Abstract
G protein-gated inwardly rectifying K+ (GIRK) channels, which are important regulators of membrane excitability both in heart and brain, appear to function as heteromultimers. GIRK1 is unique in the GIRK channel family in that although it is by itself inactive, it can associate with the other family members (GIRK2–GIRK5) to enhance their activity and alter their single-channel characteristics. By generating a series of chimeras, we identified a phenylalanine residue, F137, in the pore region of GIRK1 that critically controls channel activity. F137 is found only in GIRK1, while the remaining GIRK channels possess a conserved serine residue in the analogous position. The single-point mutant GIRK4(S143F) behaved as a GIRK1 analog, forming multimers with GIRK2, GIRK4, or GIRK5 channels that exhibited prolonged single-channel open-time duration and enhanced activity compared with that of homomultimers. Expression of the corresponding GIRK1(F137S) mutant alone resulted in appreciable channel activity with novel characteristics that was further enhanced upon coexpression with other GIRK subunits. Thus, although the F137 residue renders the GIRK1 subunit inactive, when combined with other GIRK heteromeric partners it alters their gating and contributes to their enhanced activity.
Keywords: heteromeric subunits, channel gating, P-region, F137 residue of G protein-gated inwardly rectifying K+ 1 subunit
G protein-gated inwardly rectifying K+ (GIRK) channels provide cardiac, neuronal, and endocrine cells with a rapid mechanism of inhibiting membrane excitability in response to extracellular signals. The best studied case, the atrial KACh channel, slows the heart rate in response to acetylcholine (ACh) released by the vagus nerve. ACh binds muscarinic type 2 (m2) receptors coupled to pertussis toxin-sensitive heterotrimeric G proteins, allowing the Gβγ subunits to activate KACh directly (1). Five homologous members have been identified in this ion channel subfamily (GIRK1–GIRK5) (2, 3, 4, 5, 6). Heterologous expression of any single GIRK channel has failed to produce channel activity characteristic of any known native conductance (e.g., ref. 4). While expression of GIRK1 alone does not give rise to functional homomeric channels (e.g., refs. 6 and 7), expression of each of the remaining GIRK channels has resulted mostly in functional channels but with low activity and poorly resolved openings (4, 6, 7, 8). Results from coimmunoprecipitation (4, 7), immunocytochemical (9), in situ hybridization (10, 11, 12), and functional coexpression (4, 6, 7, 8, 13, 14, 15) studies have demonstrated the heteromeric nature of GIRK channels. GIRK1 is unique among GIRK family members in that although by itself it is inactive, when coexpressed with the other GIRK channels, it greatly enhances their activity and alters their single-channel properties (4, 6, 7, 8, 13, 14, 15). The objective of our study was to identify GIRK1 structural determinants involved in the dramatic changes in activity obtained upon coexpression of GIRK1 with its heteromeric partners.
MATERIALS AND METHODS
Construction of Chimeric and Point Mutant Channels.
All cDNA constructs were produced using the “splicing by overlapping extension” method (17). Human homologs of GIRK1 and GIRK4 (7) were each subcloned in pGEMHE (16) and were used as templates for making all the chimeras listed in Fig. 1a. PCR using Vent DNA polymerase were performed for 20 cycles to reduce errors introduced during amplification. PCR products were always gel purified before used in subsequent steps as templates for PCR. Whenever possible, T7 (or SP6 or both) were used as flanking primers. Therefore, each linear full-length chimeric construct was equipped with a promoter site upstream and a poly(A) sequence downstream (amplified from the pGEMHE vector) and was used directly as a template for in vitro transcription (Ambion, Austin, TX). Each construct was subsequently subcloned back into pGEMHE and four positive clones were selected randomly and sequenced (18) (Amersham) to confirm the region near the chimeric junction of each construct. The point mutants shown in Fig. 1d, were similarly produced with the exception that the template used was the linear full-length G41–186G1181–501 chimeric DNA. GIRK1(F137S) and GIRK4(S143F) were similarly constructed and the mutations were confirmed by sequencing. Plasmid DNA was produced, linearized with NheI and transcribed in vitro using the “message machine” kit (Ambion). cRNAs were electrophoresed on formaldehyde gels and concentrations were estimated from two dilutions using RNA marker (GIBCO) as a standard.
Figure 1.
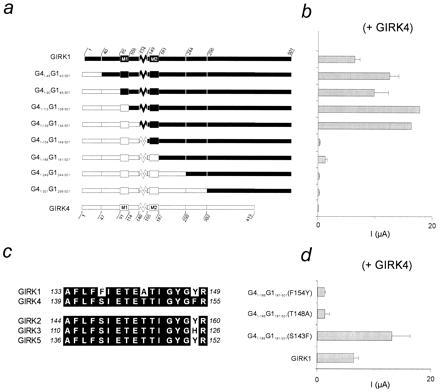
Localization of F137 in GIRK1, a critical determinant of activity of GIRK1/GIRK4 heteromultimers. (a) Schematic diagram of wild-type GIRK1 (Upper) and GIRK4 (Lower) channels and chimeric constructs between them. The amino acid positions shown for the wild-type channels denote positions where chimeric junctions were generated and are shown as subscript numbers preceded by G1 (for GIRK1) or G4 (for GIRK4). (b) Bar graph of Ba2+-sensitive currents at −80 mV from oocytes coinjected with the corresponding channels or constructs shown in part a with GIRK4. Values represent overall averages (±SEM) from different oocyte batches (N) (ranging in number from two to five per species shown, except GIRK1/GIRK4: N = 28, GIRK4: N = 15, G41–113G1108–501: N = 1, and G41–139G1134–501: N = 1). Each oocyte batch was represented by the average value of several oocytes (n ≥ 3). Four nanograms of cRNA per channel species was injected in each oocyte. (c) Alignment of the GIRK1 and GIRK4 P-region amino acid sequences reveals three amino acid differences. Comparison with other GIRK channels shows that two of these three differences are unique to GIRK1. (d) Bar graph of Ba2+-sensitive currents at −80 mV from oocytes coinjected with point mutants of chimera G41–186G1181–501 and GIRK4. Values represent overall averages (±SEM) from different oocyte batches (N = 2 per chimeric species shown).
Expression in Xenopus Oocytes and Electrophysiological Studies.
Oocytes were isolated and microinjected as described (7). Channel expression was assessed by electrophysiological means two to three days following cRNA injection. Two-electrode voltage clamp on Xenopus oocytes was performed as described (7, 19). Briefly, a Gene Clamp 500 amplifier (Axon Instruments, Foster City, CA) was used and microelectrodes were filled with 3 M KCl and had resistances of 0.5–1.0 MΩ. High potassium (ND96K) bath solution was composed of 91 mM KCl, 1 mM NaCl, 1 mM MgCl2, 5 mM Hepes·KOH, and 1.8 mM CaCl2 (pH 7.4). BaCl2 (3 mM) added to ND96K solutions was used to block the GIRK current and the Ba2+-sensitive currents are shown. Niflumic acid (300 μM) was routinely added to the bath solutions to block chloride currents (Sigma). ACh (5 μM) was used when indicated. All experiments were performed at 20–25°C. About 2–4 ng of each channel subunit RNA and 1.5 ng of human m2 (hm2) receptor were injected per oocyte.
Macroscopic currents were averaged for each batch of oocytes (at least n = 3 experiments per batch). Several batches were tested and the mean number (N) ± SEM between batches is indicated.
Single-channel activity was recorded on devitellinized oocytes under the cell-attached mode of standard patch-clamp methods (21, 22) using an Axopatch 200A amplifier (Axon Instruments, CA). The pipette solution (96 mM KCl/1.8 mM CaCl2/1 mM MgCl2/10 mM Hepes, pH 7.4) was similar to that in the bath (96 mM KCl/5 mM EGTA/1 mM MgCl2/10 mM Hepes, pH 7.4). Gadolinium (100 μM) was also included in the pipette solution. Most of the single-channel recordings were performed at a holding membrane potential of −80 mV. Single-channel currents were sampled at 5–10 kHz and filtered at 1–2 kHz. The digidata 1200 interface and pclamp (version 6.01) software were used for data acquisition and analysis (Axon Instruments). Mean open time (To) represents average values for minute-long experiments, estimated using our own software (23). Single-channel properties, such as conductance and open-time duration showed no difference among batches. For single-channel experiments the number (n) of oocytes tested across batches is given.
RESULTS
F137 of GIRK1 Is a Critical Determinant of Heteromeric Channel Activity with GIRK4.
Fig. 1a shows the strategy used to localize critical GIRK1 determinants of heteromeric activity. We used human homologs of GIRK1 (99% identical to the rat clone) and GIRK4 (or KGP, 94% identical to the rat clone also known as CIR) (7), which constitute the heteromeric subunits of KACh (4). Chimeras were constructed in which progressively longer N-terminal segments of GIRK1 (G1) were replaced with the corresponding portion of GIRK4 (G4). Each chimera was coexpressed in Xenopus oocytes with the GIRK4 subunit and tested for its ability to produce the large basal whole-cell currents typical of GIRK1/GIRK4 heteromers (Fig. 1b). These data indicated that replacement of the pore region (P-region) of GIRK1 (G1134–148) with the corresponding GIRK4 sequence decreased the macroscopic current amplitudes. Comparison of the respective P-regions of the two subunits revealed three amino acid differences (Fig. 1b). Individual substitutions of each of the three GIRK4 residues with the corresponding ones from GIRK1 were carried out in the context of chimera G41–186G1181–501. Each of these substituted chimeras were then coexpressed with GIRK4 (Fig. 1d). Only the G41–186G1181–501(S143F) mutation recovered the large amplitude currents, indicating that amino acid F137 in the pore of GIRK1 was critically involved in the enhancement of currents seen in the GIRK1/GIRK4 heteromers.
GIRK4(S143F) Behaves as a GIRK1 Analog.
Is F137 of GIRK1 sufficient to alter heteromeric channel activity? Could a phenylalanine residue at the analogous position of GIRK channels other than GIRK1 endow them with the ability to regulate heteromeric channel activity? To address such questions, we introduced the phenylalanine residue in GIRK4 (at position S143 that corresponds to F137 of GIRK1). Expression of either GIRK4(S143F) or GIRK4 wild type alone did not produce appreciable basal or agonist-induced currents (Fig. 2c). In contrast, coexpression of GIRK4(S143F)/GIRK4 yielded greatly enhanced currents, when compared with expression of either channel alone (Fig. 2c), but that were smaller than those of heteromeric GIRK1/GIRK4 channels (Fig. 2 a–c). When GIRK1 was coexpressed with GIRK4(S143F), no current enhancement was obtained. Basal and agonist induced current-voltage relationships for the coexpressed GIRK4(S143F)/GIRK4 channels showed that these channels remained inwardly rectifying and were able to couple to a G protein-linked receptor (Fig. 2d). Pertussis toxin sensitivity and stimulation by Gβγ subunits was similar for GIRK4(S143F)/GIRK4 and GIRK1/GIRK4 (data not shown) and the channel retained its high selectivity for K+ over Na+ (Fig. 2e). Single-channel recording revealed that the GIRK4(S143F)/GIRK4 channel characteristics were similar to those of GIRK1/GIRK4 heteromers both in terms of conductance (γ = 28.1 ± 0.7, n = 6 vs. γ = 34.6 ± 0.8 pS, n = 10, respectively) and To (To = 1.44 ± 0.10 ms, n = 5 and To = 2.63 ± 0.26 ms, n = 12, respectively) (Fig. 3 a and b). These characteristics were clearly different from those of the poorly resolved GIRK4 channels expressed alone (γ = 15–30 pS; To = 0.31 ± 0.02 ms, n = 7) (6) (Fig. 3b). These results provided strong evidence that the introduced phenylalanine in GIRK4(S143F) was sufficient to turn GIRK4 into a GIRK1 analog, producing “KACh-like” properties upon coexpression with the wild-type GIRK4. Macroscopic GIRK4(S143F)/GIRK4 currents showed fast-activating kinetics (Fig. 2b) rather than the characteristic slow activation seen with GIRK1/GIRK4 heteromers (Fig. 2a). These results suggest that F137 of GIRK1, previously reported to be a critical component (20), may not be the sole GIRK1 determinant of the slow macroscopic activation kinetics.
Figure 2.
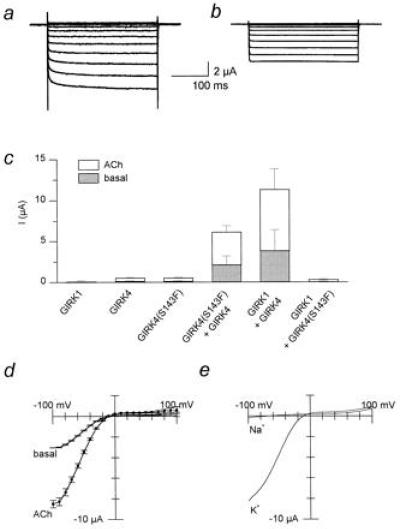
Inclusion of the phenylalanine P-region residue in GIRK4 multimers produces “GIRK1-like” enhancing activity. (a) Coexpression of GIRK1 and GIRK4. (b) GIRK4(S143F) and GIRK4 cRNAs were coinjected. Representative Ba2+-sensitive current traces in the presence of 5 μM ACh are shown for a–c. (c) Current magnitude comparison for GIRK1 (N = 3), GIRK4 (N = 4), GIRK4(S143F) (N = 4), and GIRK4(S143F)/GIRK4 (N = 5), GIRK1/GIRK4 (N = 9), and GIRK4(S143F)/GIRK1 (N = 4). (d) Current-voltage relationships of Ba2+-sensitive GIRK4(S143F)/GIRK4 currents in the absence (basal) or presence of 5 μM ACh from one representative batch of oocytes (n = 5). (e) Representative macroscopic currents (n = 3 oocytes/condition) recorded from oocytes injected with hm2 as well as GIRK4 and GIRK4(S143F) cRNAs in 96 mM KCl in the bath (K) versus 96 mM NaCl (Na) (i.e., KCl-free), in the presence of 5 μM ACh. Currents were obtained by employing a voltage ramp protocol (−100 mV to +100 mV from a holding potential of 0 mV).
Figure 3.
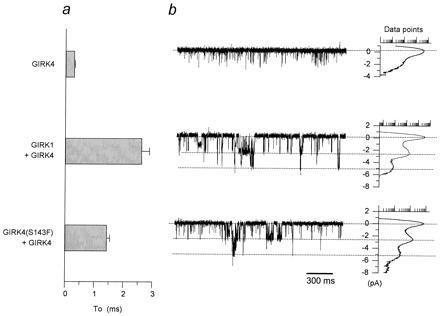
Inclusion of the phenylalanine P-region residue in GIRK4 multimers produces “GIRK1/GIRK4-like” single-channel activity. (a) “To” comparisons (calculated from oocytes from several batches) among GIRK4 (n = 7 patches), GIRK1/GIRK4 (n = 12) and GIRK4(S143F)/GIRK4 (n = 5). (b) Single-channel currents from representative cell-attached patches from Xenopus oocytes expressing GIRK4 (n ≥ 100 patches), GIRK1/GIRK4 (n ≥ 100), and GIRK4(S143F)/GIRK4 (n = 10). The membrane was held at −80 mV. An all-point histogram plot indicates the amplitudes resulting from the various activity levels ranging from closed to multiple open levels. Data points in this and other figures are shown in a logarithmic scale ranging from 1–10,000 points.
The Phenylalanine Residue in GIRK4(S143F) Alters Gating and Enhances Other Heteromeric GIRK Channel Currents As Well.
Can GIRK4(S143F) substitute for GIRK1 in the phenotypic changes seen upon coexpression of GIRK1 with other GIRK channel subunits as well? We addressed this question by coexpressing the GIRK4(S143F) or GIRK4 subunits with GIRK5 or GIRK2. Fig. 4 shows both basal and agonist-induced macroscopic current amplitudes (Fig. 4a) and representative single-channel records of basal activity along with To and amplitude histograms (Fig. 4 b and c) of the four possible combinations. Coexpression of either GIRK5 or GIRK2 with GIRK4(S143F) gave longer-lived openings (Fig. 4 b and c) and greater currents (Fig. 4a) than when coexpressed with GIRK4. The phenylalanine residue of GIRK4(S143F) was capable of altering GIRK5 and GIRK2 channel activity and single-channel characteristics, in a manner similar to that of GIRK1.
Figure 4.
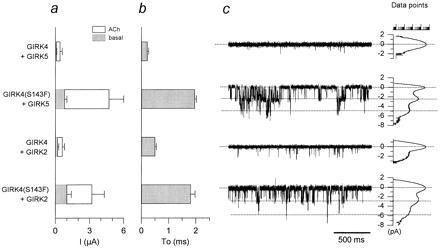
The phenylalanine residue in GIRK4(S143F) alters gating kinetics and enhances activity of heteromers with other GIRK channels. (a) Ba2+-sensitive current magnitude comparisons are shown among coinjections of GIRK5 or GIRK2 with GIRK4(S143F) or GIRK4: GIRK4/GIRK5 (N = 3); GIRK4(S143F)/GIRK5 (N = 3); GIRK4/GIRK2 (N = 4); GIRK4(S143F)/GIRK2 (N = 4). (b) “To” comparison (as in Fig. 3) from activity resulting from coinjection of GIRK5 or GIRK2 with GIRK4 or GIRK4(S143F): GIRK4/GIRK5 (n = 3 patches); GIRK4(S143F)/GIRK5 (n = 10); GIRK4/GIRK2 (n = 3); GIRK4(S143F)/GIRK2 (n = 10). (c) Single-channel currents from representative cell-attached patches from Xenopus oocytes coexpressing GIRK4/GIRK5, GIRK4(S143F)/GIRK5, GIRK4/GIRK2, GIRK4(S143F)/GIRK2, and associated all-point histogram plots are shown. The membrane was held at −80 mV. hm2 receptor (1.5 ng/oocyte) was coinjected with 2 ng/oocyte each of the channel cRNAs tested.
F137S Reveals Novel GIRK1 Channel Activity.
Since F137 of GIRK1 was a sufficient determinant, we proceeded to test whether it was also a necessary one to alter heteromeric GIRK activity. Thus, we constructed the F137S mutant in GIRK1 and tested at first the effects of a serine residue in GIRK1 functional expression. In sharp contrast to its wild-type counterpart, GIRK1(F137S) displayed large inwardly rectifying currents (at −80 mV, Fig. 5a2, compared with +80 mV, Fig. 5a1), particularly when coexpressed with hm2 receptor and induced with ACh (Fig. 5c). GIRK1(F137S) currents in oocytes showed differences in the macroscopic kinetics of activation (Fig. 5a) from those of wild-type GIRK1 (presumably forming heteromers with the endogenous Xenopus inward rectifier, XIR or GIRK5), consistent with a previous report (ref. 20 and Fig. 2a). The single-channel characteristics of GIRK1(F137S) (Fig. 5b) were distinct from those obtained from coexpression of GIRK1 with other GIRK subunits, both in terms of conductance (γ = 15.4 ± 0.9 pS, n = 6) and kinetics, with channel openings organized in clear bursts of activity separated by long closed intervals. Intraburst openings were either very long (tens to hundreds of milliseconds, Fig. 5b1) or quite short (To(sh) = 0.43 ± 0.06 ms, n = 4; Fig. 5b2). The unique characteristics of GIRK1(F137S) could not be due to heteromultimerization with endogenous XIR, since coexpression of GIRK1(F137S) with exogenous XIR yielded channels with unitary conductance characteristics distinct from those of GIRK1(F137S) alone and kinetics distinct from those of XIR alone (data not shown). These results indicated that a single-point mutation GIRK1(F137S) allowed GIRK1 activity, revealing unique properties. Thus, the presence of F137 is a contributing factor to the functional impairment of GIRK1. Moreover, the presence of phenylalanine residues at this position renders even the heteromeric channel inactive, as was the case with GIRK1/GIRK4(S143F) coexpression (see Fig. 2c).
Figure 5.
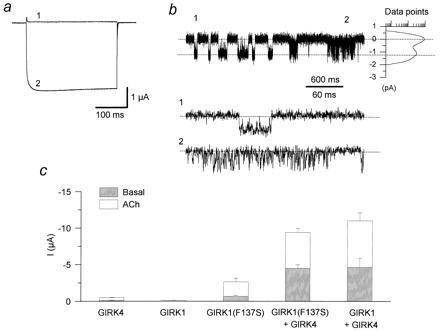
The F137S mutation reveals novel GIRK1 currents and implicates additional GIRK1 determinants for activity. hm2 (1.5 ng/oocyte) was coinjected with 2 ng each of GIRK4, GIRK1, and GIRK1(F137S) alone or in combination of GIRK4 with GIRK1(F137S) or GIRK1 cRNAs. Oocyte currents were recorded three or more days following injection of cRNAs using two-microelectrode voltage clamp. Bath solutions contained 96 mM K+ in the bath. Ba2+-sensitive currents were obtained by subtracting currents obtained in the presence from those in the absence of 3 mM BaCl2 (see Materials and Methods). (a) Experiments from oocytes expressing GIRK1(F137S) (N = 10). Ba2+-sensitive current traces at −80 mV (2) and +80 mV (1) after application of 5 μM ACh. (b) Single-channel currents from a cell-attached patch from Xenopus oocytes expressing GIRK1(F137S) (n = 6). (1) shows an example of long openings within a burst, while (2) shows brief openings within a burst. The membrane was held at −80 mV. (c) Ba2+-sensitive current magnitude comparisons (N = 2) among injections of each of GIRK4, GIRK1, and GIRK1(F137S) alone or in combination of GIRK4 with GIRK1(F137S) or GIRK1 cRNAs.
High Activity of GIRK Heteromers Involves Additional GIRK1 Determinants Other Than F137.
We finally tested whether in the absence of F137, GIRK1 was still capable of enhancing the activity of other GIRK partners. We approached this question by coexpressing GIRK4 with GIRK1(F137S), thus eliminating any effects of F137 on gating and current enhancement. Coexpression of GIRK1(F137S) with GIRK4 produced macroscopic heteromeric currents that were greatly enhanced in comparison to the homomeric controls (Fig. 5c). Thus, the current enhancement obtained in these experiments, where only serine residues were present at the specific P-region position, implicated additional GIRK1 determinants for the large current size of the GIRK1/GIRK4 heteromers, other than the F137 pore residue of GIRK1. Coinjection of GIRK1(F137S) with GIRK1 did not cause current enhancement (data not shown) as was the case for heteromers of GIRK1 or GIRK4(S143F) with GIRK2, GIRK4, or GIRK5. This result further emphasized differences between GIRK1 and the other subfamily members.
DISCUSSION
Heteromultimers of distinct but homologous subunits producing novel functional properties have been described for other channels such as voltage-gated K+ channels (24, 25, 26, 27), cyclic nucleotide-gated cation channels (28, 29), the amiloride-sensitive sodium channel (30), and inwardly rectifying K+ channels, which are not G protein sensitive (31). GIRK heteromeric channels use the inactive GIRK1 subunit as their functional partner. We have determined that F137 of GIRK1, a residue previously found to be required for the slow macroscopic current activation kinetics (20), is a critical determinant of multimeric GIRK activity and channel gating. The presence of F137 in GIRK1 does not afford this channel with functional expression. In contrast, the F137S mutation reveals large macroscopic currents with novel single-channel characteristics, suggesting formation of functional homomeric channels. Transplantation of this P-region residue into GIRK4 produced a GIRK1 analog capable of enhancing currents and altering single-channel characteristics of GIRK channels. It is not surprising that some differences between GIRK1 and the GIRK1 analog GIRK4(S143F) exist, most notably the macroscopic activation kinetics and relative current magnitudes, suggesting the involvement of additional GIRK1 structural determinants for such properties. What is rather striking however, is that even though the primary sequence of GIRK1 diverges from that of GIRK4 in many places, particularly in the ≈80-amino acid-longer C-terminal end, a crucial element of the uniqueness of GIRK1 lies in the control exerted by a single residue within the most conserved GIRK region, the P-region. Is F137 the only structural determinant of GIRK1 causing current enhancement of other GIRK currents? Since GIRK1(F137S) is capable of enhancing GIRK4 currents, it is likely that additional determinants, other than F137 of GIRK1 are involved in current enhancement. Yet, regardless of the, presence of such determinants, the exclusive presence of phenylalanine residues at this position, as with GIRK1/GIRK4(S143F) coexpression, prevented current enhancement, underscoring the crucial role of phenylalanine residues at this position in controlling GIRK channel activity.
Identification of pore residues in other K+ channels, neighboring in position to F137 of GIRK1 and showing specific gating effects, have been reported previously. Such examples include position 369 of Kv2.1 (two residues 3′ from the pore position described here) and in Shaker (Kv1.1), W434 (one residue 5′ from the pore position described here as compared to refs. 32 and 33). Intramolecular interactions with residues at these positions (resulting in C-type inactivation) have been suggested (34) but the identity of such interacting residues remains to be elucidated. Whether intramolecular interactions with residues at F137 or S143 of GIRK heteromers are the basis of the gating effects we have described is currently unknown.
We are most appreciative to Drs. David Clapham and Michel Lazdunski for sharing with us the GIRK5 and GIRK2 cDNAs, respectively. We thank Amanda Pabon and Xiaying Wu for technical assistance. We are grateful to Drs. Noelle Langan, Robert Margolskee, Marianna Max, and William B. Thornhill for critical comments on the manuscript. J.L.S. is an Aaron Diamond Fellow. The work was supported by grants from the Aaron Diamond Foundation, National Institutes of Health (HL54185) and the American Heart Association, New York City Affiliate (GIA) and National Center (96011620).
Footnotes
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. §1734 solely to indicate this fact.
Abbreviations: GIRK, G protein-gated inwardly rectifying K+ channel; P-region, pore region; ACh, acetylcholine; To, mean open time; m2, muscarinic type 2; hm2, human m2.
References
- 1.Logothetis D E, Kurachi Y, Galper J, Neer E J, Clapham D E. Nature (London) 1987;325:321–326. doi: 10.1038/325321a0. [DOI] [PubMed] [Google Scholar]
- 2.Kubo Y, Reuveny E, Slesinger P A, Jan Y N, Jan L Y. Nature (London) 1993;364:802–806. doi: 10.1038/364802a0. [DOI] [PubMed] [Google Scholar]
- 3.Dascal N, Schreibmayer W, Lim N F, Wang W, Chavkin C, DiMagno L, Labarca C, Kieffer B L, Gaveriaux-Ruff C, Trollinger D, Lester H A, Davidson N. Proc Natl Acad Sci USA. 1993;90:10235–10239. doi: 10.1073/pnas.90.21.10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krapivinsky G, Gordon E A, Wickman K, Velimirovic B, Krapivinsky L, Clapham D E. Nature (London) 1995;374:135–141. doi: 10.1038/374135a0. [DOI] [PubMed] [Google Scholar]
- 5.Lesage F, Duprat F, Fink M, Guillemare E, Coppola T, Lazdunski M, Hugnot J-P. FEBS Lett. 1994;353:37–42. doi: 10.1016/0014-5793(94)01007-2. [DOI] [PubMed] [Google Scholar]
- 6.Hedin K E, Lim N F, Clapham D E. Neuron. 1996;16:423–429. doi: 10.1016/s0896-6273(00)80060-4. [DOI] [PubMed] [Google Scholar]
- 7.Chan K W, Langan M N, Sui J, Kozak J A, Pabon A, Ladias J A A, Logothetis D E. J Gen Physiol. 1996;107:381–397. doi: 10.1085/jgp.107.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kofuji P, Davidson N, Lester H A. Proc Natl Acad Sci USA. 1995;92:6542–6546. doi: 10.1073/pnas.92.14.6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karschin C, Schreibmayer W, Dascal N, Lester H, Davidson N, Karschin A. FEBS Lett. 1994;348:139–144. doi: 10.1016/0014-5793(94)00590-7. [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi T, Ikeda K, Ichikawa T, Abe S, Togashi S, Kumanishi T. Biochem Biophys Res Commun. 1995;208:1166–1173. doi: 10.1006/bbrc.1995.1456. [DOI] [PubMed] [Google Scholar]
- 11.Ponce A, Bueno E, Vega-Saenz de Miera E, Chow L, Hillman D, Chen S, Wu M B, Wu X, Zhu L, Rudy B, Thornhill W, B. J Neurosci. 1996;16:1990–2001. doi: 10.1523/JNEUROSCI.16-06-01990.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spauschus A, Lentes K-U, Wischmeyer E, Dissmann E, Karschin C, Karschin A. J Neurosci. 1996;16:930–938. doi: 10.1523/JNEUROSCI.16-03-00930.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Velimirovich B M, Gordon E A, Lim N F, Navarro B, Clapham D E. FEBS Lett. 1996;379:31–37. doi: 10.1016/0014-5793(95)01465-9. [DOI] [PubMed] [Google Scholar]
- 14.Lesage F, Guillemare E, Fink M, Duprat F, Heurteaux C, Fosset M, Romey G, Barhanin J, Lazdunski M. J Biol Chem. 1995;270:28660–28667. doi: 10.1074/jbc.270.48.28660. [DOI] [PubMed] [Google Scholar]
- 15.Duprat F, Lesage F, Guillemare E, Fink M, Hugnot J P, Bigay J, Lazdunski M, Romey G, Barhanin J. Biochem Biophys Res Commun. 1995;212:657–663. doi: 10.1006/bbrc.1995.2019. [DOI] [PubMed] [Google Scholar]
- 16.Liman E R, Tytgat J, Hess P. Neuron. 1992;9:861–871. doi: 10.1016/0896-6273(92)90239-a. [DOI] [PubMed] [Google Scholar]
- 17.Horton R M, Hunt H D, Ho S N, Pullen J K, Pease L R. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 18.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stühmer W. Methods Enzymol. 1992;207:319–339. doi: 10.1016/0076-6879(92)07021-f. [DOI] [PubMed] [Google Scholar]
- 20.Kofuji P, Doupnik A, Davidson N, Lester H A. J Physiol (London) 1996;490.3:633–645. doi: 10.1113/jphysiol.1996.sp021173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamill O P, Marty A, Neher E, Sakmann B, Sigworth F J. Pflügers Arch. 1981;391:85–100. doi: 10.1007/BF00656997. [DOI] [PubMed] [Google Scholar]
- 22.Methfessel C, Witzemann V, Takahashi T, Mishina M, Numa S, Sakmann B. Pflügers Arch. 1986;407:577–588. doi: 10.1007/BF00582635. [DOI] [PubMed] [Google Scholar]
- 23.Sui J-L, Chan K W, Logothetis D E. J Gen Physiol. 1996;108:381–392. doi: 10.1085/jgp.108.5.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Isacoff E Y, Jan Y N, Jan L Y. Nature (London) 1990;345:530–534. doi: 10.1038/345530a0. [DOI] [PubMed] [Google Scholar]
- 25.Ruppersberg J P, Schroter K H, Sakmann B, Stocker M, Sewing S, Pongs O. Nature (London) 1990;345:535–537. doi: 10.1038/345535a0. [DOI] [PubMed] [Google Scholar]
- 26.Christie M J, North R A, Osborne P B, Douglass J, Adelman J P. Neuron. 1990;4:405–411. doi: 10.1016/0896-6273(90)90052-h. [DOI] [PubMed] [Google Scholar]
- 27.McCormack K, Lin J W, Iverson L E, Rudy B. Biochem Biophys Res Commun. 1990;171:1361–1371. doi: 10.1016/0006-291x(90)90836-c. [DOI] [PubMed] [Google Scholar]
- 28.Chen T-Y, Peng Y-W, Dhallan R S, Ahamed B, Reed R R, Yau K-W. Nature (London) 1993;362:764–767. doi: 10.1038/362764a0. [DOI] [PubMed] [Google Scholar]
- 29.Bradley J, Li J, Davidson N, Lester H A, Zinn K. Proc Natl Acad Sci USA. 1994;91:8890–8894. doi: 10.1073/pnas.91.19.8890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Canessa C M, Schild L, Buell G, Thorens B, Gautschl I, Horisberger J-D, Rossier B C. Nature (London) 1994;367:463–467. doi: 10.1038/367463a0. [DOI] [PubMed] [Google Scholar]
- 31.Pessia M, Tucker S J, Lee K, Bond C T, Adelman J P. EMBO J. 1996;15:2980–2987. [PMC free article] [PubMed] [Google Scholar]
- 32.Kirsch G E, Drewe J A, Hartmann H A, Tagliatela M, de Biasi M, Brown A M, Joho R H. Neuron. 1992;8:499–505. doi: 10.1016/0896-6273(92)90278-l. [DOI] [PubMed] [Google Scholar]
- 33.Perozo E, MacKinnon R, Bezanilla F, Stefani E. Neuron. 1993;11:353–358. doi: 10.1016/0896-6273(93)90190-3. [DOI] [PubMed] [Google Scholar]
- 34.Yan Y, Yang Y, Sigworth F, J. Biophys J. 1996;70:A190. [Google Scholar]


