examining Cox2(1-109)-HA peptide and not a degradation product of a higher molecular weight species, the Cox2(1-109)-HA peptide was selectively immunoprecipitated from extracts of mitoplasts with anti-HA antibody (Fig. 3B). The majority of the protein, precipitated from both untreated and protease-treated mitoplasts by virtue of its C-terminal epitope, corresponded to unprocessed pre-Cox2(1-109)-HA. A small fraction of peptide representing N-terminally processed Cox2(1-109)-HA was shortened by treatment with protease, indicating very inefficient N-tail export of the protein synthesized in isolated mitochondria.
FIG. 3.
Cox2(1-109)-HA remains N-terminally unprocessed following translation in isolated mitochondria. (A) Mitochondrial translation products were radiolabeled with [35S]methionine in mitochondria purified from strain HLF3-60 (lane 1). Labeled mitochondria (lanes 2 and 3) or mitoplasts created through osmotic shock treatment (lanes 4 and 5) were mock treated (lanes 2 and 4) or treated with protease (lanes 3 and 5) (see Materials and Methods). The samples were separated by a 14 to 20% gradient SDS-PAGE, visualized by autoradiography, and probed with antibodies raised against cytochrome b2 (IMS localized) and citrate synthase (matrix localized) as controls for mitoplasting efficiency. A fraction of the total sample was treated with detergent (1% octylglucoside [OG]) and protease (lane 6), as indicated. (B) The samples in lanes 7 and 8 were obtained from the fractions analyzed in lanes 4 and 5, respectively, by immunoprecipitation with anti-HA coupled to beads. The major species corresponded to N-terminally unprocessed peptide (unfilled arrow). A smaller amount of processed Cox2(1-109)-HA (filled arrows) was shortened during protease treatment. (C) Radiolabeled Cox2(1-109)-HA from IMP1 strain HLF3-60 following translation in cycloheximide-treated cells (lane 9) and in isolated organelles (lane 11) was compared to that synthesized in cycloheximide-treated cells from imp1Δ strain HLF4-73 (lane 10) following separation by 14 to 20% gradient SDS-PAGE. The products analyzed in lanes 9 to 11 were immunoprecipitated with anti-HA antibody coupled to beads (in lanes 12 to 14, respectively) to verify the identity of Cox2(1-109)-HA species. While the N terminus of Cox2(1-109)-HA was processed by Imp1 in cycloheximide-treated cells, it remained unprocessed in the imp1Δ mutant and during translation in isolated mitochondria.
These results contrasted sharply with the efficient N-tail translocation observed in vivo after labeling of cycloheximide-treated cells (Fig. 2). We therefore directly compared translocation of Cox2(1-109)-HA following synthesis in isolated organelles with that in whole cells. Following mitochondrial translation in isolated mitochondria and in cycloheximide-treated cells, newly synthesized Cox2(1-109)-HA was selectively immunoprecipitated with anti-HA antibodies (Fig. 3C). Cox2(1-109)-HA newly synthesized in cycloheximide-treated cells was N-terminally processed by Imp1 in the IMS, whereas Cox2(1-109)-HA newly synthesized in isolated mitochondria remained largely unprocessed. The failure of Cox2(1-109)-HA N-tail export in isolated mitochondria precluded studies of newly synthesized C-tail export under these conditions. We therefore attempted to examine C-tail export of protein newly synthesized in cycloheximide-treated cells by protease protection experiments on highly purified labeled mitochondria. However, Cox2(1-109)-HA proved too unstable during the mitochondrial isolation procedure. Thus, we have been unable to examine by pulse-labeling the behavior of the C terminus of this truncated protein immediately after synthesis.
Insertion of both transmembrane domains in the absence of the C-tail.
The tagged C terminus of full-length Cox2-HA is translocated across the inner membrane and is accessible to exogenously added proteases in mitoplasts (46). To ask whether the C-terminal epitope of the Cox2(1-109)-HA truncation was also translocated across the inner membrane, we examined the topology of accumulated, steady-state protein in mitochondria from unlabeled growing cells. Mitoplasts were prepared and exposed to exogenously added protease to digest epitope residues that had been exported to the IMS. Western analysis of the mitoplasts containing Cox2(1-109)-HA revealed that the HA tag was accessible to added proteases (Fig. 4), demonstrating that the C terminus of the accumulated truncated protein was exposed on the IMS side of the inner membrane. This behavior, identical to that of the full-length protein (reference 46 and data not shown), suggests that the C-tail domain of Cox2 is not required for the insertion of both the first and second transmembrane domains nor for translocation of the small hydrophilic epitope.
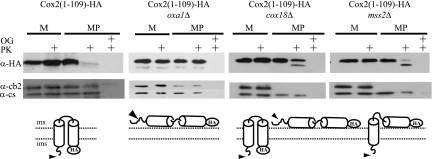
FIG. 4.
The C-terminal domain of Cox2 is not required for integration of the transmembrane domains nor translocation of an HA epitope. Mitochondria (M) carrying full-length Cox2-HA (50 μg) or Cox2(1-109)-HA (200 μg) from strains with wild-type, oxa1Δ, cox18Δ, or mss2Δ nuclear backgrounds was purified from strains HLF3-60, HLF4-17, HLF4-30, and HLF4-63, respectively, and converted to mitoplasts (MP) in the absence or presence of proteinase K (PK). The mitochondrial proteins were resolved by 12% SDS-PAGE, immunoblotted, and decorated with anti-HA antibodies. The size difference between Imp1-processed and unprocessed Cox2(1-109)-HA was not resolved under these electrophoretic conditions. To control for mitoplasting effectiveness, similar blots (containing 5 μg mitochondria) were probed with antibodies against cytochrome b2 and citrate synthase. Solubilization of mitochondrial membranes in 1% octyl glucoside (OG) demonstrated effective protease activity.
We next asked whether translocation of the C-terminal epitope of the Cox2(1-109)-HA protein was affected by the absence of Oxa1, Cox18, or Mss2. Protease protection experiments were performed on mitochondria and mitoplasts from oxa1Δ, cox18Δ, and mss2Δ strains harboring Cox2(1-109)-HA (Fig. 4). In the absence of Oxa1, the Cox2(1-109)-HA C-terminal epitope remained on the matrix side of the inner membrane, protected from exogenous protease. As expected, the protein was not shortened by protease treatment, indicating that the N terminus of Cox2(1-109)-HA also remained on the matrix side.
Since Cox18 and Mss2 have roles in translocation of the C-tail domain of Cox2, we expected that cox18Δ and mss2Δ mutations might not affect translocation of the N or C termini of Cox2(1-109)-HA, allowing export of the epitope. Consistent with this idea, most of the epitope on accumulated Cox2(1-109)-HA in mitoplasts disappeared after protease treatment (Fig. 4), indicating a substantial level of export in both cox18Δ and mss2Δ mutants. However, mitoplasts from a cox18Δ strain also accumulated a significant amount of peptide whose N and C termini remained protected. Mitoplasts from the mss2Δ mutant accumulated Cox2(1-109)-HA whose N terminus was translocated across the membrane (as judged by the shortening of the immunoreactive peptide) but whose C terminus remained protected. The accumulation of some untranslocated N-tail of Cox2(1-109)-HA in cox18Δ and mss2Δ mitochondria despite the apparently normal N-tail export of newly synthesized protein in these mutants suggests that there is a subtle slowing of N-tail translocation in these mutants.
Sequences within the Cox2 C-tail domain affect C-tail translocation.
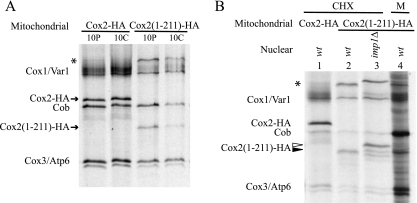
To ask whether the C-tail domain might contain sequence or structural information necessary to direct its own membrane translocation, we created another cox2 truncation mutant, cox2(1-211)::3xHA. This altered allele encodes a C-terminally tagged truncated form of Cox2 lacking the last 40 residues of the 144-residue C-tail domain. The stability of newly synthesized Cox2(1-211)-HA was evaluated by [35S]methionine pulse-labeling of mitochondrial translation products in cycloheximide-treated cells followed by an unlabeled chase. Like Cox2(1-109)-HA, newly synthesized Cox2(1-211)-HA was degraded during the chase, in contrast to the wild-type protein (Fig. 5A). Its steady-state level was also reduced relative to full-length Cox2-HA (not shown).
We evaluated Cox2(1-211)-HA N-tail translocation by asking whether the N terminus was processed by the Imp1 protease after synthesis in cycloheximide-treated cells or in isolated mitochondria (Fig. 5B). In both cases, newly synthesized Cox2(1-211)-HA was processed by Imp1. An unidentified high-molecular-weight mitochondrial translation product, apparently also processed by Imp1, appeared in the cox2(1-211)::3xHA mitochondria (Fig. 5A and B). It is likely to be an aggregated form of Cox2(1-211)-HA. No corresponding band appears in Western blots of cox2(1-211)::3xHA mitochondria probed with anti-HA or anti-Cox2 antibodies, suggesting that this species is unstable.
We evaluated the location of accumulated Cox2(1-211)-HA C-tail in mitochondria by Western analysis to detect the HA epitope (Fig. 6A). Addition of exogenous protease to mitoplasts shortened the protein but left the epitope intact, indicating that the N-tail was exposed on the IMS side of the inner membrane but the C-tail remained protected in the matrix. Thus, removal of the last 40 amino acids of the C-tail prevented its translocation.
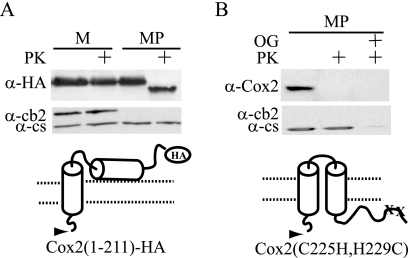
FIG. 6.
Truncation of 40 residues from the C-terminal domain of Cox2 prevents C-tail translocation. (A) A 200-μg aliquot of purified mitochondria (M) containing Cox2(1-211)-HA (strain HLF4-13) was converted to mitoplasts (MP) in the absence or presence of proteinase K (PK). The mitochondrial proteins were resolved by 12% SDS-PAGE, immunoblotted, and decorated with anti-HA antibodies. (B) A 200-μg aliquot of purified mitochondria containing the copper-binding-deficient Cox2-C225H, H229C (49) (strain HLF5-22), was converted to mitoplasts (MP) in the absence or presence of proteinase K (PK). Control mitoplasts were solubilized with 1% octyl glucoside (OG). The mitochondrial proteins were resolved by 12% SDS-PAGE, immunoblotted, and decorated with anti-Cox2 monoclonal CCO6, which recognizes a C-tail epitope (21).
The amino acids deleted from Cox2(1-211)-HA contain three of the four copper-ligating residues in the binuclear CuA site of Cox2. To test whether copper binding could be a requirement for C-tail translocation, a strain carrying missense substitutions of two of these copper-binding residues (C225H and H229C) (49) was created. The C-tail domain of this copper-binding-defective protein was sensitive to protease treatment of mitoplasts, demonstrating that the C-tail was fully translocated (Fig. 6B). Thus, copper binding is not a requirement for C-tail translocation.
Interaction of Mss2 with newly synthesized Cox2 requires the C-tail.
Mss2 is a hydrophilic protein located on the inner surface of the inner membrane and could therefore interact directly with newly synthesized Cox2 to carry out its role in the export process (6). To test this idea, we asked whether immunoprecipitation of Mss2 would coprecipitate newly synthesized Cox2 labeled in isolated mitochondria. Radiolabeling of mitochondrial translation products was carried out in mitochondria purified from MSS2 or MSS2::HA strains. The mitochondria were then solubilized in digitonin and immunoprecipitated using anti-HA antibody coupled to beads (Fig. 7A). Mature Cox2 was selectively immunoprecipitated with Mss2-HA but not Mss2, suggesting that Mss2 interacts physically with newly synthesized Cox2 molecules whose N termini have been exported and processed. In addition, we found that Cox2-HA was selectively coimmunoprecipitated with Mss2 bearing a MYC epitope (Mss2-MYC) (Fig. 7B). Finally, we asked whether Cox18, which has been shown to coimmunoprecipitate with Mss2 (46), would also pull down newly synthesized Cox2. Precipitation of Cox18-MYC from solubilized mitochondria with anti-MYC antibody selectively enriched newly synthesized Cox2-HA (Fig. 7C).
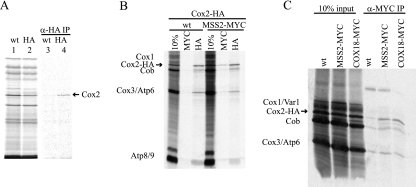
FIG. 7.
Mss2 and Cox18 immunoprecipitate newly synthesized Cox2. (A) [35S]methionine radiolabeling was performed in mitochondria purified from MSS2 (TF215) or MSS2-HA (SB19A) strains containing wild-type mtDNA (lanes 1 and 2). Translation products analyzed in lanes 1 and 2 were solubilized in 1% digitonin (see Materials and Methods), cleared by centrifugation, and immunoprecipitated with anti-HA affinity resin (lanes 3 and 4, respectively). The mitochondrial translation products were resolved by 16% SDS-PAGE and visualized by autoradiography. Lanes 1 and 2 contain 10% of the total input used for the immunoprecipitates in lanes 3 and 4. (B) Mitochondria containing full-length Cox2-HA were purified from MSS2 (HLF5-13) or MSS2-MYC (HLF5-14) strains. Translation in the isolated mitochondria and solubilization were carried out as for panel A. Equal aliquots were incubated with either anti-MYC or anti-HA affinity resin. A 10% portion of each extract and the immune precipitates were resolved by 14 to 20% gradient SDS-PAGE and visualized by autoradiography. (C) Mitochondria containing full-length Cox-HA were purified from strains HLF5-13 (wt), HLF5-14 (MSS2-MYC), and HLF5-65 (COX18-MYC). Translation in the isolated mitochondria and solubilization were carried out as for panel A, followed by immune precipitations with anti-MYC affinity resin.
To ask whether the interaction between Cox2 and Mss2 might occur through the Cox2p C-tail domain, similar immunoprecipitation experiments were performed in mitochondria from strains containing Mss2-MYC and either Cox2(1-109)-HA or Cox2(1-211)-HA (Fig. 8). Cox2(1-211)-HA, lacking the last 40 residues of the C-tail, coprecipitated with Mss2-MYC. However, Cox2(1-109)-HA, lacking the entire C-tail, was not coimmunoprecipitated with Mss2-MYC. To ensure that HA-tagged forms of Cox2 were indeed present as labeled species in the solubilized mitochondrial extracts, we subjected them to direct immunoprecipitation with anti-HA. Both truncated variants of HA-tagged Cox2 were detected in these direct precipitates (Fig. 7B and 8). However, some N-terminal proteolysis of Cox2(1-109)-HA was evidenced by the presence of shorter species that were selectively enriched by immunoprecipitation. Surprisingly, the Cox2(1-211)-HA directly precipitated with anti-HA-coupled beads was longer than the Cox2(1-211)-HA coprecipitated with Mss2-MYC using anti-MYC-coupled resin. This probably reflects proteolysis of the triply repeated HA epitope on Cox2(1-211)-HA in the extract, which is prevented by the protection afforded by the anti-HA antibody. This proteolysis was not inhibited by the addition of protease inhibitors (not shown).
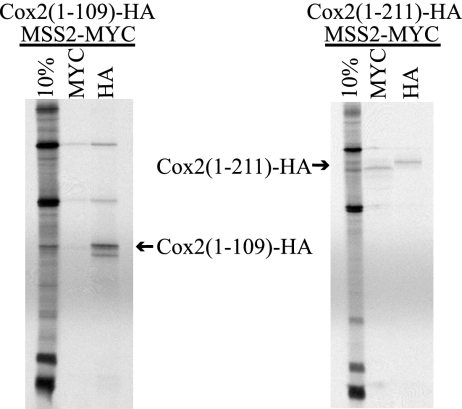
FIG. 8.
Coimmunoprecipitation of truncated Cox2 variants with Mss2-MYC. Mitochondria whose genomes encode Cox2(1-109)-HA or Cox2(1-211)-HA were purified from strains whose nuclei encode Mss2-Myc (HLF5-17 and HLF5-32). Mitochondrial translation products were labeled, and solubilized extracts were subjected to immunoprecipitations as described in the legend for Fig. 7, using anti-MYC and anti-HA affinity resins as indicated. The relevant bands are indicated by labeled arrows.
Taken together, these data indicate that the interaction between Mss2 and newly synthesized Cox2 requires sequences or structures formed by residues 109 to 211 of the C-tail but that this interaction alone is not sufficient to cause C-tail translocation.
DISCUSSION
Translocation of the hydrophilic N-tail and C-tail domains of yeast Cox2 from the mitochondrial matrix to the IMS is carried out by different mechanisms. N-tail export of pre-Cox2 is strictly dependent upon the integral membrane protein Oxa1 but does not require a potential difference across the inner membrane (21). While export of the C-tail depends upon Oxa1, it also requires the integral membrane protein Cox18, the peripheral membrane protein Mss2 (Pnt1 assists these proteins but is dispensable), and a membrane potential (6, 21-23, 46). While Oxa1 appears to have a role in the membrane insertion of several inner membrane proteins (24, 29), the interacting proteins Cox18 and Mss2 appear to be specific for Cox2 C-tail translocation (6, 22, 46). Yeast mitochondria do not possess an identifiable translocation system homologous to the bacterial Sec apparatus (17). However, the Cox2 C-tail translocation system can export a large passenger protein, Arg8m, fused to the Cox2 C terminus (21).
Oxa1 appears to have two functional domains (41). Membrane protein insertion is carried out by the N-terminal polytypic integral membrane segment of Oxa1, which is homologous to bacterial YidC as well as yeast Cox18. The C-terminal domain of Oxa1 is exposed on the inner surface of the mitochondrial membrane and interacts with mitochondrial ribosomes, apparently to promote cotranslational targeting of nascent proteins (28, 50). Both Cox18, which is functionally conserved among eukaryotes (14), and YidC lack this C-terminal domain. Fusion of this domain of Oxa1 to the C terminus of bacterial YidC generates a chimeric protein that can partially compensate for the absence of normal Oxa1 in yeast, while unmodified YidC can partially compensate for the absence of Cox18 (41). These observations suggest that direct interaction with the ribosome presents the N terminus of pre-Cox2 to the translocase domain of Oxa1 but that other functions may be required to present the Cox2 C-terminal domain to Cox18.
Mss2 is exposed on the inner surface of the inner membrane and thus could function to recognize the Cox2 C-tail prior to translocation by Cox18 (6). Consistent with this idea, Mss2 has a tetratricopeptide motif (6) similar to one in the mitochondrial import receptor Tom70, which binds cytoplasmic chaperone-associated translocation substrates (9, 60). We observed here that newly synthesized full-length Cox2 could be coimmunoprecipitated with Mss2 from solubilized mitochondria, consistent with the idea that Mss2 recognizes the Cox2 C-tail prior to export. The coprecipitated Cox2 was processed at its N terminus, demonstrating that N-tail export can occur while Cox2 is still bound, directly or indirectly, to Mss2 on the inner surface of the inner membrane. Thus, C-tail translocation may be a posttranslational event that can occur subsequent to cotranslational N-tail export and N-terminal processing. Mss2-Cox2 coprecipitation was not observed when the entire C-tail domain was completely deleted, consistent with the possibility that Mss2 might bind to the C-tail. If so, Mss2 could hold the C-tail domain of Cox2 in an unfolded conformation, or in an intermediate folded conformation (51), required for translocation after translation termination. (The N-tail domain of truncated Cox2 lacking the C-tail was not processed during synthesis in isolated mitochondria, making it formally possible that interaction with Mss2 depends on N-terminal processing rather than the absence of the C-tail per se.)
To explore the features of Cox2 that are required for membrane insertion and translocation, we studied the behavior of truncated Cox2 variants bearing an epitope tag at their C termini, encoded in mtDNA. The N-tail of a protein lacking the entire C-tail domain, Cox2(1-109)-HA, was efficiently translocated across the inner membrane during pulse-labeling of whole cells in a reaction that was dependent upon Oxa1 but not Cox18 or Mss2. Interestingly, N-tail export of the Cox2(1-109)-HA N-tail did not occur during pulse-labeling of isolated mitochondria, demonstrating that the isolated organelles are deficient in some feature of this process relative to intact cells.
Unlabeled Cox2(1-109)-HA accumulated (albeit in greatly diminished amounts relative to wild-type Cox2) with both its N terminus and epitope-tagged C terminus on the outside of the inner membrane. Thus, insertion of the two transmembrane domains of Cox2, and export of the C-terminal epitope, can occur in the absence of the C-tail. Both the N and C termini of Cox2(1-109)-HA accumulated in the matrix in an oxa1Δ mutant. Since previous work indicated that Cox18 and Mss2 are required only for C-tail translocation, we expected export of the Cox2(1-109)-HA N-tail to be unaffected in cox18Δ and mss2Δ mutants. However, both mutations caused accumulation of detectable levels of Cox2(1-109)-HA, whose N-tail was in the matrix (cox18Δ more so than mss2Δ). This could indicate either a direct but minor role for Cox18 and Mss2 in N-tail export or a general inefficiency of a larger translocation complex lacking either protein, in either case leading to accumulation of species not detected during pulse-labeling. Similarly, while most of the C-terminal epitope of Cox2(1-109)-HA was exported in both mutants, demonstrating that Cox18 and Mss2 are not required for insertion of the second transmembrane domain, both mutations clearly reduced export of the C terminus relative to wild type. In this context it is important to note that the addition of the 33-residue hydrophilic epitope tag to the C terminus of the second transmembrane domain at residue 109 may reduce the efficiency of its insertion. This insertion is completely blocked by the presence of a 402-residue hydrophilic passenger protein moiety (Arg8m) fused at the same position (21).
We also examined features of the 144-residue Cox2 C-tail domain that could contribute to its recognition as a translocation substrate. A truncated variant of Cox2 lacking the 40 C-terminal residues of this domain [Cox2(1-211)-HA] accumulated in mitochondria with its N-tail exported to the IMS and its C terminus remaining in the matrix. Thus, it is clear that sequence and/or structural features of the C-tail domain itself are recognized by its translocation apparatus independently of N-tail translocation. Recognition of these features in the C-tail can also lead to translocation of a fused passenger protein (21). Despite the fact that the truncated C-tail of Cox2(1-211)-HA is not translocated, this variant protein nevertheless did coimmunoprecipitate with Mss2. Thus, while the Cox2-Mss2 interaction is highly likely to be necessary for C-tail export, recognition of the C-tail as a translocation substrate must involve other steps as well.
Three of the four copper-ligating residues of the cytochrome c oxidase CuA site are absent from the truncated protein Cox2(1-211)-HA. While copper insertion is thought to occur in the IMS after export (7), the mitochondrial matrix has an accessible nonproteinaceous copper pool (10) that could provide metal ions to the C-tail domain. However, we found that point mutations altering the copper-binding residues C225 and H229 (corresponding to C200 and H204 from the bovine Cox2 sequence) (49) are not required for efficient C-tail translocation. Furthermore, Sco1, a mitochondrial protein required for copper delivery to Cox2 (37, 42), is not required for C-tail translocation (45). Thus, a fully folded C-tail domain containing copper ions is not required for C-tail translocation.
Both the exported 22-residue N-tail and 144-residue C-tail domains of Cox2 are negatively charged, with predicted pI values of 3.8 and 4.4, respectively. Thus, these translocation systems adhere to the “positive-inside rule” (15, 20, 43). However, while charge may play a role in driving the Cox2 C-tail across the membrane in a potential-dependent fashion, neither acidity nor the charged residues immediately flanking the transmembrane domains are sufficient to direct this process, since the acidic C-tail domain of the truncated variant Cox2(1-211)-HA (pI 4.0) is not translocated.
The molecular mechanism(s) of bacterial YidC has been studied extensively but remains to be precisely defined (reviewed in reference 32). YidC is at least partially associated with the bacterial Sec translocase, although it is not required for most Sec-dependent reactions. YidC is necessary for the assembly of the Escherichia coli respiratory chain (54). Plasma membrane insertion of the E. coli protein CyoA, a subunit of the bacterial cytochrome bo oxidase that is similar to mitochondrial Cox2, also depends upon distinct mechanisms for insertion of its N- and C-terminal domains into the membrane (8, 12, 53). In this case, the Oxa1/Cox18 homolog YidC is required for translocation of the N-terminal domain, while the C-terminal domain is translocated by the Sec translocase. While cells depleted for YidC do not translocate either domain of the wild-type protein, this is due to the fact that prior YidC-dependent translocation of the N-terminal sequences and insertion of the transmembrane domains are a prerequisite for Sec-dependent translocation of the C-terminal domain (8). Interestingly, both translocation reactions are independent of membrane potential when carried out in vitro in proteoliposomes (12). Thus, there are both similarities and clear differences between the yeast mitochondrial and bacterial systems: in both cases the early events are catalyzed by Oxa1/YidC, while in mitochondria the Cox2-specific functions of Cox18/YidC and Mss2 have taken the place of the Sec translocon.
Acknowledgments
We thank L. Briggs for technical assistance, as well as S. Saracco, S. Shabalina, N. Bsat, C. Martin, T. L. Mason, and G. Schatz for gifts of biological materials and reagents.
This study was supported by a postdoctoral fellowship to H.L.F. (GM65664) and a research grant to T.D.F. (GM29362) from the U.S. National Institutes of Health.
Footnotes
Published ahead of print on 23 April 2007.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1987. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, NY.
- 2.Bonnefoy, N., N. Bsat, and T. D. Fox. 2001. Mitochondrial translation of Saccharomyces cerevisiae COX2 mRNA is controlled by the nucleotide sequence specifying the pre-Cox2p leader peptide. Mol. Cell. Biol. 21:2359-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonnefoy, N., F. Chalvet, P. Hamel, P. P. Slonimski, and G. Dujardin. 1994. OXA1, a Saccharomyces cerevisiae nuclear gene whose sequence is conserved from prokaryotes to eukaryotes controls cytochrome oxidase biogenesis. J. Mol. Biol. 239:201-212. [DOI] [PubMed] [Google Scholar]
- 4.Bonnefoy, N., and T. D. Fox. 2001. Genetic transformation of Saccharomyces cerevisiae mitochondria. Methods Cell Biol. 65:381-396. [DOI] [PubMed] [Google Scholar]
- 5.Bonnefoy, N., and T. D. Fox. 2000. In vivo analysis of mutated initiation codons in the mitochondrial COX2 gene of Saccharomyces cerevisiae fused to the reporter gene ARG8m reveals lack of downstream reinitiation. Mol. Gen. Genet. 262:1036-1046. [DOI] [PubMed] [Google Scholar]
- 6.Broadley, S. A., C. M. Demlow, and T. D. Fox. 2001. Peripheral mitochondrial inner membrane protein, Mss2p, required for export of the mitochondrially coded Cox2p C tail in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:7663-7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carr, H. S., and D. R. Winge. 2003. Assembly of cytochrome c oxidase within the mitochondrion. Acc. Chem. Res. 36:309-316. [DOI] [PubMed] [Google Scholar]
- 8.Celebi, N., L. Yi, S. J. Facey, A. Kuhn, and R. E. Dalbey. 2006. Membrane biogenesis of subunit II of cytochrome bo oxidase: contrasting requirements for insertion of N-terminal and C-terminal domains. J. Mol. Biol. 357:1428-1436. [DOI] [PubMed] [Google Scholar]
- 9.Chan, N. C., V. A. Likic, R. F. Waller, T. D. Mulhern, and T. Lithgow. 2006. The C-terminal TPR domain of Tom70 defines a family of mitochondrial protein import receptors found only in animals and fungi. J. Mol. Biol. 358:1010-1022. [DOI] [PubMed] [Google Scholar]
- 10.Cobine, P. A., L. D. Ojeda, K. M. Rigby, and D. R. Winge. 2004. Yeast contain a non-proteinaceous pool of copper in the mitochondrial matrix. J. Biol. Chem. 279:14447-14455. [DOI] [PubMed] [Google Scholar]
- 11.Diekert, K., A. I. de Kroon, G. Kispal, and R. Lill. 2001. Isolation and subfractionation of mitochondria from the yeast Saccharomyces cerevisiae. Methods Cell Biol. 65:37-51. [DOI] [PubMed] [Google Scholar]
- 12.du Plessis, D. J. F., N. Nouwen, and A. J. M. Driessen. 2006. Subunit a of cytochrome o oxidase requires both YidC and SecYEG for membrane insertion. J. Biol. Chem. 281:12248-12252. [DOI] [PubMed] [Google Scholar]
- 13.Fontanesi, F., I. C. Soto, D. Horn, and A. Barrientos. 2006. Assembly of mitochondrial cytochrome c oxidase, a complicated and highly regulated cellular process. Am. J. Physiol. Cell Physiol. 291:C1129-C1147. [DOI] [PubMed] [Google Scholar]
- 14.Gaisne, M., and N. Bonnefoy. 2006. The COX18 gene, involved in mitochondrial biogenesis, is functionally conserved and tightly regulated in humans and fission yeast. FEMS Yeast Res. 6:869-882. [DOI] [PubMed] [Google Scholar]
- 15.Gavel, Y., and G. von Heijne. 1992. The distribution of charged amino acids in mitochondrial inner-membrane proteins suggests different modes of membrane integration for nuclearly and mitochondrially encoded proteins. Eur. J. Biochem. 205:1207-1215. [DOI] [PubMed] [Google Scholar]
- 16.Glick, B. S., and L. A. Pon. 1995. Isolation of highly purified mitochondria from Saccharomyces cerevisiae. Methods Enzymol. 260:213-223. [DOI] [PubMed] [Google Scholar]
- 17.Glick, B. S., and G. Von Heijne. 1996. Saccharomyces cerevisiae mitochondria lack a bacterial-type sec machinery. Protein Sci. 5:2651-2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Green-Willms, N. S., C. A. Butler, H. M. Dunstan, and T. D. Fox. 2001. Pet111p, an inner membrane-bound translational activator that limits expression of the Saccharomyces cerevisiae mitochondrial gene COX2. J. Biol. Chem. 276:6392-6397. [DOI] [PubMed] [Google Scholar]
- 19.Guthrie, C., and G. R. Fink. 1991. Guide to yeast genetics and molecular biology. Methods Enzymol. 194:1-863. [PubMed] [Google Scholar]
- 20.Harley, C. A., J. A. Holt, R. Turner, and D. J. Tipper. 1998. Transmembrane protein insertion orientation in yeast depends on the charge difference across transmembrane segments, their total hydrophobicity, and its distribution. J. Biol. Chem. 273:24963-24971. [DOI] [PubMed] [Google Scholar]
- 21.He, S., and T. D. Fox. 1997. Membrane translocation of mitochondrially coded Cox2p: distinct requirements for export of N and C termini and dependence on the conserved protein Oxa1p. Mol. Biol. Cell 8:1449-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He, S., and T. D. Fox. 1999. Mutations affecting a yeast mitochondrial inner membrane protein, Pnt1p, block export of a mitochondrially synthesized fusion protein from the matrix. Mol. Cell. Biol. 19:6598-6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hell, K., J. Herrmann, E. Pratje, W. Neupert, and R. A. Stuart. 1997. Oxa1p mediates the export of the N- and C-termini of pCoxII from the mitochondrial matrix to the intermembrane space. FEBS Lett. 418:367-370. [DOI] [PubMed] [Google Scholar]
- 24.Hell, K., W. Neupert, and R. A. Stuart. 2001. Oxa1p acts as a general membrane insertion machinery for proteins encoded by mitochondrial DNA. EMBO J. 20:1281-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hell, K., A. Tzagoloff, W. Neupert, and R. A. Stuart. 2000. Identification of Cox20p, a novel protein involved in the maturation and assembly of cytochrome oxidase subunit 2. J. Biol. Chem. 275:4571-4578. [DOI] [PubMed] [Google Scholar]
- 26.Herrmann, J. M., and S. Funes. 2005. Biogenesis of cytochrome oxidase-sophisticated assembly lines in the mitochondrial inner membrane. Gene 354:43-52. [DOI] [PubMed] [Google Scholar]
- 27.Jan, P. S., K. Esser, E. Pratje, and G. Michaelis. 2000. Som1, a third component of the yeast mitochondrial inner membrane peptidase complex that contains Imp1 and Imp2. Mol. Gen. Genet. 263:483-491. [DOI] [PubMed] [Google Scholar]
- 28.Jia, L., M. Dienhart, M. Schramp, M. McCauley, K. Hell, and R. A. Stuart. 2003. Yeast Oxa1 interacts with mitochondrial ribosomes: the importance of the C-terminal region of Oxa1. EMBO J. 22:6438-6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kermorgant, M., N. Bonnefoy, and G. Dujardin. 1997. Oxa1p, which is required for cytochrome c oxidase and ATP synthase complex formation, is embedded in the mitochondrial inner membrane. Curr. Genet. 31:302-307. [DOI] [PubMed] [Google Scholar]
- 30.Leonhard, K., B. Guiard, G. Pellecchia, A. Tzagoloff, W. Neupert, and T. Langer. 2000. Membrane protein degradation by AAA proteases in mitochondria: extraction of substrates from either membrane surface. Mol. Cell 5:629-638. [DOI] [PubMed] [Google Scholar]
- 31.Leonhard, K., J. M. Herrmann, R. A. Stuart, G. Mannhaupt, W. Neupert, and T. Langer. 1996. AAA proteases with catalytic sites on opposite membrane surfaces comprise a proteolytic system for the ATP-dependent degradation of inner membrane proteins in mitochondria. EMBO J. 15:4218-4229. [PMC free article] [PubMed] [Google Scholar]
- 32.Luirink, J., G. von Heijne, E. N. Houben, and J. W. Gier. 2005. Biogenesis of inner membrane proteins in Escherichia coli. Annu. Rev. Microbiol. 59:329-355. [DOI] [PubMed] [Google Scholar]
- 33.Mulero, J. J., and T. D. Fox. 1993. PET111 acts in the 5′-leader of the Saccharomyces cerevisiae mitochondrial COX2 mRNA to promote its translation. Genetics 133:509-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Naithani, S., S. A. Saracco, C. A. Butler, and T. D. Fox. 2003. Interactions among COX1, COX2, and COX3 mRNA-specific translational activator proteins on the inner surface of the mitochondrial inner membrane of Saccharomyces cerevisiae. Mol. Biol. Cell 14:324-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakai, T., T. Yasuhara, Y. Fujiki, and A. Ohashi. 1995. Multiple genes, including a member of the AAA family, are essential for degradation of unassembled subunit 2 of cytochrome c oxidase in yeast mitochondria. Mol. Cell. Biol. 15:4441-4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neff, N. F., J. H. Thomas, P. Grisafi, and D. Botstein. 1983. Isolation of the beta-tubulin gene from yeast and demonstration of its essential function in vivo. Cell 33:211-219. [DOI] [PubMed] [Google Scholar]
- 37.Nittis, T., G. N. George, and D. R. Winge. 2001. Yeast Sco1, a protein essential for cytochrome c oxidase function is a Cu(I)-binding protein. J. Biol. Chem. 276:42520-42526. [DOI] [PubMed] [Google Scholar]
- 38.Nunnari, J., T. D. Fox, and P. Walter. 1993. A mitochondrial protease with two catalytic subunits of nonoverlapping specificities. Science 262:1997-2004. [DOI] [PubMed] [Google Scholar]
- 39.Pearce, D. A., and F. Sherman. 1995. Degradation of cytochrome oxidase subunits in mutants of yeast lacking cytochrome c and suppression of the degradation by mutation of YME1. J. Biol. Chem. 270:20879-20882. [DOI] [PubMed] [Google Scholar]
- 40.Perez-Martinez, X., S. A. Broadley, and T. D. Fox. 2003. Mss51p promotes mitochondrial Cox1p synthesis and interacts with newly synthesized Cox1p. EMBO J. 22:5951-5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Preuss, M., M. Ott, S. Funes, J. Luirink, and J. M. Herrmann. 2005. Evolution of mitochondrial Oxa proteins from bacterial YidC. Inherited and acquired functions of a conserved protein insertion machinery. J. Biol. Chem. 280:13004-13011. [DOI] [PubMed] [Google Scholar]
- 42.Rentzsch, A., G. Krummeck-Weiss, A. Hofer, A. Bartuschka, K. Ostermann, and G. Rodel. 1999. Mitochondrial copper metabolism in yeast: mutational analysis of Sco1p involved in the biogenesis of cytochrome c oxidase. Curr. Genet. 35:103-108. [DOI] [PubMed] [Google Scholar]
- 43.Rojo, E. E., B. Guiard, W. Neupert, and R. A. Stuart. 1999. N-terminal tail export from the mitochondrial matrix. Adherence to the prokaryotic “positive-inside” rule of membrane protein topology. J. Biol. Chem. 274:19617-19622. [DOI] [PubMed] [Google Scholar]
- 44.Sanchirico, M. E., T. D. Fox, and T. L. Mason. 1998. Accumulation of mitochondrially synthesized Saccharomyces cerevisiae Cox2p and Cox3p depends on targeting information in untranslated portions of their mRNAs. EMBO J. 17:5796-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saracco, S. A. 2003. Analysis of the export of Saccharomyces cerevisiae Cox2p from the mitochondrial matrix. Ph.D. thesis. Cornell University, Ithaca, NY.
- 46.Saracco, S. A., and T. D. Fox. 2002. Cox18p is required for export of the mitochondrially encoded Saccharomyces cerevisiae Cox2p C-tail and interacts with Pnt1p and Mss2p in the inner membrane. Mol. Biol. Cell 13:1122-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schneider, B. L., W. Seufert, B. Steiner, Q. H. Yang, and A. B. Futcher. 1995. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast 11:1265-1274. [DOI] [PubMed] [Google Scholar]
- 48.Souza, R. L., N. S. Green-Willms, T. D. Fox, A. Tzagoloff, and F. G. Nobrega. 2000. Cloning and characterization of COX18, a Saccharomyces cerevisiae PET gene required for the assembly of cytochrome oxidase. J. Biol. Chem. 275:14898-15902. [DOI] [PubMed] [Google Scholar]
- 49.Speno, H., M. R. Taheri, D. Sieburth, and C. T. Martin. 1995. Identification of essential amino acids within the proposed CuA binding site in subunit II of Cytochrome c oxidase. J. Biol. Chem. 270:25363-25369. [DOI] [PubMed] [Google Scholar]
- 50.Szyrach, G., M. Ott, N. Bonnefoy, W. Neupert, and J. M. Herrmann. 2003. Ribosome binding to the Oxa1 complex facilitates co-translational protein insertion in mitochondria. EMBO J. 22:6448-6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Teter, S. A., and D. J. Klionsky. 1999. How to get a folded protein across a membrane. Trends Cell Biol. 9:428-431. [DOI] [PubMed] [Google Scholar]
- 52.Thorsness, P. E., K. H. White, and T. D. Fox. 1993. Inactivation of YME1, a member of the ftsH-SEC18-PAS1-CDC48 family of putative ATPase-encoding genes, causes increased escape of DNA from mitochondria in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:5418-5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Bloois, E., G. J. Haan, J. W. de Gier, B. Oudega, and J. Luirink. 2006. Distinct requirements for translocation of the N-tail and C-tail of the Escherichia coli inner membrane protein CyoA. J. Biol. Chem. 281:10002-10009. [DOI] [PubMed] [Google Scholar]
- 54.Van Der Laan, M., M. L. Urbanus, C. M. Ten Hagen-Jongman, N. Nouwen, B. Oudega, N. Harms, A. J. Driessen, and J. Luirink. 2003. A conserved function of YidC in the biogenesis of respiratory chain complexes. Proc. Natl. Acad. Sci. USA 100:5801-5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wach, A. 1996. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12:259-265. [DOI] [PubMed] [Google Scholar]
- 56.Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 57.Weber, E. R., T. Hanekamp, and P. E. Thorsness. 1996. Biochemical and functional analysis of the YME1 gene product, an ATP and zinc-dependent mitochondrial protease from S. cerevisiae. Mol. Biol. Cell 7:307-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Westermann, B., J. M. Herrmann, and W. Neupert. 2001. Analysis of mitochondrial translation products in vivo and in organello in yeast. Methods Cell Biol. 65:429-438. [DOI] [PubMed] [Google Scholar]
- 59.Yi, L., and R. E. Dalbey. 2005. Oxa1/Alb3/YidC system for insertion of membrane proteins in mitochondria, chloroplasts and bacteria (review). Mol. Membr. Biol. 22:101-111. [DOI] [PubMed] [Google Scholar]
- 60.Young, J. C., N. J. Hoogenraad, and F. U. Hartl. 2003. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell 112:41-50. [DOI] [PubMed] [Google Scholar]