Abstract
Chromatin domain boundary elements demarcate independently regulated domains of eukaryotic genomes. While a few such boundary sequences have been studied in detail, only a small number of proteins that interact with them have been identified. One such protein is the boundary element-associated factor (BEAF), which binds to the scs′ boundary element of Drosophila melanogaster. It is not clear, however, how boundary elements function. In this report we show that BEAF is associated with the nuclear matrix and map the domain required for matrix association to the middle region of the protein. This region contains a predicted coiled-coil domain with several potential sites for posttranslational modification. We demonstrate that the DNA sequences that bind to BEAF in vivo are also associated with the nuclear matrix and colocalize with BEAF. These results suggest that boundary elements may function by tethering chromatin to nuclear architectural components and thereby provide a structural basis for compartmentalization of the genome into functionally independent domains.
Genomes of higher eukaryotes are organized into distinct functional domains that are defined by boundary elements. These boundaries restrict the action of regulatory elements to the appropriate genomic targets (13, 30). Boundaries are also postulated to act as barriers against the spread of inactive heterochromatin into euchromatic regions. Sequences that act as boundaries have been identified and studied in organisms from yeasts to mammals (2, 13, 39). In vertebrates, the most extensively studied boundaries are those of the β-globin cluster (36) and the chicken lysozyme gene (4). In Drosophila melanogaster, several boundaries have been characterized, including those from the regulatory regions of the homeotic gene complexes, the gypsy retrotransposon element, and the 87A7 heat shock locus (14). The fission yeast boundary elements have been isolated from the regions flanking the silent mating loci (34), and these elements separate euchromatin from heterochromatin. In budding yeast, boundary activity has been characterized in regions bordering telomeres, the ribosomal DNA locus, and the silent mating-type locus (10, 35). All these elements have been characterized by their ability to either insulate a reporter gene from chromosomal position effects or prevent the action of an enhancer on an adjacent promoter. No significant sequence similarity has been detected among the known boundaries, suggesting that there are many sequences that function as boundary elements. However, it has recently been shown that CTCF, a vertebrate boundary element binding protein, is conserved in Drosophila and is one of the components of the well-characterized boundary sequence Fab-8 (32). This finding raises the possibility that different nucleoprotein complexes at the boundary elements may participate to create distinct domains.
The importance of boundary elements in transcriptional regulation and imprinting during development is well documented (2, 28, 30, 39). However, the physical and molecular basis of these boundaries is still unclear. Two models have been proposed to explain their action. In one model, the boundary elements, along with their binding proteins, are suggested to form a physical barrier for the interaction of regulatory elements of neighboring domains. The second model posits that the interaction of boundary elements with one another and with a nuclear substructure leads to tethering of the ends, creating a topologically independent looped domain. Thus, the functional boundary that separates independent transcriptional units is thought to coincide with the physical boundary of the loops that are generated when chromatin is folded into higher-order structures for compaction within the nuclear space (14, 30, 39).
scs and scs′ (for specialized chromatin structures as characterized by DNaseI mapping) were among the first boundary sequences to be identified. These elements bracket the heat shock puff at the 87A7 locus of Drosophila and function not only to block enhancer action but also to insulate from position effect (22, 23). Proteins that bind to specific sequences within scs and scs′ fragments have been identified. The CGATA motifs in scs′ are bound by boundary element-associated factor (BEAF), a complex of two proteins, BEAF-32A and BEAF-32B. These two proteins are derived from the same gene and differ only in the N-terminal 80 amino acids (aa). In addition to scs′, BEAF-32A and BEAF-32B bind to numerous other interbands on the polytene chromosome. Thus, there are several targets for BEAF-32A and BEAF-32B, some of which have been identified and shown to possess boundary activity (9). Similarly, the ZW5 protein binds to specific sequences in the scs boundary element and ZW5 mutants show compromised scs-dependent boundary activity in transgenic assays (11). BEAF and ZW5 interact with each other in vitro and in vivo (3). Further, the scs and scs′ boundaries, although physically separated by 15 kb, interact with each other in vivo, suggesting that the intervening chromatin is looped out into a distinct domain.
Structural and biochemical studies suggest a highly organized arrangement of chromosomes within the nucleus (7). The limited movement of chromosomes within the nuclear space, the existence of chromosome-based subnuclear compartments such as the nucleolus, and peripheral localization of heterochromatin suggest the existence of tethering sites for chromosomes (27). The internal network of nuclear lamina that is a component of nuclear matrix along with other proteins and RNA that forms the internal architecture of the nucleus are some of the possible tethering sites. Several proteins such as the lamins, nucleolar proteins, and topoisomerase II have been localized to the matrix. The procedures used for the isolation of nuclear matrix involve DNase I and RNase A digestions to remove the nucleic acids. Further, treatment under very-high-salt conditions removes any loosely associated proteins and allows the retention of only very tightly associated proteins, along with bound nucleic acids that are protected from nuclease action. The structure that remains after these treatments has been operationally defined as the nuclear matrix. Although the term is descriptive, some electron microscopic data do show the nuclear matrix to be composed of fibrillar structures. However, the exact nature of this structure remains unresolved and its molecular composition is not known. Many nuclear activities such as transcription, DNA repair (1), etc., are known to occur within discrete nuclear spaces, and these compartments may be assembled transiently. Indeed, nuclear compartmentalization may also play a role in boundary function, by bringing together regions of similar activity (12, 25).
Recently, several groups have reported links between the operationally defined nuclear matrix fraction and chromatin elements that confer boundary or insulator activity (5, 19, 33, 40). In order to investigate this relationship more directly, we have attempted to molecularly characterize the nuclear matrix. Using Drosophila embryos as the primary source, we have isolated proteins of the nuclear matrix and identified them through matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) tandem mass spectrometry (MS-MS) (unpublished data). During this analysis we identified BEAF-32 as one of the components of the nuclear matrix. Subsequent analysis showed that 25% of the total BEAF protein pool is associated with the nuclear matrix. We also mapped the domain required for matrix association. The middle region of BEAF contains a predicted coiled-coil domain and several potential sites for posttranslational modifications, including O glycosylation, and is critical for its association with the nuclear matrix. Boundary sequences that are in vivo targets of BEAF-32 are also enriched in the nuclear matrix. Taken together these results provide a structural basis for the mechanism of boundary element function in vivo.
MATERIALS AND METHODS
Plasmids.
The plasmid pFPc19 was obtained from R. E. Kingston. It is a pCaSpeR4-based vector, carrying an 845-bp Pc promoter fragment, a PCR-generated sequence encoding the Flag tag (MDYKDDDK), and a cDNA encoding aa 1 to 390 of Pc. The Pc coding fragment was replaced with the PCR-amplified fragments encoding the full-length BEAF or its different domains (N terminus, middle region, C terminus, coiled coil plus C terminus, middle region without coiled coil, and middle region 2). Expression of these constructs was checked by Western blot assays using anti-Flag antibody from Sigma-Aldrich.
Embryo collection and nuclear matrix preparation.
Embryos (0 to 16 h old) were obtained from a laboratory population of D. melanogaster (Canton-S) maintained at 25°C. Embryos were washed extensively with water, dechorionated by being stirred in 50% Clorox for 1 min, and then washed again with several volumes of distilled water to remove any traces of Clorox. Nuclei were isolated, and nuclear matrix was prepared according to published procedures (6, 26, 38) with a few modifications as described below. Dechorionated embryos were homogenized into a 10% homogenate in buffer A plus 0.25 M sucrose on ice (buffer A is 15 mM Tris [pH 7.4], 40 mM KCl, 1 mM EDTA, 0.1 mM EGTA, 0.1 mM phenylmethylsulfonyl fluoride [PMSF]). The homogenate was filtered through Mira cloth (Calbiochem), and the filtrate was centrifuged for 2 min at 600 × g. The supernatant was centrifuged at 3,000 × g for 10 min to pellet the crude nuclei. The crude nuclear pellet was suspended in buffer A plus 1 M sucrose and centrifuged at 6,000 × g for 10 min to obtain a pure nuclear pellet, which was washed three times in buffer A plus 0.25 M sucrose. At this step, an aliquot of nuclei was lysed in 0.5% sodium dodecyl sulfate (SDS) and its absorbance was measured at 260 nm. Nuclei were suspended at a concentration of 10 OD260 (optical density at 260 nm) units/ml.
For the nuclear matrix preparation, nuclei were stabilized by incubation at 37°C for 20 min. Extraction was carried out at a concentration of 10 OD260 units/ml in a buffer containing 0.4 M NaCl, 5 mM HEPES (pH 7.5), 2 mM KCl, 2 mM EDTA, 0.5% Triton X-100, and 0.1 mM PMSF. After 5 min at room temperature, the nuclei were further diluted to a concentration of 1 OD260 unit/ml and the NaCl concentration was increased to 2 M. Extraction was carried out for another 5 min at room temperature with gentle shaking. The resulting nuclear halos were centrifuged to obtain a gelatinous pellet that was washed three times in DNase I digestion buffer (20 mM Tris [pH 7.4], 20 mM KCl, 70 mM NaCl, 10 mM MgCl2, 0.5% Triton X-100, and 0.1 mM PMSF). DNase I digestion was carried out in the same buffer with 200 μg/ml DNase I (Sigma) at 4°C for 1 h. The resulting nuclear matrix preparations were used for Western blotting, two-dimensional (2D) gel electrophoresis, or extracting out matrix-associated region (MAR) DNA for quantitative PCR. For quantitative PCR experiments, the DNase I extract and the salt extracts were also saved for extraction of DNA.
The procedure described above extracts most of the soluble proteins. The sequential increase in salt as opposed to direct exposure to high salt levels prevents the association of proteins nonspecifically with the chromatin or nuclear matrix. Digestion with DNase I was carried out after the salt extraction. We found that this method, where a graded salt extraction precedes DNase I digestion, gives a reproducible protein profile. We considered retention of 0.92% (±0.11%) DNA, 45% (±3%) RNA, and 10% (±3%) proteins in the nuclear matrix preparations compared to the total nuclear DNA, RNA, and protein, respectively, as quality control parameters. Also, on silver-stained SDS gels, matrix preparations meeting these quality control standards showed enrichment of proteins of >40 kDa in size, while smaller proteins were not retained and histones, in particular, were absent.
Nuclear matrix preparation from S2 cells.
Nuclear matrix from S2 cells was prepared according to a published protocol (17). Cells were washed in phosphate-buffered saline and extracted in cytoskeleton buffer (10 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], pH 6.8, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA, 1 mM PMSF, and 0.5% Triton X-100). After 5 min on ice, the cytoskeleton frameworks were separated from soluble protein by centrifugation at 600 × g for 3 min. Chromatin was removed with 200 μg/ml DNase I at 4°C for 1 h in digestion buffer (same as that for embryos). Further salt extractions were also carried out as described above for embryos. The resulting nuclear matrix preparation was used for Western blotting.
For immunofluorescence experiments, nuclear matrix was prepared in situ on slides as described previously (5). Cells were centrifuged onto slides and then extracted with 2 M NaCl and treated with DNase I. These slides were then processed for immunofluorescence or immunofluorescent in situ hybridization (immuno-FISH). Briefly, cells were fixed with 4% paraformaldehyde for 15 min, washed extensively, blocked with bovine serum albumin, and then incubated with primary antibody at 4°C overnight, followed by secondary antibody. 4′,6′-Diamidino-2-phenylindole (DAPI) was routinely added to the mounting medium for visualization of DNA. For immuno-FISH, the antibody-treated cells were further fixed with 4% formaldehyde and then hybridized to probes synthesized using digoxigenin-dUTP (Roche) and detected by fluorescently labeled antidigoxigenin antibodies.
The matrix preparations labeled for BE28 and BEAF were examined with a Zeiss LSM510 META confocal microscope equipped with argon (488 nm)/HeNe (543 nm) lasers and a 100× numerical-aperture-1.4 PlanApo objective. Serial optical sections were collected in steps of 0.3 μm in the multitrack mode to eliminate emission cross talk between the fluorescein isothiocyanate and Cy3 channels. Black level, gain, and laser intensity were set to ensure that there was no background signal compared to secondary antibody control. The pinhole aperture used was 1 aperture unit. Two or three optical sections were projected and checked for colocalization in the Zeiss software LSM-FCS. The image region of interest was drawn around the regions of probe hybridization, and using the cross-hair function, the weighted colocalization coefficient was calculated. This calculation yields the sum of the intensities of the colocalized pixels in the two channels compared to the overall sum of the pixel intensities above threshold in each channel.
Phosphatase treatment of the nuclear matrix proteins.
Ten OD260 units of nuclear matrix was used for each treatment. Matrix preparations were suspended in 100 μl of 1× protein phosphatase 1 (PP1) buffer or 1× T-cell protein tyrosine phosphatase (TC-PTP) buffer (New England Biolabs) with or without enzyme and incubated at 25°C for 1 h. For PP1 inhibitor treatment, the matrix preparations were preincubated for 10 min with the inhibitor protein phosphatase inhibitor 2 (I-2), prior to the addition of PP1, as recommended by the manufacturer's protocol. After the incubation, the matrix was collected by centrifugation and the respective supernatants were dried in a Speed-Vac. The treated matrix preparations and the dried supernatants were reconstituted in 1× Laemmli buffer, run on an SDS-polyacrylamide gel, and analyzed by Western blotting.
Isolation of glycoproteins from nuclear extracts.
Nuclear extracts were loaded onto the affinity column containing concanavalin A and wheat germ agglutinin (WGA) with use of the Qproteome total glycoprotein kit (QIAGEN) to isolate glycoproteins. Eluted fraction and flowthrough fractions were stored in SDS gel loading buffer. Volume equivalents of nuclear extract, eluted fraction, and flowthrough were used for Western blotting with BEAF and HP1 antibodies. Under conditions when the nuclear extract was limiting, we observed all BEAF being bound to the column and no BEAF in the flowthrough. To test the glycosylation state of BEAF in the nuclear matrix, the matrix preparation was dissolved in 6 M guanidinium chloride-containing buffer and was dialyzed against PBS overnight. The soluble components that contain most of the BEAF were loaded on the WGA affinity beads, and bound proteins were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) Western blotting.
Gel electrophoresis and Western blotting.
Nuclear matrix proteins were analyzed by standard one-dimensional or 2D SDS-PAGE, transferred to an Immobilon membrane, and probed with antibodies to BEAF and ZW5 (gifts from Paul Schedl and Uli Laemmli), histone H3 and UBX (Santa Cruz Biotechnologies), ABD-B (a gift from Susan Celnicker), Flag (Sigma), and Batman (raised in our laboratory). The blots were developed using the chemiluminescence kit from NEN Life Sciences Products per the manufacturer's protocol.
2D gel electrophoresis and identification of protein spots.
2D gel electrophoresis was carried out using the Protean II electrophoresis setup (Bio-Rad). One hundred micrograms of nuclear matrix protein was loaded on the first-dimension gel. The second dimension was run on a 10% SDS-polyacrylamide slab gel. Gels were stained with Coomassie brilliant blue R-250. In-gel digestion of the excised spots was carried out with trypsin (sequencing grade; Bio-Rad), and the peptides were extracted in 5 μl of 50% acetonitrile-0.1% trifluoroacetic acid. An 0.8-μl amount of the extracted peptides and 0.8 μl of the matrix (α-cyanohydroxycinnamic acid) were mixed and spotted on the target plate. MS fingerprint analysis was performed on a PerSeptive Biosystems Voyager STR MALDI-TOF instrument. Each spectrum was processed using Data Explorer (Applied Biosystems). Postacquisition calibration was performed using a standard calibration mix supplied by Applied Biosystems. Protein identification was done using MASCOT online software (Matrix Science).
Quantitative real-time PCR analysis of matrix attachment.
For the in vivo MAR assay, DNA was prepared by two different methods. The first method uses DNase I for cleaving the chromatin, and the second method uses restriction enzymes for the purpose. DNA was extracted out of the nuclei, the nuclear matrix, and the different fractions by proteinase K digestion and phenol-chloroform extraction. The target DNA sequences were quantified by real-time PCR with a SYBR green mix in the ABI Prism 7900HT sequence detection system from Applied Biosystems (software version, SDS Enterprise Database 2.1). All the reactions were routinely performed in triplicate. Melting curve assays were systematically performed to check for specific amplifications. For both the methods, DNA was taken from equal quantities of nuclei, nuclear matrix, or supernatant fractions. The primer sequences are available on request.
RESULTS
The chromatin domain boundary protein BEAF-32B is a component of nuclear matrix.
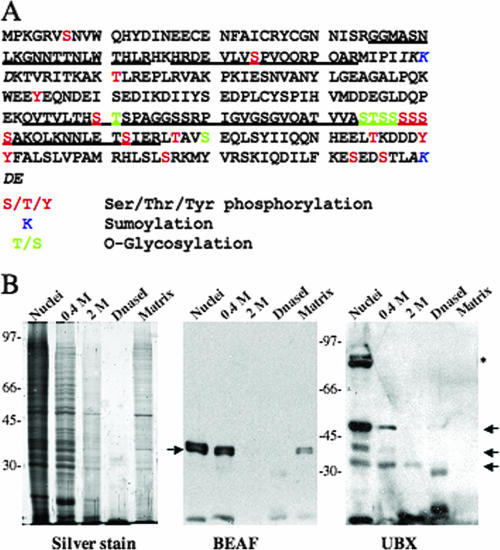
We prepared nuclear matrix from a Drosophila embryo following published protocols but incorporated several modifications which included stepwise salt extraction in the presence of mild detergent (see Materials and Methods for details). Proteins of the nuclear matrix were resolved by 2D gel electrophoresis and processed for MALDI-TOF MS-MS. Several proteins were identified in this way, including many known components of the nuclear matrix such as lamin, actin, and heat shock proteins. One of the moderately abundant protein spots with a molecular mass of ∼42 kDa and a pI of ∼6.0 was identified as BEAF-32B (Fig. 1A). Three of the six tryptic peptides that matched were from the N-terminal portion of the protein (Fig. 1A), showing that the matrix-bound protein is BEAF-32B. We analyzed the distribution of proteins in different fractions during matrix preparation and directly tested for the presence of BEAF in the matrix fraction using Western blotting. The results show that a significant proportion of BEAF is retained in the matrix while UBX, a known transcription factor, is not retained (Fig. 1B).
FIG. 1.
Identification of BEAF-32 in nuclear matrix from Drosophila embryo. (A) Amino acid sequence of BEAF-32B; underlined regions were covered in the MALDI-TOF MS-MS analysis. Various posttranslational modification sites are also marked by indicated color coding. Underlined stretches were covered by MALDI. (B) Nuclear BEAF exists in soluble and insoluble fractions. Nuclei were digested with DNase I and extracted with a stepwise increase in salt concentration. Release of BEAF protein was monitored by Western analysis and compared to that of a known soluble protein, UBX. The left panel shows the silver-stained protein profiles of different fractions, the middle panel is a Western blot probed with anti-BEAF antibody, and the right panel is the same blot stripped and reprobed with anti-UBX antibody. The loading pattern for all panels is as follows: total nuclear proteins, 0.4 M plus 0.5% Triton X-100 extract, 2 M NaCl extract, DNase I extract, and nuclear matrix fraction. Numbers are molecular masses in kilodaltons. Arrows indicate that BEAF is retained in the matrix fraction but UBX is not.
BEAF-32 exists in at least two isoforms, termed 32A and 32B, differing in the amino-terminal 80 aa (16). We could not detect any tryptic peptides representing the amino-terminal portion of BEAF-32A, suggesting that either it is absent or it is present in a form or amount that is undetectable. It is known that BEAF-32B is at least four times more abundant than BEAF-32A (16). The monoclonal antibody used in our experiments does not distinguish the two isoforms, and hence, we cannot formally rule out the presence of the 32A isoform in the matrix. Furthermore, as shown below, the region of BEAF-32B that is necessary for matrix association is common to the two isoforms of BEAF, suggesting that BEAF-32A has the potential to be present in the nuclear matrix. Therefore, we refer to this protein as BEAF throughout this report.
BEAF has previously been purified from nuclear extracts (42). In our extraction procedure, we observed that while a large fraction of BEAF was soluble in different extraction buffers, 25% of the nuclear BEAF remained matrix bound. An identical extraction procedure removes all of the nuclear UBX protein, and it is totally absent from the nuclear matrix fraction (Fig. 1B). Earlier studies have suggested that some components of the nuclear matrix are released upon RNase A digestion (17) of the matrix. We treated the matrix preparations with RNase A and observed that this treatment does not release BEAF from the nuclear matrix (data not shown). In this respect, BEAF behaves like CTCF, the mammalian boundary protein that is associated with the nuclear matrix but is not released upon RNase treatment.
Nuclear matrix-associated BEAF is posttranslationally modified.
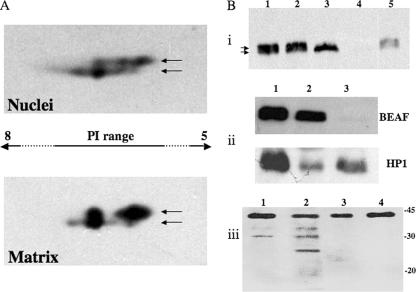
Many nuclear proteins, including histones, show a rich variety of posttranslational modifications that play an important role in their function. BEAF has been earlier shown to be phosphorylated as evidenced by the upper band of the doublet seen in Western blots (16). BEAF also contains several potential sites for other posttranscriptional modifications (Fig. 1A; see also Fig. 4A). In order to investigate the nature of these modifications and their possible relation to matrix association, we performed Western blot assays using narrow-pH-range 2D gels (Fig. 2A). The nuclear BEAF resolves into six spots. The three upper spots shift towards a more acidic pH. Phosphorylation, myristoylation, and methylation render proteins more acidic whereas esterification makes them more basic. Some modifications such as glycosylation and prenylation alter the molecular weight of the protein but not its pI. While all forms of the BEAF protein are seen in the soluble fraction, the protein form with a higher molecular weight and a more-acidic pI is enriched in the matrix fraction (Fig. 2A), suggesting extensive posttranslational modifications.
FIG. 4.
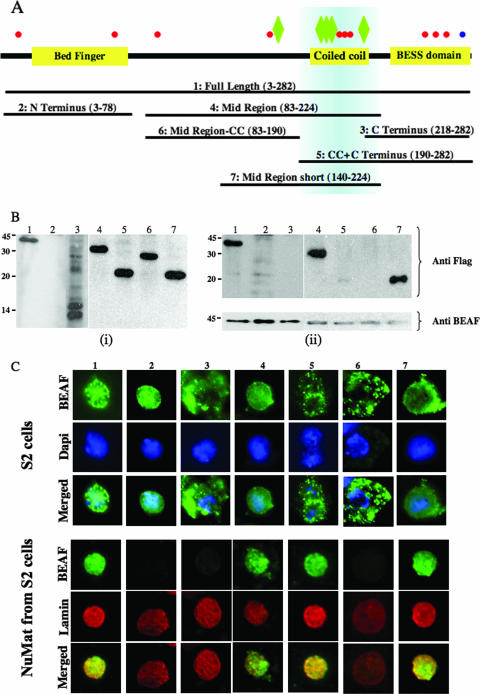
The middle region of BEAF is essential for nuclear matrix association. (A) Schematic diagram showing the various domains in BEAF, potential posttranslational modification sites, and the different constructs of BEAF fused with Flag tag that were used in this study. Red circles denote potential sites for phosphorylation, blue circles denote sumoylation, and green diamonds denote O-glycosylation sites. Blue shading across the map shows the region of the protein that is necessary for nuclear matrix association. (B) S2 cells were transfected with the indicated constructs. After 36 to 48 h, cells were lysed and nuclear matrix was prepared. The matrix preparations and total cell extracts were analyzed by Western blotting with anti-Flag antibody. (i) Total cell lysate of the transfected cells showing expression of all constructs. (ii) Nuclear matrix preparations from the same set of cells. Blots were stripped and reprobed with anti-BEAF antibody, bottom panel. Lanes 1, full-length BEAF-32B (aa 3 to 282); lanes 2, N terminus of the protein (aa 3 to 78); lanes 3, C terminus (aa 218 to 282); lanes 4, middle region (aa 83 to 224); lanes 5, C terminus with coiled coil (aa 190 to 282); lanes 6, middle region without coiled coil (aa 83 to 190); lanes 7, middle region, shorter version (aa 140 to 224). Numbers at left of each panel are molecular masses in kilodaltons. (C) Immunofluorescence for Flag-BEAF and its subfragments in transfected S2 cells. The upper panel shows whole cells stained for BEAF (top row) and DNA (middle row) and the merged images (third row). The lower panel shows nuclear matrix prepared from transfected S2 cells immunostained with anti-Flag antibody (green, top row) and anti-lamin Dm0 antibody (red, middle row). The third row shows the merged images. Lanes: 1, full-length BEAF-32B; 2, N terminus of the protein; 3, C terminus; 4, middle region; 5, C terminus with coiled coil; 6, middle region without coiled coil; 7, middle region, shorter version.
FIG. 2.
BEAF is posttranslationally modified. (A) Total nuclear proteins and the nuclear matrix proteins were electrofocused on a pH 5 to 8 strip and then resolved on an 8% polyacrylamide gel. The respective Western blots were probed with anti-BEAF antibody. BEAF protein in the matrix as well as the soluble fraction has several posttranslationally modified isoforms, marked by arrows. The spots marked by the upper arrow are more enriched in the matrix fraction. (B) BEAF is both phosphorylated and O glycosylated. (i) Both phosphorylated and dephosphorylated BEAF are retained in the nuclear matrix. Nuclear matrix preparations were treated with phosphatase alone (PP1) or with phosphatase in the presence of the specific inhibitor (I-2) and resolved on a 6 to 10% gradient polyacrylamide gel. The matrix association status of BEAF was analyzed by Western blot assays. Lane 1, untreated nuclear matrix; lane 2, matrix mock treated with 1× buffer for PP1; lane 3, matrix treated with PP1; lane 4, supernatant of PP1-treated matrix; lane 5, matrix treated with PP1 plus I-2. (ii) BEAF binds to WGA. Nuclear extracts were loaded on the affinity matrix column containing covalently attached WGA. Bound and unbound fractions in equivalent proportions were separated by SDS-PAGE, and the Western blots were probed with anti-BEAF and anti-HP1 antibodies as indicated. Lane 1, input; lane 2, bound fraction; lane 3, unbound fraction. BEAF but not HP1 is enriched in the WGA-bound fraction. (iii) Effect of alloxan treatment on BEAF. S2 cells were grown for 12 h in medium supplemented with alloxan, which is an inhibitor of the enzyme O-linked GlcNAc transferase. Indicated samples were separated by SDS-PAGE, transferred to nitrocellulose membranes, and probed with anti-BEAF antibody. Lanes 1 and 2, lysates of cells treated with 10 mM and 20 mM alloxan, respectively; lanes 3 and 4, nuclear matrixes prepared from cells treated with 10 mM and 20 mM alloxan, respectively. Numbers at right are molecular masses in kilodaltons.
To directly test if phosphorylation of BEAF is necessary for its association with the nuclear matrix, we treated the nuclear matrix preparation with PP1. PP1 is a Mn2+-dependent phosphatase with activity towards phosphoserine/threonine residues. Its activity is inhibited by I-2, which specifically interacts with the catalytic subunit of PP1. When the preparation was analyzed on a 6 to 10% gradient gel (Fig. 2Bi), which resolved the BEAF doublet, we observed that the upper band of the doublet disappeared upon phosphatase treatment. However, the intensity of the lower band increased, and it remained quantitatively associated with the matrix. This shift is not observed in the presence of PP1-specific inhibitor I-2, implying that the upper band is a phosphorylated form of BEAF that is dephosphorylated by PP1. It is interesting that both the phosphorylated and dephosphorylated forms are components of the nuclear matrix. We also tested TC-PTP, a phosphotyrosine-specific protein phosphatase, to check for tyrosine-specific phosphorylation. Treatment with TC-PTP did not affect BEAF mobility or levels (data not shown), suggesting that BEAF is not phosphorylated on tyrosine residues.
BEAF contains several potential glycosylation sites, particularly clustered in the middle region of the protein (Fig. 1A; see also Fig. 4A). We used WGA binding to check if BEAF is glycosylated. WGA is known to specifically bind terminal N-acetylglucosamine (GlcNAc) moieties and GlcNAc oligomers and has been extensively used to isolate glycosylated proteins (18, 31). We find that both the unphosphorylated and phosphorylated forms of BEAF from nuclear extracts are retained on the WGA column, showing that BEAF is indeed glycosylated, irrespective of its phosphorylation status (Fig. 2Bii). We also solubilized the nuclear matrix by adding denaturant to the matrix preparation, dialyzed the soluble proteins, and looked for binding to WGA-Sepharose beads. BEAF, which does not bind to Sepharose alone, was found to bind WGA-Sepharose (data not shown), indicating that both free and matrix-bound forms of BEAF are glycosylated. To further explore any link between the glycosylation and matrix association, we interfered with the endogenous glycosylation of proteins using alloxan, an inhibitor of O-GlcNAc transferase (24), in S2 cells. The cell lysate and nuclear matrix preparation of alloxan-treated cells were analyzed by Western blot assays with anti-BEAF antibody (Fig. 2Biii). With increasing concentrations of alloxan, several smaller bands of BEAF appear in the Western blot analysis (lanes 1 and 2). Since bacterially expressed BEAF that is likely to be unmodified shows the same mobility as does nuclear BEAF (15, 16), the smaller peptides may be degradation products of BEAF. It is possible that lack of glycosylation destabilizes BEAF, leading to smaller peptides. Importantly, these peptides are absent in the nuclear matrix prepared from alloxan-treated cells (Fig. 2Biii, lanes 3 and 4), indicating that unglycosylated BEAF is unstable and does not bind to nuclear matrix. However, these results do not rule out the possibility that unglycosylated BEAF that is still bound to the matrix is more stable due to its association with matrix components.
BEAF and ZW5 colocalize in nuclear matrix preparations.
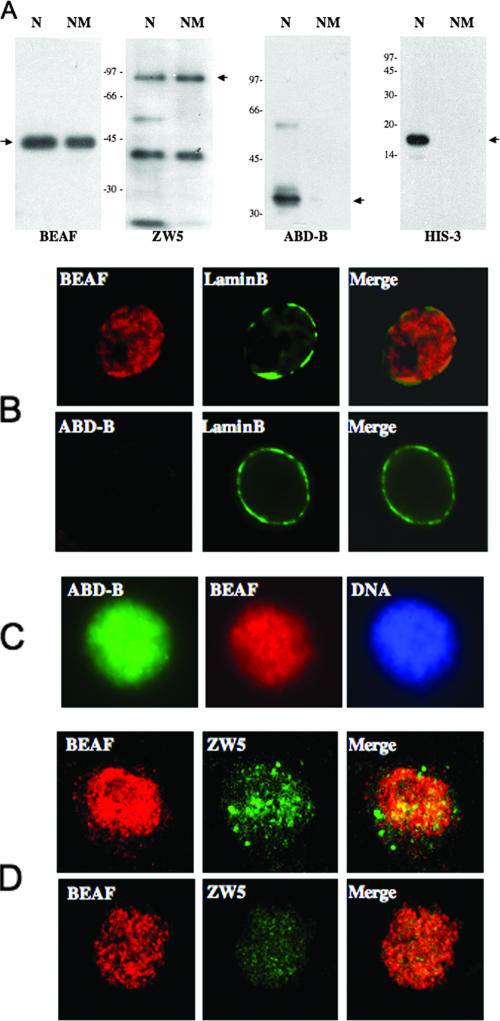
To further analyze the association of BEAF with the nuclear matrix and to ascertain whether a known partner of BEAF, ZW5, is also present in the matrix, we carried out a Western blot analysis of total nuclear protein and the nuclear matrix fraction. Equal amounts of protein from nuclei and nuclear matrix were loaded onto an SDS-polyacrylamide gel, blotted, and probed with monoclonal anti-BEAF and anti-ZW5 antibodies (Fig. 3A). ZW5 is the boundary-interacting protein that binds to scs, and like BEAF, ZW5 was also found in the matrix fraction, while several other nuclear proteins such as histone H3 and ABD-B were absent. Similar results were obtained when matrix was prepared from the embryos or S2 cells.
FIG. 3.
BEAF and ZW5 are associated with nuclear matrix. (A) Equal amounts of protein from nuclei (N) and nuclear matrix (NM) were analyzed by Western blotting using antibodies to BEAF, ZW5, ABD-B, and histone H3. BEAF and ZW5 are retained in the matrix, but histone and ABD-B are not. (B) Nuclear matrix was prepared from S2 cells in situ on slides and used for immunofluorescence. In the upper panels nuclear matrix was stained with antibodies to BEAF (red) and lamin B (green). Lamin stains the nuclear rim, and BEAF stains multiple sites in the interior of the nucleus. In the lower panels nuclear matrix is stained with antibodies to ABD-B (green) and lamin B, showing that no ABD-B is retained in the nuclear matrix. (C) Whole cells (nuclei) were stained for ABD-B, BEAF, and DNA. (D) Immunofluorescence staining of BEAF and ZW5 shown as projections of three optical sections. The top panels show BEAF (red) and ZW5 (green) in S2 cell nuclei. Regions of yellow show colocalization of the two signals. The lower panel shows the localization of BEAF (red) and ZW5 (green) in nuclear matrix. ZW5 is poorly retained in the matrix, but almost all ZW5 sites in the matrix also contain BEAF.
In order to study the distribution of BEAF and ZW5 in the nucleus, S2 cells were spun onto glass slides and extracted to reveal the nuclear matrix. These in situ matrix preparations were stained with antibodies to BEAF, ZW5, lamin, and ABD-B. As expected, lamin showed a perinuclear rim staining that was retained in the salt-extracted matrix preparations, while ABD-B was lost during salt extractions (Fig. 3B). Prior to extraction, in the nucleus as such (Fig. 3C), both ABD-B and BEAF are present. As shown in Fig. 3D, both BEAF and ZW5 were retained in the nuclear matrix. While ZW5 appears to be less abundant in the matrix, most of it colocalizes with BEAF. We estimated that about 50% of BEAF and of ZW5 colocalize in the nucleus. However, in the nuclear matrix preparations, almost 95% of ZW5 colocalizes with BEAF, consistent with the earlier observation that these two proteins interact with one another in vivo (3). These data clearly show that BEAF-32 and ZW5 are associated in the context of the nuclear matrix. Our immunofluorescence data also demonstrate that these proteins are unevenly distributed in the matrix and are not associated with prominent structures such as the nuclear lamina or nucleolus.
The middle region of BEAF is responsible for its nuclear matrix binding.
BEAF has three distinct domains (Fig. 4A): the amino-terminal BED finger domain (amino acids 27 to 77, with DNA binding activity), the carboxy-terminal BESS domain (amino acids 237 to 276, with protein-protein interaction and trimerization activity [16]), and the middle region coiled-coil domain (amino acids 203 to 223). To identify the region of the protein required for association with the nuclear matrix, full-length protein, the three distinct domains of BEAF-32B, and three other overlapping fragments were individually tagged with a Flag epitope (Fig. 4A) and were expressed as N-terminal Flag fusions under the control of the Polycomb promoter in S2 cells (37). The expression of the recombinant fusion proteins was confirmed in S2 cell lysate by Western blot assays using anti-Flag antibody (Fig. 4Bi). All the constructs were found to be expressed at relatively equal levels except for the amino-terminal fusion, which could not be detected on Western blots, possibly due to its small size of 75 aa. However, the N-terminal fusion was visible in immunofluorescence experiments (see below).
Nuclear matrix was prepared from cells transfected with each of these constructs. Western blot analysis with anti-Flag antibody reveals that the full-length protein and the middle-region (aa 83 to 224) constructs are retained in the nuclear matrix (Fig. 4Bii). The endogenous BEAF was retained in matrix preparations from all samples (Fig. 4Bii, lower panel). It is interesting that this middle region contains the coiled-coil domain (aa 203 to 223) and a shorter middle-region construct (lane 7, middle region-short, aa 140 to 224) that also contains this domain is matrix bound (Fig. 4Bii). The construct with the coiled-coil domain and the C terminus (lane 5, CC + C terminus, aa 190 to 282) is also matrix associated, although with much lower efficiency. Taken together, it appears that aa 140 to aa 224 are sufficient for targeting BEAF-32 to the nuclear matrix.
We also immunostained the transfected S2 cells and matrix prepared from them with anti-Flag antibody. As seen in Fig. 4C, full-length protein, N terminus, middle region, and middle region 2 are localized in the nucleus, whereas the C terminus, the C terminus with the coiled coil, and the middle region without the coiled coil are cytosolic. Thus, the coiled-coil region alone is not sufficient for nuclear localization but may be necessary because the middle region without the coiled coil is also cytosolic. Immunostaining confirms the findings of the Western blot analysis and shows that the region corresponding to aa 140 to 224 is responsible for nuclear matrix targeting (Fig. 4C, lower panel). The N terminus (the DNA binding domain) is also independently capable of localizing to the nucleus but is not retained in the matrix. If the coiled-coil region is included with the C terminus, the protein is unable to localize into the nucleus efficiently but the small amount of protein that does enter the nucleus associates with nuclear matrix (Fig. 4C). These results are summarized in Table 1.
TABLE 1.
Nuclear localization and matrix association properties of different Flag-tagged constructs of BEAF
| Construct no. | Construct name | Amino acid range | Nuclear localization | Matrix association |
|---|---|---|---|---|
| 1 | Full length | 3-282 | Yes | Yes |
| 2 | N terminus | 3-78 | Yes | No |
| 3 | C terminus | 218-282 | No | No |
| 4 | Middle region | 83-224 | Yes | Yes |
| 5 | Coiled coil plus C terminus | 190-282 | Partial | Yes |
| 6 | Middle region without coiled coil | 83-190 | No | No |
| 7 | Middle region (short) | 140-224 | Partial | Yes |
Interestingly, the region of BEAF that appears to play a key role in its localization to the nuclear matrix contains most of the serine/threonine residues that have a high potential to become phosphorylated as well as glycosylated, based on analysis done using web-based servers NetPhos 2.0 (http://www.cbs.dtu.dk/services/NetPhos/) and YinOYang 1.2 (http://www.cbs.dtu.dk/services/YinOYang/). When expressed separately as a Flag fusion, this middle region also shows lower mobility, indicating that it may be glycosylated. It is likely that these posttranslational modifications are important in the matrix association of BEAF. Further, this region is common to the two isoforms of BEAF, suggesting that BEAF-32A also has the potential to be a nuclear matrix component, even though we did not detect this isoform in our proteomic analysis.
BEAF target sequences associate with nuclear matrix.
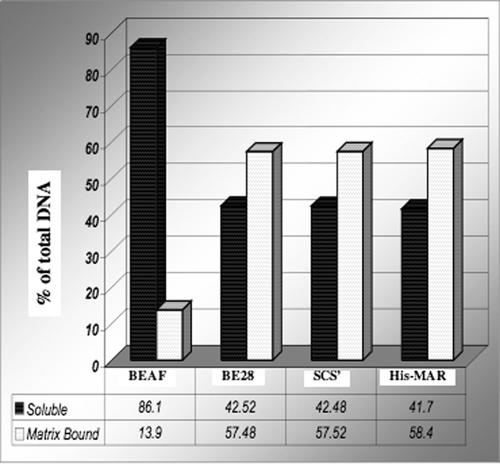
The results described above show that BEAF is associated with the nuclear matrix. Since BEAF binds to several sequences that have boundary activity, we tested the hypothesis that these target sequences are also matrix bound. We determined the presence of four different loci from the Drosophila genome in nuclear matrix preparations by quantitative PCR and immuno-FISH. Two different sequences known to be targets of BEAF were tested. First, a 111-bp amplicon from BE28, a moderately repeated 1.2-kb DNA sequence that functions as a BEAF-dependent boundary element in transgenic flies, was chosen (8). This amplicon encompasses the BEAF binding sites. The second BEAF target is a 97-bp amplicon from the scs′ boundary element including the BEAF binding sites. A 105-bp amplicon from an exon of the BEAF protein coding region with no predicted MAR was chosen as a negative control. Finally, a 192-bp amplicon from the well-characterized MAR from the histone gene cluster was chosen as a positive control (29).
We prepared nuclear matrix using EcoRI and HindIII in place of DNase I for digestion prior to extraction. The DNA fragments (operationally defined MARs) obtained from the nuclear matrix preparation were used for quantitative real-time PCR. The results show that ∼10% of the BEAF coding region amplicon is retained in the matrix, reflecting a background-level presence or transient matrix association due to transcriptional activity (which may involve indirect association with the matrix) (Fig. 5). In contrast, >50% of the BEAF target sequences (BE28 as well as scs′) and the positive-control His-MAR are present in the matrix under these conditions (Fig. 5).
FIG. 5.
Distribution of BEAF target sequences in different nuclear fractions. Nuclear matrix was prepared using restriction enzymes (EcoRI and HindIII) followed by stepwise extraction with salt and mild detergent. The graph shows the relative levels of target sequences in different fractions determined by quantitative PCR. The BEAF coding region was used as a nonmatrix control, and known MAR from the histone locus was used as a positive control. BEAF binding regions, BE28, and scs′ are enriched in nuclear matrix preparations.
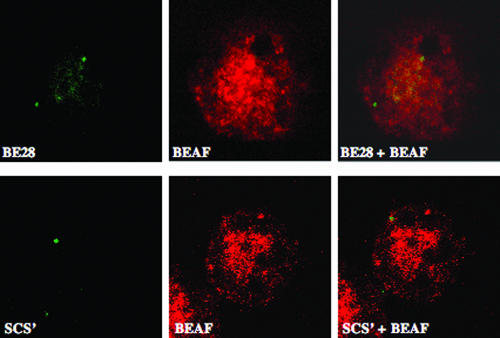
Retention of a high proportion of the BEAF target sequences (equivalent to a known MAR) suggests specific association with the nuclear matrix. If the BEAF target sequences are MARs and BEAF itself is a matrix component, they would be predicted to colocalize in nuclear matrix preparations. We performed immuno-FISH on matrix preparations of S2 cells with BEAF antibody and BE28/scs′ fluorescently labeled probes. As seen in Fig. 6, intensely staining spots of BE28 probe are clearly retained in the matrix. BEAF protein, in contrast, shows a more elaborate staining. Using optical sectioning, we could confirm that the BE28 spots are located in the interior of the nucleus. As shown in Fig. 6, scs′ too is detected in the matrix preparation. Matrix preparations doubly stained with BE28 and BEAF were examined by confocal microscopy for colocalization of the two signals using the cross-hair function. The weighted colocalization coefficient was calculated for selected regions where the BE28 signal was detected. We observed that >70% of the BE28 signal and >75% of the scs′ signal colocalize with the BEAF. The data suggest that all BE28 sites overlap with BEAF signals but that BEAF has several other target sites and gives a more disperse pattern.
FIG. 6.
BEAF target sequences BE28 (upper panel) and scs′ (lower panel) are associated with the nuclear matrix and colocalize with BEAF. S2 cells were spun onto glass slides, extracted in situ with salt, and digested with DNase I. The cell skeletons obtained after extraction and digestion were stained with antibody to BEAF (red) and hybridized fluorescent DNA probes (green) for detection of BE28/scs′ sequences.
DISCUSSION
In higher eukaryotes, regulatory elements, such as enhancers and silencers, often act from a long distance and generally have poor or no promoter specificity. One mechanism to restrict the action of specific regulatory sequences to the appropriate target gene is based on the idea that genomes are organized into functionally independent domains, which are insulated from the effects of neighboring domains by insulators or boundary elements. These insulators are operationally defined elements based on the assays that have been used to characterize them. Two distinct assays have been used extensively to study boundaries. In the enhancer blocker assay, the ability of a sequence inserted between the enhancer and promoter to insulate the promoter from the action of the enhancer is tested (23). Such sequences are called enhancer blockers. In the second assay, the ability of a sequence inserted adjacent to a reporter gene to protect reporter gene activity from chromosomal position effects is tested (22). These sequences are called insulators or barriers. Many sequences from yeasts to mammals have been demonstrated to have either or both of these activities, and a few proteins that bind to these sequences and contribute to this property have been identified. As is expected for a function as basic as organization of chromatin domains, this mechanism is highly conserved, with sequences and proteins isolated from one organism able to function as a boundary in another organism (20). Intriguingly, however, there is no significant sequence similarity among the various boundary elements known in any one organism, except for a few small common motifs. Further, the mechanism by which these sequences and their associated nucleoprotein complexes function to establish boundaries is not understood.
Two distinct models have been proposed for boundary action. The barrier model suggests that boundaries act as a physical barrier and prevent the action of neighboring enhancers or silencers. Alternatively, it is thought that these boundary sequences, both physically and functionally, subdivide the chromosome into topologically independent domains of action. This model predicts the existence of tethers to which the boundaries of the domains would attach, allowing the intervening chromatin to loop out and create independent domains. The two models are not mutually exclusive, and it is likely that the two kinds of boundaries coexist. Our results provide a possible structural basis for this tethering function. A recent study showing that scs and scs′ sequences are in close proximity in vivo also demonstrated that BEAF-32 and ZW5 interact with each other, leading to the looping out of the intervening sequences flanked by the two interacting boundaries (3). Similarly, the looping out of intervening sequences between the adjacent endogenous target sequences of Su(HW) has also been demonstrated elsewhere (5). Taken in this context our study suggests that an underlying nuclear matrix facilitates this type of looping.
Our results show that a fraction of BEAF-32 is associated with the nuclear matrix. Indeed, BEAF-32 is found at many sites in the matrix, consistent with the numerous binding sites for BEAF in the Drosophila genome (9, 42). As ZW5 and BEAF interact in vivo, we asked if the association of BEAF and ZW5 persists in the matrix preparations in vitro. Nuclear matrix stained for ZW5 yielded a very faint signal, indicating that only a small proportion of this protein is retained in the matrix. Colocalization of BEAF and ZW5 in the nucleus shows that the two proteins colocalize at over 50% of the sites. In the matrix, however, we find that over 95% of the ZW5 sites overlap with BEAF-32 but less than 70% of BEAF is localized with ZW5. This observation suggests that since almost all of ZW5 association with the matrix is in the context of BEAF, ZW5 may be associated with the matrix via BEAF. Alternatively, those boundaries where the two proteins work together may have matrix association properties.
The middle region of BEAF-32 that contains the coiled-coil region and several sites for posttranslational modification contains the signal for matrix targeting/association. We also find that BEAF target sequences that are known to function as boundaries are enriched in the matrix. These results suggest that boundary sequences and their associated proteins may function through interaction with the nuclear matrix. It is possible that BEAF does not directly interact with the nuclear matrix but is associated with the matrix through its interaction with other proteins. For example, it is possible that the sequences in scs′ or BE28 may also contain MARs that bind to matrix proteins and that BEAF-32 interacts with these proteins and hence is found in association with the nuclear matrix. Indeed, BEAF interacts with D1 chromosomal protein, which also binds to the AT-rich sequences in BE28 (8). However, in either case, it is evident that matrix association may be a property of BEAF-32-dependent boundary elements and may be an important, if not the sole, mode of action of these boundaries.
Recently, two insulator proteins have been shown to be matrix associated. One of them, Su(HW), is a Zn finger-containing protein required for the function of the gypsy insulator. Su(HW) interacts with Mod(mdg4), a BTB domain protein, and both are localized to “insulator bodies” at the nuclear periphery (12). The endogenous target sites have also been shown to be associated with these insulator bodies, and indeed, two adjacent target sites of Su(HW) seem to associate with a single insulator body, resulting in the looping out of the intervening chromosomal domain. This looping is thought to be essential for the insulator function of gypsy. Similarly, CTCF, a vertebrate insulator protein, was also shown to be associated with the matrix, specifically with the nucleolar region, and to copurify with nucleolar proteins (41). The target DNA sequences of CTCF were also shown to be matrix associated (40), and this association appears to be dependent upon the DNA sequences bound by CTCF.
Although the experiments described above show that the two insulator proteins Su(HW) and CTCF are found in two different compartments of the nucleus, namely, the nuclear periphery and the nucleolus, respectively, there are common themes. First, individual proteins seem to associate with a particular part of the nucleus, which can be thought of as their tethering site. Second, these subnuclear sites also contain the target DNA sequences, suggesting tethering of target DNA to these subnuclear locations via the insulator proteins. In both cases, indirect evidence suggests that the presence of intact insulator protein binding DNA sequences is essential for matrix association. Tethering to different nuclear components may be a common theme for insulator/boundary protein function. For example, the boundary function of Saccharomyces cerevisiae requires tethering to nuclear pore complexes to establish barrier activity (19).
Ectopically expressed BEAF-32 acts as a boundary/barrier for heterochromatinization in yeast (20). Although yeast does not have BEAF protein and there are no BEAF binding sites in the characterized boundaries of yeast, when BEAF was artificially targeted to sites where boundary activity can be assayed, it functions as a boundary protein. This finding implies that the basic mechanism of boundary function is conserved at least from yeast to flies. Interestingly, the region of BEAF that was shown to contain the boundary activity (20) overlaps with the region that we have shown to be required for matrix targeting. It is therefore possible that the tethering mechanism operating in yeast for boundary function is similar to the matrix association that we observe in Drosophila, and this physical association is the mechanistic basis for boundary function. Our results are consistent with the idea that boundary sequences along with their protein partners directly or indirectly interact with the nuclear matrix and create a topologically independent domain.
We find that the region of BEAF-32 that is essential for matrix association contains a cluster of potential sites for glycosylation. Taken together with our observation that unmodified BEAF is unstable and the earlier report that BEAF is also phosphorylated, this finding raises the possibility that posttranslational modifications are important for BEAF function. It is interesting that a variety of proteins, including several chromatin-associated proteins and transcription factors such as Sp1, estrogen receptor, and RNA polymerase II (18, 21), are known to posses these modifications. In particular, Sp1 has also been demonstrated to have boundary activity (20). In the light of these observations, it seems likely that posttranslational modifications play an important regulatory role in chromatin structural proteins. Our results show that BEAF present in the soluble fraction is also glycosylated, indicating that this modification per se is not sufficient to target BEAF to the matrix. It is possible that glycosylation and perhaps other modifications enable these chromatin proteins to interact with appropriate partners, which allows them to participate in different functions. The precise role of the GlcNAc modification of BEAF in the context of its nuclear matrix association and function needs further investigation.
Considering that an increasing number of boundary/insulator proteins are found to be associated with some kind of nuclear tether, these interactions between components of the nuclear architecture and boundary proteins warrant extensive studies on the nature of these interactions, the components of the nuclear architecture that mediate these interactions, and their possible regulation by external signaling molecules. Several studies have raised the interesting possibility that the nuclear matrix is not merely a static structure but a dynamic one with tissue- and temporally specific protein composition that is amenable to regulation. Our studies support such a model where dynamic association of factors with nuclear matrix may have regulatory consequences.
Acknowledgments
We thank Susan Celnicker, U. K. Laemmli, and P. Schedl for antibodies and R. Kingston for the Flag-Pc plasmid. We also thank Jyotsna Dhawan for critical reading of the manuscript and suggestions. We acknowledge the help of the CCMB proteomics facility in identification of 2D gel spots.
This work was supported by a Young Investigators grant (RGY0316/2001-M) from the Human Frontier Science Program to R.K.M.
Footnotes
Published ahead of print on 7 May 2007.
REFERENCES
- 1.Aten, J. A., J. Stap, P. M. Krawczyk, C. H. van Oven, R. A. Hoebe, J. Essers, and R. Kanaar. 2004. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science 303:92-95. [DOI] [PubMed] [Google Scholar]
- 2.Bell, A. C., A. G. West, and G. Felsenfeld. 2001. Insulators and boundaries: versatile regulatory elements in the eukaryotic. Science 291:447-450. [DOI] [PubMed] [Google Scholar]
- 3.Blanton, J., M. Gaszner, and P. Schedl. 2003. Protein:protein interactions and the pairing of boundary elements in vivo. Genes Dev. 17:664-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonifer, C., U. Jagle, and M. C. Huber. 1997. The chicken lysozyme locus as a paradigm for the complex developmental regulation of eukaryotic gene loci. J. Biol. Chem. 272:26075-26078. [DOI] [PubMed] [Google Scholar]
- 5.Byrd, K., and V. G. Corces. 2003. Visualization of chromatin domains created by the gypsy insulator of Drosophila. J. Cell Biol. 162:565-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cockerill, P. N., and W. T. Garrard. 1986. Chromosomal loop anchorage sites appear to be evolutionarily conserved. FEBS Lett. 204:5-7. [DOI] [PubMed] [Google Scholar]
- 7.Cremer, T., and C. Cremer. 2001. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2:292-301. [DOI] [PubMed] [Google Scholar]
- 8.Cuvier, O., C. M. Hart, E. Kas, and U. K. Laemmli. 2002. Identification of a multicopy chromatin boundary element at the borders of silenced chromosomal domains. Chromosoma 110:519-531. [DOI] [PubMed] [Google Scholar]
- 9.Cuvier, O., C. M. Hart, and U. K. Laemmli. 1998. Identification of a class of chromatin boundary elements. Mol. Cell. Biol. 18:7478-7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donze, D., and R. T. Kamakaka. 2002. Braking the silence: how heterochromatic gene repression is stopped in its tracks. Bioessays 24:344-349. [DOI] [PubMed] [Google Scholar]
- 11.Gaszner, M., J. Vazquez, and P. Schedl. 1999. The Zw5 protein, a component of the scs chromatin domain boundary, is able to block enhancer-promoter interaction. Genes Dev. 13:2098-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerasimova, T. I., K. Byrd, and V. G. Corces. 2000. A chromatin insulator determines the nuclear localization of DNA. Mol. Cell 6:1025-1035. [DOI] [PubMed] [Google Scholar]
- 13.Gerasimova, T. I., and V. G. Corces. 1996. Boundary and insulator elements in chromosomes. Curr. Opin. Genet. Dev. 6:185-192. [DOI] [PubMed] [Google Scholar]
- 14.Gerasimova, T. I., and V. G. Corces. 2001. Chromatin insulators and boundaries: effects on transcription and nuclear organization. Annu. Rev. Genet. 35:193-208. [DOI] [PubMed] [Google Scholar]
- 15.Gilbert, M. K., Y. Y. Tan, and C. M. Hart. 2006. The Drosophila boundary element-associated factors BEAF-32A and BEAF-32B affect chromatin structure. Genetics 173:1365-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hart, C. M., K. Zhao, and U. K. Laemmli. 1997. The scs′ boundary element: characterization of boundary element-associated factors. Mol. Cell. Biol. 17:999-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He, D. C., J. A. Nickerson, and S. Penman. 1990. Core filaments of the nuclear matrix. J. Cell Biol. 110:569-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hiromura, M., C. H. Choi, N. A. Sabourin, H. Jones, D. Bachvarov, and A. Usheva. 2003. YY1 is regulated by O-linked N-acetylglucosaminylation (O-glcNAcylation). J. Biol. Chem. 278:14046-14052. [DOI] [PubMed] [Google Scholar]
- 19.Ishii, K., G. Arib, C. Lin, G. Van Houwe, and U. K. Laemmli. 2002. Chromatin boundaries in budding yeast: the nuclear pore connection. Cell 109:551-562. [DOI] [PubMed] [Google Scholar]
- 20.Ishii, K., and U. K. Laemmli. 2003. Structural and dynamic functions establish chromatin domains. Mol. Cell 11:237-248. [DOI] [PubMed] [Google Scholar]
- 21.Jackson, S. P., and R. Tjian. 1988. O-glycosylation of eukaryotic transcription factors: implications for mechanisms of transcriptional regulation. Cell 55:125-133. [DOI] [PubMed] [Google Scholar]
- 22.Kellum, R., and P. Schedl. 1991. A position-effect assay for boundaries of higher order chromosomal domains. Cell 64:941-950. [DOI] [PubMed] [Google Scholar]
- 23.Kellum, R., and P. Schedl. 1992. A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol. Cell. Biol. 12:2424-2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Konrad, R. J., F. Zhang, J. E. Hale, M. D. Knierman, G. W. Becker, and J. E. Kudlow. 2002. Alloxan is an inhibitor of the enzyme O-linked N-acetylglucosamine transferase. Biochem. Biophys. Res. Commun. 293:207-212. [DOI] [PubMed] [Google Scholar]
- 25.Labrador, M., and V. G. Corces. 2002. Setting the boundaries of chromatin domains and nuclear organization. Cell 111:151-154. [DOI] [PubMed] [Google Scholar]
- 26.Loc, P. V., and W. H. Stratling. 1988. The matrix attachment regions of the chicken lysozyme gene co-map with the boundaries of the chromatin domain. EMBO J. 7:655-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marshall, W. F. 2002. Order and disorder in the nucleus. Curr. Biol. 12:R185-R192. [DOI] [PubMed] [Google Scholar]
- 28.Mihaly, J., I. Hogga, S. Barges, M. Galloni, R. K. Mishra, K. Hagstrom, M. Muller, P. Schedl, L. Sipos, J. Gausz, H. Gyurkovics, and F. Karch. 1998. Chromatin domain boundaries in the Bithorax complex. Cell. Mol. Life Sci. 54:60-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mirkovitch, J., M. E. Mirault, and U. K. Laemmli. 1984. Organization of the higher-order chromatin loop: specific DNA attachment sites on nuclear scaffold. Cell 39:223-232. [DOI] [PubMed] [Google Scholar]
- 30.Mishra, R. K., and F. Karch. 1999. Boundaries that demarcate structural and functional domains of chromatin. J. Biosci. 24:377-399. [Google Scholar]
- 31.Monsigny, M., A. C. Roche, C. Sene, R. Maget-Dana, and F. Delmotte. 1980. Sugar-lectin interactions: how does wheat-germ agglutinin bind sialoglycoconjugates? Eur. J. Biochem. 104:147-153. [DOI] [PubMed] [Google Scholar]
- 32.Moon, H., G. Filippova, D. Loukinov, E. Pugacheva, Q. Chen, S. T. Smith, A. Munhall, B. Grewe, M. Bartkuhn, R. Arnold, L. J. Burke, R. Renkawitz-Pohl, R. Ohlsson, J. Zhou, R. Renkawitz, and V. Lobanenkov. 2005. CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Rep. 6:165-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nabirochkin, S., M. Ossokina, and T. Heidmann. 1998. A nuclear matrix/scaffold attachment region co-localizes with the gypsy retrotransposon insulator sequence. J. Biol. Chem. 273:2473-2479. [DOI] [PubMed] [Google Scholar]
- 34.Noma, K., C. D. Allis, and S. I. Grewal. 2001. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science 293:1150-1155. [DOI] [PubMed] [Google Scholar]
- 35.Oki, M., and R. T. Kamakaka. 2002. Blockers and barriers to transcription: competing activities? Curr. Opin. Cell Biol. 14:299-304. [DOI] [PubMed] [Google Scholar]
- 36.Saitoh, N., A. C. Bell, F. Recillas-Targa, A. G. West, M. Simpson, M. Pikaart, and G. Felsenfeld. 2000. Structural and functional conservation at the boundaries of the chicken beta-globin domain. EMBO J. 19:2315-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shao, Z., F. Raible, R. Mollaaghababa, J. R. Guyon, C. T. Wu, W. Bender, and R. E. Kingston. 1999. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell 98:37-46. [DOI] [PubMed] [Google Scholar]
- 38.Shermoen, A. W., and S. K. Beckendorf. 1982. A complex of interacting DNAase I-hypersensitive sites near the Drosophila glue protein gene, Sgs4. Cell 29:601-607. [DOI] [PubMed] [Google Scholar]
- 39.West, A. G., M. Gaszner, and G. Felsenfeld. 2002. Insulators: many functions, many mechanisms. Genes Dev. 16:271-288. [DOI] [PubMed] [Google Scholar]
- 40.Yusufzai, T. M., and G. Felsenfeld. 2004. The 5′-HS4 chicken beta-globin insulator is a CTCF-dependent nuclear matrix-associated element. Proc. Natl. Acad. Sci. USA 101:8620-8624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yusufzai, T. M., H. Tagami, Y. Nakatani, and G. Felsenfeld. 2004. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol. Cell 13:291-298. [DOI] [PubMed] [Google Scholar]
- 42.Zhao, K., C. M. Hart, and U. K. Laemmli. 1995. Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell 81:879-889. [DOI] [PubMed] [Google Scholar]