Abstract
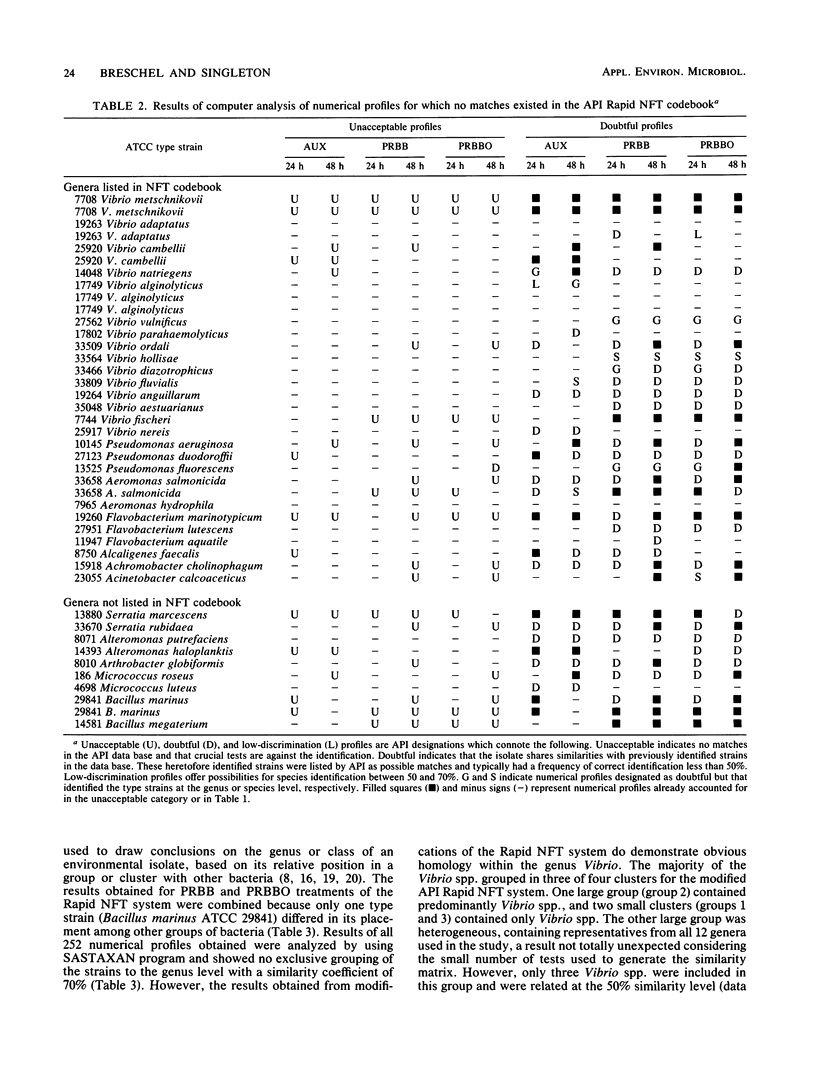
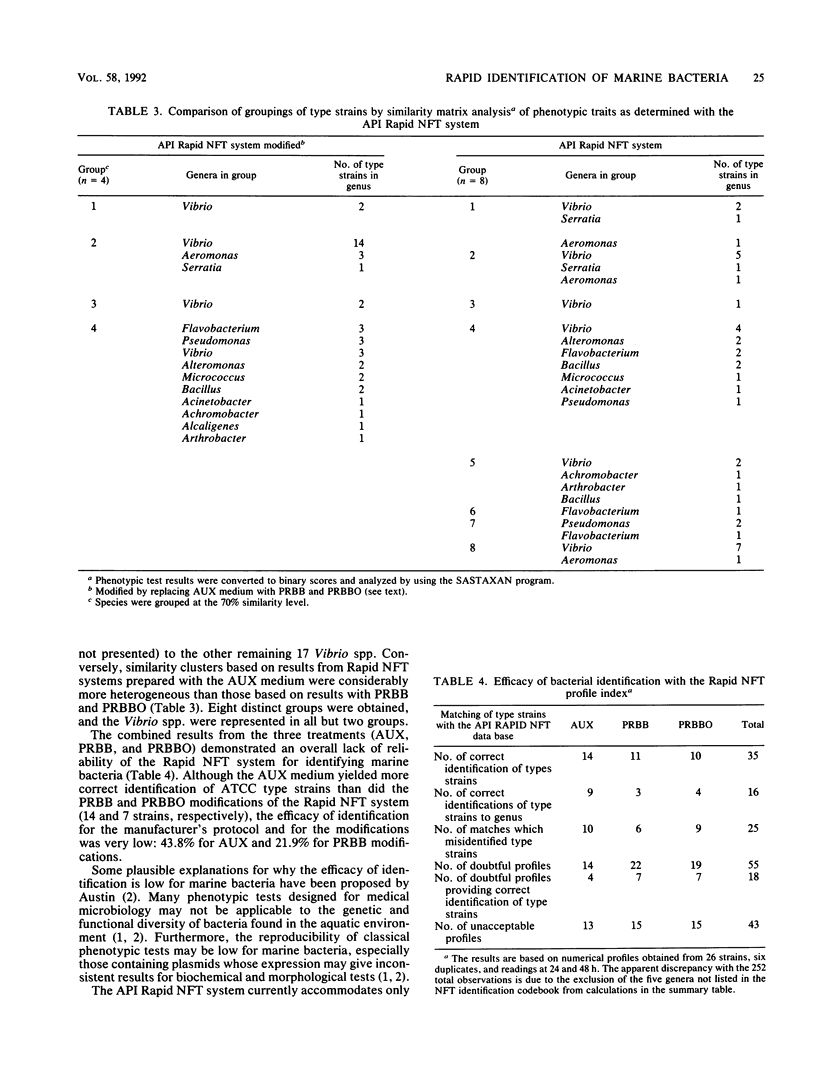
Thirty-five American Type Culture Collection type strains of marine bacteria were used to evaluate the Rapid NFT system (API Analab Products, Plainview, N.Y.) for use in identifying heterotrophic marine bacteria. The 21 biochemical and assimilation tests on the Rapid NFT test strips were treated according to the manufacturer's protocol, which included use of AUX medium (provided with the Rapid NFT system) for preparing assimilation tests, and by substituting phenol red broth base (BBL Microbiology Systems, Cockeysville, Md.) with and without an oil overlay for the AUX medium. A seven-digit numerical profile was obtained for each NFT test strip from each of the three procedures and matched to its corresponding number in the Rapid NFT identification codebook. Also, all biochemical and assimilation test results were analyzed with SASTAXAN and SAS/GRAPH programs (SAS Institute, Inc., Cary, N.C.); similarity matrices were computed for all 35 strains. For comparison purposes, bacterial strains were grouped at a similarity level of 70%. The results indicated a low efficacy of identification for all three procedures. In addition, similarity matrix analysis showed more cohesive grouping based on results of phenol red broth base-treated strains than for the AUX medium provided by the manufacturer. However, none of the three treatments provided exclusive grouping of type strains at the genus level. Thus, the reliability of the data obtained from the NFT system and modifications thereof should be evaluated carefully when environmental isolates are characterized.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bryant T. N., Lee J. V., West P. A., Colwell R. R. Numerical classification of species of Vibrio and related genera. J Appl Bacteriol. 1986 Nov;61(5):437–467. doi: 10.1111/j.1365-2672.1986.tb04308.x. [DOI] [PubMed] [Google Scholar]
- Glenville M., Broughton R., Wing A. M., Wilkinson R. T. Effects of sleep deprivation on short duration performance measures compared to the Wilkinson auditory vigilance task. Sleep. 1978 Winter;1(2):169–176. doi: 10.1093/sleep/1.2.169. [DOI] [PubMed] [Google Scholar]
- Grimes D. J., Singleton F. L., Colwell R. R. Allogenic succession of marine bacterial communities in response to pharmaceutical waste. J Appl Bacteriol. 1984 Oct;57(2):247–261. doi: 10.1111/j.1365-2672.1984.tb01389.x. [DOI] [PubMed] [Google Scholar]
- MacDonell M. T., Singleton F. L., Hood M. A. Diluent composition for use of API 20E in characterizing marine and estuarine bacteria. Appl Environ Microbiol. 1982 Aug;44(2):423–427. doi: 10.1128/aem.44.2.423-427.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R., Siavoshi F., McDougal D. L. Comparison of Rapid NFT system and conventional methods for identification of nonsaccharolytic gram-negative bacteria. J Clin Microbiol. 1986 Dec;24(6):1089–1092. doi: 10.1128/jcm.24.6.1089-1092.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhofer T. R. Comparison of the API 20E and Oxi/Ferm systems in identification of nonfermentative and oxidase-positive fermentative bacteria. J Clin Microbiol. 1979 Feb;9(2):220–226. doi: 10.1128/jcm.9.2.220-226.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto L. A., Pickett M. J. Rapid method for identification of gram-negative, nonfermentative bacilli. J Clin Microbiol. 1976 Jun;3(6):566–575. doi: 10.1128/jcm.3.6.566-575.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overman T. L., Kessler J. F., Seabolt J. P. Comparison of API 20E, API rapid E, and API rapid NFT for identification of members of the family Vibrionaceae. J Clin Microbiol. 1985 Nov;22(5):778–781. doi: 10.1128/jcm.22.5.778-781.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri M. J., Carito S. L., Meyer R. F. Comparison of rapid NFT and API 20E with conventional methods for identification of gram-negative nonfermentative bacilli from pharmaceuticals and cosmetics. Appl Environ Microbiol. 1988 Nov;54(11):2838–2841. doi: 10.1128/aem.54.11.2838-2841.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peele E. R., Singleton F. L., Deming J. W., Cavari B., Colwell R. R. Effects of pharmaceutical wastes on microbial populations in surface waters at the puerto rico dump site in the atlantic ocean. Appl Environ Microbiol. 1981 Apr;41(4):873–879. doi: 10.1128/aem.41.4.873-879.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santavy D. L., Willenz P., Colwell R. R. Phenotypic study of bacteria associated with the caribbean sclerosponge, Ceratoporella nicholsoni. Appl Environ Microbiol. 1990 Jun;56(6):1750–1762. doi: 10.1128/aem.56.6.1750-1762.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Søgaard P., Gahrn-Hansen B., Zhou H. P., Frederiksen W. An investigation of three commercial methods for rapid identification of non-enteric gram-negative rods. Application on Pseudomonas paucimobilis and some other Pseudomonas species. Acta Pathol Microbiol Immunol Scand B. 1986 Oct;94(5):357–363. doi: 10.1111/j.1699-0463.1986.tb03067.x. [DOI] [PubMed] [Google Scholar]



