Abstract
The preemptive quality control (pQC) pathway protects cells from acute endoplasmic reticulum (ER) stress by attenuating translocation of nascent proteins despite their targeting to translocons at the ER membrane. Here, we investigate the hypothesis that the DnaJ protein p58IPK plays an essential role in this process via HSP70 recruitment to the cytosolic face of translocons for extraction of translocationally attenuated nascent chains. Our analyses revealed that the heightened stress sensitivity of p58−/− cells was not due to an impairment of the pQC pathway or elevated ER substrate burden during acute stress. Instead, the lesion was in the protein processing capacity of the ER lumen, where p58IPK was found to normally reside in association with BiP. ER lumenal p58IPK could be coimmunoprecipitated with a newly synthesized secretory protein in vitro and stimulated protein maturation upon overexpression in cells. These results identify a previously unanticipated location for p58IPK in the ER lumen where its putative function as a cochaperone explains the stress-sensitivity phenotype of knockout cells and mice.
INTRODUCTION
Impairment or saturation of the protein maturation machinery of the endoplasmic reticulum (ER) causes protein misfolding stress in the ER lumen. The unfolded protein response (UPR) consists principally of two broad strategies for dealing with ER stress: increased clearance of misfolded proteins from the ER lumen and reduction of new protein influx into the ER. These two strategies are temporally separate phases of the UPR. Reduced influx occurs rapidly and transiently to prevent exacerbation of an already perturbed ER, whereas improved protein processing capacity in the ER is dependent on transcriptional up-regulation of maturation and degradation machinery. Over time, homeostasis is reestablished to restore protein maturation and substrate flux through the ER (Rutkowski and Kaufman, 2004).
The different responses of the UPR are initiated principally by the ER-resident stress sensors ATF6, IRE1, and PERK (Schröder and Kaufman, 2005). Although the pathways leading from activation of these sensors to transcriptional induction are fairly well described, the pathways that suppress ER protein influx are less clear. The best characterized of these latter pathways is translational attenuation by PERK, a kinase that phosphorylates the α subunit of the eukaryotic translation initiation factor eIF2. This phosphorylation leads to rapid inhibition of protein synthesis without any obvious specificity for substrates destined for the ER. Recently, two new pathways for ER-selective reduction of substrate burden have been described. One involves the selective degradation of ER-associated mRNAs by the endoribonuclease activity of activated IRE1 (Hollien and Weissman, 2006). The other pathway, termed preemptive quality control (pQC), triages nascent polypeptides for rapid cytosolic proteasomal degradation before their entry into a stressed ER (Kang et al., 2006).
Although the mechanistic basis of pQC is not yet fully elucidated, the first step is mediated by signal sequence-dependent attenuation of protein translocation after nascent polypeptides have been targeted to the ER translocon. This process involves yet unidentified lumenal factor(s), possibly including BiP, whose titration during acute ER stress leads to reversible inhibition of the initiation of translocation (Kang et al., 2006). The idea that the availability of lumenal proteins could influence translocation is consistent with previous observations demonstrating that lumenal chaperones can either facilitate forward translocation or prevent backward slipping of translocating nascent chains (Nicchitta and Blobel, 1993; Brodsky et al., 1995; Matlack et al., 1999; Tyedmers et al., 2003). Thus, the initial commitment to ER translocation may be linked in a substrate-selective manner to chaperone availability in the lumen, providing a means to match substrate flux with maturation capacity.
The next step in the pQC pathway is presumably the release of translocationally attenuated nascent chains from the translocon for subsequent delivery to the proteasome. Although the mechanism of this release is poorly understood, it might involve the DnaJ family protein p58IPK. Best known as an inhibitor of the eIF2α protein kinases PKR (Lee et al., 1994) and PERK (Yan et al., 2002; van Huizen et al., 2003), p58IPK was recently proposed to have an independent function in protection from ER stress (Oyadomari et al., 2006). In that study, p58IPK was proposed to interact with both ER translocons and cytosolic HSP70, thereby recruiting cytosolic chaperone and degradation machinery to the translocon for the clearance of translocationally stalled nascent proteins. Because this p58-dependent extraction of nascent chains would be necessary to prevent ongoing translocation into the stressed ER, p58IPK was proposed to be an important factor in reducing substrate influx into the ER during acute stress. Indeed, p58−/− cells accumulate higher levels of misfolded proteins upon prolonged stress (Oyadomari et al., 2006) and p58−/− mice show a mild diabetic phenotype (Ladiges et al., 2005) similar to mice compromised in other UPR-mediated aspects of ER protection (Harding et al., 2001; Scheuner et al., 2001, 2005).
Although it is clear that p58IPK is transcriptionally induced as part of the ER stress response, several aspects of its putative function in translocational attenuation and pQC remain puzzling. First, the PERK-inhibitory function of p58IPK would act at cross-purposes to its putative role in reducing substrate burden on the ER. During stress, p58IPK would seem to simultaneously increase substrate generation (by preventing PERK-dependent eIF2α phosphorylation) while putatively acting to reduce substrate entry into the ER. Second, based on the available cryoelectron microscopic structures of native mammalian ribosome–translocon complexes (Menetret et al., 2005), it is unclear how a large protein complex composed minimally of p58IPK and HSP70 could have access to the cytosolic face of an engaged translocon. And finally, no substrates were directly demonstrated to be reduced in their translocation in a p58-dependent manner during either normal or ER-stressed conditions (Oyadomari et al., 2006). These and other considerations led us to investigate the relationship between p58IPK and translocational attenuation during ER stress. Our analysis shows that p58IPK is localized in the ER lumen where it interacts with BiP and may function as a cochaperone. Because p58IPK cannot recruit chaperones to the cytosolic side of the translocon from this location, it proved dispensable for translocational attenuation during pQC. We therefore propose that the principal role of p58IPK in protecting the stressed ER is from its lumenal cochaperone function, and not any putative function in stress-dependent translocational attenuation and degradation.
MATERIALS AND METHODS
Materials
p58+/+ and p58−/− mouse embryonic fibroblasts (MEFs) were prepared as described previously (Ladiges et al., 2005). Endogenous p58IPK detected by immunoblot used monoclonal antibody 9F10 (Barber et al., 1994). Rabbit p58-specific antiserum used for immunoprecipitation was raised against a KLH-conjugated synthetic peptide encoding residues 496–504 of mouse p58IPK. Specificity was confirmed by immunoblot and immunoprecipitation by using p58+/+ and p58−/− MEFs. Polyclonal BiP antiserum for immunoprecipitations was a generous gift of Peter Arvan (University of Michigan). Antibodies against translocon-associated protein (TRAP)α, SEC61α, and SEC61β have been described previously (Fons et al., 2003). Other antibodies were obtained commercially: protein disulfide isomerase (PDI) (Assay Designs, Ann Arbor, MI), heat-shock complex (Hsc)/HSP70 (SPA-820; Assay Designs), BiP (BD Biosciences, San Jose, CA), SRP54 (BD Biosciences), transferrin receptor (TfR) (Zymed Laboratories, South San Francisco, CA), α-actin (MP Biomedicals, Irvine, CA), 80K-H (BD Biosciences), and calreticulin (Assay Designs). Tunicamycin (TM) and thapsigargin (TG) were from EMD Biosciences (San Diego, CA), and they were dissolved in dimethyl sulfoxide (DMSO). Dithiothreitol (DTT) was from Roche Diagnostics (Indianapolis, IN), and it was dissolved in water. Digitonin was from Calbiochem (“high-purity” grade). Because of its isolation from natural sources, digitonin is at most 80% pure, and it requires additional purification for reliable behavior in membrane solubilization and biochemical fractionation (Görlich and Rapoport, 1993; Fons et al., 2003). In brief, solid digitonin was dissolved in boiling water (6 g/100 ml), cooled to room temperature, and equilibrated at 4°C. Precipitates were removed by centrifugation, filtered, and passed successively through cation and anion exchange resins at pH 7 (Fons et al., 2003). The reversible lysine-reactive cross-linker dithiobis[succinimindylpropionate] (DSP) from Pierce Chemical (Rockford, IL) was dissolved in DMSO immediately before use. Reagents for in vitro transcription, translation, and translocation assays were prepared and used as described previously (Fons et al., 2003, and references therein). Hun-7293 was obtained from D. Boger (The Scripps Research Institute), and it was tested in translocation assays of vascular cell adhesion molecule (VCAM)1 (our unpublished data) to have the same properties as those previously published for Cotransin and CAM741 (Besemer et al., 2005; Garrison et al., 2005). The p58IPK sequence was analyzed using SignalP 3.0 (Bendtsen et al., 2004).
Plasmid Constructions
Constructs were made using standard methods. p58IPK was cloned by reverse transcription-polymerase chain reaction (RT-PCR) amplification of RNA isolated from MEFs. A C-terminal hemagglutinin (HA) tag and either one (after Asn504 immediately before the HA tag) or three acceptor sites (the aforementioned plus two inserted after Pro496) for N-linked glycosylation near the C terminus were added by PCR. The final amino acid sequence for p583CHO constructs (with added amino acids underlined and glycosylation acceptor sites bold) was … QGFNPFSSGGPNGSNGSGPFRFKFHFNGTYPYDVPDYA-Stop. Because of their juxtaposition, all three sites might not be used on any one polypeptide chain. For creation of Δs.s./p58, amino acids 2–31 (VAP… AEC) of p58IPK were deleted; and for Prl/p58, these residues were replaced by amino acids 2–30 of bovine preprolactin. Deletion of the p58 J-domain was made by fusion PCR by using the HA-tagged glycosylatable p58IPK construct. Signal sequences were cloned onto the A120L variant of the prion protein (PrP) mature domain as described previously (Kim et al., 2001). All constructs were cloned into pcDNA 3.1 Zeo (+) (Invitrogen, Carlsbad, CA), and they were verified by sequencing. Constructs coding for VCAM1 and HA-tagged Crt have been described previously (Garrison et al., 2005; Shaffer et al., 2005).
Measurement of Translocational Inhibition and Ratio of Glycoproteins to Cytosolic Proteins (GCR)
Measurement of protein synthesis efficiency, isolation of cytosolic and glycoprotein fractions, quantification by phosphorimaging, and determination of GCR were carried out as described previously (Kang et al., 2006). In brief, cells were pretreated with stressors or vehicle for 30 min in methionine-free media, and then [35S]methionine was added to 150 μCi/ml. After 15 min, cells were harvested in one of two ways. Total cell lysates were prepared in 1% SDS and 0.1 M Tris, pH 8. For fractionation, cytosolic proteins were selectively extracted on ice for 5 min with 1 ml/35-mm dish of 100 μg/ml digitonin in KHM buffer (110 mM KAc, 20 mM HEPES, pH 7.2, and 2 mM MgAc2). The remaining cellular material was solubilized in immunoprecipitation (IP) buffer (1% Triton X-100, 50 mM HEPES, pH 7.4, and 100 mM NaCl), clarified by centrifugation, and bound to immobilized concanavalin-A (ConA) for 2–4 h at 4°C. After washing the ConA beads three times in IP buffer, glycoproteins were selectively eluted for 1 h at 25°C with competitive sugar (0.2 M α-d-methyl-mannopyrannoside in IP buffer). Selective elution was critical for accurate results due to the nonspecific proteins that were eluted if SDS was used. After separation of samples by SDS-polyacrylamide gel electrophoresis (PAGE), all gels were stained with Coomassie blue to confirm equal recoveries and loading of all samples before further processing by phosphorimaging.
Cell Culture, Transfection, and Analysis
MEF, NIH 3T3, and HeLa cells were cultured as described previously (Kang et al., 2006; Rutkowski et al., 2006). Transfections were carried out using FuGENE 6 (Roche Diagnostics) or Effectene (QIAGEN, Valencia, CA). Endoglycosidase H (EndoH) and peptide-N-glycosidase F (PNGaseF) (New England Biolabs, Ipswich, MA) digestions were carried out according to the manufacturer's instructions on total cell lysates harvested in 1% SDS and 0.1 M Tris, pH 7.5. For protease protection assays, cell lysates were made by dounce homogenization in hypotonic buffer (0.25 M sucrose and 10 mM Tris-Cl, pH 7.5) followed by centrifugation at 2500 rpm for 10 min in a microfuge to remove nuclei and cell debris. The lysates were incubated for 30 min on ice with 0.5 mg/ml proteinase K (PK) in the absence or presence of 1% Triton X-100. Reactions were terminated with 5 mM phenylmethylsulfonyl fluoride (PMSF) and transfer to boiling 1% SDS, 0.1 M Tris, pH 8.8. Xbp1 mRNA splicing was assessed as described previously (Rutkowski et al., 2006). Pulse-chase analyses of PrP synthesis and maturation were performed on transiently transfected HeLa cells as described previously (Kang et al., 2006). Pulse labeling times were for 10 min, and chase times were from 10 to 60 min as indicated in the figure legends.
Tissue Preparation, Fractionation, and Topology Analysis
Mice were injected intraperitoneally with TM or vehicle exactly as described previously (Zinszner et al., 1998), and livers were isolated 16 h after injection. Rough microsomes and cytosol were purified from liver by modification of previously described methods for pancreatic microsomes (Walter and Blobel, 1983). Briefly, tissue was homogenized in 50 mM HEPES, pH 7.4, 6 mM MgAc2, 1 mM EDTA, 0.25 M sucrose, 1 mM DTT, and 0.5 mM PMSF by using a glass-on-glass homogenizer, and the tissue was centrifuged twice at 15,000 rpm for 10 min in a microfuge. This postmitochondrial supernatant was layered over a cushion of 0.8 M sucrose and centrifuged in a 70.1 Ti rotor for 3 h at 57,000 rpm. Cytosol was collected from the top of the tube, whereas the microsomal pellet was resuspended in 0.25 M sucrose, 50 mM HEPES, pH 7.4, and adjusted to a concentration corresponding to A260 of 50 (the same as is customary for pancreatic microsomes). Analysis by extraction (Figure 3C) was performed by diluting microsomes fivefold into various buffers at 0°C: physiological salt buffer (PSB) contained 100 mM KAc, 50 mM HEPES, pH 7.4, and 2 mM MgAc2; high salt buffer was PSB containing 500 mM KAc; and detergent extractions were in PSB. After 5 min on ice, the samples were centrifuged at 70,000 rpm for 15 min in a TLA100.3 (Beckman Coulter, Fullerton, CA). Equal aliquots of the supernatant and pellet were analyzed. We note that alkaline extraction using NaCO3, pH 11.5, was not included in our analysis, because it cannot distinguish between lumenal and peripheral proteins. Indeed, extraction at any pH above 9 will remove lumenal contents (including p58IPK; data not shown) and is a common method for lumenal protein depletion (Nicchitta and Blobel, 1993). Protease protection assays were performed in PSB by using 0.5 mg/ml PK at 0°C for 1 h as described previously (Fons et al., 2003). For sucrose velocity sedimentation, microsomes were adjusted to 1% digitonin in PSB and layered over a 2-ml 10–50% continuous sucrose gradient prepared in 1% digitonin/PSB. After centrifugation at 55,000 rpm for 1 h in a TLS-55 rotor (Beckman Coulter), fractions were removed from the top and analyzed by SDS-PAGE and immunoblotting.
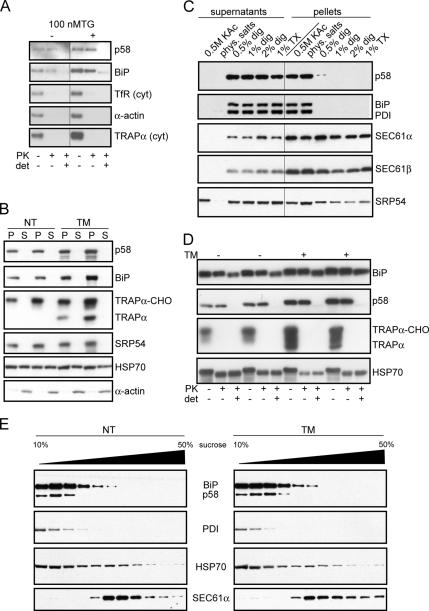
Figure 3.
Endogenous p58IPK is a component of the ER lumen. (A) Postnuclear hypotonic lysates from NIH 3T3 cells were treated with PK in the presence or absence of Triton X-100 (det) and probed by immunoblot against the indicated proteins. Antibodies against the transmembrane proteins TRAPα and TfR were directed against cytosolic epitopes. Note that BiP contains a protease-resistant core that is not digested fully even in the presence of detergent. (B) Postmitochondrial supernatants from homogenized livers of untreated or TM-injected mice (16 h) were sedimented over a cushion of 0.8 M sucrose. The microsomal pellets (P) and cytosolic supernatants (S) were probed by immunoblot against the indicated proteins. Efficacy of the TM was reflected in inhibition of TRAPα glycosylation and increased expression of p58IPK and BiP, which are both up-regulated by ER stress. (C) Canine pancreatic microsomes were extracted using high salt, physiological salt, or varying concentrations of digitonin (dig) or Triton X-100 (TX). Microsomes were then separated by centrifugation into extracted (supernatants) and nonextracted (pellets) fractions, followed by immunoblot as indicated. (D) Murine liver microsomes prepared as described in B were treated with PK in the presence or absence of detergent (1% digitonin) and analyzed by immunoblot against the indicated proteins to assess their respective topologies. Note the complete protection from PK digestion of both BiP and p58IPK in the absence of detergent. By contrast, TRAPα (detected using an antibody against its cytosolic tail) and HSP70 are equally accessible in the absence or presence of detergent. Note that both BiP and HSP70 contain protease-resistant cores that are not digested fully even in the presence of detergent. (E) Murine liver microsomes from either nontreated or TM-treated mice were solubilized in a physiological salt buffer containing 1% digitonin. The samples were separated by velocity sedimentation through a continuous sucrose gradient and individual fractions probed by immunoblot against the indicated proteins.
In Vitro Assays
In vitro transcription, translation, and protease protection assays were essentially as described previously with modifications as indicated below and in the figure legends (Fons et al., 2003; Garrison et al., 2005). For coimmunoprecipitation analyses, translation reactions were performed for 30 min at 32°C before carrying out the remaining steps at 4°C. In some experiments, the microsomes were sedimented (at 70,000 rpm for 15 min in a TL100.3 rotor) and resuspended in PSB, whereas other experiments used the translation products directly. For native immunoprecipitations, the sample was adjusted to 1% Triton X-100, incubated with the respective antibodies for 90 min, diluted to 1 ml with IP buffer, and incubated with a mixture of protein A- and protein G-Sepharose for 1 h to recover the immune complexes, washed three times in IP buffer, and eluted in SDS-PAGE sample buffer. For denaturing immunoprecipitations, translation reactions were adjusted to 1% SDS from a 10% stock, incubated for 10 min at 37°C, diluted 10-fold in IP buffer, and immunoprecipitated as described above.
Immunoprecipitation of Cell Lysates
Samples diluted into IP buffer were incubated with antibodies for 1–2 h, followed by recovery of antibodies by addition of protein A-Sepharose (Pierce Chemical) for 1 h. The samples were washed three to five times in IP buffer before elution of the immune complexes with SDS-PAGE sample buffer. For in situ cross-linking, DSP was dissolved at 20 mM in DMSO, diluted to 2 mM in PBS, and incubated with PBS-washed adherent cells at room temperature for 30 min. Cross-linker was quenched with 20 mM Tris-Cl, pH 7.5, the cells were lysed in 1% SDS as described above, and then they were solubilized in immunoprecipitation buffer. After immunoprecipitation as described above, the cross-linker was reversed in SDS-PAGE sample buffer (containing 100 mM DTT).
RESULTS AND DISCUSSION
Analysis of Protein Biosynthesis and Translocation in p58+/+ and p58−/− Cells
A general role for p58IPK in reducing substrate burden via cotranslocational degradation was proposed based on the idea that p58IPK extracts nascent chains whose translocation is temporarily delayed at the translocon during ER stress. Hence, in the absence of p58IPK, forward translocation would proceed despite any transient delays, resulting in excessive substrate burden in the ER lumen (Oyadomari et al., 2006). Therefore, during ER stress, protein translocation into the lumen should proceed at a higher efficiency in p58−/− cells compared with p58+/+ cells. We tested this idea directly by measuring the relative rates of protein synthesis and ER translocation during normal and acutely stressed conditions in p58+/+ and p58−/− cells (Figure 1A).
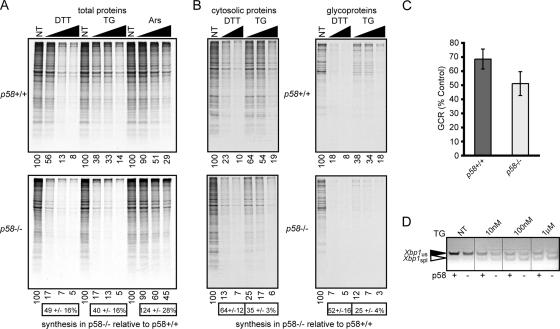
Figure 1.
p58−/− cells exhibit a lower ER substrate burden than wild-type cells. (A) Wild-type (top) and p58−/− (bottom) MEFs were treated with varying concentrations of DTT (0.1, 0.3, and 1 mM), TG (0.1, 1, and 10 μM), or arsenite (Ars; 10, 30, and 100 μM) for 30 min and labeled for 15 min with [35S]methionine before analysis of total cell lysates by SDS-PAGE and autoradiography. Relative levels of [35S]incorporation (quantitated using a phosphorimager) are shown below individual lanes of each panel. The level of synthesis in p58−/− cells relative to wild-type cells for each stressor (mean ± SD; n = 3) is shown below the respective bottom panels. Note that for both DTT and TG (but not Ars), p58−/− cells show about half the level of overall protein synthesis relative to p58+/+ cells. In this and subsequent experiments, Coomassie blue staining before autoradiography confirmed equal total protein recovery and loading (data not shown). (B) After treatment with DTT (1 and 10 mM) or TG (0.1, 1, and 10 μM) and [35S]methionine labeling as described in A, cells were fractionated into cytosolic proteins (left) and glycoproteins (right) that were each analyzed by SDS-PAGE and quantified by phosphorimaging. The amount of 35S incorporation in each lane relative to the untreated condition in that panel is indicated below the autoradiographs. Note that for both p58−/− and p58+/+ cells, glycoproteins are consistently attenuated to a slightly greater degree during stress than cytosolic proteins. Quantitation of the average 35S incorporation into either cytosolic proteins or glycoproteins in p58−/− cells relative to wild-type cells was tabulated as described in A, and it is shown below the respective bottom panels. Note that although both cytosolic proteins and glycoproteins are made to lower relative amounts in p58−/− cells, the glycoproteins are affected to a greater extent. (C) The radiolabeled GCR was quantitated for all five ER stress conditions in B and normalized against the respective GCRs from nontreated cells for both genotypes. Plotted are the overall changes in GCR (mean ± SD) observed during ER stress in p58−/− and p58+/+ cells. (D) Wild-type and p58−/− MEFs were treated with the indicated concentrations of TG for 30 min, followed by RT-PCR to detect both spliced (spl) and unspliced (us) Xbp1 mRNA. Note the slight but consistently higher proportion of spliced Xbp1 seen in p58−/− cells at each stress condition. The image is presented in black-and-white inverted form for greater visual clarity.
Analysis of [35S]methionine incorporation into total cellular proteins during both ER stress (TG or DTT treatments) and non-ER stress (arsenite treatment) showed a dose-dependent inhibition of protein synthesis in p58+/+ and p58−/− cells. Whereas the profile of arsenite-dependent inhibition was similar in both cell types, the knockout cells consistently showed ∼40–50% less protein synthesis than wild-type cells during either TG- or DTT-mediated ER stress. This increased translational inhibition in p58−/− cells is consistent with the originally characterized role of p58IPK as an eIF2α kinase inhibitor (in this case, PERK).
To assess the newly proposed role for p58IPK in cotranslocational degradation (Oyadomari et al., 2006), we combined pulse labeling with fractionation of cells into cytosolic proteins and N-linked glycoproteins (the latter, a surrogate for the nascent ER-translocated fraction). The aim was to directly quantify the substrate burden on the ER during stress in p58−/− cells relative to matched p58+/+ cells. As expected, wild-type cells showed a greater reduction in glycoprotein synthesis than cytosolic protein synthesis during each ER stress condition (Figure 1B, compare top left with top right). This finding demonstrates that during acute stress, the generation of ER substrates (monitored using the glycoprotein fraction) is reduced more than what might have been expected based on the degree of global translational attenuation alone (monitored using the cytosolic fraction). Thus, the ratio of glycoproteins to cytosolic proteins (a parameter termed GCR) decreases during stress and reflects, at least in part, reduced translocation of substrates into the ER (Kang et al., 2006). When all of the stress conditions (n = 5) were tabulated together, the average GCR for wild-type cells during acute ER stress was ∼70% of the unstressed value (Figure 1C), suggesting up to ∼30% overall reduction in ER protein translocation. Contrary to expectations, the GCR in p58−/− cells under the same conditions was even lower (∼50%) than in p58+/+ cells. Thus, the overall efficiency of protein influx into the stressed ER of p58−/− cells is not higher than in p58+/+ cells, and in fact it seems to be slightly lower.
These results suggest that in the absence of p58IPK, the ER is not subjected to an increased substrate burden under either normal or stressed conditions. This is seen in at least two ways. First, the absolute amount of glycoproteins generated during ER stress in p58−/− cells (Figure 1B, bottom right) is consistently lower than in p58+/+ cells (Figure 1B, top right) regardless of the nature or severity of the stressor. Second, even when adjusting for the lower rate of overall protein synthesis during stress in p58−/− cells, the relative translocation efficiency (as judged by the GCR) is lower (Figure 1C). Despite this overall lower substrate burden on the ER lumen during stress, p58−/− cells are nonetheless more sensitive to ER stress as judged by at least two independent parameters. Not only are the cultured cells and certain tissues more prone to ER protein misfolding (Oyadomari et al., 2006) but also the stress sensor IRE1 is consistently activated to a slightly greater degree as judged by Xbp1 mRNA splicing (Figure 1D). Considered together, these results demonstrate that p58−/− cells are more sensitive to ER stress despite a lower substrate burden (due to both decreased translation and decreased translocation) than in p58+/+ cells. Hence, a role for p58IPK in the reduction of substrate burden on the ER lumen during stress seems unlikely. Instead, the decreased capacity of p58−/− cells to cope with misfolded substrates that are generated during stress suggests a role for p58IPK in protein processing or maturation.
p58IPK Has an N-Terminal Signal Sequence for ER Targeting and Translocation
Given that substrate folding and maturation are initiated only upon entry into the ER, the principal lesion in p58−/− cells would seem to be in the lumenal environment. We therefore reexamined the subcellular localization of p58IPK, which had previously been proposed to be a peripheral protein of the ER membrane (Yan et al., 2002; Oyadomari et al., 2006). p58IPK from all species tested, including human, rodent, nematode, and plant, contains a hydrophobic N-terminal region predicted to be a cleavable ER signal sequence (Figure 2A; data not shown). In vitro-synthesized p58IPK was translocated into ER-derived microsomes (as judged by a protease protection assay) with an efficiency of ∼50%. This was comparable in efficiency to the translocation of the ER lumenal chaperone calerticulin in this same assay system. Most (but not all) of the in vitro translocated p58IPK seemed to be processed by signal peptidase; hence, it migrated slightly faster than nontranslocated p58. Translocation was abrogated by deletion of the 31 residues constituting the putative p58IPK signal sequence (Δs.s./p58), whereas replacement of this region with the signal sequence from the secretory hormone prolactin (Prl) conferred efficient translocation (Figure 2B).
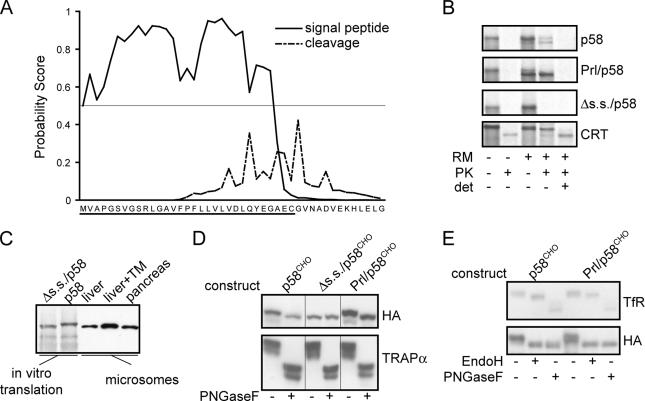
Figure 2.
p58IPK contains a functional N-terminal signal sequence. (A) SignalP3.0 analysis of the murine p58IPK N terminus predicts a likely ER targeting signal (solid line) and cleavage site (broken line). In this analysis, the probability of each residue being part of a targeting signal (based on an artificial neural network trained on known eukaryotic signal sequences) or suitable site for signal peptidase cleavage is plotted (Bendtsen et al., 2004). A horizontal line at the 0.5 probability value for the targeting signal plot is indicated, showing that nearly the entire putative signal sequence of p58IPK (underlined) is above this cut-off. (B) Transcripts encoding the indicated constructs were translated in vitro in the presence or absence of ER-derived rough microsomes (RM). After synthesis, samples were divided into equal aliquots and treated as indicated with 0.5 mg/ml PK in the presence or absence of 1% Triton-X100 (det) to assess translocation into the lumen of microsomes. Note that calreticulin (Crt) contains a core domain that is resistant to digestion by PK. (C) In vitro translations of full-length murine p58 and Δs.s./p58 were analyzed adjacent to microsomes isolated from mouse liver, TM-treated murine liver, and dog pancreas. Detection of p58 was performed by immunoblotting. (D) NIH3T3 cells were transfected with constructs encoding p58CHO, Δs.s./p58CHO, or Prl/p58CHO, each of which is appended at its C-terminus with an HA epitope tag and contains an inserted potential N-linked glycosylation site near the C terminus. Cell lysates were incubated in the absence or presence of PNGaseF before immunoblot with antibodies against the HA epitope or the ER resident glycoprotein TRAPα. (E) Lysates of NIH 3T3 cells transfected as indicated were treated with PNGaseF or EndoH followed by immunoblot against the HA epitope or the cell surface TfR. Resistance to EndoH digestion is indicative of glycan modifications that accompany trafficking of proteins past the cis-Golgi.
Fusion of the predicted p58IPK signal sequence to an unrelated protein (the mature domain of PrP) was sufficient to mediate PrP translocation as evidenced by both glycosylation and protease protection (Supplemental Figure S1). The translocation efficiency of p58-PrP was within the range of previously characterized signal sequence fusions to PrP (Kim et al., 2002), being more efficient than the interferon-γ signal and slightly less efficient than the native PrP signal. As expected, fusion of the N-terminal region of the cytosolic protein globin to PrP yielded no translocation. Finally, endogenous cellular p58IPK from multiple sources comigrated with in vitro-translated Δs.s./p58 rather than with full-length p58IPK (Figure 2C; data not shown). Collectively, these data suggested that p58IPK contains a functional cleavable ER signal sequence.
Consistent with this conclusion, p58IPK engineered to contain a C-terminal glycosylation site (p58CHO) was quantitatively modified by an N-linked glycan upon expression in NIH3T3 cells (Figure 2D). Prl/p58CHO was identically glycosylated, whereas Δs.s./p58CHO was not glycosylated (Figure 2D). The glycans on both p58CHO and Prl/p58CHO were fully sensitive to digestion by endoglycosidase H, whereas those of the plasma membrane-resident transferrin receptor were resistant (Figure 2E). This result suggests that p58IPK is retained in the ER, consistent with previous immunofluorescence localization studies (Yan et al., 2002), proteomic analysis of the secretory pathway (Gilchrist et al., 2006), and our own observations using green fluorescent protein (GFP)-tagged p58IPK constructs (data not shown). Based on these analyses with chimeric constructs and exogenously expressed protein, we conclude that p58IPK can be targeted to and translocated across the ER membrane in vitro and in vivo, where it may be a resident of the ER lumen.
Endogenous p58IPK Is an ER-Resident Lumenal Protein
Analysis of the subcellular localization of endogenous p58IPK in cell lysates by a protease protection assay corroborated the results obtained for the exogenously expressed protein. We observed that p58IPK was protected from proteolytic digestion to an extent comparable with other ER lumenal proteins, including BiP and HSP47, whereas cytosolic proteins and cytosolic domains of membrane proteins were protease accessible (Figure 3A; data not shown). As expected, all of these proteins became protease accessible in the presence of detergent (Figure 3A). ER stress did not alter the electrophoretic migration or protease protection of p58IPK (Figure 3A), suggesting that there is no substantial relocalization of endogenous p58IPK during ER stress.
In support of these conclusions based on experiments in cultured cells, p58IPK was isolated quantitatively with the rough microsomal fraction, and it was absent in the cytosolic fraction of murine liver homogenates prepared from both nontreated and TM-injected animals (Figure 3B). p58IPK and other lumenal proteins could be quantitatively extracted from microsomes by even low concentrations of nondenaturing detergents (including digitonin and Triton X-100), but not by high salt treatment that strips microsomal membranes of the peripherally associated targeting protein SRP54 (Figure 3C). It should be noted that the alkaline extraction conditions previously used to conclude a peripheral location for p58IPK (Oyadomari et al., 2006) also efficiently extract lumenal proteins of ER (Nicchitta and Blobel, 1993) and are thus not contradictory to our results. As in total cell lysates (Figure 3A), protease protection analysis of isolated liver microsomes (Figure 3D and Supplemental Figure S2) revealed that p58IPK, like known ER lumenal proteins, was fully protected from digestion. By contrast, peripherally associated cytosolic proteins and cytosolic domains of transmembrane proteins were sensitive to digestion (Figure 3D and Supplemental Figure S2). The same results were obtained from pancreatic microsomes (Supplemental Figure S2). Importantly, the microsomally associated fraction of the cytosolic HSP70 chaperone was quantitatively accessible to protease (Figure 3D). Therefore, p58IPK is topologically separated from HSP70 and other cytosolic proteins in liver microsomes under both normal and ER-stressed conditions.
Fractionation of digitonin-solubilized microsomes by velocity sedimentation showed p58IPK migrating exclusively in the low-molecular-weight fractions, along with other ER lumenal proteins such as BiP and PDI (Figure 3E). In contrast, the SEC61α protein was found in higher molecular weight fractions consistent with its presence in ribosome–translocon complexes (Figure 3E). Although the microsomal-associated HSP70 fractionated heterogenously (Figure 3E), we did not observe any fraction that simultaneously contained SEC61α, HSP70, and p58IPK. A similar separation of p58IPK and SEC61α was also observed in microsomes solubilized with Triton X-100 rather than digitonin (Supplemental Figure S2). Furthermore, the proportion of HSP70 that cosedimented with the ribosome-translocon complex (in fractions 5–9) was unchanged in the TM-treated microsomes. This result indicates that a marked stress-dependent recruitment of HSP70 to translocons during ER stress does not occur. Even if some subtle changes in HSP70 recruitment were occurring, it seems unlikely to directly or indirectly involve ER-associated p58IPK, because this factor is located in the lumen and does not cofractionate with the translocon.
The localization of p58IPK in the ER lumen is inconsistent with its putative association (by cross-linking) with nascent VCAM1 when VCAM1 translocation and degradation are pharmacologically inhibited (Oyadomari et al., 2006). This discrepancy can be reconciled by our observation that p58IPK translocation into the ER was itself inhibited by the same agent that inhibits VCAM1 translocation (Supplemental Figure S3). Furthermore, when translocation and proteasomal degradation were inhibited simultaneously, nontranslocated VCAM1 and nontranslocated p58IPK were both found in detergent-insoluble aggregates that are presumably in the cytosol. Thus, interactions observed under these conditions (when multiple basic cellular processes are pharmacologically perturbed) may not accurately reflect the situation in either normal or ER-stressed cells.
P58IPK Associates with BiP in the ER Lumen
The localization of p58IPK in the ER lumen (Figure 3) together with its lack of any apparent role in reducing substrate burden during acute stress (Figure 1) raised the issue of how p58IPK might protect the ER from stress (Oyadomari et al., 2006). Because DnaJ proteins interact with and regulate the function of HSP70 family members (Cheetham and Caplan, 1998), we speculated that p58IPK might associate with the lumenal HSP70 family member BiP, a central component in maintaining homeostasis of the ER folding environment. Indeed, immunoprecipitation of BiP from liver microsomes of either untreated or TM-injected animals copurified p58IPK, and vice versa, whereas neither protein was precipitated by irrelevant or preimmune sera (Figure 4A).
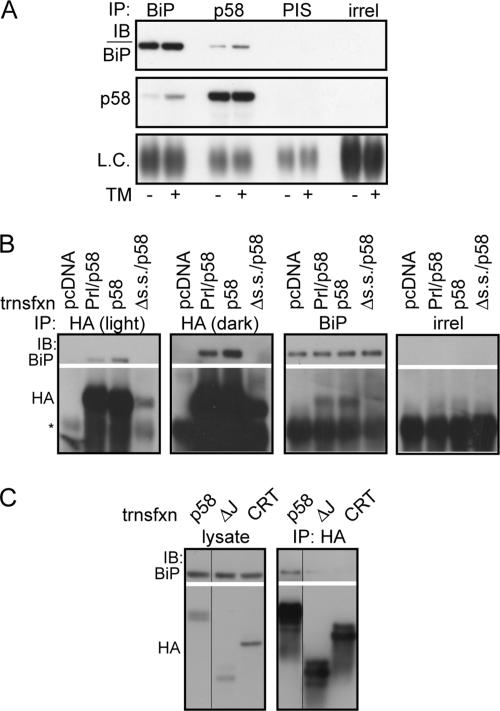
Figure 4.
p58IPK interacts with BiP in the ER lumen. (A) Liver microsomes from nontreated or TM-injected mice were solubilized in 1% Triton X-100 and immunoprecipitated with antiserum against BiP, an antiserum directed against the C terminus of p58IPK, preimmune serum (PIS), or an irrelevant antibody against interferon-γ (irrel). Immune complexes were then probed by immunoblot (IB) for BiP, p58IPK, or immunoglobulin (Ig)G light chain. (B) NIH 3T3 cells were transfected with empty vector, Prl/p58, p58, or Δs.s./p58. Cells were incubated in situ with the membrane-permeable cross-linker DSP at 2 mM, followed by lysis under denaturing conditions and immunoprecipitation with antibodies against BiP, the HA epitope, or an irrelevant antigen (interferon-γ). After cross-linker reversal, the immunoprecipitations were probed by immunoblot as indicated. Asterisk represents IgG heavy chain. Expression of Δs.s./p58 is much lower, likely due to degradation. Thus, a darker exposure is also shown to confirm the absence of an association between this protein and BiP. (C) NIH 3T3 cells were transfected with full-length p58CHO or the same construct with the J-domain deleted (ΔJ). Overexpression of HA-tagged Crt served as a negative specificity control for BiP interaction. Cell lysates were prepared under nondenaturing conditions (no cross-linker was used) and immunoprecipitated with an HA antibody, followed by immunoblot as indicated. Multiple bands for p58CHO constructs represent heterogeneity of glycosylation for the protein.
This association could be recapitulated in cultured cells using overexpressed HA-tagged p58IPK (Figure 4B). Importantly, the p58–BiP interaction was also observed with Prl/p58 (whose quantitative localization to the ER lumen was ensured by the highly efficient Prl signal sequence). In contrast, Δs.s./p58 did not coimmunoprecipitate BiP (Figure 4B), indicating that a spurious interaction after cell lysis did not occur. Identical results were also observed with p58CHO, Prl/p58CHO, and Δs.s./p58CHO (data not shown). A p58IPK construct lacking its J-domain also did not coimmunoprecipitate BiP (Figure 4C), further supporting the notion that the p58IPK interaction with BiP is specific. These results demonstrate a specific interaction between ER-lumenal p58IPK and BiP in both heterologous and endogenous contexts. Thus, by analogy to other J-domain proteins, p58IPK may function as a cochaperone to facilitate the maturation or metabolism of newly synthesized secretory or membrane proteins.
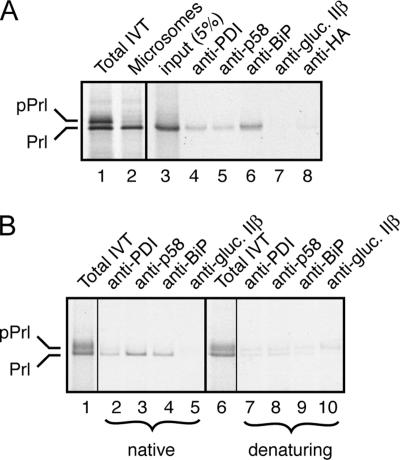
The potential role for p58IPK as a cochaperone for BiP was supported by our observation that ER lumenal p58IPK can associate with a newly translocated secretory protein. Using in vitro translation, we translocated the secretory protein Prl into ER microsomes, and, after isolation of the microsomes, we tested for Prl association with p58IPK by coimmunoprecipitation (Figure 5A). Radiolabeled Prl was coimmunoprecipitated with antibodies against p58IPK, PDI, and BiP, but not the β subunit of ER glucosidase II or the HA epitope (an irrelevant antibody control). The specificity of this interaction was further confirmed in two additional ways. First, when total translation extracts were used for the analyses, p58IPK coimmunoprecipitated only translocated Prl, and not the nontranslocated precursor (Figure 5B). Second, the interaction was substantially diminished if the samples were first treated with SDS for 10 min at 37°C before immunoprecipitation (Figure 5B). These results suggest that p58IPK, like the chaperones BiP and PDI, can interact with a newly synthesized secretory protein. Whether this interaction is direct or indirect (e.g., through BiP) remains to be determined at present.
Figure 5.
p58IPK interacts with a secretory protein in the ER lumen. (A) Preprolactin was synthesized in vitro by using rabbit reticulocyte lysate and pancreatic microsomes. Untranslocated (pPrl) and translocated, signal sequence-cleaved material (Prl) are indicated. The microsomes were isolated by centrifugation, solubilized under nondenaturating conditions, and subjected to immunoprecipitation by using antibodies against PDI, p58IPK, BiP, the β subunit of α glucosidase II (also known as 80K-H), and the HA epitope tag. Aliquots of the total translation products (lane 1), the isolated microsomes (lane 2), solubilized microsomes (lane 3), and immunoprecipitates were analyzed by SDS-PAGE and autoradiography. (B) Preprolactin was synthesized in vitro by using rabbit reticulocyte lysate and pancreatic microsomes as described above and adjusted to either 1% Triton X-100 (native conditions; lanes 1–5) or 1% SDS (denaturing conditions; lanes 6–10). After incubation of the denaturing sample at 37°C for 10 min, it was adjusted to 1% Triton X-100 and 0.1% SDS. Both samples were subjected to immunoprecipitation with the indicated antibodies. Aliquots of the input (lanes 1 and 6) and immunoprecipitates are shown.
P58IPK Overexpression Stimulates PrP Maturation in the ER Lumen
The interaction of p58IPK with BiP and its ability to coassociate (perhaps indirectly) with a secretory protein in the ER lumen suggested a potential role in either maturation or metabolism of secretory and membrane proteins. Because straightforward assays for Prl maturation were not available, we turned to the PrP. Maturation of this polypeptide requires several posttranslational events, including folding, disulfide bond formation, glycosylation, glycosylphosphatidyl inositol anchor addition, and possibly dimerization. This complexity increases the possibility that subtle changes in the overall processing capacity of the ER might be detected as a change in the rate of PrP maturation, even as an indirect consequence. Previous analyses have shown that upon completion of its maturation in the ER lumen, PrP transits to the Golgi where glycan processing causes both a mobility change and resistance to endoglycosidase H digestion (Kang et al., 2006). Using the disappearance of core-glycosylated PrP and concomitant appearance of Golgi-modified PrP as markers of maturation in a pulse-chase experiment, we analyzed the consequences of p58IPK overexpression.
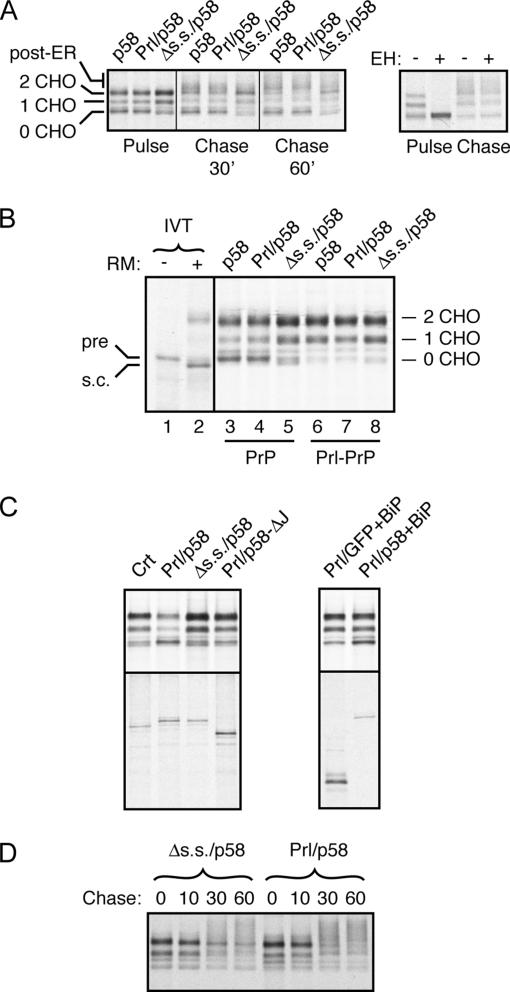
Overexpressed p58 and Prl/p58, but not Δs.s./p58, altered PrP metabolism in two detectable ways. First, there was a subtle (∼30%) but consistent decrease in fully glycosylated PrP with a concomitant increase in unglycosylated species at the pulse time point (Figure 6A). Comparison with in vitro-translated markers showed this unglycosylated band to represent precursor (Figure 6B), suggesting that it resulted from failed translocation of PrP into the ER lumen. Second, at the chase time points, we observed decreased levels of core-glycosylated PrP relative to higher-molecular-weight glycoforms (Figure 6A). The assignment of this latter species as mature, post-ER forms of PrP was confirmed by its resistance to endoglycosidase H digestion (Figure 6A). Thus, p58IPK overexpression in the ER lumen, but not the cytosol, has effects on both PrP translocation into and processing within the ER.
Figure 6.
p58IPK overexpression alters PrP metabolism. (A) HeLa cells cotransfected with PrP and the indicated p58IPK constructs were pulse labeled for 10 min and then chased for either 30 or 60 min before immunoprecipitation of PrP. The positions of immature PrP species (containing 0, 1, or 2 N-linked glycans) and post-ER PrP are indicated to the left. The post-ER species of PrP migrate as a heterogenous set of products, the most prominent of which migrates just above the doubly glycosylated immature PrP band. Right, products from pulse and 60-min chase time points (the equivalent of lanes 2 and 9) incubated overnight with either nothing or endoglycosidase H. Independent experiments showed that the expression levels of the different p58IPK constructs were equal (data not shown; see also C). (B) HeLa cells cotransfected with either PrP (lanes 3–5) or Prl-PrP (lanes 6–8), and the indicated p58IPK constructs were pulse labeled for 10 min before immunoprecipitation of PrP. In vitro translation products of PrP synthesized in the absence or presence of rough microsomes (RM) to generate markers for percursor (pre) and signal-cleaved (s.c.) species of PrP were analyzed on the same gel (but taken from a longer exposure of the autoradiograph). (C) HeLa cells cotransfected with PrP and either Crt or the indicated p58IPK constructs without (left) or with BiP (right) were pulse labeled for 10 min before immunoprecipitation of PrP (top) or the cotransfected protein (bottom). Note that in the absence of BiP cotransfection, Prl/p58 overexpression results in decreased PrP translocation (lane 2). This effect is largely offset by BiP overexpression. (D) Pulse-chase analysis of Prl–PrP in HeLa cells cotransfected with either Δs.s./p58 or Prl/p58. Note the more rapid conversion of immature PrP species to mature species in the Prl/p58-expressing cells.
When considered with the observation that p58IPK interacts with BiP (Figure 5), we interpreted these effects of p58IPK overexpression on PrP as follows. First, excess p58IPK in the ER lumen would interact with and titrate BiP away from other J-domain–containing proteins, including the translocon-associated proteins Sec63 and Mtj1. Because PrP translocation into the ER is dependent on lumenal chaperones to compensate for a weak signal sequence (Kang et al., 2006), its translocation would be diminished slightly by this titration effect. Second, the shift of BiP from its putative role in translocation at the translocon to a role in protein folding in the ER lumen would be the basis for the improved efficiency of PrP transit to the Golgi. In this view, the greater proportion of BiP (along with the J-domain protein p58IPK) devoted to posttranslocation folding events would either directly or indirectly facilitate PrP maturation.
Several additional observations supported the above-mentioned model. First, p58IPK overexpression had essentially no effect on translocation of Prl–PrP (Figure 6B). Prl–PrP, in which the PrP signal sequence is replaced with the highly efficient Prl signal sequence, exhibits lumenal protein-independent translocation (Kang et al., 2006) and would therefore not be affected by a shift in BiP functionality. Second, the inhibitory effect on PrP translocation by p58IPK overexpression in the ER lumen was nearly abolished by deletion of the J-domain (Figure 6C), a region required for interaction with BiP (Figure 4C). Third, coexpression of BiP with Prl/p58 also minimized the inhibitory effect on PrP translocation (Figure 6C), further supporting a titration-based mechanism. And finally, the maturation of Prl–PrP was faster upon overexpression of ER lumenal Prl/p58 compared with cytosolic Δss/p58 (Figure 6D). Thus, p58IPK in the ER lumen can stimulate PrP maturation independently of any effect on PrP translocation.
Considered together, these findings argue that p58IPK, via its J-domain–mediated interaction with BiP, facilitates the maturation of newly synthesized proteins in the ER lumen. It should be stressed that at present, we do not know whether p58IPK directly interacts with and influences PrP maturation. Coassociation experiments between p58IPK and PrP as shown for Prl (Figure 5) were hampered by the propensity of immature PrP to aggregate under the native immunoprecipitation conditions used in this assay. Nonetheless, the data support the conclusion that p58IPK overexpression can functionally influence the protein processing capacity of the ER lumen. Although overexpressed p58IPK also caused a selective decrease in PrP translocation, this may not be physiologically relevant, because normally, p58IPK is co-up-regulated with BiP and would therefore not have the titration effect that seems to underlie this phenomenon.
A functional role for p58IPK as a maturation factor in the ER lumen would explain the increased stress sensitivity of p58−/− cells (e.g., Figure 1) and their decreased capacity to deal with misfolded proteins in certain specialized contexts where the secretory pathway is severely taxed. Two apparent examples of this include pancreatic β cells producing mutant insulin, and hepatocytes pharmacologically perturbed in apolipoprotein B maturation (Oyadomari et al., 2006). Further work analyzing the functional importance of the p58–BiP interaction as it relates to the folding and maturation of insulin and apolipoprotein B will be needed to test this working hypothesis.
The p58IPK Signal Sequence Allows for Inefficient Translocation during ER Stress
Our localization of p58IPK to the ER lumen raises the puzzling question of how it is able to regulate the cytosolically disposed kinase activities of PERK and PKR. The simplest explanation is that slight inefficiencies in the translocation of p58IPK into the ER, either constitutively or selectively under certain conditions, generate sufficient amounts of cytosolic p58IPK to inhibit these kinases. Consistent with this idea, the p58IPK signal sequence seems to be less efficient (at least in vitro) than the canonical Prl signal sequence (Figure 2 and Supplemental Figure S1). Indeed, other slightly inefficient signal sequences have been documented to generate small but detectable cytosolic populations (Levine et al., 2005) that in some instances can have either physiological (Shaffer et al., 2005) or pathological consequences (Rane et al., 2004). We have not been able to reliably detect the presence of endogenous p58IPK in the cytosol by fractionation or protease protection assays of tissue or cultured cells under either normal or stressed conditions (Figure 2). This makes a proposed stoichiometric role for this protein on the cytosolic face of the abundantly expressed translocon very unlikely. However, given that the kinases proposed to be inhibited by p58IPK are very low-abundance proteins, even slight or transient translocational inefficiency might be sufficient to account for this alternative functional activity.
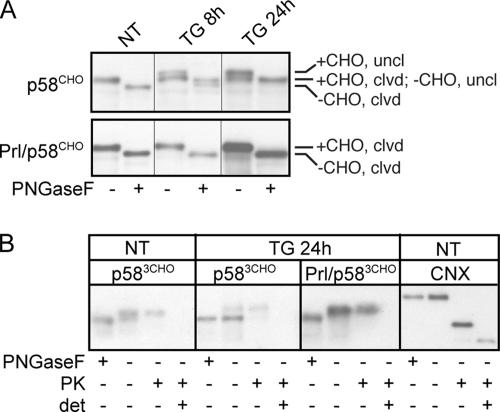
To test whether p58IPK translocation would be capable of such regulation, we used glycosylation as a reporter to compare the ER import of overexpressed p58CHO and Prl/p58CHO during ER stress. As expected by virtue of its highly efficient signal sequence, Prl/p58CHO was constitutively translocated (and thus glycosylated) in both the presence and absence of ER stress (Figure 7A). In contrast, treatment with 100 nM TG had two effects on p58CHO: signal sequence cleavage was inhibited (quantitatively so by 24 h) and glycosylation of p58CHO was blocked (∼50% by 24 h; Figure 7A). Because the glycosylation acceptor site is located very near the C terminus of p58CHO, the presence of glycosylated but signal sequence-uncleaved chains suggests that the inhibition of signal sequence cleavage exists apart from any potential effect on translocation, because these chains are almost certainly translocated. However, a protease protection assay of hypotonic cell lysates demonstrated that although all of the glycosylated chains were protected from digestion and thus translocated, the unglycosylated chains were completely sensitive, suggesting a cytosolic localization (Figure 7B). Therefore, the inefficient signal sequence of p58IPK protein is capable, at least in principle, of allowing small populations of the protein to be rerouted to the cytosol during ER stress. Hence, up-regulation of p58IPK during the late phase of the UPR could conceivably simultaneously improve ER folding capacity (via its abundant ER lumenal form) while mitigating translational attenuation by PERK inhibition (via a minor cytosolic form). This may provide a novel mechanism to match substrate synthesis with folding capacity during the recovery phase of the UPR. Future quantitative analysis of p58IPK translocation and localization during different cellular conditions will be required to confirm this possibility.
Figure 7.
ER stress reduces the efficiency of p58IPK translocation. (A) NIH 3T3 cells were transfected with p58CHO or Prl/p58CHO and treated with 100 nM TG for the indicated times. Lysates were digested with PNGaseF to resolve N-glycosylated from unmodified chains. Although ER stress does not affect either signal sequence cleavage or glycosylation of Prl/p58CHO, TG inhibits signal sequence cleavage of p58CHO, such that by 24 h all chains exist in the uncleaved (uncl) state. Of the uncleaved chains, approximately half are glycosylated (+CHO) and half unglycosylated (−CHO). (B) NIH 3T3 cells were transfected with p583CHO or Prl/p583CHO (identical to the constructs used in A except containing three C-terminal N-linked glycosylation sites to better separate glycosylated from nonglycosylated material) and treated with 100 nM TG for 24 h as indicated. Hypotonic lysates were prepared and treated with PK as described in Figure 3A, or PNGaseF as described above. p583CHO and Prl/p583CHO were identified by immunoblotting. Note that ER stress causes a substantial portion of p583CHO chains, but not Prl/p583CHO, to be unglycosylated, and these unglycosylated chains are sensitive to PK digestion, whereas glycosylated chains are resistant. Immunoblotting to detect a lumenal epitope of endogenous calnexin (CNX) confirmed the proper orientation and integrity of the membranous vesicles within the homogenate.
CONCLUSIONS
Two principal conclusions can be drawn from the results in this study. First, the increased stress sensitivity of p58−/− cells is not attributable to promiscuous or excessive protein import during acute ER stress. Second, p58IPK resides in the ER lumen, where it interacts with BiP in a J-domain–dependent manner and influences protein maturation efficiency. These results point to a new functional assignment for p58IPK in serving as a BiP cochaperone to optimize protein folding homeostasis in the ER. Because p58IPK represents one among several (at least five others) ER localized J-domain proteins that interface with BiP (Feldheim et al., 1992; Brightman et al., 1995; Shen et al., 2002; Hosoda et al., 2003; Shen and Hendershot, 2005), its absence would result in an ER lumen that is only modestly compromised in overall folding capacity. The functional consequences of this slightly lower protein maturation capacity would be obscured under all but the most taxing conditions and can explain each of the following phenotypic observations in cells and animals lacking p58IPK.
First, p58−/− cells seem to experience a greater level of stress (as judged by IRE1 activation) despite an overall lower substrate burden at any given level of exogenous stressor (Figure 1). Second, during longer term ER stress, p58−/− cells accumulate higher levels of terminally misfolded and aggregated proteins relative to p58+/+ cells (Oyadomari et al., 2006). Third, cells from p58−/− mice are essentially indistinguishable from p58+/+ cells unless the maturation of a highly expressed protein is perturbed in a highly secretory cell type (e.g., insulin in pancreatic β cells and apolipoprotein B in hepatocytes). Hence, the overall phenotype of p58−/− mice is remarkably mild relative to deletions or alterations in core UPR pathway components (such as PERK, IRE1α, XBP1, chaperones, and eIF2α). And finally, worms reduced in p58IPK show a low level of basal UPR activation and exacerbate the phenotype of IRE1 deletion. Because each of these observations is consistent with a decreased maturation capacity of the ER lumen where essentially all ER-associated p58IPK was localized under both normal and stressed conditions (Figure 3), we propose that an independent role for p58IPK in either translocational regulation, pQC, or degradation need not be invoked. Thus, the major mechanism by which p58IPK protects the stressed ER is probably through its function as a cochapeone that contributes to the overall protein processing capacity of the lumen.
Supplementary Material
ACKNOWLEDGMENTS
We thank P. Arvan for the polyclonal BiP antibody, D. Boger for Hun-7293, and J. Wu for reading the manuscript. R.J.K. is funded by the Howard Hughes Medical Institute and by National Institutes of Health grants R01 DK-042394 and R01 HL-052173. R.S.H. is funded by the Intramural Research Program of National Institute of Child Health and Human Development at the National Institutes of Health.
Abbreviations used:
- GCR
glycoprotein to cytosolic protein ratio
- pQC
preemptive quality control
- TG
thapsigargin
- TM
tunicamycin
- UPR
unfolded protein response.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-03-0272) on June 13, 2007.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Barber G. N., Thompson S., Lee T. G., Strom T., Jagus R., Darveau A., Katze M. G. The 58-kilodalton inhibitor of the interferon-induced double-stranded RNA-activated protein kinase is a tetracopeptide repeat protein with oncogenic properties. Proc. Natl. Acad. Sci. USA. 1994;91:4278–4282. doi: 10.1073/pnas.91.10.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Besemer J., Harant H., Wang S., Oberhauser B., Marquardt K., Foster C. A., Schreiner E. P., de Vries J. E., Dascher-Nadel C., Lindley I. J. Selective inhibition of cotranslational translocation of vascular cell adhesion molecule 1. Nature. 2005;436:290–293. doi: 10.1038/nature03670. [DOI] [PubMed] [Google Scholar]
- Brightman S. E., Blatch G. L., Zetter B. R. Isolation of a mouse cDNA encoding MTJ1, a new muring member of the DnaJ family of proteins. Gene. 1995;153:249–254. doi: 10.1016/0378-1119(94)00741-a. [DOI] [PubMed] [Google Scholar]
- Brodsky J. L., Goeckeler J., Schekman R. BiP and Sec63p are required for both co- and posttranslational protein translocation into the yeast endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 1995;92:9643–9646. doi: 10.1073/pnas.92.21.9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheetham M. E., Caplan A. J. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldheim D., Rothblatt J., Schekman R. Topology and functional domains of Sec63p, an endoplasmic reticulum membrane protein required for secretory protein translocation. Mol. Cell. Biol. 1992;12:3288–3296. doi: 10.1128/mcb.12.7.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fons R. D., Bogert B. A., Hegde R. S. Substrate-specific function of the translocon-associated protein complex during translocation across the ER membrane. J. Cell Biol. 2003;160:529–539. doi: 10.1083/jcb.200210095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison J. L., Kunkel E. J., Hegde R. S., Taunton J. A substrate-specific inhibitor of protein translocation into the endoplasmic reticulum. Nature. 2005;436:285–289. doi: 10.1038/nature03821. [DOI] [PubMed] [Google Scholar]
- Gilchrist A., et al. Quantitative proteomics analysis of the secretory pathway. Cell. 2006;127:1265–1281. doi: 10.1016/j.cell.2006.10.036. [DOI] [PubMed] [Google Scholar]
- Görlich D., Rapoport T. A. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell. 1993;75:615–630. doi: 10.1016/0092-8674(93)90483-7. [DOI] [PubMed] [Google Scholar]
- Harding H. P., Zeng H., Zhang Y., Jungries R., Chung P., Plesken H., Sabatini D. D., Ron D. Diabetes mellitus and exocrine pancreatic dysfunction in perk−/− mice reveals a role for translational control in secretory cell survival. Mol. Cell. 2001;7:1153–1163. doi: 10.1016/s1097-2765(01)00264-7. [DOI] [PubMed] [Google Scholar]
- Hollien J., Weissman J. S. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313:104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- Hosoda A., Kimata Y., Tsuru A., Kohno K. JPDI, a novel endoplasmic reticulum-resident protein containing both a BiP-interacting J-domain and thioredoxin-like motifs. J. Biol. Chem. 2003;278:2669–2676. doi: 10.1074/jbc.M208346200. [DOI] [PubMed] [Google Scholar]
- Kang S. W., Rane N. S., Kim S. J., Garrison J. L., Taunton J., Hegde R. S. Substrate-specific translocational attenuation during ER stress defines a pre-emptive quality control pathway. Cell. 2006;127:999–1013. doi: 10.1016/j.cell.2006.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. J., Mitra D., Salerno J. S., Hedge R. S. Signal sequences control gating of the protein translocation channel in a substrate-specific manner. Dev. Cell. 2002;2:207–217. doi: 10.1016/s1534-5807(01)00120-4. [DOI] [PubMed] [Google Scholar]
- Ladiges W. C., Knoblaugh S. E., Morton J. F., Korth M. J., Sopher B. L., Baskin C. R., Macauley A., Goodman A. G., Leboeuf R. C., Katze M. G. Pancreatic {beta}-cell failure and diabetes in mice with a deletion mutation of the endoplasmic reticulum molecular chaperone gene P58IPK. Diabetes. 2005;54:1074–1081. doi: 10.2337/diabetes.54.4.1074. [DOI] [PubMed] [Google Scholar]
- Lee T. G., Tang N., Thompson S., Miller J., Katze M. G. The 58,000-dalton cellular inhibitor of the interferon-induced double-stranded RNA-activated protein kinase (PKR) is a member of the tetratricopeptide repeat family of proteins. Mol. Cell. Biol. 1994;14:2331–2342. doi: 10.1128/mcb.14.4.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine C. G., Mitra D., Sharma A., Smith C. L., Hegde R. S. The efficiency of protein compartmentalization into the secretory pathway. Mol. Biol. Cell. 2005;16:279–291. doi: 10.1091/mbc.E04-06-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlack K.E.S., Misselwitz B., Plath K., Rapoport T. A. BiP acts as a molecular ratchet during posttranslational transport of prepro-alpha factor across the ER membrane. Cell. 1999;97:553–564. doi: 10.1016/s0092-8674(00)80767-9. [DOI] [PubMed] [Google Scholar]
- Menetret J. F., Hegde R. S., Heinrich S. U., Chandramouli P., Ludtke S. J., Rapoport T. A., Akey C. W. Architecture of the ribosome-channel complex derived from native membranes. J Mol. Biol. 2005;348:445–457. doi: 10.1016/j.jmb.2005.02.053. [DOI] [PubMed] [Google Scholar]
- Nicchitta C. V., Blobel G. Lumenal proteins of the mammalian endoplasmic reticulum are required to complete protein translocation. Cell. 1993;73:989–998. doi: 10.1016/0092-8674(93)90276-v. [DOI] [PubMed] [Google Scholar]
- Oyadomari S., et al. Cotranslocational degradation protects the stressed endoplasmic reticulum from protein overload. Cell. 2006;126:727–739. doi: 10.1016/j.cell.2006.06.051. [DOI] [PubMed] [Google Scholar]
- Rane N. S., Yonkovich J. L., Hegde R. S. Protection from cytosolic prion protein toxicity by modulation of protein translocation. EMBO J. 2004;23:4550–4559. doi: 10.1038/sj.emboj.7600462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski D. T., Arnold S. M., Miller C. N., Wu J., Li J., Gunnison K. M., Mori K., Sadighi Akha A. A., Raden D., Kaufman R. J. Adaptation to ER stress is mediated by differential stabilities of pro-survival and pro-apoptotic mRNAs and proteins. PLoS Biol. 2006;4 doi: 10.1371/journal.pbio.0040374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski D. T., Kaufman R. J. A trip to the ER: coping with stress. Trends Cell Biol. 2004;14:20–28. doi: 10.1016/j.tcb.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Scheuner D., Mierde D. V., Song B., Flamez D., Creemers J.W.M., Tasukamoto K., Ribick M., Schuit F. C., Kaufman R. J. Control of mRNA translation preserves endoplasmic reticulum function in beta cells and maintains glucose homeostasis. Nat. Med. 2005;11:757–764. doi: 10.1038/nm1259. [DOI] [PubMed] [Google Scholar]
- Scheuner D., Song B., McEwen E., Liu C., Laybutt R., Gillespie P., Saunders T., Bonner-Weir S., Kaufman R. J. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol. Cell. 2001;7:1165–1176. doi: 10.1016/s1097-2765(01)00265-9. [DOI] [PubMed] [Google Scholar]
- Schröder M., Kaufman R. J. The mammalian unfolded protein response. Annu. Rev. Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- Shaffer K. L., Sharma A., Snapp E. L., Hegde R. S. Regulation of protein compartmentalization expands the diversity of protein function. Dev. Cell. 2005;9:545–554. doi: 10.1016/j.devcel.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Shen Y., Hendershot L. ERdj3, a stress-inducible endoplasmic reticulum DnaJ homologue, serves as a coFactor for BiP's interactions with unfolded substrates. Mol. Biol. Cell. 2005;16:40–50. doi: 10.1091/mbc.E04-05-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Meunier L., Hendershot L. M. Identification and characterization of a novel endoplasmic reticulum (ER) DnaJ homologue, which stimulates ATPase activity of BiP in vitro and is induced by ER stress. J. Biol. Chem. 2002;277:15947–15956. doi: 10.1074/jbc.M112214200. [DOI] [PubMed] [Google Scholar]
- Tyedmers J., Lerner M., Widemann M., Volkmer J., Zimmermann R. Polypeptide-binding proteins mediate completion of co-translational protein translocation into the mammalian endoplasmic reticulum. EMBO Rep. 2003;4:505–510. doi: 10.1038/sj.embor.embor826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Huizen R., Martindale J. L., Gorospe M., Holbrook N. J. P58IPK, a novel endoplasmic reticulum stress-inducible protein and potential negative regulator of eIF2alpha signaling. J. Biol. Chem. 2003;278:15558–15564. doi: 10.1074/jbc.M212074200. [DOI] [PubMed] [Google Scholar]
- Walter P., Blobel G. Preparation of microsomal membranes for cotranslational protein translocation. Methods Enzymol. 1983;96:84–93. doi: 10.1016/s0076-6879(83)96010-x. [DOI] [PubMed] [Google Scholar]
- Yan W., Frank C. L., Korth M. J., Sopher B. L., Novoa I., Ron D., Katze M. G. Control of PERK eIF2alpha kinase activity by the endoplasmic reticulum stress-induced molecular chaperone P58IPK. Proc. Natl. Acad. Sci. USA. 2002;99:15920–15925. doi: 10.1073/pnas.252341799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinszner H., Kuroda M., Wang X., Batchvarova N., Lightfoot R. T., Remotti H., Stevens J. L., Ron D. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998;12:982–995. doi: 10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.