Abstract
We isolated an MgATG1 gene encoding a serine/threonine protein kinase from the rice blast fungus Magnaporthe grisea. In the ΔMgatg1 mutant, in which the MgATG1 gene had been deleted, autophagy was blocked; the mutant also showed fewer lipid droplets in its conidia, lower turgor pressure of the appressorium, and such defects in morphogenesis as delayed initiation and slower germination of conidia. As a result of lower turgor pressure of the appressorium, the ΔMgatg1 mutant lost its ability to penetrate and infect the two host plants, namely, rice and barley. However, normal values of the parameters and infective abilities were restored on reintroducing an intact copy of the MgATG1 gene into the mutant. Autophagy is thus necessary for turnover of organic matter during the formation of conidia and appressoria and for normal development and pathogenicity in M. grisea.
Magnaporthe grisea, a filamentous ascomycete fungus, is best known as the causal agent of rice blast, the most serious disease of cultivated rice throughout the world (15), and has been developed as a model organism for investigating fungus-host interactions (4, 27). The appressorium, a specialized cell necessary for infection by the rice blast fungus, generates tremendous intracellular turgor pressure (as much as 8.0 MPa) to penetrate the leaf cuticle (5, 26). Such enormous turgor in the appressorium is a consequence of accumulation of very large quantities of glycerol in the cell, and potential sources of glycerol biosynthesis are lipid, glycogen, and two sugars, trehalose and mannitol, in the conidium (21).
The process of development from the conidium to the appressorium, and then from the appressorium to the penetration peg or infectious hypha, requires a cell to undergo significant phenotypic changes accompanied by the breakdown and recycling of old cellular components in about 30 h (21). In M. grisea, appressorium formation involves autophagy, but nuclei in conidia of the ΔMgatg8 mutant were not degraded during appressorium formation, and the mutant failed to infect the plant through the appressoria (29). Also, the mutant produced far fewer conidia. Autophagy is a common and evolutionarily preserved process that degrades and recycles old proteins and organelles in all eukaryotic cells (8, 9, 18, 31). For many years, autophagy was believed to be involved in changes in cellular architecture during differentiation and development, presumably through its role in the turnover of organelles and proteins (9). Our study sought to find out whether autophagy has a role in the turnover of organic matter that enables a hypha to form a conidium, which then goes on to develop an appressorium, and in generating the turgor pressure in the appressorium required for successful infection.
An expressed sequence tag (clone s197; GenBank accession no. CK828251) for MgATG1 (autophagy-related gene 1), homologous to ATG1 of yeast in its protein sequence, was found in the appressorium of the rice blast fungus (10). Because the fungus is used as a primary model for host-pathogen interactions (27), the role of autophagy in fungal development, appressorium turgor, and pathogenicity of M. grisea through the turnover of organelles and proteins forms an attractive topic for research and is the subject of this paper, which reports that autophagy was blocked in a mutant that lacked the gene MgATG1 and that the mutant not only had fewer lipid droplets in its conidia and lower turgor pressure in its appressoria but also failed to penetrate its host.
MATERIALS AND METHODS
Fungal strains and growth conditions.
M. grisea wild-type strain Guy-11 and mutant strains were cultured on CM medium (22) at 28°C with a 12-h photo phase. Three media were used to study the growth characteristics of the fungus: MM medium (6 g NaNO3, 0.52 g KCl, 0.52 g MgSO4, 1.52 g KH2PO4, 10 g glucose, and 0.5% biotin), MM−N medium (MM medium without the nitrogen source), and MM−C medium (MM medium without the carbon source). Studies involving a cross with strain 2539 were conducted on OMA medium (30 g oat meal and 10 g agar in 1 liter of distilled water).
Cloning and sequencing of the MgATG1 gene.
A cDNA fragment containing a full opening read frame of the MgATG1 gene was cloned from the cDNA library (11) by long-distance PCR with a BD Advantage 2 PCR kit (Clontech Laboratories, California) using the forward primer 5′-CTCGAACCGATAACCACTGCCAGGAAT-3′ and the reverse primer 5′-CTTAGAAACACTCGGGCTCAACCGGG-3′. The amplified product was cloned into a T-easy vector, pUCm-T (Sangon, Shanghai, China) and sequenced on an ABI377 DNA sequencer (Invitrogen).
Complementation of the Saccharomyces cerevisiae Δatg1 mutant.
The MgATG1 open reading frame amplified from MgATG1 cDNA by PCR with primers (5′-TTGGATCCATGGCGGACCGATCAGCACGCCGTGCGCG-3′ and 5′-CCTCTAGATCAAGAGCTGTATGAAGGGACACTTCGCG-3′) was inserted into BamHI-XbaI sites of the yeast expression vector pYES2 (Invitrogen) to create pYES-ATG1. The pYES-ATG1 vector was introduced into the S. cerevisiae Δatg1 mutant (kindly provided by Yoshinori Ohsumi) using a small-scale transformation protocol (12). The uracil prototrophic transformants were tested for the presence of the vector pYES-ATG1 by PCR. The S. cerevisiae Δatg1 mutant has been reported to display a starvation-sensitive phenotype (12). When grown in a starvation medium, the Δatg1 mutant tends to lose its viability over time. The protocol to check for the complementation of the starvation-sensitive phenotype in the yeast Δatg1 mutant was as follows: yeast cells were incubated on YPD (1% yeast extract, 2% polypeptone, and 2% glucose) plates for 2 days at 30°C, multiplied on an SD(−N) plate (2% glucose and 0.17% yeast nitrogen base with neither amino acids nor ammonium sulfate), incubated for 14 days at 30°C, and subcultured on a YPD plate for 2 days at 30°C before recording the growth of cells (12).
Generation of the GFP-MgATG1 fusion construct.
The promoter fragment of the NAR gene (norsolorinic acid reductase; GenBank accession no. EF486491) was cloned from Guy-11 genetic DNA by PCR using primers (5′-CAGGATCCGGGAAGCGATTGCGTT-3′ and 5′-GGTGCCATGGTGTCGGTTGTGGTG-3′) and inserted into BamHI-NcoI sites of pEGFP (Clontech) to produce a vector, namely, pEGFP-NAR. The GFP gene coding DNA sequences (CDS) fragment without the stop codon (TAA) under the control of the NAR promoter was amplified with primers (5′-CTCCCGGGAAGCGATTGCGTTT-3′ and 5′-CAGGATCCTTGTACAGCTCGTCCATGC-3′) from pEGFP-NAR. The NAR promoter-GFP CDS fusion fragment thus obtained was then cloned into SmaI-BamHI sites of the pBS-HPH1 vector to generate a vector, namely, pBS-HPH1-NAR-GFP. pBS-HPH1 is a modified pBluescript II SK(+) vector (Stratagene) inserted in SalI-HindIII sites with a hygromycin-resistant gene (HPH) cloned from pCB1003 vector (3) using primers (HPH1, 5′-CCGTCGACCTACTCTATTCCTTTGCCCTCG-3′; HPH2, 5′-CCAAGCTTTGGAGGTCAACACATCAAT-3′). Subsequently, the coding domain of the MgATG1 fragment without the initiation codon (ATG) was amplified with primers (5′-TTGGATCCGCGGACCGATCAGCACGCCGTGCGCG-3′ and 5′-GCGCGGCCGCTCAAGAGCTGTATGAAGGGACACTTCGCG-3′) obtained from MgATG1 cDNA and inserted into BamHI-NotI sites of pBS-HPH1-NAR-GFP to generate pGFP-ATG1. The resulting vector, namely, pGFP-ATG1, linearized with SmaI, was transformed into protoplasts of Guy-11, and the resulting transformants were confirmed to have a single-copy genomic integration by Southern hybridization analysis. Fluorescence of the transformants was observed using an Olympus-BX51 microscope (Japan) with UV epifluorescence.
Construction of targeted gene deletion vector and fungal transformation.
The targeted gene deletion vector pBS-ATG1 was constructed by inserting two flanking sequences of the MgATG1 gene into two sides of the HPH gene in the pBS-HPH1 vector. A 1.6-kb XhoI-SalI upstream flanking sequence fragment, amplified from M. grisea genomic DNA using the primers ATG1up-p1 (5′-TTCTCGAGCTCACTGACTCCGCCAAACCCCACACT-3′) and ATG1up-p2 (5′-CCGTCGACGGCAACGGCGGCACCAGATACCT-3′), was first inserted into XhoI-SalI sites of the pBS-HPH1 vector to generate pBS-ATG1up. The targeted gene deletion vector pBS-ATG1 was then generated after inserting another HindIII-XbaI downstream flanking sequence fragment amplified using the primers ATG1low-p1 (5′-TTCGAGGATGCCCCGAAGCGAAGCTT-3′) and ATG1low-p2 (5′-CCTCTAGACAGGGCGTGCGACAAACCAAAGAGTA-3′) into HindIII-XbaI sites of pBS-ATG1up. The 4.6-kb XhoI-ClaI-digested targeted deletion fragment of pBS-ATG1 was transformed into protoplasts of Guy-11 cells.
The fungal protoplasts were produced and transformed with DNA as previously described (22). The mutants were screened using OCM agar (CM stabilized osmotically by adding 1 M sucrose) containing hygromycin B (200 μg/ml). To identify the mutants in which the gene had been deleted, PCR was performed using the checking primers internal to MgATG1 (yzp1, 5′-AACCGCGAATCATCATCCCCAACT-3′; yzp2, 5′-GCAACCCGCGCATCCTCGTCTA-3′). Integration of the HPH gene expression cassette was also confirmed by PCR with HPH-specific primers HPH1 and HPH2. The mutants in which the gene had been deleted were purified by culturing single conidia, and the single integration event was confirmed by Southern hybridization.
For complementation assays, the 6-kb PCR products containing 2-kb upstream sequence, full-length M. grisea MgATG1 gene coding region, and 1-kb downstream sequence were first amplified from genomic DNA using primers hb1 (5′-GTCGTTCATCAGGCGTTCTATTTG-3′) and hb2 (5′-TACTCACTCCTGCTCCTGGGCCTG-3′) and then cloned into a pCR-XL-TOPO vector (Invitrogen) to produce a pTOPO-ATG1 vector. The EcoRI-digested PCR fragment was then ligated to the EcoRI site in pBarKS1 (16) to obtain pBAR-ATG1. Resistance to glufosinate ammonium (150 μg/ml) was used as a selectable marker during transformation with XhoI-linearized pBAR-ATG1 (with pBARKS1 as control), and successful single-copy genomic integration was confirmed by Southern hybridization.
Nucleic acid manipulation and Southern blotting.
Restriction digest, gel electrophoresis, ligation reactions, and PCR were all carried out using standard procedures (19).
For Southern hybridization analysis of ΔMgatg1 mutants, genomic DNA was digested with NcoI, separated on a 0.7% agarose gel in 1× Tris-borate-EDTA buffer, and transferred onto a positively charged nylon transfer membrane. Labeled probes were amplified from genomic DNA using the primers probe1 (5′-CAGCGGTGGTGACGGAGAAGATG-3′) and probe2 (5′-CACCAAGTTGCGCCCCGTAGTC-3′). The hybridization was carried out in accordance with the manufacturer's instructions for digoxigenin high-prime DNA labeling and the detection starter kit I (Roche, Germany).
To detect MgATG1 transcripts in the transformants, a mixture of conidia and mycelia cultured for 10 days on a cellophane membrane that covered the CM medium was quickly frozen in liquid nitrogen, and total RNA was isolated from the mixture by the TRIzol method following the procedure laid down by the manufacturer (Molecular Research Center, Inc.). cDNA was reverse transcribed from 500 ng of the total RNA with random 6-mers using the SYBR ExScript reverse transcription-PCR (RT-PCR) kit (TaKaRa Bio Co. Ltd., Dalian, China) and diluted 1:1 with nuclease-free water. RT-PCR was conducted thrice with three replicates each time. The primer pairs for transcript amplifications were as follows: MgATG1, F_qRT (ATGGGCAAGCACAAGGTA)/R_qRT (TCATGGAGGGCAACAATAT) and β-tubulin gene (internal control), 2_215U20 (GTGACCCTCGCAACGGAAAG)/2_315L20 (CGACGAACTGGATGCTACGC).
Assays for vegetative growth, formation of conidia, and formation of appressorium.
Discs of mycelia, 5 mm in diameter, from 8-day-old CM plates were individually transferred to the center of 90-mm petri dishes containing MM medium, MM−N medium, and MM−C medium and cultured at 28°C with a 12-h photophase. Radial growth of vegetative mycelia was measured at 9 days and 14 days.
Fungal growth cultured on CM agar at 28°C for 8 days was scraped with a sterilized glass rod to remove the aerial mycelia and dried aseptically with a blower for 2 to 3 h (30). To induce production of conidia, the mycelium was incubated at 28°C with a 12-h photophase for 3 days. Conidia taken from three mycelial discs, each 1 cm in diameter, were suspended in sterile water and counted with a hemocytometer under a microscope.
The conidial suspension thus obtained was filtered through three layers of lens paper and resuspended in sterile distilled water to ensure 1 × 105 conidia/ml. Droplets (20 μl or 100 μl) of the conidial suspension were placed on plastic microscope coverslips and incubated in a humid environment at 25°C. The frequency of conidial germination and appressorium formation was determined at 2 h, 4 h, and 6 h by counting the number of germ tubes and/or appressoria formed from more than 150 conidia. All the experiments were repeated three times with seven replicates each time.
Measurement of appressorium turgor.
Appressorial turgor was estimated using incipient cytorrhysis (cell collapse) assays (6). Conidia in 20-μl droplets (1 × 106 conidia/ml) were induced to form appressoria on plastic microscope coverslips incubated in a humid chamber for 24 h or 48 h. Water surrounding the conidia was removed carefully and replaced with an equal volume (20 μl) of glycerol in concentrations ranging from 0.5 M to 4.0 M. The number of appressoria that had collapsed after 10 min was recorded. The experiments were replicated three times, and more than 200 appressoria were observed each time.
Nile Red assays.
Conidia, germinating conidia, and appressoria were stained directly in Nile Red solution consisting of 50 mM Tris-maleate buffer (pH 7.5), 20 mg/ml polyvinylpyrrolidone, and 2.5 μg/ml Nile Red oxazone (9-diethylamino-5H-benzo[α]phenoxazine-5-one; Sigma) (24). Within a few seconds, lipid droplets in the conidia and appressoria began to fluoresce when seen under a microscope, whereas no fluorescence was observed with the same solution without Nile Red.
Electron microscopy.
Conidia collected from 10-day-old mycelia were cultured at 28°C for 24 h in CM liquid medium continuously shaken at 150 rpm. The mycelial growth was collected, thoroughly washed in distilled water, transferred to MM−N liquid medium with 2 mM phenylmethylsulfonyl fluoride (PMSF), and incubated at 28°C for 4 h on a 150-rpm shaker. The fungal mass was then collected and fixed overnight at 4°C in modified Karmovsky's fixative containing 2% paraformaldehyde and 2.5% (vol/vol) glutaraldehyde in 0.1 M phosphate buffer (pH 7.2). The fixed samples were washed three times, for 10 min each time, with 0.1 M phosphate buffer (pH 7.2). The samples were postfixed in 1% OsO4 for 2 h at 25°C, washed three times with phosphate buffer as before, dehydrated in a graded ethanol series, embedded in Spurr resin, and stained with 2% uranyl acetate and Reynold's lead solution before sectioning. The ultrathin sections were examined under a JEM-1230 electron microscope (JEOL, Tokyo, Japan) operating at 70 kV.
Plant infection assays.
For the cut leaf assay, the third leaf in 14-day-old rice (Oryza sativa cv. CO-39) seedlings or a leaf from 8-day-old barley (Hordeum vulgare cv. ZJ-8) seedlings was used. Conidia harvested from 10-day-old cultures on CM plates were resuspended in 0.2% (wt/vol) gelatin solution to ensure 1 × 105 conidia/ml. A 20-μl droplet was deposited onto the upper side of the cut leaves maintained on 4% (wt/vol) water agar plates (distilled water was used). The leaves were observed for disease lesions after 24, 48, 72, and 96 h of incubation at 25°C. The leaves that showed disease lesions were decolored by methanol, stained with acid fuchsin (1) or 0.01% trypan blue-0.06% aniline blue in lactophenol (7), and observed under a microscope for host penetration.
For spray infection assays, 2-week-old seedlings of rice and 8-day-old seedlings of barley were used. The suspension (1 × 105 conidia/ml) was sprayed evenly onto plant leaves using an artist's airbrush (Badger Co., Illinois). Inoculated plants were placed in a dew chamber at 25°C for 48 h (rice) or 24 h (barley) in the dark and then transferred to the growth chamber with a photoperiod of 12 h achieved by using fluorescent lights. Lesion formation was examined 7 days (rice) or 4 days (barley) after inoculation. Disease severity in rice was rated using the scale developed by Bonman et al. (2), and the densities of lesions were recorded from 20 to 30 infected leaves using a 5-cm section of the most severely infected leaf from each plant. Infection assays were replicated three times using 45 plants each time.
Nucleotide sequence accession number.
The sequence of the MgATG1 cDNA-containing CDS was determined and deposited in GenBank under accession no. DQ224381.
RESULTS
Isolation of MgATG1.
An expressed sequence tag (GenBank accession no. CK828251) encoding Mgatg1, a serine/threonine kinase required for autophagy, was found in screening the genes that were highly expressed during the appressorium stage from a subtractive suppressive cDNA library using Blastx in GenBank (10). To find out the potential role of Mgatg1 during the formation and maturation of, and penetration by, the appressorium, we isolated a full-length MgATG1 gene from genomic DNA of M. grisea and an appressorium-stage cDNA library (11) by long-distance PCR. A 2,949-bp cDNA fragment containing full CDS was cloned to pUCm-T (Sangon, Shanghai, China) and then sequenced (GenBank accession no. DQ224381).
Mgatg1 in M. grisea, containing an S_TKc domain of 256 residues, is a serine/threonine protein kinase that is 982 amino acids long with a calculated molecular mass of 108 kDa. The Mgatg1 and Atg1s from other organisms show very high homology (data not shown); Mgatg1 in M. grisea, for example, is 51% identical to Atg1 (Apg1/Aut3) in S. cerevisiae (12, 20). To determine whether MgATG1 in the rice blast fungus is functionally related to the ATG1 of S. cerevisiae, a full-length MgATG1 cDNA fragment of M. grisea was introduced into S. cerevisiae strain Δatg1 (kindly provided by Yoshinori Ohsumi) using pYES2, an expression vector (Invitrogen). The expression of MgATG1 in M. grisea was confirmed in the Δatg1 mutant of S. cerevisiae under the control of a GAL1 promoter by RT-PCR (data not shown). When cultured on starvation medium for 14 days, the mutant died, but those complemented with MgATG1 survived, which shows that MgATG1 had restored the corresponding defects in these mutants, thereby ensuring their survival (12) (Fig. 1). Therefore, MgATG1 of M. grisea is presumably homologous to ATG1 of yeast in structure and function.
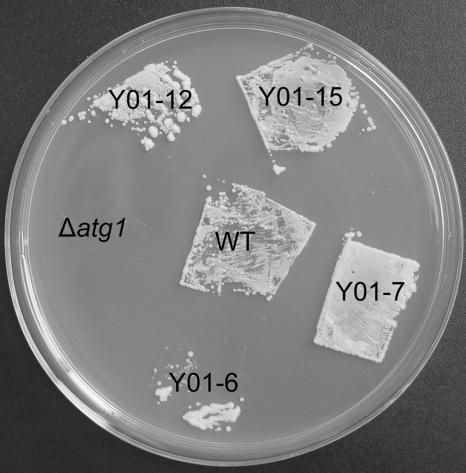
FIG. 1.
Complementation of the S. cerevisiae Δatg1 mutant by the MgATG1 gene. After incubation for 14 days at 30°C on SD−N medium, the yeast cells were grown on YPD plates for 2 days at 30°C. Cells of MgATG1-rescued yeast strains Y01-6, Y01-7, Y01-12, and Y01-15 and of the wild-type strain (WT) survived the starvation regimen, but those of Δatg1 mutant did not.
Cellular localization of MgATG1.
To investigate the localization of MgATG1 in M. grisea, we constructed a fluorescent green protein gene-MgATG1 fusion expression cassette (pGFP-ATG1). pGFP-ATG1 linearized with SmaI was introduced into the protoplasts of the wild-type strain. The transformant NGA7 was selected for further studies after Southern blot analysis, which confirmed that it contained a single-copy integration of pGFP-ATG1. Expression of GFP-ATG1 was uniformly detectable in the cytoplasm of conidia, mycelia, and appressoria of the transformant NGA7 (Fig. 2). When an autophagic phenotype for fungal cells was induced in the presence of 2 mM PMSF in the medium without nitrogen for 4 h, the fluorescence was confined to the cytoplasm. As a control, when another green fluorescent protein fusion expression vector, pGFP-MNH6 (MNH6; GenBank accession no. DQ222524), built with the same strategy as pGFP-ATG1, was introduced into the protoplasts of the wild-type strain, the fluorescence was detected in the nucleus (Fig. 2), thereby implying that in M. grisea, the Mgatg1 protein is localized to the cytoplasm.
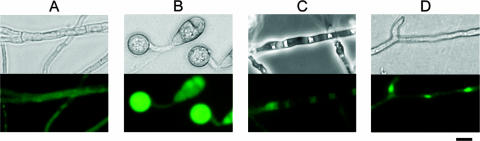
FIG. 2.
Localization of MgATG1 in cells. GFP-MgATG1 protein appeared in the cytoplasm of M. grisea strain NGA7. (A and B) The mycelia, conidia, and appressoria (at 6 h postincubation) of strain NGA7 were observed by light microscopy (top panels) and by fluorescence microscopy (bottom panels). (C) The mycelia were subjected to the presence of 2 mM PMSF in a nitrogen-limiting medium for 4 h and then observed by differential interference microscopy (top) and fluorescence microscopy (bottom). (D) The mycelia of the Guy-11 strain introduced with vector pGFP-MNH6 were observed by light microscopy (top) and by fluorescence microscopy (bottom), and GFP-MNH6 was detected in the nucleus. Bar, 10 μm.
Targeted deletion of MgATG1.
A targeted gene deletion strategy was adopted to determine the role of MgATG1 in fungal development and pathogenicity at the molecular level. The gene deletion vector pBS-ATG1 conferring resistance to hygromycin B was constructed as described earlier. The 4.6-kb XhoI-ClaI DNA fragment containing the replacement cassettes was transformed into the wild-type protoplasts. Initially, 30 transformants resistant to hygromycin B were screened by PCR with the primers internal to MgATG1, and two of these transformants, s197-4 and s197-28, were confirmed to have lost their MgATG1 gene. These two were then purified by culturing them from single conidia. In Southern hybridization analysis, a 2-kb fragment was detected in the two transformants, in contrast with a 4.1-kb fragment found in the wild-type strain (data not shown). The band shift from 4.1 kb to 2 kb showed that the MgATG1 gene had been deleted in transformants s197-4 and s197-28 and that homologous recombination had occurred at a single site.
A complementation assay was carried out to ensure that any phenotype attributed to the ΔMgatg1 mutants was indeed due to deletion of the MgATG1 gene. Plasmid pBAR-ATG1 (containing a full-length sequence of the gene MgATG1) and pBarKS1 (control) were transformed to ΔMgatg1 mutant s197-4. Among the 10 bialaphos-resistant transformants screened by Southern hybridization analysis, one strain (HB24) that carried single-copy integration of the MgATG1 gene at an ectopic site was identified (data not shown).
An RT-PCR assay indicated that the 120-bp strand had not been amplified in cDNA of the ΔMgatg1 mutant, whereas the strand was prominent in that of the wild-type strain and of the complementary mutant HB24 with the primers internal to the MgATG1 cDNA sequence (with β-tubulin as a control). These results indicated that, at the mRNA level, the MgATG1 gene had been deleted in ΔMgatg1 mutant s197-4 but rescued in the complementary mutant, HB24. Therefore, the ΔMgatg1 mutant s197-4 is referred to as a ΔMgatg1 mutant strain and the transformant HB24 as an MgATG1-rescued strain.
Blocking of autophagy in ΔMgatg1 mutants.
After the ΔMgatg1 mutants had been cultured in the nitrogen-limiting medium (MM−N) in the presence of 2 mM PMSF for 4 h, it was found by electron microscopy that they did not accumulate autophagosomes in the cytoplasm or autophagic bodies in the lumen of vacuoles in their hyphal cells (Fig. 3); differential interference microscopy also confirmed the observation about autophagic bodies not being accumulated in vacuoles. These observations suggest that the autophagic pathway in ΔMgatg1 mutants is blocked and that the MgATG1 gene is responsible for autophagy in this fungal species.
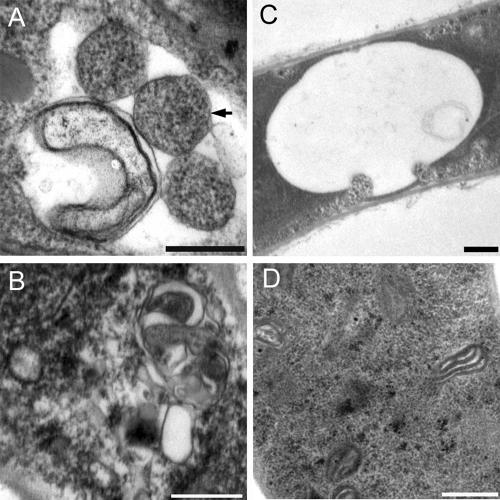
FIG. 3.
Autophagy was blocked in ΔMgatg1 mutants. The vacuole was filled with autophagic bodies (A) and the cytoplasm with autophagosomes in the wild-type Guy-11 cells (B), whereas no autophagic body was evident in the vacuole (C) and no autophagosomes were evident in the cytoplasm of ΔMgatg1 mutants (D) after culturing on MM−N medium in the presence of 2 mM PMSF for 4 h. An autophagosomes are indicated by the arrow. Bars, 0.5 μm.
Defects during growth and during the formation of conidia and appressoria in ΔMgatg1 mutants.
The ΔMgatg1 mutants cultured on both CM and MM plates had sparse aerial hyphae, in contrast to the dense aerial hyphae in the wild-type strain (Fig. 4), although growth of their colonies was not affected (P ≤ 0.01). Conidiogenesis in ΔMgatg1 mutants was reduced significantly (Table 1). The mutants produced (0.11 ± 0.02) × 104/mm2 conidia, approximately 1/15 of that in the wild-type strain. In mating experiments, ΔMgatg1 mutants did not form perithecia even after 4 weeks of incubation with the opposite mating-type strain, 2539, while the wild-type strain and the MgATG1-rescued strain produced many perithecia with viable ascospores. In the germination assay, conidia of ΔMgatg1 mutants were slower to germinate than those of the wild-type strain (Table 1). After 2 h of being placed onto plastic covers, only 72.25% of the conidia of ΔMgatg1 mutants germinated, compared to 88.60% of those of the wild-type strain. However, 4 h later, the rate of germ tube emergence was similar in both. On plastic covers resting on water agar (made using double-distilled water), appressorium formation, either from conidia or from hyphae, remained unaffected in mutants that lacked MgATG1; however, autophagy was blocked in the mutants (Table 1). On reintroducing MgATG1, these defects were completely repaired.
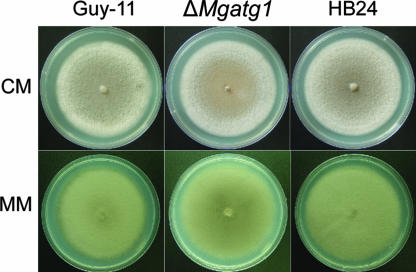
FIG. 4.
Growth characteristics of ΔMgatg1 mutants. Colonies of the wild-type Guy-11, the MgATG1-rescued strain HB24, and ΔMgatg1 mutants were cultured on CM plates (10 days) and on MM (14 days). ΔMgatg1 mutants showed sparse mycelia, in contrast to the dense ones of the wild-type and MgATG1-rescued strains.
TABLE 1.
Phenotypic analysis of ΔMgatg1 mutants of M. griseaa
| Strain | Conidiation (104/mm2)b | Conidial germination (%)c
|
Appressorium formation (%)d | Appressorium turgor (%) after exposure to glycerol concn for indicated timee
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 h | 4 h | 0.5 M
|
1 M
|
2 M
|
3 M
|
4 M
|
||||||||
| 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | |||||
| Guy-11 | 1.66 ± 0.21A | 88.60 ± 4.28A | 99.06 ± 0.65A | 98.83 ± 0.84A | 2.65 ± 0.24A | 2.12 ± 1.37A | 3.23 ± 0.71A | 4.01 ± 0.40A | 24.87 ± 4.78A | 16.88 ± 1.87A | 34.77 ± 3.37A | 46.76 ± 6.19A | 78.36 ± 7.71A | 74.32 ± 8.49A |
| ΔMgatg1 | 0.11 ± 0.02B | 72.25 ± 2.70B | 97.66 ± 0.60A | 93.42 ± 3.99A | 3.86 ± 0.90B | 6.13 ± 1.04B | 19.17 ± 7.69B | 13.22 ± 3.17B | 48.64 ± 5.04B | 37.24 ± 3.25B | 81.71 ± 4.35B | 83.12 ± 1.89B | 85.38 ± 1.08A | 87.81 ± 2.83A |
| HB24 | 1.51 ± 0.33A | 90.32 ± 1.43A | 98.53 ± 0.94A | 95.15 ± 2.30A | 2.19 ± 0.06A | 1.89 ± 1.24A | 4.56 ± 0.15A | 4.95 ± 2.09A | 19.46 ± 1.69A | 28.03 ± 2.44A | 33.93 ± 8.92A | 49.78 ± 3.77A | 79.10 ± 6.39A | 83.61 ± 1.43A |
The tested M. grisea strains were wild-type Guy-11, ΔMgatg1 mutant, and MgATG1-rescued strain HB24. The same capital letters in a column denote that the difference, as estimated by Duncan's test (shortest significant ranges) (P ≤ 0.01), is not significant.
Conidia on three 1-cm-diameter discs of mycelium were collected in water and counted with a hemocytometer (n = 3; seven replicates for each experiment).
The percentage of conidia that had elaborated a germ tube after incubation in water on plastic coverslips after 2 h or 4 h.
The percentage of conidia forming appressoria after incubation on plastic coverslips for 24 h.
Appressorium turgor was measured by incipient cytorrhysis analysis. Appressoria were allowed to form on plastic coverslips for 24 h or 48 h, and the proportion of collapsed appressoria was recorded after exposure to a series of glycerol solutions of various strengths.
The growth of the ΔMgatg1 mutants was assessed on minimal media lacking either of the two crucial nutrients, nitrogen and carbon. Colonies of ΔMgatg1 mutants spread more slowly than those of the wild-type strain in both cases (∼63% of Guy-11 in MM−N and ∼68% in MM−C), but growth of the mutant colonies and of the wild-type strain was comparable on the MM medium (P ≤ 0.01). The MgATG1-rescued strain, however, grew completely normally on MM−N medium and MM−C medium. These results indicate that the ΔMgatg1 mutant is sensitive to such starvation and that the MgATG1 gene is required for normal growth under these circumstances.
Lipid droplets in ΔMgatg1 mutant conidia.
To assess the effect of MgATG1 on the number of lipid droplets in conidia, the conidia were observed by transmission electron microscopy or, after staining with Nile Red, by light microscopy. Conidia of the ΔMgatg1 mutant contained less abundant and more highly vacuolated lipid bodies than those of the wild-type strain or of the MgATG1-rescued strain (Fig. 5); also, conidia of the mutant were stained much more weakly by Nile Red, and the number of lipid droplets stained was lower (Fig. 6). These observations imply that the mutant conidia contained smaller quantities of lipids than those of the wild-type strain and the MgATG1-rescued strain. Also, during development of the appressorium, lipid reserves localized in appressoria of the mutant were lower than in those of the wild-type strain and the MgATG1-rescued strain (Fig. 6). These results indicate that the number of lipid bodies in the mutant is influenced by deletion of the MgATG1 gene.
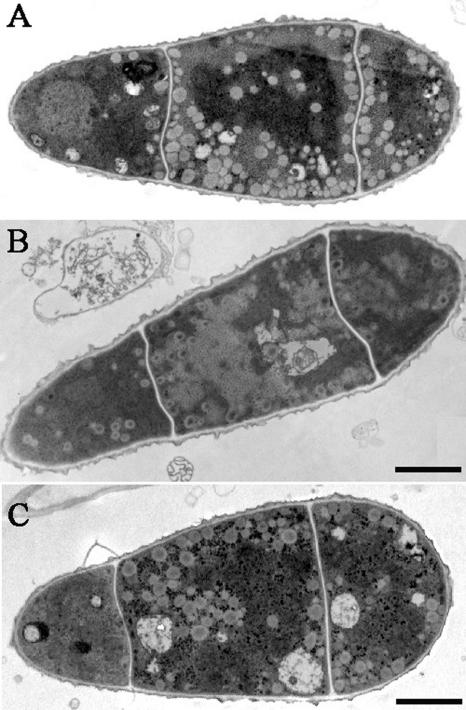
FIG. 5.
Ultrastructures of conidia of Guy-11 and ΔMgatg1 mutants. Conidia were harvested from 10-day-old CM plate cultures. Bars, 2 μm. (A) Conidia of the wild-type Guy-11, showing numerous lipid bodies. (B) Conidia of the ΔMgatg1 mutant, showing fewer and vacuolated lipid bodies. (C) Conidia of the MgATG1-rescued strain HB24, showing an adequate number of lipid bodies.
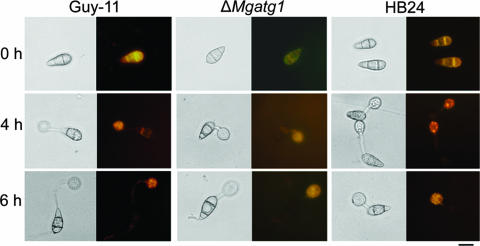
FIG. 6.
Cellular distribution of lipid droplets. Conidia of the wild-type Guy-11, ΔMgatg1 mutants, or MgATG1-rescued strain HB24 were allowed to form appressoria on plastic coverslips. Samples were stained with Nile Red in triacylglycerol and observed in the dark with UV epifluorescence, as described in Materials and Methods. Photographs were taken at 0 h, 4 h, and 6 h after inoculation. Images on the left are of the corresponding fields examined by bright-field microscopy. Bar, 10 μm.
Appressorium turgor in ΔMgatg1 mutants.
To evaluate the role of MgATG1 in appressorium turgor, we measured the turgor of the mature appressorium using the incipient cytorrhysis test described by Howard et al. (6). The number of collapsed appressoria in the series of glycerol solutions of various strengths differed significantly between ΔMgatg1 mutants and the wild-type strain or MgATG1-rescued strain (P ≤ 0.01) (Table 1). In 2 M glycerol, ∼25% of wild-type strain appressoria and ∼19% of MgATG1-rescued strain appressoria had collapsed, compared to ∼49% in appressoria of the ΔMgatg1 mutants 24 h after inoculation. The concentration of glycerol leading to the collapse of 50% of the appressoria was ∼2 M for the mutants and more than 3 M for the wild-type strain. Our study of such incipient cytorrhysis showed that appressorium turgor was reduced significantly in ΔMgatg1 mutants and restored in the MgATG1-rescued strain, suggesting that turgor of the appressorium in the rice blast fungus is regulated by the MgATG1 gene.
Pathogenicity in ΔMgatg1 mutants.
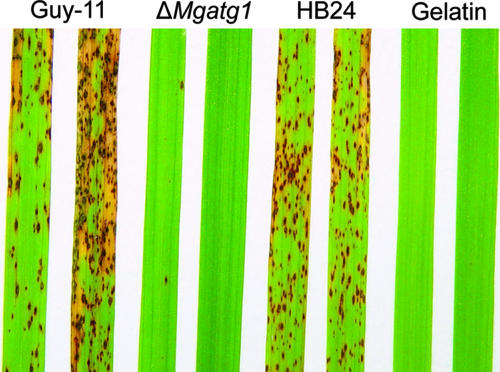
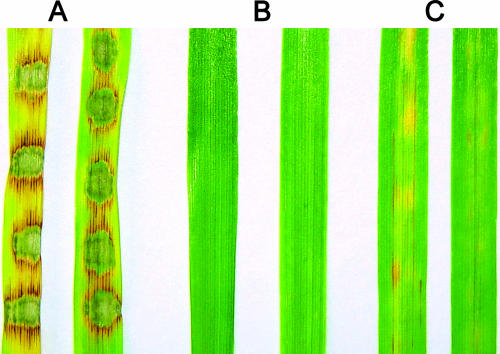
Pathogenicity of ΔMgatg1 mutants to the two susceptible hosts, barley (H. vulgare cv. ZJ-8) and rice (O. sativa cv. CO-39), was tested to assess the role of MgATG1 in pathogenicity. When sprayed on rice, the wild-type strain and MgATG1-rescued strain caused typical diamond-shaped, gray-centered, and sporulating lesions on leaves, most of which later coalesced, whereas ΔMgatg1 mutants caused only a few small and isolated lesions (Fig. 7). Severity of disease caused by ΔMgatg1 mutants was low; 57% of the plants showed no lesions and were graded 1 to 2, whereas more than 90% of plants sprayed with the wild-type strain were graded 4 to 5. On the other hand, the mean (±standard deviation) number of lesions per 5 cm of leaf was 0.87 ± 0.51 for the ΔMgatg1 mutants, 32.08 ± 5.48 for the wild-type strain, and 30.37 ± 9.09 for the MgATG1-rescued strain (P ≤ 0.01). The pattern was similar in barley: ΔMgatg1 mutants failed to produce any observable lesions on leaves, whereas the wild-type strain and MgATG1-rescued strain caused severe lesions.
FIG. 7.
Pathogenicity assays on rice leaves. Seedlings of rice (O. sativa cv. CO-39) were inoculated by spraying them with a suspension of conidia of the wild-type Guy-11, ΔMgatg1 mutants, or MgATG1-rescued strain HB24 individually with 0.2% gelatin as control. Lesion formation on leaves was observed 7 days after inoculation.
In cut leaf assays, both rice and barley leaves inoculated with a 1 × 105 conidia/ml suspension (20-μl droplets) showed severe blast 4 days after inoculation in the case of the wild-type strain or MgATG1-rescued strain, whereas ΔMgatg1 mutants failed to produce any lesions.
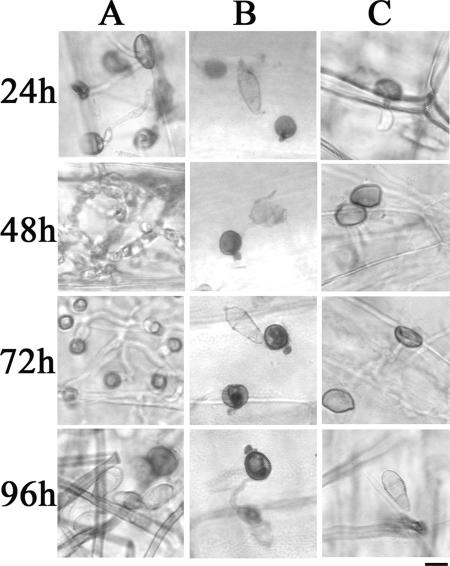
To elucidate why ΔMgatg1 mutants lose their infectivity, we carefully examined the lesions on barley leaves obtained from the cut leaf assays under a microscope. Twenty-four hours after inoculation, ∼49.7% of the appressoria of the wild-type strain had formed long penetration pegs, whereas only ∼2.8% of the appressoria of ΔMgatg1 mutants had done so, and the pegs were also much shorter. Even after a longer period, the frequency of formation of penetration pegs in ΔMgatg1 appressoria continued to be low (∼6.0% after 48 h and ∼6.2% after 72 h). Until 96 h after inoculation, penetration pegs formed by ΔMgatg1 were as short as those formed 24 h, 48 h, or 72 h after inoculation and failed to enter the epidermal cells (Fig. 8), whereas the infectious hyphae of the wild-type strain spread over the infected cells quickly (48 h), colonized the neighboring cells (72 h), and then produced secondary conidia (96 h) (Fig. 8). On wounded leaves, however, the appressoria of ΔMgatg1 mutants could produce functional penetration pegs and long infectious hyphae 24 h after inoculation (Fig. 8). As shown in Fig. 9, ΔMgatg1 mutants failed to produce lesions on intact barley leaves but did so on wounded leaves, although the lesions were far milder than those produced by the wild-type strain on intact leaves. This suggests that infectious growth of ΔMgatg1 mutants on their hosts is also affected. The mutants also fared poorly in terms of the number of conidia formed on wounded leaves, producing fewer than 10 conidia on average on each lesion compared to about 3.4 × 104 produced by the wild-type strain on intact leaves at 72 h after inoculation.
FIG. 8.
Host penetration and growth of infectious hyphae of ΔMgatg1 mutants on barley leaves. Leaves of barley were inoculated with a conidial suspension (5 × 104 conidia/ml), and growth of infectious hyphae was examined at the indicated intervals after inoculation. Bar, 10 μm. (A) Growth of infectious hyphae of wild-type Guy-11 on intact barley leaves. (B) Conidia of ΔMgatg1 mutants failed to develop infectious hyphae on intact barley leaves. (C) Growth of a few infectious hyphae of ΔMgatg1 mutants on abraded barley leaves.
FIG. 9.
Lesions on abraded barley leaves by ΔMgatg1 mutants. Leaves of 8-day-old barley seedlings were inoculated with a conidial suspension (5 × 104 conidia/ml). Disease symptoms were allowed to develop for 96 h. (A) Disease symptoms developed on intact leaves inoculated with wild-type Guy-11 conidia. (B) No disease symptoms developed on intact leaves inoculated with ΔMgatg1 mutant conidia. (C) Weak disease symptoms developed on abraded leaves inoculated with ΔMgatg1 mutant conidia.
From these results, it is clear that ΔMgatg1 mutants were unable to infect barley and rice, owing mainly to their inability to penetrate the cuticle, and that the MgATG1 gene is essential to the pathogenicity of M. grisea.
DISCUSSION
Autophagy is an evolutionary mechanism preserved in all eukaryotes from yeasts to mammals, a catabolic membrane-trafficking phenomenon that depends on signal transduction events. The mechanism is a cellular response to both extracellular and intracellular stress and allows lower organisms such as yeasts to survive nutrient starvation (8, 18). Starvation is one of the key factors that induce the differentiation of a conidium into an appressorium in M. grisea (23). In M. grisea, autophagy is essential to appressorium formation and is known to avert the death of conidia in ΔMgatg8 mutants and make them nonpathogenic to rice (29). Conidiation, appressorium formation, and appressorium-mediated penetration of plant tissue in ΔMgatg8 mutants are reported to be greatly reduced (29). In this study, we identified an MgATG1 gene in M. grisea orthologous to S. cerevisiae ATG1 and found, by studying mutants that lacked MgATG1, that autophagy is essential to fungal development, production of appressorium turgor, and pathogenicity in M. grisea.
Autophagy is necessary for fungal development.
Defective sporulation is a known effect of blocked autophagy in yeast (12, 20, 25). As in yeast, the deletion of MgATG8 in M. grisea significantly reduced the production of conidia (29). Our results show that deletion of MgATG1 also had a negative effect on the efficiency of conidiogenesis. Moreover, ΔMgatg1 mutants displayed less-dense aerial hyphae than the wild-type strain, as was reported in ΔPaATG8 and ΔPaATG1 mutants in Podospora anserine (17), and slightly delayed conidial germination. As in ΔPaATG8 and ΔPaATG1 mutants of P. anserine (17), ΔMgatg1 mutants produced no perithecia, because autophagy is required for differentiation of female reproductive organs. Thus, autophagy is essential to fungal development, although its exact role is different in different species.
Autophagy is coupled with completion of mitosis during morphogenesis of the appressorium and is necessary for infection in the rice blast fungus (29). As in other eukaryotes, autophagy is induced upon starvation or treatment with PMSF in M. grisea. Starvation is one of the key factors in inducing the tip of the germ tube to differentiate into the appressorium (23). It has been shown in many other lower eukaryotic organisms that autophagy is required for differentiation and development processes under inadequate nutrition (9, 12, 17, 20, 25). In M. grisea, hyphae and conidia of the ΔMgatg1 mutant produced appressoria under such conditions, as did those of the wild-type strain, although autophagy was blocked in the mutant. Therefore, it can be inferred that both appressorium formation and autophagy during appressorium morphogenesis are activated simultaneously, but independently, by starvation.
Autophagy is necessary for organic matter turnover during the formation of conidia and appressoria.
Autophagy is believed to be associated with changes in cellular architecture during differentiation and development (9). In M. grisea, buildup of stocks of organic substances in conidia, at least those of lipids in the form of droplets, is affected by autophagy. It was found that lipid droplets in conidia were not only fewer in ΔMgatg1 mutants but also had larger vacuoles.
Autophagy is also known to provide the nutrients to maintain metabolism, which is essential to survival during starvation (8, 28, 31). In M. grisea, ΔMgatg1 mutants are sensitive to lack of nitrogen and carbon, as reported in other Δatg1 mutants in many eukaryotes (8, 13, 14). Differentiation of the conidium or the hypha into the appressorium in M. grisea, occurring on hard hydrophobic surfaces during starvation, is known to require endogenous sources of nutrition for rebuilding cell structure and for accumulation of glycerol in the cell in very large quantities for infection. In ΔMgatg1 mutants, appressorium turgor, achieved through intracellular glycerol (5), was decreased markedly but restored on reintroduction of MgATG1, suggesting that production of glycerol in appressoria is also affected by autophagy.
Autophagy is required for pathogenicity in M. grisea.
Autophagy is necessary if the rice blast fungus is to be infective: the ΔMgatg8 mutants were inefficient in forming the penetration hyphae and failed to infect the plant (29). That ΔMgatg8 mutants can form appressoria but cannot form penetration hyphae efficiently could be explained by changes in turgor pressure in the appressorium. Deletion of MgATG1 blocks the autophagic pathway in ΔMgatg1 mutants, as has been reported in the ΔMgatg8 mutants (29). Intracellular turgor pressure is known to be the main force that enables the appressorium to penetrate the plant cuticle and form the penetration hyphae (5, 26), whereas appressorium turgor was much lower in the ΔMgatg1 mutants. Therefore, it can be inferred that the invasion of the host plant by appressoria will be severely hindered or blocked altogether in ΔMgatg1 mutants. This inference is supported by studies of plant infection in ΔMgatg1 mutants, which have yielded similar results as those described earlier in the case of ΔMgatg8 mutants (29). The frequency of formation of the penetration peg by the appressorium on intact barley leaves was reduced greatly in the ΔMgatg1 mutants. Further, normal barley leaves, when inoculated with the mutant by placing droplets of its conidial suspension on the leaves, formed no observable lesions. However, the mutant could penetrate the cuticle when the leaves had been abraded before inoculation and also produced lesions, although milder than those caused by the wild-type strain. Probably, appressoria in the mutants could exert a force adequate for some appressoria to penetrate abraded cuticles but not strong enough to penetrate intact leaf cuticles. These observations suggest that blocked autophagy in the mutants lowered appressorium turgor, which in turn lowered the ability of pegs to penetrate the cuticle, with the ultimate result being lost pathogenicity.
In summary, MgATG1 plays a significant role in autophagy of M. grisea, is responsible for fungal morphogenesis, and ensures the turnover of organic matter necessary for generating an appressorium turgor of the required strength. ΔMgatg1 mutants failed to initiate autophagy and showed reduced conidiation, conidial germination, lipid turnover, and appressorium turgor. As a result, the mutants lost their ability to penetrate and infect the two tested host plants, namely, rice and barley.
Acknowledgments
This study was supported by grants (nos. 30270049, 30470064, and 30671351) from the National Natural Science Foundation of China, from the 863 Program (no. 2002AA245041) of the Ministry of Science and Technology of China, and from a major project (no. 2004C12020-5) of the Zhejiang Science and Technology Bureau of China to F.-C. Lin.
Footnotes
Published ahead of print on 6 April 2007.
REFERENCES
- 1.Balhadère, P. V., A. J. Foster, and N. J. Talbot. 1999. Identification of pathogenicity mutants of the rice blast fungus Magnaporthe grisea using insertional mutagenesis. Mol. Plant-Microbe Interact. 12:129-142. [Google Scholar]
- 2.Bonman, J. M., D. D. T. Vergel, and M. M. Khin. 1986. Physiologic specialization of Pyricularia oryzae in the Philippines. Plant Dis. 70:767-769. [Google Scholar]
- 3.Carroll, A. M., J. A. Sweigard, and B. Valent. 1994. Improved vectors for selecting resistance to hygromycin. Fungal Genet. Newslett. 41:22. [Google Scholar]
- 4.Dean, R. A., N. J. Talbot, D. J. Ebbole, M. L. Farman, T. K. Mitchell, M. J. Orbach, et al. 2005. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434:980-986. [DOI] [PubMed] [Google Scholar]
- 5.de Jong, J. C., B. J. McCormack, N. Smirnoff, and N. J. Talbot. 1997. Glycerol generates turgor in rice blast. Nature 389:244. [Google Scholar]
- 6.Howard, R. J., M. A. Ferrari, D. H. Roach, and N. P. Money. 1991. Penetration of hard substrates by a fungus employing enormous turgor pressures. Proc. Natl. Acad. Sci. USA 88:11281-11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim, S., I. P. Ahn, H. S. Rho, and Y. H. Lee. 2005. MHP1, a Magnaporthe grisea hydrophobin gene, is required for fungal development and plant colonization. Mol. Microbiol. 57:1224-1237. [DOI] [PubMed] [Google Scholar]
- 8.Klionsky, D. J. 2005. The molecular machinery of autophagy: unanswered questions. J. Cell Sci. 118:7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levine, B., and D. J. Klionsky. 2004. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell 6:463-477. [DOI] [PubMed] [Google Scholar]
- 10.Lu, J. P., T. B. Liu, and F. C. Lin. 2005. Identification of mature appressorium-enriched transcripts in Magnaporthe grisea, the rice blast fungus, using suppression subtractive hybridization. FEMS Microbiol. Lett. 245:131-137. [DOI] [PubMed] [Google Scholar]
- 11.Lu, J. P., T. B. Liu, X. Y. Yu, and F. C. Lin. 2005. Representative appressorium stage cDNA library of Magnaporthe grisea. J. Zhejiang Univ. Sci. B 6:132-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuura, A., M. Tsukada, Y. Wada, and Y. Ohsumi. 1997. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene 192:245-250. [DOI] [PubMed] [Google Scholar]
- 13.Melendez, A., Z. Talloczy, M. Seaman, E. L. Eskelinen, D. H. Hall, and B. Levine. 2003. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 301:1387-1391. [DOI] [PubMed] [Google Scholar]
- 14.Nair, U., and D. J. Klionsky. 2005. Molecular mechanisms and regulation of specific and nonspecific autophagy pathways in yeast. J. Biol. Chem. 280:41785-41788. [DOI] [PubMed] [Google Scholar]
- 15.Ou, S. H. 1985. Rice diseases, 2nd ed. Commonwealth Mycological Institute, Kew, United Kingdom.
- 16.Pall, M. C., and P. Brunelli. 1993. A series of six compact fungal transformation vectors containing polylinkers with multiple unique restriction sites. Fungal Genet. Newsl. 40:59-62. [Google Scholar]
- 17.Pinan-Lucarre, B., A. Balguerie, and C. Clave. 2005. Accelerated cell death in Podospora autophagy mutants. Eukaryot. Cell 4:1765-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reggiori, F., and D. J. Klionsky. 2002. Autophagy in the eukaryotic cell. Eukaryot. Cell 1:11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook, J., E. F. Fritsch, and T. Maniatis. 2002. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, New York, N.Y.
- 20.Straub, M., M. Bredschneider, and M. Thumm. 1997. AUT3, a serine/threonine kinase gene, is essential for autophagocytosis in Saccharomyces cerevisiae. J. Bacteriol. 179:3875-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Talbot, N. J. 2003. On the trail of a cereal killer: exploring the biology of Magnaporthe grisea. Annu. Rev. Microbiol. 57:177-202. [DOI] [PubMed] [Google Scholar]
- 22.Talbot, N. J., D. J. Ebbole, and J. E. Hamer. 1993. Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell 5:1575-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Talbot, N. J., H. R. K. McCafferty, M. Ma, K. Moore, and J. E. Hamer. 1997. Nitrogen starvation of the rice blast fungus Magnaporthe grisea may act as an environmental cue for disease symptom expression. Physiol. Mol. Plant Pathol. 50:179-195. [Google Scholar]
- 24.Thines, E., R. W. Weber, and N. J. Talbot. 2000. MAP kinase and protein kinase A-dependent mobilization of triacylglycerol and glycogen during appressorium turgor generation by Magnaporthe grisea. Plant Cell 12:1703-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsukada, M., and Y. Ohsumi. 1993. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333:169-174. [DOI] [PubMed] [Google Scholar]
- 26.Tucker, S. L., and N. J. Talbot. 2001. Surface attachment and pre-penetration stage development by plant pathogenic fungi. Annu. Rev. Phytopathol. 39:385-417. [DOI] [PubMed] [Google Scholar]
- 27.Valent, B. 1990. Rice blast as a model system for plant pathology. Phytopathology 80:33-36. [Google Scholar]
- 28.Vellai, T., K. Takacs-Vellai, Y. Zhang, A. L. Kovacs, L. Orosz, and F. Muller. 2003. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature 426:620. [DOI] [PubMed] [Google Scholar]
- 29.Veneault-Fourrey, C., M. Barooah, M. Egan, G. Wakley, and N. J. Talbot. 2006. Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science 312:580-583. [DOI] [PubMed] [Google Scholar]
- 30.Xiao, J. Z., T. Watanabe, T. Kamakura, A. Ohshima, and I. Yamaguchi. 1994. Studies on cellular differentiation of Magnaporthe grisea. Physicochemical aspects of substratum surfaces in relation to appressorium formation. Physiol. Mol. Plant Pathol. 44:227-236. [Google Scholar]
- 31.Yorimitsu, T., and D. J. Klionsky. 2005. Autophagy: molecular machinery for self-eating. Cell Death Differ. 12(Suppl. 2):1542-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]