Abstract
Type 7 cyclic nucleotide phosphodiesterases (PDE7s) are a newly described family of enzymes having high affinity and specificity for cAMP. However, little is known about their structure, function, or regulation. We have isolated a mouse skeletal muscle cDNA representing a new alternative splice variant (PDE7A2) of the PDE7 gene. The ORF encodes a 456-amino acid protein having a predicted molecular weight of 52.4 kDa. The 5′ end of the mouse PDE7A2 is divergent from the 5′ end of the human PDE7A1 sequence and is more hydrophobic. A comparison of the 5′ ends of the two cDNA clones with human genomic sequence indicates that they represent alternate splice products rather than species variation. RNase protection analysis of several mouse tissues indicates that PDE7 is expressed widely with highest levels in skeletal muscle. HPLC fractionation and Western blot analysis of two human lymphocyte T-cell lines shows that an unknown PDE activity described by Ichimura and Kase [Ichimura, M. & Kase, H. (1993) Biochem. Biophys. Res. Commun. 193, 985–990] is most likely to be PDE7A1. A single immunoreactive band of ≈55 kDa, which comigrates with PDE7A1, is seen in fractions of the HPLC profile containing this activity suggesting that the original human PDE7A1 clone contains a full-length ORF, and is not truncated at the 5′ end as was originally postulated. In a human lymphocyte B-cell line and also in mouse skeletal muscle, a large amount of PDE7 mRNA but little PDE7 protein or activity is expressed suggesting that the translation or stability of PDE7 protein may be highly regulated in these tissues.
Cyclic nucleotides are used as second messengers by a wide range of extracellular signals. Termination of the cyclic nucleotide signal is via hydrolysis by one or more members of the cyclic nucleotide phosphodiesterase (PDE) superfamily. There are seven identified families of mammalian PDEs, differing in their substrate specificity, allosteric regulation and sensitivity to specific pharmacological agents (1). Most families contain several genes, and most of these genes can produce multiple transcripts. The PDE7 family is the most recently identified PDE family (2). However, low levels of PDE7 activity have precluded the characterization of this activity in most tissues and cell types. The initial characterization of a human PDE7A1 cDNA clone suggested that it might be truncated at the 5′ end (2), but repeated efforts using various methods to isolate the remainder of the 5′ end have been ineffective. Since Northern blot analysis showed that skeletal muscle had the highest expression of PDE7 among several human tissues, we decided to try to isolate a full-length clone from muscle.
Little is known about what PDEs are present in skeletal muscle, although surveys of PDE activity profiles using anion exchange (3, 4) or size exclusion chromatography (5, 6, 7) showed the existence of multiple isoforms. Rolipram sensitivity of PDE activity in skeletal muscle homogenates demonstrated the existence of one or more PDE4 isozymes (8), and PDE4D expression in rat skeletal myoblasts was confirmed by Northern blot analysis (9). Strangely, although muscle appears to contain high levels of PDE7 mRNA, no reports have described a high affinity cAMP-specific PDE activity in this tissue that is also insensitive to PDE4 inhibitors, as would be expected for PDE7.
However, in several human T-cell lines but not B cells, Ichimura and Kase (10) reported an unknown PDE activity in DEAE-fractions that did have a high affinity for cAMP, did not hydrolyze cGMP at 1 μM and was insensitive to the PDE4-selective inhibitor, RO 20–1724. These characteristics were similar to those seen for PDE7 expressed in Sf9 cells. Therefore, we examined these T-cell lines to determine whether there was in fact expression of a PDE7.
In this report, we describe a mouse cDNA encoding a new PDE7 splice variant that differs from the human protein only in the predicted N-terminal region. Additionally, we show that PDE7A1 is expressed in two human T-cell lines and that PDE7 translation or stability may be highly regulated. Recently, what appears to be a homologous new PDE7A2 splice variant has also been isolated independently from a human muscle cDNA library.*
EXPERIMENTAL PROCEDURES
Materials.
Radiochemicals were purchased from DuPont/NEN, except [2,8-3H]cAMP, which was from ICN. Century RNA molecular weight markers were purchased from Ambion (Austin, TX). Nucleobond AX DNA purification columns were purchased from The Nest Group (Southport, MA). Sequenase (version 2.0) was purchased from Amersham/United States Biochemical. Nitrocellulose membranes were from Schleicher & Schull. The mouse skeletal muscle cDNA library and cloning vector pCRII were obtained from Clontech. The human genomic library was a gift from Mark Hamblin (Seattle Veterans Affairs Hospital).
Library Screening.
Approximately 1 × 106 plaques from each library were screened using standard procedures (P. Han, C. F. Fletcher, N. G. Copeland, N. A. Jenkins, and T. Michaeli, personal communication) with the complete ORF of the human PDE7 clone as a probe. Plaque-purified clones were subcloned into the vector pMOB and sequenced using the TN1000 transposon method (12). Subclones were either manually sequenced with Sequenase (version 2.0) or with the Prism PCR sequencing kit from Applied Biosystems followed by fluorescent detection on an Applied Biosystems model 373A sequencer. Computer analysis of DNA was done using version 8.0 of the Genetics Computer Group (Madison, WI) software package.
Expression and Assay of PDE7 Activity.
Baculovirus constructs for expression of PDE in Sf9 insect cells were made using the shuttle vector pVL1393. Production of recombinant virus and infection of Sf9 cells for viral amplification and protein expression were by standard protocols (13). PDE activity was measured essentially as described previously using 50 mM Tris (pH 7.5), 0.1 mg/ml BSA, 10 mM MgCl2, 100,000 cpm of [2,8-3H]cAMP, and sufficient unlabeled cAMP to produce the desired concentration (14). Protein concentrations were determined by the Bradford method (15).
Cell Line Growth and Maintenance.
Hut78, Jurkat clonal line E6-1, and H33HJ-JA1 cells (T-lymphocyte-derived cell lines) and Jiyoye and Ramos cells (B-lymphocyte-derived) were obtained from the American Type Culture Collection/National Institutes of Health Repository (Rockville, MD) and grown according to the supplier’s recommendations.
HPLC Fractionation of Cell Extracts.
Cells were resuspended in four volumes of ice cold homogenization buffer [40 mM Tris (pH 7.5)/50 μM EDTA/200 μM 4-(2-aminoethyl)benzenesulfonyl fluoride/10 μM leupeptin/1 μM pepstatin] and homogenized with 25 strokes of a Dounce homogenizer using the A pestle. The homogenate was then centrifuged at 10,000 × g for 15 min and the supernatant removed to a fresh tube.
HPLC on a Pharmacia Mono-Q anion exchange column was used to fractionate the PDE activities. Samples were filtered through a 0.2 μm pore Acrodisc (Gelman) prior to injection on the column. The flow rate was 0.5 ml/min and 0.25 ml fractions were collected at 4°C. The column buffers were 50 mM Tris (pH 7.2), 1 mM EDTA (buffer A), and 50 mM Tris (pH 7.2), 1 mM EDTA, and 500 mM NaCl (buffer B). The gradient consisted of three phases: a 2-min linear increase in buffer B of 5%/min, followed by a 3-min linear increase of 10%/min and a final 35-min phase linearly increasing to 100% buffer B.
Antisera Production and Characterization.
The C-terminal peptide of HSPDE7A1, CELNSQLLTQENRLS (the N-terminal cysteine was added for crosslinking) was coupled to maleimide activated keyhole limpet hemocyanin from Pierce. Rabbits were maintained and antiserum was produced by R & R Rabbitry (Stanwood, WA). Before use antisera were affinity purified on human PDE7A1 bound to nitrocellulose. For Western blot analysis, proteins were separated on SDS/10% PAGE gels, then transferred to nitrocellulose overnight at 25 V. Blots were developed using a horseradish peroxidase-coupled goat antirabbit IgG (Bio-Rad) as the secondary antibody, and chemiluminescent detection using luminol (Pierce) as the substrate.
RNA Analysis.
Total RNA was prepared using the one step method of Chomczynski and Sacchi (16). The RPA II kit from Ambion was utilized for all RNase protection assays, and the Maxiscript kit from Ambion was used to make riboprobes. Samples were run on 5% sequencing gels and exposed to Kodak X-Omat film. For Northern blot analysis, 10 μg poly(A)+ RNA was electrophoresed in a formaldehyde gel and transferred to Hybond N+ nylon (Amersham) overnight using 20× standard saline citrate (SSC). The membrane was probed with the ORF of cDNA HSPDE7A1 (clone TM22) and washed at high stringency.
RESULTS
Isolation of Mouse PDE7 cDNA Clones.
A mouse skeletal muscle cDNA library produced 12 independent clones when screened with a probe consisting of the entire ORF of a human PDE7A1 cDNA. Several of the larger clones were chosen for sequence analysis. PCR amplification revealed that clones S15C (0.8 kb), S17C (0.8 kb), and S20D (2.7 kb) had the longest 5′ ends. None of these clones contained all of the ORF. Clone S1A (3 kb) was missing the 5′ end but did contain all of the 3′ end of the ORF, as well as a long untranslated region. To complete the sequencing of this murine PDE7, clone S1A was used in combination with clones S15C and S17C. The sequence of a consensus clone (P2A) constructed from these overlapping clones was determined and has been deposited in GenBank with the locus name of MMPDE7A2. The structure of this consensus clone was confirmed by sequence of an independent cDNA clone of 2.3 kb spanning all of the junctions.
Comparison of the Mouse Clone P2A and the Human Clone TM22.
DNA sequence analysis of the mouse P2A and the human TM22 clones show that they are completely different at the 5′ end of the coding sequence but are >90% identical downstream of HSPDE7A1 nucleotide 181. This high homology includes much of the 3′ untranslated region. The 5′ divergence was confirmed in four independent skeletal muscle cDNA clones, and by 5′ race cloning (data not shown).
To show that the differences between the 5′ ends of the mouse and human clones was not due to species variation, the region of divergence was examined at the genomic level using a human genomic DNA clone. A human genomic library was screened using the entire ORF of cDNA clone TM22 as a probe. Five clones were plaque purified and characterized by Southern blot analysis. The clones were digested and probed with different fragments of TM22 to determine which portions of the cDNA were present within each genomic clone. A 1.6-kb NotI fragment from genomic clone 1.2 contains the region representing the human cDNA 5′ end. Sequence analysis of this fragment revealed that the genomic sequence matched that of the cDNA until cDNA nucleotide 187 (Fig. 1). At this point the genomic clone contains the consensus sequence for the 5′ end of an intron and its splice donor. The presence of an intron–exon boundary at this site of divergence between the human and mouse cDNA clones strongly suggests that the two are splice variants.
Figure 1.
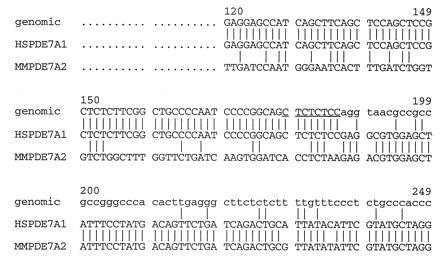
Alignment of PDE7 cDNA and genomic sequence. Alignment of the 5′ ends of human cDNA clone TM22, the mouse cDNA clone P2A and the human genomic clone 1.2 was done using the Genteics Computer Group program pileup. Uppercase letters indicate exon sequences, lowercase letters indicate intron sequence. The consensus sequence for a 5′ splice donor is underlined. The numbers correspond to clone TM22.
Fig. 2A shows an alignment of the N-terminal regions of PDE7A1 and 7A2. The sequence divergence occurs upstream of Arg-45 of HSPDE7A1. Interestingly, the same 6-amino acid motif, RRGAIS, also occurs again at positions 21–27 of the HSPDE7A1 sequence and perhaps serves some regulatory function. Fig. 2B shows a Kyte–Doolittle comparison (17) of the hydrophilicity of the murine and human PDE7A predicted N-terminal sequences. The N terminus predicted for the human HSPDE7A1 protein is longer and significantly more hydrophilic than that of the mouse PDE7A2 protein.
Figure 2.
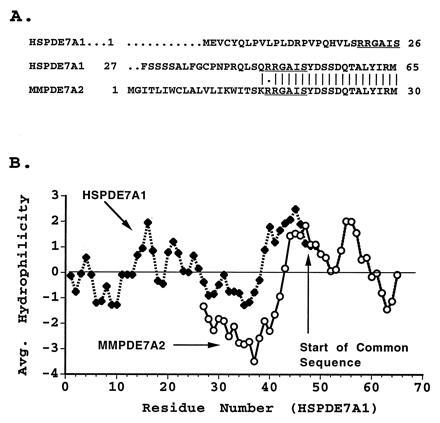
Comparison of PDE7 N termini. (A) Sequence alignment of the amino terminal region of HSPDE7A1 and MMPDE7A2. The repeated sequence, RRGAIS, is underlined. (B) The hydrophilicity of the predicted N termini of the human and mouse cDNA clones was compared using hydrophilicity index of Kyte and Doolittle (18) incorporated into the Genetics Computer Group program peptidestructure. The hydrophilicity value was averaged over a window of 7. The comparison was limited to the area ending at human PDE7A1 residue 65 and mouse PDE7A2 residue 30. Divergence in sequence occurs after residue 44 of HSPDE7A1.
Expression of Skeletal Muscle Clone P2A.
The ORF of clone P2A runs from the ATG at position 78 to the stop codon at position 1445 giving a predicted polypeptide of 456 amino acids having a molecular mass of 52,441. The enzyme expressed in Sf9 cells shows a single band of ≈52 kDa when analyzed by Western blot. In addition, a human variant of the PDE7A2, which has a slightly longer 5′ end contains an in-frame stop codon upstream of the same ATG.* The kinetic parameters of PDE7A2 activity expressed in Sf9 cells were very similar to those seen for the expressed human PDE7A1 cDNA clone (2). In brief, the activity was selective for cAMP as a substrate, with a Km for cAMP of ≈0.2 μM, and was insensitive to selective concentrations of isozyme-specific inhibitors of the PDE2, PDE3, and PDE4 families. Additionally, the Sf9-expressed mouse PDE7A2 was unaffected by inclusion of imidazole, EGTA, or calcium and calmodulin, known effectors of PDE1 (data not shown).
Localization of PDE7 in Mouse Tissues.
Ribonuclease protection analysis was used as a sensitive method to determine the distribution of PDE7 in mouse tissues. Using a probe consisting of the 3′ end from the catalytic domain of mouse PDE7A2, a protected band of the appropriate size was detected in all tissues examined (Fig. 3). The highest signal is seen in skeletal muscle, followed by spleen. Lower and roughly equivalent signals are seen in all other tissues surveyed, with the exception of testis. The signal in testis is barely detectable with a 10-day exposure, indicating an extremely low level of expression.
Figure 3.
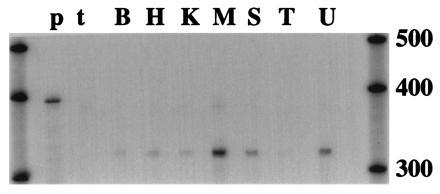
Expression of PDE7 in mouse tissues. RNase protection assays using 20 μg total RNA from the indicated tissues were performed with a riboprobe from the catalytic domain. The protected fragments were separated on a 5% sequencing gel and dried onto filter paper for exposure to film for 10 days at −70°C. The film was then scanned at 300 dpi and adjusted for brightness in photoshop 3.0. Molecular weight markers are RNA transcripts from Ambion Century kit. B, brain; H, heart; K, kidney; L, lung; M, skeletal muscle; S, spleen; T, testis; U, uterus; p, undigested probe; t, tRNA.
PDE7 in Human T-Cell Lines.
While surveying the PDE activity profile of several lymphocyte T-cell and B-cell derived lines, Ichimura and Kase (10) observed that the T-cell lines they examined contained a peak of PDE activity absent from B-cell lines. As the kinetic properties of the unknown PDE activity they described were very similar to those seen using either of the PDE7 expression constructs, we examined several T-cell lines for the expression of PDE7 using both immunochemical and molecular approaches.
The human T-cell line, Hut78, and Sf9 cells expressing PDE7A1 were analyzed by Western blot analysis with affinity purified antiserum 6858 (Fig. 4). The sera used for these studies was made to the C-terminal region of the human PDE7A1 polypeptide. For either homogenate, a single immunoreactive band was seen at ≈55 kDa (Fig. 4A, lanes 1 and 2). Addition of peptide at 20 μg/ml blocked the band (Fig. 4B, lanes 1 and 2), and antiserum 6858 purified against cell extracts from noninfected Sf9 cells showed no such band (data not shown). When the homogenate of the Hut78 cells was separated into soluble and particulate components both fractions showed a single strong band (Fig. 4C, lanes 1 and 2).
Figure 4.
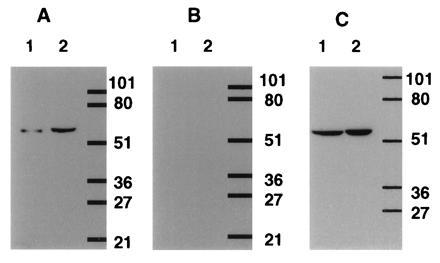
Western blot analysis of PDE7 in Sf9-cell and T-cell extracts. Antipeptide antiserum, 6858, was produced and purified as described in the text. A and B show a blot of homogenates of Hut78 T-cells (lane 1) and Sf9-expressed PDE7A1 (human cDNA TM22; lane 2). A shows a single immunoreactive band present in each homogenate, while B shows the effect of added peptide at 20 μg/ml. C shows the presence of an immunoreactive band in both the 10,000 × g supernatant (lane 1) and pellet (lane 2) of Hut78 cells. Scanning and printing were as in Fig. 3.
Soluble extracts from several human T-cell and B-cell lines were fractionated by Mono-Q HPLC anion exchange chromatography. Assay of the column profile of the Hut78 T-cell line revealed the presence of two peaks of PDE activity when using 0.1 μM cAMP as substrate (Fig. 5A). Both peaks were specific for cAMP, showing no activity with 1 μM cGMP. Neither peak was affected by the addition of EGTA, calcium/calmodulin, cGMP or the PDE3-specific inhibitor, enoximone. The second peak was inhibited by RO 20–1724 at concentrations known to be selective for PDE4 and the back portion of the first peak also showed some inhibition by RO 20–1724, suggesting the presence of an unresolved PDE4 in this region. The first peak was insensitive to all other selective inhibitors tested.
Figure 5.
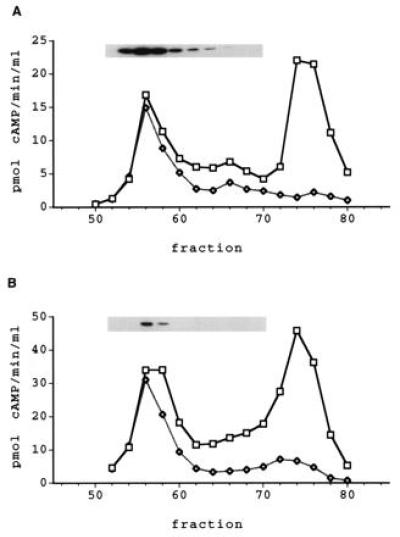
PDE7 Activity in Hut78 and H33HJ-JA1 T-cells. Soluble extract (20 mg) was fractionated using Mono-Q HPLC, as described in the text. (A) The elution profile for Hut78 cells and (B) the profile for H33HJ-JA1 cells. Squares represent total PDE activity, and diamonds indicate PDE activity insensitive to 10 μM RO 20–1724. Samples corresponding to the peaks of PDE activity were Western blotted using antiserum 6858. The insets show the immunoreactive bands from the blots overlying their respective fractions.
Western blot analysis of the Mono-Q HPLC profile of Hut78 soluble extract showed the presence of a band of ≈55 kDa in the fractions corresponding to the first peak of PDE activity (Fig. 5A). The intensity of the immunoreactive band in each fraction was proportional to the PDE activity as measured with 0.1 μM cAMP as substrate. However, fractions on the back part of the peak had a slightly lower proportional signal than fractions from the front part. This is consistent with the observation that the back side of the peak was partially inhibited by RO 20–1724 and probably contains some PDE4 activity. The immunoreactive bands were competitively blocked by the addition of immunizing peptide (data not shown).
In initial studies, the size of the immunoreactive band from Hut78 cell extracts and the Mono-Q peak appeared to be approximately the same as that seen with Sf9-expressed PDE7A1 (Fig. 4A). To confirm this, samples of Sf9-expressed TM22 and Hut78 cell extract were Western blotted side-by-side and also combined in the same sample. A single band was noted (data not shown) indicating that the native size of PDE7 in T-cells is the same as that predicted by the human cDNA clone for PDE7A1. Reverse transcription–PCR using primers based on the 5′ end of the human cDNA also showed the presence of PDE7A1 in Hut78 total RNA (data not shown).
Another T-cell line, H33HJ-JA1, also was fractionated by Mono-Q HPLC (Fig. 5B). The profile from the soluble extract of these cells also had two peaks of low Km, cAMP-specific PDE activity, eluting at the same position as the peaks from the Hut78 cell extract. As with the Hut78 peaks, the first peak was insensitive to all selective PDE inhibitors tested, as would be expected for PDE7. The second peak was inhibited by 10 μM RO 20–1724, indicating that it represents one or more PDE4s. The H33HJ-JA1 also showed and immunoreactive band on Western blot analysis that corresponded with PDE7-like activity, although the intensity of the signal was substantially lower compared with the Hut78 cells. This band also could be competed away by the peptide used to generate the antiserum.
Two B-cell lines (Jiyoye and Ramos cells) were also examined for the presence of PDE7 activity and immunoreactivity. Ichimura and Kase (10) showed that several B-cell lines contained only one peak of cAMP PDE activity after DEAE chromatography. This peak contained PDE4, based on RO 20–1724 sensitivity. In our hands, Mono-Q separation of soluble extracts from the B-cell lines Jiyoye and Ramos also showed a single broad peak of PDE activity. This peak was almost completely inhibited by 10 μM RO 20–1724. Importantly, Western blot analysis of the fractions across the peak with antiserum 6858 detected no immunoreactive bands under conditions where a band was easily seen in the HUT-78 and H33HJ-JA1 cells (data not shown).
Northern Blot Analysis of PDE7 mRNA in T-Cell and B-Cell Lines.
To determine whether PDE7 mRNA might be expressed in both lymphocyte cell types, several T cell and B cell lines were examined by Northern blot analysis. Messenger RNA was prepared from Hut78, Jurkat, and H33HJ-JA1 T-cells, and from Jiyoye B-cells. In an effort to increase the mRNA and/or activity level of PDE7, the cells were incubated for six hours with 2 μg/ml phytohemagglutinin, an activator of lymphocyte growth. This incubation was sufficient to produce a morphological change consistent with lymphocyte activation, specifically a strong clumping of the cells. Fig. 6 shows the presence of a single band of 3.8 kb in all lanes, including the B-cell lane. This band is the same size as previously shown for PDE7A1 mRNA in human astrocytoma cells (2). The T-cell lines Hut78 and H33HJ-JA1 have the highest level of expression, followed by the Jurkat T-cell and Jiyoye B-cell lines. No increase of mRNA signal was seen after phytohemagglutinin treatment of any of the cell lines and if anything a small decrease was noted (Fig. 6). There was also no change in PDE activity.
Figure 6.
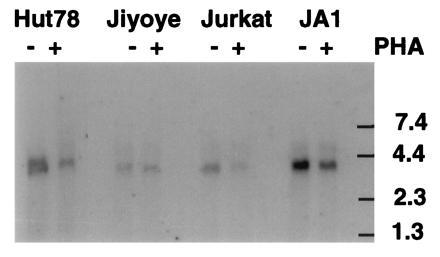
Northern blot analysis of PDE7 in lymphocyte cell lines. Poly(A)+ RNA (10 μg) from the indicated cell lines was probed as described in the text. Cells were incubated in the presence (+) or absence (−) of 2 μg/ml phytohemagglutinin for 6 hr prior to RNA isolation. This blot was exposed to film for 3 days. After probing for PDE7, the blot was stripped and a probe directed against G3PDH was used to ensure that equivalent amounts of RNA were loaded in each lane (not shown). Jiyoye is a B-cell line and the others are T-cell lines. Scanning and printing were as in Fig. 3.
DISCUSSION
A New Variant of the PDE7A Gene Has Been Isolated from a Mouse Skeletal Muscle cDNA Library.
This cDNA encodes a protein that is 90% identical to that encoded by the human clone, with the major difference corresponding to an altered N terminus of 48 amino acids. Within the domain common to both proteins, the predicted identity is 96%. The similarity of the two clones extends into the 3′ untranslated region, where there is >90% identity. A similar high degree of relatedness in untranslated regions across species also occurs with several other members of the mammalian PDE superfamily. However, it is not a universal feature of PDE genes nor does it occur with many other gene products. This high degree of 3′ sequence conservation suggests a likely regulatory role for these untranslated sequences of PDE7.
Comparison of the human PDE7A1 cDNA with corresponding human genomic sequence shows that the area of divergence between the PDE7A1 and the new PDE7A2 cDNA clones coincides with a consensus 5′ splice donor sequence in the genomic sequence. It therefore seems unlikely that the alternate 5′ end of the mouse clones is a cloning artifact. Four independent clones containing the 5′ end were isolated from the mouse cDNA library, and three independent clones with the same 5′ end were PCR amplified from mouse leg muscle cDNA. If the alternate 5′ end actually represented an unprocessed intron, it would be unusual for all of the clones isolated to have been generated from unprocessed mRNA.*
The mouse PDE7A2 cDNA was used to create a recombinant baculovirus for expression studies. PDE activity expressed in extracts of Sf9 cells infected with this recombinant virus has biochemical and pharmacological characteristics nearly identical to those seen with the human clone expressed in yeast. Both are specific for cAMP as a substrate, with a Km below 1 μM. When used at concentrations less than 50 μM, inhibitors selective for other PDE families (in this case EHNA, enoximone, RO 20–1724, and zaprinast) had little effect on the PDE activity expressed by either the human HSPDE7A1 clone (TM22) or the mouse MMPDE7A2 clone (P2A) in Sf9 cells as described (2) for the HSPDE7A1 enzyme expressed in yeast (data not shown).
Ichimura and Kase (10) demonstrated the existence of a novel PDE activity in several T-cell lines. They suggested based on kinetic criteria that this PDE might correspond to a PDE7, and that it was possibly specific to T-cells, as the B-cell lines they examined lacked the activity. The results shown here partially support this hypothesis. Mono-Q chromatography of Hut78 T-cell soluble extracts resolved two peaks of low Km, cAMP-specific PDE activity. The first of these was insensitive to the PDE4-specific inhibitor, RO 20–1724 and the PDE3-specific inhibitor, enoximone. This first peak coincided with a single immunoreactive band when Western blotted using affinity-purified antisera generated against a peptide predicted by a conserved region of the human PDE7A cDNA. Additionally, both reverse transcription–PCR and Northern blot analysis of total and poly(A)+ RNA, respectively, showed PDE7A mRNA in Hut78 cells.
The apparent molecular weight of the immunoreactive band seen in Hut78 cell extracts, and in the first peak of PDE activity in both Hut78 and H33HJ-JA1 T-cells, is the same as that predicted by the human cDNA clone strongly suggesting that it is PDE7A1. Interestingly, the original sequence analysis of the cDNA clone TM22 and comparison of the size of the clone with the size of a signal seen with Northern blots suggested that the TM22 PDE7A1 cDNA might be truncated on the 5′ end (2). However, the Western blot data seen here strongly suggest the human TM22 clone contains the entire coding region.
Large relative differences in the levels of PDE7 mRNA, PDE7 protein and PDE7 activity were noted between the T-cell and B-cell lines implying that regulation of PDE7 expression and possibly also activity may occur in these cells. For example, Northern blot analysis of PDE7 shows an mRNA signal in B-cell lines that is comparable in intensity to the signal in several of the T-cell lines, even though no PDE7 protein or activity can be detected in these extracts or in Mono-Q HPLC fractions from these cells. Similarly, a strong Northern signal can be seen in skeletal muscle but little or no PDE activity or protein having properties characteristic of PDE7 was seen. This suggests that in tissues having these properties (i.e., high mRNA and low protein and activity) either much of the mRNA is not translated or that much of the translated protein is inactive or unstable.
The presence of PDE7 activity, protein, and mRNA in the T-cell lines raises questions about what role this PDE might play in the immune response. Possible physiological role(s) of cAMP and PDEs in T-cells have been extensively examined. Elevated cAMP typically has a negative effect on the rate of cell growth, secretion of the stimulatory cytokines interleukin 1 and 2, mitogen-stimulated generation of arachidonic acid, and expression levels of various genes (reviewed in refs. 18 and 19). One aspect of current interest related to PDE function in lymphocytes is the determination of which PDEs are present in particular T-cell types, and what role(s) these PDEs may play in specific immune responses mediated by these cells. Since elevated cAMP has an immunosuppressive action, increasing cAMP levels by selective PDE inhibition appears to be a potentially promising route for blocking specific immune responses and perhaps eventually improved clinical treatment of a variety of disease states. The current lack of a specific inhibitor for PDE7 will hamper studies to answer these questions. Finally, the observation that T-cell lines and B-cell lines are able to express similar levels of PDE7 mRNA, but vastly different levels of PDE activity and immunoreactive protein suggest that some cellular regulation of this PDE exists in lymphocytes. What effect PDE7 regulation may have on T- or B-cell function is a question requiring further study.
Acknowledgments
The research presented in this manuscript was supported in part by National Institutes of Health Grant DK21723.
Footnotes
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. §1734 solely to indicate this fact.
Abbreviation: PDE, cyclic nucleotide phosphodiesterase.
Data deposition: The sequence reported in this paper has been deposited in the GenBank data base (accession no. U68171U68171).
Michaeli and colleagues reported recently at the Third International Conference on Cyclic Nucleotide Phosphodiesterases in Glasgow, Scotland on a cDNA from human muscle that appears to encode the same splice variant of PDE7A2 as reported in this manuscript. This cDNA differs from the one described here in that it has a somewhat longer 5′-untranslated sequence that includes an in frame stop codon (personal communication).
References
- 1.Beavo J A. Physiol Rev. 1995;75:725–748. doi: 10.1152/physrev.1995.75.4.725. [DOI] [PubMed] [Google Scholar]
- 2.Michaeli T, Bloom T J, Martins T, Loughney K, Ferguson K, Riggs M, Rodgers L, Beavo J A, Wigler M. J Biol Chem. 1993;268:12925–12932. [PubMed] [Google Scholar]
- 3.Kemp R G, Huang Y C. Methods Enzymol. 1974;38:240–244. doi: 10.1016/0076-6879(74)38037-8. [DOI] [PubMed] [Google Scholar]
- 4.Hidaka H, Yamaki T, Ochiai Y, Asano T, Yamabe H. Biochim Biophys Acta. 1977;484:398–407. doi: 10.1016/0005-2744(77)90095-x. [DOI] [PubMed] [Google Scholar]
- 5.Ball E H, Seth P K, Sanwal B D. J Biol Chem. 1980;255:2962–2968. [PubMed] [Google Scholar]
- 6.Seth P K, Rogers J, Narindrasorasak S, Sanwal B D. J Cell Physiol. 1983;116:336–344. doi: 10.1002/jcp.1041160311. [DOI] [PubMed] [Google Scholar]
- 7.Narindrasorasak S, Tan L U, Seth P K, Sanwal B D. J Biol Chem. 1982;257:4618–4626. [PubMed] [Google Scholar]
- 8.Nicholson C D, Wilke R. J Auton Pharmacol. 1989;9:159–165. doi: 10.1111/j.1474-8673.1989.tb00207.x. [DOI] [PubMed] [Google Scholar]
- 9.Kovala T, Lorimer I A, Brickenden A M, Ball E H, Sanwal B D. J Biol Chem. 1994;269:8680–8685. [PubMed] [Google Scholar]
- 10.Ichimura M, Kase H. Biochem Biophys Res Commun. 1993;193:985–990. doi: 10.1006/bbrc.1993.1722. [DOI] [PubMed] [Google Scholar]
- 11.Ausbel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. London: Wiley; 1995. [Google Scholar]
- 12.Strathmann M, Hamilton B A, Mayeda C A, Simon M I, Meyerowitz E M, Palazzolo M J. Proc Natl Acad Sci USA. 1991;88:1247–1250. doi: 10.1073/pnas.88.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Reilly D R, Miller L K, Luckow V A. Baculovirus Expression Vectors: A Laboratory Manual. New York: Freeman; 1992. [Google Scholar]
- 14.Martins T J, Mumby M C, Beavo J A. J Biol Chem. 1982;257:1973–1979. [PubMed] [Google Scholar]
- 15.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 16.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 17.Kyte J, Doolittle R F. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 18.Kammer G M. Immunol Today. 1988;9:222–229. doi: 10.1016/0167-5699(88)91220-0. [DOI] [PubMed] [Google Scholar]
- 19.Kuehl F A, Jr, Zanetti M E, Soderman D D, Miller D K, Ham E A. Am Rev Respir Dis. 1987;136:210–213. doi: 10.1164/ajrccm/136.1.210. [DOI] [PubMed] [Google Scholar]


