Abstract
Candida albicans is the leading cause of systemic fungal infections in immunocompromised humans. The ability to form biofilms on surfaces in the host or on implanted medical devices enhances C. albicans virulence, leading to antimicrobial resistance and providing a reservoir for infection. Biofilm formation is a complex multicellular process consisting of cell adhesion, cell growth, morphogenic switching between yeast form and filamentous states, and quorum sensing. Here we describe the role of the C. albicans EAP1 gene, which encodes a glycosylphosphatidylinositol-anchored, glucan-cross-linked cell wall protein, in adhesion and biofilm formation in vitro and in vivo. Deleting EAP1 reduced cell adhesion to polystyrene and epithelial cells in a gene dosage-dependent manner. Furthermore, EAP1 expression was required for C. albicans biofilm formation in an in vitro parallel plate flow chamber model and in an in vivo rat central venous catheter model. EAP1 expression was upregulated in biofilm-associated cells in vitro and in vivo. Our results illustrate an association between Eap1p-mediated adhesion and biofilm formation in vitro and in vivo.
Candida albicans is the leading cause of candidiasis, most often manifesting as superficial mucosal infections. Candida spp. are also major agents of systemic bloodstream infections, causing 8% of all such nosocomial infections in the United States, and are behind only coagulase-negative staphylococci, Staphylococcus aureus, and enterococci as causes of bloodstream infections (13, 25, 43). Frequently, candidemia is associated with C. albicans colonization of indwelling devices, such as catheters, endotracheal tubes, and pacemakers (12, 29, 45). In fact, C. albicans is the most commonly occurring fungus in biofilms formed on medical devices (36). In addition to providing the fungus a route through host barrier defenses, these devices provide a surface for biofilm growth and development (12). Sessile C. albicans cells exhibit distinct gene expression patterns compared to planktonic cells, including differences in protein synthesis, metabolism, cell cycle control, and signal transduction (16). Cells in biofilms are also much more resistant to antifungal agents, including amphotericin B and azoles (1, 23).
C. albicans biofilms consist of a dense network of yeast cells, pseudohyphae, and hyphae organized in an extracellular polymeric matrix in a complex three-dimensional structure containing water channels (9, 22, 31, 38, 47). These biofilms are extremely heterogenous in structure, depending on the substrate composition, environmental conditions, and C. albicans strains used (9, 16, 28, 34, 52). C. albicans biofilms formed in vitro or in vivo contain primarily yeast cells at the substrate surface and hyphae in the upper portion of the biofilm (5, 9, 22). While the yeast-hypha morphogenic transition is important for full development of the biofilm, strains that are unable to grow as yeast cells and strains that are unable to form hyphae can still form biofilms (5).
Adhesion between C. albicans cells and materials or host cells has been implicated as an early step in biofilm formation. Attachment of C. albicans cells to materials is mediated by nonspecific interactions, such as hydrophobic and electrostatic forces, as well as by specific adhesin-ligand bonds (8, 10, 26, 44, 45, 50). Cell hydrophobicity and charge also depend on cell growth morphology and cell surface structure (27). A class of adhesins, termed glycosylphosphatidylinositol-dependent cell wall proteins (GPI-CWPs), possess a common structure characterized by the presence of an N-terminal signal peptide and a C-terminal sequence containing a GPI anchor attachment site. C. albicans genes encoding GPI-CWP adhesins, including ALS1, ALS2, ALS4, ALS5 (ALA1), HWP1, and EAP1, mediate adhesion to organic and inorganic surfaces, extracellular matrix proteins, human endothelial cells, and epithelial cells (15, 17, 32, 51, 61). One or more genes in the ALS family are upregulated in biofilm-associated cells compared with planktonic cells (9). ALS3 expression was shown to be necessary for biofilm formation on silicone substrates in vitro (39, 60). While ALS3 expression was not required for biofilm formation in vivo, overexpression of ALS3 restored biofilm formation in bcr1/bcr1 strains (39). Farnesol, a quorum-sensing molecule that inhibits C. albicans biofilm formation, decreases HWP1 expression in biofilms (46). HWP1 expression was found to be required for establishment of biofilms in an in vivo catheter model (41). These studies suggest that GPI-CWP adhesins may play a mechanistic role in C. albicans biofilm formation.
The C. albicans EAP1 gene was originally identified because of its ability to mediate adhesion to polystyrene when expressed in a Saccharomyces cerevisiae flocculin-deficient strain (32). EAP1 expression also restored invasive and filamentous growth to S. cerevisiae flo11Δ mutants and enhanced attachment of S. cerevisiae to HEK293 kidney epithelial cells (32). In this study, we demonstrate that Eap1p is a glucan-cross-linked cell wall-localized protein. In addition, we found that eap1 mutants exhibited reduced adhesion to plastic surfaces and epithelial cells and that Eap1p was able to mediate adhesion to yeast cells. EAP1 expression was also required for biofilm formation under shear flow in vitro and in an in vivo central venous catheter biofilm model. Finally, we show that the expression of EAP1 was differentially regulated between planktonic and biofilm-associated cells. Our results suggest that the adhesin Eap1p plays a role in C. albicans biofilm formation.
MATERIALS AND METHODS
Strains and media.
The C. albicans and S. cerevisiae strains used in this study are listed in Tables 1 and 2. Escherichia coli strain DH5α was used for general recombination techniques according to protocols described by Sambrook et al. (48). Yeast strains were routinely cultured in YPD medium (1% yeast extract, 2% peptone, 2% glucose) or minimal defined medium (2% glucose, 0.67% yeast nitrogen base without amino acids) at 30°C. YPD medium was supplemented with 80 mg/liter uridine when Ura− C. albicans strains were cultured. Solid Spider medium (35) was used to induce C. albicans filamentous growth, as described by Bensen et al. (6). Synthetic minimal medium lacking specific nutrients was prepared as described previously (42). Galactose was added to media to replace glucose in order to express genes within plasmids containing the S. cerevisiae GAL1 promoter. S. cerevisiae cells and C. albicans cells were transformed by lithium acetate transformation as previously described (18, 57).
TABLE 1.
C. albicans strains used in this study
| Strain | Genotype or description | Source or reference |
|---|---|---|
| SC5314 | Clinical isolate | 19 |
| BWP17 | ura3::λimm434/ura3::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG | 59 |
| YJB6284 | ura3::λimm434/ura3::λimm434 HIS1::his1::hisG/his1::hisG ARG4-URA3::arg4::hisG/arg4::hisG | 6 |
| SPY313 | ura3::λimm434/ura3::λimm434 HIS1::his1::hisG/his1::hisG ARG4::arg4::hisG/arg4::hisG eap1::URA3/EAP1 | This study |
| SPY314 | ura3::λimm434/ura3::λimm434 HIS1::his1::hisG/his1::hisG arg4::hisG/arg4::hisG eap1::URA3/eap1::ARG4 | This study |
| SPY315 | ura3::λimm434/ura3::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG EAP1-HIS1::eap1::URA3/eap1::ARG4 | This study |
| SPY316 | ura3::λimm434/ura3::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG eap1::URA3/EAP1 | This study |
| SPY317 | ura3::λimm434/ura3::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG eap1::URA3/eap1::ARG4 | This study |
| SPY387 | ura3::λimm434/ura3::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG ACT1/ACT1-URA3-pACT1-HAEAP1 | This study |
| SPY388 | ura3::λimm434/ura3::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG ACT1/ACT1-URA3-pACT1-HAEAP1.NOGPI | This study |
TABLE 2.
S. cerevisiae strains used in this study
| Strain | Background and genotype | Source or reference |
|---|---|---|
| SKY760 | Σ1278b MATaura3-52 his3::hisG leu2::hisG | Our collection |
| SPY308 | Σ1278b MATaura3-52 his3::hisG leu2::hisG flo8Δ::Kanr | 32 |
| SPY415 | Σ1278b MATaura3-52 his3::hisG leu2::hisG flo8Δ::HIS3-pADH1-GFP | This study |
Plasmid construction.
A partial open reading frame (ORF) encoding the N-terminal 42 amino acids of Eap1p was amplified from C. albicans SC5314 genomic DNA with the oligonucleotides EAP1.5primer and EAP1.HA.Sig3, which contains the sequence encoding the hemagglutinin (HA) epitope of influenza virus. The PCR product was digested with PacI and SpeI and ligated into pHwp1Sig.GFP.GPI (37) to yield pEap1SigHA.GFP.Hwp1GPI. A partial ORF encoding Eap1p without the N-terminal 42 amino acids was amplified from C. albicans SC5314 genomic DNA. This PCR product was digested with SpeI and SmaI and ligated into pEap1SigHA.GFP.Hwp1GPI to generate pSigHAEap1.GPI. The ORF encoding HA-tagged Eap1p was amplified from pSigHAEap1.GPI and cloned into the XhoI and XbaI sites of pAU3 (32) to yield pHAEAP1. pHAEAP1.NOGPI is essentially identical to pHAEAP1 except that 21 amino acid residues from the C terminus of Eap1p, encoding the GPI anchor signal, were deleted. For all constructs in which fragments were generated by PCR, the final constructs were verified by DNA sequence analysis.
Expression and detection of HA-tagged Eap1p.
The pHAEAP1 and pHAEAP1.NOGPI constructs were linearized by cutting in the promoter of CaACT1 with BglII to direct integration to the CaACT1 locus. The linearized pHAEAP1 and pHAEAP1.NOGPI constructs were used to transform C. albicans BWP17. Ura+ clones were selected, and integration of the third copy of EAP1 was verified by PCR. The transformants were cultured in 3 ml SC−Ura medium overnight at 30°C (optical density at 600 nm [OD600] = 1.0). The cells were blocked with 10% goat serum in phosphate-buffered saline (PBS) and then incubated with an anti-HA monoclonal antibody (Covance, Berkeley, CA) for 30 min, followed by incubation with an anti-mouse antibody-fluorescein isothiocyanate conjugate (Zymed, San Francisco, CA) for 30 min. The cells were examined for green fluorescence by using an inverted epifluorescence Olympus IX70 microscope coupled to a Spot charge-coupled device camera (Diagnostic Instruments, Sterling Heights, MI) and MetaVue image acquisition and analysis software (Universal Imaging Corporation, Downingtown, PA).
Western analysis.
Yeast cells expressing HA-tagged Eap1p constructs were cultured as described above and harvested by centrifugation. To analyze the HA-tagged Eap1p protein in medium, the supernatant was concentrated with 100-kDa-cutoff Microcon centrifugal filters (Amicon, Billerica, MA). Yeast cell walls were isolated according to the method of Mao et al. (37). Briefly, 3 ml yeast cells (OD600 = 1.5 to 2.0) was harvested and disrupted by vortexing with glass beads. The resulting yeast debris was centrifuged to obtain supernatants and pellets. The pellets were boiled twice in 2% sodium dodecyl sulfate (SDS) for 5 min each and washed five times in 1 M NaCl and five times in water. The washed pellets were treated with laminarinase (β-1,3-glucanase from Penicillium; Sigma) (0.25 U/g wet weight) in 100 mM sodium acetate, pH 5.5, or with the buffer alone for 4 h at 37°C. The proteins released into the supernatant by enzymatic digestion or from concentrated cell-free medium were subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by Western blotting using an ECL Western blot kit (Amersham) and anti-HA monoclonal antibody.
Construction of eap1 mutant strains.
To construct C. albicans disruption strains, we used PCR-mediated gene disruption methods as previously described (20, 58). The marker cassettes used for gene disruption were amplified by primers that provide 100 bp of sequence homology corresponding to the EAP1 ORF at its 5′ or 3′ end. An eap1::URA3 PCR product was amplified with pDDB57 (58) as the DNA template and transformed into BWP17 (59) to generate the heterozygous strain SPY316. SPY316 was transformed with an eap1::ARG4 disruption fragment amplified from the DNA template pFA-ARG4 (20) to generate the eap1 null mutant strain SPY317. SPY316 and SPY317 were made prototrophic by transforming them with pGEM-HIS1 (59) linearized with NruI and/or pRS-Arg4ΔSpeI linearized with ClaI to generate strains SPY313 and SPY314, respectively. BWP17 was made prototrophic (strain YJP6284) as described by Bensen et al. (6).
To complement the eap1/eap1 mutant with EAP1, HIS1 was amplified by PCR with pGEM-HIS1 as the DNA template and cloned into the SalI and SpeI sites of pBluescript KS(+) (Stratagene) to generate pHIS1. A full-length EAP1 gene, including a 562-bp sequence upstream and a 337-bp sequence downstream of the ORF for Eap1p, was PCR amplified from C. albicans BWP17 genomic DNA. This PCR product was cloned into pHIS1 cut with SpeI and SacII to generate pHIS1-EAP1. The final construct was verified by DNA sequence analysis. This plasmid was linearized by cutting in the EAP1 promoter with SphI and was then integrated into the native EAP1 locus of strain SPY317 to generate strain SPY315.
Parallel plate flow chamber cell adhesion assay.
Yeast cell adhesion to polystyrene surfaces was quantified using a parallel plate flow chamber (Glycotech, Rockville, MD) as previously described (33). Briefly, yeast cells were cultured in YPD overnight, pelleted, and resuspended in 0.1 M sodium phosphate buffer, pH 6.0. After brief sonication to disperse cellular aggregates, the cell suspension was pumped into the flow chamber and incubated for 15 min. The detachment assay was performed by increasing the flow rate of the sodium phosphate buffer, and thus the shear stress, in a stepwise manner. For each applied shear stress, the number of cells remaining attached to the surface was identified and counted, using an inverted Olympus microscope coupled to a Spot charge-coupled device camera and MetaVue image acquisition and analysis software. The adhesion of cells was quantified as the mean fraction of cells in each of three selected fields remaining attached after exposure to an applied shear force.
S. cerevisiae adhesion to C. albicans cells.
To distinguish S. cerevisiae cells bound to a C. albicans monolayer, an S. cerevisiae flo8Δ strain constitutively expressing green fluorescent protein (GFP) under the control of the ADH1 promoter was constructed. Primers flo8pADHGFP_F (5′-GTT TAT AGA CAT AAA TAA AGA GGA AAC GCA TTC CGT GGT AGA ATG GAA TTC GAG CTC GTT TAA AC-3′) and flo8pADHGFP_R (5′-TTA AGA GTT TTT ATT TTT TAT TAT AAT ACT CAA CAC GTG ACT TCA TCT ATA TTA CCC TGT TAT CCC-3′) were designed to amplify the HIS-pADH1-GFP cassette from plasmid pFA6a-His3MX6-pADH1-GFP (54). Plasmid-specific regions of these primers are indicated in bold. The resulting DNA cassette was transformed into SKY760, and selection was carried out on minimal glucose plates lacking histidine. Replacement of the FLO8 ORF with the HIS-pADH1-GFP construct in the resulting strain (SPY415) was verified by PCR. SPY415 was then transformed with plasmid pYE-1, which contains a fragment of C. albicans DNA that carries EAP1 under the control of the GAL1 promoter (33).
C. albicans BWP17 was cultured overnight in a petri dish in YPD with mild shaking. Cells were allowed to settle to the dish surface, and the flow chamber apparatus was placed atop them. The chamber was filled with 0.1 M sodium phosphate buffer, pH 6.0, and the flow rate was increased incrementally from 0 to 30.7 dyn/cm2 to generate a “monolayer” of cells. S. cerevisiae SPY415 cells, carrying either pYE-1 (EAP1) or pYESR (vector), were cultured overnight in SG medium and then resuspended in 0.1 M sodium phosphate, pH 6.0, and introduced into the flow chamber. After 60 min, the number of GFP-expressing cells in the chamber was quantified by epifluorescence imaging. Flow was applied at 2.5 dyn/cm2 and increased incrementally to 30.7 dyn/cm2, with the flow held steady for 3 min at each rate prior to acquiring an epifluorescence image. Cell adhesion was quantified as the number of fluorescent S. cerevisiae cells remaining attached in each field. At least 200 cells in multiple fields were observed. Phase-contrast images were also acquired to ensure that C. albicans cells did not detach from the substrate.
Adhesion to human kidney epithelial cells.
Yeast adhesion to 293 cells was measured essentially as previously described (33), using confluent monolayers of 293 cells in six-well tissue culture plates. After the cells were washed in PBS containing Mg2+ and Ca2+ (PBS++) at 37°C, yeast cells (500 cells/μl) grown in YPD overnight were suspended in PBS++ and added to a confluent monolayer of 293 human kidney epithelial cells. After incubating for 30 min at 37°C, nonadherent yeast cells were rinsed off with PBS++. Next, trypsin was added to the wells, and the cells were suspended in sterile deionized water and plated on YPD agar. The number of cells added to the 293 monolayer and the number of adherent cells following rinsing were determined by colony counting, and adherence was expressed as the fraction of cells added to the 293 monolayer that remained attached after rinsing. The correlations between CFU and the numbers of cells added and recovered for each strain were determined using mock adhesion assays lacking 293 cells.
Parallel plate flow chamber in vitro biofilm assay.
C. albicans cells were cultured overnight at 30°C in YPD medium. Cells were pelleted and resuspended in 0.1 M sodium phosphate buffer (pH 6.0). The suspension was injected into the flow chamber, which was placed on the stage of a microscope enclosed in a 37°C constant-temperature box. Following a 30-min incubation to allow the cells to adhere to the surface of a polystyrene petri dish, the flow of sterile YPD medium was induced at a shear stress of 6 dyn/cm2. Images of several fields of the flow chamber surface were acquired to capture the growth of cells on the surface as time progressed.
In vivo catheter biofilm formation and imaging.
C. albicans biofilms were developed in a rat central venous catheter system and imaged using scanning electron microscopy as described by Andes et al. (2). Heparinized (100 U/ml) polyethylene tubing (Becton Dickinson Company, Franklin Lakes, NJ), mimicking the catheters used in human patients, was placed in the internal jugular veins of specific-pathogen-free female Sprague-Dawley rats weighing 400 g (Harlan Sprague-Dawley, Indianapolis, IN). The catheters were conditioned for 24 h prior to infection to allow the deposition of host proteins on the catheter surface. C. albicans cells were cultured in YPD overnight and suspended in 0.85% NaCl. The inoculum was adjusted to 106 CFU/ml and instilled in each catheter in a 500-μl volume (the entire catheter volume) for 4 h. After 24 h of development, the distal 2 cm of catheter was cut perpendicular to the catheter length into 5-mm segments. For scanning electron microscopy, the catheter segments were placed in fixative (1% [vol/vol] glutaraldehyde and 4% [vol/vol] formaldehyde) overnight. The specimens were subsequently dehydrated in a series of ethanol washes and desiccated by critical point drying (Tousimis, Rockville, MA). The specimens were then coated with gold and observed with a scanning electron microscope (Hitachi S-5700) in high-vacuum mode at 10 kV. The images were processed for display by using Adobe Photoshop software (Adobe, Mountain View, CA).
RNA isolation and quantitative reverse transcription-PCR (RT-PCR).
Total RNA from in vivo biofilm-associated cells was obtained as described by Andes et al. (2), with modifications. The distal 2 cm of catheter after 24 h of biofilm development was cut into 2-mm-thick segments. The segments were sonicated and vortexed to collect biofilm-associated cells, and total RNA was isolated using an RNeasy mini kit (QIAGEN, Valencia, CA) and treated with an RNase-free DNase set (QIAGEN, Valencia, CA). For comparative reference, cells were grown in RPMI 1640 at 37°C to an OD600 of 1.0. Extraction of RNAs from sessile cells cultured in vitro was carried out as described by Ramage et al. (46). C. albicans cells were suspended in RPMI 1640 (106 cells/ml), and 25 ml of cell suspension was incubated in 75-cm2 vent-cap tissue culture flasks for 1 h. Next, the medium was decanted to wash away nonattached cells, and 50 ml fresh RPMI 1640 was added. The flasks were gently shaken at 37°C for 24 h. The sessile cells were removed from the flask surface with a sterile scraper, and total RNA was isolated as described above. RNAs were obtained from suspension-cultured cells under identical culture conditions as a reference.
Quantitative real-time RT-PCR was performed by using a QuantiTect probe RT-PCR kit (QIAGEN, Valencia, CA) and an ABI PRISM 7700 v1.7 sequence detection system (Applied Biosystems, Foster City, CA). The comparison of mRNA abundance of EAP1 in planktonic cultured cells and biofilm-associated cells was characterized as the x-fold change normalized to the ACT1 gene of C. albicans (2). The significance of ΔCT values was determined by applying a paired t test.
RESULTS
Eap1p is a glucan-cross-linked CWP.
Sequence analysis predicted that Eap1p is a member of the family of GPI-CWPs, with an N-terminal signal sequence and a C-terminal GPI addition signal (11, 32, 53). To verify that Eap1p is a GPI-anchored CWP, the HA epitope of influenza virus was engineered into the EAP1 coding sequence C-terminal to the predicted Eap1p signal peptide. The HA-tagged Eap1 protein (HAEAP1) was expressed in C. albicans BWP17. Transformants expressing HAEAP1 exhibited localized fluorescence at the cell surface (Fig. 1A), consistent with a CWP. We next investigated if HAEAP1 was anchored to the cell surface via a GPI anchor by expressing a version of HAEAP1 lacking the GPI anchor site (HAEAP.NOGPI); this protein was not detected at the cell surface (Fig. 1A), indicating that GPI anchoring is essential for localization. We tested the cell-free medium of the HAEAP1.NOGPI transformants for secretion of HA-tagged Eap1p by Western blotting with an anti-HA monoclonal antibody. The HAEAP1.NOGPI transformants secreted a protein that was detected by the anti-HA antibody, whereas HA-tagged Eap1p was not detected in culture supernatants of the HAEAP1 transformants (Fig. 1B).
FIG. 1.
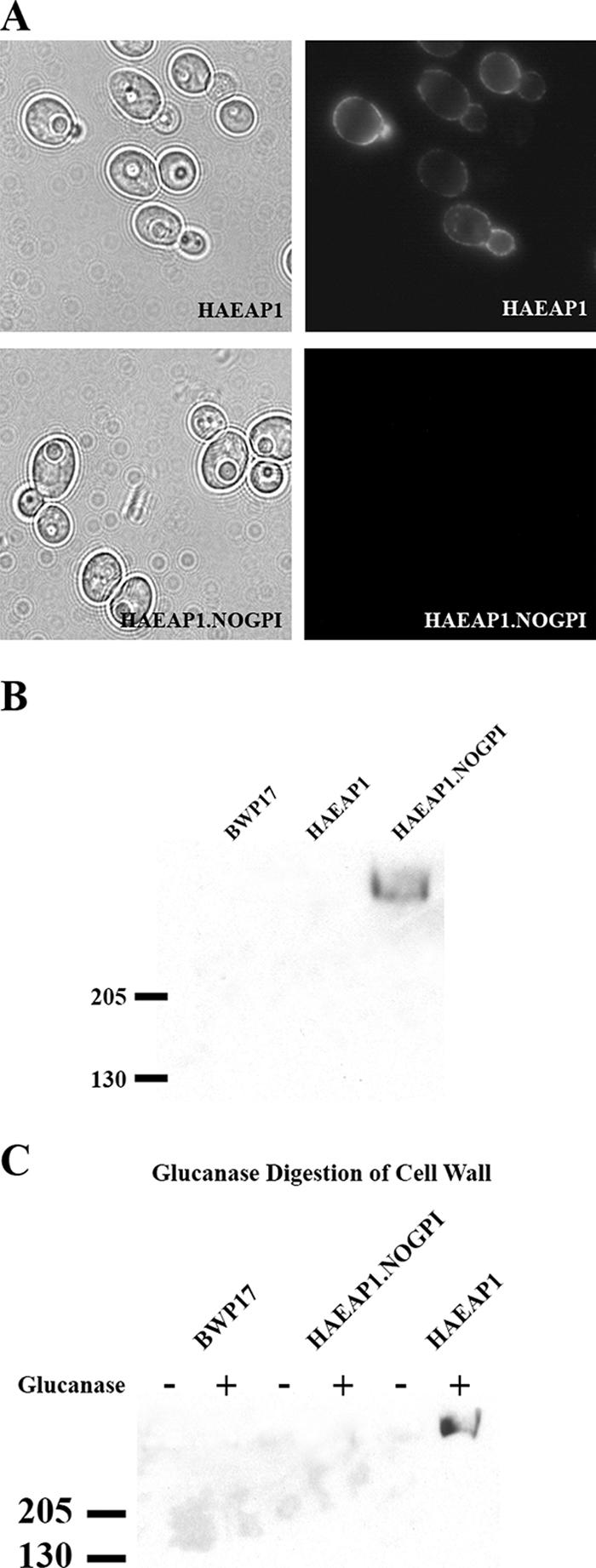
Cell wall localization of HA-tagged Eap1p. (A) Epifluorescence microscopy analysis of C. albicans strains SPY387 and SPY388. SPY387 expresses an HA-tagged Eap1p protein, and SPY388 expresses a protein identical to HA-tagged Eap1p except that 21 amino acid residues from the C terminus of Eap1p, encoding the GPI anchor signal, were deleted. Cells were cultured in SC−Ura medium overnight at 30°C prior to being imaged. (B) Analysis of cell-free supernatants from C. albicans strains BWP17, SPY387, and SPY388. The fractions were run in a 4 to 15% SDS-PAGE gradient gel and visualized by Western blotting with an anti-HA monoclonal antibody. (C) Cell wall extraction of C. albicans strains BWP17, SPY387, and SPY388. Cell walls were extracted twice in boiling SDS, digested with β-1,3-glucanase or no enzyme, and then separated into pellet and supernatant fractions. The glucanase-treated and untreated supernatant fractions were loaded into a 4 to 15% SDS-PAGE gradient gel and visualized by Western blotting with an anti-HA monoclonal antibody.
To address whether Eap1p anchors to the plasma membrane or is incorporated into the cell wall, purified cell walls from C. albicans strains transformed with HA-tagged Eap1p constructs were digested with β-glucanase. HA-tagged Eap1p was released from the cell walls of the HAEAP1 transformants and detected with anti-HA antibody (Fig. 1C). In control experiments, immunoreactive HA-tagged proteins were not found in β-glucanase-treated cell walls of cells transformed with HAEAP1.NOGPI or in samples treated with buffer alone (Fig. 1C). Based on these results, we concluded that Eap1p is a GPI-CWP.
Deletion of EAP1 reduces C. albicans adhesion.
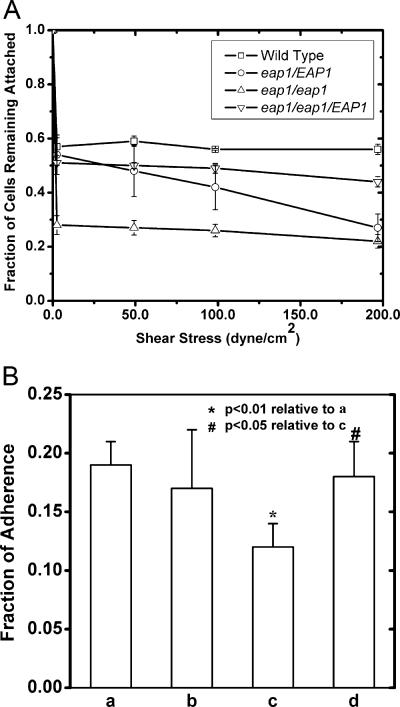
Eap1p localized to the yeast cell wall, as demonstrated in Fig. 1, and heterologous expression of EAP1 in S. cerevisiae enhanced adhesion to polystyrene and to HEK293 kidney epithelial cells (32). These findings suggest that EAP1 expression may affect adhesion of C. albicans to materials or mammalian cells. We used a parallel plate flow chamber to quantify the shear stresses required to detach wild-type and eap1 mutant strains from polystyrene surfaces. A dramatic difference existed in the ability of the strains to adhere (P < 0.001); adhesion of the eap1/eap1 strain to polystyrene was reduced approximately 50% relative to that of the wild-type strain, up to a shear stress of 200 dyn/cm2 (Fig. 2A), as defined by the number of adherent cells at any value of shear stress. In wild-type and eap1/eap1 strains, the number of cells adhered to the surface was relatively independent of shear stress over the range tested. EAP1 expression affected the number of cells that adhered to the surface, but cells that adhered appeared to have a high affinity for the surface, suggesting a very low dissociation rate constant or cooperative binding to the surface. Adhesion was not completely abolished in the eap/eap1 strain, as approximately 25 to 30% of cells were able to adhere even at high shear rates, indicating that EAP1-independent mechanisms also play a role in adhesion to polystyrene. Diminished adhesion of the heterozygous strain compared to that of the wild-type strain implies that the gene dosage of EAP1 affected the extent of adhesion to polystyrene (Fig. 2A). The eap1/eap1::EAP1 strain adhered to polystyrene to a similar extent as the heterozygous strain (Fig. 2A), indicating that the reduction in adhesion resulted from the absence of EAP1.
FIG. 2.
Eap1p mediates C. albicans adhesion to polystyrene surfaces and to 293 cells. (A) C. albicans strains YJB6284 (EAP1/EAP1), SPY313 (eap1Δ/EAP1), SPY314 (eap1Δ/eap1Δ), and SPY315 (eap1Δ/eap1Δ/EAP1) were grown in YPD and incubated on the surface of a petri dish in a parallel plate flow chamber for 10 min. Sodium phosphate buffer (0.1 M; pH 6.0) flowed through the chamber for 15 min at a controlled shear stress. The fraction of cells adhering to the plate after being subjected to the flow was determined via image analysis of three fields containing 300 to 800 cells each prior to flow exposure. Error bars represent the standard deviations for three experiments. (B) Disruption of EAP1 in C. albicans reduces adhesion to 293 human embryonic kidney cells. Bars: a, YJB6284 (EAP1/EAP1); b, SPY313 (eap1Δ/EAP1); c, SPY314 (eap1Δ/eap1Δ); and d, SPY315 (eap1Δ/eap1Δ/EAP1). Strains were grown in YPD and incubated on a confluent 293 cell monolayer for 60 min at 37°C. The cells were rinsed with PBS containing Ca2+ and Mg2+ to remove nonadherent cells. The 293 cell monolayer and associated yeast cells were detached by trypsinization and added to a YPD plate. Adhesion was quantified as the number of colonies formed on the YPD plate divided by the number of yeast cells initially added to the 293 monolayer. Error bars represent the standard deviations for three separate experiments.
To confirm that EAP1 is also involved in C. albicans adhesion to mammalian cells, we measured the adherence of strains expressing different gene dosages of EAP1 to HEK293 kidney epithelial cell monolayers. Compared with that of the wild-type strain, adhesion of the eap1/eap1 strain, measured as the fraction of cells remaining attached to the 293 monolayer following a rinse in PBS, was reduced 37% (Fig. 2B). Similar to the observations with polystyrene, the adhesion of the null mutant to epithelial cells was not eliminated (Fig. 2B), suggesting that while EAP1 expression affects C. albicans adhesion to epithelial cells, other attachment factors also contribute to this adhesion.
Eap1p mediates cell-cell adhesion.
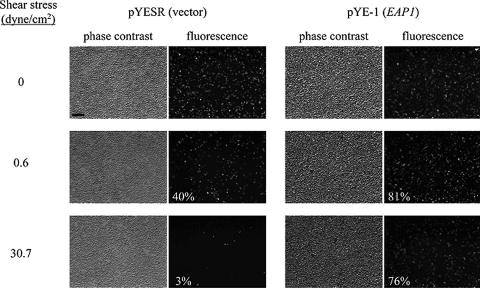
During biofilm development, sessile cells proliferate and daughter cells must remain attached to the surface-bound cells or to the polysaccharide matrix in the biofilm. To test whether Eap1p plays a role in cell-cell adhesion in addition to cell surface adhesion, we expressed EAP1 in an adhesin-deficient S. cerevisiae strain and measured the shear stress required to detach these cells from a C. albicans monolayer. A cassette encoding GFP regulated by the constitutive ADH1 promoter was integrated into the FLO8 locus, deleting the FLO8 ORF, in a haploid S. cerevisiae strain. This GFP-expressing strain was transformed with pYE-1, carrying EAP1 under control of the GAL1 promoter. A parallel plate flow chamber was assembled atop C. albicans BWP17 cells that had been grown overnight in a polystyrene petri dish, and flow was applied to generate a monolayer of cells. This monolayer remained stably attached to the surface at shear stresses of up to 31 dyn/cm2 (not shown). S. cerevisiae cells expressing GFP and carrying either a plasmid containing EAP1 or an empty vector were then introduced into the flow chamber and allowed to attach to the C. albicans monolayer. The fraction of S. cerevisiae cells remaining adherent was quantified as a function of shear stress, using fluorescence microscopy to detect GFP-expressing cells (Fig. 3). Eighty-one percent of S. cerevisiae cells expressing EAP1 were able to adhere to the C. albicans monolayer under very low flow (0.6 dyn/cm2), while only 40% of cells containing the empty vector were adherent. Likewise, 76% of S. cerevisiae cells expressing EAP1 resisted detachment at 31 dyn/cm2, while only 3% of cells containing the empty vector remained on the surface. These results suggest that Eap1p mediates adhesion to one or more components of the C. albicans cell surface.
FIG. 3.
Eap1p mediates adhesion to C. albicans cells. S. cerevisiae strain SPY415 (flo8Δ; constitutively expresses GFP) containing pYE-1 (EAP1) or pYESR (empty vector) was incubated on a confluent monolayer of C. albicans BWP17 cells in a parallel plate flow chamber for 1 h in 0.1 M sodium phosphate, pH 6.0. Phase-contrast and epifluorescence images were acquired. Buffer flow was increased incrementally to obtain shear stresses from 0 to 30.7 dyn/cm2, and phase-contrast and epifluorescence images were acquired after 3 min at each flow rate. The percentage of S. cerevisiae cells remaining attached at each shear stress, shown on the epifluorescence image, was determined by counting fluorescent cells in each field. Bar = 50 μm.
EAP1 expression is required for biofilm formation under flow in vitro.
EAP1 is involved in adhesion to surfaces and adhesion to other C. albicans cells. These processes play roles in biofilm formation and development (7). To determine whether EAP1 expression affects biofilm formation, we tested the ability of C. albicans eap1 mutant strains to form biofilms under shear flow in a parallel plate flow chamber. In this assay, C. albicans cells experience well-controlled shear stresses, and a previous report demonstrated that biofilms formed under flow produce more extracellular matrix than do biofilms formed under static conditions (21). C. albicans cells were cultured in YPD medium at 30°C overnight, resuspended in 0.1 M sodium phosphate, pH 6.0, and added to the parallel plate flow chamber. After 30 min of incubation at 37°C, fresh YPD was allowed to flow through the chamber at a shear stress of 6 dyn/cm2 for 20 h to allow cell growth and biofilm development.
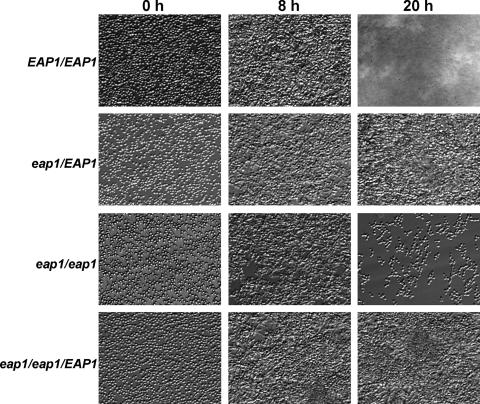
We were able to observe all stages of biofilm formation by capturing phase-contrast images at different times. Wild-type C. albicans biofilm formation proceeded in three developmental phases. During the early phase, yeast cells adhered to the uniform surface of the petri dish and began to divide to form microcolonies (Fig. 4). At 8 to 12 h, wild-type cells produced confluent multiple layers of adherent blastospores, and initial differentiation into pseudohyphae and hyphae occurred (Fig. 4). Finally, multilayer biofilms were formed at 20 h, in which C. albicans communities were completely encased within extracellular material (Fig. 4). The eap1 null mutant was able to adhere to the polystyrene surface under this shear stress and formed pseudohyphae and hyphae at 8 h (Fig. 4). However, the biofilm reached the maximum thickness of a few layers of cells at 8 h, and the number of cells retained in the chamber decreased after 8 h. Almost all eap1 null mutant cells were washed out of the chamber at 20 h, perhaps reflecting the defect in cell-cell and cell-substrate adhesion as colony size increased and the colonies experienced greater shear forces away from the chamber wall (Fig. 4). The heterozygous strain formed a confluent monolayer containing yeast and filamentous cells (Fig. 4) but was not able to form a thick biofilm, even after 48 h (not shown). Thus, it appears that EAP1 gene dosage, not solely the kinetics of biofilm formation, affects the qualitative ability of C. albicans to form biofilms in vitro. The eap1/eap1::EAP1 strain formed a biofilm to a similar extent as the heterozygous strain (Fig. 4).
FIG. 4.
EAP1 expression is required for in vitro biofilm formation under flow. C. albicans strains YJB6284 (EAP1/EAP1), SPY313 (eap1Δ/EAP1), SPY314 (eap1Δ/eap1Δ), and SPY315 (eap1Δ/eap1Δ/EAP1) were cultured on the surface of a parallel plate flow chamber with YPD at 37°C flowing at 6 dyn/cm2. Cells were allowed to incubate on the surface in sodium phosphate buffer for 30 min prior to the initiation of flow.
EAP1 expression is required for C. albicans biofilm formation in vivo.
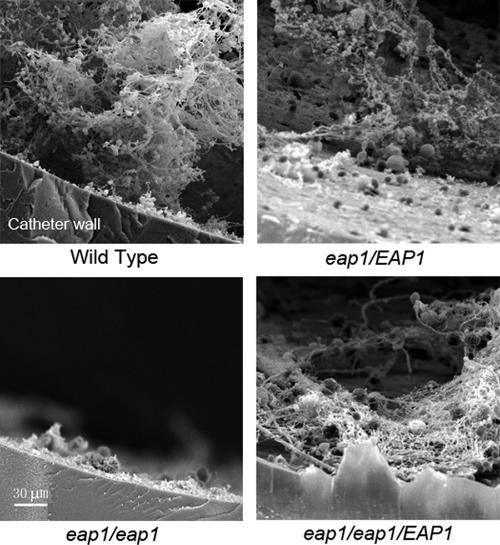
While an in vitro biofilm assay provides a regulatable environment and permits monitoring of biofilm formation in real time, it lacks certain features that are important for biofilm formation in vivo, such as the host immune response and surface fouling with serum and extracellular matrix components. To determine if EAP1 expression affects biofilm formation in vivo, we tested wild-type and eap1 mutant C. albicans strains in a rat model that simulates a central venous catheter biofilm infection (2). Cells were grown in YPD, resuspended in PBS (pH 7.0), and instilled in catheters that had been placed in the internal jugular veins of rats and conditioned for 24 h prior to infection. After 24 h, the catheters were removed from the rats, cut into 5-mm transverse segments, and examined by scanning electron microscopy. Wild-type cells formed a mature biofilm consisting of C. albicans cells embedded in a network of extracellular matrix strands that covered virtually the entire surface of the catheter (Fig. 5). In contrast, the eap1 null mutant was not able to form a biofilm on the catheter surface (Fig. 5). The heterozygous strain formed a thinner biofilm that contained fewer cells and was sparsely distributed over the catheter surface (Fig. 5). The eap1/eap1::EAP1 strain formed a biofilm to a level similar to that of the heterozygous strain (Fig. 5). Together, the observations shown in Fig. 4 and 5 illustrate the necessity of EAP1 expression for biofilm formation and development under flow in vitro and on a catheter surface in vivo.
FIG. 5.
EAP1 expression affects biofilm formation in a rat central venous catheter model. Scanning electron microscopy images of C. albicans strain YJB6284 (EAP1/EAP1), SPY313 (eap1Δ/EAP1), SPY314 (eap1Δ/eap1Δ), and SPY315 (eap1Δ/eap1Δ/EAP1) biofilm cross sections formed in a rat central venous catheter model. Catheters were conditioned for 24 h, 5 × 105 CFU were instilled into the catheters, and biofilms were allowed to develop for 24 h. Catheter segments were fixed overnight and dehydrated by a series of ethanol washes. Samples were observed in the high-vacuum mode at 10 kV. Bar = 30 μm.
EAP1 expression is upregulated in biofilms.
Total RNA was isolated from C. albicans YJB6284 cells associated with in vivo biofilms developed in catheters or with in vitro biofilms grown in RPMI 1640 in tissue culture flasks undergoing agitation. RNA was also obtained from suspension-cultured cells under identical culture conditions as a reference. No variations in the morphology of planktonic and in vitro sessile cultures were observed at different time points up to 24 h; both yeast and filamentous cells were observed (not shown). The change in abundance of EAP1 mRNA extracted from the biofilm relative to that obtained from planktonic cells, measured by quantitative real-time RT-PCR, is shown in Fig. 6. In both cases, the EAP1 mRNA abundance in the biofilm state was nearly twice that found in the corresponding planktonic cells (P < 0.01). These differential patterns of EAP1 expression in sessile cells compared with their planktonic counterparts further suggest a role for EAP1 in C. albicans biofilm formation. However, it is not clear whether cells with higher levels of EAP1 expression are more likely to adhere to the surface and participate in biofilm formation or whether growth in a sessile state induces EAP1 expression.
FIG. 6.
EAP1 mRNA abundance for in vivo biofilm-associated or in vitro sessile C. albicans YJB6284 cells compared to that for suspension-grown cells. Five hundred nanograms of RNA harvested from biofilm-associated C. albicans cells isolated from the rat central venous catheter model, from sessile cells cultured in RPMI 1640, or from suspension-grown cells was used in separate quantitative RT-PCRs. ACT1 expression was used as an internal control. Data represent the x-fold changes in EAP1 expression in sessile versus planktonic cells. Error bars indicate the standard deviations for two independent experiments.
DISCUSSION
We report that expression of the C. albicans EAP1 gene affects cell adhesion and biofilm formation in vitro and in vivo. The primary amino acid sequence of Eap1p suggests that it is a member of the family of GPI-CWPs. We demonstrated that the C-terminal sequence of Eap1p was required to target HA-tagged Eap1p to the surfaces of C. albicans cells. In the absence of the GPI anchor signal, tagged Eap1p did not cross-link with the cell wall, similar to what was observed for C-terminal deletion of the Candida glabrata CWP Epa1p (14). In addition, HA-Eap1p was released from the cell wall by glucanase treatment. These results strongly suggest that Eap1p is indeed a GPI-CWP.
The availability of the C. albicans genome provides a strategy for predicting ORFs that potentially encode GPI-CWPs based on their common sequence features. Fifty-four ORFs in assembly 6 of the C. albicans genome have been predicted to encode GPI-CWPs (53), and 104 putative GPI-anchored protein ORFs have been identified in assembly 19 of the C. albicans genome (11). Most of these proteins have not been characterized functionally, although several adhesins have been identified and at least partially characterized, including Eap1p, Hwp1p, and members of the Als family of proteins (24, 55). The existence of multiple putative adhesins in C. albicans may account for the diversity of substrates, from human tissues to plastic prostheses, to which C. albicans cells are able to attach. Furthermore, genetic recombination and epigenetic regulation have been suggested to provide a reservoir of adhesins with new functions in S. cerevisiae (56) and may also have contributed to evolution of adhesin function in C. albicans. In this work, we demonstrated that EAP1 expression confers adhesion of C. albicans to polystyrene, human epithelial cells, and C. albicans cells, although it is not the sole contributor to adhesion to these surfaces. Understanding the nature of Eap1p-mediated adhesion, as well as the contributions of other adhesins, to different materials will presumably increase our ability to design strategies to prevent such adhesion and resulting infections.
Many Candida infections involve the formation of biofilms on implanted devices, such as central venous catheters. These biofilms exhibit resistance to a variety of antifungal agents currently in clinical use, including amphotericin B and fluconazole (1, 4, 23). Adhesion to the surface is the initial step in biofilm formation, and cell-cell interactions may be important in the hierarchical organization of cells within the biofilm, so presumably adhesin expression and activity can regulate the biofilm formation rate and perhaps the biofilm structure. We found that the EAP1 mRNA abundance in the biofilm state was nearly twice that in the corresponding planktonic cells, and eap1 null mutants were defective in C. albicans biofilm development under shear flow in vitro and in an in vivo central venous catheter biofilm model. This increase in EAP1 expression may be the result of a selection process in which cells containing higher surface densities of Eap1p are more likely to attach to surfaces and to form biofilms. Alternatively, surface binding may directly induce EAP1 expression; the mitogen-activated protein kinase Mkc1p is activated by cell surface binding and enhances hyphal growth and biofilm formation (30).
Als3p and Hwp1p have also been implicated, directly and indirectly, in biofilm formation (39, 41, 60). The transcription factor Bcr1p contributes to biofilm formation and induces expression of the adhesins Als3p and Hwp1p, among other CWPs (40). While microarray data did not identify changes in EAP1 transcription upon BCR1 deletion (40), alternative transcriptional pathways may affect biofilm formation via EAP1 expression. Eap1p and Hwp1p both appear to be necessary for biofilm formation in the rat central venous catheter model, while Als3p is not strictly required (39, 41). The functional redundancy of adhesins and mechanisms of their regulation in biofilm formation are not yet clear.
No significant effect of EAP1 expression on C. albicans morphogenesis was observed. Hypha formation by the eap1/eap1 strain was similar to that of the wild-type strain in liquid culture in response to 0.1% to 1% fetal bovine serum and under conditions used to assess biofilm formation (Fig. 4). Thus, it is likely that the effect of EAP1 on adhesion and biofilm formation is independent of filamentation and that adhesion and filamentation contribute to biofilm development separately. Segregation of adhesion and filamentation was also observed in Als3p-mediated biofilm formation on silicone by an efg1/efg1 strain, which formed biofilms lacking hyphal cells (60). EAP1 is expressed in both yeast and hyphal cells (32), in contrast to ALS3 and HWP1, which are specifically expressed during hyphal development (3, 49).
The observation of biofilm growth in vitro indicated that eap1 null mutants exhibited sufficient adhesion to permit microcolony formation on a polystyrene surface, but as these colonies grew larger they were unable to maintain sufficient adhesion, prohibiting full biofilm development. The nature of the requirement of Eap1p-mediated adhesion during biofilm formation in the rat central venous catheter model is unknown. One likely hypothesis is that eap1 mutants failed to adhere to the catheter surface, as very few cells were observed on the catheter surface. Other possibilities include Eap1p involvement in attaching C. albicans cells to the exopolymeric matrix of an immature biofilm and the possibility that exopolymeric matrix synthesis requires Eap1p.
In summary, these data demonstrate that C. albicans EAP1 encodes a GPI-dependent, glucan-cross-linked CWP that mediates adhesion to polystyrene, C. albicans cells, and epithelial cells. EAP1 expression is required for C. albicans biofilm development under shear stress in vitro and in an in vivo central venous catheter biofilm model.
Acknowledgments
We thank Eric Benson and Judith Berman for providing the C. albicans strain YJB6284, Aaron Mitchell for providing the C. albicans strain BWP17, Jurgen Wendland for providing plasmids for PCR-based gene disruption in C. albicans, and Yuxin Mao and Brian Wong for GFP expression plasmids.
This work was supported by a Whitaker Foundation grant (RG-01-0421) to S.P.P. and was partially supported by the NSF-funded University of Wisconsin Nanoscale Science and Engineering Center (UW-NSEC). M.J.S. was supported by the NIH Biotechnology Training Program (GM08349).
Footnotes
Published ahead of print on 6 April 2007.
REFERENCES
- 1.Al-Fattani, M. A., and L. J. Douglas. 2004. Penetration of Candida biofilms by antifungal agents. Antimicrob. Agents Chemother. 48:3291-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andes, D., J. Nett, P. Oschel, R. Albrecht, K. Marchillo, and A. Pitula. 2004. Development and characterization of an in vivo central venous catheter Candida albicans biofilm model. Infect. Immun. 72:6023-6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Argimon, S., J. A. Wishart, R. Leng, S. Macaskill, A. Mavor, T. Alexandris, S. Nicholls, A. W. Knight, B. Enjalbert, R. Walmsley, F. C. Odds, N. A. Gow, and A. J. Brown. 2007. Developmental regulation of an adhesin gene during cellular morphogenesis in the fungal pathogen Candida albicans. Eukaryot. Cell 6:682-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bachmann, S. P., K. VandeWalle, G. Ramage, T. F. Patterson, B. L. Wickes, J. R. Graybill, and J. L. Lopez-Ribot. 2002. In vitro activity of caspofungin against Candida albicans biofilms. Antimicrob. Agents Chemother. 46:3591-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baillie, G. S., and L. J. Douglas. 1999. Candida biofilms and their susceptibility to antifungal agents. Methods Enzymol. 310:644-656. [DOI] [PubMed] [Google Scholar]
- 6.Bensen, E. S., S. G. Filler, and J. Berman. 2002. A forkhead transcription factor is important for true hyphal as well as yeast morphogenesis in Candida albicans. Eukaryot. Cell 1:787-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blankenship, J. R., and A. P. Mitchell. 2006. How to build a biofilm: a fungal perspective. Curr. Opin. Microbiol. 9:588-594. [DOI] [PubMed] [Google Scholar]
- 8.Chaffin, W. L., J. L. Lopez-Ribot, M. Casanova, D. Gozalbo, and J. P. Martinez. 1998. Cell wall and secreted proteins of Candida albicans: identification, function, and expression. Microbiol. Mol. Biol. Rev. 62:130-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandra, J., D. M. Kuhn, P. K. Mukherjee, L. L. Hoyer, T. McCormick, and M. A. Ghannoum. 2001. Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J. Bacteriol. 183:5385-5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandra, J., J. D. Patel, J. Li, G. Zhou, P. K. Mukherjee, T. S. McCormick, J. M. Anderson, and M. A. Ghannoum. 2005. Modification of surface properties of biomaterials influences the ability of Candida albicans to form biofilms. Appl. Environ. Microbiol. 71:8795-8801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Groot, P. W., A. D. de Boer, J. Cunningham, H. L. Dekker, L. de Jong, K. J. Hellingwerf, C. de Koster, and F. M. Klis. 2004. Proteomic analysis of Candida albicans cell walls reveals covalently bound carbohydrate-active enzymes and adhesins. Eukaryot. Cell 3:955-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Douglas, L. J. 2003. Candida biofilms and their role in infection. Trends Microbiol. 11:30-36. [DOI] [PubMed] [Google Scholar]
- 13.Edmond, M. B., S. E. Wallace, D. K. McClish, M. A. Pfaller, R. N. Jones, and R. P. Wenzel. 1999. Nosocomial bloodstream infections in United States hospitals: a three-year analysis. Clin. Infect. Dis. 29:239-244. [DOI] [PubMed] [Google Scholar]
- 14.Frieman, M. B., J. M. McCaffery, and B. P. Cormack. 2002. Modular domain structure in the Candida glabrata adhesin Epa1p, a beta1,6 glucan-cross-linked cell wall protein. Mol. Microbiol. 46:479-492. [DOI] [PubMed] [Google Scholar]
- 15.Fu, Y., A. S. Ibrahim, D. C. Sheppard, Y. C. Chen, S. W. French, J. E. Cutler, S. G. Filler, and J. E. Edwards, Jr. 2002. Candida albicans Als1p: an adhesin that is a downstream effector of the EFG1 filamentation pathway. Mol. Microbiol. 44:61-72. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Sanchez, S., S. Aubert, I. Iraqui, G. Janbon, J. M. Ghigo, and C. d'Enfert. 2004. Candida albicans biofilms: a developmental state associated with specific and stable gene expression patterns. Eukaryot. Cell 3:536-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaur, N. K., and S. A. Klotz. 1997. Expression, cloning, and characterization of a Candida albicans gene, ALA1, that confers adherence properties upon Saccharomyces cerevisiae for extracellular matrix proteins. Infect. Immun. 65:5289-5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gietz, D., A. St. Jean, R. A. Woods, and R. H. Schiestl. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20:1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gillum, A. M., E. Y. Tsay, and D. R. Kirsch. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198:179-182. [DOI] [PubMed] [Google Scholar]
- 20.Gola, S., R. Martin, A. Walther, A. Dunkler, and J. Wendland. 2003. New modules for PCR-based gene targeting in Candida albicans: rapid and efficient gene targeting using 100 bp of flanking homology region. Yeast 20:1339-1347. [DOI] [PubMed] [Google Scholar]
- 21.Hawser, S. P., G. S. Baillie, and L. J. Douglas. 1998. Production of extracellular matrix by Candida albicans biofilms. J. Med. Microbiol. 47:253-256. [DOI] [PubMed] [Google Scholar]
- 22.Hawser, S. P., and L. J. Douglas. 1994. Biofilm formation by Candida species on the surface of catheter materials in vitro. Infect. Immun. 62:915-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hawser, S. P., and L. J. Douglas. 1995. Resistance of Candida albicans biofilms to antifungal agents in vitro. Antimicrob. Agents Chemother. 39:2128-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoyer, L. L. 2001. The ALS gene family of Candida albicans. Trends Microbiol. 9:176-180. [DOI] [PubMed] [Google Scholar]
- 25.Jarvis, W. R. 1995. Epidemiology of nosocomial fungal infections, with emphasis on Candida species. Clin. Infect. Dis. 20:1526-1530. [DOI] [PubMed] [Google Scholar]
- 26.Jeng, H. W., A. R. Holmes, and R. D. Cannon. 2005. Characterization of two Candida albicans surface mannoprotein adhesins that bind immobilized saliva components. Med. Mycol. 43:209-217. [DOI] [PubMed] [Google Scholar]
- 27.Kriznik, A., M. Bouillot, J. Coulon, and F. Gaboriaud. 2005. Morphological specificity of yeast and filamentous Candida albicans forms on surface properties. C. R. Biol. 328:928-935. [DOI] [PubMed] [Google Scholar]
- 28.Kuhn, D. M., J. Chandra, P. K. Mukherjee, and M. A. Ghannoum. 2002. Comparison of biofilms formed by Candida albicans and Candida parapsilosis on bioprosthetic surfaces. Infect. Immun. 70:878-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumamoto, C. A. 2002. Candida biofilms. Curr. Opin. Microbiol. 5:608-611. [DOI] [PubMed] [Google Scholar]
- 30.Kumamoto, C. A. 2005. A contact-activated kinase signals Candida albicans invasive growth and biofilm development. Proc. Natl. Acad. Sci. USA 102:5576-5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumamoto, C. A., and M. D. Vinces. 2005. Alternative Candida albicans lifestyles: growth on surfaces. Annu. Rev. Microbiol. 59:113-133. [DOI] [PubMed] [Google Scholar]
- 32.Li, F., and S. P. Palecek. 2003. EAP1, a Candida albicans gene involved in binding human epithelial cells. Eukaryot. Cell 2:1266-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, F., and S. P. Palecek. 2005. Identification of Candida albicans genes that induce Saccharomyces cerevisiae cell adhesion and morphogenesis. Biotechnol. Prog. 21:1601-1609. [DOI] [PubMed] [Google Scholar]
- 34.Li, X., Z. Yan, and J. Xu. 2003. Quantitative variation of biofilms among strains in natural populations of Candida albicans. Microbiology 149:353-362. [DOI] [PubMed] [Google Scholar]
- 35.Liu, H., J. Kohler, and G. R. Fink. 1994. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266:1723-1726. [DOI] [PubMed] [Google Scholar]
- 36.Lopez-Ribot, J. L. 2005. Candida albicans biofilms: more than filamentation. Curr. Biol. 15:R453-R455. [DOI] [PubMed] [Google Scholar]
- 37.Mao, Y., Z. Zhang, and B. Wong. 2003. Use of green fluorescent protein fusions to analyse the N- and C-terminal signal peptides of GPI-anchored cell wall proteins in Candida albicans. Mol. Microbiol. 50:1617-1628. [DOI] [PubMed] [Google Scholar]
- 38.Mukherjee, P. K., G. Zhou, R. Munyon, and M. A. Ghannoum. 2005. Candida biofilm: a well-designed protected environment. Med. Mycol. 43:191-208. [DOI] [PubMed] [Google Scholar]
- 39.Nobile, C. J., D. R. Andes, J. E. Nett, F. J. Smith, F. Yue, Q. T. Phan, J. E. Edwards, S. G. Filler, and A. P. Mitchell. 2006. Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathog. 2:e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nobile, C. J., and A. P. Mitchell. 2005. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Curr. Biol. 15:1150-1155. [DOI] [PubMed] [Google Scholar]
- 41.Nobile, C. J., J. E. Nett, D. R. Andes, and A. P. Mitchell. 2006. Function of Candida albicans adhesin Hwp1 in biofilm formation. Eukaryot. Cell 5:1604-1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Palecek, S. P., A. S. Parikh, and S. J. Kron. 2000. Genetic analysis reveals that FLO11 upregulation and cell polarization independently regulate invasive growth in Saccharomyces cerevisiae. Genetics 156:1005-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pfaller, M. A., R. N. Jones, S. A. Messer, M. B. Edmond, and R. P. Wenzel. 1998. National surveillance of nosocomial blood stream infection due to Candida albicans: frequency of occurrence and antifungal susceptibility in the SCOPE program. Diagn. Microbiol. Infect. Dis. 31:327-332. [DOI] [PubMed] [Google Scholar]
- 44.Price, C. L., D. W. Williams, M. G. Waters, L. Coulthwaite, J. Verran, R. L. Taylor, D. Stickler, and M. A. Lewis. 2005. Reduced adherence of Candida to silane-treated silicone rubber. J. Biomed. Mater. Res. B 74:481-487. [DOI] [PubMed] [Google Scholar]
- 45.Ramage, G., S. P. Saville, D. P. Thomas, and J. L. Lopez-Ribot. 2005. Candida biofilms: an update. Eukaryot. Cell 4:633-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramage, G., S. P. Saville, B. L. Wickes, and J. L. Lopez-Ribot. 2002. Inhibition of Candida albicans biofilm formation by farnesol, a quorum-sensing molecule. Appl. Environ. Microbiol. 68:5459-5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramage, G., K. Vandewalle, B. L. Wickes, and J. L. Lopez-Ribot. 2001. Characteristics of biofilm formation by Candida albicans. Rev. Iberoam. Micol. 18:163-170. [PubMed] [Google Scholar]
- 48.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 49.Sharkey, L. L., M. D. McNemar, S. M. Saporito-Irwin, P. S. Sypherd, and W. A. Fonzi. 1999. HWP1 functions in the morphological development of Candida albicans downstream of EFG1, TUP1, and RBF1. J. Bacteriol. 181:5273-5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singleton, D. R., J. Masuoka, and K. C. Hazen. 2005. Surface hydrophobicity changes of two Candida albicans serotype B mnn4Δ mutants. Eukaryot. Cell 4:639-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Staab, J. F., S. D. Bradway, P. L. Fidel, and P. Sundstrom. 1999. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science 283:1535-1538. [DOI] [PubMed] [Google Scholar]
- 52.Suci, P. A., G. G. Geesey, and B. J. Tyler. 2001. Integration of Raman microscopy, differential interference contrast microscopy, and attenuated total reflection Fourier transform infrared spectroscopy to investigate chlorhexidine spatial and temporal distribution in Candida albicans biofilms. J. Microbiol. Methods 46:193-208. [DOI] [PubMed] [Google Scholar]
- 53.Sundstrom, P. 2002. Adhesion in Candida spp. Cell. Microbiol. 4:461-469. [DOI] [PubMed] [Google Scholar]
- 54.Svarovsky, M. J., and S. P. Palecek. 2005. Disruption of LRG1 inhibits mother-daughter separation in Saccharomyces cerevisiae. Yeast 22:1117-1132. [DOI] [PubMed] [Google Scholar]
- 55.Verstrepen, K. J., and F. M. Klis. 2006. Flocculation, adhesion and biofilm formation in yeasts. Mol. Microbiol. 60:5-15. [DOI] [PubMed] [Google Scholar]
- 56.Verstrepen, K. J., T. B. Reynolds, and G. R. Fink. 2004. Origins of variation in the fungal cell surface. Nat. Rev. Microbiol. 2:533-540. [DOI] [PubMed] [Google Scholar]
- 57.Walther, A., and J. Wendland. 2003. An improved transformation protocol for the human fungal pathogen Candida albicans. Curr. Genet. 42:339-343. [DOI] [PubMed] [Google Scholar]
- 58.Wilson, R. B., D. Davis, B. M. Enloe, and A. P. Mitchell. 2000. A recyclable Candida albicans URA3 cassette for PCR product-directed gene disruptions. Yeast 16:65-70. [DOI] [PubMed] [Google Scholar]
- 59.Wilson, R. B., D. Davis, and A. P. Mitchell. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao, X., K. J. Daniels, S. H. Oh, C. B. Green, K. M. Yeater, D. R. Soll, and L. L. Hoyer. 2006. Candida albicans Als3p is required for wild-type biofilm formation on silicone elastomer surfaces. Microbiology 152:2287-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao, X., S. H. Oh, K. M. Yeater, and L. L. Hoyer. 2005. Analysis of the Candida albicans Als2p and Als4p adhesins suggests the potential for compensatory function within the Als family. Microbiology 151:1619-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]