Abstract
Forkhead transcription factors of the O class (FOXOs) are important targets of the phosphatidylinositol 3-kinase (PI3-kinase)/Akt pathway. FOXOs have been implicated in the regulation of cell cycle progression, oxidative stress resistance, and apoptosis. Using DNA microarrays, we analyzed the transcriptional response to FOXO3a activation by gene expression analysis in DLD-1 colon cancer cells stably expressing a FOXO3a.A3-ER fusion protein. We found that activation of FOXO3a resulted in repression of a number of previously identified Myc target genes. Furthermore, FOXO3a activation induced expression of several members of the Mad/Mxd family of transcriptional repressors, most notably Mxi1. The induction of Mxi1 by FOXO3a was specific to the Mxi1-SRα isoform and was mediated by three highly conserved FOXO binding sites within the first intron of the gene. Activation of FOXO3a in response to inhibition of Akt also resulted in activation of Mxi1-SRα expression. Silencing of Mxi1 by small interfering RNA (siRNA) reduced FOXO3a-mediated repression of a number of Myc target genes. We also observed that FOXO3a activation induced a switch in promoter occupancy from Myc to Mxi1 on the E-box containing promoter regions of two Myc target genes, APEX and FOXM1. siRNA-mediated transient silencing of Mxi1 or all Mad/Mxd proteins reduced exit from S phase in response to FOXO3a activation, and stable silencing of Mxi1 or Mad1 reduced the growth inhibitory effect of FOXO3a. We conclude that induction of Mad/Mxd proteins contributes to the inhibition of proliferation in response to FOXO3a activation. Our results provide evidence of direct regulation of Mxi1 by FOXO3a and imply an additional mechanism through which the PI3-kinase/Akt/FOXO pathway can modulate Myc function.
Forkhead transcription factors of the O class (FOXOs) belong to a family of transcription factors that are characterized by their conserved DNA binding domain (forkhead box). Daf-16, the FOXO orthologue in Caenorhabditis elegans, has been identified as a target of insulin-like signaling through the Daf-2/AGE-1 pathway and is involved in formation of the Dauer stage (40, 53). The FOXO subgroup in mammals consists of four members, FOXO1, FOXO3a, and FOXO4 (previously termed FKHR, FKHRL1, and AFX, respectively) and FOXO6 (7, 24, 33, 62).
FOXO transcription factors bind to DNA as monomers through their winged-helix domain at a consensus motif termed DBE (for Daf-16 family member binding element) with the core sequence TTGTTAC (23). A number of FOXO target genes have been identified so far (for review, see reference 28). FOXO target genes are involved in cell cycle arrest, apoptosis, metabolism, differentiation, and the stress response. It has been shown that FOXOs induce G1 arrest through expression of p27KIP1 and p130 (36, 46, 51) and increase the duration of the G2 phase of the cell cycle by inducing cyclin G2 (43). In lymphocytes, FOXO activation induces cell death through induction of Bim, Trail, and Fas ligand (9, 48, 66). Upregulation of glucose-6-phosphatase and phosphoenolpyruvate carboxykinase by FOXOs results in induction of gluconeogenesis (5, 30). The role of FOXO transcription factors in differentiation is less well understood and is cell type dependent. In adipocytes, FOXOs inhibit differentiation most likely through induction of p21, while FOXO activation induces erythroid differentiation through induction of BTG and is required for myoblast differentiation (4, 6, 50). Finally, induction of Mn-superoxide dismutase and catalase by FOXO is involved in detoxification of reactive oxygen species (19, 34).
Mammalian FOXO transcription factors are regulated through the phosphatidylinositol 3-kinase (PI3-kinase) pathway. In the absence of growth factors, FOXOs are localized to the nucleus and transcriptionally active. Activation of Akt or SGK in response to growth factor stimulation induces phosphorylation of FOXO transcription factors at three highly conserved serine and threonine residues (9, 35). As a result, FOXO transcription factors translocate to the cytoplasm. Phosphorylation of FOXO factors by Akt or SGK induces binding of 14-3-3 proteins, which could induce nuclear export, possibly by exposing a nuclear export sequence located in the C terminus or by enhancing interaction with the nuclear export machinery. Interaction with 14-3-3 proteins could also contribute to cytoplasmic retention of FOXO factors by masking a nuclear localization signal. Akt-dependent phosphorylation can also modulate the stability of FOXO transcription factors (3, 45) by inducing interaction with the Skp2/Cul1/F-box complex and subsequent targeting to the proteasome (32, 54).
It has been shown more recently that FOXO factors are regulated by a Ral/JNK-dependent mechanism in response to oxidative stress. Phosphorylation of FOXO4 by JNK induces nuclear translocation even in the presence of growth factors (19).
The transcriptional activity of FOXO factors can be modulated by p300/CBP-dependent acetylation (22, 44, 70). The inhibitory effect of acetylation is counteracted by the action of NAD+-dependent deacetylases of the sirtuin family (10, 13, 70), and acetylation status may affect target gene specificity (25).
FOXOs are frequently inactivated in cancer cells due to the action of the PI3-kinase/Akt pathway (2). Surprisingly, no inactivating mutations in FOXO genes have been found in human tumors so far. Chromosomal translocations involving FOXO factors found in alveolar rhabdomyosarcomas or acute leukemias result in fusion proteins that retain DNA binding activity of the fusion partner, Pax3 or MLL, respectively (7, 24, 62). However, it is conceivable that loss of function of FOXO1 or FOXO4 contributes to tumorigenesis in this context (64, 65). In addition, it has been reported that FOXO1 is inactivated through direct binding of the androgen receptor in prostate cancer cells, and FOXO3a is downregulated during progression to the androgen-independent state in LNCaP cells (39, 42). These findings emphasize the importance of inactivation of the growth inhibitory functions of FOXO factors during tumorigenesis.
In order to achieve further insight into the role of FOXO factors as tumor suppressors, we investigated the transcriptional program induced by FOXO3a activation using DNA microarrays. The results of our study show that FOXO3a induces the expression of several members of the Mad/Mxd family of transcriptional repressors. We observed that induction of Mxi1 is specific to the Mxi1-SRα isoform and that FOXO3a binds directly to conserved DBEs in the first intron of the gene. Induction of Mxi1-SRα was required for efficient inhibition of Myc-dependent transcription by FOXO3a, and silencing of Mxi1 or Mad1 reduced the growth inhibitory effect of FOXO3a. Our results provide evidence of a novel mechanism through which the PI3-kinase/Akt pathway can regulate Myc function.
MATERIALS AND METHODS
Plasmids and reagents.
Expression plasmids for FOXO3a.A3-ER and a firefly luciferase reporter construct containing six DBE consensus sites (pGL3-DBE) have been described previously (36). To create expression vectors for Mxi1-SRα, SRβ, and WR, coding sequences were amplified from cDNA and inserted into pCDNA3.
A luciferase reporter construct containing eight consecutive E-boxes (MycT) and the corresponding empty vector (CT) were obtained from Panomics.
Promoter regions of the human Mxi1 gene were amplified from genomic DNA from DL23 cells by PCR and cloned into the pGL3-Basic vector (Promega).
Successive deletions of the Mxi1-SRα(−441/+1292) region were performed using PCR. Mutations in DBEs in the construct Mxi1-SRα(−441/+1292) were generated using the QuikChange site-directed mutagenesis kit (Stratagene). Primer sequences are listed in the supplemental material.
The specific Akt inhibitor triciribine was obtained from Calbiochem (Akt inhibitor V).
Cell culture.
The DL23 colon carcinoma cell line has been described elsewhere (36). DL23 and parental DLD-1 cells were grown in RPMI 1640 with 10% fetal calf serum. 4-Hydroxy-tamoxifen (4-OHT; Sigma) was dissolved in ethanol and used at a final concentration of 100 nM.
MCF-10A cells were grown in Ham's nutrient mixture F-12-Dulbecco's modified Eagle's medium (1:1) containing 5% horse serum and 10 μg/ml insulin, 20 ng/ml epidermal growth factor, 5 μg/ml hydrocortisone, and 100 ng/ml cholera toxin. The retroviral vector pBABEpuro-HA-FKHR-L1.A3-ER was packaged in GP+E cells and used to infect MCF-10A cells expressing the ecotropic retrovirus receptor. Infected cells were selected, and clone M11 was chosen for further study.
For colony formation assays, DL23 cells or stable pools expressing retroviral RNA inhibition (RNAi) vectors were seeded at a density of 104 cells per 6-cm dish. Cells were cultured for 10 to 15 days in the presence of 100 nM 4-OHT or solvent (ethanol). Cells were fixed in methanol and stained with crystal violet.
BrdU incorporation and FACS analysis.
Cell were treated with 10 μM bromodeoxyuridine (BrdU; Sigma) for 30 min prior to harvesting, fixed in ethanol, stained, and analyzed by fluorescence-activated cell sorting (FACS).
Immunoblotting.
Cells were lysed in Triton buffer (1% Triton X-100, 50 mM Tris pH 7.5, 300 mM NaCl, 1 mM EGTA, and protease inhibitor cocktail [Roche]). Plates were incubated for 20 min on ice, and lysates were cleared by centrifugation at 13,000 rpm. Lysates were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and transferred onto an Immobilon membrane (Millipore).
The following antibodies were used: anti-Mxi1 (sc-1042), anti-Mad1 (sc-222), anti-Max (sc-197), anti-cyclin A1 (sc-751), and anti-c-Myc antibody (sc-764) (all from Santa Cruz Biotechnology); anti-p27KIP1 (no. 610241; BD Transduction Laboratories); anti-c-Myc (9E10), anti-FKHRL1, and anti-phospho-FKHRL1 (Thr32) (Upstate); anti-phospho-Akt (Ser473) and anti-Akt (Cell Signaling Technology); anti-actin-horseradish peroxidase (Ac-15; Sigma); and anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH)-horseradish peroxidase (Abcam).
RNA preparation, array hybridization, and data analysis.
For gene expression profiling, total RNA was prepared using TRIzol reagent (GibcoBRL) following the manufacturer's protocol. Total RNA from control and experimental samples was labeled by oligo(dT)-primed first-strand cDNA synthesis in the presence of Cy3-dUTP or Cy5-dUTP (Amersham). Labeled cDNA was purified using AutoSeq G-50 columns and hybridized to human cDNA microarrays (Sanger human 10K version 1.2.1). All hybridizations were performed in quadruplicate using duplicate RNA samples and dye swaps. For detailed description of clone sequences, preparation of the microarrays, and protocols for array hybridization, washing, and handling, see http://www.sanger.ac.uk/Projects/Microarrays/.
Images were quantified using the adaptive circle method (QuantArray; GSI-Lumonics). Intensity values were imported into Genespring 6 (Silicon Genetics) and per spot and per chip normalizations were performed. Intensity ratios were normalized to a self-to-self comparison of the control sample. Genes significantly regulated in response to FOXO3a.A3-ER activation were identified by performing a statistical group analysis with a P value of <0.05 and by applying a restriction on change of twofold. To rule out effects of 4-OHT, RNA from DLD-1 cells treated with 100 nM 4-OHT was used in a control experiment. Probes were annotated according to the Hver1.2.1_35 annotation provided by the Sanger Microarray Facility (see http://www.sanger.ac.uk/Projects/Microarrays/informatics/annotation.shtml). All microarray data have been submitted to ArrayExpress (EBI).
Transfections and reporter assays.
DL23 cells were transiently transfected using Lipofectamine Plus reagent (Gibco-BRL) in serum-free medium (optiMEMI; Invitrogen). M11 cells were transfected using Effectene (QIAGEN) according to the manufacturer's instructions. Cells were harvested in passive lysis buffer (Promega), and luciferase activity was determined using the luciferase reporter assay kit (Promega) and a Berthold Bioluminat LB luminometer. Activity of firefly luciferase was normalized to the activity obtained from a cotransfected expression construct for Renilla luciferase (phRG-TK; Promega).
siRNA experiments.
For transient silencing of Mxi1 expression, DL23 cells were transfected with 100 nM of a small interfering RNA (siRNA) oligonucleotide specific for Mxi1 or a mixture of three Mxi1-specific sequences (33 nM each) using DharmaFECT 3 reagent (Dharmacon) in serum-free medium (optiMEMI; Invitrogen). Cells were split 24 h posttransfection and incubated for an additional 24 h prior to stimulation with 4-OHT or solvent for 16 or 24 h, as indicated.
The following siRNA oligonucleotides were used: Mxi1-1 (17300), Mxi1-2 (17207), Mxi1-3 (17113), Mad1-1 (114221), Mad1-2 (106784), Mad1-3 (106785), p27 (16104), and Silencer negative control 1 (all from Ambion) and deconvoluted SMARTpools for Mad3, Mad4, and c-Myc (Dharmacon).
Retroviral vectors expressing shRNA specific for Mxi1 or Mad1 were obtained from the NKI RNAi library. The shRNA expression cassette was subcloned into pMSCV-BLAST (NKI, Amsterdam, The Netherlands).
Reverse transcription-PCR.
Total RNA was extracted using RNeasy kits (QIAGEN). Total RNA (1 to 5 μg) was used for first-strand cDNA synthesis using oligo(dT) primers and SuperScript II reverse transcriptase (Invitrogen). Real-time PCR was performed with SYBR Green PCR master mix (Applied Biosystems) in 96-well plates using the Chromo 4 system (MJ Research). All reactions were performed in duplicate, and experiments were repeated at least three times. The relative amount of mRNA was calculated using the comparative CT method after normalization to GAPDH. Primer sequences are listed in the supplemental material.
Chromatin immunoprecipitation (ChIP) assay.
Immunoprecipitation of FOXO3a bound to chromatin has been described previously (18). Cells were fixed in 1% (wt/vol) formaldehyde for 10 min followed by addition of 0.136 M glycine and incubation for a further 10 min. Cells were washed and sonicated five times for 10 seconds each in 400 μl sonication buffer (Upstate). The lysate was cleared by centrifugation at 13,000 × g and diluted 10 times with diluent buffer (Upstate). The chromatin solution was precleared with 2 mg of sonicated herring sperm DNA (Sigma) and 45 μl of a 50% (vol/vol) slurry of protein A or G-Sepharose (Pharmacia). The supernatant was either incubated with an anti-FOXO3a antibody raised against the C-terminal region of the protein, an anti-Mxi-1 antibody (sc-1042X; Santa Cruz), an anti-Myc antibody (sc-764X), or an isotype control antibody (Babco) for 16 h at 4°C with rotation and an additional 2 h in the presence of 2 mg of sonicated herring sperm DNA and 45 μl of a 50% (vol/vol) slurry of protein A- or G-Sepharose. The beads were then washed and DNA was extracted with 350 μl of 1% SDS-1.1 M NaHCO3 and incubated at 65°C for 16 h with 0.2 M NaCl to reverse cross-linking followed by 1 h of proteinase K digestion at 45°C. DNA was purified using the QIAGEN PCR purification kit. PCR was performed in the presence of 2.5 mM MgCl2, and the annealing temperature was 55°C.
We used the following primers: Mxi1A forward (5′-CACACTGGTTCCCTTCTTCCC-3′) and reverse (5′-CGGAATGGAAAAAAATACCTGATG-3′), Mxi1B forward (5′-TGCTAAAGATGTAACAAAGACGGG-3′) and reverse (5′-GCCTGAGAAGGCTCCGGAAA-3′), and Mxi1C forward (5′-CAAGTGTTGTTTTTCTGTTG-3′) and reverse (5′-CCGTGGAGAGCTGTTTG-3′), all for 28 cycles. APEX forward (5′-GGGGACCTAAGTGTCC-3′) and reverse (5′-GAGCAACCCCGTATCTG-3′) were used for 27 cycles. FOXM1 forward (5′-CGGAATGCCGAGACAAGG-3′) and reverse (5′-TCCGCTGTTTGAAATTGGC-3′) were used for 30 cycles.
Microarray data accession number.
The microarray data in this study have been submitted to ArrayExpress and assigned accession number E-MEXP-721.
RESULTS
The transcriptional program induced by FOXO3a activation reveals repression of Myc target genes.
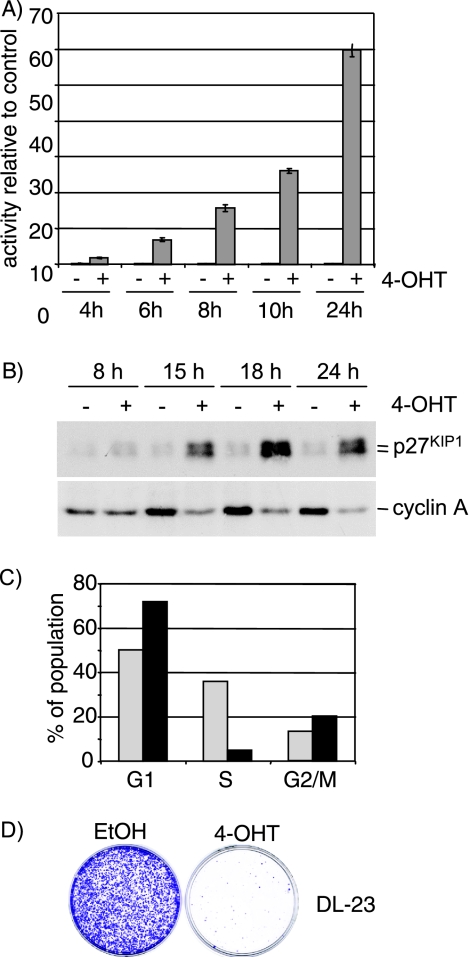
In order to elucidate how FOXOs affect diverse cellular processes, such as cell cycle progression, the stress response, and transformation, we made use of an inducible version of the FOXO3a protein fused to the hormone binding domain of the human estrogen receptor (FOXO3a.A3-ER), in which all three Akt phosphorylation sites have been mutated to alanine. FOXO3a.A3-ER was stably expressed in the human colon carcinoma cell line DLD-1 (36). Stimulation of FOXO3a.A3-ER-expressing cells (DL23) with the estrogen analogue 4-OHT induced a rapid and sustained activation of FOXO-dependent transcription indicated by activation of a DBE reporter construct (Fig. 1A). As previously described for these cells (36), activation of FOXO3a.A3-ER led to induction of p27KIP1 (Fig. 1B). We also observed a reduction of cyclin A expression, as well as a reduced proportion of cells incorporating BrdU, indicating cell cycle arrest (Fig. 1B and C). Furthermore, long-term activation of FOXO3a.A3-ER resulted in substantial inhibition of cell proliferation (Fig. 1D).
FIG. 1.
Activation of FOXO3a.A3-ER induces cell cycle arrest in DL23 cells. (A) Time course of induction of DBE-dependent transcription in response to FOXO3a activation. DL23 cells were transfected with 500 ng DBE-luc. At 24 h posttransfection, cells were treated with 100 nM 4-OHT or solvent (ethanol) for the indicated times. Values represent firefly luciferase activities relative to solvent-treated cells and are normalized to the activity of a cotransfected Renilla luciferase construct. (B) Time course of induction of cell cycle regulators. DL23 cells were treated with 100 nM 4-OHT or solvent (ethanol) for the indicated times, and total cell lysates were analyzed for the expression of p27KIP1 or cyclin A1 by immunoblotting. (C) DL23 cells were treated with solvent (light bars) or 100 nm 4-OHT (black bars) for 16 h prior to addition of 10 mM BrdU for 30 min. Proportions of BrdU-positive cells were determined by FACS analysis. (D) Colony formation assay. DL23 cells were seeded at clonal density and grown in the presence of 100 nM 4-OHT or solvent for 10 days. Cells were fixed and stained with crystal violet.
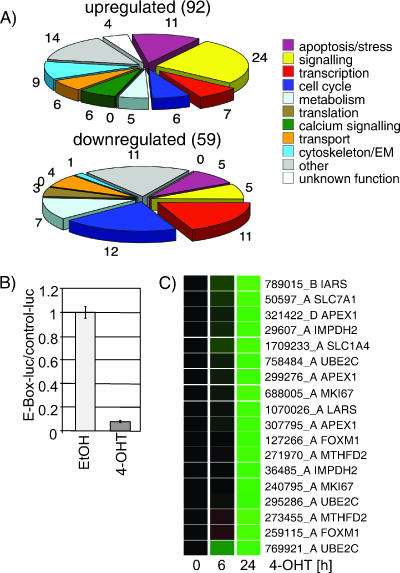
We analyzed gene expression profiles from DL23 or parental DLD-1 cells, after 6 or 24 h of 4-OHT treatment, using cDNA microarrays. A list of 151 probes that show significant changes in signal intensity after FOXO3a activation can be found in Table S1 in the supplemental material. Figure 2A shows pie charts representing the number of genes in each category found to be up- or downregulated in response to FOXO3a activation. We observed that a significant number of cell cycle regulators are downregulated by FOXO, i.e., 12 downregulated probes mapping to seven genes, compared to 6 upregulated probes mapping to two genes (Fig. 2A). This is consistent with its role in inhibition of proliferation. We also noticed that a number of genes involved in transcriptional regulation are affected by FOXO3a activation.
FIG. 2.
Activation of FOXO3a.A3-ER in DL23 cells results in inhibition of Myc-dependent transcription. (A) DL23 or DLD-1 cells were treated with 100 nM 4-OHT or solvent for 6 or 24 h. Relative mRNA abundance was measured by comparative hybridization to cDNA microarrays. Genes regulated in response to FOXO3a activation were grouped according to their GO annotations. The pie charts represent the number of genes in each class found to be induced or repressed in response to FOXO3a activation. (B) Repression of E-box-dependent transcription in response to FOXO3a activation. DL23 cells were transfected with 500 ng of a reporter construct containing a series of eight E-boxes (MycT-luc) or the corresponding control vector (CT-luc) prior to treatment with 4-OHT or solvent for 24 h. Values represent the relative activities of MycT-luc normalized to the activity of CT-luc and a cotransfected Renilla luciferase reporter. (C) Expression profile of Myc target genes in DL23 cells. The panel shows expression profiles of 18 cDNA clones which were found to be downregulated in response to FOXO3a.A3-ER activation and mapped to 10 Myc target genes. Green indicates downregulation relative to solvent-treated control cells. Results have been reproduced by qPCR.
It has previously been postulated that FOXO factors might repress Myc-dependent transcription by inhibiting formation of the preinitiation complex and loading of polymerase II (8). Figure 2B shows that activation of FOXO3a for 24 h resulted in a 10-fold reduction of activity of an E-box-containing reporter construct in DL23 cells. We compared our data set of genes regulated in response to FOXO3a activation to a public database of Myc target genes (www.myc-cancer-gene.org). We found that 18 out of 59 probes showed a reduction in signal intensity after FOXO3a activation code for genes that have previously been identified as Myc target genes. Expression profiles of these 18 probes (coding for 11 genes) following FOXO3a.A3-ER activation in DL-23 cells are shown in Fig. 2C. We noticed that downregulation of most genes was only apparent after 24 h of FOXO3a activation, suggesting that it might be an indirect event requiring synthesis of intermediate factors (Fig. 2C). To confirm that these potential Myc target genes are regulated by Myc in the cell system used here, we quantified mRNA levels following transfection of siRNA oligonucleotides targeting c-myc expression. Partial silencing of c-myc resulted in reduced expression of the majority of these potential Myc target genes (see Fig. S1 in the supplemental material).
FOXO3a induces expression of transcriptional repressors of the Mad/Mxd family.
Among the regulators of transcription found to be upregulated in response to FOXO3a activation were Mad1 and Mxi1, two members of the Mad/Mxd family of transcriptional repressors, as well as the Myc binding partner Max. Mad/Mxd proteins antagonize Myc-dependent transcription by heterodimerization with Max, binding to E-boxes and recruiting the Sin3/HDAC histone deacetylase activity (41). We therefore set out to investigate whether induction of Mad/Mxd proteins by FOXO3a could be involved in repression of Myc-dependent transcription.
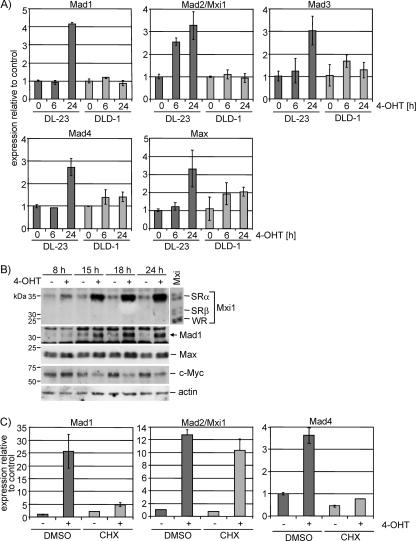
We then analyzed expression of all four Mad/Mxd family members, Mad1, Mxi1 (also termed Mad2), Mad3, and Mad4, in DL23 and parental DLD1 cells. Activation of FOXO3a.A3-ER by 4-OHT resulted in a two- to fourfold increase in mRNA abundance of all Mad/Mxd family members in DL23 cells, while the same treatment had no effect in parental cells (Fig. 3A). While the induction of Mad1, Mad3, and Mad4 mRNA was only apparent after 24 h, Mxi1 expression was already induced after 6 h of FOXO3a activation. Likewise, induction of Mxi1 protein was readily detectable after 8 h of FOXO3a activation, while Mad1 protein started to appear only after 15 h (Fig. 3B). We did not observe any changes in expression of Mnt or Miz1 (data not shown), and 4-OHT treatment had no effect in DLD1 cells (see Fig. S2 in the supplemental material).
FIG. 3.
Changes in expression of members of the Myc/Max/Mxd network in response to FOXO3a activation. (A) DL23 and DLD-1 cells were treated with 100 nM 4-OHT or solvent for 6 or 24 h. Expression of Mad1, Mad2/Mxi1, Mad3, Mad4, and Max was determined by qPCR. It should be noted that CT values for Mad3 were very high, suggesting low abundance of the transcript. (B) DL23 cells were treated with 100 nM 4-OHT or solvent for 8, 10, 15, and 24 h, and total cell lysates were used to detect expression of Mxi1, Mad1, Max, and Myc by immunoblotting. Lysates from cells transfected with expression vectors for Mxi1-SRα, -SRβ, and -WR were loaded as size controls, and actin is shown as a loading control. (C) Mxi1 but not Mad1 or Mad4 is a direct target of FOXO3a. DL23 cells were stimulated with 100 nM 4-OHT in the presence of 2 μg/ml cycloheximide or dimethyl sulfoxide (DMSO) for 24 h. Expression of Mad1, Mad2/Mxi1, and Mad4 was determined by qPCR.
Induction of Mxi1 by FOXO3a was also observed in a mammary epithelial cell line (MCF-10A) expressing the FOXO3a.A3-ER fusion protein (clone M11) (see Fig. S3 and S5B in the supplemental material). To exclude potential gain-of-function activity of strong activation of FOXO3a, we performed a dose-response experiment using low concentrations of 4-OHT (see Fig. S4 in the supplemental material). Mxi1 expression was readily induced by FOXO3a activation using only 10 nM 4-OHT.
Activation of FOXO3a for 24 h also led to a threefold increase in Max expression, thus confirming our microarray result (Fig. 3A). However, the observed increase in mRNA resulted in only a minor change in Max protein levels (Fig. 3B). Expression of c-Myc protein was reduced by about 50% after 24 h of FOXO3a activation (Fig. 3B). Thus, we concluded that activation of FOXO3a resulted in a concerted induction of Myc antagonists while simultaneously reducing c-Myc protein levels.
We next asked whether the temporal differences in induction of Mxi1 and other Mad/Mxd proteins are due to differences in the mechanism of activation. Stimulation of DL23 cells with 4-OHT in the presence or absence of cycloheximide revealed that induction of Mxi1 mRNA, but not Mad1 or Mad4 mRNA, is independent of de novo protein synthesis (Fig. 3C).
FOXO3a induces Mxi1-SRα expression through binding to conserved DBEs within the first intron.
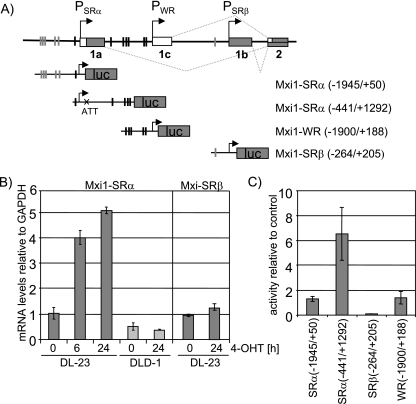
As induction of the Mxi1 transcript in response to FOXO3a activation was independent of de novo protein synthesis, we analyzed the human Mxi1 locus for the presence of FOXO3a binding sites. The human Mxi1 gene has been mapped to chromosome 10q24-25 (63, 72). The Mxi1 locus codes for three individual transcripts (Mxi1-SRα, Mxi1-SRβ, and Mxi1-WR), which differ in their first exons (15, 17). We identified a cluster of highly conserved DBEs, depicted in Fig. 4A. Sequences with similarity to DBEs but poorer conservation were found elsewhere in the region. Using primers specific for Mxi1-SRα and Mxi1-SRβ, we were able to show that the SRα transcript is induced upon FOXO3a activation, while the transcript specific for Mxi1-SRβ shows no significant change in expression (Fig. 4B). We were unable to generate quantitative PCR (qPCR) primers specific for Mxi1-WR due to high sequence homology with the other isoforms.
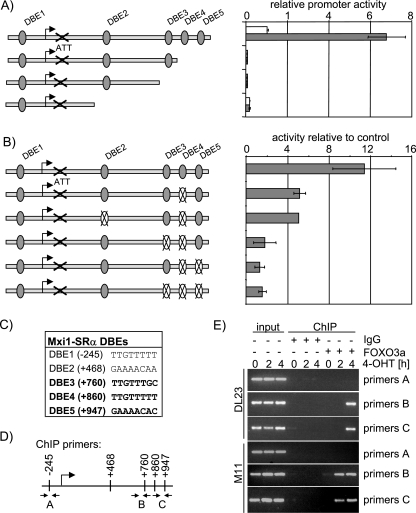
FIG. 4.
FOXO3a activation induces expression of Mxi1-SRα. (A) Schematic overview of the human Mxi1 locus. The human Mxi1 locus contains three transcriptional start sites that give rise to three transcripts (Mxi1-SRα, Mxi1-SRβ, and Mxi1-WR) that differ in their first exon. The locations of highly conserved DBEs are indicated by black bars. Sequences with similarity to DBEs but poorer conservation are indicated by gray bars. Luciferase reporter constructs used in panel C are depicted. (B) cDNA from DL23 or DLD-1 cells treated with 4-OHT for 6 or 24 h was used to perform isoform-specific qPCR. Values represent expression levels relative to solvent-treated controls. (C) DL23 cells were transfected with the luciferase reporter constructs indicated in panel A and treated with 100 nM 4-OHT or solvent for 24 h. Values represent luciferase activities relative to solvent-treated control and are normalized to the activity of a cotransfected Renilla luciferase construct.
The Mxi1-SRα transcript encodes a 295-amino-acid polypeptide of approximately 33 kDa (15, 17). This is consistent with the molecular mass of a single Mxi1 protein band detected by immunoblotting after FOXO3a activation (Fig. 3B). Taken together these data indicate that only Mxi1-SRα is induced after FOXO3a activation.
To further analyze regulation of the Mxi1 gene by FOXO3a, we generated luciferase reporter constructs containing sequences upstream of the transcriptional start sites of the three Mxi1 transcripts [Mxi1-SRα(−1945/+50), Mxi1-SRβ(−264/+205), and Mxi1-WR(−1900/+188)]. Transient transfection in DL23 cells showed that these constructs were not induced in response to FOXO3a activation (Fig. 4C). However, when we used a reporter construct including sequences from the first intron of the Mxi1-SRα gene [Mxi1-SRα(−441/+1292)], we observed a five- to eightfold induction of luciferase activity in response to FOXO3a activation (Fig. 4C). Serial deletion of the promoter revealed that a region containing a cluster of three DBEs (DBE3, -4, and -5, depicted in Fig. 5C) is required for induction of Mxi1-SRα expression by FOXO3a (Fig. 5A). Mutation of DBE4 (located at position +860) reduced FOXO3a-dependent activation of the Mxi1-SRα promoter twofold, and mutation of two or all three sites completely abolished the induction (Fig. 5B). Mutation of DBE2 (located at position +468) did not have any effect on induction of the promoter by FOXO3a (Fig. 5B). As DBE1 was also present in the luciferase construct containing sequences upstream of PSRα [Mxi1-SRα(−1945/+50)] which did not respond to FOXO3a activation, we conclude that DBE1 does not contribute to induction of Mxi1-SRα by FOXO3a. Similar results were obtained in MCF-10A cells expressing the FOXO3a.A3-ER fusion protein (clone M11) (data not shown). These results indicate that induction of Mxi1-SRα expression by FOXO3a is mediated through three highly conserved DBEs located in the first intron.
FIG. 5.
FOXO3a activates Mxi1-SRα through conserved DBEs located in the first intron. (A) Deletion analysis of the Mxi1-SRα promoter. Luciferase reporter constructs representing a series of deletions of the Mxi1-SRα downstream promoter were transfected into DL23 cells prior to induction with 4-OHT (dark bars) or solvent (light bars) for 24 h. Values represent luciferase activity relative to the activity of the full-length construct in solvent-treated cells and are normalized to the activity of a cotransfected Renilla luciferase reporter. (B) Mutation analysis of Mxi1-SRα downstream promoter. Luciferase reporter constructs in which the indicated DBEs were disrupted by mutation were transfected into DL23 cells prior to induction with 4-OHT for 24 h. Values represent luciferase activities relative to solvent-treated controls and are normalized to the activity of a cotransfected Renilla luciferase reporter. (C) Sequence of the DBEs located in the first intron of the Mxi1-SRα gene. Positions are given relative to the putative transcriptional start site. (D) Schematic representation of the regions amplified by primer pairs used in panel E. (E) FOXO binding to the DBE-containing region of the Mxi1 promoter. DL23 or MCF-10A cells expressing the FOXO3a.A3-ER fusion protein (M11) were treated with 100 nM 4-OHT for 2 or 4 h. Chromatin immunoprecipitations using a FOXO3a-specific antibody or unspecific serum (immunoglobulin G [IgG]) were analyzed using primer pairs depicted in panel D.
To confirm direct involvement of FOXO3a in Mxi1-SRα induction, we performed ChIP experiments using a FOXO3a-specific antibody or matched control immunoglobulins. Chromatin precipitates from DL23 or M11 cells treated with 4-OHT for 2 or 4 h were analyzed for the presence of sequences from the Mxi1-SRα promoter by PCR using primer pairs complementary to different regions of the Mxi1 promoter (depicted in Fig. 5D). Figure 5E shows that 4-OHT treatment induces specific binding of FOXO3a to regions flanked by primer pairs B or C, while a region upstream of the transcriptional start site, flanked by primer pair A, shows no binding. Furthermore, binding of FOXO3a to the Mxi1-SRα promoter can be detected in DL23 cells only after 4 h of 4-OHT treatment, while it is already observed after 2 h in M11 cells. This is consistent with different kinetics of Mxi1-SRα induction in the two different cell lines. The Mxi1-SRα transcript is already strongly induced after 4 h of 4-OHT treatment in M11 cells (see Fig. S5 in the supplemental material).
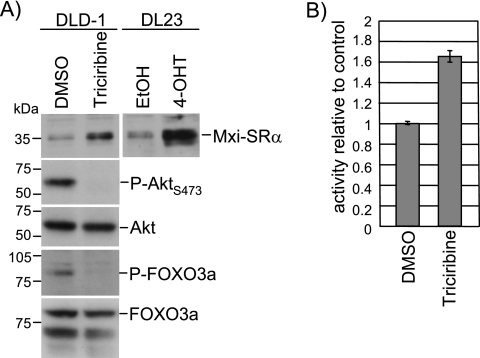
Activation of endogenous FOXO proteins in response to Akt inhibition induces Mxi1-SRα expression.
We next asked whether activation of endogenous FOXO proteins also activates Mxi1-SRα expression. We used the specific inhibitor triciribine, which has been shown to prevent phosphorylation and activation of Akt (74). Treatment of DLD-1 cells with triciribine for 24 h resulted in complete loss of serine 473 phosphorylation of Akt as well as threonine 32 phosphorylation of FOXO3a (Fig. 6A). Expression of Mxi1-SRα was induced in triciribine-treated DLD-1 cells, albeit to a lesser extent than following FOXO3a.A3-ER activation in DL23 cells (Fig. 6A). Activity of the Mxi1-SRα(−441/+1292) luciferase reporter construct was also increased following triciribine treatment of DLD-1 cells (Fig. 6B). These results indicate that activation of endogenous FOXO proteins induces Mxi1-SRα expression.
FIG. 6.
Chemical inhibition of Akt induces expression of Mxi1-SRα in DLD-1 cells. (A) DLD-1 cells were treated with 1 μg/ml triciribine or solvent (dimethyl sulfoxide [DMSO]) for 24 h and analyzed for expression of Mxi1-SRα, Akt, and FOXO3a by immunoblotting. Phosphorylation of Akt and FOXO3a was investigated using phospho-specific antibodies. (B) DL23 cells were transfected with a luciferase construct containing sequences of the Mxi1-SRα promoter, Mxi1-SRα(−441/+1292), and treated with 1 μg/ml triciribine or solvent (DMSO) for 24 h. Values were normalized to the activity of a cotransfected Renilla luciferase reporter.
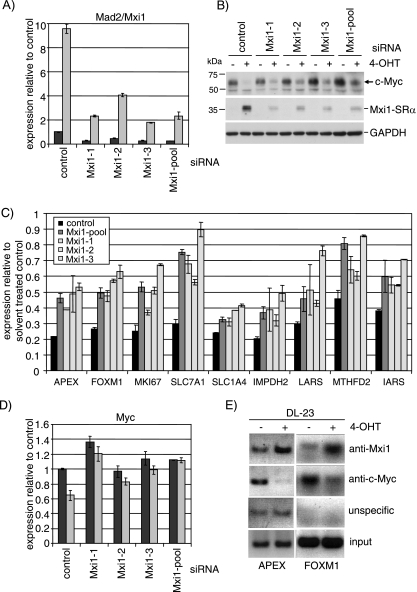
Induction of Mxi1-SRα contributes to repression of Myc-dependent gene expression by FOXO3a.
We next asked whether Mxi1-SRα could be involved in the repression of Myc-dependent transcription observed in response to FOXO3a activation in DL23 cells (Fig. 2B and C). To address this question, we employed RNA interference to silence Mxi1 expression in these cells. Figure 7A shows a quantitation of Mxi1 mRNA by qPCR after transfection of siRNA oligonucleotides specific to Mxi1 or an unspecific control. Induction of Mxi1 mRNA by FOXO3a is reduced by 60 to 90% following transfection of either 100 nM of the individual oligonucleotides or a combination of three Mxi1-specific RNAi oligonucleotides at a concentration of 33 nM each. Silencing of the Mxi1 transcript by siRNA is also accompanied by a significant reduction in Mxi1-SRα protein (Fig. 7B, middle panel).
FIG. 7.
Induction of Mxi1 contributes to repression of Myc-dependent transcription in response to FOXO3a activation. (A) Silencing of Mxi1 expression using siRNA. DL23 cells were transfected with 100 nM of individual RNA oligonucleotides targeting Mxi1 (Mxi1-1, Mxi1-2, or Mxi1-3), a mixture of the three sequences (Mxi1-pool), or scrambled control prior to stimulation with 4-OHT for 24 h. Expression of Mxi1 after 24 h of 4-OHT (gray bars) or solvent (ethanol) treatment (black bars) was detected by qPCR. (B) Cells transfected with Mxi1 or scrambled control oligonucleotides were treated as for panel A. Expression levels of Mxi1-SRα and c-Myc were detected by immunoblotting. GAPDH is shown as a loading control. (C) Silencing of Mxi1 rescues repression of Myc target gene expression by FOXO3a. The same RNA samples shown in panel A were analyzed for expression of the same nine genes shown in Fig. 2C by qPCR. Values represent expression levels relative to solvent-treated control after transfection of scrambled control (back bars), a pool of three oligonucleotides targeting Mxi1 (dark gray bars), or individual oligonucleotides (light gray bars). (D) Expression of c-Myc mRNA in DL23 cells after silencing of Mxi1 by siRNA and subsequent activation of FOXO3a.A3-ER. Values represent relative mRNA abundance in 4-OHT-treated cells (gray bars) compared to solvent controls (black bars). (E) DL23 cells were treated with 100 nM 4-OHT for 24 h. Chromatin immunoprecipitations using antibodies specific for Mxi1 (top panel), c-Myc (second panel), or unspecific serum (immunoglobulin G [unspecific]) were analyzed by PCR using primers derived from the human APEX and FOXM1 promoters.
We next asked whether blocking induction of Mxi1-SRα had any effect on FOXO3a-dependent inhibition of Myc-dependent transcription and went on to analyze expression of FOXO3a-regulated Myc target genes that had been identified in the microarray analysis (Fig. 2C). Silencing of Mxi1 (individual siRNA oligonucleotides or a combination of all three sequences) substantially increased expression of all genes in the presence of 4-OHT (Fig. 7C). Furthermore, the level of derepression of Myc target genes in individual siRNA transfections correlated well with the efficiency of Mxi1 silencing as determined by qPCR (compare Mxi1-3 in Fig. 7A and C).
We conclude that Mxi1 contributes to repression of Myc target genes after FOXO3a activation. However, individual Myc target genes seem to differ in their response to Mxi1 silencing. Further investigation is required to elucidate whether this difference can be explained by variations in E-box sequences or other regulatory elements in the promoter regions of these genes.
Since we observed inhibition of expression of c-Myc following FOXO3a.A3-ER activation (Fig. 3B), we asked whether ablation of Mxi1 would affect Myc expression under the conditions used here. It had previously been shown that Mxi1 represses expression of the c-myc gene through inhibiting activation of the P2 core promoter by USF (38). Figure 7D shows a 40% reduction in c-myc mRNA levels following FOXO3a.A3-ER activation in DL-23 cells. Mxi1 silencing significantly reduced downregulation of c-myc mRNA expression and resulted in increased c-Myc protein levels after FOXO activation (Fig. 7B, top panel). These results indicate that Mxi1 contributes to repression of c-myc expression by FOXO3a.
We next asked whether Mxi1 could also directly inhibit expression of Myc target genes by binding to E-box sequences within their promoters. We chose the APEX and FOXM1 genes, as high-affinity E-boxes have been identified in the promoter sequences of these genes (20). Figure 7E shows increased binding of Mxi1 to the APEX and FOXM1 promoter in DL-23 cells after 24 h of 4-OHT treatment. Conversely, binding of c-Myc to the same promoter regions is reduced in response to FOXO3a activation (Fig. 7E, anti-c-Myc). This result strongly suggests that binding of Mxi1 and displacement of Myc from its target gene promoters contribute to repression of gene expression in response to FOXO3a activation.
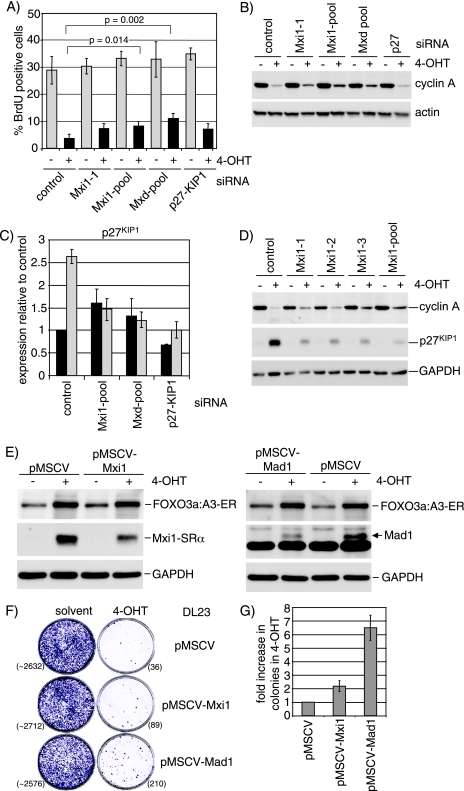
Silencing of Mad/Mxd family members reduces cell cycle arrest and growth inhibition by FOXO3a.
It has been shown previously that activation of FOXO3a and FOXO4 leads to induction of p27KIP1 expression and cell cycle arrest (46). However, subsequent studies found that FOXO4 was still able to induce cell cycle arrest in p27−/− mouse embryo fibroblasts due to induction of p130 as well as repression of cyclins D1 and D2 (36, 58). In order to study the role of induction of Mad/Mxd family proteins on FOXO3a-mediated inhibition of cell proliferation, we determined the proportion of cells in S phase after FOXO3a.A3-ER activation by BrdU labeling and FACS analysis. The data in Fig. 8A show an eightfold reduction in cells in S phase in response to 4-OHT treatment in DL-23 cells. Silencing of Mxi1 (Mxi1-1 and Mxi1-pool) or simultaneous knockdown of all Mad/Mxd family members (Mxd-pool) resulted in a significant increase in the percentage of cells in S phase (3.7% in the control compared to 7.1% and 11.1% in Mxi1-pool or Mxd-pool, respectively). Similar results were also obtained using different combinations of individual oligonucleotides targeting all Mad/Mxd family members (see Fig. S6 in the supplemental material). Interestingly, silencing of p27KIP1 did not have a significant effect on cell cycle arrest in response to FOXO3a activation (Fig. 8A) and could not further increase the number of cells in S phase when combined with knockdown of Mxi1 (data not shown).
FIG. 8.
Induction of Mxi1-SRα and other Mad/Mxd proteins contributes to inhibition of proliferation by FOXO3a. (A) DL23 cells were transfected with siRNA oligonucleotides targeting Mxi1 (Mxi1-1 and Mxi1-pool), a mixture of oligonucleotides targeting all Mad/Mxd family members simultaneously (Mxd-pool), or p27KIP1. At 48 h posttransfection, cells were reseeded at lower density and treated with 100 nM 4-OHT (black bars) or solvent (gray bars) for 16 h. DNA synthesis was detected by incorporation of BrdU and FACS analysis. Values represent the means and standard deviations of three independent experiments. P values were determined by performing unpaired t tests with equal variance. (B) Cells were treated as for panel A and analyzed for expression of cyclin A by immunoblotting. Actin is shown as a loading control. (C) DL23 cells were transfected with the indicated siRNA oligonucleotides as for panel A and treated with 100 nM 4-OHT (gray bars) or solvent (black bars) for 24 h. Expression of p27KIP1 mRNA was quantified by qPCR. (D) DL23 cells were transfected with three different siRNA oligonucleotides targeting Mxi1 expression (Mxi1-1, Mxi1-2, or Mxi1-3) or a pool of all three sequences (Mxi1-pool). At 48 h posttransfection, cells were treated with 100 nM 4-OHT or solvent for 24 h. Expression of cyclin A and p27KIP1 was analyzed by immunoblotting. GAPDH is shown as a loading control. (E) DL23 cells stably expressing pMSCV-Mxi1 (left panel), pMSCV-Mad1 (right panel), or empty vector (pMSCV) were treated with 100 nM 4-OHT (+) or solvent (−) for 24 h. Expression of the FOXO3a.A3-ER fusion protein as well as Mxi1 or Mad was analyzed by immunoblotting. GAPDH is shown as a loading control. (F) DL23 cells stably expressing pMSCV-Mxi1, pMSCV-Mad1, or empty vector were seeded at clonal density and grown in the presence of 100 nM 4-OHT or solvent (ethanol) for 14 days. Cells were fixed and stained with crystal violet. Numbers of colonies are given in brackets. Note that colony number of the solvent-treated cultures was estimated by counting three segments of the plate. (G) Quantification of colony formation experiments, performed as for panel B. The values are normalized to the empty vector control and represent the results of two independent experiments.
The efficiency of silencing of Mxi1, Mad1, Mad4, and p27KIP1 was quantified by qPCR and immunoblotting (Fig. 8C; see also Fig. S7 in the supplemental material). While expression of the mRNAs for Mxi1, Mad4, and p27KIP1 was reduced by at least 70% following siRNA transfection, we only observed a modest decrease in Mad1 expression (see Fig. S7A). We were unable to improve efficiency of Mad1 silencing using six different oligonucleotide sequences (data not shown). Analysis of Mad3 expression failed due to low abundance of this mRNA (data not shown).
The observed reduction in FOXO3a-dependent cell cycle arrest following silencing of Mxi1 or all Mad/Mxd proteins was accompanied by an increase in cyclin A expression (Fig. 8B and D). It has previously been shown that expression of p27KIP1 is repressed by c-Myc and that overexpression of c-Myc can abrogate induction of p27KIP1 in WEHI 231 cells (12, 76). We observed that ablation of Mxi1 by siRNA prevented efficient induction of p27KIP1 mRNA in response to FOXO3a activation (Fig. 8C) and resulted in a significant reduction in p27KIP1 protein (Fig. 8D).
Taken together, these data indicate that induction of Mxi1 and other Mad/Mxd proteins contributes to the induction of cell cycle arrest in response to FOXO3a activation in DL-23 cells.
To investigate the effect of stable ablation of Mad/Mxd family proteins on FOXO-mediated inhibition of cell growth, we generated DL-23 cells in which Mxi1 or Mad1 expression was stably silenced by RNA interference using a retroviral expression system. DL-23 cells stably expressing pMSCV-Mxi1 or pMSCV-Mad1 showed reduced induction of Mxi1 or Mad1 in response to FOXO3a activation, while expression of the FOXO3a.A3-ER fusion protein was similar between the different cell lines (Fig. 8E, top and middle panels). Both cell lines showed a small increase in the number of surviving colonies observed after 10 days of 4-OHT treatment (twofold for pMSCV-Mxi1 and sixfold for pMSCV-Mad1) (Fig. 8F and G). This result indicates that induction of Mad/Mxd family proteins can contribute to growth inhibition by FOXO3a. However, we still observed a strong reduction in colony number after FOXO3a activation compared to solvent-treated controls (Fig. 8F). Incomplete silencing, as well as induction of the remaining Mad/Mxd family members or other FOXO target genes (i.e., p27KIP1 and p130Rb2), could be responsible for the observed growth inhibition.
DISCUSSION
In this study, we investigated changes in gene expression in response to FOXO3a activation. We showed that FOXO3a induces expression of all four members of the Mad/Mxd family of transcriptional repressors. Moreover, FOXO3a induced Mxi1-SRα expression by binding to a cluster of conserved DBEs within the first intron of the gene. Activation of FOXO3a in response to chemical inhibition of Akt also resulted in increased expression of Mxi1-SRα. Silencing of Mxi1-SRα by RNAi prevented downregulation of c-myc by FOXO3a activation and partially restored expression of a number of Myc target genes. We also found increased binding of Mxi1 to the promoter regions of two previously identified Myc target genes (APEX and FOXM1) in response to FOXO3a activation. In addition, binding of Myc to the same regions was decreased, indicating a switch in promoter occupancy. Finally, abrogation of Mxi1 or all Mad/Mxd proteins significantly reduced cell cycle exit in response to FOXO3a activation, and stable silencing of Mxi1 or Mad1 increased the number of colonies after long-term FOXO3a activation, indicating that induction of Mad/Mxd proteins is involved in FOXO3a-dependent inhibition of proliferation. Taken together, these results provide evidence of an additional mechanism for the regulation of Myc activity by the PI3-kinase/Akt/FOXO pathway.
Previous studies have analyzed changes in gene expression induced by FOXO factors in renal carcinoma cells (FOXO1 [56]), prostate carcinoma cells (FOXO1 and FOXO3a [48]), or endothelial cells (FOXO1 [14]). Although these studies were performed using different cell lines and experimental systems, we observed some overlap between published data sets and our results (see Table S1 in the supplemental material). Xuan and Zhang (75) have identified Mxi1 as a potential FOXO target gene by searching for conserved DBE motifs in mammalian promoters. In addition, Mxi1 was found upregulated in response to FOXO1 expression in renal carcinoma cells (56). This induction seemed to be independent of DNA binding, since a mutant of FOXO1, in which the DNA binding domain had been altered by point mutation, was still able to induce Mxi1 expression (56). Our results, however, show that FOXO3a binds to conserved sequences in the first intron of the Mxi1-SRα gene. This discrepancy could be explained by differences in DNA binding specificity of different FOXO proteins or cell-type-specific differences. It is also possible that the FOXO1 mutant used in the above study has residual binding activity towards DBE sequences embedded in chromatin.
In our study, expression of all four members of the Mad/Mxd family was induced in response to FOXO3a activation. However, we found that only Mxi1 is a direct target. It seems unlikely that induction of Mxi1 by FOXO3a is involved in the regulation of other Mad/Mxd family members, since silencing of Mxi1 by siRNA did not block FOXO3a-induced upregulation of Mad1 expression (data not shown). The mechanism responsible for induction of other Mad/Mxd family proteins by FOXO3a remains to be elucidated.
Mad/Mxd proteins form heterodimers with Max proteins and compete with Myc/Max heterodimers for binding to E-box sequences (78). They recruit Sin3/histone deacetylases through an N-terminal Sin3 interaction domain (SID) and inhibit transcription through induction of chromatin remodelling (60). While Mad1, Mad3, and Mad4 all contain SIDs, Mxi1 isoforms differ in their ability to bind Sin3. Both Mxi1-SR isoforms (SR, strong repressor) bind Sin3, but Mxi1-WR (WR, weak repressor) lacks a SID and inhibits transcription only by competing with Myc (15, 17, 59). The Mxi1-SRα isoform (also termed Mxi1-0) has been identified recently by two independent studies (15, 17). While both studies showed binding of Mxi1-SRα to Sin3, they differed in their results regarding the function of this isoform. Engstrom et al. showed that Mxi1-0 localizes to the cytoplasm and is unable to inhibit Myc-dependent transcription. In contrast, Dugast-Darzacq and coworkers concluded that Mxi1-SRα shows increased affinity to Sin3 as well as enhanced repression in a transient-reporter assay compared to the Mxi1-SRβ isoform (15).
It is intriguing that FOXO3a specifically induces expression of the Mxi1-SRα isoform. Our results show that FOXO3a does not induce the Mxi1-WR gene despite several conserved DBEs upstream of the promoter (Fig. 3B and 4C). Further investigation is required to analyze whether sequences downstream of the cluster of DBEs are responsible for repression of the Mxi1-WR promoter.
We observed regulation of the Mxi1-SRα promoter through a cluster of highly conserved DBEs located in the first intron of the gene. Binding of FOXO3a to this region correlates with activation of Mxi1-SRα expression in two different cells lines (Fig. 5E; see also Fig. S5 in the supplemental material). The FOXO binding sites of most target genes are located upstream of their transcriptional start sites. However, the FOXO binding element within the p130RB2 gene has been mapped within the first intron (36), and a DBE has been identified at an exon-intron boundary of the rat bim promoter (26). Regulation of gene expression though sequences located in the first intron has also been described for other forkhead transcription factors (71).
It was shown previously that FOXO factors inhibit Myc-dependent transcription of the cyclin D2 gene by preventing the formation of the preinitiation complex and loading of RNA polymerase (8). In that study, Bouchard et al. showed binding of FOXO3a to a non-DBE-containing region of the cyclin D2 promoter in response to inhibition of PI3-kinase while binding of Myc-ER to the E-box was not affected.
Our results suggest that FOXO3a inhibits Myc-dependent transcription through induction of expression of Mad/Mxd family proteins, particularly Mxi1-SRα. Abrogation of Mxi1 expression by RNA interference significantly reduced FOXO3a-dependent inhibition of the panel of Myc target genes (Fig. 7C). Furthermore, we observed that silencing of Mxi1 reduced inhibition of c-Myc expression by FOXO3a activation (Fig. 7B and D). Indeed, regulation of the c-myc gene by Mxi1 has been described before (38). Mxi1 was found to block activation of the P2 promoter of c-Myc by USF through sequences located in the core promoter. Interestingly, inhibition of c-Myc expression did not require the Sin3 binding domain of Mxi1. It will be interesting to see whether inhibition of c-Myc expression in response to FOXO3a activation is mediated by Mxi1-SRα through the same mechanism.
It is possible that the increased expression of c-Myc alone could account for the observed derepression of Myc target genes. However, we only observed a moderate increase in c-Myc protein levels after Mxi1 silencing, indicating that c-Myc might be regulated by FOXO3a through posttranscriptional mechanisms (Fig. 7B). Furthermore, we observed direct binding of Mxi1 to the promoter sequences of two Myc target genes, APEX and FOXM1 (Fig. 7E). The gene for the APEX nuclease, a multifunctional DNA repair enzyme, has been identified as a Myc target gene by microarray and SAGE analysis (29, 47) and contains three high-affinity Myc binding sites in its promoter (20). The gene for the forkhead box protein M1 (FOXM1) contains a single high-affinity Myc binding site (20). We found that binding of Mxi1 to both promoters was increased upon FOXO3a activation while binding of c-Myc decreased, indicating a switch in promoter occupancy.
The observed derepression of Myc target gene expression by Mxi1 silencing, although significant, was only partial and showed variation between the different genes tested here (Fig. 7C). This is most likely due to incomplete silencing of Mxi1 or compensation by other Mad/Mxd family members. It seems likely that different E-box sequences have different affinities to Myc/Max, Mxi1/Max, or Mad/Max complexes. Indeed, sequence-specific gene regulation among the Myc/Mad family has been studied (52). A more detailed analysis of the E-box sequences present in different Myc target genes will be required to elucidate Mxi1- and Mad/Mxd-specific effects on gene expression.
The results presented in this study raise questions about the role of induction of Mxi1-SRα and other Mad/Mxd family members by FOXO3a. Mxi1-SRβ has been shown to inhibit proliferation of prostate carcinoma and glioblastoma cell lines when overexpressed (67, 73). Similarly, Mad1 overexpression inhibits proliferation in different cell lines (31, 57) and blocks Myc/Ras-dependent tumor formation (11).
We observed a significant increase in the percentage of cells undergoing DNA synthesis following silencing of Mxi1 or simultaneous silencing of all Mad/Mxd family members (Fig. 8A; see also Fig. S6 in the supplemental material). This was accompanied by increased expression of cyclin A (Fig. 8B), while induction of p27KIP1 mRNA and protein expression by FOXO3a were greatly diminished (Fig. 8C and D). It has previously been shown that c-Myc regulates expression of p27 through an initiator element within its promoter (76) and that overexpression of c-Myc abrogates FOXO-dependent induction of p27KIP1 in response to inhibition of PI3-kinase or engagement of the B-cell receptor in WEHI 231 cells (12). It seems possible, therefore, that derepression of c-Myc contributes to the reduction in FOXO3a-induced p27KIP1 expression following Mxi1 silencing. However, since reduction of p27KIP1 protein levels was more pronounced than changes in mRNA levels, involvement of posttranscriptional mechanisms regulating p27KIP1 protein stability seems likely.
We also observed a small but significant effect on FOXO3a-induced repression of colony formation after stable silencing of Mxi1 or Mad1 in DL-23 cells (Fig. 8F and G). The failure to obtain a more pronounced increase in colony number is most likely due to incomplete silencing as well as compensation by other Mad/Mxd family proteins.
Loss of function of the mxi1 gene has been implicated in the development of prostate cancer (16, 27, 55), although its role is still somewhat controversial (37). Interestingly, mxi1−/− mice exhibit hyperplasia in several tissues, including prostate, and show increased sensitivity to chemical carcinogenesis (61), while deletion of the mad1 gene in mice had no obvious effect, most likely due to redundancy between different Mad/Mxd family members (21). Downregulation of FOXO factors has been observed during progression to the androgen-independent state in LnCap prostate cancer cells (39, 42). It is thus possible that induction of Mxi1-SRα by FOXO factors contributes to tumor suppression through the PTEN/PI3-kinase pathway.
Our results also highlight an intriguing parallel between daf-16 in Caenorhabditis elegans and mammalian FOXO factors. Mdl-1, the worm orthologue of Mad, has been identified as a daf-16 target gene in a longevity screen (49). Mdl-1 binds DNA as a heterodimer with the C. elegans Max orthologue Mxl-1, and it is able to substitute for mammalian Mad in repressing Myc-dependent transcription as well as transformation by Myc and Ras in mammalian cells (77). Abrogation of Mdl-1 expression by RNAi resulted in a ∼10% decrease in life span (49). Several other daf-16 target genes that increase longevity in C. elegans, like SOD or catalase, are involved in stress response mechanisms in mammalian cells (19, 34). Aberrant activation of c-Myc has been shown to induce DNA damage and increase oxidative stress, possibly by deregulating mitochondrial oxidative metabolism, resulting in production of reactive oxygen species (1, 69). Therefore, induction of expression of Mad/Mxd proteins may be a mechanism by which FOXO factors limit activation of Myc-dependent gene expression to prevent excessive reactive oxygen species production.
Recent work has established a role for FOXO proteins in the maintenance of hematopoietic stem cells. Conditional deletion of three members of the FOXO family resulted in a decrease in specific hematopoietic stem cell lineages and decreased resistance to oxidative stress (68). It will be interesting to see whether regulation of the Myc/Max/Mxd network by FOXO factors contributes to stem cell maintenance.
Taken together, our results provide evidence for a novel mechanism by which FOXO3a can repress the expression of Myc target genes. Induction of Mad/Mxd proteins contributes to repression of Myc target genes and is required for efficient cell cycle arrest in response to FOXO3a. Inhibition of Mad/Mxd protein expression through inactivation of FOXO3a by the PI3-kinase/Akt pathway may thus be required for the expression of Myc target genes and could contribute to cell transformation.
Supplementary Material
Acknowledgments
We thank Subham Basu, Gordon Peters, Bernhard Luescher, Claudio Santos, Bruno Amati, and Martin Eilers for helpful discussions and/or reagents and Selma El Messaoudi for her help and encouragement with ChIP experiments. We also thank the Microarray Facility at the Sanger Institute for production of cDNA microarrays and Gavin Kelly from the CRUK Bioinformatics and Biostatistics Service for advice. We thank the CRUK LRI Equipment Park and FACS Laboratory staff for valuable assistance.
This work was funded by Cancer Research UK.
Footnotes
Published ahead of print on 23 April 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adhikary, S., and M. Eilers. 2005. Transcriptional regulation and transformation by Myc proteins. Nat. Rev. Mol. Cell Biol. 6:635-645. [DOI] [PubMed] [Google Scholar]
- 2.Altomare, D. A., and J. R. Testa. 2005. Perturbations of the AKT signaling pathway in human cancer. Oncogene 24:7455-7464. [DOI] [PubMed] [Google Scholar]
- 3.Aoki, M., H. Jiang, and P. K. Vogt. 2004. Proteasomal degradation of the FoxO1 transcriptional regulator in cells transformed by the P3k and Akt oncoproteins. Proc. Natl. Acad. Sci. USA 101:13613-13617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bakker, W. J., M. Blazquez-Domingo, A. Kolbus, J. Besooyen, P. Steinlein, H. Beug, P. J. Coffer, B. Lowenberg, M. von Lindern, and T. B. van Dijk. 2004. FoxO3a regulates erythroid differentiation and induces BTG1, an activator of protein arginine methyl transferase 1. J. Cell Biol. 164:175-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barthel, A., D. Schmoll, K. D. Kruger, G. Bahrenberg, R. Walther, R. A. Roth, and H. G. Joost. 2001. Differential regulation of endogenous glucose-6-phosphatase and phosphoenolpyruvate carboxykinase gene expression by the forkhead transcription factor FKHR in H4IIE-hepatoma cells. Biochem. Biophys. Res. Commun. 285:897-902. [DOI] [PubMed] [Google Scholar]
- 6.Bois, P. R., and G. C. Grosveld. 2003. FKHR (FOXO1a) is required for myotube fusion of primary mouse myoblasts. EMBO J. 22:1147-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borkhardt, A., R. Repp, O. A. Haas, T. Leis, J. Harbott, J. Kreuder, J. Hammermann, T. Henn, and F. Lampert. 1997. Cloning and characterization of AFX, the gene that fuses to MLL in acute leukemias with a t(X;11)(q13;q23). Oncogene 14:195-202. [DOI] [PubMed] [Google Scholar]
- 8.Bouchard, C., J. Marquardt, A. Bras, R. H. Medema, and M. Eilers. 2004. Myc-induced proliferation and transformation require Akt-mediated phosphorylation of FoxO proteins. EMBO J. 23:2830-2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brunet, A., A. Bonni, M. J. Zigmond, M. Z. Lin, P. Juo, L. S. Hu, M. J. Anderson, K. C. Arden, J. Blenis, and M. E. Greenberg. 1999. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96:857-868. [DOI] [PubMed] [Google Scholar]
- 10.Brunet, A., L. B. Sweeney, J. F. Sturgill, K. F. Chua, P. L. Greer, Y. Lin, H. Tran, S. E. Ross, R. Mostoslavsky, H. Y. Cohen, L. S. Hu, H. L. Cheng, M. P. Jedrychowski, S. P. Gygi, D. A. Sinclair, F. W. Alt, and M. E. Greenberg. 2004. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 303:2011-2015. [DOI] [PubMed] [Google Scholar]
- 11.Cerni, C., B. Skrzypek, N. Popov, S. Sasgary, G. Schmidt, L. G. Larsson, B. Luscher, and M. Henriksson. 2002. Repression of in vivo growth of Myc/Ras transformed tumor cells by Mad1. Oncogene 21:447-459. [DOI] [PubMed] [Google Scholar]
- 12.Chandramohan, V., S. Jeay, S. Pianetti, and G. E. Sonenshein. 2004. Reciprocal control of Forkhead box O 3a and c-Myc via the phosphatidylinositol 3-kinase pathway coordinately regulates p27Kip1 levels. J. Immunol. 172:5522-5527. [DOI] [PubMed] [Google Scholar]
- 13.Daitoku, H., M. Hatta, H. Matsuzaki, S. Aratani, T. Ohshima, M. Miyagishi, T. Nakajima, and A. Fukamizu. 2004. Silent information regulator 2 potentiates FoxO1-mediated transcription through its deacetylase activity. Proc. Natl. Acad. Sci. USA 101:10042-10047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daly, C., V. Wong, E. Burova, Y. Wei, S. Zabski, J. Griffiths, K. M. Lai, H. C. Lin, E. Ioffe, G. D. Yancopoulos, and J. S. Rudge. 2004. Angiopoietin-1 modulates endothelial cell function and gene expression via the transcription factor FKHR (FOXO1). Genes Dev. 18:1060-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dugast-Darzacq, C., M. Pirity, J. K. Blanck, A. Scherl, and N. Schreiber-Agus. 2004. Mxi1-SRα: a novel Mxi1 isoform with enhanced transcriptional repression potential. Oncogene 23:8887-8899. [DOI] [PubMed] [Google Scholar]
- 16.Eagle, L. R., X. Yin, A. R. Brothman, B. J. Williams, N. B. Atkin, and E. V. Prochownik. 1995. Mutation of the MXI1 gene in prostate cancer. Nat. Genet. 9:249-255. [DOI] [PubMed] [Google Scholar]
- 17.Engstrom, L. D., A. S. Youkilis, J. L. Gorelick, D. Zheng, V. Ackley, C. A. Petroff, L. Q. Benson, M. R. Coon, X. Zhu, S. M. Hanash, and D. S. Wechsler. 2004. Mxi1-0, an alternatively transcribed Mxi1 isoform, is overexpressed in glioblastomas. Neoplasia 6:660-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Essafi, A., S. Fernandez de Mattos, Y. A. Hassen, I. Soeiro, G. J. Mufti, N. S. Thomas, R. H. Medema, and E. W. Lam. 2005. Direct transcriptional regulation of Bim by FoxO3a mediates STI571-induced apoptosis in Bcr-Abl-expressing cells. Oncogene 24:2317-2329. [DOI] [PubMed] [Google Scholar]
- 19.Essers, M. A., S. Weijzen, A. M. de Vries-Smits, I. Saarloos, N. D. de Ruiter, J. L. Bos, and B. M. Burgering. 2004. FOXO transcription factor activation by oxidative stress mediated by the small GTPase Ral and JNK. EMBO J. 23:4802-4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernandez, P. C., S. R. Frank, L. Wang, M. Schroeder, S. Liu, J. Greene, A. Cocito, and B. Amati. 2003. Genomic targets of the human c-Myc protein. Genes Dev. 17:1115-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foley, K. P., and R. N. Eisenman. 1999. Two MAD tails: what the recent knockouts of Mad1 and Mxi1 tell us about the MYC/MAX/MAD network. Biochim. Biophys. Acta 1423:M37-M47. [DOI] [PubMed] [Google Scholar]
- 22.Fukuoka, M., H. Daitoku, M. Hatta, H. Matsuzaki, S. Umemura, and A. Fukamizu. 2003. Negative regulation of forkhead transcription factor AFX (Foxo4) by CBP-induced acetylation. Int. J. Mol. Med. 12:503-508. [PubMed] [Google Scholar]
- 23.Furuyama, T., T. Nakazawa, I. Nakano, and N. Mori. 2000. Identification of the differential distribution patterns of mRNAs and consensus binding sequences for mouse DAF-16 homologues. Biochem. J. 349:629-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galili, N., R. J. Davis, W. J. Fredericks, S. Mukhopadhyay, F. J. Rauscher III, B. S. Emanuel, G. Rovera, and F. G. Barr. 1993. Fusion of a forkhead domain gene to PAX3 in the solid tumour alveolar rhabdomyosarcoma. Nat. Genet. 5:230-235. [DOI] [PubMed] [Google Scholar]
- 25.Giannakou, M. E., and L. Partridge. 2004. The interaction between FOXO and SIRT1: tipping the balance towards survival. Trends Cell Biol. 14:408-412. [DOI] [PubMed] [Google Scholar]
- 26.Gilley, J., P. J. Coffer, and J. Ham. 2003. FOXO transcription factors directly activate bim gene expression and promote apoptosis in sympathetic neurons. J. Cell Biol. 162:613-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gray, I. C., S. M. Phillips, S. J. Lee, J. P. Neoptolemos, J. Weissenbach, and N. K. Spurr. 1995. Loss of the chromosomal region 10q23-25 in prostate cancer. Cancer Res. 55:4800-4803. [PubMed] [Google Scholar]
- 28.Greer, E. L., and A. Brunet. 2005. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene 24:7410-7425. [DOI] [PubMed] [Google Scholar]
- 29.Guo, Q. M., R. L. Malek, S. Kim, C. Chiao, M. He, M. Ruffy, K. Sanka, N. H. Lee, C. V. Dang, and E. T. Liu. 2000. Identification of c-myc responsive genes using rat cDNA microarray. Cancer Res. 60:5922-5928. [PubMed] [Google Scholar]
- 30.Hall, R. K., T. Yamasaki, T. Kucera, M. Waltner-Law, R. O'Brien, and D. K. Granner. 2000. Regulation of phosphoenolpyruvate carboxykinase and insulin-like growth factor-binding protein-1 gene expression by insulin. The role of winged helix/forkhead proteins. J. Biol. Chem. 275:30169-30175. [DOI] [PubMed] [Google Scholar]
- 31.Holzel, M., F. Kohlhuber, I. Schlosser, D. Holzel, B. Luscher, and D. Eick. 2001. Myc/Max/Mad regulate the frequency but not the duration of productive cell cycles. EMBO Rep. 2:1125-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang, H., K. M. Regan, F. Wang, D. Wang, D. I. Smith, J. M. van Deursen, and D. J. Tindall. 2005. Skp2 inhibits FOXO1 in tumor suppression through ubiquitin-mediated degradation. Proc. Natl. Acad. Sci. USA 102:1649-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacobs, F. M., L. P. van der Heide, P. J. Wijchers, J. P. Burbach, M. F. Hoekman, and M. P. Smidt. 2003. FoxO6, a novel member of the FoxO class of transcription factors with distinct shuttling dynamics. J. Biol. Chem. 278:35959-35967. [DOI] [PubMed] [Google Scholar]
- 34.Kops, G. J., T. B. Dansen, P. E. Polderman, I. Saarloos, K. W. Wirtz, P. J. Coffer, T. T. Huang, J. L. Bos, R. H. Medema, and B. M. Burgering. 2002. Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature 419:316-321. [DOI] [PubMed] [Google Scholar]
- 35.Kops, G. J., N. D. de Ruiter, A. M. De Vries-Smits, D. R. Powell, J. L. Bos, and B. M. Burgering. 1999. Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature 398:630-634. [DOI] [PubMed] [Google Scholar]
- 36.Kops, G. J., R. H. Medema, J. Glassford, M. A. Essers, P. F. Dijkers, P. J. Coffer, E. W. Lam, and B. M. Burgering. 2002. Control of cell cycle exit and entry by protein kinase B-regulated forkhead transcription factors. Mol. Cell. Biol. 22:2025-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuczyk, M. A., J. Serth, C. Bokemeyer, J. Schwede, R. Herrmann, S. Machtens, V. Grunewald, K. Hofner, and U. Jonas. 1998. The MXI1 tumor suppressor gene is not mutated in primary prostate cancer. Oncol. Rep. 5:213-216. [PubMed] [Google Scholar]
- 38.Lee, T. C., and E. B. Ziff. 1999. Mxi1 is a repressor of the c-Myc promoter and reverses activation by USF. J. Biol. Chem. 274:595-606. [DOI] [PubMed] [Google Scholar]
- 39.Li, P., H. Lee, S. Guo, T. G. Unterman, G. Jenster, and W. Bai. 2003. AKT-independent protection of prostate cancer cells from apoptosis mediated through complex formation between the androgen receptor and FKHR. Mol. Cell. Biol. 23:104-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin, K., J. B. Dorman, A. Rodan, and C. Kenyon. 1997. daf-16: an HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science 278:1319-1322. [DOI] [PubMed] [Google Scholar]
- 41.Luscher, B. 2001. Function and regulation of the transcription factors of the Myc/Max/Mad network. Gene 277:1-14. [DOI] [PubMed] [Google Scholar]
- 42.Lynch, R. L., B. W. Konicek, A. M. McNulty, K. R. Hanna, J. E. Lewis, B. L. Neubauer, and J. R. Graff. 2005. The progression of LNCaP human prostate cancer cells to androgen independence involves decreased FOXO3a expression and reduced p27KIP1 promoter transactivation. Mol. Cancer Res. 3:163-169. [DOI] [PubMed] [Google Scholar]
- 43.Martinez-Gac, L., M. Marques, Z. Garcia, M. R. Campanero, and A. C. Carrera. 2004. Control of cyclin G2 mRNA expression by forkhead transcription factors: novel mechanism for cell cycle control by phosphoinositide 3-kinase and forkhead. Mol. Cell. Biol. 24:2181-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsuzaki, H., H. Daitoku, M. Hatta, H. Aoyama, K. Yoshimochi, and A. Fukamizu. 2005. Acetylation of Foxo1 alters its DNA-binding ability and sensitivity to phosphorylation. Proc. Natl. Acad. Sci. USA 102:11278-11283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsuzaki, H., H. Daitoku, M. Hatta, K. Tanaka, and A. Fukamizu. 2003. Insulin-induced phosphorylation of FKHR (Foxo1) targets to proteasomal degradation. Proc. Natl. Acad. Sci. USA 100:11285-11290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Medema, R. H., G. J. Kops, J. L. Bos, and B. M. Burgering. 2000. AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature 404:782-787. [DOI] [PubMed] [Google Scholar]
- 47.Menssen, A., and H. Hermeking. 2002. Characterization of the c-MYC-regulated transcriptome by SAGE: identification and analysis of c-MYC target genes. Proc. Natl. Acad. Sci. USA 99:6274-6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Modur, V., R. Nagarajan, B. M. Evers, and J. Milbrandt. 2002. FOXO proteins regulate tumor necrosis factor-related apoptosis inducing ligand expression. Implications for PTEN mutation in prostate cancer. J. Biol. Chem. 277:47928-47937. [DOI] [PubMed] [Google Scholar]
- 49.Murphy, C. T., S. A. McCarroll, C. I. Bargmann, A. Fraser, R. S. Kamath, J. Ahringer, H. Li, and C. Kenyon. 2003. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424:277-283. [DOI] [PubMed] [Google Scholar]
- 50.Nakae, J., T. Kitamura, Y. Kitamura, W. H. Biggs III, K. C. Arden, and D. Accili. 2003. The forkhead transcription factor Foxo1 regulates adipocyte differentiation. Dev. Cell 4:119-129. [DOI] [PubMed] [Google Scholar]
- 51.Nakamura, N., S. Ramaswamy, F. Vazquez, S. Signoretti, M. Loda, and W. R. Sellers. 2000. Forkhead transcription factors are critical effectors of cell death and cell cycle arrest downstream of PTEN. Mol. Cell. Biol. 20:8969-8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O'Hagan, R. C., N. Schreiber-Agus, K. Chen, G. David, J. A. Engelman, R. Schwab, L. Alland, C. Thomson, D. R. Ronning, J. C. Sacchettini, P. Meltzer, and R. A. DePinho. 2000. Gene-target recognition among members of the myc superfamily and implications for oncogenesis. Nat. Genet. 24:113-119. [DOI] [PubMed] [Google Scholar]
- 53.Ogg, S., S. Paradis, S. Gottlieb, G. I. Patterson, L. Lee, H. A. Tissenbaum, and G. Ruvkun. 1997. The Forkhead transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389:994-999. [DOI] [PubMed] [Google Scholar]
- 54.Plas, D. R., and C. B. Thompson. 2003. Akt activation promotes degradation of tuberin and FOXO3a via the proteasome. J. Biol. Chem. 278:12361-12366. [DOI] [PubMed] [Google Scholar]
- 55.Prochownik, E. V., L. Eagle Grove, D. Deubler, X. L. Zhu, R. A. Stephenson, L. R. Rohr, X. Yin, and A. R. Brothman. 1998. Commonly occurring loss and mutation of the MXI1 gene in prostate cancer. Genes Chromosomes Cancer 22:295-304. [PubMed] [Google Scholar]
- 56.Ramaswamy, S., N. Nakamura, I. Sansal, L. Bergeron, and W. R. Sellers. 2002. A novel mechanism of gene regulation and tumor suppression by the transcription factor FKHR. Cancer Cell 2:81-91. [DOI] [PubMed] [Google Scholar]
- 57.Roussel, M. F., R. A. Ashmun, C. J. Sherr, R. N. Eisenman, and D. E. Ayer. 1996. Inhibition of cell proliferation by the Mad1 transcriptional repressor. Mol. Cell. Biol. 16:2796-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmidt, M., S. Fernandez de Mattos, A. van der Horst, R. Klompmaker, G. J. Kops, E. W. Lam, B. M. Burgering, and R. H. Medema. 2002. Cell cycle inhibition by FoxO forkhead transcription factors involves downregulation of cyclin D. Mol. Cell. Biol. 22:7842-7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schreiber-Agus, N., L. Chin, K. Chen, R. Torres, G. Rao, P. Guida, A. I. Skoultchi, and R. A. DePinho. 1995. An amino-terminal domain of Mxi1 mediates anti-Myc oncogenic activity and interacts with a homolog of the yeast transcriptional repressor SIN3. Cell 80:777-786. [DOI] [PubMed] [Google Scholar]
- 60.Schreiber-Agus, N., and R. A. DePinho. 1998. Repression by the Mad(Mxi1)-Sin3 complex. Bioessays 20:808-818. [DOI] [PubMed] [Google Scholar]
- 61.Schreiber-Agus, N., Y. Meng, T. Hoang, H. Hou, Jr., K. Chen, R. Greenberg, C. Cordon-Cardo, H. W. Lee, and R. A. DePinho. 1998. Role of Mxi1 in ageing organ systems and the regulation of normal and neoplastic growth. Nature 393:483-487. [DOI] [PubMed] [Google Scholar]
- 62.Shapiro, D. N., J. E. Sublett, B. Li, J. R. Downing, and C. W. Naeve. 1993. Fusion of PAX3 to a member of the forkhead family of transcription factors in human alveolar rhabdomyosarcoma. Cancer Res. 53:5108-5112. [PubMed] [Google Scholar]
- 63.Shapiro, D. N., V. Valentine, L. Eagle, X. Yin, S. W. Morris, and E. V. Prochownik. 1994. Assignment of the human MAD and MXI1 genes to chromosomes 2p12-p13 and 10q24-q25. Genomics 23:282-285. [DOI] [PubMed] [Google Scholar]
- 64.So, C. W., and M. L. Cleary. 2002. MLL-AFX requires the transcriptional effector domains of AFX to transform myeloid progenitors and transdominantly interfere with forkhead protein function. Mol. Cell. Biol. 22:6542-6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.So, C. W., and M. L. Cleary. 2003. Common mechanism for oncogenic activation of MLL by forkhead family proteins. Blood 101:633-639. [DOI] [PubMed] [Google Scholar]
- 66.Stahl, M., P. F. Dijkers, G. J. Kops, S. M. Lens, P. J. Coffer, B. M. Burgering, and R. H. Medema. 2002. The forkhead transcription factor FoxO regulates transcription of p27Kip1 and Bim in response to IL-2. J. Immunol. 168:5024-5031. [DOI] [PubMed] [Google Scholar]
- 67.Taj, M. M., R. J. Tawil, L. D. Engstrom, Z. Zeng, C. Hwang, M. G. Sanda, and D. S. Wechsler. 2001. Mxi1, a Myc antagonist, suppresses proliferation of DU145 human prostate cells. Prostate 47:194-204. [DOI] [PubMed] [Google Scholar]
- 68.Tothova, Z., R. Kollipara, B. J. Huntly, B. H. Lee, D. H. Castrillon, D. E. Cullen, E. P. McDowell, S. Lazo-Kallanian, I. R. Williams, C. Sears, S. A. Armstrong, E. Passegue, R. A. Depinho, and D. G. Gilliland. 2007. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell 128:325-339. [DOI] [PubMed] [Google Scholar]
- 69.Vafa, O., M. Wade, S. Kern, M. Beeche, T. K. Pandita, G. M. Hampton, and G. M. Wahl. 2002. c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: a mechanism for oncogene-induced genetic instability. Mol. Cell 9:1031-1044. [DOI] [PubMed] [Google Scholar]
- 70.van der Horst, A., L. G. Tertoolen, L. M. de Vries-Smits, R. A. Frye, R. H. Medema, and B. M. Burgering. 2004. FOXO4 is acetylated upon peroxide stress and deacetylated by the longevity protein hSir2SIRT1. J. Biol. Chem. 279:28873-28879. [DOI] [PubMed] [Google Scholar]
- 71.Watanabe, M., M. L. Rebbert, M. Andreazzoli, N. Takahashi, R. Toyama, S. Zimmerman, M. Whitman, and I. B. Dawid. 2002. Regulation of the Lim-1 gene is mediated through conserved FAST-1/FoxH1 sites in the first intron. Dev. Dyn. 225:448-456. [DOI] [PubMed] [Google Scholar]
- 72.Wechsler, D. S., A. L. Hawkins, X. Li, E. W. Jabs, C. A. Griffin, and C. V. Dang. 1994. Localization of the human Mxi1 transcription factor gene (MXI1) to chromosome 10q24-q25. Genomics 21:669-672. [DOI] [PubMed] [Google Scholar]
- 73.Wechsler, D. S., C. A. Shelly, C. A. Petroff, and C. V. Dang. 1997. MXI1, a putative tumor suppressor gene, suppresses growth of human glioblastoma cells. Cancer Res 57:4905-4912. [PubMed] [Google Scholar]
- 74.Whyte, D. B., and S. L. Holbeck. 2006. Correlation of PIK3Ca mutations with gene expression and drug sensitivity in NCI-60 cell lines. Biochem. Biophys. Res. Commun. 340:469-475. [DOI] [PubMed] [Google Scholar]
- 75.Xuan, Z., and M. Q. Zhang. 2005. From worm to human: bioinformatics approaches to identify FOXO target genes. Mech. Ageing Dev. 126:209-215. [DOI] [PubMed] [Google Scholar]
- 76.Yang, W., J. Shen, M. Wu, M. Arsura, M. FitzGerald, Z. Suldan, D. W. Kim, C. S. Hofmann, S. Pianetti, R. Romieu-Mourez, L. P. Freedman, and G. E. Sonenshein. 2001. Repression of transcription of the p27Kip1 cyclin-dependent kinase inhibitor gene by c-Myc. Oncogene 20:1688-1702. [DOI] [PubMed] [Google Scholar]
- 77.Yuan, J., R. S. Tirabassi, A. B. Bush, and M. D. Cole. 1998. The C. elegans MDL-1 and MXL-1 proteins can functionally substitute for vertebrate MAD and MAX. Oncogene 17:1109-1118. [DOI] [PubMed] [Google Scholar]
- 78.Zervos, A. S., J. Gyuris, and R. Brent. 1993. Mxi1, a protein that specifically interacts with Max to bind Myc-Max recognition sites. Cell 72:223-232. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.