Abstract
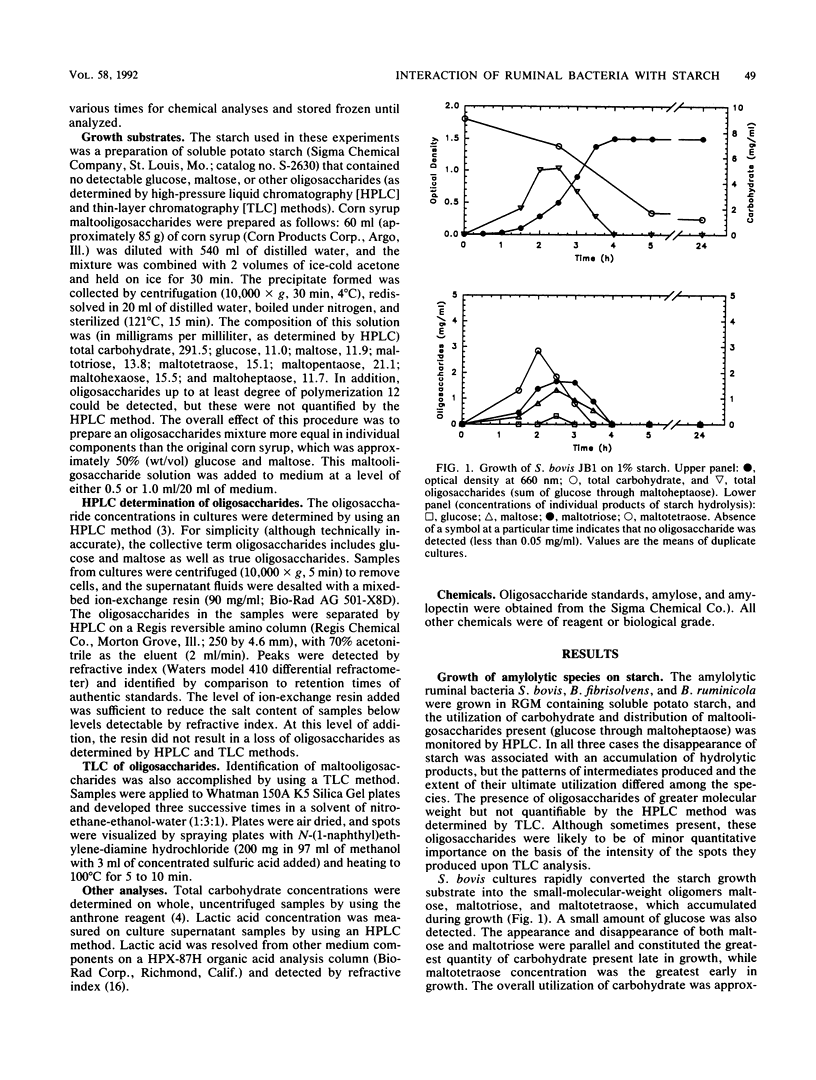
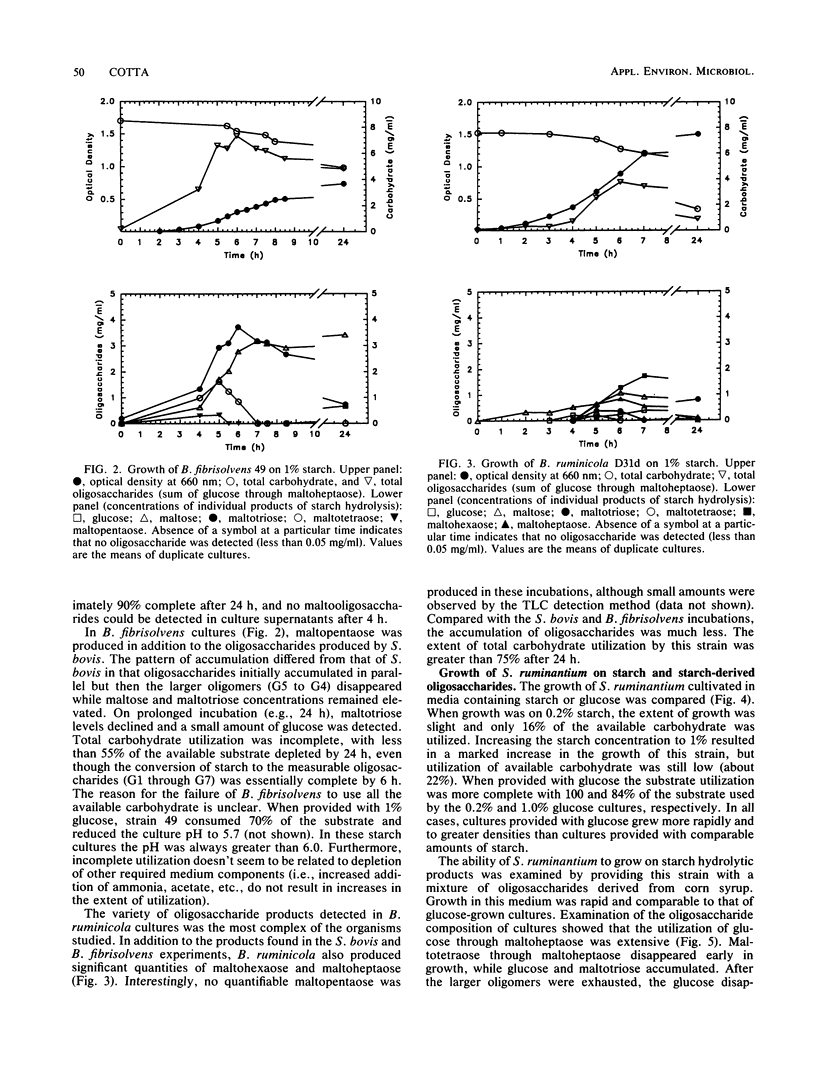
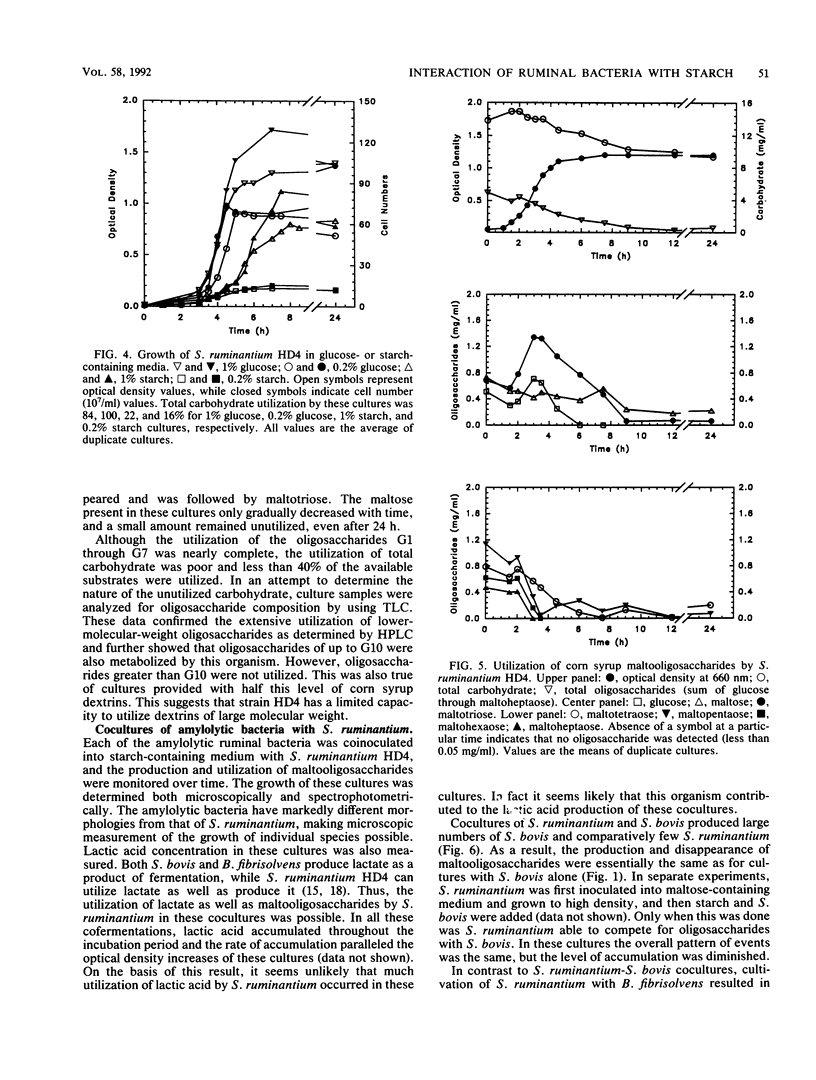
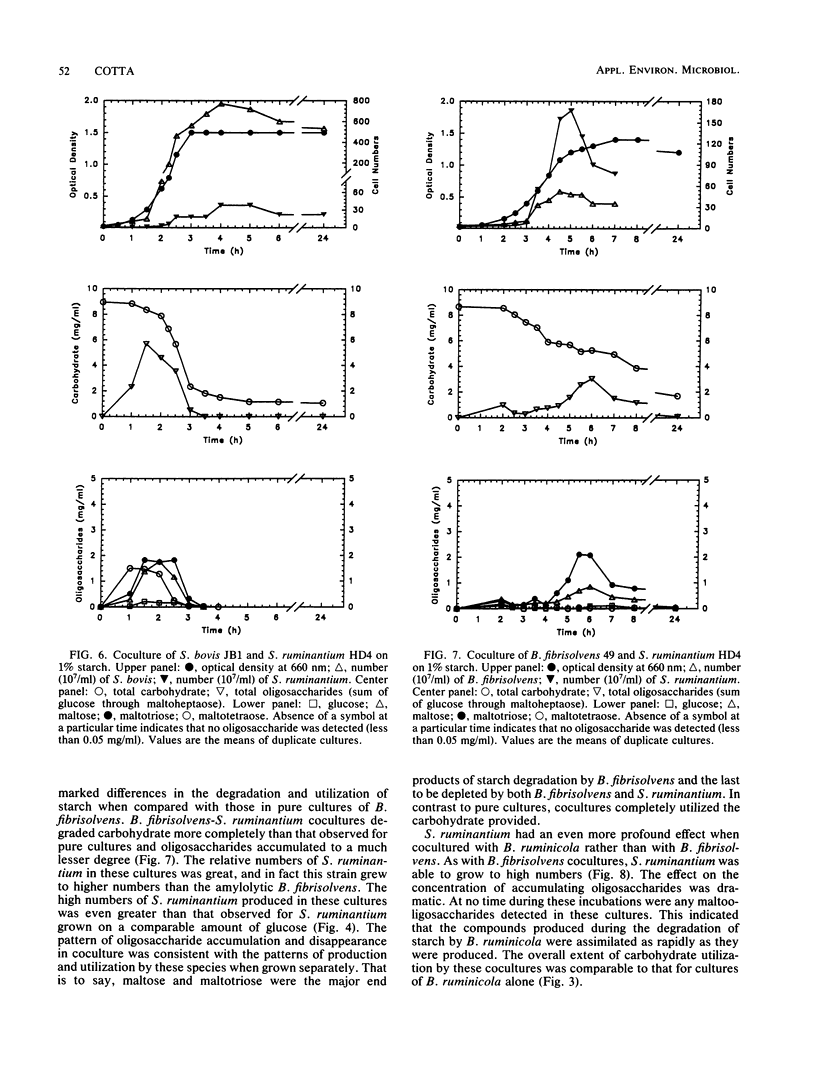
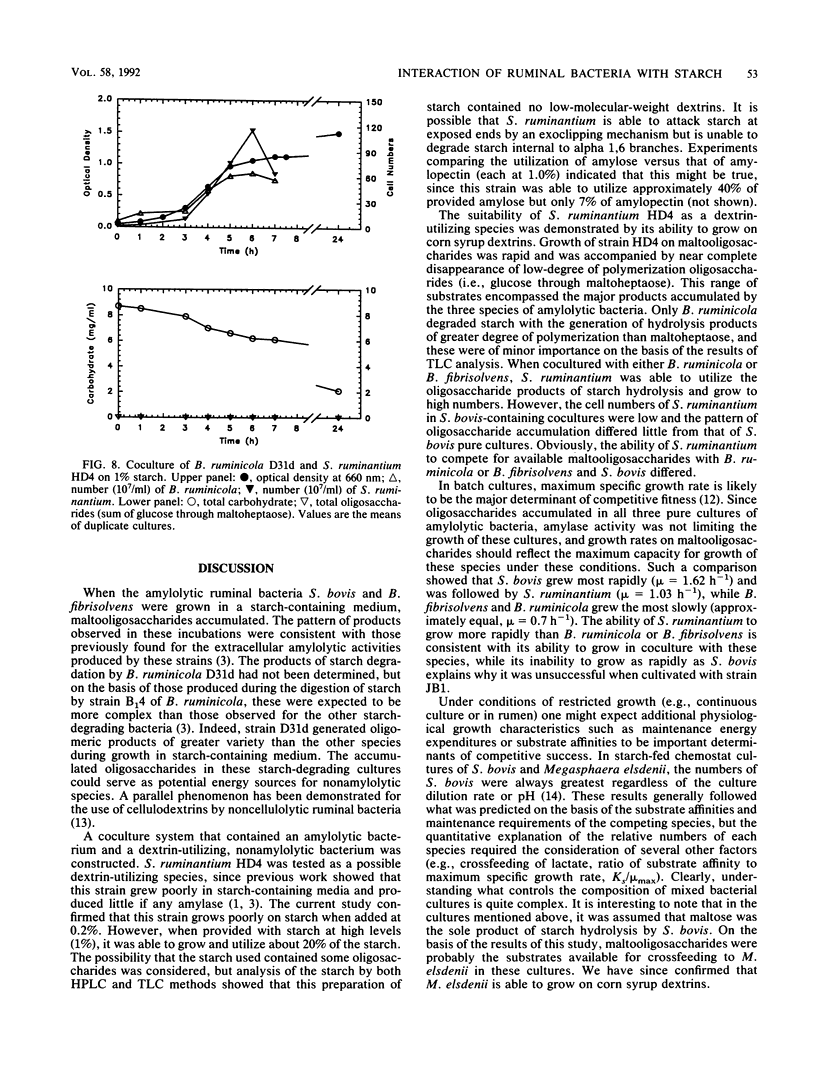
The degradation and utilization of starch by three amylolytic and one nonamylolytic species of ruminal bacteria were studied. Pure cultures of Streptococcus bovis JB1, Butyrivibrio fibrisolvens 49, and Bacteroides ruminicola D31d rapidly hydrolyzed starch and maltooligosaccharides accumulated. The major starch hydrolytic products detected in S. bovis cultures were glucose, maltose, maltotriose, and maltotetraose. In addition to these oligosaccharides, B. fibrisolvens cultures produced maltopentaose. The products of starch hydrolysis by B. ruminicola were even more complex, yielding glucose through maltotetraose, maltohexaose, and maltoheptaose but little maltopentaose. Selenomonas ruminantium HD4 grew poorly on starch, digested only a small portion of the available substrate, and generated no detectable oligosaccharides as a result of cultivation in starch containing medium. S. ruminantium was able to grow on a mixture of maltooligosaccharides and utilize those of lower degree (less than 10) of polymerization. A coculture system containing S. ruminantium as a dextrin-utilizing species and each of the three amylolytic bacteria was developed to test whether the products of starch hydrolysis were available for crossfeeding to another ruminal bacterium. Cocultures of S. ruminantium and S. bovis contained large numbers of S. bovis but relatively few S. ruminantium and exhibited little change in the pattern of maltooligosaccharides observed for pure cultures of S. bovis. In contrast, S. ruminantium was able to compete with B. fibrisolvens and B. ruminicola for these growth substrates. When grown with B. fibrisolvens, S. ruminantium grew to high numbers and maltooligosaccharides accumulated to a much lesser degree than in cultures of B. fibrisolvens alone. S. ruminantium-B. ruminicola cultures contained large numbers of both species, and maltooligosaccharides never accumulated in these cocultures.(ABSTRACT TRUNCATED AT 250 WORDS)
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BRYANT M. P. The characteristics of strains of Selenomonas isolated from bovine rumen contents. J Bacteriol. 1956 Aug;72(2):162–167. doi: 10.1128/jb.72.2.162-167.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotta M. A. Amylolytic activity of selected species of ruminal bacteria. Appl Environ Microbiol. 1988 Mar;54(3):772–776. doi: 10.1128/aem.54.3.772-776.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hespell R. B., Wolf R., Bothast R. J. Fermentation of xylans by Butyrivibrio fibrisolvens and other ruminal bacteria. Appl Environ Microbiol. 1987 Dec;53(12):2849–2853. doi: 10.1128/aem.53.12.2849-2853.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo H., Cheng K. J., Costerton J. W. Interactions between Treponema bryantii and cellulolytic bacteria in the in vitro degradation of straw cellulose. Can J Microbiol. 1987 Mar;33(3):244–248. doi: 10.1139/m87-041. [DOI] [PubMed] [Google Scholar]
- Leedle J. A., Barsuhn K., Hespell R. B. Postprandial trends in estimated ruminal digesta polysaccharides and their relation to changes in bacterial groups and ruminal fluid characteristics. J Anim Sci. 1986 Mar;62(3):789–803. doi: 10.2527/jas1986.623789x. [DOI] [PubMed] [Google Scholar]
- Miura H., Horiguchi M., Ogimoto K., Matsumoto T. Nutritional interdependence among rumen bacteria during cellulose digestion in vitro. Appl Environ Microbiol. 1983 Feb;45(2):726–729. doi: 10.1128/aem.45.2.726-729.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relationship of lactate dehydrogenase specificity and growth rate to lactate metabolism by Selenomonas ruminantium. Appl Microbiol. 1975 Dec;30(6):916–921. doi: 10.1128/am.30.6.916-921.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell J. B., Cotta M. A., Dombrowski D. B. Rumen Bacterial Competition in Continuous Culture: Streptococcus bovis Versus Megasphaera elsdenii. Appl Environ Microbiol. 1981 Jun;41(6):1394–1399. doi: 10.1128/aem.41.6.1394-1399.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell J. B. Fermentation of cellodextrins by cellulolytic and noncellulolytic rumen bacteria. Appl Environ Microbiol. 1985 Mar;49(3):572–576. doi: 10.1128/aem.49.3.572-576.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton T. B., Canale-Parola E. Treponema bryantii sp. nov., a rumen spirochete that interacts with cellulolytic bacteria. Arch Microbiol. 1980 Sep;127(2):145–156. doi: 10.1007/BF00428018. [DOI] [PubMed] [Google Scholar]


