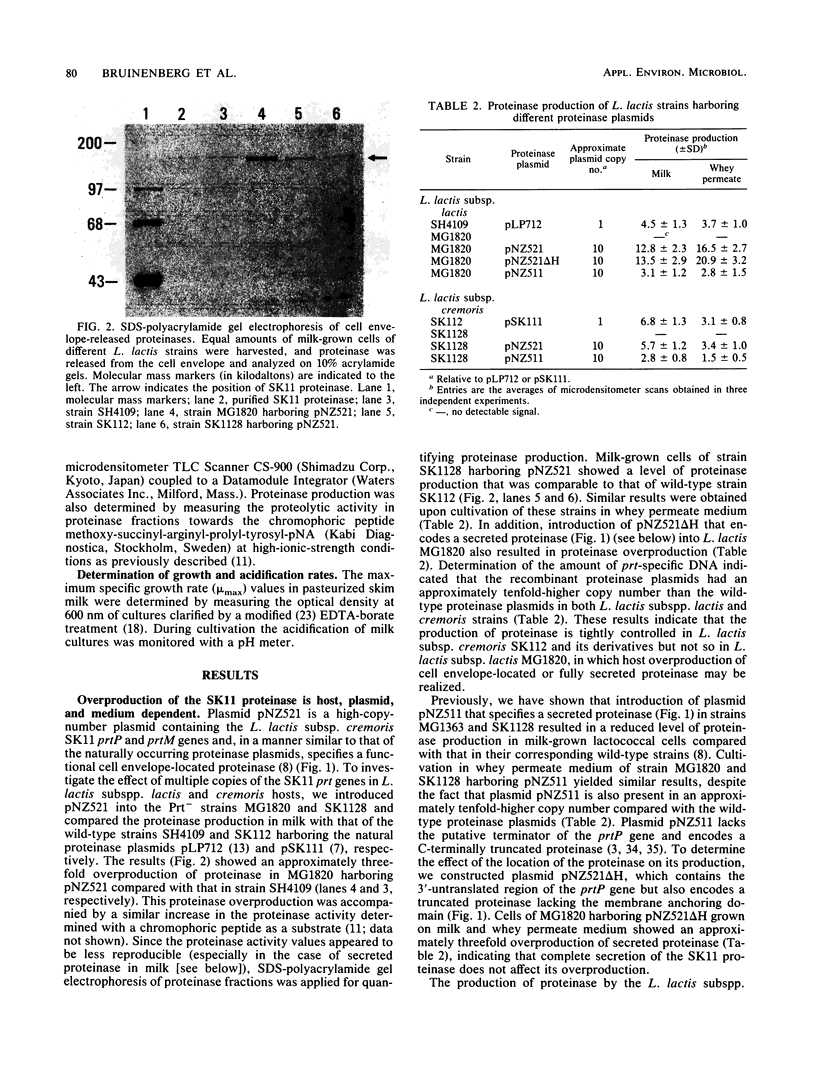
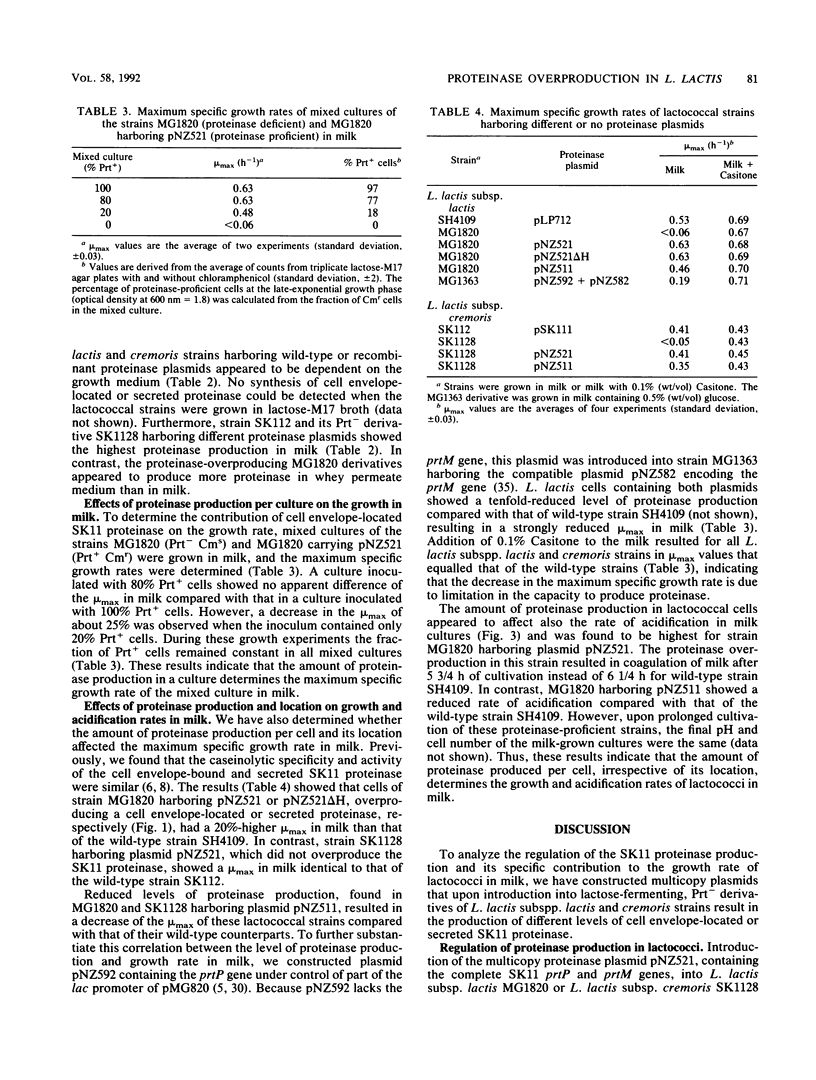
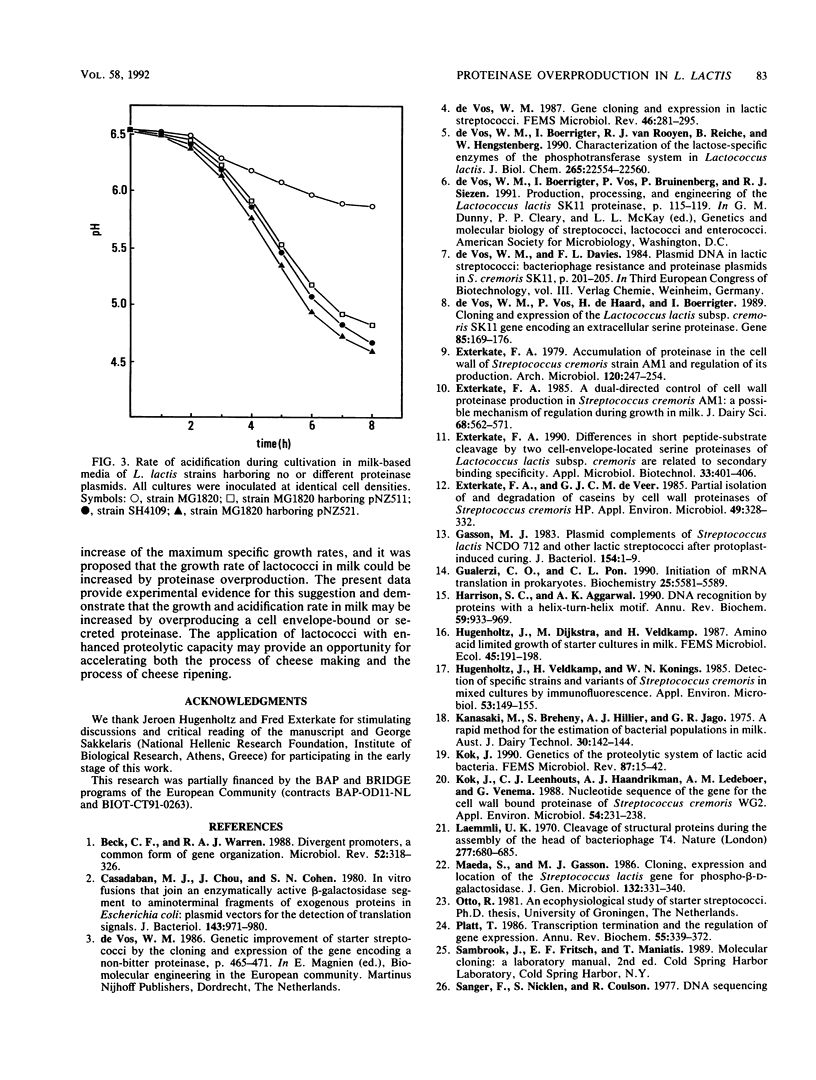
Abstract
Multicopy plasmids that contained the complete of 3'-deleted forms of the proteinase (prtP) gene of Lactococcus lactis subsp. cremoris SK11 under the control of different promoters were constructed and introduced into Prt- lactococcal strains. The production and location of the SK11 proteinase was determined in different hosts grown in industrial and laboratory media. In spite of the 10-fold-higher copy number of the prt genes, no overproduction of proteinase was observed in strain SK1128, a Prt- derivative of L. lactis subsp. cremoris SK112. In contrast, an approximately threefold overproduction of the cell envelope-located or fully secreted proteinase was found in strain MG1820 compared with that of its parental strain L. lactis subsp. lactis SH4109. In all strains proteinase production appeared to be regulated by the medium composition. Highest proteinase production of the SK11 derivatives was found in milk, in contrast to derivatives of SH4109 that produced most proteinase in whey permeate medium. Analysis of single strains with different levels of proteinase production or mixed cultures containing various ratios of Prt+ and Prt- cells indicated that the amount of proteinase produced per cell or culture determines the specific growth rate in milk. Overproduction of cell envelope-located or secreted proteinase in strain MG1820 resulted in a 20%-higher specific growth and acidification rate in milk compared with that in the wild-type strain SH4109. These results indicate that the growth of lactococci in milk is limited by the caseinolytic activity of the proteinase.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beck C. F., Warren R. A. Divergent promoters, a common form of gene organization. Microbiol Rev. 1988 Sep;52(3):318–326. doi: 10.1128/mr.52.3.318-326.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Chou J., Cohen S. N. In vitro gene fusions that join an enzymatically active beta-galactosidase segment to amino-terminal fragments of exogenous proteins: Escherichia coli plasmid vectors for the detection and cloning of translational initiation signals. J Bacteriol. 1980 Aug;143(2):971–980. doi: 10.1128/jb.143.2.971-980.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exterkate F. A. Differences in short peptide-substrate cleavage by two cell-envelope-located serine proteinases of Lactococcus lactis subsp. cremoris are related to secondary binding specificity. Appl Microbiol Biotechnol. 1990 Jul;33(4):401–406. doi: 10.1007/BF00176654. [DOI] [PubMed] [Google Scholar]
- Exterkate F. A., de Veer G. J. Partial Isolation and Degradation of Caseins by Cell Wall Proteinase(s) of Streptococcus cremoris HP. Appl Environ Microbiol. 1985 Feb;49(2):328–332. doi: 10.1128/aem.49.2.328-332.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasson M. J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983 Apr;154(1):1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison S. C., Aggarwal A. K. DNA recognition by proteins with the helix-turn-helix motif. Annu Rev Biochem. 1990;59:933–969. doi: 10.1146/annurev.bi.59.070190.004441. [DOI] [PubMed] [Google Scholar]
- Hugenholtz J., Veldkamp H., Konings W. N. Detection of Specific Strains and Variants of Streptococcus cremoris in Mixed Cultures by Immunofluorescence. Appl Environ Microbiol. 1987 Jan;53(1):149–155. doi: 10.1128/aem.53.1.149-155.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok J. Genetics of the proteolytic system of lactic acid bacteria. FEMS Microbiol Rev. 1990 Sep;7(1-2):15–42. doi: 10.1111/j.1574-6968.1990.tb04877.x. [DOI] [PubMed] [Google Scholar]
- Kok J., Leenhouts K. J., Haandrikman A. J., Ledeboer A. M., Venema G. Nucleotide sequence of the cell wall proteinase gene of Streptococcus cremoris Wg2. Appl Environ Microbiol. 1988 Jan;54(1):231–238. doi: 10.1128/aem.54.1.231-238.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Maeda S., Gasson M. J. Cloning, expression and location of the Streptococcus lactis gene for phospho-beta-D-galactosidase. J Gen Microbiol. 1986 Feb;132(2):331–340. doi: 10.1099/00221287-132-2-331. [DOI] [PubMed] [Google Scholar]
- Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- Schmeissner U., McKenney K., Rosenberg M., Court D. Removal of a terminator structure by RNA processing regulates int gene expression. J Mol Biol. 1984 Jun 15;176(1):39–53. doi: 10.1016/0022-2836(84)90381-4. [DOI] [PubMed] [Google Scholar]
- Visser S., Exterkate F. A., Slangen C. J., de Veer G. J. Comparative Study of Action of Cell Wall Proteinases from Various Strains of Streptococcus cremoris on Bovine alpha(s1)-, beta-, and kappa-Casein. Appl Environ Microbiol. 1986 Nov;52(5):1162–1166. doi: 10.1128/aem.52.5.1162-1166.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos P., Boerrigter I. J., Buist G., Haandrikman A. J., Nijhuis M., de Reuver M. B., Siezen R. J., Venema G., de Vos W. M., Kok J. Engineering of the Lactococcus lactis serine proteinase by construction of hybrid enzymes. Protein Eng. 1991 Apr;4(4):479–484. doi: 10.1093/protein/4.4.479. [DOI] [PubMed] [Google Scholar]
- Vos P., Simons G., Siezen R. J., de Vos W. M. Primary structure and organization of the gene for a procaryotic, cell envelope-located serine proteinase. J Biol Chem. 1989 Aug 15;264(23):13579–13585. [PubMed] [Google Scholar]
- Vos P., van Asseldonk M., van Jeveren F., Siezen R., Simons G., de Vos W. M. A maturation protein is essential for production of active forms of Lactococcus lactis SK11 serine proteinase located in or secreted from the cell envelope. J Bacteriol. 1989 May;171(5):2795–2802. doi: 10.1128/jb.171.5.2795-2802.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vos W. M., Boerrigter I., van Rooyen R. J., Reiche B., Hengstenberg W. Characterization of the lactose-specific enzymes of the phosphotransferase system in Lactococcus lactis. J Biol Chem. 1990 Dec 25;265(36):22554–22560. [PubMed] [Google Scholar]
- de Vos W. M., Vos P., de Haard H., Boerrigter I. Cloning and expression of the Lactococcus lactis subsp. cremoris SK11 gene encoding an extracellular serine proteinase. Gene. 1989 Dec 21;85(1):169–176. doi: 10.1016/0378-1119(89)90477-0. [DOI] [PubMed] [Google Scholar]
- van Rooijen R. J., de Vos W. M. Molecular cloning, transcriptional analysis, and nucleotide sequence of lacR, a gene encoding the repressor of the lactose phosphotransferase system of Lactococcus lactis. J Biol Chem. 1990 Oct 25;265(30):18499–18503. [PubMed] [Google Scholar]