Abstract
It is not known how the volume of the cell nucleus is set, nor how the ratio of nuclear volume to cell volume (N/C) is determined. Here, we have measured the size of the nucleus in growing cells of the budding yeast Saccharomyces cerevisiae. Analysis of mutant yeast strains spanning a range of cell sizes revealed that the ratio of average nuclear volume to average cell volume was quite consistent, with nuclear volume being ∼7% that of cell volume. At the single cell level, nuclear and cell size were strongly correlated in growing wild-type cells, as determined by three different microscopic approaches. Even in G1-phase, nuclear volume grew, although it did not grow quite as fast as overall cell volume. DNA content did not appear to have any immediate, direct influence on nuclear size, in that nuclear size did not increase sharply during S-phase. The maintenance of nuclear size did not require continuous growth or ribosome biogenesis, as starvation and rapamycin treatment had little immediate impact on nuclear size. Blocking the nuclear export of new ribosomal subunits, among other proteins and RNAs, with leptomycin B also had no obvious effect on nuclear size. Nuclear expansion must now be factored into conceptual and mathematical models of budding yeast growth and division. These results raise questions as to the unknown force(s) that expand the nucleus as yeast cells grow.
INTRODUCTION
It has long been appreciated that the size of a cell and its nucleus are intertwined (Gulliver, 1875; Wilson, 1925). In a great range of eukaryotes, from yeast to sea urchins to mice, it has been found that changing ploidy alters the volume of the cell, such that tetraploid cells are twice the size of diploids, which are in turn twice the size of haploids (Wilson, 1925; Fankhauser, 1945; Mortimer, 1958; Henery et al., 1992). More than a hundred years ago, such observations led Hertwig, Boveri, and their colleagues to propose that eukaryotic cells actively maintain a ratio of nuclear volume to cell volume, the “karyoplasmic” ratio.
Furthermore, there is a striking correlation between cell volume and nuclear DNA content across vast stretches of evolutionary space. The correlation is upheld in the face of greater than 100,000-fold variation in DNA content among eukaryotes (Cavalier-Smith, 2005; Gregory, 2005). Explanations for the consistent relationship between DNA content and cell size during eukaryotic evolution have been proposed. Cavalier-Smith (2005) has argued that there is natural selection for optimal cell size and an underlying, optimal karyoplasmic ratio. Selection for optimum cell size leads to selection for the matching optimum nuclear size and because in this theory nuclear size depends on DNA content, this leads to selection for larger or smaller amounts of DNA. This adjustment of DNA need not be for protein-coding genes, perhaps thus accounting for the overwhelming majority of noncoding DNA in large genomes (Cavalier-Smith, 2005).
The karyoplasmic ratio is important in several theories of cell cycle control. In many single-celled eukaryotes, the cell division cycle is under size control, such that cells do not commit to division until they have grown to a certain critical size. The ultimate mechanistic basis for this size control is still unknown, but some theories propose that the cell is somehow measuring the relative volumes of a growing cytoplasm versus a constant nucleus; that is, when the cytoplasm has grown sufficiently large, so that the ratio of cytoplasmic to nuclear volume is sufficiently large, cell division is triggered (Futcher, 1996; Csikasz-Nagy et al., 2006). A requirement for these theories is that the nucleus must stay relatively constant in volume throughout G1-phase. To our knowledge, the relationship of nuclear volume to the cell cycle, and in particular G1-phase, has not been definitively reported in any cell type. It has been reported that budding yeast in S-phase have larger nuclei than cells in G1-phase, and cells in early mitosis have larger nuclei than cells in S-phase (Winey et al., 1997).
Cell size control is best characterized in yeast (Rupes, 2002; Jorgensen and Tyers, 2004). In the budding yeast Saccharomyces cerevisiae, cell cycle progression is coupled to cell growth at Start, a transition period between G1- and S-phase. Start occurs when yeast have reached a critical cell size (Johnston et al., 1977), although it is still not clear how yeast measure size. It is presumed that some parameter that correlates with size, such as the overall protein synthesis rate, is monitored. In the predominant model, cell size is reported by the cyclin Cln3, which complexes with the cyclin-dependent kinase Cdk1 to activate Start by phosphorylating and thereby inactivating the Start repressor Whi5 (Jorgensen et al., 2002; Costanzo et al., 2003; de Bruin et al., 2004). Cln3 is a highly unstable protein expressed from an mRNA present at fairly consistent levels during G1-phase (Tyers et al., 1992, 1993; Cross and Blake, 1993), so its absolute abundance is expected to be highly sensitive to its rate of synthesis and thereby reflect the current translation rate. As Cln3 locates predominantly to the nucleus (Miller and Cross, 2000; Edgington and Futcher, 2001), it was proposed that Cln3 nuclear concentration increases as cells grow and increase their absolute translational capacity (Futcher, 1996). Again, this theory requires that the volume of the nucleus must stay relatively constant through G1-phase.
An important reason for exploring the relationship between nuclear volume and the cell cycle is that cancer cells often have large nuclear to cell volume (N/C) ratios. For instance, in cervical carcinoma, a high N/C ratio is a well-accepted cytological indicator of an aggressive tumor and a poor prognosis (Slater et al., 2005). Despite hundreds of articles documenting correlations between nuclear size or shape and the progression of malignancies (Zink et al., 2004), there is no understanding of the mechanism by which the N/C ratio is set or why some cancer cells might have a larger N/C ratio than other cells.
MATERIALS AND METHODS
Yeast Strains, Culture, and Drugs
Yeast culture, including size analysis with a Coulter particle analyzer, fluorescence-activated cell sorting (FACS) analysis, and cell size selection by centrifugal elutriation has been described (Futcher, 1999; Jorgensen et al., 2002). Yeast strains are listed in Table 1. Alleles of bud21::BUD21YFP-his5+ and sec63::SEC63CFP-kanR were generated by genomic integration of standard C-terminal tagging cassettes that were PCR-amplified with integration site-specific primers. Template plasmids for cyan and yellow fluorescent protein (CFP and YFP, respectively) fusions were obtained from the Yeast Resource Center (University of Washington, Seattle). whi and CLN3 alleles have been described previously (Jorgensen et al., 2002). Synthetic complete medium is 0.2% amino acid mix, 0.17% yeast nitrogen base without amino acids and ammonium sulfate, 0.5% ammonium sulfate supplemented with 3% ethanol, 2% glucose, 2% raffinose, or 2% galactose, as indicated. YEP medium is 2% peptone, 1% yeast extract supplemented with 3% ethanol or 2% glucose, as indicated. Rapamycin and leptomycin B (LMB) were obtained from Sigma and were used at working concentrations of 0.2 μg/ml and 100 ng/ml, respectively.
Table 1.
Strains used in this study
| Strain | Genotype | MAT | Background | Reference |
|---|---|---|---|---|
| yMT1448 | ura3Δ leu2Δ his3Δ | a | S288c | Brachmann et al. (1998) |
| yMT3040 | bud21::BUD21YFP-his5+ sec63::SEC63CFP-kanR ura3Δ leu2Δ his3Δ | a | S288c | This study |
| yMT3121 | sfp1Δ::URA3 bud21::BUD21YFP-his5+ sec63::SEC63CFP-kanR ura3Δ leu2Δ his3Δ | α | S288c | This study |
| yMT3131 | sch9Δ::natR bud21::BUD21YFP-his5+ sec63::SEC63CFP-kanR ura3Δ leu2Δ his3Δ | α | S288c | This study |
| yMT3160 | whi5Δ::natR sec63::SEC63CFP-kanR ura3Δ leu2Δ his3Δ | a | S288c | This study |
| yMT3182 | cln3Δ::URA3-GAL1-CLN3-1 bud21::BUD21YFP-his5+ sec63::SEC63CFP-kanR ura3Δ leu2Δ his3Δ | α | S288c | This study |
| yMT3985 | sec63::SEC63CFP-kanR crm1Δ::kanR [pDC-crm1(T539C)-LEU2/CEN] | a | W303/S288c | This study |
| yMT3981 | bud21::BUD21YFP-his5/+ sec63::SEC63CFP-kanR/+ ura3Δ leu2Δ his3Δ | a/α | S288c | |
| N-419 | ura3Δ leu2Δ his3Δ lys2Δ | a | S288c | This study |
| W303 | ade2-1 trpl-1 leu2-3,112 his3-11,15 ura3 can1-100 | a | W303 | R. Rothstein |
| BS100 | cln1::LEU2-GALpr-CLN1-HA3 cln2Δ::TRP1 cln3Δ::HIS3 ade2-1 trpl-1 leu2-3,112 his3-11,15 ura3 | a | W303 | Schneider et al. (1996) |
Microscopy and Morphometry
Sec63CFP Reporter.
All experiments in Figures 1–3 and 6 were performed with log phase cells at 30°C with OD600 <0.5 and cell concentration <3 × 107 cells/ml to prevent inadvertent nutrient depletion. Log phase cultures in synthetic media were rapidly concentrated by centrifugation. Medium, 2.5 μl, with concentrated live cells was mounted at room temperature and immediately visualized. Cell fields were imaged within 5 min of being removed from the 30°C incubator.
Figure 1.
Nuclear and nucleolar size is altered in yeast with abnormal cell size. Yeast cells bearing a CLN3-1 allele under the control of the GAL1 promoter were grown in media that generates very low CLN3 activity (top panels, glucose medium) or very high CLN3 activity (bottom panels, galactose medium). The cells expressed the nuclear envelope/ER marker Sec63CFP and the nucleolar marker Bud21YFP from the endogenous loci. All images are equally magnified; scale bar, 10 μm.
Figure 2.
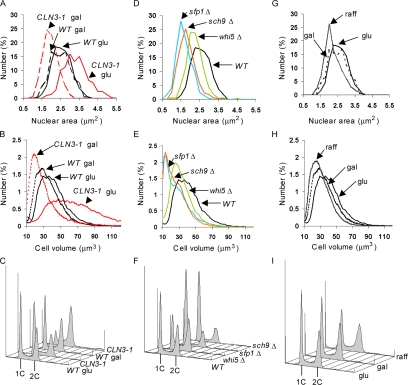
Nuclear size correlates with cell size in different mutant backgrounds and nutrient conditions. (A) CLN3 affects nuclear size. Wild-type and cln3::GAL1pr-CLN3-1 strains expressing Sec63CFP were propagated to log phase in synthetic glucose (glu) or galactose (gal) medium, cross-sectional nuclear areas were measured from micrographs, and the distributions were plotted. (B and C) Representative cell volume and DNA content distributions of cultures in A. (D) whi mutants have small nuclei. Strains of the indicated genotype and expressing Sec63CFP were propagated in synthetic glucose medium to log phase, and the distribution of cross-sectional nuclear areas was determined. (E and F) Representative cell volume and DNA content distributions of cultures in D. (G) Carbon source influences nuclear size. Wild-type cells expressing Sec63CFP were measured as in A, but were propagated in glucose, galactose, or raffinose (raff) medium. (H and I) Representative cell volume and DNA content distributions of cultures in G. Live cell volume was measured with a Coulter particle analyzer. DNA content was measured by flow cytometry. Each nuclear size histogram is composed of 185–539 nuclei (see Table 2 for exact numbers). The Glu and Gal data for wild-type cells presented in A–C is the same data presented for Glu and Gal cells in G–I.
Figure 3.
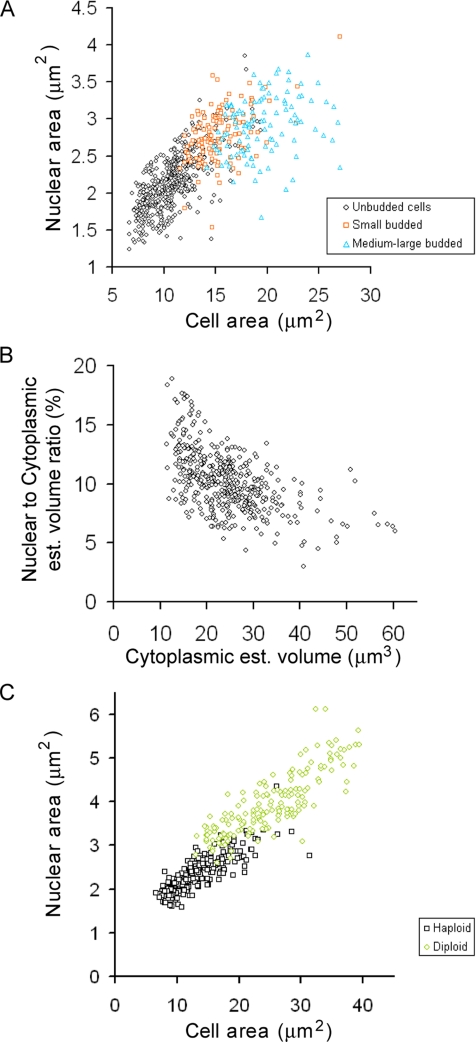
Nuclear size increases as cells grow. (A) Nuclear size correlates with cell size. Wild-type cells expressing Sec63CFP were propagated to log phase in synthetic raffinose medium and subjected to epifluorescence microscopy and morphometry. Nuclear and cell cross-sectional areas were plotted for unbudded cells (G1-phase, n = 454 cells, ◇), cells with small buds (S-phase, n = 132 cells, □), and cells with medium-to-large buds (late S/G2-phase, n = 108 cells, ▵). (B) The N/Cyt volume ratio decreases slightly as cells grow during G1-phase. Cross-sectional nuclear and cell areas of the unbudded cells from A were used to estimate nuclear and cell volume, assuming the nuclei and cells are spheres. Cytoplasmic volume was estimated to be the difference between cell and nuclear volume. Cells begin budding at ∼30 μm3 (data not shown). (C) Nuclear size correlates with cell size in both haploid (□, n = 207 cells) and diploid cells (○, n = 166 cells). Wild-type cells expressing Sec63CFP were propagated in synthetic glucose medium and imaged. Unlike A and B, each cell field was imaged as a small z-series and the maximal cross-sectional area for both the nucleus and the cell were determined before morphometry.
For Cells in Figures 1, 2, and 3, A and B.
Each cell field was sequentially imaged by differential interference contrast (DIC) and epifluorescent microscopy with an Eclipse E600FN microscope (100× objective, Plan Apo, Nikon, Melville, NY) and an Orca II CCD camera (Hamamatsu, Bridgewater, NJ). Sec63CFP signal delineated the nuclear and cell envelopes. Metamorph (MDS Analytical Technologies, St. Laurent, QC, Canada) tools were used to individually outline nuclear, cell, and bud cross-sectional areas.
For Cells in Figures 3C and 6.
Figure 6.
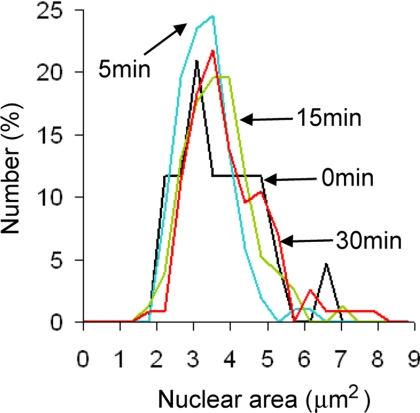
Blocking Crm1-mediated nuclear export has no effect on nuclear size. Cells bearing SEC63-CFP and crm1(T539C) alleles were propagated to log phase in synthetic glucose medium, and an aliquot of culture was pelleted and imaged (0 min, black line, n = 43 cells). LMB was added to a final concentration of 100 ng/ml and aliquots of cells were pelleted and imaged 5 (blue line, n = 102 cells), 15 (green line, n = 153 cells), and 30 (red line, n = 115 cells) min later. Nuclear size was quantified for 43-153 cells at each time point. As in Figure 3C, each cell field was imaged as a small z-series and the maximal cross-sectional area for both the nucleus and the cell were determined before morphometry.
Each cell field was imaged as a small z-series, with five steps separated by 0.4 μm each. Sec63CFP signal delineated the nuclear and cell envelopes. For each nucleus and cell, the largest cross-sectional area was determined from the z-series in Photoshop (Adobe Systems, San Jose, CA) and the cells and nuclei were converted to two separate binary masks that were analyzed in ImageJ (http://rsb.info.nih.gov/ij/) using the “Particles 8 Plus” plugin (http://rsb.info.nih.gov/ij/plugins/index.html) in the Morphology package developed by Gabriel Landini (http://rsb.info.nih.gov/ij/plugins/index.html).
For Cells in Figures 1–3 and 6.
All cells in a field were quantified unless they were undergoing anaphase or cytokinesis, that is, if they possessed an elongated nucleus or two separated nuclei. For all experiments, multiple cell fields were imaged. The resulting data were analyzed in Microsoft Excel (Redmond, CA) and in Matlab (MathWorks, Natick, MA). For volume estimates, it was assumed that nuclei and G1 cells were spherical and that measured areas were cross-sections through the centers of these spheres.
NLSGFP Reporter.
Wild-type yeast (N-419) carrying the plasmid pMGGLA (ARS/CEN, LEU2, MET3pr-GFP-NLS-A) were cultured in synthetic -Met-Leu-Trp 3% ethanol medium. pMGGLA has been described previously (Edgington and Futcher, 2001). To enrich for small cells, the culture was elutriated, and 12 fractions of incrementally larger cells were obtained and kept on ice. Cells in fractions 1, 3, 4, and 10 were mounted. Slides were prepared with a small amount of molten agar mixed with minimal medium lacking a carbon source in microwells. Cells, 1 μl, were spotted onto the cooled, solidified medium, and a coverslip was applied. The coverslip edges were sealed with nail polish, the cells were examined with a microscope (Olympus) and photographed with a 1.3 megapixel Axiocam camera (Zeiss, Thornwood, NY), and morphometry was executed with Openlab software v.2 (Improvision, Lexington, MA). The perimeter of the nucleus was delineated by the edge of the green fluorescent protein (GFP) fluorescent signal, and the cell edge was obtained from DIC images.
Electron Microscopy
To generate a range of cell sizes, cells were collected from three different sources. All cells were in the W303 genetic background, whereas in the prior two approaches, all strains were from the S288c background. To measure small G1-phase cells, unbudded cells from a log phase, YEP 3% ethanol wild-type W303 culture were obtained by elutriation. Fifty-seven cell fields were analyzed by electron microscopy (EM) from these ethanol fractions. To measure moderately sized G1-phase cells, unbudded cells from a log phase, YEP 2% glucose wild-type W303 culture were obtained by elutriation. Twelve cell fields were analyzed by EM from these glucose fractions. To measure large G1-phase cells, conditional G1-cyclin depletion was performed by first propagating BS100 (cln1::LEU2-GAL-CLN1-HA3 cln2Δ::TRP1 cln3Δ::HIS3) to log phase in YEP 1% raffinose, 1% galactose. Cells were pelleted, washed twice in YEP without sugar, and then resuspended in YEP 2% glucose for 5 h. From these experiments, 16 BS100 cell fields were analyzed by EM. All samples were processed and photographed by Tamara Howard, a technician in Dr. David Spector's laboratory, Cold Spring Harbor Laboratory. Photomicrographs were scanned and analyzed using Image Reader 1.0 software from FujiFilm Science Lab for Windows (Stamford, CT). In each field of 30–50 cells, the cell with the largest cross-sectional nuclear area was chosen for further analysis, compensating for the fact that most sections were not taken through the center of the nucleus. It should be noted that our fluorescent studies indicate that there is not a strong relationship between nuclear size variability and cell size, so this method should not generate artifactual correlations between nuclear and cell size for that reason. For the chosen cell, nuclear and cell cross-sectional areas were measured.
RESULTS
Nuclear Size Is Altered in Cell Size Mutants
The model of Start entry described in the Introduction predicts that budding yeast mutations that alter cell size control will have no affect on nuclear size. That is, cells bearing Start mutations should have altered karyoplasmic ratios. Cells bearing the hypermorphic mutation CLN3-1 or the deletion mutation whi5Δ pass Start precociously and form populations with small cell size (the Whi phenotype in S. cerevisiae). The Start repressors Sfp1 and Sch9 function in a parallel pathway related to nutrient conditions (Jorgensen et al., 2004). sfp1Δ and sch9Δ mutants exhibit the strongest known Whi phenotypes (Jorgensen et al., 2002). To observe nuclear and nucleolar compartments, we fused CFP and YFP coding sequences to the genomic coding sequences of SEC63 and BUD21. Sec63 is an integral membrane protein that localizes to the endoplasmic reticulum (ER). The ER is contiguous with the outer bilayer of the nuclear envelope and, in budding yeast, a peripheral ER lies beneath the cell membrane (Feldheim et al., 1992). Bud21 is a nonessential nucleolar protein (Dragon et al., 2002). Neither C-terminal fusion caused observable cell size or growth defects (data not shown).
To determine if cell size had an impact on nuclear size, we examined Sec63CFP and Bud21YFP in a strain whose CLN3 allele had been replaced with the hyperactive CLN3-1 allele under control of the GAL1 promoter. When this strain is grown in galactose medium, CLN3-1 overexpression produces populations with small cell size. When this strain is grown in glucose medium, GAL1pr-CLN3-1 is repressed, and the cells are as large as those of a cln3 null strain (data not shown). We found that the large cells had large nuclei and large nucleoli, whereas the small cells had small nuclei and small nucleoli (Figure 1).
To quantitate these differences in nuclear size, the nuclear areas bounded by Sec63CFP in focused optical sections of GAL1pr-CLN3-1 and wild-type cells were determined. Only cells with a single round nucleus were measured, thereby avoiding cells undergoing anaphase and cytokinesis. As each cell field was visualized in only one plane, such two-dimensional measurements are subject to uncertainty caused by the nucleus not being perfectly centered in the plane of focus. We reasoned that, even with such error, measuring several hundred cells from multiple cell fields would allow us to observe differences between the nuclear size distributions. Indeed, the cross-sectional nuclear areas measured in the small cell populations, and the large cell populations generated by CLN3 manipulation were significantly different from the nuclear areas measured in wild-type cells in the same medium (Figure 2A, Table 2, Student's t tests, p < 10−15). The differences in the nuclear areas appeared to be roughly proportional to the differences in cell volume measured with a Coulter particle analyzer (Figure 2B). To estimate nuclear volumes, we assumed that the nuclei were spherical, which is true or nearly true (Winey et al., 1997). The N/C ratio determined from population averages was quite constant, such that nuclear volume was 6–8% of cell volume, even though these alterations in CLN3 activity generated twofold differences in average cell volume (Table 2).
Table 2.
Summary of nuclear and cell size distributions in asynchronous cultures
| Genotype | Carbon source | n | Measured nuclear A (μm2) |
Estimated nuclear V (μm3) |
Measured cell V (μm3) |
N/C volume ratio (%) | |||
|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | ||||
| Wild-type | Glucose | 271 | 2.44 | 0.48 | 2.91 | 0.85 | 41.8 | 16.5 | 7.0 |
| Galactose | 185 | 2.37 | 0.54 | 2.80 | 0.95 | 35.7 | 15 | 7.9 | |
| Raffinose | 287 | 2.09a | 0.51 | 2.33 | 0.86 | 30.9 | 12.9 | 7.5 | |
| GAL1-CLN3-1 | Glucose | 419 | 3.17a | 0.67 | 4.31 | 1.36 | 63 | 27.2 | 6.8 |
| Galactose | 539 | 1.83b | 0.39 | 1.89 | 0.61 | 30.4 | 18.1 | 6.2 | |
| whi5Δ | Glucose | 369 | 2.12a | 0.43 | 2.35 | 0.73 | 33.3 | 13.8 | 7.1 |
| sch9Δ | Glucose | 357 | 1.70a | 0.45 | 1.72 | 0.69 | 26 | 13.1 | 6.6 |
| sfp1Δ | Glucose | 367 | 1.54a | 0.39 | 1.47 | 0.56 | 27.4 | 16 | 5.4 |
Strains were propagated to log phase in synthetic medium with the indicated carbon source, and the area (A) of a nuclear cross-section was determined for n cells. All strains were in the S288c background and were congenic except at the noted alleles. Nuclear volume (V) was estimated under the assumption that each nucleus was spherical. Cell volume distributions were directly measured with a Coulter particle analyzer and a representative distribution was analyzed. The mean and SD of each distribution were calculated.
a The mean nuclear A measurement is significantly different from that for wild-type cells in glucose medium (Student's t test, p < 10−15).
b The mean nuclear A measurement is significantly different from that for wild-type cells in galactose medium (Student's t test, p < 10−27). The difference between the mean nuclear area of wild-type cells in glucose and galactose was not significant (Student's t test, p > 0.05).
We asked if other Whi mutants exhibited decreased nuclear volume. The distributions of nuclear area for the sfp1Δ, sch9Δ, and whi5Δ mutants were significantly different from that of a congenic wild-type strain (Figure 2D, Student's t tests, p < 10−17). The N/C ratio determined from population averages was also between 6 and 8% in wild-type, whi5Δ, and sch9Δ strains, but was somewhat decreased in the sfp1Δ strain (5.4%, Table 2). Carbon source also influences budding yeast cell size (Johnston et al., 1979; Lorincz and Carter, 1979; Tyson et al., 1979). There was a significant decrease in the size of nuclei when wild-type cells were cultured in raffinose as opposed to glucose medium (Figure 2, G and H, Student's t test, p < 10−15), although we did not observe a significant change in nuclear size between glucose and galactose medium (Table 2).
Despite having much smaller average nuclear size than the wild-type control, populations of cells overexpressing CLN3-1 or bearing the whi5Δ mutation were enriched for cells with replicated DNA (Figure 2, C and F), as expected for these types of Whi mutants. This result suggests that DNA content is not, on its own, the determining factor for nuclear volume.
Nuclear Size Increases as Cells Grow
We asked if the correlation between nuclear and cell size we observed with populations of cell size mutants was maintained at the single cell level over the course of a wild-type cell cycle. There are two issues of particular interest. First, the commitment of cells to division during G1-phase might be dependent on the karyoplasmic ratio, so we were interested to see whether or not the nucleus grew during G1-phase. Second, the DNA content of the cell doubles abruptly during S-phase, and we were interested to see whether a sharp increase in nuclear volume accompanied DNA replication.
We quantified the cell and nuclear volume of individual cells growing exponentially. Three different methods were used to mark the nucleus: the nuclear envelope was highlighted by Sec63CFP (as above), GFP was directed to the nucleoplasm by a powerful nuclear localization signal (NLS) to mark the interior of the nucleus, and EM directly visualized the nuclear envelope.
We measured three categories of Sec63CFP-expressing cells cultured in synthetic raffinose medium: unbudded cells, small budded cells (bud was <¼ the size of the mother cell), and medium-to-large budded cells (bud was >¼ the size of the mother cell). These three categories correspond to cells in G1-, S-, and late S/G2-phases of the cell cycle. Nuclear and cell area measurements were obtained from micrographs of live cells, as described above for the cell size mutant strains. There was a strong positive correlation between a cell's size and the size of its nucleus (Figure 3A; r = 0.77, n = 694). There was no observable discontinuity in cell and nuclear size between unbudded cells in late G1-phase and budded cells in S/G2-phase; i.e., there was no evidence that S-phase caused an abrupt increase in nuclear volume.
It was clear that nuclear size increased during G1-phase, a result that confounds a model of how cell size triggers the Start transition (see Introduction). We wanted to determine if the ratio between nuclear and cell volume declined at all during G1-phase. Because the nuclear volume is ∼6–8% of cell volume and we in fact wanted to test if the ratio of nuclear to cytoplasmic volume was declining during G1-phase, in this one instance we estimated cytoplasmic volume for each cell by subtracting the nuclear volume from the cell volume. More accurate estimates of cytoplasmic volume, as opposed to cell volume, are challenging to obtain (see Discussion). The estimated nuclear to cytoplasmic (N/Cyt) volume ratio was plotted against the estimated cytoplasmic volume for unbudded, G1-phase cells (Figure 3B). There was a significant negative correlation in G1-phase cells between these two parameters (r = −0.55, n = 454, Student's t test, p < 0.001). Linear regression of the data for unbudded cells gave the relationship: N/Cyt volume ratio = −0.17%(Vcytoplasm) + 14.39%. Thus, the nucleus grows during G1-phase, but it does not grow quite as fast as the cytoplasm. In our data set, the smallest daughter cells (11–15 μm3 estimated cytoplasmic volume, n = 42) had an average N/Cyt volume ratio of 13%, whereas cells in late G1-phase (27–30 μm3 estimated cytoplasmic volume, n = 67) had an average ratio of 10%.
Multiple z-Section Approach.
We worried that our use of single optical sections might impose some unforeseen bias on the measurements of cell and nuclear size. We therefore subjected fields of cells to small z-series that extended over a 2-μm range, which is about half the width of a haploid yeast cell and chose maximal cell and nuclear cross-sectional areas from these optical sections. This more onerous approach improved the correlation between cell and nuclear cross-sectional areas (Figure 3C, r = 0.82, n = 207 haploid cells, vs. r = 0.77 by the single section method). However, because the qualitative result was unchanged, and the improvement in the r was so small, we concluded that the use of a single optical section was not likely to be an important source of error.
We used the multiple z-section approach, we compared haploid and diploid cells. We found that nuclear and cell size are also highly correlated in diploids and that the haploid and diploid correlations are fairly continuous (Figure 3C). However, in the region where large (presumably 2C) haploid cells and small diploid cells overlapped (∼15–22 μm2), the diploid cells tended to have slightly larger nuclei. This observation is consistent with the aforementioned observation that early G1-phase cells have a higher N/C ratio (Figure 3B). Alternatively, this very slight shift in the curve between large haploid cells and small diploid cells in Figure 3C between 15 and 22 μm2 may reflect different sphericality in these populations. Large haploid cells are predominantly budded (Figure 3A), whereas small diploids are predominantly unbudded daughters. Assuming that the haploid and diploid cells in this size range have nuclei with equal sphericality, it would be expected that the cross-sectional area of budded haploid cells would overrepresent the volume of those cells relative to the more spherical unbudded diploid cells. Therefore, the actual nuclear versus cell volume distributions for haploid and diploid cells may in fact be perfectly continuous. Our measurements cannot resolve such subtle points.
GFP-NLS.
As a second independent method of measuring the nucleus, cells were transformed with a plasmid expressing GFP fused to two SV40 nuclear localization signals (Edgington and Futcher, 2001). This GFP-NLS was efficiently localized to the nucleus, and the disk of GFP seen by fluorescent microscopy gave an estimated cross-sectional area of the nucleus. Fractions of small unbudded cells and fractions of budded cells were obtained by elutriation of an exponential phase, ethanol culture and live cells were examined with differential interference contrast (DIC) and epifluorescence microscopy. Three categories of cells were defined. Class I cells were unbudded with a single nucleus (G1-phase). Class II cells were budded with a single nucleus (S/G2/M-phase). Class III cells were budded with two nuclei (telophase).
The GFP-NLS reporter confirmed the results garnered with the Sec63CFP reporter. Nuclear size expanded through G1-phase, because there was a significant positive correlation between cell and nuclear cross-sectional areas in class I cells (r = 0.65, n = 60, Student's t test, p < 0.001, Figure 4A). Again, there was no obvious jump in nuclear volume within the size range of cells initiating and undergoing DNA replication (∼10–15 μm2, where class I and class II cells overlap).
Figure 4.
Two independent microscopic methods confirm the correlation between cell and nuclear size. (A) Cells cultured in synthetic, 3% ethanol media and expressing a nuclear localized GFP were elutriated into different cell size fractions. Cells from a range of size fractions were imaged, and the cell and nuclear cross-sectional area measured. The nuclear boundary was defined by the edge of the green fluorescence, whereas the cell boundary was determined from a DIC image. ○, unbudded cells (G1-phase, n = 60 cells); ■, budded cells with a single nucleus (S/G2/M-phase, n = 17 cells). (B) The cell and nuclear areas of 17 cells in telophase plotted as horizontal bars. Telophase cells had two nuclei, one in the mother and one in the daughter cell. The proportion of the combined cell or nuclear cross-sectional area that is derived from the mother compartment (■) and the daughter compartment (□) is shown. (C) EM micrographs of representative G1-phase cells. The smaller cell was derived from elutriation of cells cultured in ethanol medium, and the larger cell was arrested by G1 cyclin depletion in glucose medium. (D) The cross-sectional area of the nucleus and the cell was determined from EM micrographs. Area is reported in arbitrary units. Fifty-seven cells from an elutriation in ethanol medium (black circles with gray interior), 12 cells from elutriation in glucose medium (■), and 16 cells from an arrest caused by G1 cyclin depletion (▵) were measured. All cells were in G1-phase.
We asked if nuclear volume partitions symmetrically or asymmetrically at mitosis with the class III cells. When cultured in ethanol medium, the daughter compartment is always much smaller than the mother compartment at cytokinesis and so can be unambiguously identified. In these telophase cells, the size of the daughter nucleus was, in most cases, also much smaller than that of the mother (Figure 4B).
EM.
Finally, as a third method for measuring nuclear size, we used EM to look at the nuclear envelope directly. Cells were fixed, sectioned, and imaged by transmission electron microscopy (Figure 4C). In this method, the section is necessarily taken at a random position with respect to the center of the cell. Therefore, to choose a section that happened to be taken close to the center of some nucleus, we inspected fields of 30–50 cells with sampled nuclei and chose the cell with the largest cross-sectional nuclear area for further analysis. For the cell so chosen, we measured both cross-sectional nuclear area and cross-sectional cell area. By simple geometry, it can be estimated that this method should yield a nuclear cross-section that is within 2% of the maximal cross-sectional area of the average nucleus in the field more than 95% of the time. Given that the nucleus is not necessarily in the center of the cell and that yeast cells are not exactly spherical, it is expected that the corresponding cellular cross-sectional areas will display a fairly broad distribution. Still, we anticipated that by measuring many fields of cells the technique would suffice to determine if nuclear and cell size are correlated.
Three kinds of G1-phase yeast cells were prepared in order to generate a broad range of cell sizes: small G1 cells were isolated by elutriation from exponential, ethanol cultures; medium size G1 cells were isolated by elutriation from exponential, glucose cultures; and very large G1 cells were generated by depletion of G1 cyclins (see Materials and Methods). The largest cells quantified were ∼10-fold larger in volume than the smallest cells; examples are shown in Figure 4C. Cross-sectional cell and nuclear area was measured for cells of all three types, and once again, it was apparent that nuclear area increased with cell area, even though all cells were in G1-phase (Figure 4D, r = 0.77, n = 85, Student's t test, p < 0.001. Although there are theoretical reasons to think the EM method could give less accurate estimates of cell size than other methods (see above), nevertheless the r for the EM method (0.77) was exactly the same as the r for the single optical slice/Sec63CFP method and was quite similar to the r for the z-series method (r = 0.82). This result could suggest that our measurement errors may be relatively small compared with real biological variation in cell and nuclear size.
Continual Cell Growth Is Not Required To Maintain Nuclear Volume
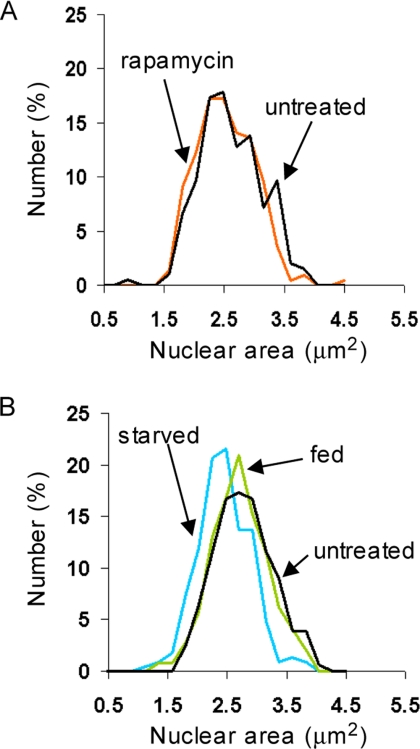
The mechanisms that cause nuclear and cell size to be so well correlated remain to be discovered. Because the absolute rate of cell growth increases with cell size in growing budding yeast (Elliott and McLaughlin, 1978), we wondered if nuclear size might reflect the absolute growth rate of the cell. To investigate this possibility, we asked if continual cell growth was required to maintain the large nuclear size exhibited by wild-type cells growing in glucose medium. We challenged such cells with two conditions that rapidly abrogate cell growth: repression of the TOR kinase by rapamycin, and carbon starvation. In the short-term (36–60 min after addition), rapamycin had no effect on the distribution of nuclear sizes (Figure 5A), but noticeably decreased nucleolar size (data not shown), as was previously reported (Tsang et al., 2003). In the longer term (∼120 min after addition), rapamycin did decrease nuclear size, but this decline accompanied the expected accumulation of small G1-phase cells in the culture (data not shown). After 50 min of carbon starvation, there was a slight but significant decrease in nuclear size (Figure 5B; Student's t test, p < 10−7). This decrease in nuclear size was accompanied, however, by a ∼10% reduction in average cell volume, as measured with a Coulter particle analyzer (data not shown), presumably due to starved cells arresting in G1-phase. In short, neither rapamycin nor carbon starvation strongly reduced nuclear size. The changes we did observe in nuclear size could not be dissociated from approximately proportional changes in cell size. Continual cell growth is apparently not required to maintain a large nuclear size. It is still possible, however, that the nucleus does expand in proportion to the rate of cellular growth, as opposed to cell volume per se, but that it is incapable of compression, at least over the time periods examined here.
Figure 5.
Blocking cell growth and repressing ribosome biogenesis does not strongly influence nuclear size. (A) Rapamycin treatment has no impact on nuclear size in the short-term. Wild type cells expressing Sec63CFP were propagated to log phase in synthetic glucose medium, treated with 0.2 μg/ml rapamycin, and visualized 36–60 min later. DMSO carrier had no effect on nuclear size (data not shown). Rapamycin, n = 220 cells; untreated, n = 196 cells. (B) Carbon starvation causes small decreases in nuclear size. Wild-type cells expressing Sec63CFP were propagated to log phase in synthetic glucose medium at 30°C. Aliquots of cells were then washed and resuspended in prewarmed 30°C synthetic medium with (fed) or without (starved) glucose. Starved cells were visualized 47–52 min after washing (n = 227 cells), and glucose-fed cells were visualized 54–58 min after washing (n = 244 cells). Untreated, n = 156 cells.
Accumulation of Proteins in the Nucleus Does Not Change Nuclear Size
We wondered if protein accumulation in the nucleus would suffice to expand nuclear size. Binding to the nuclear export karyopherin Crm1 is the predominant conduit for nuclear protein export in yeast. Although S. cerevisiae is naturally resistant to LMB, replacement of the endogenous allele of CRM1 with the crm1-T539C allele creates LMB sensitivity. In crm1-T539C cells, Crm1-mediated protein export and cell viability are sensitive to LMB (Neville and Rosbash, 1999). Within 15 min of LMB addition to the medium of crm1-T539C mutant yeast, nuclear export of the large and small ribosomal subunits stalled (Gadal et al., 2001; Moy and Silver, 2002). Crm1 also mediates the export of many additional proteins in yeast (Cullen, 2003), but LMB treatment had no obvious effect on the nuclear size of crm1-T539C mutant yeast expressing Sec63CFP after 5, 15, or 30 min (Figure 6).
The mechanisms that do couple cell and nuclear size remain to be characterized.
DISCUSSION
It was noted by Theodor Boveri a century ago that “The constant, which we must accept as something given and not at present further analyzable, is the fixed proportion between nuclear volume and protoplasmic volume, namely, the karyoplasmic ratio.” (Wilson, 1925). This statement was echoed recently by Cavalier-Smith (2005), who wrote that “The invariant karyoplasmic ratio is a basic fact of cell biology established for more than a century (but forgotten by two generations of textbooks, which are insufficiently quantitative) and a necessary consequence of the optimization of growth processes by selection for rapid and efficient cell reproduction.”
In this study of the budding yeast nucleus, we have discovered that the karyoplasmic ratio is actively, although not perfectly, maintained in budding yeast. In cell size mutants, nuclear size is altered in rough proportion with cell size. Similarly, as wild-type cells grow, nuclear size increases. The close relationship between cell and nuclear size appeared to overwhelm any direct, immediate nuclear expansions caused by replicating the genome in S-phase, at least at the level of precision afforded by this analysis. A previous analysis of nuclear volume during the S. cerevisiae cell cycle could not have arrived at this conclusion, as the authors did not monitor cell size (Winey et al., 1997). This previous study could only conclude that cells in early mitosis had larger nuclei than cells in S-phase, whereas cells in S-phase had larger nuclei than cells in G1-phase. If DNA content were an important and direct factor in setting nuclear volume, a step-like increase in nuclear volume would be expected during S-phase, and we saw no sign of such an increase in either haploid or diploid cells. That nuclear size is more closely correlated with cell size than DNA content is further demonstrated by the small average nuclear size of whi5Δ cells and cells overexpressing CLN3-1. FACS analysis demonstrates that asynchronous populations of both strains are enriched for cells with replicated genomes (Figure 2, C and F).
In one model for how budding yeast measure the critical cell size for cell cycle entry, the highly unstable Cln3 protein increases in concentration in a constant volume nucleus as cells grow and increase their absolute translational capacity (Futcher, 1996). In this model, a constant G1 nuclear volume acts as the metric against which ribosome content and hence cell volume are measured. But we have demonstrated that nuclear volume increases during G1-phase. This observation renders the model very unlikely, although the fact that the nucleus does not grow quite as quickly as the cytoplasm means the model cannot be completely ruled out. Our observations accommodate the possibility that increased nuclear size is what triggers the Start transition, as previously suggested for S-phase entry in mammalian cells (Yen and Pardee, 1979). Alternatively, some aspect of DNA quantity and not nuclear volume may be the metric against which cell volume is gauged before Start, because the critical cell size requirement is nearly twice as large in diploid budding yeast (Lorincz and Carter, 1979).
Although we estimated cytoplasmic volume by subtracting estimated nuclear volume from estimated cell volume in Figure 3B, accurately measuring the volume of the cytoplasm is much more complicated. Even before any measurements, it would need to be decided which organelles to include in the definition of cytoplasm. One would almost certainly want to subtract vacuolar volume, which is comparable to the nuclear volume (Figure 4C). Measuring vacuolar volume would be complicated by the often eccentric shape of this organelle. In addition, the volume of the vacuole may not increase proportionately with cell size. It has been previously noted that cln3Δ mutant cells, which are large relative to wild-type, have disproportionate increases in total vacuolar volume (Han et al., 2003). The allometric relationship between vacuole size and cell size remains to be further characterized, as does the possible influence of vacuolar volume on cell size control at Start.
It is generally presumed that the size of the nucleus is set directly by the physical bulk of the chromatin. As the nucleus must house the chromatin, DNA content must directly limit the minimal nuclear size. But even in the smallest yeast, we saw no flattening of the correlation between cell and nuclear size, suggesting that this minimal size has not been reached. Indeed, all of our evidence suggests that DNA content does not directly control nuclear size in growing budding yeast. We therefore anticipate that changes in cell ploidy impact nuclear size indirectly in yeast. By raising the critical cell size threshold at Start (Lorincz and Carter, 1979), increased DNA content will increase nuclear size due to the unknown mechanisms that maintain the N/C ratio, not because of the greater space occupied by the larger genome. DNA content may also affect nuclear size indirectly in other organisms.
We were unable to obtain any clues as to the mechanism that coordinates nuclear and cell size. Blocking cell growth had little effect in the short-term (<60 min) on nuclear size (Figure 5). In terms of sheer magnitude, ribosome biogenesis is the dominant process in the yeast cellular economy (Warner, 1999). Rapamycin treatment rapidly represses ribosome synthesis rates and shrinks the nucleolus (Warner, 1999; Tsang et al., 2003), but had no such effects on nuclear size (Figure 5A). In the inverse experiment, causing ribosomal subunits to accumulate in the nucleus by blocking Crm1-mediated nuclear export also had no effect on nuclear size (Figure 6). As yeast growing under these conditions double every ∼90 min, in 15 min a yeast will synthesize ∼⅙ of the total ribosomes synthesized each cell cycle. If nuclear export is completely blocked, then ∼⅙ of the total ribosomes synthesized each cell cycle will have accumulated in a space that is the volume of the cell (Table 2), resulting in nuclear ribosomal concentrations twice that of the cytoplasm. These considerations assume no negative feedback to ribosome synthesis rates as well as full blockage of ribosome export during the 15 min. Despite these caveats, it seems reasonable to suppose that these brief LMB treatments strongly increased nuclear protein concentration, and yet there was no effect on nuclear size.
How then might the yeast nucleus expand? Are there dedicated mechanisms that actively maintain the N/C ratio by controlling the density of the chromatin or the structure of the nuclear envelope? Notably, yeast lack the lamin-based meshwork that underlies the nuclear envelope in metazoan cells (Taddei et al., 2004). Recently, three genes whose loss leads to rampant nuclear membrane expansion have been characterized in budding yeast. All three gene products appear to repress the transcription of enzymes involved in phospholipid biosynthesis (Santos-Rosa et al., 2005). Remarkably, the nuclear membrane in these cells is no longer round, but displays long extensions that are filled with nucleolar material (Campbell et al., 2006). It is not clear whether overall nuclear or cell volume is markedly increased in these mutant cells. Mutations that alter nuclear volume without altering cell volume could be very informative.
There are many more interesting questions about nuclear size. Budding yeast do not break down the nuclear envelope during mitosis. But after this closed mitosis, daughter cells clearly have smaller nuclei than mother cells (Figure 4B and noted by Gasser, 2002). Is the nuclear volume segregated differentially at anaphase? Or is the nuclear mass segregated equally followed by rapid adjustments in nuclear volume? On a related note, can the nucleus shrink in volume without cell division? It has been demonstrated that parts of the nucleus are engulfed by the vacuole in a process termed piecemeal microautophagy of the nucleus (Roberts et al., 2003).
Finally, how is nuclear volume related to cell growth in other organisms? Nuclear volume in metazoan cells can change without alterations in cellular DNA content. A handful of studies with mammalian cells have suggested, but not convincingly demonstrated, that the nucleus may increase in volume before S-phase (Maul et al., 1972; Yen and Pardee, 1979; Fidorra et al., 1981). It is evident in many animal embryos that nuclear volume shrinks during embryonic cleavage cycles as cell volume decreases (Wilson, 1925; Gerhart, 1980; Sulston et al., 1983). Inversely, injecting the nuclei of HeLa cells into much larger frog oocytes can lead to 10-fold or greater increases in nuclear volume, while fusing hen erythrocytes with much larger HeLa cells leads to expansion of the erythrocyte nucleus (Harris, 1967; Gurdon, 1976). In the latter case, the expansion was shown to occur independently of DNA replication (Harris, 1967). Pathologists have used aspects of nuclear morphology, including size, to diagnose and grade cancers for many decades (Zink et al., 2004). These and many other studies illustrate that nuclear volume is dynamic, but the mechanisms causing nuclear volume alterations remain largely unknown.
ACKNOWLEDGMENTS
We thank Daniel Durocher (Samuel Lunenfeld Research Institute, Mount Sinai Hospital, Toronto, Ontario, Canada) for providing the LMB-sensitive strain. We thank Tamara Howard, David Spector, and other members of the Cold Spring Harbor EM facility, as well as Marc Angeli, for technical assistance. This work was supported by a Canadian Institutes of Health Research (CIHR) grant to M.T., a Canada Research Chair to M.T., a National Institutes of Health (NIH) Grant GM077874 and a grant from The Ted Nash Long Life Foundation to B.L.S., and by a NIH award, RO1 GM039978, to B.F.
Abbreviations used:
- CFP
cyan fluorescent protein
- DIC
differential interference contrast
- EM
electron microscopy
- ER
endoplasmic reticulum
- GFP
green fluorescent protein
- N/C
nuclear to cell volume ratio
- YFP
yellow fluorescent protein.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-10-0973) on June 27, 2007.
REFERENCES
- Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., Boeke J. D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Campbell J. L., Lorenz A., Witkin K. L., Hays T., Loidl J., Cohen-Fix O. Yeast nuclear envelope subdomains with distinct abilities to resist membrane expansion. Mol. Biol. Cell. 2006;17:1768–1778. doi: 10.1091/mbc.E05-09-0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. Economy, speed and size matter: evolutionary forces driving nuclear genome miniaturization and expansion. Ann. Bot. (Lond.) 2005;95:147–175. doi: 10.1093/aob/mci010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M., Schub O., Andrews B. G1 transcription factors are differentially regulated in Saccharomyces cerevisiae by the Swi6-binding protein Stb1. Mol. Cell. Biol. 2003;23:5064–5077. doi: 10.1128/MCB.23.14.5064-5077.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross F. R., Blake C. M. The yeast Cln3 protein is an unstable activator of Cdc28. Mol. Cell. Biol. 1993;13:3266–3271. doi: 10.1128/mcb.13.6.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csikasz-Nagy A., Battogtokh D., Chen K. C., Novak B., Tyson J. J. Analysis of a generic model of eukaryotic cell-cycle regulation. Biophys. J. 2006;90:4361–4379. doi: 10.1529/biophysj.106.081240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen B. R. Nuclear RNA export. J. Cell Sci. 2003;116:587–597. doi: 10.1242/jcs.00268. [DOI] [PubMed] [Google Scholar]
- de Bruin R.A.M., McDonald W. H., Kalashnikova T. I., Yates J., III, Wittenberg C. Cln3 activates G1-specific transcription via phosphorylation of the SBF-bound repressor Whi5. Cell. 2004;117:887–898. doi: 10.1016/j.cell.2004.05.025. [DOI] [PubMed] [Google Scholar]
- Dragon F., et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002;417:967–970. doi: 10.1038/nature00769. [DOI] [PubMed] [Google Scholar]
- Edgington N. P., Futcher A. B. Relationship between the function and the location of G1 cyclins in S. cerevisiae. J. Cell Sci. 2001;114:4599–4611. doi: 10.1242/jcs.114.24.4599. [DOI] [PubMed] [Google Scholar]
- Elliott S. G., McLaughlin C. S. Rate of macromolecular synthesis through the cell cycle of the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 1978;75:4384–4388. doi: 10.1073/pnas.75.9.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser G. Maintenance of normal structure in heteroploid salamander larvae through compensation of changes in cell size by adjustment of cell number and cell shape. J. Exp. Zool. 1945;100:445–455. doi: 10.1002/jez.1401000310. [DOI] [PubMed] [Google Scholar]
- Feldheim D., Rothblatt J., Schekman R. Topology and functional domains of Sec63p, an endoplasmic reticulum membrane protein required for secretory protein translocation. Mol. Cell. Biol. 1992;12:3288–3296. doi: 10.1128/mcb.12.7.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidorra J., Mielke T., Booz J., Feinendegen L. E. Cellular and nuclear volume of human cells during the cell cycle. Radiat. Environ. Biophys. 1981;19:205–214. doi: 10.1007/BF01324188. [DOI] [PubMed] [Google Scholar]
- Futcher B. Cyclins and the wiring of the yeast cell cycle. Yeast. 1996;12:1635–1646. doi: 10.1002/(SICI)1097-0061(199612)12:16%3C1635::AID-YEA83%3E3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Futcher B. Cell cycle synchronization. Methods Cell Sci. 1999;21:79–86. doi: 10.1023/a:1009872403440. [DOI] [PubMed] [Google Scholar]
- Gadal O., Strauss D., Kessl J., Trumpower B., Tollervey D., Hurt E. Nuclear export of 60s ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol. Cell. Biol. 2001;21:3405–3415. [Google Scholar]
- Gasser S. M. Visualizing chromatin dynamics in interphase nuclei. Science. 2002;296:1412–1416. doi: 10.1126/science.1067703. [DOI] [PubMed] [Google Scholar]
- Gerhart J. C. Mechanisms regulating pattern formation in the amphibian egg and early embryo. In: Goldberger R. F., editor. Biological Regulation and Development, Molecular Organization and Cell Function. Vol. 2. New York: Plenum Press; 1980. pp. 133–316. [Google Scholar]
- Gregory T. R. Genome size evolution in animals. In: Gregory T. R., editor. The Evolution of the Genome. New York: Elsevier; 2005. pp. 3–87. [Google Scholar]
- Gulliver G. Observations on the sizes and shapes of the red corpuscles of the blood of vertebrates, with drawings of them to a uniform scale, and extended and revised tables of measurements. Proc. Zool. Soc. Lond. 1875;1875:474–495. [Google Scholar]
- Gurdon J. B. Injected nuclei in frog oocytes: fate, enlargement, and chromatin dispersal. J. Embryol. Exp. Morphol. 1976;36:523–540. [PubMed] [Google Scholar]
- Han B. K., Aramayo R., Polymenis M. The G1 cyclin Cln3p controls vacuolar biogenesis in Saccharomyces cerevisiae. Genetics. 2003;165:467–476. doi: 10.1093/genetics/165.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris H. The reactivation of the red cell nucleus. J. Cell Sci. 1967;2:23–32. doi: 10.1242/jcs.2.1.23. [DOI] [PubMed] [Google Scholar]
- Henery C. C., Bard J. B., Kaufman M. H. Tetraploidy in mice, embryonic cell number, and the grain of the developmental map. Dev. Biol. 1992;152:233–241. doi: 10.1016/0012-1606(92)90131-y. [DOI] [PubMed] [Google Scholar]
- Johnston G. C., Ehrhardt C. W., Lorincz A., Carter B.L.A. Regulation of cell size in the yeast Saccharomyces cerevisiae. J. Bacteriol. 1979;137:1–5. doi: 10.1128/jb.137.1.1-5.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston G. C., Pringle J. R., Hartwell L. H. Coordination of growth with cell division in the yeast Saccharomyces cerevisiae. Exp. Cell Res. 1977;105:79–98. doi: 10.1016/0014-4827(77)90154-9. [DOI] [PubMed] [Google Scholar]
- Jorgensen P., Nishikawa J. L., Breitkreutz B. J., Tyers M. Systematic identification of pathways that couple cell growth and division in yeast. Science. 2002;297:395–400. doi: 10.1126/science.1070850. [DOI] [PubMed] [Google Scholar]
- Jorgensen P., Rupes I., Sharom J. R., Schneper L., Broach J. R., Tyers M. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004;18:2491–2505. doi: 10.1101/gad.1228804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P., Tyers M. How cells coordinate growth and division. Curr. Biol. 2004;14:R1014–R1027. doi: 10.1016/j.cub.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Lorincz A., Carter B.L.A. Control of cell size at bud initiation in Saccharomyces cerevisiae. J. Gen. Microbiol. 1979;113:287–295. [Google Scholar]
- Maul G. G., Maul H. M., Scogna J. E., Lieberman M. W., Stein G. S., Hsu B. Y., Borun T. W. Time sequence of nuclear pore formation in phytohemagglutinin-stimulated lymphocytes and in HeLa cells during the cell cycle. J. Cell Biol. 1972;55:433–447. doi: 10.1083/jcb.55.2.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. E., Cross F. R. Distinct subcellular localization patterns contribute to functional specificity of the Cln2 and Cln3 cyclins of Saccharomyces cerevisiae. Mol. Cell. Biol. 2000;20:542–555. doi: 10.1128/mcb.20.2.542-555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer R. K. Radiobiological and genetic studies on a polyploid series (haploid to hexaploid) of Saccharomyces cerevisiae. Rad. Res. 1958;9:312–326. [PubMed] [Google Scholar]
- Moy T. I., Silver P. A. Requirements for the nuclear export of the small ribosomal subunit. J. Cell Sci. 2002;115:2985–2995. [Google Scholar]
- Neville M., Rosbash M. The NES-Crm1p export pathway is not a major mRNA export route in Saccharomyces cerevisiae. EMBO J. 1999;18:3746–3756. doi: 10.1093/emboj/18.13.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts P., Moshitch-Moshkovitz S., Kvam E., O'Toole E., Winey M., Goldfarb D. S. Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol. Biol. Cell. 2003;14:129–141. doi: 10.1091/mbc.E02-08-0483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupes I. Checking cell size in yeast. Trends Genet. 2002;18:479–485. doi: 10.1016/s0168-9525(02)02745-2. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H., Leung J., Grimsey N., Peak-Chew S., Siniossoglou S. The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J. 2005;24:1931–1941. doi: 10.1038/sj.emboj.7600672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider B. L., Yang Q. H., Futcher A. B. Linkage of replication to start by the Cdk inhibitor Sic1. Science. 1996;272:560–562. doi: 10.1126/science.272.5261.560. [DOI] [PubMed] [Google Scholar]
- Slater D. N., Rice S., Stewart R., Melling S. E., Hewer E. M., Smith J. H. Proposed Sheffield quantitative criteria in cervical cytology to assist the grading of squamous cell dyskaryosis, as the British Society for Clinical Cytology definitions require amendment. Cytopathology. 2005;16:179–192. doi: 10.1111/j.1365-2303.2005.00271.x. [DOI] [PubMed] [Google Scholar]
- Sulston J. E., Schierenberg E., White J. G., Thomson J. N. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Taddei A., Hediger F., Neumann F. R., Gasser S. M. The function of nuclear architecture: a genetic approach. Annu. Rev. Genet. 2004;38:305–345. doi: 10.1146/annurev.genet.37.110801.142705. [DOI] [PubMed] [Google Scholar]
- Tsang C. K., Bertram P. G., Ai W., Drenan R., Zheng X. F. Chromatin-mediated regulation of nucleolar structure and RNA Pol I localization by TOR. EMBO J. 2003;22:6045–6056. doi: 10.1093/emboj/cdg578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyers M., Tokiwa G., Futcher B. Comparison of the Saccharomyces cerevisiae G1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO J. 1993;12:1955–1968. doi: 10.1002/j.1460-2075.1993.tb05845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyers M., Tokiwa G., Nash R., Futcher B. The Cln3-Cdc28 kinase complex of S. cerevisiae is regulated by proteolysis and phosphorylation. EMBO J. 1992;11:1773–1784. doi: 10.1002/j.1460-2075.1992.tb05229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson C. B., Lord P. G., Wheals A. E. Dependency of size of Saccharomyces cerevisiae cells on growth rate. J. Bacteriol. 1979;138:92–98. doi: 10.1128/jb.138.1.92-98.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner J. R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- Wilson E. B. New York: Macmillan; 1925. The Cell in Development and Heredity. [Google Scholar]
- Winey M., Yarar D., Giddings T. H., Mastonarde D. N. Nuclear pore complex number and distribution throughout the Saccharomyces cerevisiae cell cycle by three-dimensional reconstruction from electron micrographs of nuclear envelopes. Mol. Biol. Cell. 1997;8:2119–2132. doi: 10.1091/mbc.8.11.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen A., Pardee A. B. Role of nuclear size in cell growth initiation. Science. 1979;204:1315–1317. doi: 10.1126/science.451539. [DOI] [PubMed] [Google Scholar]
- Zink D., Fischer A. H., Nickerson J. A. Nuclear structure in cancer cells. Nat. Rev. Cancer. 2004;4:677–687. doi: 10.1038/nrc1430. [DOI] [PubMed] [Google Scholar]