Abstract
Centrin is a calcium-binding centrosome/basal body–associated protein involved in duplication and segregation of these organelles in eukaryotes. We had shown that disruption of one of the centrin genes (centrin1) in Leishmania amastigotes resulted in failure of both basal body duplication and cytokinesis. Here, we undertook to define the role of centrin1 (TbCen1) in the duplication and segregation of basal body and its associated organelles kinetoplast and Golgi, as well as its role in cytokinesis of the procyclic form of Trypanosoma brucei by depleting its protein using RNA inhibition methodology. TbCen1-depleted cells showed significant reduction in growth compared with control cells. Morphological analysis of these cells showed they were large and pleomorphic with multiple detached flagella. Both immunofluorescence assays using organelle-specific antibodies and electron microscopic analysis showed that TbCen1-deficient cells contained multiple basal bodies, kinetoplasts, Golgi, and nuclei. These multiple organelles were, however, closely clustered together, indicating duplication without segregation in the absence of centrin. This failure in organelle segregation may be the likely cause of inhibition of cytokinesis, suggesting for the first time a new and unique role for centrin in the segregation of organelles without affecting their multiplication in the procyclic form of T. brucei.
INTRODUCTION
Centrin is a calcium-binding protein associated with microtubule-organizing centers (MTOC), such as the centrosome of nonciliated cells, basal bodies of ciliated or flagellated cells, and spindle pole body of yeast cells (Salisbury et al., 1988; Spang et al., 1993; Salisbury, 1995; Wolfrum and Salisbury, 1998; Klink and Wolniak, 2001). It is part of the filaments within and attached to such organelles that can contract in response to changes in Ca2+ concentration as in Tetraselmis striata and Chlamydomonas reinhardtii (Salisbury et al., 1984; Wright et al., 1985). Thereby centrin is involved in duplication and segregation of centrosome/basal bodies and is important for cell division in eukaryotes (Errabolu et al., 1994; Salisbury, 1995). However, the basic mechanism of such centrin function is not known. There are several centrin genes identified in different eukaryotes: one in Chlamydomonas (CrCen) and Saccharomyces cerevisiae (CDC31), four in mice (MmCen1-4), and three in humans (HsCen1-3; Huang et al., 1988; Errabolu et al., 1994; Salisbury, 1995; Gavet et al., 2003). The recently completed genome database for two trypanosomatids, i.e., Trypanosoma brucei, causative agent of African sleeping sickness and Leishmania major that causes cutaneous leishmaniasis disease in human (Berriman et al., 2005; Ivens et al., 2005) and our own studies with L. donovani, causative agent of visceral leishmaniasis in human, have revealed that there are five putative centrin genes in this group of organisms (Selvapandiyan et al., 2001, 2004).
Some of the centrins have been assigned particular functions. For example, one group of centrins, which includes human centrins (HsCen1 and 2), mouse centrins (MmCen1 and 2), C. reinhardtii centrin (CrCen) and Paramecium centrins (PtCen2 and 3) is involved in centrosome/basal body segregation (Middendorp et al., 1997; Koblenz et al., 2003; Ruiz et al., 2005). The other group containing human centrin3 (HsCen3), mouse centrin3 (MmCen3), yeast centrin (CDC31), and Leishmania centrin1 (LdCen1) plays a role in centrosome/basal body duplication (Middendorp et al., 1997; Khalfan et al., 2000; Gavet et al., 2003; Selvapandiyan et al., 2004). Mouse centrin4 (MmCen4) is involved in the basal body assembly in the brain ependymal and choroidal ciliated cells (Gavet et al., 2003). Knockdown of centrin2 by ribonucleic acid inhibition (RNAi) in human cells and a centrin gene deletion in Tetrahymena thermophila yielded defects in centrosome/basal-body duplication and cell cycle progression (Salisbury et al., 2002; Stemm-Wolf et al., 2005; Tsang et al., 2006), whereas centrin disruption in Chlamydomonas led to aberrant numbers of basal bodies that interfered with cytokinesis (Koblenz et al., 2003). Centrins have also been found involved in other cellular processes such as maintenance of membrane integrity and cell morphology in yeast (yeast centrin, CDC31; Ivanovska and Rose, 2001), homologous recombination and nucleotide excision repair in Arabidopsis (centrin2) and humans (HsCen2; Molinier et al., 2004; Nishi et al., 2005), nuclear mRNA export in yeast (CDC31; Fischer et al., 2004), genomic instability via increased chromosome loss in C. reinhardtii (CrCen; Zamora and Marshall, 2005), Golgi duplication in T. brucei (TbCen2; He et al., 2005) and in defining the geometry of basal body duplication in Paramecium (PtCen2 and 3; Ruiz et al., 2005).
Our earlier studies with Leishmania donovani have shown that when the N-terminally (1-28 amino acids) deleted L. donovani centrin protein (LdCen1) was overexpressed in the parasite, it had a dominant negative effect on the growth of the parasite, resulting in an enrichment of cells in the G2/M stage (Selvapandiyan et al., 2001). Further, a deletion of this gene in L. donovani resulted in specific growth arrest in the amastigote form (a stage that proliferates intracellularly in human macrophages) in vitro as well as ex vivo in human macrophages (Selvapandiyan et al., 2004). The growth arrested amastigotes were defective in basal body duplication followed by a failure in cytokinesis resulting in large multinucleated cells (Selvapandiyan et al., 2004).
The eukaryotic cytoskeleton regulates important cellular functions, including the control of cell shape, cell movement, intracellular transport, and cell division. Centrosome/basal body nucleate microtubules necessary for postmitotic changes, which include organelle repositioning and cytokinesis. The protozoan parasite T. brucei has unique and precisely ordered microtubule-mediated cytoskeletal structures. These include the subpellicular corset of microtubules that maintain cell shape, the single flagellum that exits the cell body through the flagellar pocket, the mitochondrial DNA (kinetoplast), the basal body, and the single nucleus (Robinson et al., 1995). In addition basal bodies in T. brucei play a role, probably through microtubules, in determining the site of the new Golgi and endoplasmic reticulum export site (He et al., 2004). The multiplication and segregation of these structures are developmentally coordinated with great precision (Robinson et al., 1995). They offer a great opportunity for studying the relationship between the organelles, and the proteins associated with them that are involved in their function.
A genomic sequence search in the T. brucei database (Berriman et al., 2005) revealed the presence of five putative centrin genes in these organisms (TbCen1-5). The functions of two of these centrins (TbCen2 and 3) have been recently reported (He et al., 2005). TbCen2, which is localized at the basal body and a bilobed structure close to the Golgi, is responsible for the duplication of both basal body and the Golgi. TbCen3, which is localized at the basal body of the parasite, is also responsible for the duplication of the basal body in T. brucei (He et al., 2005). Here, we characterize the localization and function of TbCen1 in T. brucei. Our studies reveal that TbCen1 is localized at the basal body and a bilobed structure close to the Golgi. TbCen1 depletion affects cell growth and results in impairment of the appropriate segregation of basal bodies and nucleus, along with the arrest in the movement of the other basal body–associated organelles, viz., kinetoplasts and Golgi. In addition, we observed early on detachment of flagella, which is part of basal body complex. The outcome of such a defect leads to loss of cytokinesis, resulting in enlarged cell size. Our studies also revealed that depletion in TbCen1 does not affect the duplication of basal body, kinetoplast, Golgi, and nucleus, unlike TbCen2. This suggests that TbCen1 may have a unique role in overall organelle segregation and not in organelle duplication.
MATERIALS AND METHODS
In Vitro Culture of Parasites
T. brucei procyclic form strain 29-13 that harbors integrated genes for T7 RNA polymerase and tetracycline repressor (Wirtz et al., 1999) was used. The parasites were grown in SDM-79 medium supplemented with 10% fetal bovine serum during normal growth and 15% during transfection and clonal selection. G418 (15 μg/ml) and hygromycin B (50 μg/ml) were added in the medium to preserve T7 RNA polymerase and tetracycline repressor gene constructs within the cells. The parasites were grown and harvested as described previously (Morris et al., 2004).
Gene Cloning and Transfection of Parasites for RNAi
Gene characterization via cognate mRNA degradation (RNAi) and arrest of translation is a routinely followed molecular technique as described in certain eukaryotes (Ullu et al., 2004). This is achieved through plasmid construct carrying a unique portion of the gene, which after transfection in cell generates double-stranded RNA of the gene upon induction (currently with tetracycline), which then targets the cognate native mRNA for degradation. Specifically in this study to amplify PCR fragment of TbCen1 gene for developing RNAi construct, gene-specific forward and reverse primers were designed utilizing the putative centrin sequence from the T. brucei genome sequence databank (Berriman et al., 2005). Oligos were designed (Supplementary Table S1) in which the amplicon constitutes a portion from the 5′ untranslated region (from −69 base pairs) of the genes into approximately the middle of the open reading frame (ORF; +278 base pairs) with HindIII and XhoI restriction sites added to the termini of the PCR fragments. The PCR amplified fragment was a 347-base pair unique sequence that had no significant sequence identity with the rest of the T. brucei genome sequences. The fragment was subcloned into the HindIII and XhoI sites of the pZJM vector (Wang et al., 2000). The recombinant plasmid was linearized with NotI and DNA transfection carried out on the procyclic form of T. brucei using a published procedure (Morris et al., 2004) using a BTX ECM 630 Electroporator (BTX, San Diego, CA, 500 V and 275 μF). Transfectants were selected in the presence of 2.5 μg/ml phleomycin and a clonal cell line was obtained by limiting dilutions. For induction of RNAi, the cloned stable transfectant that reached a constant growth rate after at least three regular subpassages was cultured in the presence of 1.0 μg/ml tetracycline. For monitoring cell growth, the cells were counted at different time intervals using a Coulter Z1 particle counter (Beckman Coulter, Fullerton, CA) with a starting culture of 1 × 105 cells/ml. To include counting of larger cells that appear during RNAi, an aperture setting between 6.5 and 30 μm was used.
Isolation of RNA and Northern Blot Analysis
To examine the specific reduction in centrin transcription during RNAi induction, the cloned stable transfectant, either uninduced or induced with tetracycline for 3 d, was analyzed for TbCen1 mRNA level using Northern blot analysis. The membrane was rehybridized with α-tubulin gene-specific probe as loading control, and with TbCen2-5 gene-specific probes to confirm the specific inhibition of TbCen1 transcript. Probes were designed (Supplementary Table S1) in such a way that the amplicon constitute a portion from the middle of the ORF, not including the region selected for RNAi. The signal intensity was quantitated using a Phosphor Imager system (Molecular Dynamics, Amersham Pharmacia Biotech, Piscataway, NJ) as described previously (Selvapandiyan et al., 2001).
Ectopic Expression of Centrins in T. brucei
In the absence of specific antibodies expression of a gene is routinely monitored by the localization of an ectopic expression of that protein through an expression plasmid (Selvapandiyan et al., 2001). Such proteins are usually expressed with tag sequences that help in the specific identification of protein's localization. To ectopically express TbCen1 in the procyclic-form T. brucei, the TbCen1 gene ORF was amplified using the primers described in Supplementary Table S2 and cloned into pLEW100 (Wirtz et al., 1999) in frame with either three concatameric hemagglutinin (HA) tag sequences (Pati, 1992; pLEW100-HA) or a cyan fluorescent protein (CFP; Chudakov et al., 2004) tag sequence (pLEW100-CFP) at the 3′-end. The authenticity of the PCR products was confirmed by DNA sequencing. The constructs were transfected into the T. brucei 29-13 for integration in the genome as described above (Wirtz et al., 1999). The proteins overexpressed by addition of 0.5 μg/ml tetracycline were analyzed by Western blot analysis (Selvapandiyan et al., 2001).
Flow Cytometry
Procyclics inoculated at 1 × 105 cells/ml were allowed to grow and the cultures were harvested at 0, 3, and 5 d, fixed with paraformaldehyde, stained with propidium iodide (PI), and analyzed by flow cytometry according to the procedure described previously (Tu and Wang, 2004; Raslova et al., 2006). Cells were analyzed for populations with 2C, 4C, and >4C as a measure of relative DNA content. The stained cell samples were also examined with an Olympus phase-contrast and fluorescence microscope (Olympus, Melville, NY) to count the number of nuclei and kinetoplasts in individual cells from a population of 200 cells.
Immunofluorescence Analysis
For the immunofluorescence experiments, cells were prepared and analyzed under a fluorescence microscope following the procedure described previously (Kumar and Wang, 2006). Paraformaldehyde fixed midlog T. brucei cells were stained with anti-LdCen1 (rabbit polyclonal antibody [pAb] against L. donovani LdCen1; Selvapandiyan et al., 2001; 1:300 dilution) for staining TbCen1; YL1/2 (rat mAb against yeast tyrosinated-α-tubulin from Chemicon, Temecula, CA; 1:400 dilution; Kilmartin et al., 1982) for staining the basal body; L8C4 (anti-paraflagellar rod antibodies from Dr. Keith Gull, Oxford University; 1:4 dilution; Kohl et al., 1999) to stain the flagella; GRASP (pAb against Golgi reassembly stacking protein from Dr. Graham Warren, Yale University School of medicine; 1:500 dilution; He et al., 2004) to stain Golgi; anti-HA-Tag (from Santa Cruz Biotechnology, Santa Cruz, CA, sc-7392; 1:500 dilution) to stain ectopically expressed HA-tagged TbCen1 proteins. Appropriate secondary antibodies (anti-rat Alexa Flour 488 A-21208, anti-mouse Alexa Flour 488 A-21422, anti-mouse Alexa Flour 588 A-21422, and anti-rabbit Alexa Flour 488 A11008; all from Molecular Probes [Invitrogen, Carlsbad, CA]) were used at 1:500 dilutions.
Electron Microscopy
Parasites harvested at appropriate time periods from culture were prepared and examined by electron microscopy as described previously (Lingle et al., 1998; Lee et al., 2002).
RESULTS
Sequence Analysis and Nomenclature of T. brucei Centrins
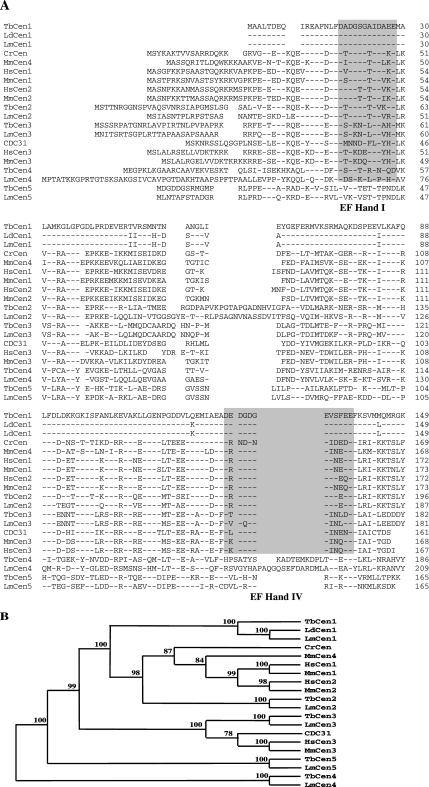
The five putative T. brucei centrin ORF sequences obtained from GeneDB were compared with the characterized three human, four mouse, one yeast, and one Chlamydomonas centrin genes by ClustalW (Figure 1A). For comparison we have also included the Leishmania centrin counterparts (L. donovani centrin [Selvapandiyan et al., 2001] and five putative L. major centrins [www.genedb.org]) in the analysis. Based on the sequence similarity and clustal analysis, the centrins from L. donovani (AAL01153), L. major (LmjF22.1410), and T. brucei (XP_845964) that share 97–100% sequence similarity (Supplementary Data: Supplementary Table S3) fall in the same cluster with HsCen1, HsCen2, MmCen1, MmCen2, and CrCen in the phylogenetic tree (Figure 1B) were designated as LdCen1, LmCen1, and TbCen1, respectively. Centrin1 sequences from the trypanosomatid family are smaller in size, in comparison to all the other centrins in the eukaryotes, because of a shorter N terminal region (Figure 1A). Similarly, the L. major and T. brucei centrins (LmjF07.0710; XP_824555) that share 71% similarities to both HsCen2 and MmCen2 were designated as LmCen2 and TbCen2, respectively. Leishmania centrin (LmjF34.2390) and T. brucei centrin (XP_825248), which have 67–71% similarity with HsCen3 and MmCen3 and coexist with HsCen3, MmCen3, and CDC31 in the phylogeny, were designated as LmCen3 and TbCen3, respectively. Recently, He et al. (2005) named the five centrins of T. brucei based on their homology to only Chlamydomonas centrin. According to their nomenclature TbCen1 is homologous to human or mouse centrin 3. This nomenclature is at odds with the known nomenclature for centrins from different organisms; therefore, in this report we have used centrin nomenclature that is consistent with all other organisms, based on the above-mentioned extensive sequence and phylogenetic analysis. Thus, we referred centrin1 of He et al. (2005) as TbCen3 in the present study. Both, Leishmania and T. brucei centrins 1-3, similar to HsCen1-3, also have EF-hands I and IV as two putative calcium-binding sites predicted by the motif analysis program (http://molbio.info.nih.gov/molbio/; Figure 1A). The Leishmania and T. brucei centrins (LmjF32.0660; XP_829446) with a single putative calcium-binding site (EF-hand I) were designated as LmCen4 and TbCen4, respectively, whereas, the Leishmania and T. brucei centrins (LmjF36.6; XP_823096), with no predicted calcium-binding site, were designated as LiCen5 and TbCen5, respectively (Figure 1B). The mouse has a unique centrin MmCen4, which is more closely related to mammalian centrin2 and 3 but is expressed exclusively in the basal bodies of the ependymal and choroidal ciliated cells (Gavet et al., 2003). However none of the kinetoplastid members have such a centrin homolog. The use of such a uniform nomenclature will be ultimately beneficial in comparing the functions of centrins across different organisms.
Figure 1.
(A) Multiple alignment of centrin protein sequences of T. brucei (TbCen), L. donovani (LdCen), L. major (LmCen), Chlamydomonas (CrCen), yeast (CDC31), mouse (MmCen), and human (HsCen). The sequence names with the accession numbers are as follows: TbCen1 (XP_824270), TbCen2 (XP_824555), TbCen3 (XP_825248), TbCen4 (XP_829446), TbCen5 (XP_823096), LdCen1 (AAL01153), LmCen1 (LmjF22.1410), LmCen2 (LmjF07.0710), LmCen3 (LmjF34.2390), LmCen4 (LmjF32.0660), LmCen5 (LmjF36.6), CrCen (P05434), CDC31 (NP-014 900), MmCen1 (NP_031619), MmCen2 (NP_062278), MmCen3 (AAH54097), MmCen4 (CAI26236), HsCen1 (AAH29515), HsCen2 (NP_004335), and HsCen3 (O15182). The accession numbers of T. brucei, L. donovani, Chlamydomonas, yeast, and human are from GenPept data bank. The accession numbers of L. major are from www.genedb.org. The alignment was generated utilizing ClustalW of MacVector 7.2.2 program. Amino acids are listed in the standard one-letter code, and residues identical to TbCen1 are indicated by dashes. The gray boxes (EF-hands I and IV) are the putative Ca2+-binding domains. (B) Dendrogram of complete protein sequences of centrins mentioned above were generated in the MacVector 7.2.2 program, utilizing the systematic bootstrap unweighted pair group method with arithmetic mean. The numbers on the nodes indicate the percent of times the respective centrins grouped together in 1000 bootstrap samples in the program.
RNAi-induced Reduction of Specific TbCen1 mRNA
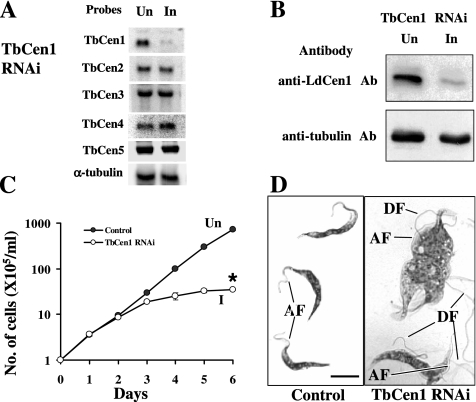
To characterize TbCen1 function for the first time, we knocked down the mRNA level of this centrin using RNAi methodology in the procyclic stage of the parasite. Northern blot analysis of RNA obtained from the tetracycline-induced culture on day three revealed the reduction of the cognate mRNA level (Figure 2A). As a loading control, the membrane was also reprobed with a α-tubulin gene fragment, and its mRNA level was quantitated. After normalizing the centrin band intensity with the intensity of the α-tubulin mRNA control, the reduction in the amount of centrin mRNAs in the tetracycline-induced culture compared with the uninduced culture was ∼92%. Rehybridizing the membrane with probes of other centrin genes (TbCen2-5) showed that the mRNA levels of these genes were not affected. As an additional confirmation, based on the observation that LdCen1 antibody, cross-reacts with TbCen1 (Supplementary Figure S1), we measured the level of TbCen1 protein in the uninduced and the induced cells. A significant reduction in the level of TbCen1 protein was also observed in the induced cells (Figure 2B).
Figure 2.
(A) Northern blots of RNA from induced and uninduced TbCen1 culture. Membrane containing 12 μg total RNA from 3-d uninduced (Un) and induced (I) cells was used. Membrane for TbCen1, in addition to hybridization with either specific centrin DNA or α-tubulin (Ngo et al., 1998) probes was reprobed with probes of TbCen2-5 to confirm on the specific inhibition of only the TbCen1 transcript. (B) Western blots on the parasite lysates showing reduction in the protein level of TbCen1 during RNAi induction by anti-LdCen1 Ab (top) and the loading control showing the level of α-tubulin by anti-tubulin Ab (bottom). Un, uninduced; In, induced. In each lane 20 μg of total extracted proteins were loaded. (C) The effect of TbCen1 knockdown on the in vitro growth of T. brucei procyclics. The cells were grown with (I) or without (Un) tetracycline. The data represent the means of ± SD of three independent experiments. *p < 0.04. (D) Geimsa-stained images of the control and TbCen1-depleted cells, 4 d after RNAi induction. Scale bar for both images, 5 μm. AF, attached flagella; DF, detached flagella.
TbCen1 Is Essential for the Growth of the Parasite
The effect of reduction in TbCen1 level on the growth of the TbCen1 RNAi cells was monitored by counting cell number daily for 6 d after RNAi induction (Figure 2C). The tetracycline-induced parasites started showing the growth defect on day 3 compared with uninduced control cells. Cell density on day 5 was 11% of the control for TbCen1-depleted cells. There was no increase in the cell numbers of the RNAi-induced TbCen1 parasites after day 5, whereas the uninduced culture continued to grow throughout the experiment. Thus, TbCen1 RNAi-expressing cells showed a reduced growth rate after induction compared with the uninduced cells. Geimsa-stained TbCen1-depleted cells at day 4 in culture, under microscope, showed that the cells were large, pleomorphic in shape and had multidetached flagella, unlike the uninduced cells that were uniform in shape with one or two attached flagellum, depending on the cell cycle stage (Figure 2D and see Figure 9).
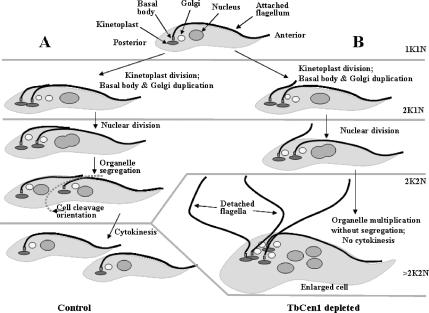
Figure 9.
Schematic diagram of the segregation of the organelles and its failure in the absence of TbCen1 in T. brucei procyclic form. (A) Normal cell division. (B) The arrest of organelle segregation and cytokinesis upon depletion of TbCen1.
TbCen1 Depletion Arrests Cytokinesis Resulting in Large Cells with Multiple Organelles
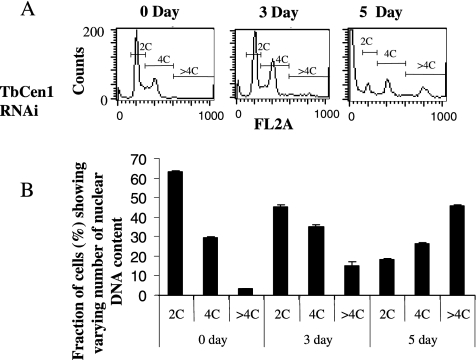
To determine the cause of growth arrest, the RNAi-induced TbCen1 cells along with uninduced cells as control were stained with PI and subjected to flow cytometry to analyze for relative DNA content (C) at different time intervals in culture (Figure 3A). The percent of cells having 2C, 4C, and >4C were calculated and the data are shown in Figure 3B. The analysis revealed that the TbCen1 RNAi cultures displayed gradual increase in the number of cells with >4C (Figure 3, A and B). Approximately 15% of TbCen1 RNAi-induced cells had >4C at day 3 compared with 3% of the cells at day 0 and the percentage of >4C cells increased to up to 45% at day 5 (Figure 3B). The increase in the >4C cell population coincided with the simultaneous decrease in 2C cells with progression of TbCen1 depletion (Figure 3, A and B). In Figure 3A the 5-d time point the far left-hand edge of the figure shows cells having less than 2C, which usually represent dying cells, as was observed previously in LdCen1-depleted L. donovani amastigotes (Selvapandiyan et al., 2004).
Figure 3.
Fluorescence-activated cell sorting analysis for relative DNA content of TbCen1-depleted cells. The percentage of cells that contain two times the DNA content (2C), four times (4C), and more than 4 times (>4C) were measured at each time point and are shown. (A) The corresponding cell frequency units obtained directly from the flow cytometry analysis are shown. (B) Histograms with the quantitated values of the cell populations are shown. Data represent the means of ± SD of three independent experiments.
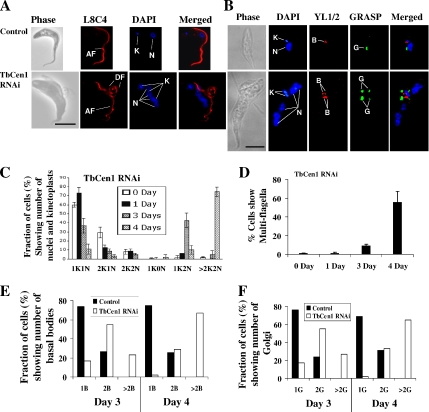
As a first step to analyze the organization of various organelles of both control and TbCen1-depleted cells, the cells at day 4 after induction were stained with DAPI to identify the nuclei and kinetoplasts and with L8C4 antibody (antibodies to paraflagellar rod, which is a complex lattice-like structure running alongside the axoneme in the flagellum of T. brucei; Bastin et al., 1998) to visualize the flagella. The stained cells were observed under a fluorescence microscope for organelle position. The majority of the uninduced or wild-type cells showed a single kinetoplast (the mitochondrial genome), a single nucleus with one flagellum (Figure 4A, top). However, TbCen1-depleted cells have multinuclei, multikinetoplasts, and multiflagella in each cell (Figure 4A, bottom). Such cells were also large and highly pleomorphic (Figure 4A, bottom), as opposed to the control cells that were small and twisted (Figure 4A, top). With such large cells, we noticed multiple detached flagella along with the attached flagellum (Figure 4A, bottom). We presume that such an attached flagellum could be the initial flagellum of the cells before the induction of RNAi. To monitor the progression of organelle replication, we counted the number of nuclei, kinetoplasts, basal bodies, Golgi, and flagella in centrin-depleted cells until day 4 after induction. A gradual increase in the number of cells with multiple nuclei and multiple kinetoplasts (>2K2N) was observed from day 3 onward in the centrin-depleted cultures, coupled with a concomitant decrease in cells with one nucleus and one kinetoplast (1K1N; Figure 4C). We also observed cells with two kinetoplasts and one nucleus (2K1N), two kinetoplasts and two nuclei (2K2N), one kinetoplast and no nucleus (1K0N: zoid) and one kinetoplast and two nuclei (1K2N) as intermediate populations before finally the majority (>70%) of them on day 4 became >2K2N cells (Figure 4C).
Figure 4.
(A and B) Microscopic images of control and TbCen1-depleted procyclic cells from day 4 cultures. In each cell, DAPI stained kinetoplasts (K) and nuclei (N), the L8C4 stained flagella (F), YL1/2 stained basal bodies (B), and GRASP stained Golgi (G). AF, attached flagellum; DF, detached flagellum. All the images are the same scale (scale bars, 5 μm). (C) Bar graph showing increase in the proportion of multinucleated and multikinetoplast cells with days after RNAi knockdown of TbCen1. Cells after different days of TbCen1 RNAi induction were stained with PI and examined by fluorescence microscope for tabulation of the cells with different number of kinetoplasts (K) and nuclei (N). (D) Percentage of TbCen1-depleted cells with multiflagella. Cells after different days of TbCen1 RNAi induction were observed under microscope under bright field, and the cells that contained more than two flagella were counted. For both B and C at least 250 total cells were observed for the quantitation at each time point. The data represent the means of ± SD of three independent experiments. (E and F) Percent of cells on day 3 and 4, which show varying number of basal bodies (E) and Golgi (F). The data only shows the number of mature basal bodies that were stained with YL1/2 Ab and were easily visible than the probasal bodies and GRASP that stains the Golgi as in B. In each case more than 125 cells were analyzed.
We also observed that the TbCen1-depleted cells in culture continue to grow in size, with continuous replication of kinetoplasts and nuclei until the nutrients are depleted in the medium (Figures 2D and 4C). Because the TbCen1 RNAi cells have multiflagella, we quantitated them in a separate analysis. Almost 60% of the TbCen1-depleted cells on day 4 had more than two flagella (Figure 4D). Previously, we had observed that knockdown of LdCen1 expression in L. donovani amastigotes affected basal body duplication. Further, coordinated biogenesis of basal body and Golgi has been recently shown in T. brucei (Field et al., 2000; He et al., 2004). To determine the fate of basal bodies and Golgi in the TbCen1-depleted T. brucei cells, an immunofluorescence analysis was conducted on such cells using YL1/2 antibody that stains the basal bodies and GRASP antibody that stains the Golgi. Most of the uninduced cells show a pair of basal bodies and single Golgi (Figure 4B, top). However, majority of TbCen1-depleted cells displayed multibasal bodies and multi-Golgi (Figure 4B, bottom). Careful quantification of the basal body and Golgi number in the centrin-depleted cells on day 3 and 4 revealed the existence of cells with one, two, or more than two mature basal bodies and Golgi. Approximately 60% of cells with more than two basal bodies and Golgi on day 4 after RNAi induction were observed with concomitant decrease in the cells with two basal bodies and two Golgi (Figure 4, E and F).
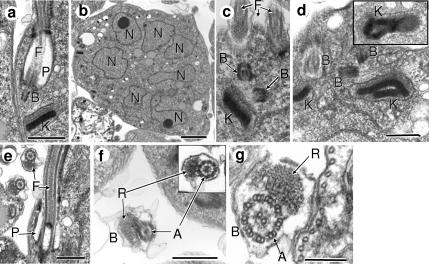
To further confirm the abnormal morphological characteristics of TbCen1-depleted cells, we examined such cells by electron microscopy (EM). The features of multibasal bodies, multikinetoplasts, multiflagella, and multinuclei observed in the TbCen1-depleted cells by immunofluorescence microscopy were confirmed by the EM studies (Figure 5, B–D). The uninduced cells mostly had a single nucleus, kinetoplast and basal body (Figure 5A). In a limited number of observations the EM morphology of the flagellar pocket in the TbCen1-depleted cells appears to be normal (Figure 5E). Although the newly formed flagella are detached type in TbCen1 RNAi cells (Figure 6A), the 9 + 2 arrangement of the microtubules of the axoneme and paraflagellar rod (Bastin et al., 2000) is not affected (Figure 5F) and appears similar to the attached flagellum (Figure 5G). All these observations clearly demonstrate the failure of cell division in the TbCen1-depleted T. brucei cells, which resulted in increase in number of the cellular organelles with concomitant increase in cell size.
Figure 5.
Electron microscopy of centrin-depleted cells. (a) A nondividing control cell shows single flagellum, basal body, and kinetoplast. (b–d) TbCen1-depleted cells with multinuclei, multibasal bodies, and multikinetoplasts are sequentially displayed in images. Inset in d is more representation of divided kinetoplasts. (e) Similar cell showing the unaffected flagellar pocket with the old attached flagellum and a cross section of a newly formed detached flagellum. (f) The cross section of detached flagella from TbCen1-depleted cells showing the paraflagellar rod and the unaffected 9 + 2 microtubule structure in the flagellar axoneme (inset). (g) The cross section of an attached flagellum of a centrin-depleted cell showing the typical regions of paraflagellar rod and the axoneme that shows the 9 + 2 arrangement of microtubules. F, flagellum; P, flagellar pocket; B, basal body; K, kinetoplast; N, nucleus; R, paraflagellar rod; A, axoneme. Cells in all experiments were from 5 d after RNAi induction, except in d, where the inset of f and g are from 2 d after RNAi induction. Scale bars, (a and c–f) 500 nm; (b), 2 μm; and (g) 250 nm.
Figure 6.
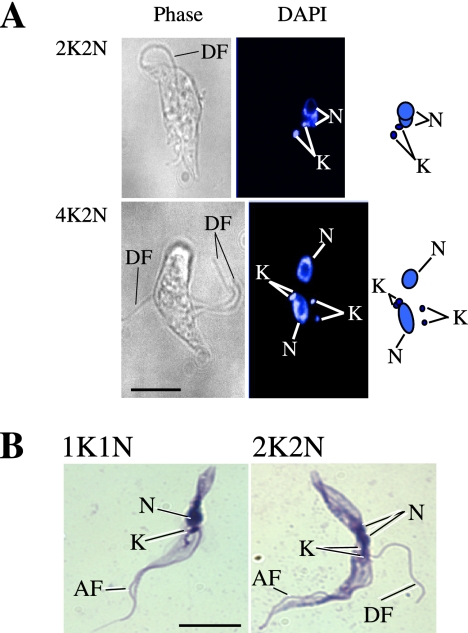
Effect on the attachment of the newly formed flagella in the TbCen1 RNAi cells. (A) DAPI-stained cells; (B) Geimsa-stained cells. More than 40 cells were used for the analysis. AF, attached flagella; DF, detached flagella; K, kinetoplast; N, nucleus. Scale bars, 5 μm.
TbCen1 Depletion Affects Flagellar Attachment
Centrin-depleted cells showed newly formed flagella that were of detached type although the original flagellum in such cells was still seen as attached (Figure 2D). To find out whether the detachment of the flagella in the centrin RNAi cells is either a primary or a secondary effect of centrin depletion, we analyzed cells at early stages of RNAi induction. Careful observation of 2K2N cells on day 2 of RNAi induction revealed that the very first new flagella of more than 90% of such cells, where the kinetoplasts did not segregate, showed detached flagellum in addition to the old attached flagellum (Figure 6A, top, and B, right). Similarly in 4K2N centrin RNAi cells, we observed three new detached flagella (Figure 6A, bottom). These results suggest that flagellar detachment is an early event in the TbCen1-depleted cells and occurs concomitantly with the failure of segregation of basal-bodies, flagellum, and kinetoplast.
TbCen1 Is Associated with the Basal Bodies
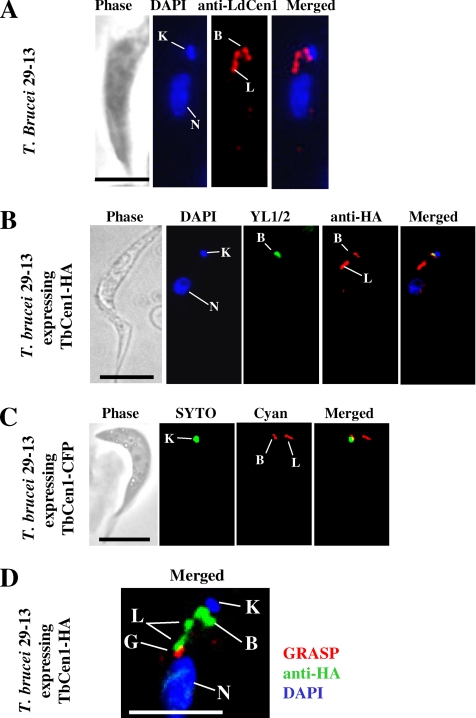
To ascertain the localization of TbCen1 in T. brucei, we carried out immunofluorescence analysis. Using anti-LdCen1 Ab (Selvapandiyan et al., 2001), which cross-reacts specifically to TbCen1 protein in a Western blot analysis (Supplementary Figure S1), we noticed staining at the basal body region (Figure 7A). However, in addition to the basal body, the anti-LdCen1 Ab also stained an additional area (a bilobed structure) next to the basal body. To confirm that the localization of TbCen1 in the bilobed structure was not an aberrant observation, we ectopically expressed 1) TbCen1 fused with an HA tag (TbCen1-HA) using a pLEW100-HA vector and 2) TbCen1 fused with CFP using a pLEW100-CFP vector in the parasite. The parasites that express either TbCen1-HA or TbCen1-CFP showed normal growth compared with control cells, and the expression of both the fusion proteins was observed in over 90% of the cells using immunofluorescence analysis (data not shown). The anti-HA Ab-stained TbCen1 at both the basal body and the bilobed structure (Figure 7B). The fluorescent TbCen1-CFP also similarly localized at the basal body and the bilobed structure (Figure 7C). The TbCen1-CFP localization was monitored in the live cells by fluorescence microscope. Using three different immunolocalization approaches for TbCen1 clearly suggesting that TbCen1 localizes both in the basal body region and to a bilobed structure. In addition, it also suggests that TbCen1 localization to bilobed structure is not due to overexpression of the protein because such localization was found in nonoverexpressing cells (Figure 7A). Previously, it was reported that TbCen2 is also localized to the both basal body and a bilobed structure that is in close proximity with the Golgi apparatus (He et al., 2005). Using both GRASP, an antibody that stains Golgi, and anti-HA Ab, which localizes TbCen1, we confirmed that the bilobed structure is also in close proximity to Golgi in the transfected T. brucei cells (Figure 7D). However, the reason for the localization of TbCen1 (in this study) and TbCen2 (in He et al., 2005) at the bilobed structure along with basal body and its relevance to the function of these proteins is not known at this time.
Figure 7.
(A) Immunolocalization of TbCen1 in the procyclics of T. brucei 29-13 using anti-LdCen1 Ab (stains LdCen1 at the basal body in L. donovani (Selvapandiyan et al., 2004) stains here the basal body and the bilobe. (B) Localization of TbCen1-HA proteins in T. brucei 29-13 procyclic cells transformed with pLEW100-TbCen1. Staining with anti-HA Ab stains the basal body and the bilobe, and YL1/2 stains the basal body. (C) Fluorescent images of a live T. brucei cell ectopically expressing TbCen-CFP are stained with cyan, and kinetoplast was stained with SYTO Green-Fluorescent Nucleic Acid Stain (Molecular Probes, Invitrogen). (D) Localization of TbCen1 with respect to Golgi. Cells were stained with GRASP to stain the Golgi and anti-HA Ab to stain TbCen1-HA at the basal body and the bilobe. In all, the parasites obtained were from midlog culture and also stained with DAPI for the nuclei and the kinetoplasts. K, kinetoplast; N, nucleus; B, basal body; G, Golgi; L, bilobe. Scale bar in all, 5 μm.
TbCen1 Is Involved in Segregation of Organelles Necessary for Completion of Cytokinesis
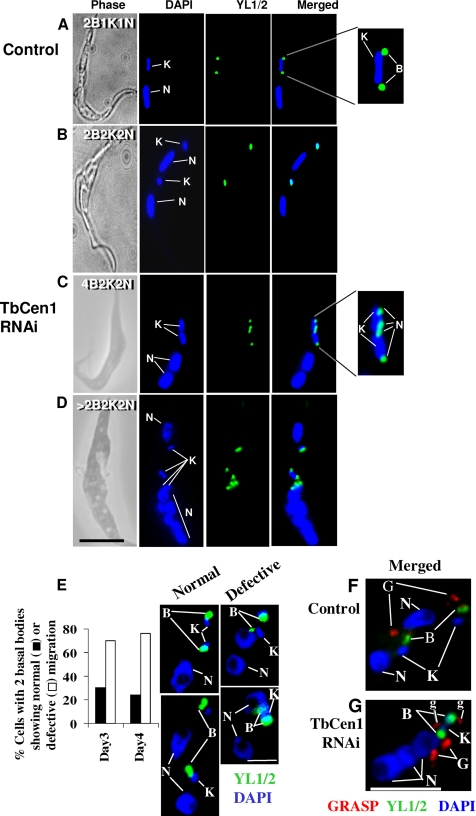
Previously we showed that disruption of a L. donovani centrin (LdCen1) affected basal body duplication, which in turn affected cytokinesis in the amastigote form of L. donovani (Selvapandiyan et al., 2004). In this study we explored the effect of TbCen1 depletion on the basal body duplication and segregation, along with the segregation of other organelles, such as kinetoplasts, nuclei, and Golgi in the procyclic form of TbCen1-depleted cells. Immunofluorescence analysis was carried out using YL1/2 and GRASP antibodies for identifying basal bodies and Golgi, respectively. Control cells displayed a normal segregation pattern of the duplicated basal bodies and the divided kinetoplasts (Figure 8A, magnified portion in 8A and 8B, and Figure 9A), which is a prerequisite for the initiation of cytokinesis (Robinson and Gull, 1991). Careful observation of TbCen1 RNAi cells revealed the absence of segregation of the duplicated basal bodies and kinetoplast complex (Figures 8, C and D, and 9B). The magnified portion in Figure 8C shows the basal bodies that have gone through a second round of duplication without segregation. High-resolution EM of TbCen1-depleted cells also showed multiple number of basal bodies and kinetoplasts grouped in one area (Figure 5, c and d). Basal-body migration pattern in the normal T. brucei cells was described previously (Robinson et al., 1995). On the basis of that study we have measured the migration of basal bodies in TbCen1 RNAi cells at the two-nucleated stage. At day 3 after RNAi induction the quantitation revealed that ∼75% of the cells contained the basal bodies that were close to each other and not separated as in the uninduced cells (Figure 8E). The two nucleated cells with multiple nonseparated basal bodies continued to increase from day 3 to day 4. These results clearly indicate a defect in basal body segregation after centrin depletion. Using a Golgi-specific antibody, GRASP, we found normal duplication of Golgi similar to other organelles in the TbCen1-depleted cells (Figure 8, F and G). We have also noticed more than one new Golgi structures stained with GRASP in both the control (data not shown) and the centrin-depleted cells (Figure 8G marked as g), as observed by He et al. (2004) during the normal T. brucei cell cycle. However, in TbCen1-depleted cells, there was an abnormal segregation of Golgi as was observed with other organelles in such cells compared with control (Figure 8, F and G). In conclusion, depletion of TbCen1 did not affect organelle duplication but interfered with their segregation.
Figure 8.
Effects on the duplication and segregation of basal bodies, kinetoplasts, Golgi, and nuclei in TbCen1-depleted cells. The cells were stained with DAPI for nuclei and kinetoplasts, GRASP for Golgi, and YL1/2 for basal bodies. (A–D) In A and C the separate enlarged areas at right show the number and position of the duplicated basal bodies and the divided kinetoplasts. Note that the divided basal bodies and the kinetoplasts in image C are not as well separated compared with the cell in B. (E) Histogram showing the percent of TbCen1 RNAi cells with two basal bodies that are either normal or defective in their separation were analyzed on days 3 and 4. The images show the type of the cells used in the analysis. Note the cells that show basal bodies defective in separation have already two nuclei in them. More than 40 cells were used for the analysis. (F and G) Effects on the duplication and segregation of the Golgi in TbCen1-depleted parasite (G) compared with control (F). Note that the divided two Golgi in image G are not as well separated as the two Golgi seen in the F. The time point at which the cells analyzed after induction was day 2 for A–C and F and day 3 for D and I. B, basal body; K, kinetoplast; G, Golgi; N, nucleus; g, additional Golgi structures as described (He et al., 2004). Scale bars, 5 μm.
DISCUSSION
Trypanosomatids Have Unique Centrin Genes
Eukaryotic cells are reported to have a varying numbers of centrin genes, for example, one in Chlamydomonas and yeast, three in humans, and four in mice (Wolfrum and Salisbury, 1998; Gavet et al., 2003). The genomic sequences of T. brucei and Leishmania reveal the presence of five putative centrins in their genome. The protein sequence homology among the five genes within trypanosomatids is very low (35–54% similarity). However, each gene displays significant similarity to its homologue among other species of trypanosomatids, suggesting diversification of this centrin family early in evolution. All five genes in trypanosomatids have variable amino-terminal extensions. The amino-terminal regions are thought to confer functional diversity to centrins (Salisbury, 1995; Wiech et al., 1996). Shorter amino-terminal extension seen either in TbCen1, LdCen1, or LmCen1, seems unique to the Trypanosomatid family among the eukaryotes. Similarly, TbCen4 (and its Leishmania counterpart), which has one putative calcium-binding site, and TbCen5, which has no putative calcium-binding site, branched off early in the phylogenetic tree from a common centrin ancestor, are unique to this family. Further in the phylogenetic analysis the centrins that possess two putative calcium-binding sites branch into two clusters. One cluster comprises all the centrins 1 and 2 and MmCen4. The other cluster includes all the centrins 3 and CdC31. These observations suggest that the unique structure of each of centrin gene might play different functional roles. Therefore, further characterization of each T. brucei centrin is important for understanding trypanosome biology.
Localization of T. brucei TbCen1
Centrins have been localized to the basal-body region in the flagellated eukaryotes, such as Chlamydomonas (CrCen; Salisbury, 1995), Leishmania (LdCen1; Selvapandiyan et al., 2001), Paramecium (PtCen2 and 3; Ruiz et al., 2005), and T. brucei (TbCen2; He et al., 2005). We have similarly found that TbCen1 is localized to the basal body region of T. brucei. In addition, we observed TbCen1 is also localized at a bilobed structure close to the Golgi. The additional localization of Tbcen1 to a bilobed structure in close proximity to the Golgi has also been observed with TbCen2 but not for TbCen3 (He et al., 2005). At present little is known about the localization of TbCen4 and TbCen5. The functional significance of some of the T. brucei centrins localizing at the bilobed structure in addition to the basal body is not known and needs further investigation.
TbCen1 Depletion Leads to Pleomorphic Cell Shape and Detached Flagella
The pleomorphic shape observed in LdCen1 deleted Leishmania (Selvapandiyan et al., 2004) and TbCen1-depleted T. brucei in the current study has also been observed in S. cerevisiae. For example, mutations in yeast centrin (CdC31p) have generated cells defective in spindle pole body biogenesis with large-budded cell morphology (Sullivan et al., 1998; Ivanovska and Rose, 2001). Other structural proteins besides centrin have also been shown to be important for the cell morphology of T. brucei. For example, depletion of a flagellar adhesion protein (Fla1) seen at the flagellum attachment zone (LaCount et al., 2002) or the cytoskeletal protein α-tubulin (Ngo et al., 1998) causes parasites to lose their shape because of failure in cell division. Whether the loss of shape in the centrin-depleted protozoan parasites (Leishmania and T. brucei) is a direct effect from loss of centrin or an indirect effect due to the uncontrolled enlargement of cell size needs to be further examined.
TbCen1-depleted T. brucei cells also displayed detached flagella in addition to the original attached flagellum. A similar phenotype was observed in Fla1 deficient T. brucei cells (LaCount et al., 2002). However, in the present study, the Fla1 level was unaffected in the TbCen1-depleted cells with detached flagellum (Supplementary Figure S2). Because Fla1 is apparently not affected in TbCen1-depleted T. brucei cells and TbCen1 is not localized between flagellum and flagellum attachment zone (FAZ) like Fla1 (LaCount et al., 2002), the flagellar detachment due to TbCen1 depletion could be an indirect effect. However, it has been shown that the FAZ filament originates close to basal body area and occurs around the time, when the probasal body matures and new axoneme extends and is essential in determining the process of cytokinesis in T. brucei (Kohl et al., 1999; LaCount et al., 2002). In the present study depletion of TbCen1 in T. brucei resulted in the detachment of new flagellum at an early stage (day 2 after RNAi induction) of failure in segregation of the basal body suggesting the role of centrin in flagellum attachment and its requirement in cytokinesis. Role of flagellum in basal body and kinetoplast segregation has also been previously demonstrated by either altering the expression of intraflagellar transport proteins resulting in either short or lost flagella (Kohl et al., 2003) or by ablation of axonemal proteins which alter flagellar mobility (Dawe et al., 2005).
It is also known that the centrin associated centrosome/basal body regulates the number, spatial arrangement, and stability of microtubules responsible for cell polarity and shape in other eukaryotes (Lingle and Salisbury, 1999). The flagellum's position and polarity along with the subpellicular cytoskeleton enables the morphogenesis of the distinct flagellar pocket (Gull, 2003). However in our limited EM studies to analyze flagellar pocket morphology, we were not able to conclusively determine whether TbCen1 depletion had any effect on the overall structure of the flagellar pocket in such cells.
TbCen1 Is Involved in Organelle Segregation
TbCen1 depletion by RNAi revealed that it is important for the growth of T. brucei procyclic form. The parasites depleted for TbCen1 stopped dividing and the cells became enlarged and highly pleomorphic with multiple detached flagella. Such parasites had multiple basal bodies, kinetoplasts, Golgi, and nuclei, suggesting that there was no effect on organelle multiplication but a defect in cytokinesis. The fate of large cells eventually led to cell death, as indicated by the increased number of cells with less than 2C DNA content. Similar phenomenon was observed in L. donovani centrin disrupted amastigotes (Selvapandiyan et al., 2004).
Organelles like the flagellum, basal bodies, mitochondrion, Golgi, and nucleus must duplicate and segregate during each cell division in the protozoan parasite (Robinson et al., 1995; Gull, 1999). The sequence of events that occur during cell division in the procyclic form of T. brucei are maturation and duplication of the basal bodies, division of kinetoplast and nucleus, assembly and emergence of a new flagellum followed by segregation of the linked organelles (flagellum, basal bodies, and kinetoplast), a prerequisite for cytokinesis (Robinson et al., 1995; Kohl and Gull, 1998; Gull, 1999), and are depicted in Figure 9. In the centrin-depleted T. brucei cells, we have observed no effect on the multiplication of the linked-organelles, as indicated by a majority of cells (∼80%) with multikinetoplasts and multinuclei (>2K2N) on day 4, but a significant defect in their segregation leading to loss of cytokinesis. Interestingly, we have noticed a significant population (40%) of cells with 1K2N only at day 3 and not at day 4. The increase in the cell population with only single kinetoplast (in 1K2N) observed by DAPI is most probably due to a lack of enough segregation of the divided kinetoplasts at day 3 because DAPI staining is not sufficiently sensitive to distinguish a single kinetoplast from a duplicated kinetoplast. However, at day 4 cells the duplicated kinetoplasts are separated enough to be counted as >2K2N (Figure 4C). A similar increase in the 1K2N population was observed because of the failure of nuclear segregation, leading to both the nuclei remaining in one daughter cell and leaving the second daughter cell with one kinetoplasts and no nucleus (1K0N; Ploubidou et al., 1999). However, we did not observe an increase in 1K0N population in TbCen1-depleted cells. Our observation that the divided kinetoplasts often staying close together without separation in the TbCen1-depleted cells was further substantiated by the EM analysis in Figure 5D. Therefore, the increase in number of cells with multibasal bodies, multiflagella, and multi-Golgi as the RNAi induction proceeds reinforces the idea that there is no defect in the duplication of these organelles after depletion of TbCen1.
A coordination in the biogenesis of Golgi and basal bodies has been previously suggested (Field et al., 2000; He et al., 2004). For example, in T. brucei, duplication of the basal bodies was followed by the formation of a new Golgi near to the new basal body. An inhibition of the separation of the basal bodies and kinetoplasts by anistomicin-p, which inhibits the microtubule morphogenesis, also inhibited separation of the old and new Golgi in T. brucei (He et al., 2004). From the above-mentioned observations it is apparent that segregation of various organelles is interdependent and the basal body plays a control role in this process. Further, the failure of segregation that we observed with both the duplicated basal bodies and kinetoplasts soon after the induction of RNAi is probably the cause of the defect in the initiation of cytokinesis. Involvement of centrins in the segregation of basal bodies/centrosome is known in human, mouse, Chlamydomonas, and Paramecium (Middendorp et al., 1997; Koblenz et al., 2003; Ruiz et al., 2005). However, in the present study the role of TbCen1 in the segregation of other organelles (kinetoplast and Golgi) besides basal body has been demonstrated in T. brucei. Hence from our studies the localization of TbCen1 to the basal body and its depletion affecting the segregation of all the associated organelles suggest the importance of this protein in the completion of cell cycle in T. brucei.
We observed that nuclear division occurs despite arrest in cytokinesis in centrin-depleted T. brucei cells. This confirms previous observations by others that nuclear division and cell division in both T. brucei and Leishmania are not linked processes (Ploubidou et al., 1999; Selvapandiyan et al., 2004; Kumar and Wang, 2006). It is known that the nuclear division in Trypanosomes is due to the spindles that are intranuclear in origin and not due to the basal body nucleating microtubules (Ogbadoyi et al., 2000). The divided nuclei, which were not repositioned in the TbCen1-depleted cells as they were in the uninduced control cells but remained as a cluster in the centrin-depleted cells, suggest that the repositioning of the divided nuclei might require a distinct unknown mechanism that needs the presence of TbCen1.
It has been shown that the position of the spindle pole body (basal body counterpart in yeast) after segregation determines the division cleavage plane and cytokinesis (Hoyt, 2000). Postanaphase repositioning of the centrosome in HeLa cells controls the release of central microtubules from the midbody and the completion of cell division (Piel et al., 2001). The involvement of the spindle pole body nucleating microtubules in the nuclear migration and repositioning needed for cell division in Aspergillus nidulans has been also shown (Oakley et al., 1990; Xiang and Fischer, 2004). Centrin depletion in Chlamydomonas cells resulted in aberrant numbers of basal bodies, which disturbed the microtubular cytoskeleton independent of the mitotic spindle and arrested cytokinesis (Koblenz et al., 2003). In addition, a precedent for centrin-associated fibers interacting with microtubules has been established in Chlamydomonas (Sanders and Salisbury, 1989, 1994). These studies describe the association between centrin and microtubules and may explain the observed failure in organelle segregation in the T. brucei centrin-depleted cells. Therefore, in the future studies, it will be of interest to determine the effect on microtubule assembly in the absence of centrin and its effect on trypanosomatid organelle segregation.
Differential Function of Centrins in Trypanosomatids
TbCen1-depleted cells showed continued duplication of basal bodies and kinetoplasts in the absence of cytokinesis, whereas, the TbCen2- and 3-depleted cells showed defects both in the duplication of basal bodies and kinetoplast (He et al., 2005). In the L. donovani amastigote form, another member of the trypanosomatid family, there is a failure in the duplication of basal bodies without affecting kinetoplast division in the absence of LdCen1 (Selvapandiyan et al., 2004). There are additional differences between T. brucei and L. donovani. For example, the knockout for LdCen1 in L. donovani only affected the growth of amastigote stage parasites and not the in vitro growth of the insect form promastigote (Selvapandiyan et al., 2004), whereas, in T. brucei we see a distinct change in the phenotype of the insect stage procyclic form upon depletion of centrins. At this time the function of centrins in the bloodstream form of T. brucei is not known. It is possible that the differences in function among centrins in trypanosomatids could be due to structural differences. The variable amino-terminus, which is thought to confer functional diversity to centrins (Salisbury, 1995; Wiech et al., 1996), may be involved in the functional differences. Therefore, it will be of interest to find how differences in centrins among T. brucei and Leishmania, which cause various diseases, result in the functional differences.
The results in this study indicate that the primary event that occurs in the absence of TbCen1 could be the failure of the segregation of the linked organelles (basal bodies, flagellum, kinetoplasts, and Golgi). The detachment of new flagella, arrest in cytokinesis, and the enlargement of cell size due to repeated organelle multiplication could be the associated events and may be occurring simultaneously. In conclusion, the present study describes a novel function for TbCen1, which is the involvement in the spatial positioning of multiple organelles that is required for the initiation of cytokinesis in T. brucei. Such a centrin function may be unique in T. brucei among the eukaryotes, probably due to a primitive centrin-dependent mechanism of coordination in the biogenesis of the organelles seen in this evolutionarily ancient protozoan parasite.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Paul T. Englund (Johns Hopkins School of Medicine, Baltimore, MD) for providing plasmid vector pZJM; and George A.M. Cross (The Rockefeller University, New York, NY), Graham Warren (Yale University School of Medicine, New Haven, CT), and Keith Gull (University of Oxford, Oxford, United Kingdom) for providing anti-flagellum adhesion protein (Fla1), anti-Golgi reassembly stacking protein (GRASP), and anti-paraflagellar rod (L8C4) antibodies, respectively. We also acknowledge Robert Duncan and Alain Debrabant (Division of Emerging and Transfusion and Transmitted Diseases, Center for Biologics Evaluation and Research, Food and Drug Administration, Bethesda, MD) for critical reading of the manuscript.
Abbreviations used:
- HA
hemagglutinin
- DAPI
4′6-diamidino-2-phenylindole
- ORF
open reading frame
- pAb
polyclonal antibodies
- EM
electron microscopy.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-01-0022) on June 13, 2007.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Bastin P., Pullen T. J., Moreira-Leite F. F., Gull K. Inside and outside of the trypanosome flagellum: a multifunctional organelle. Microbes. Infect. 2000;2:1865–1874. doi: 10.1016/s1286-4579(00)01344-7. [DOI] [PubMed] [Google Scholar]
- Bastin P., Sherwin T., Gull K. Paraflagellar rod is vital for trypanosome motility. Nature. 1998;391:548. doi: 10.1038/35300. [DOI] [PubMed] [Google Scholar]
- Berriman, et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- Chudakov D. M., Verkhusha V. V., Staroverov D. B., Souslova E. A., Lukyanov S., Lukyanov K. A. Photoswitchable cyan fluorescent protein for protein tracking. Nat. Biotechnol. 2004;22:1435–1439. doi: 10.1038/nbt1025. [DOI] [PubMed] [Google Scholar]
- Dawe H. R., Farr H., Portman N., Shaw M. K., Gull K. The Parkin co-regulated gene product, PACRG, is an evolutionarily conserved axonemal protein that functions in outer-doublet microtubule morphogenesis. J. Cell Sci. 2005;118:5421–5430. doi: 10.1242/jcs.02659. [DOI] [PubMed] [Google Scholar]
- Errabolu R., Sanders M. A., Salisbury J. L. Cloning of a cDNA encoding human centrin, an EF-hand protein of centrosomes and mitotic spindle poles. J. Cell Sci. 1994;107(Pt 1):9–16. doi: 10.1242/jcs.107.1.9. [DOI] [PubMed] [Google Scholar]
- Field H., Sherwin T., Smith A. C., Gull K., Field M. C. Cell-cycle and developmental regulation of TbRAB31 localisation, a GTP-locked Rab protein from Trypanosoma brucei. Mol. Biochem. Parasitol. 2000;106:21–35. doi: 10.1016/s0166-6851(99)00192-9. [DOI] [PubMed] [Google Scholar]
- Fischer T., Rodriguez-Navarro S., Pereira G., Racz A., Schiebel E., Hurt E. Yeast centrin Cdc31 is linked to the nuclear mRNA export machinery. Nat. Cell Biol. 2004;6:840–848. doi: 10.1038/ncb1163. [DOI] [PubMed] [Google Scholar]
- Gavet O., Alvarez C., Gaspar P., Bornens M. Centrin4p, a novel mammalian centrin specifically expressed in ciliated cells. Mol. Biol. Cell. 2003;14:1818–1834. doi: 10.1091/mbc.E02-11-0709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gull K. The cytoskeleton of trypanosomatid parasites. Annu. Rev. Microbiol. 1999;53:629–655. doi: 10.1146/annurev.micro.53.1.629. [DOI] [PubMed] [Google Scholar]
- Gull K. Host-parasite interactions and trypanosome morphogenesis: a flagellar pocketful of goodies. Curr. Opin. Microbiol. 2003;6:365–370. doi: 10.1016/s1369-5274(03)00092-4. [DOI] [PubMed] [Google Scholar]
- He C. Y., Ho H. H., Malsam J., Chalouni C., West C. M., Ullu E., Toomre D., Warren G. Golgi duplication in Trypanosoma brucei. J. Cell Biol. 2004;165:313–321. doi: 10.1083/jcb.200311076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C. Y., Pypaert M., Warren G. Golgi duplication in Trypanosoma brucei requires Centrin2. Science. 2005;310:1196–1198. doi: 10.1126/science.1119969. [DOI] [PubMed] [Google Scholar]
- Hoyt M. A. Exit from mitosis: spindle pole power. Cell. 2000;102:267–270. doi: 10.1016/s0092-8674(00)00031-3. [DOI] [PubMed] [Google Scholar]
- Huang B., Mengersen A., Lee V. D. Molecular cloning of cDNA for caltractin, a basal body-associated Ca2+-binding protein: homology in its protein sequence with calmodulin and the yeast CDC31 gene product. J. Cell Biol. 1988;107:133–140. doi: 10.1083/jcb.107.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanovska I., Rose M. D. Fine structure analysis of the yeast centrin, Cdc31p, identifies residues specific for cell morphology and spindle pole body duplication. Genetics. 2001;157:503–518. doi: 10.1093/genetics/157.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivens, et al. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309:436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalfan W., Ivanovska I., Rose M. D. Functional interaction between the PKC1 pathway and CDC31 network of SPB duplication genes. Genetics. 2000;155:1543–1559. doi: 10.1093/genetics/155.4.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin J. V., Wright B., Milstein C. Rat monoclonal antitubulin antibodies derived by using a new nonsecreting rat cell line. J. Cell Biol. 1982;93:576–582. doi: 10.1083/jcb.93.3.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klink V. P., Wolniak S. M. Centrin is necessary for the formation of the motile apparatus in spermatids of Marsilea. Mol. Biol. Cell. 2001;12:761–776. doi: 10.1091/mbc.12.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koblenz B., Schoppmeier J., Grunow A., Lechtreck K. F. Centrin deficiency in Chlamydomonas causes defects in basal body replication, segregation and maturation. J. Cell Sci. 2003;116:2635–2646. doi: 10.1242/jcs.00497. [DOI] [PubMed] [Google Scholar]
- Kohl L., Gull K. Molecular architecture of the trypanosome cytoskeleton. Mol. Biochem. Parasitol. 1998;93:1–9. doi: 10.1016/s0166-6851(98)00014-0. [DOI] [PubMed] [Google Scholar]
- Kohl L., Robinson D., Bastin P. Novel roles for the flagellum in cell morphogenesis and cytokinesis of trypanosomes. EMBO J. 2003;22:5336–5346. doi: 10.1093/emboj/cdg518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl L., Sherwin T., Gull K. Assembly of the paraflagellar rod and the flagellum attachment zone complex during the Trypanosoma brucei cell cycle. J. Eukaryot. Microbiol. 1999;46:105–109. doi: 10.1111/j.1550-7408.1999.tb04592.x. [DOI] [PubMed] [Google Scholar]
- Kumar P., Wang C. C. Dissociation of cytokinesis initiation from mitotic control in a eukaryote. Eukaryot. Cell. 2006;5:92–102. doi: 10.1128/EC.5.1.92-102.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCount D. J., Barrett B., Donelson J. E. Trypanosoma brucei FLA1 is required for flagellum attachment and cytokinesis. J. Biol. Chem. 2002;277:17580–17588. doi: 10.1074/jbc.M200873200. [DOI] [PubMed] [Google Scholar]
- Lee N., Bertholet S., Debrabant A., Muller J., Duncan R., Nakhasi H. L. Programmed cell death in the unicellular protozoan parasite Leishmania. Cell Death Differ. 2002;9:53–64. doi: 10.1038/sj.cdd.4400952. [DOI] [PubMed] [Google Scholar]
- Lingle W. L., Lutz W. H., Ingle J. N., Maihle N. J., Salisbury J. L. Centrosome hypertrophy in human breast tumors: implications for genomic stability and cell polarity. Proc. Natl. Acad. Sci. USA. 1998;95:2950–2955. doi: 10.1073/pnas.95.6.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingle W. L., Salisbury J. L. Altered centrosome structure is associated with abnormal mitoses in human breast tumors. Am. J. Pathol. 1999;155:1941–1951. doi: 10.1016/S0002-9440(10)65513-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middendorp S., Paoletti A., Schiebel E., Bornens M. Identification of a new mammalian centrin gene, more closely related to Saccharomyces cerevisiae CDC31 gene. Proc. Natl. Acad. Sci. USA. 1997;94:9141–9146. doi: 10.1073/pnas.94.17.9141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinier J., Ramos C., Fritsch O., Hohn B. CENTRIN2 modulates homologous recombination and nucleotide excision repair in Arabidopsis. Plant Cell. 2004;16:1633–1643. doi: 10.1105/tpc.021378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J. C., Wang Z., Motyka S. A., Drew M. E., Englund P. T. Boca Raton, FL: CRC Press; 2004. An RNAi-based Genomic Library for Forward Genetics in the African Trypanosome; pp. 241–258. [Google Scholar]
- Ngo H., Tschudi C., Gull K., Ullu E. Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc. Natl. Acad. Sci. USA. 1998;95:14687–14692. doi: 10.1073/pnas.95.25.14687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi R., Okuda Y., Watanabe E., Mori T., Iwai S., Masutani C., Sugasawa K., Hanaoka F. Centrin 2 stimulates nucleotide excision repair by interacting with xeroderma pigmentosum group C protein. Mol. Cell. Biol. 2005;25:5664–5674. doi: 10.1128/MCB.25.13.5664-5674.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley B. R., Oakley C. E., Yoon Y., Jung M. K. Gamma-tubulin is a component of the spindle pole body that is essential for microtubule function in Aspergillus nidulans. Cell. 1990;61:1289–1301. doi: 10.1016/0092-8674(90)90693-9. [DOI] [PubMed] [Google Scholar]
- Ogbadoyi E., Ersfeld K., Robinson D., Sherwin T., Gull K. Architecture of the Trypanosoma brucei nucleus during interphase and mitosis. Chromosoma. 2000;108:501–513. doi: 10.1007/s004120050402. [DOI] [PubMed] [Google Scholar]
- Pati U. K. Novel vectors for expression of cDNA encoding epitope-tagged proteins in mammalian cells. Gene. 1992;114:285–288. doi: 10.1016/0378-1119(92)90589-h. [DOI] [PubMed] [Google Scholar]
- Piel M., Nordberg J., Euteneuer U., Bornens M. Centrosome-dependent exit of cytokinesis in animal cells. Science. 2001;291:1550–1553. doi: 10.1126/science.1057330. [DOI] [PubMed] [Google Scholar]
- Ploubidou A., Robinson D. R., Docherty R. C., Ogbadoyi E. O., Gull K. Evidence for novel cell cycle checkpoints in trypanosomes: kinetoplast segregation and cytokinesis in the absence of mitosis. J. Cell Sci. 1999;112(Pt 24):4641–4650. doi: 10.1242/jcs.112.24.4641. [DOI] [PubMed] [Google Scholar]
- Raslova H., Baccini V., Loussaief L., Comba B., Larghero J., Debili N., Vainchenker W. Mammalian target of rapamycin (mTOR) regulates both proliferation of megakaryocyte progenitors and late stages of megakaryocyte differentiation. Blood. 2006;107:2303–2310. doi: 10.1182/blood-2005-07-3005. [DOI] [PubMed] [Google Scholar]
- Robinson D. R., Gull K. Basal body movements as a mechanism for mitochondrial genome segregation in the trypanosome cell cycle. Nature. 1991;352:731–733. doi: 10.1038/352731a0. [DOI] [PubMed] [Google Scholar]
- Robinson D. R., Sherwin T., Ploubidou A., Byard E. H., Gull K. Microtubule polarity and dynamics in the control of organelle positioning, segregation, and cytokinesis in the trypanosome cell cycle. J. Cell Biol. 1995;128:1163–1172. doi: 10.1083/jcb.128.6.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz F., Garreau de Loubresse N., Klotz C., Beisson J., Koll F. Centrin deficiency in paramecium affects the geometry of basal-body duplication. Curr. Biol. 2005;15:2097–2106. doi: 10.1016/j.cub.2005.11.038. [DOI] [PubMed] [Google Scholar]
- Salisbury J. L. Centrin, centrosomes, and mitotic spindle poles. Curr. Opin. Cell Biol. 1995;7:39–45. doi: 10.1016/0955-0674(95)80043-3. [DOI] [PubMed] [Google Scholar]
- Salisbury J. L., Baron A., Surek B., Melkonian M. Striated flagellar roots: isolation and partial characterization of a calcium-modulated contractile organelle. J. Cell Biol. 1984;99:962–970. doi: 10.1083/jcb.99.3.962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury J. L., Baron A. T., Sanders M. A. The centrin-based cytoskeleton of Chlamydomonas reinhardtii: distribution in interphase and mitotic cells. J. Cell Biol. 1988;107:635–641. doi: 10.1083/jcb.107.2.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury J. L., Suino K. M., Busby R., Springett M. Centrin-2 is required for centriole duplication in mammalian cells. Curr. Biol. 2002;12:1287–1292. doi: 10.1016/s0960-9822(02)01019-9. [DOI] [PubMed] [Google Scholar]
- Sanders M. A., Salisbury J. L. Centrin-mediated microtubule severing during flagellar excision in Chlamydomonas reinhardtii. J. Cell Biol. 1989;108:1751–1760. doi: 10.1083/jcb.108.5.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders M. A., Salisbury J. L. Centrin plays an essential role in microtubule severing during flagellar excision in Chlamydomonas reinhardtii. J. Cell Biol. 1994;124:795–805. doi: 10.1083/jcb.124.5.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvapandiyan A., Debrabant A., Duncan R., Muller J., Salotra P., Sreenivas G., Salisbury J. L., Nakhasi H. L. Centrin gene disruption impairs stage-specific basal body duplication and cell cycle progression in Leishmania. J. Biol. Chem. 2004;279:25703–25710. doi: 10.1074/jbc.M402794200. [DOI] [PubMed] [Google Scholar]
- Selvapandiyan A., Duncan R., Debrabant A., Bertholet S., Sreenivas G., Negi N. S., Salotra P., Nakhasi H. L. Expression of a mutant form of Leishmania donovani centrin reduces the growth of the parasite. J. Biol. Chem. 2001;276:43253–43261. doi: 10.1074/jbc.M106806200. [DOI] [PubMed] [Google Scholar]
- Spang A., Courtney I., Fackler U., Matzner M., Schiebel E. The calcium-binding protein cell division cycle 31 of Saccharomyces cerevisiae is a component of the half bridge of the spindle pole body. J. Cell Biol. 1993;123:405–416. doi: 10.1083/jcb.123.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemm-Wolf A. J., Morgan G., Giddings T. H., Jr, White E. A., Marchione R., McDonald H. B., Winey M. Basal body duplication and maintenance require one member of the Tetrahymena thermophila centrin gene family. Mol. Biol. Cell. 2005;16:3606–3619. doi: 10.1091/mbc.E04-10-0919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan D. S., Biggins S., Rose M. D. The yeast centrin, cdc31p, and the interacting protein kinase, Kic1p, are required for cell integrity. J. Cell Biol. 1998;143:751–765. doi: 10.1083/jcb.143.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang W. Y., Spektor A., Luciano D. J., Indjeian V. B., Chen Z., Salisbury J. L., Sanchez I., Dynlacht B. D. CP110 cooperates with two calcium-binding proteins to regulate cytokinesis and genome stability. Mol. Biol. Cell. 2006;17:3423–3434. doi: 10.1091/mbc.E06-04-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu X., Wang C. C. The involvement of two cdc2-related kinases (CRKs) in Trypanosoma brucei cell cycle regulation and the distinctive stage-specific phenotypes caused by CRK3 depletion. J. Biol. Chem. 2004;279:20519–20528. doi: 10.1074/jbc.M312862200. [DOI] [PubMed] [Google Scholar]
- Ullu E., Tschudi C., Chakraborty T. RNA interference in protozoan parasites. Cell Microbiol. 2004;6:509–519. doi: 10.1111/j.1462-5822.2004.00399.x. [DOI] [PubMed] [Google Scholar]
- Wang Z., Morris J. C., Drew M. E., Englund P. T. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 2000;275:40174–40179. doi: 10.1074/jbc.M008405200. [DOI] [PubMed] [Google Scholar]
- Wiech H., Geier B. M., Paschke T., Spang A., Grein K., Steinkotter J., Melkonian M., Schiebel E. Characterization of green alga, yeast, and human centrins. Specific subdomain features determine functional diversity. J. Biol. Chem. 1996;271:22453–22461. doi: 10.1074/jbc.271.37.22453. [DOI] [PubMed] [Google Scholar]
- Wirtz E., Leal S., Ochatt C., Cross G. A. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- Wolfrum U., Salisbury J. L. Expression of centrin isoforms in the mammalian retina. Exp. Cell Res. 1998;242:10–17. doi: 10.1006/excr.1998.4038. [DOI] [PubMed] [Google Scholar]
- Wright R. L., Salisbury J., Jarvik J. W. A nucleus-basal body connector in Chlamydomonas reinhardtii that may function in basal body localization or segregation. J. Cell Biol. 1985;101:1903–1912. doi: 10.1083/jcb.101.5.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang X., Fischer R. Nuclear migration and positioning in filamentous fungi. Fungal Genet. Biol. 2004;41:411–419. doi: 10.1016/j.fgb.2003.11.010. [DOI] [PubMed] [Google Scholar]
- Zamora I., Marshall W. F. A mutation in the centriole-associated protein centrin causes genomic instability via increased chromosome loss in Chlamydomonas reinhardtii. BMC Biol. 2005;3:15. doi: 10.1186/1741-7007-3-15. http://www.biomedcentral.com/1741-7007/3. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.