Abstract
We expressed a cDNA encoding the Arabidopsis thaliana defense-related protein ELI3-2 in Escherichia coli to determine its biochemical function. Based on a protein database search, this protein was recently predicted to be a mannitol dehydrogenase [Williamson, J. D., Stoop, J. M. H., Massel, M. O., Conkling, M. A. & Pharr, D. M. (1995) Proc. Natl. Acad. Sci. USA 92, 7148–7152]. Studies on the substrate specificity now revealed that ELI3-2 is an aromatic alcohol: NADP+ oxidoreductase (benzyl alcohol dehydrogenase). The enzyme showed a strong preference for various aromatic aldehydes as opposed to the corresponding alcohols. Highest substrate affinities were observed for 2-methoxybenzaldehyde, 3-methoxybenzaldehyde, salicylaldehyde, and benzaldehyde, in this order, whereas mannitol dehydrogenase activity could not be detected. These and previous results support the notion that ELI3-2 has an important role in resistance-related aromatic acid-derived metabolism.
Keywords: benzyl alcohol dehydrogenase, disease resistance, fungal elicitor
Plant defense toward potential pathogens encompasses a wide array of mechanisms, some leading to the rapid reinforcement of preexisting structural barriers, others to the de novo synthesis of a large diversity of defense-related compounds via transcriptional activation of the corresponding genes. In recent years, numerous plant genes potentially involved in the pathogen defense response have been isolated. However, many of them were detected by various differential screening approaches solely on the basis of enhanced expression levels without knowledge of the biochemical functions of the encoded proteins (1, 2, 3). In several cases, functional identification was subsequently achieved, for example, by expression in Escherichia coli or yeast, by the use of specific antibodies or by genetic complementation studies. Often, however, inference of function was merely based on deduced amino acid sequence similarity to known proteins, a valuable but not unequivocal means of identification.
The eli3 gene was originally identified as part of the defense response in parsley [Petroselinum crispum (Pc)] (4). Expression of this gene was shown to be rapidly and transiently stimulated in cultured parsley cells upon treatment with a cell wall preparation (elicitor) from the phytopathogenic fungus Phytophthora sojae, and histochemical studies revealed local and rapid accumulation of ELI3 mRNA around fungal infection sites in parsley leaves (5). The presence of this gene in other plant species, such as potato (Solanum tuberosum), alfalfa (Medicago sativa), and Arabidopsis thaliana (At) was also demonstrated (6). Subsequently, two sequence-related counterparts of eli3 in parsley were isolated from A. thaliana (Ateli3-1 and Ateli3-2), and their expression was shown to be induced by fungal elicitor in cultured A. thaliana cells (6). Additional evidence for an important role of the eli3 gene product in plant disease resistance came from genetic studies demonstrating that eli3 expression was dependent on the presence of the RPMI resistance gene in A. thaliana (7). The RPM1 locus confers resistance to Pseudomonas syringae strains carrying the corresponding avirulence (avr) gene avrRpm1 (8).
The ELI3 cDNAs from parsley and A. thaliana share 67% nucleotide and 70% deduced amino acid sequence identity (7). At the time of their isolation, no related sequences were found in the various data bases. Meanwhile, several plant cinnamyl-alcohol dehydrogenases (CAD) with similarity to the deduced ELI3 proteins have been reported (9). Based on this similarity, it was possible that the eli3 gene encodes a CAD. Recently, however, Williamson et al. (10) suggested that ELI3 is a mannitol dehydrogenase (MTD). Again, this was based on sequence similarity to an MTD from celery [Apium graveolens (Ag), AgMTD]. This proposal prompted speculation as to the functional relevance of eli3 gene expression (10), particularly in an accompanying commentary (11), which, in view of our present data, is no longer tenable. We now provide evidence that ELI3-2 from A. thaliana is a novel type of aromatic (preferably benzyl) alcohol dehydrogenase with substrate specificity distinct from both CAD and MTD.
MATERIALS AND METHODS
Materials.
Actinobacillus mannitol dehydrogenase was purchased from Sigma; NAD+, NADH, NADP+, and NADPH were from Boehringer Mannheim. The following aldehydes and alcohols were obtained from Aldrich: cinnamaldehyde, cinnamyl alcohol, sinapaldehyde, coniferaldehyde, coniferyl alcohol, 3,5-dimethoxy-4-hydroxybenzaldehyde, 2-methoxybenzyl alcohol, 3-methoxybenzyl alcohol, salicylaldehyde β-O-glucoside, and 4-coumaryl alcohol. Acetaldehyde, d-mannitol, benzaldehyde, benzyl alcohol, salicylaldehyde, 2-hydroxybenzyl alcohol, 2-methoxybenzaldehyde, 3-methoxybenzaldehyde, 4-methoxybenzaldehyde, 4-hydroxybenzaldehyde, 3,4-dimethoxybenzaldehyde, and vanillin were from Merck, and capronaldehyde was from Fluka. 4-Coumaraldehyde was a kind gift from J. Grima-Pettenati (Toulouse, France) and W. Heller (Neuherberg, Germany). All substrates were dissolved in methoxyethanol.
Expression in E. coli.
One of two established, closely related A. thaliana ELI3 cDNAs, AtELI3-2, containing the entire coding region (7), was cloned into the expression vector pQE50 (Qiagen, Düsseldorf, Germany), and the resulting construct, pQE50-ELI3-2, was introduced into the E. coli host strain SURE. The transformed bacterial cells were grown at 37°C in Luria–Bertani medium containing 1 mM ZnCl2 and 50 μg/ml ampicillin until they reached an OD600 of 0.8–1.0. The cells were then cooled to 15°C, and 1 mM isopropyl β-d-thiogalactoside (IPTG) was added to induce ELI3-2 protein synthesis. The bacteria were grown for another 16 h and then harvested by centrifugation. As controls, E. coli SURE cells containing the pQE50 expression vector and E. coli BL21 cells expressing a CAD cDNA (EuCAD2) from Eucalyptus gunnii (Eu; ref. 12) were treated and processed in the same manner. Preparation of crude extracts from E. coli transformants was performed according to Lauvergeat et al. (13). Briefly, the harvested cells were suspended in lysis buffer (20 mM Tris·HCl, pH 7.5/10% glycerol/5 mM DTT/0.1% Nonidet P-40/1 mM phenylmethylsulfonyl fluoride/1 mM EDTA/5 μg/ml leupeptin). Lysozyme was added to a final concentration of 2 mg/ml, and the cells were incubated at 4°C until lysis occurred. Nuclease (Benzonase, Eurogentec, Brussels, Belgium) was added to a final concentration of 50 units/ml, and the mixture was incubated at 4°C for about 15 min. Cell debris was pelleted by centrifugation, and the supernatant was tested for enzyme activity. For the analysis of total bacterial proteins, aliquots were pelleted in a microcentrifuge, boiled in SDS-lysis buffer [0.1 M Tris·HCl, pH 6.8/1.6% (vol/vol) glycerol/0.008% bromphenol blue/4 mM EDTA/10 mM DTT/3% (wt/vol) SDS] for 3 min at 95°C and loaded onto a 12% SDS-polyacrylamide gel. Proteins were visualized by Coomassie blue staining. Protein concentrations were determined spectrophotometrically by the Bradford assay (14).
Assay for MTD Activity.
MTD activity was measured by monitoring the reduction of NAD+ spectrophotometrically at 340 nm according to Stoop et al. (15). The assay mixture contained 100 mM 1,3-bis[tris(hydroxymethyl)methylamino]propane, pH 9.0, 2 mM NAD+, 150 mM d-mannitol, and pQE50-ELI3 bacterial extract in a total volume of 1 ml.
Assay for Aromatic Alcohol Dehydrogenase Activity.
Recombinant bacterial extracts were assayed spectrophotometrically for aromatic alcohol dehydrogenase activity by both the oxidation of the aromatic alcohols and the reduction of the corresponding aldehydes, as described for CAD activity (16). The assay was carried out at 30°C in 1 ml of reaction mixture containing, for the aldehyde substrates, 200 μM KH2PO4/Na2HPO4, pH 6.5, 34 μM aldehyde substrate, 200 μM NADPH or NADH, and 1–60 μl of protein extract, and for the alcohol substrates, 100 mM Tris·HCl (pH 8.8), 100 μM alcohol substrate, 200 μM NADP+ or NAD+, and 1–100 μl of protein extract. The molar extinction coefficients (ɛ340) used were 18.5 × 103 M−1 cm−1 at pH 8.8 for coniferylaldehyde, 15.8 × 103 M−1 cm−1 at pH 6.5 for sinapaldehyde, 23.5 × 103 M−1 cm−1 at pH 6.5 for 4-coumaraldehyde, and 6.3 × 103 M−1 cm−1 at pH 6.5 for NADH and NADPH. For detailed kinetic analyses, only the aldehyde substrates were used, because oxidation of the respective alcohol substrates was 50–100 times slower.
RESULTS
Expression of AtELI3-2 in E. coli.
IPTG-induced expression of the AtELI3-2 cDNA in E. coli at various temperatures ranging from 25 to 37°C resulted in the exclusive accumulation of ELI3-2 protein in inclusion bodies, as analyzed by SDS/PAGE. This problem was partly overcome by reducing the temperature to 15°C before the addition of IPTG, as described by Lauvergeat et al. (13). An appreciable amount of the 45-kDa ELI3-2 protein remained under these conditions in the soluble fraction. This fraction was used for all subsequent experiments, in parallel with identically treated control extracts derived from bacteria containing either the pQE50 vector or expressing the EuCAD2 cDNA.
Assay for MTD Activity.
Considering the recently reported sequence similarity of AgMTD with AtELI3 and PcELI3 (10), we first tested whether the recombinant AtELI3-2 protein showed MTD activity. Fig. 1 illustrates that no NAD+-dependent MTD activity was observed with the ELI3-2-containing bacterial extracts. This applies to various substrate concentrations analyzed. Use of 0.2 units of a commercially available MTD demonstrated the functionality of the assay. Furthermore, addition of authentic MTD to the crude AtELI3-2 preparation confirmed the NAD+ dependency of the reaction and excluded the presence of inhibitory factors in the enzyme assay. In all cases, NADP+ could not substitute for NAD+ as cofactor (data not shown).
Figure 1.
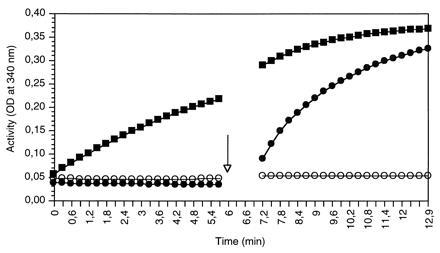
Spectrophotometric assay for MTD activity in protein extracts from ELI3-2-expressing E. coli cells. Crude extract (242 μg of protein) was incubated in MTD assay buffer with (circles) or without (open circles) the cofactor NAD+. The arrow indicates the time point of addition of 0.2 units of authentic Actinobacillus MTD (solid squares).
Assay for CAD Activity.
Sequence similarity of ELI3 to various plant CADs has also been observed (9). To address this point, the AtELI3-2 preparation was tested for CAD activity using various cinnamyl aldehydes (bottom line in Fig. 2) and alcohols. As illustrated in Fig. 3A, low but measurable activity was detected for cinnamaldehyde, 4-coumaraldehyde, coniferaldehyde, and sinapaldehyde. However, large amounts (60 μg) of protein extract were required to detect this activity. Therefore, additional aldehydes (Fig. 2) were tested as possible substrates. These assays revealed that several benzaldehyde derivatives, including 2-methoxybenzaldehyde, 3-methoxybenzaldehyde, salicylaldehyde, and benzaldehyde, were used more efficiently than the corresponding cinnamaldehyde derivatives. Fig. 3B depicts the activities observed with these substrates using only 5 μg of the bacterial protein extract.
Figure 2.
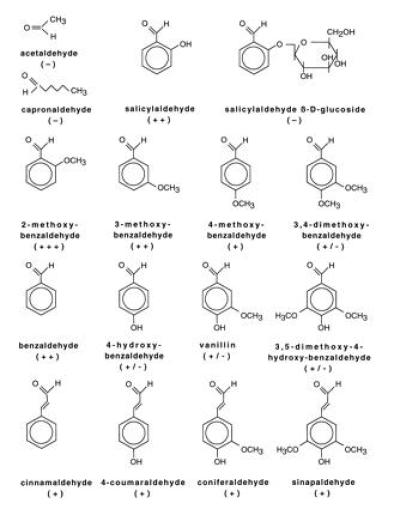
Chemical structures of various aldehydes tested as potential substrates of AtELI3-2. Symbols indicate high (+++), intermediate (++), low (+ or +/−), and undetectable (−) rates of conversion.
Figure 3.
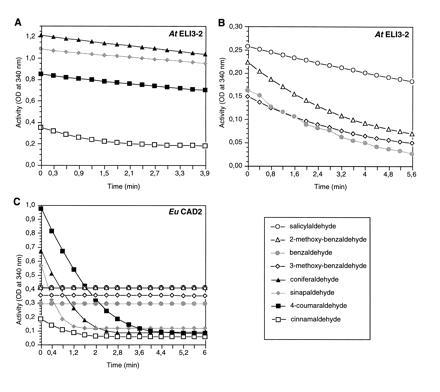
Relative conversion rates of various aldehydes using 60 μg (A) or 5 μg (B) of a crude bacterial AtELI3-2 extract, or 3.4 μg (C) of an analogous, crude bacterial EuCAD2 extract. Symbols are explained in D.
Various other aromatic aldehydes, such as 4-hydroxybenzaldehyde, 4-methoxybenzaldehyde, vanillin (3-methoxy-4-hydroxybenzaldehyde), 3,4-dimethoxybenzaldehyde, and 3,5-dimethoxy-4-hydroxybenzaldehyde, were converted even less efficiently than the cinnamaldehyde derivatives, whereas salicylaldehyde β-O-glucoside and two aliphatic aldehydes, acetaldehyde and capronaldehyde, were not converted at all (Fig. 2). Oxidation of the corresponding alcohols was also observed, albeit at 50- to 100-fold lower rates. In contrast to AtELI3-2, authentic EuCAD2 from analogous bacterial extracts showed a strong preference for cinnamyl aldehydes/alcohols, with no conversion of the four major AtELI3-2 substrates detectable even at high protein concentrations (Fig. 3C). Extracts from IPTG-stimulated bacteria harboring the pQE50-vector construct showed no detectable aromatic aldehyde reductase activity.
Thus, biochemical analysis of the recombinant AtELI3-2 protein revealed its function as an aromatic aldehyde reductase. The enzyme had an absolute requirement for NADPH as cofactor; NADH could not substitute for NADPH. Quantitative analysis by high performance liquid chromatography confirmed the stoichiometric conversion of 2-methoxybenzaldehyde to 2-methoxybenzyl alcohol.
Kinetic Parameters.
To estimate the relative affinities for some of the most efficient aldehyde substrates, the apparent Km and Vmax values were calculated from Lineweaver–Burk and Hanes plots (Table 1) using the recombinant enzyme in crude bacterial extracts. The results demonstrate that (i) 2-methoxybenzaldehyde is the most efficient of all AtELI3-2 substrates tested; (ii) various unsubstituted and substituted benzaldehydes are more efficient substrates than unsubstituted and substituted cinnamaldehydes; and (iii) the substrate specificity of AtELI3-2 differs greatly from that of an authentic CAD. We determined the apparent Km of EuCAD2 for coniferyl aldehyde under our conditions (5 μM) and found it to be in good agreement with the reported value of 4.5 μM (12). The apparent Km values for NADPH using the four most efficient benzaldehyde derivatives (Table 1) as substrates for AtELI3-2 were in the range of 9–32 μM.
Table 1.
Affinity of AtELI3-2 for selected substrates (for structures, see Fig. 2)
| Substrate | Km, μM | Vmax, nkat·mg−1 | Vmax/Km, nkat·mg−1·μM−1 |
|---|---|---|---|
| 2-Methoxybenzaldehyde | 1.3 | 24.8 | 19.1 |
| 3-Methoxybenzaldehyde | 6 | 17.5 | 2.9 |
| Salicylaldehyde | 12.5 | 13.8 | 1.1 |
| Benzaldehyde | 25 | 26.2 | 1.1 |
| Cinnamaldehyde | 24 | 17.5 | 0.7 |
| 4-Coumaraldehyde | 35 | 1.9 | 0.05 |
The values given were determined from Lineweaver–Burk and Hanes plots using 5–18 μg protein from the transformed E. coli strain pQE50-ELI3 and varying concentrations of the indicated substrates.
DISCUSSION
These results demonstrate that the eli3-2 gene from A. thaliana encodes an aromatic alcohol:NADP+ oxidoreductase and not, as recently predicted, an MTD. Although the precise function of the enzyme in vivo is not entirely clear from our present data, it is most likely involved in the generation of benzyl alcohol derivatives. In any case, the apparent substrate specificity is distinct from that of any known NAD+- or NADP+-dependent oxidoreductase, including MTD and CAD. Considering the strong preference for various benzaldehydes among the substrates tested, as well as the analogy to CAD, which preferentially accepts cinnamyl aldehyde derivatives as substrates, we propose the general name BAD for AtELI3-2. Functionally related enzymes, likewise exhibiting higher specificity for benzyl alcohols compared with coniferyl alcohol, may have been detected previously in E. gunnii (17) and Phaseolus vulgaris (18) but have not been further characterized.
Although biochemical evidence of the functions of AtELI3-1 (7) and PcELI3 (4) is not yet available, sequence comparison with AtELI3-2 (85 and 70% amino acid identity, respectively) suggests that they too may be BADs. Moreover, the established tandem location of the eli3-1 and eli3-2 genes on chromosome 4 of A. thaliana, separated by only ≈2 kb (7), suggests, apart from functional similarity, a common evolutionary origin for these two AtELI3 isoforms. Even more equivocal is the function of two A. thaliana proteins (deduced from GenBank accessions AtP42734 and AtZ31715), which were recently proposed to be CADs solely on the basis of their sequence similarity with the authentic enzyme. Although AtZ31715 may indeed be a CAD (76% identity with EuCAD2), AtP42734 shares greater similarity with AtELI3-2 (63% identity compared with 52% identity with EuCAD2) and thus may rather be a BAD. However, from the obvious functional dissimilarity of AtELI3-2 and AgMTD, despite their large sequence similarity (68% identity), we conclude that one has to be extremely cautious with such functional assignments, even though numerous successful cases exist.
The functional assignment of ELI3 is particularly interesting in view of its close association with both local defense gene expression at pathogen infection sites (5) and genetically determined disease resistance (7, 19), even though the genetic link was not always observed (20, 21). It is possible that the putative involvement of BAD in the conversion of benzaldehyde derivatives to the corresponding benzyl alcohols is associated either with the incorporation of phenolic defense materials into the cell wall or with the metabolism of soluble compounds, or both. Although, to our knowledge the accumulation of wall-bound benzyl alcohols has not been described in relation to pathogen defense, two benzaldehydes, 4-hydroxybenzaldehyde and vanillin (Fig. 2), along with unidentified wall components, have been isolated from fungal elicitor-treated, cultured P. crispum (22) and S. tuberosum cells (23), as well as from fungus-infected S. tuberosum leaves (23), and syringaldehyde was found to accumulate in fungus-infected lettuce leaves (24). Further analysis in this direction may reveal additional, structurally related compounds, possibly including the products of BAD activity.
If BAD generates soluble benzyl alcohols in vivo, these may have a role in the signaling process during infection, as suggested by the close structural relationship of the most efficient substrates of AtELI3-2 with salicylic acid, an established signal molecule in plants (25) that occurs in various structural variants (26). However, similar to the occurrence of multiple CAD (9, 27) and other alcohol dehydrogenase (28) isoforms with distinct substrate specificities, functionally distinct BADs may exist to fulfill different physiological roles. In this regard, surprising observations may lie ahead, analogous to the recent finding that the gene TASSELSEED2 encodes an alcohol dehydrogenase required for sex determination in Zea mays (29).
Acknowledgments
We thank Drs. J. Grima-Pettenati and A. Boudet (Université Paul Sabatier/ Centre National de la Recherche Scientifique, Toulouse, France) for information concerning low temperature IPTG induction of recombinant protein in E. coli, for the substrate 4-coumaraldehyde, and for the E. coli strain expressing the EuCAD2 cDNA; Dr. W. Heller (Forschungszentrum für Umwelt und Gesundheit, Neuherberg, Germany) for 4-coumaraldehyde; Dr. G. Lange, (Bayer, Leverkusen, Germany) for help with product identification; Drs. E. Kombrink and W. Knogge for valuable experimental advice; and Dr. P. Rushton (all of this institution) for critical reading of this manuscript.
Footnotes
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. §1734 solely to indicate this fact.
Abbreviations: Pc, Petroselinum crispum; At, Arabidopsis thaliana; Ag, Apium graveolens; CAD, cinnamyl-alcohol dehydrogenase; BAD, benzyl alcohol dehydrogenase; Eu, Eucalyptus gunnii; MTD, mannitol dehydrogenase; IPTG, isopropyl β-d-thiogalactoside.
References
- 1.Collinge D B, Gregersen P L, Thordal-Christensen H. In: The Induction of Gene Expression in Response to Pathogenic Microbes. Basra A S, editor. New York: Dekker; 1994. pp. 391–433. [Google Scholar]
- 2.Dixon R A, Harrison M J. Adv Genet. 1990;28:165–234. doi: 10.1016/s0065-2660(08)60527-1. [DOI] [PubMed] [Google Scholar]
- 3.Kombrink E, Somssich I E. Adv Bot Res. 1995;21:1–34. [Google Scholar]
- 4.Somssich I E, Bollmann J, Hahlbrock K, Kombrink E, Schulz W. Plant Mol Biol. 1989;12:227–234. doi: 10.1007/BF00020507. [DOI] [PubMed] [Google Scholar]
- 5.Schmelzer E, Krüger-Lebus S, Hahlbrock K. Plant Cell. 1989;1:993–1001. doi: 10.1105/tpc.1.10.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trezzini G F, Horrichs A, Somssich I E. Plant Mol Biol. 1993;21:385–389. doi: 10.1007/BF00019954. [DOI] [PubMed] [Google Scholar]
- 7.Kiedrowski S, Kawalleck P, Hahlbrock K, Somssich I E, Dangl J L. EMBO J. 1992;11:4677–4684. doi: 10.1002/j.1460-2075.1992.tb05572.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Debener T, Lehnackers H, Arnold H, Dangl J L. Plant J. 1991;1:289–302. doi: 10.1046/j.1365-313X.1991.t01-7-00999.x. [DOI] [PubMed] [Google Scholar]
- 9.Boudet A M, Lapierre C, Grima-Pettenati J. New Phytol. 1995;129:203–236. doi: 10.1111/j.1469-8137.1995.tb04292.x. [DOI] [PubMed] [Google Scholar]
- 10.Williamson J D, Stoop J M H, Massel M O, Conkling M A, Pharr D M. Proc Natl Acad Sci USA. 1995;92:7148–7152. doi: 10.1073/pnas.92.16.7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong X. Proc Natl Acad Sci USA. 1995;92:7137–7139. doi: 10.1073/pnas.92.16.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grima-Pettenati J, Feuillet C, Goffner D, Bordereis G, Boudet A M. Plant Mol Biol. 1993;21:1085–1095. doi: 10.1007/BF00023605. [DOI] [PubMed] [Google Scholar]
- 13.Lauvergeat V, Kennedy K, Feuillet C, McKie J H, Gorrichon L, Baltas M, Boudet A M, Grima-Pettenati J, Douglas K T. Biochemistry. 1995;34:12426–12434. doi: 10.1021/bi00038a041. [DOI] [PubMed] [Google Scholar]
- 14.Bradford M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 15.Stoop J M H, Williamson J D, Conkling M A, Pharr D M. Plant Physiol. 1995;108:1219–1225. doi: 10.1104/pp.108.3.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wyrambik D, Grisebach H. Eur J Biochem. 1975;59:9–15. doi: 10.1111/j.1432-1033.1975.tb02418.x. [DOI] [PubMed] [Google Scholar]
- 17.Hawkins S W, Boudet A M. Plant Physiol. 1994;104:75–84. doi: 10.1104/pp.104.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grima-Pettenati J, Campargue C, Boudet A, Boudet A M. Phytochemistry. 1994;37:941–947. doi: 10.1016/s0031-9422(00)89508-4. [DOI] [PubMed] [Google Scholar]
- 19.Mittal S, Davis K R. Mol Plant-Microbe Interact. 1995;8:165–171. doi: 10.1094/mpmi-8-0165. [DOI] [PubMed] [Google Scholar]
- 20.Reuber T L, Ausubel F M. Plant Cell. 1996;8:241–249. doi: 10.1105/tpc.8.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ritter C, Dangl J L. Plant Cell. 1996;8:251–257. doi: 10.1105/tpc.8.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kauss H, Franke R, Krause K, Conrath U, Jeblick W, Grimmig B, Matern U. Plant Physiol. 1993;102:459–466. doi: 10.1104/pp.102.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keller H, Hohlfeld H, Wray V, Hahlbrock K, Scheel D, Strack D. Phytochemistry. 1996;42:389–396. [Google Scholar]
- 24.Bennett M, Gallagher M, Fagg J, Bestwick C, Paul T, Beale M, Mansfield J. Plant J. 1996;9:101–115. [Google Scholar]
- 25.Klessig D F, Malamy J. Plant Mol Biol. 1994;26:1439–1458. doi: 10.1007/BF00016484. [DOI] [PubMed] [Google Scholar]
- 26.Pierpoint W S. Adv Bot Res. 1994;20:163–235. [Google Scholar]
- 27.Campbell M M, Sederoff R R. Plant Physiol. 1996;110:3–13. doi: 10.1104/pp.110.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jörnvall H, Persson B, Jeffery J. Eur J Biochem. 1987;167:195–201. doi: 10.1111/j.1432-1033.1987.tb13323.x. [DOI] [PubMed] [Google Scholar]
- 29.DeLong A, Calderon-Urrea A, Dellaporta S L. Cell. 1993;74:757–768. doi: 10.1016/0092-8674(93)90522-r. [DOI] [PubMed] [Google Scholar]


