Abstract
Identifying and characterizing transcriptional regulatory networks is important for guiding experimental tests on gene function. The characterization of regulatory networks allows comparisons among both closely and distantly related species, providing insight into network evolution, which is predicted to correlate with the adaptation of different species to particular environmental niches. One of the most intensely studied regulatory factors in the yeast Saccharomyces cerevisiae is the bZIP transcription factor Gcn4p. Gcn4p is essential for a global transcriptional response when S. cerevisiae experiences amino acid starvation. In the filamentous ascomycete Neurospora crassa, the ortholog of GCN4 is called the cross pathway control-1 (cpc-1) gene; it is required for the ability of N. crassa to induce a number of amino acid biosynthetic genes in response to amino acid starvation. Here, we deciphered the CPC1 regulon by profiling transcription in wild-type and cpc-1 mutant strains with full-genome N. crassa 70-mer oligonucleotide microarrays. We observed that at least 443 genes were direct or indirect CPC1 targets; these included 67 amino acid biosynthetic genes, 16 tRNA synthetase genes, and 13 vitamin-related genes. Comparison among the N. crassa CPC1 transcriptional profiling data set and the Gcn4/CaGcn4 data sets from S. cerevisiae and Candida albicans revealed a conserved regulon of 32 genes, 10 of which are predicted to be directly regulated by Gcn4p/CPC1. The 32-gene conserved regulon comprises mostly amino acid biosynthetic genes. The comparison of regulatory networks in species with clear orthology among genes sheds light on how gene interaction networks evolve.
A major means of responding to stress and environmental perturbations is to alter patterns of gene expression. The advent of full-genome microarrays enables the measurement of mRNA levels in response to these alterations. In the yeast Saccharomyces cerevisiae, the bZIP transcription factor Gcn4p is the major regulator of genes whose expression changes in response to amino acid starvation (26, 28). Gcn4p activates the transcription of more than 30 amino acid biosynthetic genes in 12 different biosynthetic pathways; mutations in GCN4 abolish transcriptional responses associated with amino acid starvation (27, 49). Starvation for single amino acids leads to increased synthesis of enzymes associated with amino acid biosynthesis (25). Microarray analysis has further determined that Gcn4p affects the transcription of a large number of genes (∼1,000) in response to amino acid starvation (34, 45).
The production of Gcn4p itself is induced during amino acid starvation via translational regulation; four short upstream open reading frames (uORFs) encoding products of 2 to 4 amino acids affect the ribosome scanning of GCN4 mRNA under conditions of stress (24, 44, 61). The translational regulation of GCN4 is associated with the kinase activity of Gcn2p, which is stimulated by the binding of deacylated tRNAs that accumulate under conditions of amino acid starvation. The phosphorylation target of Gcn2p is the α subunit of eukaryotic translation initiation factor 2 (eIF2). In addition to amino acid starvation, the Gcn4p pathway is induced by starvation for purines (50), glucose limitation (71), and exposure to DNA-damaging agents, such as methyl methanesulfonate (45) and rapamycin (67); it is not clear whether the patterns of translational regulation of GCN4 are identical under all these stress conditions.
Gcn4p binds to 5′TGACTCA3′ sequences in the promoters of target genes, such as HIS4, HIS3, ILV1, and ILV2 (1). By chromatin immunoprecipitation (ChIP) experiments, the promoters of 207 genes were found to be bound by Gcn4p (P < 0.001) (22, 36); the transcriptional profiling and ChIP data sets show an overlap of 95 genes. Gcn4p was also shown to interact physically with numerous other proteins that are predicted to affect Gcn4p DNA binding and function in vivo (22, 33, 60).
The filamentous fungus Neurospora crassa diverged from S. cerevisiae approximately 350 million years ago (16). Previously, it was shown by genetic analyses that cross pathway or general amino acid control is evolutionarily conserved between S. cerevisiae and N. crassa (reviewed in reference 53). A mutation identified in N. crassa confers a phenotype similar to that of gcn4 mutants of S. cerevisiae; the cross pathway control (cpc-1) mutant failed to induce the expression of a number of amino acid biosynthetic genes upon amino acid starvation (3, 4, 18, 37, 56). CPC1 is required for the global regulatory response observed during starvation for amino acids (9-11, 46). The cpc-1 locus encodes a bZIP transcription factor with sequence similarity to Gcn4 (46). The cpc-1 mRNA comprises a long leader segment with two uORFs of 2 and 41 codons (53). As that of GCN4, the translation of cpc-1 mRNA is regulated by amino acid starvation; the uORFs play a regulatory role (53). An ortholog of the GCN2 eIF2α kinase gene, cpc-3, is important for the regulation of cpc-1 function in response to amino acid starvation (55).
The level of amino acid identity between CPC1 and Gcn4p is only 25% (similarity, 42%). The carboxy terminus of CPC1 contains the DNA binding and dimerization domain (57 amino acid residues) (46), and regions associated with DNA binding and transcriptional activation in Gcn4p and CPC1 are more similar in primary sequence (30 amino acid residues involved in DNA binding are 70% identical) than other regions. The introduction of GCN4 into a cpc-1 mutant of N. crassa did not restore cross pathway control (48). Only constructs in which the DNA binding domain of cpc-1 was replaced by that of GCN4 complemented the cpc-1 mutant phenotype.
In N. crassa, cpc-1 is regulated throughout the asexual life cycle (14, 31, 54). The relative levels of DNA binding activity in cell extracts correlate with CPC1 binding activity (14). Binding studies of the trp-3 and arg-2 promoter regions show that CPC1 binds to a DNA sequence identical to that bound by Gcn4p (5′TGACTC3′). However, the size of the bound complex is larger than the predicted CPC1 homodimer, indicating that additional unknown proteins also bind to these regions. As many as 20% of the total detectable mRNA species in N. crassa are influenced directly or indirectly by the presence of a functional cpc-1 gene during amino acid starvation (17), including the cpc-1 mRNA itself; CPC1 regulates it own expression (14). The synthesis of many polypeptides also increases in response to arginine limitation, which is dependent on functional CPC1 (17, 58). These data indicate that CPC1 controls a large number of N. crassa genes, similar in magnitude to the number reported to be controlled by Gcn4p in S. cerevisiae (45).
In this study, we determined the regulon of CPC1 by profiling transcription in a wild-type (WT) strain and a cpc-1 mutant under normal growth conditions and under amino acid starvation. As with the GCN4 regulon, the transcription of a large number of genes was affected by conditions of amino acid starvation and was also dependent upon functional CPC1. At least 443 genes were predicted to be either direct or indirect CPC1 targets, including 67 amino acid biosynthesis genes, 16 tRNA synthetase genes, and 13 vitamin metabolism-related genes. In addition, a large number of genes showed CPC1-dependent repression. We compared our results to those obtained in a similar study of both WT strains and gcn4 mutants of S. cerevisiae and Candida albicans (62). We identified conserved and divergent elements in the response to amino acid starvation among these three fungi. A comparison of these regulons showed that the regulation of only 32 orthologous genes is conserved among all three fungi. Of these 32 genes, only 10 retain the conserved cis element bound by Gcn4p/CPC1.
MATERIALS AND METHODS
N. crassa genome 70-mer oligonucleotide microarray design and synthesis.
We developed oligonucleotide microarrays for the Neurospora research community as part of NIH program project grant GM068087. In a previous transcriptional profiling study assessing gene expression patterns associated with asexual spore germination (31), we used an oligonucleotide array comprised of 3,366 predicted N. crassa genes. Based on the results of that study, we revised our design to construct a 70-mer oligonucleotide set corresponding to the predicted 10,526 ORFs of N. crassa (Broad Institute [http://www.broad.mit.edu/annotation/fungi/neurospora_crassa_7/index.html] and the Munich Information Center for Protein Sequences [MIPS; http://mips.gsf.de/genre/proj/ncrassa/) by using the bioinformatics tool ArrayOligoSelector (6, 31). ArrayOligoSelector identifies a unique 70-bp segment to represent each ORF, avoiding self-annealing structures and repetitive sequences. In addition, 384 70-mer oligonucleotides corresponding to intergenic or telomeric regions were included, along with Ambion oligonucleotides for normalization procedures (31). A total of 10,910 70-mer oligonucleotides were synthesized (Illumina, Inc., San Diego). We printed the 10,910 70-mers onto gamma amino propyl silane slides at the University of California—San Francisco Center for Advanced Technology (http://cat.UCSF.edu/). N. crassa microarray slides are available to the research community from the Fungal Genetics Stock Center (http://www.fgsc.net/). Information on the oligonucleotide gene set is available at the Neurospora functional genomics database (http://www.yale.edu/townsend/Links/ffdatabase/introduction.htm).
Strains and culture conditions.
The cpc-1 mutant (FGSC 4264) and the WT sequenced strain (FGSC 2489) were obtained from the Fungal Genetics Stock Center. The cpc-1 CD15 allele mutation is a single-base-pair deletion in codon 93 resulting in a nonfunctional, truncated polypeptide (47). Strains were inoculated onto slants containing Vogel's (69) minimal medium and grown at 30°C for 2 days, followed by incubation at 25°C under constant light for 7 days. Conidia were harvested with water and inoculated into 50 ml of Bird's medium (43) in 250-ml Erlenmeyer flasks at a final concentration of 106 conidia/ml. The cells were grown under constant light at 30°C for 12 h in a gyratory shaker at 300 rpm. An inhibitor of imidazole glycerol phosphate dehydratase, 3-aminotriazole (3-AT), was then added to the cultures of the WT and cpc-1 strains (FGSC 2489 and FGSC 4264, respectively) to a final concentration of 6 or 30 mM. The addition of 3-AT to N. crassa cultures inhibits histidine biosynthesis and causes imidazole glycerol phosphate to accumulate. Control cultures received no 3-AT. All cultures were incubated under identical conditions for a further 2 h. Each sample was represented by duplicate cultures. The mycelium was harvested by filtration, immediately immersed in liquid nitrogen, and stored at −70°C.
In N. crassa, amino acid starvation has been experimentally induced by the addition of 6 mM 3-AT to hyphal cultures (14, 54). To determine the appropriate concentration for a transcriptional profiling analysis of amino acid starvation in N. crassa, we performed profiling experiments with 12-h-old hyphal cultures grown in liquid medium and exposed or not to 6 or 30 mM 3-AT for 30 min or 2 h. Approximately 300 genes showed differential regulation after a 2-h exposure to 3-AT at both concentrations (6 and 30 mM); expression levels were more affected by the duration of 3-AT treatment than by the 3-AT concentration. To minimize the possible side effects of using too high a concentration of 3-AT, for subsequent analyses we exposed 12-h-old hyphae to 6 mM 3-AT for 2 h.
RNA isolation and cDNA labeling.
RNA isolation was accomplished using TRIzol (Invitrogen Life Technologies), and RNA was subsequently purified using the RNAeasy kit (QIAGEN) according to the manufacturer's protocols. For cDNA synthesis and labeling, the Pronto kit (catalog no. 40076; Corning) was used according to the manufacturer's specifications. Briefly, cDNA was synthesized from a mixture containing 10 μg total RNA and oligo(dT) primer, ChipShot reverse transcriptase, and aminoallyl-deoxynucleoside triphosphate and incubated at 42°C for 2 h. The cDNA was purified by using a ChipShot membrane column. The dyes Cy3 and Cy5 (Amersham; catalog no. RPN5661) were incorporated into cDNA by adding Cy3 or Cy5 monofunctional N-hydroxysuccinimide ester dye to the cDNA solution for 1 h at 22°C. The cDNA was subsequently cleaned by using a ChipShot membrane column and dried under vacuum and was subsequently used for hybridization.
Hybridization and image acquisition.
Slides were prehybridized and hybridized at 42°C according to the instructions of the manufacturer of the Pronto kit (Corning; catalog no. 40076). Briefly, the presoak solution was prewarmed (42°C) for 30 min, and then a 1% volume of sodium borohydride was added to the presoak solution and the solution was mixed well. The microarray slide was then added to the solution and incubated at 42°C for 20 min. Slides were washed, transferred to prewarmed prehybridization solution (42°C) for 15 min, and then rewashed. The presoak and prehybridization steps reduced slide background. Following slide prehybridization, labeled cDNA was resuspended in 30 μl of hybridization solution (Pronto kit) and the suspension was heated at 95°C for 5 min and subsequently pipetted into the space between a microarray slide and a LifterSlip cover glass (Erie Scientific, Portsmouth, NH). Hybridization was carried out for 16 h at 42°C, and unbound DNA was washed off according to the manufacturer's instructions. A GenePix 4000B scanner (Axon Instruments, CA) was used to acquire images, and GenePix Pro6 software was used to quantify hybridization signals. Low-quality spots were flagged automatically by GenePix software, and subsequently each slide was inspected manually.
Experimental design and data analysis.
For the transcriptional profiling of the WT and the cpc-1 mutant under conditions of amino acid starvation, we chose to use a closed-circuit design for microarray comparisons (Fig. S1 in the supplemental material). Circuit designs for microarrays are statistically robust and improve resolution in identifying differentially regulated genes compared to designs for microarrays that use a universal reference (32, 63, 64, 68, 72).
Hybridized spots with a mean fluorescence intensity for at least one of the Cy3 and Cy5 dyes that was greater than the mean background intensity plus three standard deviations were scored for further analysis if less than 0.02% of the pixels were saturated. Normalized ratio data were analyzed using Bayesian Analysis of Gene Expression Levels software, with which we inferred a relative expression level and a credible interval for each gene in each sample (65). These inferred levels of gene expression were then clustered (15) using Hierarchical Clustering Explorer 2.0 (57), in which similarity in expression patterns between genes is measured as Pearson's correlation coefficient and the closest two genes or clusters are successively joined. The functional catalog FunCat created by MIPS (19, 52) was mined to associate functional annotations with Neurospora genes (http://mips.gsf.de/genre/proj/ncrassa/Search/Catalogs/catalog.jsp). The statistically significant overrepresentation of gene groups in functional categories relative to the whole genome was determined with the MIPS online FunCat system using hypergeometric distribution for P value calculation (http://mips.gsf.de/proj/funcatDB/help_p-value.html).
cis-element discovery and enrichment analysis.
Enrichment of motifs (P < 0.001) was assessed using Fisher's exact test performed by the Fisher test function implemented in the R 1.9 program (http://bioconductor.org). A motif logo illustrating the consensus sequence was obtained using the WebLogo program (http://weblogo.berkeley.edu). For the identification of the CPC1 DNA binding motif, the total number of putative targets was evaluated by MIPS functional category analysis. Motif searches were conducted using three programs, BioProspector (42), MDscan (38), and MEME (2), on segments of 500 bases upstream of predicted translational start sites, which were downloaded from the Broad Institute N. crassa database release version 7 (http://www.broad.mit.edu/annotation/fungi/neurospora_crassa_7/). The different predictions were compared and inspected manually. The CPC1 DNA binding cis-element matrix was built based on the MDscan prediction with the default parameter. With the results from the initial MDscan step, the conserved 7-bp region in the middle of the element was used to make the initial matrix from motif 1 (10 bp in width). By using the initial matrix, the promoter regions of the 67 amino acid genes were evaluated by the PATSER program (23). Fifty-seven genes were retained when known CPC1 targets were used to determine the cutoff value. The predicted cis-elements in the promoter regions of these 57 genes were used to build the CPC1 DNA binding matrix and logo. To identify additional CPC1 target genes in the N. crassa and S. cerevisiae genomes, predicted promoter regions were scanned using the CPC1/Gcn4p matrix (23). Enrichment analysis results were binned into a group of genes with perfect matches to the CPC1/Gcn4p consensus and a second bin reflecting a match score of 70% of the full score.
Identification of orthologs and regulogs in N. crassa and other fungi.
The sequences of predicted ORFs in the N. crassa genome were downloaded from Broad Institute N. crassa database release version 7 (http://www.broad.mit.edu/annotation/genome/neurospora/Downloads.html). Sequences of S. cerevisiae ORFs and upstream 500-bp promoter regions were downloaded from the Saccharomyces Genome Database (ftp://genome-ftp.stanford.edu/pub/yeast/data_download/sequence/genomic_sequence/orf_dna/archive/), and sequences of C. albicans ORFs and upstream regions were downloaded from http://www.candidagenome.org/download/sequence/genomic_sequence/. Orthologous genes were identified as best bidirectional hits by using BLASTp with a cutoff value of 1e−10. If orthologs were CPC1, Gcn4p, or CaGcn4p targets, they were defined as regulogs in a common regulon. The term regulog extends the concept of a protein-DNA interolog as defined by Yu et al. (73). Targets can be direct (genes involved in Gcn4p/CPC1-DNA interactions) or indirect (genes exhibiting a transcriptional response affected by mutations in cpc-1/gcn4/Cagcn4 that are not direct DNA targets). The Gcn4p target gene data set includes ChIP data (http://jura.wi.mit.edu/young_public/regulatory_code/GWLD.html; 22, 36) and transcriptional profiling data (34, 45) that were obtained from haploid and diploid cells, respectively. Transcriptional profiling data for WT strains and gcn4 mutants of S. cerevisiae and C. albicans were kindly provided by Alan G. Hinnebusch (NIH NICHD) and Alistair Brown (University of Aberdeen), respectively. We reanalyzed the CaGcn4 profiling data using the Bayesian Analysis of Gene Expression Levels software (65) and normalized expression data. Genes were defined as CaGcn4p targets if they were induced in the WT by 3-AT treatment, showed a statistically significant change in expression level upon 3-AT exposure, and were repressed in the Cagcn4 mutant but not in the WT. Sequences of promoter regions and predicted ORFs from the Candida tropicalis, Aspergillus nidulans, and Magnaporthe grisea genomes were downloaded from the Broad Institute (http://www.broad.mit.edu/annotation/fgi/).
Supplemental material and the Neurospora functional genomics microarray database.
The complete data set generated in this study is available in the supplemental material and in the Neurospora functional genomics microarray database (http://www.yale.edu/townsend/Links/ffdatabase/introduction.htm).
RESULTS
Transcriptional profile of WT N. crassa exposed to 3-AT.
To investigate the similarities between N. crassa cross pathway control and S. cerevisiae general amino acid control, we performed transcriptional profiling of the N. crassa WT and the cpc-1 mutant under conditions of amino acid starvation. In N. crassa, amino acid starvation has been experimentally induced by the addition of 3-AT to hyphal cultures (14, 54). Preliminary experiments indicated that the exposure of a 12-h culture to 6 mM 3-AT for 2 h was sufficient to induce expression levels of known targets of CPC1 (see Materials and Methods). For microarray analyses, we used a 70-mer oligonucleotide set corresponding to the 10,526 predicted N. crassa genes, with an additional set of 384 70-mer oligonucleotides corresponding to intergenic or telomeric regions and Ambion oligonucleotides for normalization procedures (see Materials and Methods). During the course of this experiment, and for all of the data sets discussed here, a total of 5,865 spots on the Neurospora array had expression data.
A large number of genes showed differential expression levels in cultures of WT N. crassa exposed to 6 mM 3-AT for 2 h relative to those in unexposed control cultures; the expression levels of 334 genes showed a statistically significant increase in the 3-AT-exposed culture, while the expression levels of 280 genes decreased in the 3-AT-exposed culture relative to those in the unexposed WT culture (Table 1). The genes identified in the increased- and decreased-expression-level data sets were subjected to functional category representation analysis (FunCat; http://mips.gsf.de/genre/proj/ncrassa/Search/Catalogs/catalog.jsp) (Table S1 in the supplemental material). Of the 334 genes in the WT whose expression levels were significantly increased by 3-AT treatment, 120 genes were classified in the metabolism functional category. Of these, 54 genes were involved in amino acid metabolism (P = 1.44e−34) and included known targets of CPC1, such as trp-1 (NCU00200.2, encoding anthranilate synthase component II), arg-12 (NCU01667.2, encoding ornithine carbamoyltransferase), his-3 (NCU03139.2, encoding a histidine biosynthesis trifunctional protein), trp-3 (NCU08409.2, encoding tryptophan synthase), and leu-6 (NCU09463.2, encoding leucine-tRNA synthetase) as well as cpc-1 itself (NCU04050.2). Another known target of CPC1, arg-2 (NCU07732.2, encoding the arginine-specific carbamoylphosphate synthetase small subunit), was slightly induced, consistent with previously published data (54), but did not pass the statistical significance test. The relative level of transcripts of the for gene (NCU02274.2, encoding cytosolic serine hydroxymethyltransferase) was not elevated after 2 h of exposure to 3-AT, although previously published Northern analysis data indicated that for gene expression increases after a 1-h exposure to 3-AT (41). We assessed for gene expression levels from both the 30-min and 2-h microarray data sets (see Materials and Methods); for expression levels were transient (detectable in the 30-min but not the 2-h data set).
TABLE 1.
Summary of transcriptional profiling results
| Type of change in gene expression | Amt of change (n-fold) | No. of genes with indicated change in comparison of: |
|||
|---|---|---|---|---|---|
| WT exposed to 3-AT vs unexposed WT | WT exposed to 3-AT vs cpc-1 mutant exposed to 3-AT | cpc-1 mutant exposed to 3-AT vs unexposed mutant | Unexposed WT vs unexposed cpc-1 mutant | ||
| Increase | >3 | 94 | 48 | 164 | 4 |
| >2 | 224 | 143 | 385 | 65 | |
| >1.5 | 334 | 255 | 669 | 290 | |
| Decrease | >3 | 43 | 65 | 91 | 72 |
| >2 | 177 | 190 | 270 | 190 | |
| >1.5 | 280 | 346 | 510 | 317 | |
A significant number of genes (280) showed reductions in expression levels when the WT was exposed to 3-AT (Table 1). A functional category analysis of these genes (Table S1 in the supplemental material) showed a distribution of functional categories different from that among genes induced by 3-AT. The set of genes repressed by 3-AT was most enriched with genes associated with ribosome biosynthesis (P = 2e−97). Within the down-regulated-gene set, 89 genes were associated with ribosome biosynthesis, with 67 of these genes encoding predicted ribosomal proteins. In S. cerevisiae, the expression levels of more than 90 transcripts for ribosome-related proteins are reduced upon amino acid starvation (43). In addition to genes associated with ribosome biogenesis, 45 N. crassa nucleus-encoded mRNAs specifying products with mitochondrial functions showed reduced expression levels after 3-AT exposure, also consistent with observations for S. cerevisiae (8). Only 47 genes (17%) whose expression levels were statistically significantly reduced by 3-AT treatment belonged to the metabolism functional category, a category not significantly enriched as a responding class.
Transcriptional profile of the N. crassa cpc-1 mutant.
The cpc-1 mutant grows like the WT in minimal medium (3), and basal levels of transcripts of the amino acid biosynthesis genes, such as his-3, trp-3, cpc-1, and arg-2, are similar between the WT and the cpc-1 mutant (FGSC 4264) (53). However, enzyme assay data showed that mutations in cpc-1 affect basal levels of activity of some amino acid biosynthetic enzymes (3). When the cpc-1 mutant was grown in minimal medium, 290 genes showed higher levels of expression than those in the WT under identical conditions, while 317 genes showed lower levels of expression (Table 1). Functional category analysis of the up-regulated-gene set revealed a statistically significant overrepresentation of genes annotated to be involved in protein synthesis (Table S1 in the supplemental material). Fifty genes (P = 1.24e−37) were predicted to be involved in ribosome biosynthesis; 37 of these 50 genes encode ribosomal proteins (P = 1.53e−30), and 7 are predicted rRNA genes (P = 7.00e-06). This gene set was also enriched with energy-related genes, including 45 genes with mitochondrial functions (P = 1.41e−19), 8 genes predicted to be involved in aerobic respiration (P = 1e−06), and 12 electron transport genes (P = 6e−8). The set was not enriched with genes predicted to be involved in amino acid biosynthesis, although five genes involved in amino acid transport overrepresented this functional category (P = 0.0004) (Table S1 in the supplemental material).
In contrast to the set of genes showing increased expression in the cpc-1 mutant, the set of down-regulated genes revealed no significant enrichment with genes of any specific functional category upon a similar analysis of gene functions (Table S1 in the supplemental material). Only 12 genes identified in the down-regulated-gene set were related to amino acid metabolism, and among these, only 3 were predicted to be involved in amino acid biosynthesis: a 2-methylcitrate dehydratase gene (NCU00680.2) and a gene for a homoaconitase precursor (NCU08898.2), both involved in lysine biosynthesis, and a methionine synthase gene (NCU06512.2).
Comparison of transcriptional profiles of the WT and the cpc-1 mutant exposed to 3-AT.
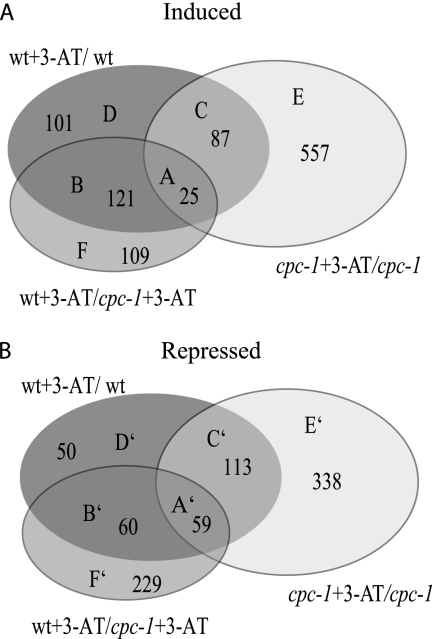
When the cpc-1 mutant was exposed to 3-AT for 2 h, it showed significant differences in the levels of expression of a large number of genes in comparison to the cpc-1 mutant grown in minimal medium; 669 genes showed statistically significantly increased expression levels, while 510 genes showed decreased expression levels (Table 1). Changes in the levels of expression of a large number of genes in the cpc-1 mutant may reflect increased stress associated with amino acid limitation. A similar transcriptional response occurs in the S. cerevisiae gcn4 mutant when it is exposed to 3-AT (45). To identify genes whose responses to amino acid starvation are dependent upon CPC1, we compared expression profiles of the WT versus the cpc-1 mutant in response to 3-AT; 255 genes showed statistically significant increases in expression levels, while 346 genes showed decreases in expression levels (Table 1). A Venn diagram showing the overlap of data sets of up-regulated genes from the WT and the cpc-1 mutant grown in minimal medium and minimal medium plus 3-AT is presented in Fig. 1A. A total of 443 genes were identified as potential CPC1 targets. This data set included 121 genes in set B, whose expression levels in the WT but not in the cpc-1 mutant were increased by treatment with 3-AT; 25 genes in set A, whose expression levels in both the WT and the cpc-1 mutant, but to a greater extent in the WT, were increased by treatment with 3-AT; 109 genes in set F, which required functional cpc-1 for appropriate expression levels in the WT; and 101 genes in set D, whose expression levels in the WT only increased upon exposure to 3-AT (Fig. S2A in the supplemental material). In addition, the 87 genes within set C were included in the CPC1 target gene set because the expression levels of 37 of these genes increased to a greater degree in the WT than in the cpc-1 mutant upon exposure to 3-AT. Genes within set E (557) were induced only in the cpc-1 mutant upon exposure to 3-AT and were therefore considered not to be CPC1 targets.
FIG. 1.
Venn diagram of genes with differences in expression levels between the WT and the cpc-1 mutant. (A) Venn diagram showing overlap among genes that exhibit statistically significantly increased or altered expression levels in the WT (FGSC 2489) versus the cpc-1 mutant (FGSC 4264) in minimal medium after exposure to 3-AT. The 443 genes comprising set B, set D, set A, set F, and set C are defined as the CPC1 target gene set (regulon) (see the text for details). A functional category analysis of these 443 genes is given in Table S1 in the supplemental material. (B) Venn diagram showing overlap among genes whose expression levels in the WT (FGSC 2489) and the cpc-1 (FGSC 4264) mutant were reduced in minimal medium upon exposure to 3-AT. A functional category analysis of genes that showed reduced expression levels is given in Table S1 in the supplemental material.
CPC1 regulatory cis-element analysis.
By biochemical analysis, CPC1 has been shown to bind the cis-element TGACTC as a core sequence (14). We assessed whether the set of 443 CPC1 target genes was enriched with the cis element identified by biochemical analysis. An initial subgroup of putative CPC1 targets composed of 500-bp regions upstream from 67 amino acid metabolism genes were analyzed by MEME, MDscan, and BioProspector (see Materials and Methods). All three analyses recovered the same cis-element, TGACTCA (Fig. 2). Interesting in this regard, biochemical studies also showed that an A in position 7 is necessary for high-affinity binding (14). Of the 443 CPC1 target genes, 87 genes contain a perfect match to this CPC1 consensus motif in their predicted promoter regions (P = 2.70e−45). An enrichment of this 87-gene set with genes involved in amino acid metabolism (24 genes; P = 1.9e−21) and aminoacyl-tRNA (aa-tRNA) synthetase genes (15 genes; P = 1.3e−22) was detected. We next expanded the search to identify genes within the 443-gene set that had less stringently defined matches to the CPC1 consensus (70% of the full score); 214 promoter regions (48%) contained a CPC1 consensus match with a score of at least 70% (P = 1.82e−13) (Table 2). This value is very similar to that obtained for the 3-AT-responsive Gcn4 target gene set of S. cerevisiae (235 out of 539, or 44%) (45).
FIG. 2.
Computational identification of cis-element enrichment in predicted promoter regions of genes within the CPC1 regulon. The 500-bp upstream regions of the predicted translational start sites of 67 amino acid metabolism genes from the CPC1 target gene set were retrieved and evaluated by MDscan (38), BioProspector (42), and MEME (2) for motif prediction. By using the initial CPC1 matrix, the promoter regions of the 67 amino acid genes were evaluated by the PATSER program (23). To identify additional CPC1 target genes in the N. crassa target gene set and the genome, predicted promoter regions were scanned using the CPC1 matrix. The logo using the CPC1 matrix was built using WebLogo (http://weblogo.berkeley.edu/logo.cgi).
TABLE 2.
Analysis of cis-element enrichment within CPC1 targets
| % Identity cutoff value (score)a | No. of genes (P value)b with a cis-element match among: |
||
|---|---|---|---|
| 67 amino acid metabolism genes | 443 target genes | Whole genome | |
| 100 (8.15) | 24 (3.74e−19) | 87 (2.70e−45) | 351 |
| 70 (5.70) | 43 (9.1e−8) | 214 (1.82e−13) | 3,468 |
Score obtained by PATSER program (23). The score for a full match to the CPC1 cis-element matrix is 8.15.
P value obtained from Fisher test (http://bioconductor.org) for results of comparison to whole-genome data.
An analysis of the predicted promoter regions of all 10,526 N. crassa genes showed that 351 predicted genes have a perfect match to the CPC1 consensus. When the 87 putative direct CPC1 targets were removed from this 351-gene data set (leaving 264 genes), no enrichment with genes of any specific functional category was detected. When the computational search was expanded to any N. crassa genes that contained a 70% match to the CPC1 consensus in the upstream region, 3,468 genes were identified. It is unlikely that this number of genes is directly regulated by CPC1.
Description of genes within functional categories overrepresented in the 443-gene set of putative CPC1 targets.
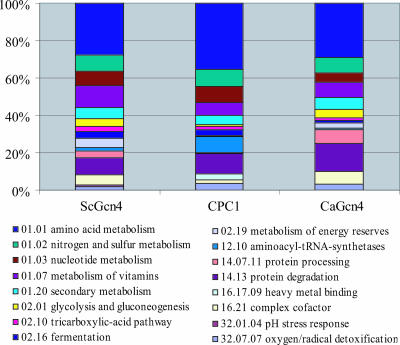
Our profiling data indicated that, similar to Gcn4p, CPC1 is a major transcriptional regulator in N. crassa. The 443 CPC1 target genes were evaluated to identify functional categories by using the MIPS FunCat system (52) (Fig. 3 and Table S1 in the supplemental material). Sixty-seven of the predicted 195 amino acid metabolism genes in FunCat were present in the 443-gene CPC1 target set, and these 67 genes represented the largest FunCat category (P = 3.71e−41); 50 genes encoding amino acid biosynthetic enzymes were identified (Table S2 in the supplemental material). Increased or WT expression levels of genes corresponding to 19 of the 20 predicted amino acid biosynthetic pathways were dependent upon functional cpc-1 in response to amino acid starvation; only genes in the alanine biosynthetic pathway were absent (Table S2 in the supplemental material). The CPC1 target gene set was also enriched with other functional categories, in particular, cytosolic aa-tRNA synthetase genes (P = 7e−13); nitrogen and sulfur metabolism genes (P = 7e−8); nucleotide metabolism genes (P = 7e−6); genes involved in the metabolism of vitamins, cofactors, and prosthetic groups (P = 4e−5); alcohol fermentation-related genes (P = 2e−5); protein degradation genes (P = 8e−6); and oxygen response and free-radical detoxification genes (P = 5.80e−06) (Fig. 3 and Table S2 in the supplemental material). Slightly more than 50% (65 of 122) of the genes listed in Table S2 in the supplemental material have N. crassa gene designations; the remaining genes have not been characterized.
FIG. 3.
Functional category analysis of Gcn4p, CaGcn4p, and CPC1 targets. Target gene sets derived from S. cerevisiae Gcn4p transcriptional profiling and ChIP data (22, 34, 36, 45) were combined to generate a set of 747 Gcn4p target genes. A statistical analysis of C. albicans Gcn4p transcriptional profiling data (62) retrieved a set of 483 CaGcn4p target genes. The S. cerevisiae Gcn4p and C. albicans Gcn4p data sets were subjected to functional category analysis using FunCat (52). The functional category analyses of the S. cerevisiae Gcn4p and C. albicans Gcn4p data sets were then compared to the functional category analysis of the set of 443 CPC1 target genes.
Within the 443-gene CPC1 target set, three genes known or likely to be involved in the translational regulation of cpc-1 were identified, including cpc-3, a GCN2 ortholog (NCU01187.2, encoding a protein kinase), and genes for the eIF2B δ subunit (NCU01468.2) and the eIF2B ɛ subunit (NCU02414.2). The promoters of the two latter genes contain a perfect match to the CPC1 cis-element consensus. In S. cerevisiae, GCN2 (YDR283C) and the eIF2B δ and ɛ subunit genes (YGR083c and YDR211w) are not induced by treatment with 3-AT (45), nor do they contain a Gcn4p consensus cis element in their promoter regions.
In cross pathway control, deacylated tRNA is the signal for amino acid starvation (26, 28, 53). Analogous to the S. cerevisiae regulatory system, N. crassa CPC3, a protein kinase (S. cerevisiae GCN2 ortholog), is believed to be activated directly by deacylated tRNA to phosphorylate the α subunit of eIF2, reducing translation at uORFs in the cpc-1 mRNA and thus increasing the translation of CPC1 (55). Sixteen of 21 predicted aa-tRNA synthetase genes (corresponding to 20 aa-tRNA synthetases, as phenylalanyl-tRNA synthetase is composed of two subunits) were identified in the CPC1 target gene set (Fig. S2B in the supplemental material). Among the 21 predicted aa-tRNA synthetase genes, 19 have a perfect cis-element match to the CPC1 consensus, including the phenylalanyl-tRNA synthetase β subunit gene (Table 3). The one aa-tRNA synthetase gene in this set that lacks a perfect cis-element, NCU07755.2 (encoding tyrosyl-tRNA synthetase), has a related element TGACTCT; this variant element can bind weakly to CPC1 in vitro (14). In contrast to the cytosolic tRNA synthetase genes, none of the nine genes annotated as mitochondrial tRNA synthetases were identified as CPC1 targets. In addition, five genes annotated as tRNA synthetase related, NCU00466.2 (related to NCU08894.2, encoding glutamyl-tRNA synthetase), NCU00920.2 (related to NCU03575.2, encoding isoleucyl-tRNA synthetase), NCU00931.2 (related to NCU04020.2, encoding lysine-tRNA ligase), NCU07082.2 (related to NCU00915.2, encoding aspartyl-tRNA synthetase), and NCU09892.2 (hypothetical; related to NCU04449.2, encoding prolyl-tRNA synthetase), were not identified as CPC1 targets, and all lack a CPC1 consensus cis-element.
TABLE 3.
cis-element analysis of predicted promoter regions of aa-tRNA synthetase genes in N. crassa, C. albicans, and S. cerevisiae
| N. crassa aa-tRNA synthetase gene | cis-elementa | Scoreb | C. albicans ortholog | cis-elementa | Scoreb | S. cerevisiae ortholog | cis-elementa | Scoreb |
|---|---|---|---|---|---|---|---|---|
| NCU00405.2 | cgTGACTCAct | 8.15 | orf19.437 | gaTGACTCTtt | 4.43 | YBR121C | taTGATTCAtg | 3.16 |
| NCU00915.2 | gtTGACTCAca | 8.15 | orf19.2407 | tgTTAGTCAta | 5.03 | YLL018C | aaTGCGTCAta | 6.72 |
| NCU01443.2 | tgTGACTCAac | 8.15 | orf19.269 | aaTGAGACAaa | 4.99 | YDR023W | atCGAGTCAaa | 4.69 |
| NCU01965.2 | ttTGACTCAtg | 8.15 | orf19.1295 | atTGACTCAtc | 8.15 | YGR094W | agTGAGACAat | 4.99 |
| NCU02380.2 | tgTGACTCAct | 8.15 | orf19.5685 | taTGACTCTtc | 4.43 | YIL078W | ttTGCGTCAgc | 6.72 |
| NCU02566.2 | aaTGACTCAag | 8.15 | orf19.5746 | taTGACTCAaa | 8.15 | YOR335C | ttTTACTCAtc | 5.06 |
| NCU03575.2 | ccTGACTCAcc | 8.15 | orf19.2138 | gaTGACTGAgc | 6.64 | YBL076C | gaTGACTCTta | 4.43 |
| NCU04020.2 | acTGACTCAcc | 8.15 | orf19.6749 | agTTACTCAtc | 5.06 | YDR037W | aaTGACTCTta | 4.43 |
| NCU04449.2 | acTGACTCAcc | 8.15 | orf19.6701 | atTGACTCAcg | 8.15 | YHR020W | agTGACTCAag | 8.15 |
| NCU05095.2 | ttTGACTCAca | 8.15 | orf19.2960 | gaTGAATCAtt | 3.16 | YFL022C | gtTTACTGAaa | 3.55 |
| NCU05642.2 | ctTGACTCAtt | 8.15 | orf19.4931 | ttTGACTCAct | 8.15 | YNL247W | ttTGACTCTtc | 4.43 |
| NCU06457.2 | ggTGACTCAcc | 8.15 | orf19.6702 | atTGACTCAcg | 8.15 | YHR019C | agTGACTCAag | 8.15 |
| NCU06722.2 | agTGACTCAag | 8.15 | orf19.5226 | aaTGAGTGAga | 6.6 | YOL097C | ttTTAGTCAtt | 5.03 |
| NCU06914.2 | ttTGACTCAgt | 8.15 | orf19.4051 | agCGAGTCAac | 4.69 | YPR033C | ctTGAGTGAtc | 6.6 |
| NCU07451.2 | tcTGACTCAcc | 8.15 | orf19.3955 | gtTGACTCAta | 8.15 | YGR264C | tcTGCCTCCga | 4.35 |
| NCU07755.2 | ggTGACTCTtc | 4.43 | orf19.2694 | taTAAGTCAtt | 4.66 | YGR185C | aaTTAGTCAtt | 5.03 |
| NCU07926.2 | ccTGACTCAcc | 8.15 | orf19.7064 | gcTCCCTCAga | 4.32 | YOR168W | ttAGACTGAaa | 2.89 |
| NCU08195.2 | ctTGACTCAgt | 8.15 | orf19.3341 | aatcTGACTCAa | 8.15 | YDR341C | acTGACTCAtt | 8.15 |
| NCU08894.2 | ctTGACTCAat | 8.15 | orf19.7057 | aagtTTAGTCAg | 5.03 | YGL245W | tagtTTCGTCAtt | 3.64 |
| NCU09463.2 | tcTGACTCAat | 8.15 | orf19.2560 | agaaTGACTCA | 8.15 | YPL160W | tgggTGACTGA | 6.64 |
Lowercase letters indicate sequences flanking a consensus cis-element.
Score obtained by PATSER program (23). The score for a full match to the CPC1 cis-element matrix is 8.15.
The cpc-1 mutant is more sensitive to purine and pyrimidine analogs (12, 53), suggesting a connection between the control of nucleotide biosynthesis and the control of amino acid biosynthesis in N. crassa. In S. cerevisiae, the Gcn4 regulon includes some purine and pyrimidine metabolism genes (45). Consistent with these data, nucleotide metabolism genes were overrepresented in the CPC1 regulon (P = 7.77e−06) (Table S2 in the supplemental material). An overrepresentation of genes related to oxygen/radical detoxification in the CPC1 target set was also identified (seven genes; P = 5.80e−6) (Table S2 in the supplemental material), which included two of the four predicted catalase genes, cat-3 (NCU00355.2) and cat-2 (NCU05770.2). In S. cerevisiae, genes known to be induced by treatment with hydrogen peroxide, including CTA1 (catalase A) and CTT1 (catalase T), are also induced by 3-AT treatment but are Gcn4p independent (45). However, unlike S. cerevisiae, in which genes for 26 transcription factors and 11 protein kinases are part of the Gcn4 regulon (34, 45), N. crassa had only three genes encoding predicted DNA binding proteins identified as CPC1 targets (Table S2 in the supplemental material). None of these predicted transcription factors have an ortholog in S. cerevisiae. In addition to cpc-3, only one gene predicted to specify a protein kinase, NCU06230.2 (out of a total ∼70 in the genome), was identified in the CPC1 data set.
Within the 443-gene CPC1 data set, 169 genes (∼40%) encode hypothetical or conserved hypothetical proteins (Table S1 in the supplemental material). Of these 169 genes, 27 (16%) have a perfect CPC1 cis-element match (TGACTCA) in their 5′ regions, a value similar to that of genes in the 443-gene target set whose functions have been annotated (20%). Of these 169 genes of unknown function, 27 have orthologs in S. cerevisiae. However, only 4 among these 27 have been identified as part of the Gcn4 regulon (45) (YIL164C, a predicted nitrilase gene, orthologous to NCU05387.2; YCR023C, orthologous to NCU03107.2; YIR035C, orthologous to NCU02018.2; and YBR147W, orthologous to NCU09195.2).
Functional categorization of genes dependent upon functional CPC1 for reduced expression levels.
Consistent with our findings that CPC1 is a major regulator in N. crassa, a large number of genes in the WT showed reduced expression levels upon exposure to 3-AT (Table 1), many (119) of which required functional CPC1 (Fig. 1B; Table S1 in the supplemental material). However, only 2 of the 119 genes contain a CPC1 consensus cis element in their predicted promoter regions. Among these 119 genes, 53 specify proteins involved in ribosomal biosynthesis, with 37 of these specifying ribosomal proteins (Fig. S2C in the supplemental material). Analysis of the 5′ regions of the 37 ribosomal protein genes led to the identification of a conserved a cis-element, AGCCCTAA, which is identical to that previously identified as a potential regulatory site (21, 31).
Characterization of CPC1-independent genes.
The expression levels of a large number of genes (557) increased only in the cpc-1 mutant when it was exposed to 3-AT; changes in the levels of expression of these genes were therefore CPC1 independent (Fig. 1A, set E). In contrast to the CPC1 target gene set, the set of CPC1-independent genes was not significantly enriched with any functional category. The slight enrichment included a group of genes encoding nucleic acid binding proteins (P = 5e−6), including predicted transcription factors and chromosomal remodeling proteins, plus eight predicted protein kinases (Table S1 in the supplemental material).
Comparative analysis of N. crassa CPC1, S. cerevisiae Gcn4, and C. albicans Gcn4 pathways.
In S. cerevisiae, Gcn4p influences the transcriptional response of many genes (635) to amino acid starvation (34, 45). Based on ChIP data, 207 genes are directly bound by Gcn4p (binding P value, <0.001) (22, 36). The Gcn4p transcriptional profiling and ChIP data sets have an overlap of 95 genes. We combined the S. cerevisiae transcriptional profiling and ChIP data to define a set of 747 S. cerevisiae genes as the Gcn4 regulon. Functional categorization of these 747 genes (Fig. 3 and Table S3 in the supplemental material) showed enrichment with the same functional categories as the CPC1 regulon, including genes involved in amino acid biosynthesis, nitrogen and sulfur metabolism, and the metabolism of vitamins. However, the Gcn4 and CPC1 regulons also showed differences, most notably in aa-tRNA synthetase genes, of which only the CPC1 data set was enriched, and genes involved in complex cofactor (NAD/NADP) binding, of which only the Gcn4 data set was enriched. Since CPC1 and Gcn4p show similarity in their DNA binding domains and the cis-element sequences bound by CPC1 and Gcn4p are identical (TGACTCA), these data suggest that cis-elements in the promoter regions of some genes within the CPC1 and Gcn4 regulons have diverged.
To further evaluate the evolution of the Gcn4 and CPC1 regulons, we analyzed an available expression profile data set for C. albicans WT strains and gcn4 mutants exposed or not to 40 mM 3-AT (62). We identified genes in the C. albicans data set whose expression levels increased or decreased to statistically significant degrees to enable comparison to our analyses of the N. crassa and S. cerevisiae data sets (see Materials and Methods). A total of 483 genes were defined as the CaGcn4 regulon, including 399 genes whose increased expression levels and 84 genes whose reduced expression levels upon exposure to 3-AT were dependent upon functional CaGCN4. Functional category analyses showed that the N. crassa, S. cerevisiae, and C. albicans data sets showed enrichment with genes within identical functional categories, including amino acid, sulfur, and vitamin metabolism (Fig. 3 and Table 3), although in some cases the data set of one or two species was specifically enriched with a category. For example, the S. cerevisiae and C. albicans data sets, but not the N. crassa data set, were significantly enriched with the complex cofactor (NAD/NADP) functional category. The C. albicans and N. crassa data sets, but not the S. cerevisiae data set, were enriched with genes within the protein degradation and oxygen detoxification functional categories, and the N. crassa and S. cerevisiae data sets, but not the C. albicans data set, were enriched with genes within the fermentation functional category.
In N. crassa, many cytosolic aa-tRNA synthetase genes were members of the CPC1 regulon and contain a perfect CPC1 consensus site. In S. cerevisiae, only three aa-tRNA synthetase genes have a perfect match to the Gcn4p consensus TGAC/GTCA (Table 3). Gcn4p ChIP data support the cis-element function in these three aa-tRNA synthetase genes (YHR020W, encoding a product related to prolyl-tRNA synthetase; YHR019C, encoding asparaginyl-tRNA synthetase; and YDR341C, encoding arginyl-tRNA synthetase) (22, 36). Of the predicted 20 cytosolic aa-tRNA synthetase genes in C. albicans, 10 have at least a 70% match to the Gcn4p cis-element in the 500-bp region upstream of the coding sequence, including 8 with a full match. Although the CaGcn4 data set was not significantly enriched with aa-tRNA synthetase genes, six of eight of the genes with a full cis-element match were induced from 1.2- to 2.4-fold upon exposure to 3-AT (62); their increase in expression levels was dependent upon functional CaGCN4 (Table 3). These data suggest a loss of regulation of aa-tRNA synthetase genes by Gcn4 orthologs in the evolution of the hemiascomycete clade.
Computational comparison of levels of ortholog and regulog identity among the CPC1, Gcn4, and CaGcn4 data sets.
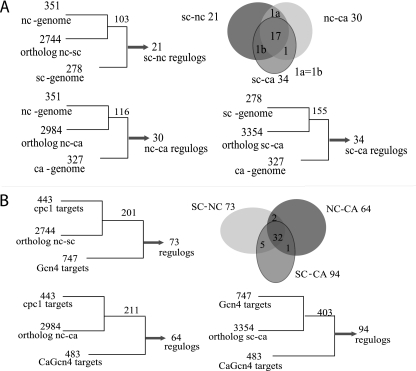
Regulogs consist of orthologous genes that have maintained their regulatory network through evolution (73). Some regulogs reflect direct interaction via an orthologous regulator (direct targets). We wished to compare the identification of Gcn4p/CaGcn4p/CPC1 targets using purely computational methods to the identification of predicted Gcn4p/CaGcn4p/CPC1 targets by transcriptional profiling methods. For the computational approach, we used the 351 genes in the N. crassa genome recovered using the CPC1 cis-element matrix. Of these 351 genes, 103 corresponded to orthologs identified in S. cerevisiae, although only 21 of the orthologs contain a Gcn4p cis-element (Fig. 4A). Similarly, for the 351 N. crassa genes, 116 orthologous genes in C. albicans were identified, but only 30 have a CaGcn4p cis-element. To compare S. cerevisiae and C. albicans, we used the CPC1 cis-element matrix to scan the promoters of S. cerevisiae genes to identify 278 that contain a perfect match to the Gcn4p cis-element; 123 of these were detected as Gcn4p targets in both ChIP and profiling data (22, 36). Out of these 278 genes, 155 had an ortholog in C. albicans, but only 34 of the C. albicans orthologs have a TGACTCA cis-element in the promoter region (Fig. 4A). When all three of these data sets were compared (S. cerevisiae to N. crassa, S. cerevisiae to C. albicans, and N. crassa to C. albicans), the promoters of only 17 orthologous genes were found to contain the Gcn4p/CaGcn4p/CPC1 cis-element (Fig. 4A; Table 4). Twelve of these genes are involved in amino acid biosynthesis, three encode aa-tRNA synthetases, one gene is required for adenine biosynthesis, and the last gene, SNZ, is involved in vitamin B6 biosynthesis.
FIG. 4.
Identification of common regulogs in the S. cerevisiae (sc), C. albicans (ca), and N. crassa (nc) genomes by computational analysis using cis-element scans compared to identification of common regulogs by transcriptional profiling. (A) In N. crassa, predicted promoter regions of 351 genes that contain the CPC1 cis-element were identified. Of these 351 genes, 103 have orthologs in S. cerevisiae, and of these 103 orthologs, 21 have a Gcn4p cis-element consensus in the promoter region (regulogs). Orthologs of 116 of the 351 N. crassa genes were identified in C. albicans. Of these 116 C. albicans genes, 30 have a CaGcn4p consensus cis-element in the predicted promoter region. In S. cerevisiae, 278 genes that contain a Gcn4p consensus cis-element in their promoter regions were identified. Of these 278 genes, 155 had orthologs identified in C. albicans, and of the 155 orthologs, 34 have a predicted CaGcn4p cis-element in predicted promoter regions. The overlap of all three data sets identified 17 orthologous gene pairs that maintained the conserved Gcn4p/CaGcn4p/CPC1 cis-element. (B) By transcriptional profiling, 443 genes were identified as CPC1 targets. The overlap between the 747-gene S. cerevisiae and N. crassa CPC1 data sets revealed 73 orthologous gene pairs (regulogs). The overlap between the N. crassa and C. albicans data sets identified 64 orthologous gene pairs, and the overlap between the S. cerevisiae and C. albicans data sets revealed 94 orthologous gene pairs. The overlap of all three data sets identified a conserved regulon of 32 orthologous genes present in all three species as determined by transcriptional profiling and ChIP.
TABLE 4.
Direct regulogs among N. crassa, C. albicans, and S. cerevisiae identified by cis element computational prediction
| N. crassa gene designation | C. albicans gene designation | S. cerevisiae gene designation | ChIP data available for S. cerevisiae | S. cerevisiae gene name | Predicted product |
|---|---|---|---|---|---|
| NCU00200.2 | orf19.5243 | YKL211C | Yes | TRP3 | Anthranilate synthase component II |
| NCU00554.2 | orf19.1559 | YDR158W | HOM2 | Aspartate-semialdehyde dehydrogenase | |
| NCU01412.2 | orf19.6779 | YOR323C | PRO2 | Gamma-glutamyl phosphate reductase | |
| NCU01667.2 | orf19.5610 | YJL088W | Yes | ARG3 | Ornithine carbamoyltransferase |
| NCU02677.2 | orf19.3221 | YJR109C | Yes | CPA2 | Arginine-specific carbamoylphosphate synthetase |
| NCU03118.2 | orf19.1789 | YIR034C | Yes | LYS1 | Saccharopine dehydrogenase |
| NCU04118.2 | orf19.1235 | YER052C | Yes | HOM3 | Aspartokinase |
| NCU04303.2 | orf19.198 | YGR124W | ASN2 | Asparagine synthetase | |
| NCU05129.2 | orf19.2546 | YER090W | Yes | TRP2 | Anthranilate synthase component I |
| NCU07156.2 | orf19.5505 | YBR248C | Yes | HIS7 | Imidazole glycerol phosphate synthase |
| NCU07987.2 | orf19.2092 | YGL184C | Yes | STR3 | Cystathionine beta-lyase |
| NCU08162.2 | orf19.6689 | YHR018C | Yes | ARG4 | Arginosuccinate lyase |
| NCU04449.2 | orf19.6701 | YHR020W | Yes | Glutamyl-prolyl-tRNA synthetase | |
| NCU06457.2 | orf19.6702 | YHR019C | Yes | DED81 | Asparaginyl-tRNA synthetase |
| NCU08195.2 | orf19.3341 | YDR341C | Yes | RRS1 | Arginyl-tRNA synthetase |
| NCU06550.2 | orf19.2947 | YMR096W | Yes | SNZ1 | Vitamin B6 biosynthesis protein |
| NCU09789.2 | orf19.4827 | YNL220W | ADE12 | Adenylosuccinate synthetase |
Transcriptional profile comparisons of orthologs and regulogs of the CPC1 and Gcn4p pathways.
We then used the transcriptional profiling data from S. cerevisiae, C. albicans, and N. crassa WT strains and gcn4/Cagcn4/cpc-1 mutants in response to amino acid starvation to compare to our computational analyses of Gcn4/CPC1 regulons based only on predicted cis-elements. We expect to identify both conserved direct targets of Gcn4p/CaGcn4p/CPC1 (direct regulogs) and conserved indirect target genes (indirect regulogs).
Of the 443 genes in the N. crassa CPC1 regulon, 201 had orthologs in S. cerevisiae, a higher percentage than that obtained in a total-genome comparison (26%). However, only 73 of these 201 genes were included in the Gcn4 regulon (Fig. 4B); 34 of these 73 genes contain the Gcn4p consensus cis-element. The binding of Gcn4p to the promoter regions of 25 of these 34 genes has been observed previously (22, 36). However, only 18 of the 73 orthologous gene pairs in S. cerevisiae and N. crassa have a full match to the Gcn4p/CPC1 cis-element consensus sequence. In C. albicans, although 211 genes were orthologs of genes in the 443-gene CPC1 target set, only 64 of these were within the CaGcn4 regulon. Only 14 of the 64 orthologous gene pairs in C. albicans and N. crassa have a predicted CaGcn4p/CPC1 cis-element.
We also compared the S. cerevisiae Gcn4 data set to the C. albicans Gcn4 data set. For the 747 genes in the Gcn4 regulon, 403 orthologs in C. albicans were identified. However, only 94 of the 403 genes in C. albicans were identified as part of the CaGcn4 regulon, and of these 94 genes, only 20 contain the CaGcn4p cis-element. A comparison among all three data sets showed that 32 regulogs are conserved among all three fungi (Fig. 4B), which includes both direct and indirect targets. Of these 32 orthologous genes, 22 encode amino acid biosynthetic enzymes, one encodes a tRNA synthetase, two encode enzymes involved in adenine biosynthesis, two encode enzymes involved in oxygen/radical detoxification, one is predicted to be involved in vitamin B6 biosynthesis, three are involved in carbon metabolism, and one produces an enzyme of unknown function which localizes to purified mitochondria (Table 5). Of these 32 genes, 10 contain a Gcn4p/CPC1/CaGcn4p consensus sequence and 18 have been confirmed by ChIP experiments to be bound by Gen4p in S. cerevisiae (22, 36).
TABLE 5.
Common regulogs among N. crassa, C. albicans, and S. cerevisiae identified by transcriptional profiling data
| N. crassa gene designation | C. albicans gene designation | S. cerevisiae gene designation | ChIP data available for S. cerevisiae | S. cerevisiae gene name | Predicted product |
|---|---|---|---|---|---|
| NCU00200.2 | orf19.5243 | YKL211C | Yes | TRP3 | Anthranilate synthase/indole glycerol phosphate synthase |
| NCU00554.2 | orf19.1559 | YDR158W | HOM2 | Aspartate-semialdehyde dehydrogenase | |
| NCU00567.2 | orf19.4788 | YER069W | Yes | ARG5,6 | Acetylglutamate kinase/acetylglutamyl-phosphate reductase |
| NCU01412.2 | orf19.6779 | YOR323C | PRO2 | Gamma-glutamyl phosphate reductase | |
| NCU01667.2 | orf19.5610 | YJL088W | Yes | ARG3 | Ornithine carbamoyltransferase |
| NCU02430.2 | orf19.1033 | YJR130C | STR2 | Cystathionine gamma-synthase | |
| NCU02639.2 | orf19.7469 | YOL058W | Yes | ARG1 | Argininosuccinate synthetase |
| NCU02677.2 | orf19.3221 | YJR109C | Yes | CPA2 | Arginine-specific carbamoylphosphate synthetase |
| NCU03010.2 | orf19.2970 | YBR115C | Yes | LYS2 | Alpha-aminoadipate reductase |
| NCU03139.2 | orf19.5639 | YCL030C | Yes | HIS4 | Histidinol dehydrogenase |
| NCU03166.2 | orf19.7484 | YAR015W | ADE1 | Phosphoribosyl-amidoimidazole succinocarboxamidesynthetase | |
| NCU04118.2 | orf19.1235 | YER052C | Yes | HOM3 | Aspartokinase |
| NCU04230.2 | orf19.6844 | YER065C | ICL1 | Isocitrate lyase | |
| NCU05238.2 | orf19.4099 | YJR137C | ECM17 | Sulfite reductase beta subunit | |
| NCU05548.2 | orf19.4060 | YBR249C | Yes | ARO4 | 3-Dehydro-deoxyphosphoheptonate aldolase |
| NCU06360.2 | orf19.4177 | YIL116W | HIS5 | Histidinol-phosphate aminotransferase | |
| NCU05526.2 | orf19.4506 | YDL131W | LYS21 | Homocitrate synthase | |
| NCU07156.2 | orf19.5505 | YBR248C | Yes | HIS7 | Imidazole glycerol phosphate synthase |
| NCU07982.2 | orf19.1613 | YMR108W | ILV2 | Acetolactate synthase | |
| NCU07987.2 | orf19.2092 | YGL184C | Yes | STR3 | Cystathionine beta-lyase |
| NCU09116.2 | orf19.2098 | YGL202W | Yes | ARO8 | Aromatic amino acid aminotransferase |
| NCU09320.2 | orf19.4026 | YER055C | Yes | HIS1 | ATP phosphoribosyltransferase |
| NCU09789.2 | orf19.4827 | YNL220W | ADE12 | Adenylosuccinate synthetase | |
| NCU10180.2 | orf19.5480 | YER086W | Yes | ILV1 | Threonine deaminase |
| NCU10468.2 | orf19.6500 | YMR062C | Yes | ECM40 | Acetylornithine acetyltransferase |
| NCU03575.2 | orf19.2138 | YBL076C | Yes | ILS1 | Isoleucyl-tRNA synthetase |
| NCU02133.2 | orf19.2770 | YJR104C | Yes | SOD1 | Cytosolic superoxide dismutase |
| NCU03151.2 | orf19.2762 | YLR109W | AHP1 | Alkyl hydroperoxide reductase | |
| NCU04415.2 | orf19.7310 | YML128C | MSC1 | Protein of unknown function, possibly involved in meiotic recombination | |
| NCU04510.2 | orf19.6757 | YOR120W | GCY1 | Aldo/keto reductase | |
| NCU06550.2 | orf19.2947 | YMR096W | Yes | SNZ1 | Vitamin B6 biosynthesis protein |
| NCU07027.2 | orf19.7021 | YPR160W | GPH1 | Glycogen phosphorylase |
DISCUSSION
CPC1 is a major transcriptional regulator in Neurospora.
In this study, we identified 443 genes that require functional cpc-1 for correct expression patterns when N. crassa is exposed to amino acid starvation. This gene set was enriched with the CPC1 cis-element compared to the entire genome. The functional categorization of these 443 genes showed the highest level of enrichment with genes associated with amino acid metabolism. The set was also enriched with genes corresponding to a number of other functional categories, including nitrogen, sulfur, and nucleotide metabolism; fermentation; protein degradation; and oxygen/radical detoxification. The functional categories of the vast majority of the genes identified have not been experimentally validated for N. crassa; our transcriptional profiling data and comparative analysis provide support for the hypothesis that the predicted biochemical functions of these genes are correct. In addition, of the 443 genes, ∼40% encode hypothetical or conserved hypothetical proteins. Genes that encode proteins involved in a common cellular pathway are often coregulated, and the patterns of expression of clusters of genes that perform related cellular functions are often correlated (15, 29). Our profiling data provide a guide for future phenotypic screening of mutants with knockouts of these genes (13).
Conservation and divergence of the Gcn4/CaGcn4/CPC1 regulons: identification of a core regulon.
Comparative microarray studies have usually analyzed evolutionarily distant species for which data were obtained under diverse experimental conditions (5). Our examination of expression patterns in these three diverse fungal species under a common set of experimental conditions (exposure to 3-AT in liquid medium) and in strains with a mutation in an orthologous transcriptional regulator (GCN4/CaGCN4/cpc-1) revealed a conserved regulon of direct and indirect targets. Thirty-two orthologous gene pairs maintained their regulation patterns in these three fungi under these conditions, and 10 of these genes are predicted to be directly regulated by Gcn4p/CaGcn4p/CPC1 based on computational analyses. These 32 orthologous genes form the core of the amino acid starvation response; 22 encode amino acid biosynthetic enzymes, and 1 encodes an aa-tRNA synthetase.
Although more than 15% of the Gcn4/CaGcn4/CPC1 targets were within the amino acid synthesis functional category, the direct regulation of most of these genes by Gcn4p/CaGcn4p/CPC1, as evaluated by the presence of cis-elements, was not conserved. These data indicate that, in some cases, orthologous genes in S. cerevisiae, C. albicans, and N. crassa maintain their regulation by either Gcn4, CaGcn4 or CPC1 but that regulation may change from direct to indirect or vice versa. For example, LYS2 was identified as a Gcn4p target in S. cerevisiae both by transcriptional profiling and by ChIP (22, 36, 45). However, its ortholog in N. crassa, NCU03010.2, was identified as a target by transcriptional profiling but lacks the CPC1 cis-element, and the score for matching to the consensus was less than 50%. Similarly, NCU07982.2 has a full match to the CPC1 consensus and was identified as a target by transcriptional profiling, but its ortholog in S. cerevisiae, ILV2, is an indirect target of Gcn4p (detected by transcriptional profiling but not by ChIP).
Regulatory network evolution by cis-element gain or loss.
The evolution of gene expression patterns can affect phenotypic plasticity and is thought to play an important role in the adaptation of species to a particular environmental niche (5, 59). In C. albicans, responses to amino acid availability are linked with pseudohyphal development, biofilm formation, and phagocytosis by human neutrophils (20, 51, 66). Similarly, the GCN4 ortholog in Aspergillus fumigatus, cpcA, is required for virulence in human pulmonary infections (7, 35). These observations suggest that the CaGcn4/CpcA regulons in these two species have evolved additional functions. It is not clear how multiple promoters of coordinately expressed genes coordinately evolve their cis-elements to become members of a regulon (39, 70). For example, the promoters of ribosomal protein genes in N. crassa contain a cis-element that is an exact match to a cis-element identified in C. albicans (21); a similar cis-element is not found in ribosomal protein genes in S. cerevisiae. However, as shown in this study, the coordinate repression of ribosomal protein gene expression upon amino acid starvation is a conserved process in N. crassa, S. cerevisiae, and C. albicans and is dependent upon functional GCN4/CaGCN4/cpc-1.
In the hemiascomycete clade, which includes S. cerevisiae and C. albicans, the loss of the coordinate regulation of cytoplasmic ribosomal protein genes or rRNA processing genes with mitochondrial ribosomal protein genes is correlated with a whole-genome duplication event and the massive loss of a conserved cis-regulatory element (30). Promoter evolution can also be more gradual, as observed for the tRNA synthetase genes described in this study. In N. crassa, 19 of the 20 cytosolic tRNA synthetase genes have an exact CPC1 cis-element located in their promoter regions; 16 of these genes were identified in the CPC1 regulon. In the filamentous ascomycete species Magnaporthe grisea and Aspergillus nidulans, the predicted aa-tRNA synthetase genes also contain the predicted CPC1 cis-element (21). In Candida tropicalis, similar to C. albicans, 8 of 16 aa-tRNA synthetase orthologs have the TGACTCA consensus. Profiling data and ChIP data show that most of the aa-tRNA synthetase genes in S. cerevisiae are not regulated by Gcn4p, nor are they part of the Gcn4 regulon. This loss of regulation of aa-tRNA synthetase genes in the hemiascomycete clade is not associated with any known genome rearrangements but may be associated with adaptation and speciation.
Regulatory network evolution.
Species’ genomes evolve over time, especially via gene or genome duplication and gene or segment loss, thus making it difficult to maintain the regulation of individual components of an entire regulon. However, networks show a buffering capacity whereby the identities of networks can be conserved through evolution, although mechanistic aspects of individual gene regulation may diverge. For example, although the patterns of temporal regulation of eve stripe expression are almost identical among species of Drosophila, this similarity in overall regulation patterns is not reflected by patterns of sequence conservation in regulatory regions (39, 40). Similarly, a comparison of expression data from six species (S. cerevisiae, Escherichia coli, Caenorhabditis elegans, Arabidopsis thaliana, humans, and Drosophila melanogaster) showed that while functionally related genes are often coexpressed and coregulated, the modular components of each transcriptional program vary significantly among organisms (5). Although our analysis showed that the number of conserved Gcn4/CaGcn4/CPC1 regulogs was only a fraction of the total number of target genes identified by transcriptional profiling, the functional categorization analyses of Gcn4/CaGcn4/CPC1 targets were remarkably similar, with the conservation of overall pathways of response to amino acid starvation.
Supplementary Material
Acknowledgments
We thank the Broad Institute and MIPS for making N. crassa gene and intergenic genomic data available for oligonucleotide prediction. We thank Audrey Gasch, Mike Eisen, Jing Zhu, and Joe DeRisi for valuable discussions on the development of oligonucleotide microarrays for N. crassa. We thank Betty Gilbert, Sarah Brown, and Anna Simonin for help printing the Neurospora microarrays. We thank Heather Hood for careful reading of the manuscript. We thank Jeff Townsend (Yale University) for reviewing our manuscript, providing very helpful suggestions, and developing the Neurospora microarray database.
This work was funded by a National Institutes of Health multi-institutional program project grant (GM068087) to N.L.G. (Core III: Transcriptional Profiling) and M.S.S. (Core II: Functional Annotation).
Footnotes
Published ahead of print on 20 April 2007.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Arndt, K., and G. R. Fink. 1986. GCN4 protein, a positive transcription factor in yeast, binds general control promoters at all 5′ TGACTC 3′ sequences. Proc. Natl. Acad. Sci. USA 83:8516-8520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey, T. L., and C. Elkan. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymer, p. 28-36. In Russ B. Altman, Douglas L. Brutlag, Peter D. Karp, Richard H. Lathrop, and David B. Searls (ed.), Proceedings of the 2nd International Conference on Intelligent Systems for Molecular Biology. Association for the Advancement of Artificial Intelligence, Menlo Park, CA. [PubMed]
- 3.Barthelmess, I. B. 1982. Mutants affecting amino acid cross-pathway control in Neurospora crassa. Genet. Res. 39:169-185. [DOI] [PubMed] [Google Scholar]
- 4.Barthelmess, I. B. 1986. Regulation of amino acid synthetic enzymes in Neurospora crassa in the presence of high concentrations of amino acids. Mol. Gen. Genet. 203:533-537. [DOI] [PubMed] [Google Scholar]
- 5.Bergmann, S., J. Ihmels, and N. Barkai. 2004. Similarities and differences in genome-wide expression data of six organisms. PLoS Biol. 2:E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bozdech, Z., J. Zhu, M. P. Joachimiak, F. E. Cohen, B. Pulliam, and J. L. DeRisi. 2003. Expression profiling of the schizont and trophozoite stages of Plasmodium falciparum with a long-oligonucleotide microarray. Genome Biol. 4:R9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braus, G. H., C. Sasse, and S. Krappmann. 2006. Amino acid acquisition, cross-pathway control, and virulence in Aspergillus. Med. Mycol. 44:91-94. [DOI] [PubMed] [Google Scholar]
- 8.Cantwell, R., C. M. McEntee, and A. P. Hudson. 1992. Regulation of mitochondrial transcription during the stringent response in yeast. Curr. Genet. 21:241-247. [DOI] [PubMed] [Google Scholar]
- 9.Carsiotis, M., and R. F. Jones. 1974. Cross-pathway regulation: tryptophan-mediated control of histidine and arginine biosynthetic enzymes in Neurospora crassa. J. Bacteriol. 119:889-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carsiotis, M., R. F. Jones, and A. C. Wesseling. 1974. Cross-pathway regulation: histidine-mediated control of histidine, tryptophan, and arginine biosynthetic enzymes in Neurospora crassa. J. Bacteriol. 119:893-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carsiotis, M., and A. M. Lacy. 1965. Increased activity of tryptophan biosynthetic enzymes in histidine mutants of Neurospora crassa. J. Bacteriol. 89:1472-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Catcheside, D. E. A. 1978. A pleiotropic mutation in Neurospora conferring sensitivity to analogues of amino acids, purines and pyrimidines. Neurospora Newsl. 25:17-18. [Google Scholar]
- 13.Colot, H. V., G. Park, G. E. Turner, C. Ringelberg, C. M. Crew, L. Litvinkova, R. L. Weiss, K. A. Borkovich, and J. C. Dunlap. 2006. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA 103:10352-10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ebbole, D. J., J. L. Paluh, M. Plamann, M. S. Sachs, and C. Yanofsky. 1991. cpc-1, the general regulatory gene for genes of amino acid biosynthesis in Neurospora crassa, is differentially expressed during the asexual life cycle. Mol. Cell. Biol. 11:928-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fitzpatrick, D. A., M. E. Logue, J. E. Stajich, and G. Butler. 2006. A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol. Biol. 22:6-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flint, H. J. 1985. Changes in gene expression elicited by amino acid limitation in Neurospora crassa strains having normal or mutant cross-pathway amino acid control. Mol. Gen. Genet. 200:283-290. [DOI] [PubMed] [Google Scholar]
- 18.Flint, H. J., and J. Wilkening. 1986. Cloning of the arg-12 gene of Neurospora crassa and regulation of its transcript via cross-pathway amino acid control. Mol. Gen. Genet. 203:110-116. [DOI] [PubMed] [Google Scholar]
- 19.Frishman, D., K. Albermann, J. Hani, K. Heumann, A. Metanomski, A. Zollner, and H.-W. Mewes. 2001. Functional and structural genomics using PEDANT. Bioinformatics 17:44-57. [DOI] [PubMed] [Google Scholar]
- 20.Garcia-Sanchez, S., S. Aubert, I. Iraqui, G. Janbon, J.-M. Ghigo, and C. d'Enfert. 2004. Candida albicans biofilms: a developmental state associated with specific and stable gene expression patterns. Eukaryot. Cell 3:536-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gasch, A. P., A. M. Moses, D. Y. Chiang, H. B. Fraser, M. Berardini, and M. B. Eisen. 2004. Conservation and evolution of cis-regulatory systems in ascomycete fungi. PLoS Biol. 2:e398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harbison, C. T., D. B. Gordon, T. I. Lee, N. J. Rinaldi, K. D. Macisaac, T. W. Danford, N. M. Hannett, J. B. Tagne, D. B. Reynolds, J. Yoo, E. G. Jennings, J. Zeitlinger, D. K. Pokholok, M. Kellis, P. A. Rolfe, K. T. Takusagawa, E. S. Lander, D. K. Gifford, E. Fraenkel, and R. A. Young. 2004. Transcriptional regulatory code of a eukaryotic genome. Nature 431:99-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hertz, G. Z., and G. D. Stormo. 1999. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics 15:563-577. [DOI] [PubMed] [Google Scholar]
- 24.Hinnebusch, A. G. 1984. Evidence for translational regulation of the activator of general amino acid control in yeast. Proc. Natl. Acad. Sci. USA 81:6442-6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hinnebusch, A. G. 1988. Mechanisms of gene regulation in the general control of amino acid biosynthesis in Saccharomyces cerevisiae. Microbiol. Rev. 52:248-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hinnebusch, A. G. 2005. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 59:407-450. [DOI] [PubMed] [Google Scholar]
- 27.Hinnebusch, A. G., and G. R. Fink. 1983. Positive regulation in the general amino acid control of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 80:5374-5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hinnebusch, A. G., and K. Natarajan. 2002. Gcn4p, a master regulator of gene expression, is controlled at multiple levels by diverse signals of starvation and stress. Eukaryot. Cell 1:22-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ihmels, J., S. Bergmann, and N. Barkai. 2004. Defining transcription modules using large-scale gene expression data. Bioinformatics 20:1993-2003. [DOI] [PubMed] [Google Scholar]
- 30.Ihmels, J., S. Bergmann, M. Gerami-Nejad, I. Yanai, M. McClellan, J. Berman, and N. Barkai. 2005. Rewiring of the yeast transcriptional network through the evolution of motif usage. Science 309:938-940. [DOI] [PubMed] [Google Scholar]
- 31.Kasuga, T., J. P. Townsend, C. Tian, L. B. Gilbert, G. Mannhaupt, J. W. Taylor, and N. L. Glass. 2005. Long-oligomer microarray profiling in Neurospora crassa reveals the transcriptional program underlying biochemical and physiological events of conidial germination. Nucleic Acids Res. 33:6469-6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kerr, M. K., and G. A. Churchill. 2001. Statistical design and the analysis of gene expression microarray data. Genet. Res. 77:123-128. [DOI] [PubMed] [Google Scholar]
- 33.Kim, S.-J., M. J. Swanson, H. Qiu, C. K. Govind, and A. G. Hinnebusch. 2005. Activator Gcn4p and Cyc8p/Tup1p are interdependent for promoter occupancy at ARG1 in vivo. Mol. Cell. Biol. 25:11171-11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kleinschmidt, M., O. Grundmann, N. Bluthgen, H. U. Mosch, and G. H. Braus. 2005. Transcriptional profiling of Saccharomyces cerevisiae cells under adhesion-inducing conditions. Mol. Genet. Genomics 273:382-393. [DOI] [PubMed] [Google Scholar]
- 35.Krappmann, S., E. M. Bignell, U. Reichard, T. Rogers, K. Haynes, and G. H. Braus. 2004. The Aspergillus fumigatus transcriptional activator CpcA contributes significantly to the virulence of this fungal pathogen. Mol. Microbiol. 52:785-799. [DOI] [PubMed] [Google Scholar]
- 36.Lee, T. I., N. J. Rinaldi, F. Robert, D. T. Odom, Z. Bar-Joseph, G. K. Gerber, N. M. Hannett, C. T. Harbison, C. M. Thompson, I. Simon, J. Zeitlinger, E. G. Jennings, H. L. Murray, D. B. Gordon, B. Ren, J. J. Wyrick, J. B. Tagne, T. L. Volkert, E. Fraenkel, D. K. Gifford, and R. A. Young. 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298:799-804. [DOI] [PubMed] [Google Scholar]
- 37.Legerton, T. L., and C. Yanofsky. 1985. Cloning and characterization of the multifunctional his-3 gene of Neurospora crassa. Gene 39:129-140. [DOI] [PubMed] [Google Scholar]
- 38.Liu, X. S., D. L. Brutlag, and J. S. Liu. 2002. An algorithm for finding protein-DNA binding sites with applications to chromatin-immunoprecipitation microarray experiments. Nat. Biotechnol. 20:835-839. [DOI] [PubMed] [Google Scholar]
- 39.Ludwig, M. Z., A. Palsson, E. Alekseeva, C. M. Bergman, J. Nathan, and M. Kreitman. 2005. Functional evolution of a cis-regulatory module. PLoS Biol. 3:e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ludwig, M. Z., N. H. Patel, and M. Kreitman. 1998. Functional analysis of eve stripe 2 enhancer evolution in Drosophila: rules governing conservation and change. Development 125:949-958. [DOI] [PubMed] [Google Scholar]
- 41.McClung, C. R., C. R. Davis, K. M. Page, and S. A. Denome. 1992. Characterization of the formate (for) locus, which encodes the cytosolic serine hydroxymethyltransferase of Neurospora crassa. Mol. Cell. Biol. 12:1412-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McGuire, A. M., J. D. Hughes, and G. M. Church. 2000. Conservation of DNA regulatory motifs and discovery of new motifs in microbial genomes. Genome Res. 10:744-757. [DOI] [PubMed] [Google Scholar]
- 43.Metzenberg, R. L. 2004. Bird medium: an alternative to Vogel medium. Fungal Genet. Newsl. 51:19-20. [Google Scholar]
- 44.Mueller, P. P., and A. G. Hinnebusch. 1986. Multiple upstream AUG codons mediate translational control of GCN4. Cell 45:201-207. [DOI] [PubMed] [Google Scholar]
- 45.Natarajan, K., M. R. Meyer, B. M. Jackson, D. Slade, C. Roberts, A. G. Hinnebusch, and M. J. Marton. 2001. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell. Biol. 21:4347-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paluh, J. L., M. J. Orbach, T. L. Legerton, and C. Yanofsky. 1988. The cross-pathway control gene of Neurospora crassa, cpc-1, encodes a protein similar to GCN4 of yeast and the DNA-binding domain of the oncogene v-jun-encoded protein. Proc. Natl. Acad. Sci. USA 85:3728-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paluh, J. L., M. Plamann, D. Kruger, I. B. Barthelmess, C. Yanofsky, and D. D. Perkins. 1990. Determination of the inactivating alterations in two mutant alleles of the Neurospora crassa cross-pathway control gene cpc-1. Genetics 124:599-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paluh, J. L., and C. Yanofsky. 1991. Characterization of Neurospora CPC1, a bZIP DNA-binding protein that does not require aligned heptad leucines for dimerization. Mol. Cell. Biol. 11:935-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Penn, M. D., B. Galgoci, and H. Greer. 1983. Identification of AAS genes and their regulatory role in general control of amino acid biosynthesis in yeast. Proc. Natl. Acad. Sci. USA 80:2704-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rolfes, R. J., and A. G. Hinnebusch. 1993. Translation of the yeast transcriptional activator GCN4 is stimulated by purine limitation: implications for activation of the protein kinase GCN2. Mol. Cell. Biol. 13:5099-5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rubin-Bejerano, I., I. Fraser, P. Grisafi, and G. R. Fink. 2003. Phagocytosis by neutrophils induces an amino acid deprivation response in Saccharomyces cerevisiae and Candida albicans. Proc. Natl. Acad. Sci. USA 100:11007-11012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruepp, A., A. Zollner, D. Maier, K. Albermann, J. Hani, M. Mokrejs, I. Tetko, U. Guldener, G. Mannhaupt, M. Munsterkotter, and H. W. Mewes. 2004. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 32:5539-5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sachs, M. S. 1996. General and cross-pathway controls of amino acid biosynthesis, p. 315-345. In R. Brambl and G. A. Marzluf (ed.), The Mycota, vol. 3. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 54.Sachs, M. S., and C. Yanofsky. 1991. Developmental expression of genes involved in conidiation and amino acid biosynthesis in Neurospora crassa. Dev. Biol. 148:117-128. [DOI] [PubMed] [Google Scholar]
- 55.Sattlegger, E., A. G. Hinnebusch, and I. B. Barthelmess. 1998. cpc-3, the Neurospora crassa homologue of yeast GCN2, encodes a polypeptide with juxtaposed eIF2alpha kinase and histidyl-tRNA synthetase-related domains required for general amino acid control. J. Biol. Chem. 273:20404-20416. [DOI] [PubMed] [Google Scholar]
- 56.Schechtman, M. G., and C. Yanofsky. 1983. Structure of the trifunctional trp-1 gene from Neurospora crassa and its aberrant expression in Escherichia coli. J. Mol. Appl. Genet. 2:83-99. [PubMed] [Google Scholar]
- 57.Seo, J., M. Bakay, Y. W. Chen, S. Hilmer, B. Shneiderman, and E. P. Hoffman. 2004. Interactively optimizing signal-to-noise ratios in expression profiling: project-specific algorithm selection and detection p-value weighting in Affymetrix microarrays. Bioinformatics 20:2534-2544. [DOI] [PubMed] [Google Scholar]
- 58.Spurgeon, S. L., and W. H. Matchett. 1977. Inhibition of aminoacyl-transfer ribonucleic acid synthetases and the regulation of amino acid biosynthetic enzymes in Neurospora crassa. J. Bacteriol. 129:1303-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stuart, J. M., E. Segal, D. Koller, and S. K. Kim. 2003. A gene-coexpression network for global discovery of conserved genetic modules. Science 302:249-255. [DOI] [PubMed] [Google Scholar]
- 60.Swanson, M. J., H. Qiu, L. Sumibcay, A. Krueger, S. J. Kim, K. Natarajan, S. Yoon, and A. G. Hinnebusch. 2003. A multiplicity of coactivators is required by Gcn4p at individual promoters in vivo. Mol. Cell. Biol. 23:2800-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thireos, G., M. D. Penn, and H. Greer. 1984. 5′ untranslated sequences are required for the translational control of a yeast regulatory gene. Proc. Natl. Acad. Sci. USA 81:5096-5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tournu, H., G. Tripathi, G. Bertram, S. Macaskill, A. Mavor, L. Walker, F. C. Odds, N. A. Gow, and A. J. Brown. 2005. Global role of the protein kinase Gcn2 in the human pathogen Candida albicans. Eukaryot. Cell 4:1687-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Townsend, J. P. 2003. Multifactorial experimental design and the transitivity of ratios with spotted DNA microarrays. BMC Genomics 4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Townsend, J. P. 2004. Resolution of large and small differences in gene expression using models for the Bayesian analysis of gene expression levels and spotted DNA microarrays. BMC Bioinformatics 5:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Townsend, J. P., and D. L. Hartl. 2002. Bayesian analysis of gene expression levels: statistical quantification of relative mRNA level across multiple strains or treatments. Genome Biol. 3:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tripathi, G., C. Wiltshire, S. Macaskill, H. Tournu, S. Budge, and A. J. Brown. 2002. Gcn4 co-ordinates morphogenetic and metabolic responses to amino acid starvation in Candida albicans. EMBO J. 21:5448-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Valenzuela, L., C. Aranda, and A. Gonzalez. 2001. TOR modulates GCN4-dependent expression of genes turned on by nitrogen limitation. J. Bacteriol. 183:2331-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vinciotti, V., R. Khanin, D. D'Alimonte, X. Liu, N. Cattini, G. Hotchkiss, G. Bucca, O. de Jesus, J. Rasaiyaah, C. P. Smith, P. Kellam, and E. Wit. 2005. An experimental evaluation of a loop versus a reference design for two-channel microarrays. Bioinformatics 21:492-501. [DOI] [PubMed] [Google Scholar]
- 69.Vogel, H. J. 1964. Distribution of lysine pathways among fungi: evolutionary implications. Am. Nat. 98:435-446. [Google Scholar]
- 70.Wray, G. A., M. W. Hahn, E. Abouheif, J. P. Balhoff, M. Pizer, M. V. Rockman, and L. A. Romano. 2003. The evolution of transcriptional regulation in eukaryotes. Mol. Biol. Evol. 20:1377-1419. [DOI] [PubMed] [Google Scholar]
- 71.Yang, R., S. A. Wek, and R. C. Wek. 2000. Glucose limitation induces GCN4 translation by activation of Gcn2 protein kinase. Mol. Cell. Biol. 20:2706-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang, Y. H., and T. Speed. 2002. Design issues for cDNA microarray experiments. Nat. Rev. Genet. 3:579-588. [DOI] [PubMed] [Google Scholar]
- 73.Yu, H., N. M. Luscombe, H. X. Lu, X. Zhu, Y. Xia, J. D. Han, N. Bertin, S. Chung, M. Vidal, and M. Gerstein. 2004. Annotation transfer between genomes: protein-protein interologs and protein-DNA regulogs. Genome Res. 14:1107-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.