Abstract
The Saccharomyces cerevisiae essential gene YNL158w/PGA1 encodes an endoplasmic reticulum (ER)-localized membrane protein. We constructed temperature-sensitive alleles of PGA1 by error-prone polymerase chain reaction mutagenesis to explore its biological role. Pulse-chase experiments revealed that the pga1ts mutants accumulated the ER-form precursor of Gas1 protein at the restrictive temperature. Transport of invertase and carboxypeptidase Y were not affected. Triton X-114 phase separation and [3H]inositol labeling indicated that the glycosylphosphatidylinositol (GPI)-anchoring was defective in the pga1ts mutants, suggesting that Pga1 is involved in GPI synthesis or its transfer to target proteins. We found GPI18, which was recently reported to encode GPI-mannosyltransferase II (GPI-MT II), as a high-copy suppressor of the temperature sensitivity of pga1ts. Both Gpi18 and Pga1 were detected in the ER by immunofluorescence, and they were coprecipitated from the Triton X-100–solubilized membrane. The gpi18ts and pga1ts mutants accumulated the same GPI synthetic intermediate at the restrictive temperature. From these results, we concluded that Pga1 is an additional essential component of the yeast GPI-MT II.
INTRODUCTION
Many proteins synthesized at the surface of the endoplasmic reticulum (ER) are transported through the secretory pathway to proper locations in the cell where they function (Palade, 1975; Mellman and Warren, 2000). The ER is an organelle where the major secretory pathway starts, and the folding status of the proteins are monitored to pass this pathway (Kleizen and Braakman, 2004). In the ER, several modifications are performed on the proteins, such as N-glycosylation, O-glycosylation, and glycosylphosphatidylinositol (GPI)-anchoring (Sipos and Fuller, 2002). The GPI is a complex glycolipid with a core structure, phosphoethanolamine (EtN-P)-6-mannose (Man)-α1,2-Man-α1,6-Man-α1,4-glucosamine (GlcN)-α1,6-inositol-phospholipid, that is conserved in all eukaryotic cells (Kinoshita and Inoue, 2000) and functions as a membrane anchor for many cell surface proteins (Englund, 1993). In budding yeast Saccharomyces cerevisiae, GPI synthesis and GPI-anchoring are carried out by >20 gene products, and most of the genes are essential for cell viability (Pittet and Conzelmann, 2006). Many GPI-anchored proteins lose lipid moiety at a certain stage of maturation, and they become attached to the cell wall β1,6-glucan through the remaining mannose residues, contributing to the cell wall integrity (Lu et al., 1995). Construction and attachment to the target protein of GPI are carried out on the ER membrane. The GPI is synthesized by a stepwise addition of sugars, an acyl chain, and EtN-P to phosphatidylinositol (PI). The GPI transamidase complex removes the C-terminal GPI attachment signal peptide of the target protein and links the newly generated C-terminal end to the EtN-P of the complete GPI precursor lipid (Eisenhaber et al., 2003). All of the genes that are known to be involved in GPI synthesis and GPI-anchoring in S. cerevisiae have homologues in mammals. Moreover, GPI is essential for the viability of some protozoan species, such as Leishmania mexicana and the bloodstream form of Trypanosoma brucei. It has been suggested that the pathway of GPI synthesis is mostly conserved among the species; thus, S. cerevisiae can contribute to GPI research as a model organism.
The GPI of S. cerevisiae contains four Man residues. The fourth Man has essential roles because SMP3, which encodes the enzyme to add the fourth Man, is essential for viability (Grimme et al., 2001), although the addition of fourth Man is not essential and the GPIs contain only three Man residues in other organisms (Taron et al., 2004). The enzymes that add Man to GPI precursors are called GPI-mannosyltransferase (GPI-MT) I, -II, -III, and -IV, according to the order of mannose addition. Each GPI-MT consists of at least one protein with several transmembrane domains that has homology to other enzymes responsible for glycosylation (Takahashi et al., 1996; Grimme et al., 2001; Maeda et al., 2001; Ashida et al., 2005; Fabre et al., 2005; Kang et al., 2005). So far, GPI-MT I has been the only enzyme that is found to consist of multisubunits. Gpi18 is recently reported to be required for the addition of a second Man to the GPI precursor, and its mammalian homologue PIG-V acts as GPI-MT II in Chinese hamster ovary (CHO) cells. PIG-V could partially rescue the lethality of Δgpi18 cells (Fabre et al., 2005; Kang et al., 2005).
Recently, large-scale analyses of the essential genes of S. cerevisiae were reported from several laboratories (Hazbun et al., 2003; Mnaimneh et al., 2004; Davierwala et al., 2005; Schuldiner et al., 2005). In their analyses, target genes were placed under the control of regulatable promoters, and the physiological roles of encoded proteins were predicted by the phenotypes they showed under the expression shut-off, and, in some cases, by the synthetic sickness or lethality with other genes that encode proteins with known functions. Although these approaches proved to be very useful for dealing with many genes, and they suggested possible involvement of the given gene product in a certain biological pathway, the functional and mechanistic information obtained for each gene product seemed to be sometimes limited. So, we took a different approach to find proteins that execute undefined but essential roles in the ER. We created the temperature-sensitive (ts) mutants of essential genes that encode the ER-localized proteins by error-prone polymerase chain reaction (PCR). The advantage of using ts mutants is that they often display the phenotypes immediately after the temperature shift; they sometimes display allele-specific phenotypes useful for regional analyses; and they are suitable for screening genetic interactions, especially genes that function as multicopy suppressors, which often provide with very valuable clues to characterize the protein function. The ER-localized proteins were selected from membrane proteins encoded by essential genes by observing the localization of the green fluorescent protein (GFP)-fusion proteins. One of them, YNL158w/PGA1, encodes a protein that has no similarity with mammalian proteins, and it was reported to be required for the proper processing of Gas1 (a GPI-anchored protein of the plasma membrane) and Pho8 (an alkaline phosphatase in the vacuole); therefore, Pga1 has been suggested to be important for normal ER function (Davierwala et al., 2005). Here, we show that Pga1 is an essential component of GPI-MT II. We identified GPI18 as a high-copy suppressor of pga1ts, and further characterization indicated that Pga1 is responsible for the second mannose addition to GPI precursors as a partner of Gpi18. This is the first report to show that a gene with no mammalian homologue is involved in GPI synthesis in S. cerevisiae.
MATERIALS AND METHODS
Strains and Media
S. cerevisiae strains used in this study are listed in Table 1. They were grown in YPD (1% Bacto yeast extract [BD Biosciences, Franklin Lakes, NJ], 2% Bacto peptone [BD Biosciences], and 2% glucose) or SD (0.17% yeast nitrogen base without amino acids [BD Biosciences], 0.5% ammonium sulfate, 2% glucose, and appropriate supplements) medium at 30°C unless other temperatures were indicated. An SG (2% galactose instead of glucose in SD) medium was used to induce genes under GAL1 promoter. Ammonium sulfate was replaced with ammonium chloride in the low-sulfate SD medium. Solid media were made with 1.5% agar; 20 μg/ml each of histidine, tryptophan, and uracil and 0.1% 5-fluoroorotic acid (Sigma-Aldrich, St. Louis, MO) were added in SD to make a 5-fluoroorotic acid (5-FOA) plate for Ura3+ counterselection, and 20 μg/ml phloxine B (Sigma-Aldrich) was added in YPD to make a phloxine plate (Tsukada and Ohsumi, 1993) for selection of temperature-sensitive mutants. Escherichia coli DH5α (F−, supE44 ΔlacU169φ80lacZΔM15 hsdR17 recA1 endA1 gyrA96 thi-1 relA1) was used in plasmid propagation. E. coli was grown in an LB (1% Bacto tryptone [BD Biosciences], 0.5% Bacto yeast extract [BD Biosciences]) and 0.5% NaCl) medium.
Table 1.
S. cerevisiae strains used in this study
| Strain | Genotype and plasmid | Origin |
|---|---|---|
| KA31a | MATa, his3Δ leu2Δ trp1Δ ura3Δ | Laboratory stock |
| BY4741 | MATa, his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Euroscarf |
| BY4743 | MATa/MATa, his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 met15Δ0/MET15 LYS2/lys2Δ0 ura3Δ0/ura3Δ0 | |
| KSY99 | as BY4741, MET15 | This study |
| KSY33 | as BY4741, pga1Δ::kanMX4, pKS5 (CEN, HIS3 PGA1-GFP) | This study |
| KSY63 | as BY4741, pga1Δ::kanMX4, pKS22 (CEN, HIS3 PGA1-6myc) | This study |
| KSY216 | as BY4741, pga1Δ::kanMX4, pKS38 (CEN, HIS3 PGA1), pKS128 (CEN, GFP-GPI18) | This study |
| KSY217 | as BY4741, pga1Δ::kanMX4, pKS110 (CEN, HIS3 PGA1-6myc), pKS128 (CEN, GFP-GPI18) | This study |
| KSY182 | as KSY99, pga1Δ::pga1-3 LEU2 | This study |
| AKY19 | as KSY99, pga1Δ::pga1-1 LEU2 | This study |
| KSY274 | as KSY99, pga1Δ::gpi18-3 LEU2 | This study |
| KSY277 | as KSY99, pga1Δ::gpi18-6 LEU2 | This study |
| KSY279 | as KSY99, gpi18Δ::PGAL1-GPI18 LEU2 | This study |
| KSY307 | as KSY99, pKS128 (CEN, GFP-GPI18) | This study |
| KSY308 | as KSY99, gpi18Δ::PGAL1-PGA1 LEU2,pKS128 (CEN, GFP-GPI18) | This study |
| KSY309 | as KSY99, gpi18Δ::PGAL1-PGA1 LEU2,pKS128 (CEN, GFP-GPI18) | This study |
| KSY312 | as AKY19, pKS128 (CEN, GFP-GPI18) | This study |
| KSY313 | as KSY277, pKS128 (CEN, GFP-GPI18) | This study |
| YKI59 | as KA31a, erd1Δ | Imai et al. (2005) |
| YNY309 | as KA31a, sec12ts | Inadome et al. (2005) |
| YNY401 | as KA31a, emp24Δ | This study |
| KSY269 | as BY4741, gpi7Δ::kanMX4 | Euroscarf |
| CAY341 | as BY4743, pga1Δ::kanMX4/PGA1 | Euroscarf |
| KSY223 | as BY4743, gpi18Δ::kanMX4/GPI18 | Euroscarf |
Plasmid Construction
Primers used in this study are listed in Table 2. The nucleotide sequence of each amplified fragment was confirmed.
Table 2.
Primers used in this study
| Primer | Gene | Restriction site | Direction | Sequence |
|---|---|---|---|---|
| CA159 | PGA1 | BamHI | Forward | 5′-CGGGATCCTTATCGTCTCTTTCTGTCC-3′ |
| CA160 | PGA1 | XhoI | Reverse | 5′-CCCTCGAGTTGGGCGAGAATTTGCTG-3′ |
| CA186 | PGA1 | BamHI | Forward | 5′-CGGGATCCATGGTCCGGCCTCAGAAT-3′ |
| YN209 | PGA1 | XhoI | Reverse | 5′-CTCGAGAAAATTTCAGCAAAAGATC-3′ |
| KS5 | PGA1 | EcoRI | Reverse | 5′-GGGAATTCGAACCTATGTTGGCTTAG-3′ |
| KS6 | PGA1 | EcoRI | Forward | 5′-GGGAATTCTTTGGCTGGATAGCTTGG-3′ |
| KS52 | GPI18 | BamHI | Forward | 5′-CGGGATCCACTGCCATGACGACGATG-3′ |
| KS53 | GPI18 | XhoI | Reverse | 5′-CCCTCGAGTCAGAGAGGGGTGATGTT-3′ |
| KS54 | PGA1 | SacI | Forward | 5′-GCGAGCTCCTCATATTCCTCCTAAGG-3′ |
| KS57 | GPI10 | BamHI | Forward | 5′-CGGGATCCCGAGTCCTATTCCCTAAA-3′ |
| KS58 | GPI10 | XhoI | Reverse | 5′-CCCTCGAGCGCTCTGTATTTATCCTC-3′ |
| KS59 | GPI14 | BamHI | Forward | 5′-CGGGATCCTCCCAAGGCCTAGGTCTT-3′ |
| KS60 | GPI14 | XhoI | Reverse | 5′-CCCTCGAGCACATCACATCAAGTATC-3′ |
| KS61 | PBN1 | BamHI | Forward | 5′-CGGGATCCCGGATTAACAGTATGCTC-3′ |
| KS62 | PBN1 | XhoI | Reverse | 5′-CCCTCGAGAGAATGAACAAAGAGCCC-3′ |
| KS63 | SMP3 | BamHI | Forward | 5′-CGGGATCCGGTACGTGCAATATAGAG-3′ |
| KS64 | SMP3 | XhoI | Reverse | 5′-CCCTCGAGTGAGCTGAGTACCGAGTT-3′ |
| KS65 | GPI18 | BamHI | Forward | 5′-CGGGATCCATGATTGTGGGGTTGACA-3′ |
| KS83 | GPI18 | XbaI | Forward | 5′-GCTCTAGAGCTTGGTGAACTACAGCA-3′ |
| KS85 | GPI18 | EcoRI | Reverse | 5′-GGGAATTCTCTCTCGAAGCCTCTCTT-3′ |
| KS86 | GPI18 | EcoRI | Forward | 5′-GGGAATTCGCCTGATCCTTAATGCTT-3′ |
Low-copy PGA1 expression plasmid (pCA148 or pKS38) was made by ligation of the PCR amplification fragment from the genomic DNA by using the primers CA177 and CA178 into the plasmid pRS316 or pRS313, respectively. Low-copy GPI18 expression plasmid (pKS71) was made by ligation of the PCR amplification fragment from the genomic DNA by using the primers KS52 and KS53 into the plasmid pRS316. High-copy GPI18 expression plasmid (pKS72) was made by replacing the plasmid vector with pRS426. High-copy GPI14, PBN1, GPI10, or SMP3 expression plasmid (pKS97, pKS98, pKS96, or pKS99, respectively) was made by ligation of the PCR amplification fragment from the genomic DNA by using the primers (KS59 and KS60, KS61 and KS62, KS57 and KS58, or KS63 and KS64, respectively) into the plasmid pRS426. The C-terminal tagging of GFP to Pga1 (pKS5) was made by ligation of the PCR amplification fragment from the genomic DNA by using the primers CA159 and YN209 into the plasmid pRS316-GFP-TDH1 terminator (pCA42).
The C-terminal fusion of 6myc to Pga1 (pKS22 or pKS110) was made by replacing the vector of the plasmid pKS5 with pNT125-2 (pRS316-6myc-TDH1 terminator) or pKS55 (pRS313-6myc-TDH1 terminator), respectively. The N-terminal fusion of GFP to Gpi18 (pKS128) was made by ligation of the PCR amplification fragment from the genomic DNA by using the primers KS65 and KS53 into the plasmid pRS316-YPT1 promoter-GFP (pCA42). The plasmid pKS213 for gap repair used to construct pga1ts mutants was made as follows: the PCR amplification fragment from the genomic DNA using primers CA159 and KS5 was ligated into pRS313, and then another PCR amplification fragment from the genomic DNA using primers KS6 and CA160 was ligated into the newly generated plasmid. The plasmid pKS170 for gap repair used to construct gpi18ts mutants was made as follows: the PCR amplification fragment from the genomic DNA using the primers KS52 and KS85 was ligated into pRS313 and then another PCR amplification fragment from the genomic DNA using the primers KS86 and KS53 was ligated into the newly generated plasmid. Plasmid pAK35 or pKS81 used for the integration of pga1ts alleles to the yeast chromosome was made by insertion of the DNA fragment containing the encoding ORF and its downstream sequence from pKSTS16 or pKSTS15 to pTY23 (pBluescript SK+-LEU2) and following insertion of the PCR amplification fragment from the genomic DNA using the primers KS54 and CA160. Plasmid pKS188 or pKS195 for the integration of the gpi18ts allele to the yeast chromosome was made by insertion of the PCR amplification fragment from the genomic DNA using the primers KS83 and KS85 into pTY23 and following insertion of the DNA fragment containing encoding open reading frame (ORF) and its downstream sequence from pKSTS113 or pKSTS114. The plasmid pKS173 for integration of GPI18 expressing under the control of GAL1 promoter was made as follows. A DNA fragment containing GAL1 promoter from pYES2 was inserted into pTY23, and the PCR amplification fragment from the genomic DNA using the primers KS65 and KS53 was ligated into the newly generated plasmid behind the LEU2 gene. The plasmid pKS189 for the integration of PGA1 expressing under the control of GAL1 promoter to the chromosome was made as follows: a DNA fragment containing GAL1 promoter from pYES2 was inserted into pTY23, and the PCR amplification fragment from the genomic DNA using the primers CA186 and CA160 was ligated into the newly generated plasmid behind the LEU2 gene.
Antibodies
Antisera against Scs2 and Kar2 were kindly provided by Dr. S. Kagiwada (Nara Woman's University, Japan) and Dr. Masao Tokunaga (Kagoshima University, Japan), respectively. Anti-3-phosphoglycerate kinase mouse (anti-PGK, 22C5; Invitrogen, Carlsbad, CA), anti-myc mouse (9E10; Berkeley Antibody, Richmond, CA), anti-alkaline phosphatase (ALP) mouse (1D3; Invitrogen) and monoclonal antibodies (mAbs), and anti-carboxypeptidase Y rabbit (anti-CPY; Rockland, Gilbertsville, PA) polyclonal antibody were purchased. The anti-GFP antiserum was prepared by immunizing rabbits with the glutathione S-transferase-GFP fusion protein as the antigen. For Western blotting, these antisera were diluted at 1:1000.
Construction of Temperature-sensitive Mutant Alleles of PGA1 and GPI18
The DNA fragment containing the whole PGA1 gene with the authentic promoter and terminator was amplified using primers KS159 and KS160 under an error-prone PCR condition (Muhlrad et al., 1992). KSY33 was transformed with the mixture of the amplified fragment and the EcoRI-cleaved pKS213 to integrate fragments into the plasmid by gap repair. The His+ transformants were replicated to 5-FOA plates to remove pKS5, and then they were replicated to phloxine B plates and incubated at 37°C for 2 d. The cells that formed pink colonies were tested for single colony formation at 25 and 37°C. After confirming that the temperature sensitivity was due to the plasmid, the nucleotide sequences of the mutant pga1 were determined. Construction of the ts mutant alleles of GPI18 was performed almost the same way as described above, except that primers KS52 and KS53 were used, the gap repair plasmid was the SmaI-cleaved pKS170, and the transformed strain was KSY239.
Subcellular Fractionation and Solubility Test of Membrane Proteins
Cells were converted to spheroplasts and burst in B88 (20 mM HEPES, pH 6.8, 150 mM potassium acetate, 5 mM magnesium acetate, and 200 mM sorbitol) containing protease inhibitors (1 μg/ml each of chymostatin, aprotinin, leupeptin, pepstatin A, antipain, and 1 mM phenylmethylsulfonyl fluoride). Unbroken cells were removed by centrifugation at 1000 × g for 5 min. For subcellular fractionation, the cleared lysate was sequentially centrifuged to generate 10,000 × g pellet (P10), 100,000 × g pellet (P100), and 100,000 × g supernatant (S100). Each fraction was adjusted to the original volume of the lysate and the same amount was applied for SDS-polyacrylamide gel electrophoresis (PAGE) and Western blotting. As a solubility test, a portion of lysate was mixed with the same amount of B88 containing 2% Triton X-100, 2 M NaCl, 0.2 M Na2CO3, or 4 M urea, and the mixture was kept on ice for 15 min. After removing a sample as the total, the mixture was centrifuged at 100,000 × g for 60 min. The precipitate was suspended in the same volume of the same buffer as the supernatant.
BiP Secretion Blots
The secretion of lumenal ER proteins was analyzed as described previously (Lewis et al., 1997).
Triton X-114 Phase Separation
The Triton X-114 phase separation was performed essentially as described by Doering and Schekman (1996) with some modifications. In brief, the cells were incubated at 34°C for 12 h, collected, and disrupted with glass beads. After centrifugation at 1000 × g for 5 min, the supernatant was incubated in B88 containing 1% Triton X-114 on ice for 30 min. After the extract was centrifuged at 10,000 × g for 4 min at 4°C, the supernatant was kept at 32°C for 5 min and centrifuged at 13,000 × g for 30 s at room temperature to separate the detergent and aqueous phases. Each phase was re-extracted with either 1% Triton X-114 (final concentration) or buffer as appropriate. Each re-extracted phase was subjected to SDS-PAGE, and Gas1, Scs2, or Pgk1 was detected by Western blotting.
Indirect Immunofluorescence
Fixation and permeabilization of the yeast cells for indirect immunofluorescence was performed as described by Wooding and Pelham (1998), and subsequent steps were performed as described by Vashist et al. (2002) with some modifications; a description of the procedure is as follows. Log-phase cells grown at 30°C were fixed by adding 2.5 ml of fresh 10% paraformaldehyde to 7.5 ml of yeast culture and pelleted by centrifugation. They were resuspended in 3.2 ml of PP (0.1 M potassium phosphate, pH 7.5), 1.8 ml of paraformaldehyde solution was added, and fixation was continued for an additional 15 min. Cells were then washed four times in PP and resuspended in 1 ml of SPP (PP with 1.2 M sorbitol) containing 100 mM dithiothreitol. Ten microliters of lyticase was added, and incubation was continued at 30°C. The spheroplasts were harvested by centrifugation and resuspended in 50 mM NH4Cl in SPP, and then in SPP before being transferred to polylysine-coated slides. The slides were immersed in methanol for 6 min and acetone for 30 s, both at −20°C, and then air-dried. The anti-myc mouse mAb and the anti-Kar2 rabbit polyclonal antibody were diluted to 1/40, 1/100 each in 1% skim milk, 0.1% bovine serum albumin, and 0.05% Tween 20 in Tris-buffered saline (TBS; 50 mM Tris, pH 7.4, and 150 mM NaCl), and incubations were carried out overnight at 4°C. Slides were washed and incubated in the secondary antibodies (Alexa 488-conjugated goat antibody to mouse immunoglobulin G and Texas Red-conjugated goat antibody to rabbit immunoglobulin G) for 2 h at room temperature, washed with phosphate-buffered saline, and mounted as described by Kilmartin and Adams (1984). Images were obtained using an FV500 confocal laser-scanning microscope (Olympus, Tokyo, Japan).
Immunoprecipitation
Cells were grown at 30°C in SD medium to an OD600 nm = 1.0, and 50 OD units of cells were converted to spheroplasts and burst in 1 ml of B88 containing protease inhibitors. Unbroken cells were removed by centrifugation at 1000 × g for 5 min. Then, 990 μl of the cleared lysate was mixed with 110 μl of 10% Triton X-100 and kept on ice for 15 min. The mixture was centrifuged at 100,000 × g for 60 min. The supernatant was recovered as the fraction “input.” The rest of the supernatant was mixed with anti-myc monoclonal (9E10) antibody and kept gently rotating at 4°C for 1 h. Protein A-Sepharose beads (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) washed with B88 containing 1% Triton X-100 were added and incubation was continued at 4°C for 1 h further. The mixture was centrifuged at 1000 × g for 5 min, and the supernatant was recovered as the fraction “unbound.” The beads were washed 4 times with B88 containing 1% Triton X-100 and then washed with 1 ml of 5 mM ammonium acetate. The bound proteins were eluted with 50 μl of 0.5 M acetic acid/ammonium acetate, pH 3.4, and recovered as the fraction “bound.” The same volume of each fraction was subjected to the SDS-PAGE, and the indicated proteins were detected by Western blotting.
Invertase Detection
Cells were incubated in YPD to OD600 nm = 1.0 at 25°C, and 10 OD unit of cells were shifted to 25 or 37°C, and incubation was continued for 30 min. Cells were washed in YPS (2% sucrose instead of glucose in YPD) and incubated in YPS for 3 h at the same temperature as they were incubated before, after which cells were collected and resuspended in solution A (25 mM Tris, pH 8.0, 0.6 M sorbitol, 2.5 mM sodium azide, 0.1% β-mercaptoethanol, and 0.25 mg/ml Zymolyase [Zymo Research, Orange, CA]). After 37°C incubation for 30 min, the mixture was centrifuged at 13,400 × g for 10 min at 4°C, and the supernatant was subjected to native PAGE. Secretory invertase in the gel was detected by activity staining as described by Ballou (1990).
Pulse-Chase Experiments of CPY and Gas1
Yeast strains were grown at 25°C in low-sulfate SD to OD600 nm = 1.0. Two and a half OD units of cells were harvested by centrifugation and resuspended in 100 μl of SD lacking sulfate. Pulse labeling was initiated 30 min after the shift to the restrictive temperature of 37°C by the addition of 50 μCi of Tran[35S]-label (MP Biomedicals, Irvine, CA). After 5 min of pulse labeling, the chase period was initiated by the addition of prewarmed 500 μl of chase solution (SD with 0.5% casamino acids [BD Biosciences]) and terminated by transferring the cells to a tube on ice containing 100 μl of 60 mM sodium azide. After a minimum of 10 min on ice, the cells were collected and broken by vortexing with glass beads in TBS containing 1% SDS and protease inhibitors, and then they were boiled for 5 min. The boiled mixture was added with 4 volumes of TBS containing 2% Triton X-100, and centrifuged at 15,000 × g for 5 min. The supernatant was added with anti-CPY or anti-Gas1 antibody, and then it was rotated overnight. Protein A-Sepharose washed with B88 containing 1% Triton X-100 was added to the mixture and rotated for 1 h. Recovered protein A-Sepharose was washed in TBS containing 1% Triton X-100 5 times. Immunoprecipitated proteins were eluted by boiling in SDS-PAGE sample buffer for 5 min and resolved by SDS-PAGE through 7.5% gel. 35S-labeled CPY or Gas1 was detected by autoradiography and analyzed by BAS-3000 (Fuji Film, Tokyo, Japan).
Radiolabeling of Lipids with myo-2-[3H]Inositol
For myo-2-[3H]inositol labeling of temperature-sensitive strains, cultures of each strain were grown at 25°C in SD medium to an OD600 nm = 1.0, and 10 OD units of cells were washed twice in SD-inositol and then resuspended in 1 ml of SD-inositol. After incubation at 25°C for 30 min, these cultures were shifted to 37°C for 20 min or maintained at 25°C before 15 μCi of [3H]inositol (American Radiolabeled Chemicals, St. Louis, MO) was added to them, and radiolabeling was continued for 90 min at the same temperature. For [3H]inositol labeling of gpi18::LEU2-PGAL1-GPI18 strains, cells were maintained on SG medium and then shifted to fresh SG or SD for 20 h to OD600 of ∼1. Ten OD cells were washed twice in SG-inositol or SD-inositol, resuspended in 1 ml of SG-inositol or SD-inositol, and then incubated for 50 min at 30°C. After the incubation, 15 μCi of [3H]inositol was added to them, and radiolabeling was continued for 90 min at 30°C. To stop [3H]inositol labeling, NaN3 was added to a 10 mM final concentration each, and cells were placed on ice. Cells were collected by centrifugation, washed twice in 10 mM NaN3, and resuspended in 500 μl of CM extraction solvent (CHCl3/CH3OH, 1:1, vol/vol). Glass beads (0.5 g) were added to the suspension, and cells were lysed by vortexing four times for 1 min each. The lysate was spun at 15,000 × g for 5 min, and the supernatant was collected and transferred to a new tube. Three hundred microliters of CMW (CHCl3/CH3OH/H2O, 10:10:3, vol/vol) was added to the remaining glass beads and cell debris, and the mixture was vortexed for 1 min. After centrifugation, supernatant was added to the first lipid extract. Pooled lipids were dried in a speed-vac. The resulting pellet was resuspended with 150 μl of H2O-saturated 1-butanol and extracted with 75 μl of 1-butanol-saturated water. The organic phase was collected, and the aqueous phase was backextracted with an additional 75 μl of H2O-saturated 1-butanol. The pooled organic phases were dried in a SpeedVac and resuspended in 30 μl of CMW. All amounts of samples were loaded onto thin layer chromatography (TLC) plates (Kieselgel 60; Merck, Darmstadt, Germany), developed in CHCl3/CH3OH/H2O (5:5:1, vol/vol). TLC-separated lipids were detected by autoradiography.
Radiolabeling of Proteins with myo-2-[3H]Inositol
Labeling of cells was performed as described above except for labeling with 120 μCi of [3H]inositol for 2 h. Washed cells were resuspended in 200 μl of lysis buffer (50 mM Tris-Cl, pH 6.8, 5 mM EDTA, pH 8.0, 1% SDS, 1% 2-melchaptoethanol, and protease inhibitors), and glass beads (0.2 g) were added to the suspension. Cells were lysed by vortexing four times for 1 min each, placing the cells on ice between each vortexing cycle, and then boiled for 5 min. Four milliliters of Con A buffer (50 mM Tris-Cl, pH 7.5, 1% Triton X-100, 500 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, and 1 mM MnCl2) was added to the boiled suspension, and the mixture was centrifuged at 15,000 × g. Con A-Sepharose (GE Healthcare) washed twice in Con A buffer was added to the supernatant and rotated overnight. Proteins were recovered by boiling Con A-Sepharose in SDS-PAGE sample buffer for 5 min, separated by SDS-PAGE through a 4–20% gradient gel (Daiichi, Tokyo, Japan), and then detected by autoradiography.
RESULTS
Previously Uncharacterized Pga1 Protein Is an ER-resident Membrane Glycoprotein
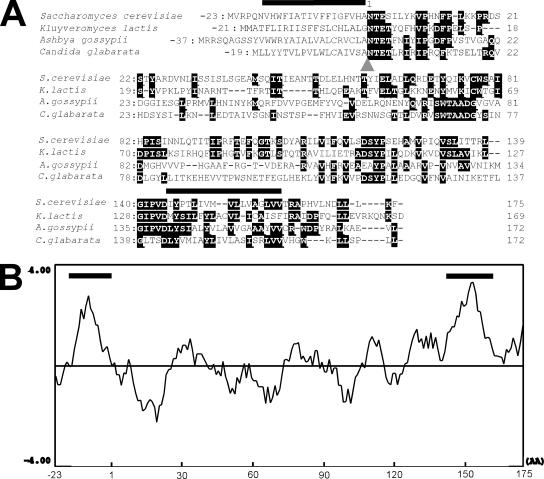
We searched the Saccharomyces Genome Database (SGD) for essential proteins with predicted transmembrane domain(s) to find proteins that execute essential functions in the ER. We introduced plasmids constructed to produce the GFP-fusion protein to each diploid strain in which one of the alleles was replaced by kanMX4. A haploid deletion strain producing GFP-fusion protein was obtained by tetrad analysis, and the localization of GFP was observed by fluorescent microscopy. We found that Ynl158w-GFP showed the localization around the nucleus and at the cell periphery, which is a pattern common to the ER proteins (Figure 2A). YNL158w/PGA1 is an essential gene and its gene disruptants were reported to be inviable even under the laboratory culture conditions (Duenas et al., 1999). According to SGD, it encodes a 198 amino-acid polypeptide of molecular weight = 22,383 and pI = 6.63, the biochemical function of which remains unknown. Homologous sequences were found in several fungal genome databases by a FASTAP search (Figure 1A), but no homologues were found in the database of higher eukaryotes. The hydropathy plot indicated that there are two hydrophobic segments at the N and C termini (Figure 1B). SignalP predicted the N-terminal segment functions as the signal peptide. The sequence 23Ala-Asn is the most probable cleavage site, and this cleavage finally generates a polypeptide of 175 amino-acid residues.
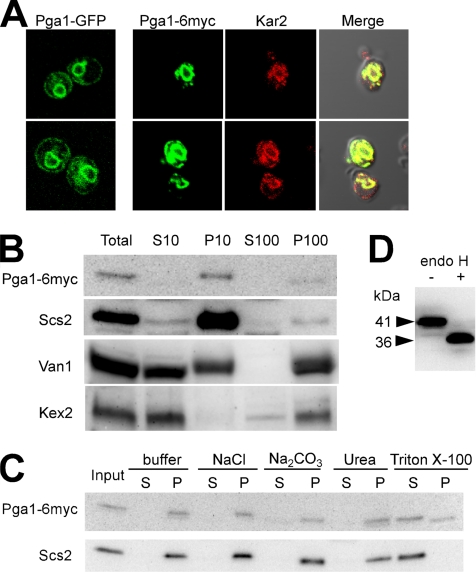
Figure 2.
Pga1 localizes in the ER as a glycosylated integral membrane protein. Pga1-GFP or Pga1-6myc was produced from CEN plasmid in Δpga1 cells, KSY33 or KSY63. (A) Confocal microscopic observation of Pga1-GFP and Pga1-6myc. The immunofluorescence images of the ER-marker Kar2 protein and the merged images with Pga1-6myc are also shown. (B) Subcellular fractionation of Pga1-6myc, the ER-marker Scs2, the early Golgi marker Van1, and the late Golgi marker Kex2. (C) Pga1-6myc and the ER membrane protein Scs2 were subjected to the solubilization test by using 1 M NaCl, 0.1 M Na2CO3, 2 M urea, and 1% Triton X-100. (D) The molecular mass of Pga1p-6myc before (−) and after (+) endoglycosidase H treatment was determined by SDS-PAGE and Western blotting.
Figure 1.
Sequence alignment of Pga1 protein and its homologues and the hydropathy profile of Pga1 protein. (A) Amino acid sequence alignment of the Pga1 homologues. Two hydrophobic segments are indicated by bars over the sequences, and the predicted signal peptide cleavage site is shown by an arrowhead. Nine terminal amino acids of the A. gossypii sequence were omitted to adjust the amino-acid alignment of the predicted signal peptides. The identical amino acids were highlighted with reverse letters. (B) The hydrophobicity index was calculated according to Kyte and Doolittle (1982), with a window size of 10 amino acids. Bold lines indicate the hydrophobic domains.
To confirm its localization to the ER, we fused 6myc at the C terminus of Pga1 and expressed it in Δpga1 cells, then we compared its localization with the ER-marker Kar2 by indirect fluorescence with confocal microscopy. The Pga1-6myc showed same pattern as Pga1-GFP, and it colocalized with Kar2 (Figure 2A). Next, we performed biochemical analysis of the Pga1-6myc. The Pga1-6myc was detected by immunoblotting, by using the anti-myc antibody as a main band of 41 kDa. By differential centrifugation of the cell lysate, Pga1-6myc was recovered mainly in the P10 fraction and some in the P100 fraction, but it was not detected in the soluble S100 fraction (Figure 2B). The ER membrane protein Scs2 was similarly recovered as Pga1, whereas the early Golgi marker Van1 was recovered almost evenly in P10 and P100 fraction, and the late Golgi marker Kex2 was mainly recovered in P100 fraction. Together with the results of microscopy observation, Pga1 should be the ER-localized protein. Next, we tested whether Pga1 is actually integral to the ER membrane. As shown in Figure 2C, Pga1-6myc was recovered in the precipitate even in the presence of 1 M sodium chloride, 0.1 M sodium carbonate, pH 11, or 2 M urea, but it became soluble in the presence of 1% Triton X-100. Therefore, we concluded that Pga1 is integral to the ER membrane.
There are two potential N-glycosylation sites, 72Asn-Thr-Thr and 80Asn-Thr-Thr, in the hydrophilic region of Pga1 polypeptide. To obtain topological information, we examined the effect of endoglycosidase H (endoH) treatment on Pga1-6myc. As shown in Figure 2D, a 41-kDa band disappeared and a 36-kDa band appeared after endoH digestion. Therefore, Pga1 accepted N-glycosylation, which indicates that the hydrophilic domain localizes in the lumen of the ER. Cells expressing Pga1 tagged with 6myc at its N terminus (6myc-Pga1) accumulated 6myc-Pga1 whose mobility of SDS-PAGE was not affected by endoH treatment (data not shown), which may indicate that N-terminal tagging to Pga1 disturbed translocation to the ER lumen. This is consistent with the SignalP prediction that the N-terminal hydrophobic segment of Pga1 functions as a signal peptide.
Isolation of Mutants with Temperature-sensitive Alleles of PGA1
Next, we sought to create temperature-sensitive alleles of PGA1 by error-prone PCR mutagenesis to investigate the biological role of Pga1. We obtained three mutant PGA1 plasmids [pKSTS16 (pga1-1), pKSTS13 (pga1-2), and pKSTS15 (pga1-3)], which supported growth of the Δpga1 haploid strain at 25°C, but not at 37°C. Nucleotide sequences indicated that a number of amino acid substitutions occurred in each of the mutant alleles. The amino acid substitutions in the pga1-1 mutation were F20I, M64T, T74P, D75G, I107F, N111S, T126A, Y144H, P152S, and Y169H; those in pga1-2 were F36Y, I57T, Y95C, C100Y, and N110S; and those in pga1-3 were T74A, D92G, W101R, I107V, Q113R, T121A, T136A, N192S, L196P, and K197E. These substitutions collectively contributed to the mutant phenotype, because the reversion of some residues resulted in the loss of temperature sensitivity, but these residues alone did not give temperature sensitivity. The chromosomal wild-type PGA1 allele was replaced with the mutant allele pga1-1 or pga1-3 by homologous recombination for further analysis.
Characterization of pga1ts Mutants
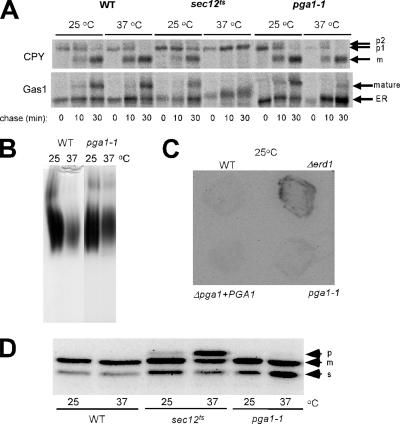
Localization in the ER suggested that Pga1 has an essential role involved in the phenomenon that specifically occurs in the ER. Therefore, we examined whether translocation, modification, and/or transport of secretory proteins was affected in these pga1 mutants. The process of protein transport was analyzed by pulse-chase experiments of Gas1 (Nuoffer at al., 1993; Popolo and Vai, 1999) and CPY (Van Den Hazel et al., 1996). A GPI-anchored plasma membrane protein Gas1 enters in the ER accompanied with the cleavage of the N-terminal secretion signal peptide, and core glycosylation of its N- and O-carbohydrate modification sites, and it becomes the 105-kDa ER form proGas1 (Kodukula et al., 1993). Then, the proGas1 C-terminal hydrophobic signal peptide is replaced with the GPI-anchor. This GPI-anchored proGas1 is transported to the plasma membrane through the Golgi where it receives the further addition of carbohydrates to the glycosyl chains, and becomes the 125-kDa mature form. ProCPY receives core N-glycosylation in the ER (form p1), additional glycosylation in the Golgi (form p2), and finally it becomes the mature enzyme (form m) by the removal of propeptide in the vacuole. As shown in Figure 3A, the 105-kDa precursor of Gas1 was largely converted to the mature 125-kDa form during a 30-min chase in the wild type both at 25 and 37°C. In the pga1-1 mutant, the mature form appeared at 25°C but not at 37°C. A large amount of the ER-form Gas1 precursor accumulated at 37°C and only a faint signal of the mature form appeared after the 30-min chase. In the sec12-ts mutant defective in the formation of COPII transport vesicles also failed to produce the mature-form Gas1 at 37°C and accumulated a precursor with somewhat reduced electrophoretic mobility. In contrast, the transport of CPY normally occurred in the pga1-1 mutant both at 25 and 37°C, whereas the sec12-ts mutant accumulated the ER-form precursor of CPY (Novick et al., 1980). These results suggest that Pga1 has an essential role in the processing or transport of a GPI-anchored membrane protein Gas1 but not in those of a soluble vacuolar protein CPY. Secretion and N-glycosylation of the periplasmic invertase were normal in the pga1-1 mutant (Figure 3B; Novick et al., 1980). The O-glycosylation of the extracellular Kre9 (Lussier et al., 1995) was also normal in the pga1-1 mutant (data not shown). The retention system of the ER lumenal proteins (Lewis et al., 1997) was normal, because no leakage of Kar2 in the medium was detected (Figure 3C). ProALP is also transported through the Golgi to the vacuole but without passing the endosome, and the propeptide is removed in the vacuole (Klinosky and Emr, 1989). An accumulation of soluble-form ALP was found in the pga1-1 mutant (Figure 3D), which is consistent with the observation in the previous report (Davierwala et al., 2005). Because the sec12ts cells also showed similar accumulation of soluble-form ALP at the permissive temperature, this might be a consequence of some aberrant condition in the ER.
Figure 3.
The secretion-related characteristics of the temperature-sensitive pga1-1 mutant. (A) The transport of CPY and Gas1 from the ER in the pga1-1 cells was analyzed by a pulse-chase experiment at either 25 or 37°C. Proteins were pulse labeled with Tran[35S]-label for 5 min and chased for indicated times. The immunoprecipitated CPY and Gas1 were detected by SDS-PAGE and autoradiography. ER, the ER-form proGas1(105 kDa); Golgi, the Golgi-form Gas1(125 kDa); p1, precursor CPY in the ER; p2, precursor CPY in the Golgi; m, mature CPY in the vacuole. (B) The N-glycosylation of secretory invertase in the pga1-1 cells at either 25 or 37°C was analyzed by native PAGE and activity staining. (C) The leakage of Kar2 protein (BiP) to the medium in the wild-type (WT), Δerd1, Δpga1/PGA1 and pga1-1 cells was analyzed at 25°C by membrane filter trapping and immunological detection of BiP. (D) The processing of ALP precursor in the WT, sec12ts, and pga1-1 cells at either 25 or 37°C was analyzed by SDS-PAGE and Western blotting. p, precursor (ER) form; m, mature (vacuolar) form; s, soluble form of ALP.
The proGas1 Protein Accumulated in pga1ts Mutants Is Not GPI-Anchored
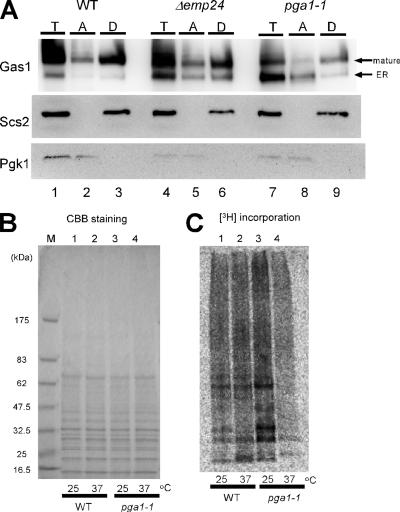
Accumulation of proGas1 in the ER might result from either of two deficiencies. First, because GPI-anchor attachment is required for the exit of the GPI target proteins from the ER (Nuoffer et al., 1993; Doering and Schekman, 1996), a failure to produce or attach GPI would result in proGas1 accumulation. Second, mutants specifically defective in the transport of GPI-anchored proteins also accumulate 105-kDa Gas1. Because the GPI-anchored 105-kDa Gas1 protein is transported to the Golgi via COPII vesicle with the assistance of p24-family proteins Emp24 and Erv25 (Muniz et al., 2000; Belden and Barlowe, 2001), in the Δemp24 mutant cell, the exit of Gas1 from the ER is delayed and the 105-kDa Gas1 precursor accumulates. To determine which deficiency caused the defect of Gas1 processing observed in the pga1-1 mutant, the proteins were subjected to Triton X-114 phase separation that distinguishes hydrophobicity (Doering and Schekman, 1996). Cells were grown at the semipermissive temperature for the pga1-1 mutant (34°C) for 12 h. The 105-kDa band, which represents the precursor form of Gas1 in the ER was only faintly detected in the total sample of the wild-type cell (Figure 4A, lane 1), but the amount significantly increased in the Δemp24 and pga1-1 mutant cells (Figure 4A, lanes 4 and 7). In the pga1-1 mutant cells, a significant amount of 125-kDa band that represents the mature form of Gas1 was detected, probably because the mutant Pga1-1 was not completely inactive in this condition; however, the 105-kDa band was more abundant than the 125-kDa band. GPI attachment increases the hydrophobicity of target proteins and is known to cause an increase of the amount of target proteins recovered in the detergent phase when separated with Triton X-114. Consistent with our prediction, the 125-kDa Gas1 in each cell and the 105-kDa Gas1 accumulated in the Δemp24 mutant was mainly detected in detergent phase, indicating these Gas1 were GPI-anchored (Figure 4A, lanes 3, 6, and 9). In contrast, the ER-form of Gas1 accumulated in pga1-1 cells was mainly detected in the aqueous phase (Figure 4A, lane 8). So, the 105-kDa Gas1 in the pga1-1 mutant cell was suggested to represent the Gas1 that was not GPI-anchored.
Figure 4.
The pga1-1 mutant showed temperature-sensitive defects in the GPI-anchoring of Gas1 and other GPI-anchored proteins. (A) Cleared lysate (total, T) of the WT, Δemp24, and pga1-1 cells was treated with 1% Triton X-114 and separated into the aqueous (A) and detergent (D) phases. Gas1, integral ER membrane protein Scs2 and the cytosolic 3-phosphoglycerate kinase (Pgk1) were detected by SDS-PAGE and Western blotting. (B) Cells were metabolically labeled with [3H]inositol and glycoproteins were collected using Con A-Sepharose. The wild-type (lanes 1 and 2) and pga1-1 (lanes 3 and 4) cells were grown at either 25°C (lanes 1 and 3) or 37°C (lanes 2 and 4). The concentrated glycoproteins were resolved by SDS-PAGE on 4–20% gradient gel and detected by Coomassie Brilliant Blue stain to confirm that the same amount of proteins was loaded on each lane. Lane M, molecular weight markers. (C) [3H]inositol incorporated into the glycoproteins was detected by autoradiography of the same gel. Lanes were the same as in B.
Because all detectable protein-linked inositol in yeast is present in the GPI-anchors, we can distinguish whether the GPIs were attached to its target proteins by checking the inositol incorporation to proteins. Therefore, we metabolically labeled the cell with [3H]inositol, and we analyzed the total protein for modification of the GPI-anchor to examine whether the pga1-1 mutant cell is actually defective in the GPI anchoring. Because all major GPI-anchored proteins are glycoproteins, almost all inositol-incorporated proteins can be recovered with Con A-Sepharose (Guillas et al., 2000). As shown in Figure 4C, [3H]inositol was incorporated in a number of proteins in the wild type, but a very small amount of [3H]inositol was detected in the pga1-1 mutant cells at 37°C. The equivalence of applied proteins was confirmed by Coomassie Brilliant Blue staining (Figure 4B). These results indicated that pga1-1 was defective in GPI synthesis or its anchoring to the target protein.
pga1ts Mutants Display the Phenotypes Related to GPI-anchoring Defect
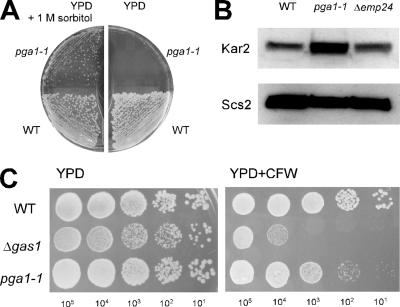
The mutants defective in GPI-anchoring generally show cell wall defects that are partially suppressed by increasing the osmolarity by adding 1 M sorbitol into the growth medium. Similarly, the pga1-1 mutant did not form colonies on the YPD plate at 36°C for 2 d, but the temperature sensitivity was partially suppressed by adding 1 M sorbitol to YPD (Figure 5A). At a sublethal temperature (34°C), a significant increase of the amount of Kar2p protein in the ER lumen was found in the pga1-1 mutant (Figure 5B). This indicates the pga1 mutant induced the unfolded protein response (UPR), which is commonly observed in GPI mutants (Ng et al., 2000, Davydenko et al., 2005). In contrast, the Δemp24 mutant that accumulates the ER-form Gas1 precursor due to the defect in transport of GPI-anchored proteins from the ER does not show UPR induction, suggesting that the UPR is induced by the accumulation of the GPI precursor or the target protein without GPI-anchoring, but not by the accumulation of GPI-anchored proteins. Mutants defective in GPI-anchoring are typically hypersensitive to the chitin-binding dye Calcofluor White (CFW). At a permissive temperature (30°C), the pga1-1 mutant showed a slightly increased sensitivity to CFW (Figure 5C). The Δgas1 mutant showed higher sensitivity. These results strongly support the likeliness that Pga1 is actually defective in GPI anchoring.
Figure 5.
The pga1-1 mutant displayed the phenotypes related to the defect of GPI-anchoring. (A) The temperature sensitivity of pga1-1 was partially suppressed by high osmolarity. The WT and pga1-1 mutant were streaked on YPD with or without 1 M sorbitol and grown 2 d at 36°C. (B) Unfolded protein response was induced in pga1-1 mutant cells. The amount of Kar2 protein (BiP) in the WT, pga1-1 mutant and Δemp24 mutant grown at 34°C was analyzed by SDS-PAGE and Western blotting, by using the ER membrane protein Scs2 as the loading control. (C) The pga1-1 mutant showed increased sensitivity to Calcofluor white. Ten microliters of 10-fold serially diluted suspension of the WT, Δgas1, and pga1-1 mutant cells were spotted on SD agar with or without 40 μg/ml Calcofluor White, and then the cells were incubated for 2 d at 30°C.
Interaction of Pga1 with Gpi18, an Essential Glycosylphosphatidylinositol-Mannosyltransferase II Component
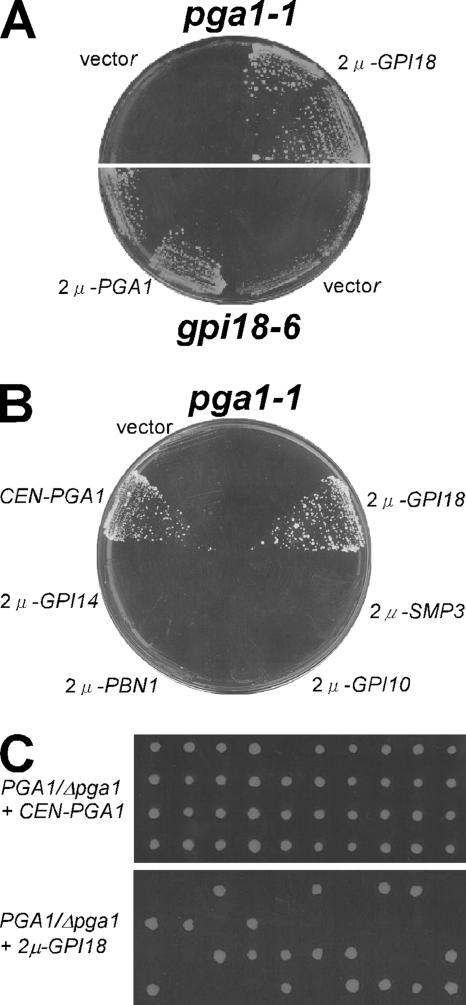
The defect of GPI anchoring in the pga1-1 mutant strongly suggested the involvement of PGA1 in GPI synthesis or anchoring process, but there are many steps in GPI synthesis and its anchoring to the target proteins. To determine where Pga1 functions in the pathway, we screened for the multicopy suppressor genes of the temperature sensitivity of the pga1-1 mutant, because it often becomes an effective clue to elucidate the functional role of uncharacterized gene products. We introduced the 2μ plasmid library to the pga1-1 mutant cells and sought the colonies whose viability at 37°C were dependent on the introduced plasmids. As a result, we found that the GPI18 gene could recover the growth defect of the pga1-1 mutant at 37°C by its introduction on 2μ plasmid (Figure 6A). GPI18 was recently reported to encode GPI-MT-II (Fabre et al., 2005; Kang et al., 2005). GPI-MT-II transfers the second Man from dolichol-P-Man to the Man-GlcN-PI precursor in the GPI anchor, so we supposed that Pga1 would have a function related to mannosylation in GPI synthesis. To test the possibility that Pga1 has genetic interaction with other GPI-MTs, we introduced GPI14, PBN1, GPI10, or SMP3 on 2μ plasmid. GPI14 and PBN1 encode subunits of GPI-MT-I (Maeda et al., 2001; Ashida et al., 2005), GPI10 encodes the third α1,2-mannosyltransferase, and SMP3 encodes the last α1,2-mannosyltransferase (Grimme et al., 2001). However, multicopy expression of each gene could not restore the growth of the pga1-1 mutant at 37°C (Figure 6B). The multicopy GPI18 could not suppress the lethality of the Δpga1 null mutant (Figure 6C), indicating that Pga1 has an indispensable role related to GPI18.
Figure 6.
Temperature sensitivity of the mutants of PGA1 and GPI18 was suppressed by introducing the mutual multicopy gene. (A) The colony-forming activity at 37°C of pga1-1 mutant was recovered by the GPI18 gene on the 2μ plasmid and that of gpi18–6 mutant was recovered by the PGA1 gene on the 2μ plasmid. (B) The high-copy introduction of GPI14, PBN1, GPI10, or SMP3 genes responsible for mannosylation in the yeast GPI synthesis did not suppress temperature sensitivity of the pga1-1 mutant at 37°C. (C) The heterozygous PGA1/Δpga1::kanMX4 diploid cells were transformed with the CEN-PGA1 (top) or 2μ-GPI18 (bottom) plasmid, and then they were sporulated. The spores were dissected to assess the complementation by tetrad analysis.
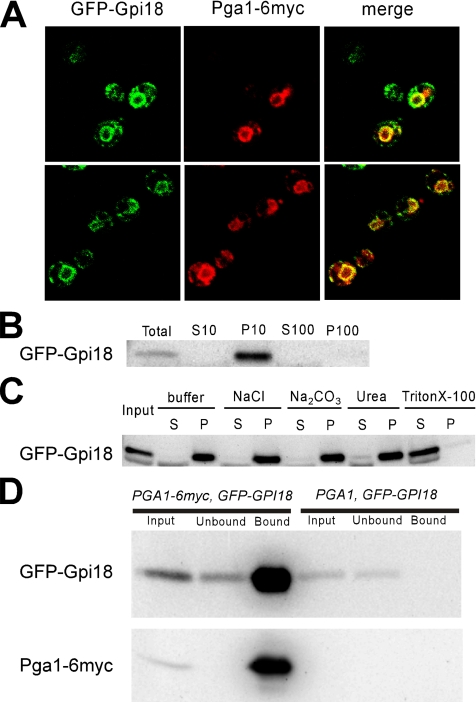
Because only GPI18 could rescue the growth defects of pga1-1 mutant cells, but other genes encoding GPI-MTs could not, the specific association between PGA1 and GPI18 was indicated. Therefore, we investigated the interaction of the two proteins in yeast cells. For this purpose, PGA1-6myc and GFP-GPI18 on low-copy plasmids were expressed in Δpga1 cells. The fluorescent imaging using GFP-tagged Gpi18 and Pga1-6myc indicated that both proteins mainly localized to the ER, and the merged images indicated that the two proteins were in the same compartments in the ER (Figure 7A). GFP-Gpi18 was mainly recovered in the P10 fraction by subcellular fractionation (Figure 7B), and it was only solubilized by Triton X-100 by our solubilization test (Figure 7C), which indicates that Gpi18 is an integral ER membrane protein. To examine whether there is a physical protein–protein interaction between Gpi18 and Pga1, we conducted immunoprecipitation experiments. The membranes in the cell lysate were solubilized by the addition of 1% Triton X-100, and Pga1-6myc was precipitated using the anti-myc mAb from the cleared lysate. The GFP-tagged Gpi18 was clearly detected in the precipitate fraction by immunoblotting (Figure 7D). Therefore, Gpi18 and Pga1 not only colocalized in the ER but also had a physical interaction in vivo.
Figure 7.
Pga1 and Gpi18 proteins colocalized in the ER and were found in a complex in the Triton X-100–solubilized lysate. (A) The fluorescent image of GFP-Gpi18 and the indirect immunofluorescent staining image of Pga1-6myc were overlapping (merge). (B) Subcellular fractionation of GFP-Gpi18 was performed as described in Figure 2B. (C) Solubilization test of GFP-Gpi18 was performed as in Figure 2C. (D) The cleared lysate of the cells having either Pga1-6myc or untagged Pga1 in addition to GFP-Gpi18 was subjected to immunoprecipitation using anti-myc mAb, and GFP-Gpi18 and Pga1-6myc were detected by SDS-PAGE and Western blotting.
Pga1 is a Novel Subunit of Glycosylphosphatidylinositol-Mannosyltransferase II
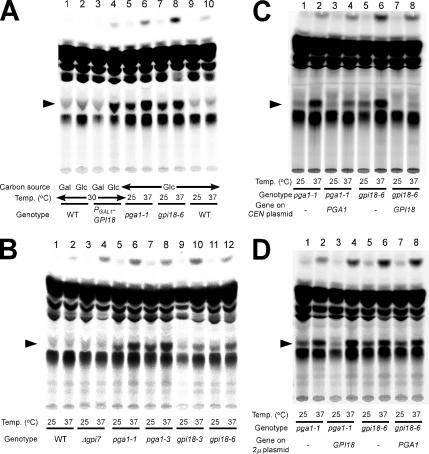
Because the pga1-1 mutant cells showed defects in the modification of the GPI anchor to the Gas1 precursor in the ER and other potential GPI-anchored proteins at 37°C, its temperature-sensitive growth defect was suppressed by the introduction of multicopy GPI18 gene, and Pga1 and Gpi18 were ER-membrane protein and found in a complex in the Triton X-100-solubilized lysate, we suspected that Pga1 and Gpi18 might cooperate in functioning as the enzyme to produce the GPI anchor. It was reported that the GPI18-depleted cell accumulated the GPI precursor “lipid 004-1,” which was identified as Man[EtN-P]-GlcN-PI (Fabre et al., 2005); thus, we tested whether the pga1 mutant accumulates the same GPI precursor. For this purpose, we constructed the PGAL1-GPI18 strain and gpi18ts mutants. Screening for gpi18ts mutants was performed by the same method by which the pga1ts mutants were constructed. As a result, we obtained 7 gpi18ts alleles and replaced the GPI18 gene on the chromosome of BY4741 with gpi18-6 for the further analysis. To analyze for accumulation of GPI anchor precursors, the cells were metabolically labeled with [3H]inositol, after which lipids were extracted from the cells and separated by TLC. [3H]Inositol-containing lipids on the TLC were detected by autoradiography. As shown in Figure 8, the pga1ts mutants and the gpi18ts mutants accumulated a spot of the same RF at 37°C (Figure 8A, lanes 6 and 8, and B, lanes 6, 8, 10, and 12). This spot was also detected in the sample of the transcription arrested cells of the PGAL1-GPI18 promoter shut-off mutant in SD (Figure 8A, lane 4); so, this corresponds to the “lipid 004-1”, namely, Man[EtN-P]-GlcN-PI. However, this spot was not detected in the wild type or Δgpi7 at 25 or 37°C (Figure 8A, lanes 9 and 10, and B, lanes 1–4), and moderate accumulation was detected in the pga1ts or the gpi18ts mutant cells at 25°C (Figure 8A, lanes 5 and 7, and B, lanes 5, 7, 10, and 11). Therefore, the functional Pga1 was necessary for the enzyme activity of GPI-MT II. We concluded that Pga1 is an essential subunit of GPI-MT II. The accumulation of the lipid 004-1 in pga1-1 and gpi18-6 mutants was complemented by the introduction of the wild-type gene on CEN plasmid (Figure 8C). The presence of a small amount of the lipid 004-1 in pga1-1/PGA1 may be caused by codominance of the pga1-1 mutation (Figure 8C, lane 4). On the contrary, the introduction of the high-copy-suppressor genes on 2μ plasmid did not diminish the accumulation (Figure 8D).
Figure 8.
The temperature-sensitive pga1 mutants accumulated the same GPI anchor precursor that was accumulated in the Gpi18-deprived cells. (A) The myo-2-[3H]inositol-labeled lipids were extracted, developed on a TLC plate in CHCl3/CH3OH/H2O (5:5:1, vol/vol), and detected by autoradiography. The wild-type (lanes 1 and 2) or PGAL1-GPI18 (lanes 3 and 4) cells were grown at 30°C in SG (lanes 1 and 3) or SD (lanes 2 and 4). An intermediate lipid (lipid 004-1, indicated by the arrowhead) accumulated in the sample of lane 4 in which the expression of GPI18 was repressed for 20 h in the glucose-containing medium. The wild-type (lanes 9 and 10), pga1-1 (lanes 5 and 6), or gpi18-6 (lanes 7 and 8) ts mutant cells were labeled at either 25°C (lanes 5, 7, and 9) or 37°C (lanes 6, 8, and 10). An accumulation of the indicated GPI biosynthetic precursor was found in lanes 5, 6, 7, and 8. (B) The wild-type (WT, lanes 1 and 2), Δgpi7 (lanes 3 and 4), pga1-1 (lanes 5 and 6), pga1-3 (lanes 7 and 8), gpi18-3 (lanes 9 and 10), or gpi18-6 (lanes 11 and 12) cells were grown at either 25°C (lanes 1, 3, 5, 7, 9, and 11) or 37°C (lanes 2, 4, 6, 8, 10, and 12), and labeled with myo-2-[3H]inositol, and the accumulation of the GPI biosynthetic precursor was analyzed as in described in A. The intermediate lipid 004-1 is indicated by the arrowhead. (C) The complementation of the lipid 004-1 (arrowhead) accumulation in the pga1-1 (lanes 1–4) or gpi18-6 (lanes 5–8) cells by introducing PGA1 (lanes 3 and 4) or GPI18 (lanes 7 and 8) on CEN plasmid was examined. The mutants transformed with pRS316 were used as the controls (lanes 1, 2, 5, and 6). (D) The suppression of the lipid 004-1 (arrowhead) accumulation in pga1-1 (lanes 1–4) or gpi18-6 (lanes 5–8) cells by introducing GPI18 (lanes 3 and 4) or PGA1 (lanes 7 and 8) on 2μ plasmid was examined. The mutants transformed with pRS426 were used as the controls (lanes 1, 2, 5, and 6).
Gpi18 Is Stable Irrespective of Pga1
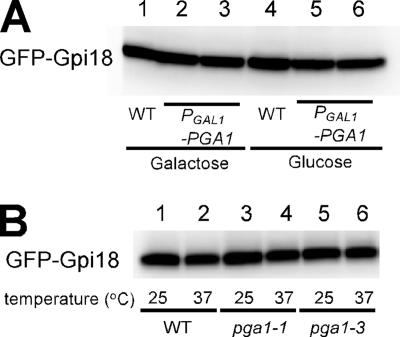
GPI-MT I also consists of two proteins in mammalian cells, PIG-M and PIG-X (Ashida et al., 2005). PIG-M is a multimembrane-spanning protein like Gpi18, and PIG-X is a protein with a single transmembrane domain like Pga1. Because PIG-X was reported to stabilize PIG-M, we speculated that the loss of Pga1 or 37°C incubation of ts allele of Pga1 would cause the destabilization of Gpi18. To test this possibility, GFP-Gpi18 was expressed in PGAL1-PGA1 mutant or pga1-1 mutant cells. Even after PGAL1-PGA1 was incubated in SD medium for 20 h when the lipid 004-1 significantly accumulated (data not shown), no reduction in the amount of GFP-Gpi18 was observed (Figure 9A). Similarly, incubation at 37°C for 2 h did not affect the stability of GFP-Gpi18 in pga1ts mutants (Figure 9B), although the 105-kDa proGas1 was significantly accumulated (data not shown). Therefore, the stability of Gpi18 is independent of Pga1.
Figure 9.
The stability of Gpi18 was not affected in pga1 mutants. (A) GFP-Gpi18 was expressed in the WT (lanes 1 and 4) or PGAL1-PGA1 (KSY308, lanes 2 and 5; KSY309, lanes 3 and 6) cells. After 20-h incubation in SG (lanes 1–3) or SD medium (lanes 4–6), GFP-Gpi18 was detected by SDS-PAGE and Western blotting. KSY308 and KSY309 are independent isolates of the same construction. (B) GFP-Gpi18 was expressed in the WT (lanes 1 and 2) or in the pga1ts mutants (pga1-1, lanes 3 and 4; pga1-2, lanes 5 and 6). After 2-h incubation at 25°C (lanes 1, 3, and 5) or at 37°C (lanes 2, 4, and 6), GFP-Gpi18 was detected as described in A.
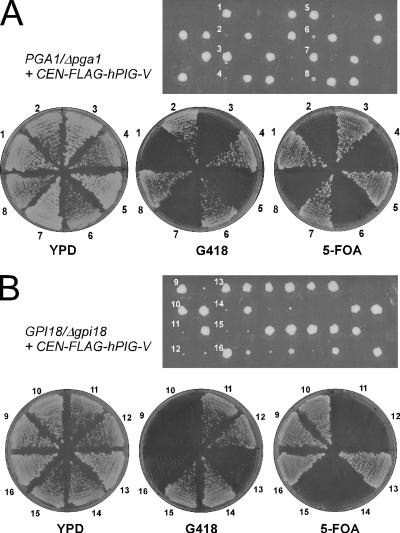
Human PIG-V Rescues the Viability of Δpga1 Cells
It is curious that PGA1 has homologues only in fungi but not in higher eukaryotes including mammals, because all S. cerevisiae genes identified so far to be involved in GPI synthesis or anchoring have their mammalian homologues. So, it is natural to assume the existence of a counterpart of Pga1 in mammalian cells. If the Pga1 counterpart exists, two probable reasons why there is no apparent PGA1 homologue were concerned. The first is that the sequence similarity between the two proteins is not high enough to be detected by homology search, although they are functionally homologous. The other is that the function of mammalian PIG-V contains not only that of Gpi18 but also that of Pga1. To discern which possibility is likely, we introduced the FLAG-human PIG-V on pRS316 (pFLAG-hPIG-V, a generous gift from Dr. Taroh Kinoshita, Research Institute for Microbial Disease; Osaka; Kang et al., 2005) to the heterozygous PGA1/Δpga1::kanMX4 diploid cells. The spores of the transformants were dissected to assess the complementation by tetrad analysis. As shown in Figure 10A, most tetrads from PGA1/Δpga1::kanNMX4 diploid cells harboring pFLAG-hPIG-V generated four viable haploid progenies with two of normal growth and two of slow growth. The slow growth cells were Δpga1 alleles whose viability was dependent on the pFLAG-hPIG-V because they were resistant to Geneticin (G418; Invitrogen) and lethal on 5-FOA plate, and the normal growth cells were wild-type cells because they were sensitive to G418 and grew normally on the 5-FOA plate. The same result was obtained when the pFLAG-hPIG-V was introduced to the GPI18/Δgpi18::kanMX4 diploid; two normal growth haploids sensitive to G418 and two slow growth haploids resistant to G418 were observed in most tetrads (Figure 10B). These results indicate that human PIG-V rescues the viability of not only Δgpi18 but also Δpga1. Furthermore, the introduction of GPI18 to Δpga1 even on 2μ plasmid could not rescue the viability, indicating that the function of Gpi18 is completely distinct from that of Pga1. Thus, human PIG-V is strongly suggested to possess the functions of both Gpi18 and Pga1 of S. cerevisiae.
Figure 10.
Human PIG-V rescues the viability of both Δgpi18 and Δpga1. (A) The heterozygous PGA1/Δpga1:: kanMX4 diploid cells were transformed with pFLAG-hPIG-V and were sporulated. The spores were dissected to assess the complementation by tetrad analysis. The genotypes of the numbered progenies (1–8) were checked by streaking the indicated cells on the G418 plate and the 5-FOA plate. (B) The heterozygous GPI18/Δgpi18::kanMX4 diploid cells were transformed with pFLAG-hPIG-V and were sporulated. The genotypes of the numbered progenies (9–16) were checked as described in A.
DISCUSSION
Enormous efforts have been made to identify the genes required for the yeast GPI biosynthesis. Two major strategies were generally taken in S. cerevisiae by most researchers; one was to screen the mutants defective in inositol incorporation, and the other was to find homologues with mammalian proteins that are proven to be involved in GPI biosynthesis. Despite these intensive studies, a flippase to move the GPI intermediate across the membrane and a GPI remodelase to introduce a ceramide to GPI that already attached to the target proteins have not been identified so far. In contrast, because an in vitro reaction of GPI synthesis using purified enzymes and substrates has not been demonstrated yet, it is not possible to exclude an undiscovered participant in any step of GPI biosynthesis.
We focused on the characterization of the ER-localized essential proteins whose functions remain unknown even after the enormous comprehensive studies of S. cerevisiae. Our strategy uses the construction of temperature-sensitive mutants suitable for both phenotypic and genetic studies. The temperature-sensitive pga1 mutants showed defects in the processing and transport of the GPI-anchored Gas1 protein (Figure 3A) and biochemical analysis indicated that GPI anchor synthesis was defective (Figure 4). Genetic analysis identified the GPI18 gene as a multicopy suppressor of the growth defect of pga1-1 (Figure 6A). Gpi18 was reported to be GPI-MT II, which adds Man to Man-GlcN-PI, and the mutants in pga1 as well as gpi18 accumulated the same intermediate, Man[EtN-P]-GlcN-PI (Figure 8). Gpi18 and Pga1 not only colocalized in the ER membrane but also were found in a complex after Triton X-100 solubilization of the cell lysate (Figure 7). From all these results, we concluded that Pga1 is a novel subunit of GPI-MT II that collaborates with another subunit Gpi18 (Figure 10).
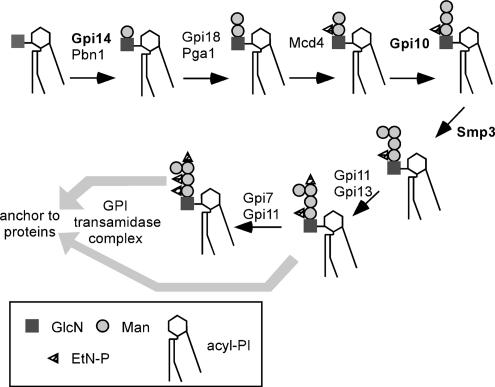
As shown in Figure 11, several steps in GPI biosynthesis are reported to be carried out by the participation of multiple proteins (Kinoshita and Inoue, 2000; Pittet and Conzelmann, 2006). Both mammalian and budding yeast GPI-MT I are a complex of a multimembrane-spanning protein (PIG-M/Gpi14) and a single membrane-spanning protein (PIG-X/Pbn1). Most GPI-MT polypeptides have a DXD motif, which is generally required for the activity of glycosyltransferase. Because PIG-M and Gpi14 have a DXD sequence, the multimembrane-spanning subunit probably represents the catalytic subunit. The molecular function of PIG-X and Pbn1 is unknown, but PIG-X was reported to be required for stability of PIG-M (Ashida et al., 2005). So, the single membrane-spanning subunit may play a structural role for the partner. However, as for Pga1, neither repression of PGA1 by the GAL1 promoter shut-off nor incubation of the pga1-1 mutant at 37°C affected the amount of Gpi18 (Figure 9), suggesting that Pga1 is not required for the stability of its partner. The overexpression of GPI18 could not rescue the lethality of Δpga1. It only rescued the temperature sensitivity of pga1ts. Conversely, PGA1 overexpression suppressed the temperature sensitivity of gpi18ts (Figure 6A). We immunoprecipitated Pga1-6myc from the digitonin-solubilized membrane fraction with the anti-myc antibody, but we could not detect any other interacting protein except Gpi18 in the coprecipitated proteins (data not shown). These data indicate that both Pga1 and Gpi18 would be required to generate the overall structure necessary for the substrate recognition and enzymatic activity of yeast GPI-MT II.
Figure 11.
The mannosylation process in GPI anchor synthesis. Pga1 is a partner of Gpi18. GPI-MT I and GPI-MT II are composed of protein complex. The GPI-MT subunits containing a DXD motif are shown in bold.
The sequences of polypeptides of GPI-MT II do not contain a DXD motif, which is commonly present in glycosyltransferases. Instead, Gpi18 and PIG-V has a structural feature common with Pmt2-family proteins (Kang et al., 2005). The Pmt2-family proteins are O-mannosyltransferases, which have multiple transmembrane domains but no DXD motif. They have two conserved hydrophilic loops that face the ER lumen. One loop is between the first and second transmembrane domains and the other is between the fifth and sixth. The first loop of each Pmt2-family protein has a Trp-Asp motif as well as Gpi18 and PIG-V. In CHO PIG-V mutants, single amino-acid substitutions of W66L or D67A in the conserved Trp-Asp motif were responsible for the mutant phenotype (Kang et al., 2005). Considering this result and structural features, Gpi18 is likely to be a catalytic subunit of the yeast GPI-MT II, but it requires the partner Pga1 for the activity.
No obvious Pga1 homologue was found in mammalian cells. There are several possible explanations why Pga1 is required only in fungi. First, the Pga1 function is necessary only in yeast cells and completely specific to yeast. Second, there is a functional homologue with very low similarity. Third, PIG-V possesses the functions of both Gpi18 and Pga1. We supposed that the first possibility was unlikely because the GPI biosynthetic pathway is conserved between the yeast and the mammalian cell very well so intermediates and enzymes in the pathway should be common. The second possibility seemed likely; however, the third possibility was most likely because the expression of the human PIG-V gene in both Δgpi18 and Δpga1 cells could suppress their lethality (Figure 10). So, it is natural to suppose that the two yeast proteins compose the GPI-MT II complex, which is functionally homologous to the single human PIG-V protein. Compared with human PIG-V, the hydrophilic segments at the both sides of the seventh transmembrane domain are missing in yeast Gpi18 (Figure 4 of Kang et al., 2005). It is possible that Pga1 compensates for the function of these missing segments by forming a complex with Gpi18 in the membrane. Because the PIG-V homologue in fruit fly has similar missing stretches, it may require a homologue of Pga1 for its activity.
Considering that both GPI-MT I and GPI-MT II are composed of (at least) two subunits in S. cerevisiae, it is natural to ask whether GPI-MT III or IV forms a similar complex, because they are using substrates with structures similar to those used by GPI-MT I and GPI-MT II. The proteins that have been proved to be responsible for the mannosylation in GPI-MT III and IV until now are Gpi10 and Smp3, respectively, both of which are multimembrane-spanning proteins like Gpi14 and Gpi18. It would be very interesting to perform the multicopy suppressor screening of gpi10 or smp3 temperature-sensitive mutants to see whether there would be any partner proteins to them.
ACKNOWLEDGMENTS
We thank Drs. S. Kagiwada and M. Tokunaga for kindly providing antisera; Dr. Taroh Kinoshita for kindly providing the human PIG-V expression plasmids; Chihiro Akimoto and Atsushi Kamiya for help in preliminary characterization of PGA1; Mariko Umemura and Dr. Takehiko Yoko-o for valuable advices in the analysis of GPI precursors; Dr. Hironori Inadome, Dr. Shah Alam Bhuiyan and Dr. Hiroyuki Adachi for helpful discussions. This work was supported by a Research Fellowship for Young Scientists from the Japan Society for the Promotion of Science (to K.S.), grants-in-aid for Scientific Research from the Japan Society for the Promotion of Science (to Y.N.), and grants for “Bioarchitect Research” from the Institute of Physical and Chemical Research (RIKEN) of Japan (to K.Y.) and a grant from the Noda Institute of Scientific Research (to K.Y.).
Abbreviations used:
- ALP
alkaline phosphatase
- CPY
carboxypeptidase Y
- endoH
endoglycosidase H
- ER
endoplasmic reticulum
- EtN-P
phosphoethanolamine
- GFP
green fluorescent protein
- GlcN
glucosamine
- GPI
glycosylphosphatidylinositol
- GPI-MT
GPI-mannosyltransferase
- Man
mannose
- PI
phosphatidylinositol
- SGD
Saccharomyces Genome Database.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-03-0258) on July 5, 2007.
REFERENCES
- Ashida H., Hong Y., Murakami Y., Shishioh N., Sugimoto N., Kim Y. U., Maeda Y., Kinoshita T. Mammalian PIG-X and yeast Pbn1p are the essential components of glycosylphosphatidylinositol-mannosyltransferase I. Mol. Biol. Cell. 2005;16:1439–1448. doi: 10.1091/mbc.E04-09-0802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballou C. E. Isolation, characterization, and properties of Saccharomyces cerevisiae mnn mutants with nonconditional protein glycosylation defects. Methods Enzymol. 1990;185:440–470. doi: 10.1016/0076-6879(90)85038-p. [DOI] [PubMed] [Google Scholar]
- Belden W. J., Barlowe C. Deletion of yeast p24 genes activates the unfolded protein response. Mol. Biol. Cell. 2001;12:957–969. doi: 10.1091/mbc.12.4.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davierwala A. P., et al. The synthetic genetic interaction spectrum of essential genes. Nat. Genet. 2005;37:1147–1152. doi: 10.1038/ng1640. [DOI] [PubMed] [Google Scholar]
- Davydenko S. G., Feng D., Jäntti J., Keränen S. Characterization of GPI14/YJR013w mutation that induces cell wall integrity signaling pathway and results in increased protein production in Saccharomyces cerevisiae. Yeast. 2005;22:993–1009. doi: 10.1002/yea.1286. [DOI] [PubMed] [Google Scholar]
- Doering T. L., Schekman R. GPI anchor attachment is required for Gas1p transport from the endoplasmic reticulum in COP II vesicles. EMBO J. 1996;15:182–191. [PMC free article] [PubMed] [Google Scholar]
- Duenas E., Revuelta J. L., del Rey F., Vazquez de Aldana C. R. Disruption and basic phenotypic analysis of six novel genes from the left arm of chromosome XIV of Saccharomyces cerevisiae. Yeast. 1999;15:63–72. doi: 10.1002/(SICI)1097-0061(19990115)15:1<63::AID-YEA338>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Eisenhaber B., Maurer-Stroh S., Novatchkova M., Schneider G., Eisenhaber F. Enzymes and auxiliary factors for GPI lipid anchor biosynthesis and post-translational transfer to proteins. Bioessays. 2003;25:367–385. doi: 10.1002/bies.10254. [DOI] [PubMed] [Google Scholar]
- Englund P. T. The structure and biosynthesis of glycosyl phosphatidylinositol protein anchors. Annu. Rev. Biochem. 1993;62:121–138. doi: 10.1146/annurev.bi.62.070193.001005. [DOI] [PubMed] [Google Scholar]
- Fabre A. L., Orlean P., Taron C. H. Saccharomyces cerevisiae Ybr004c and its human homologue are required for addition of the second mannose during glycosylphosphatidylinositol precursor assembly. FEBS J. 2005;272:1160–1168. doi: 10.1111/j.1742-4658.2005.04551.x. [DOI] [PubMed] [Google Scholar]
- Grimme S. J., Westfall B. A., Wiedman J. M., Taron C. H., Orlean P. The essential Smp3 protein is required for addition of the side-branching fourth mannose during assembly of yeast glycosylphosphatidylinositols. J. Biol. Chem. 2001;276:27731–27739. doi: 10.1074/jbc.M101986200. [DOI] [PubMed] [Google Scholar]
- Guillas I., Pfefferli M., Conzelmann A. Analysis of ceramides present in glycosylphosphatidylinositol anchored proteins of Saccharomyces cerevisiae. Methods Enzymol. 2000;312:506–515. doi: 10.1016/s0076-6879(00)12935-0. [DOI] [PubMed] [Google Scholar]
- Hazbun T. R., et al. Assigning function to yeast proteins by integration of technologies. Mol. Cell. 2003;12:1353–1365. doi: 10.1016/s1097-2765(03)00476-3. [DOI] [PubMed] [Google Scholar]
- Imai K., Noda Y., Adachi H., Yoda K. A novel endoplasmic reticulum membrane protein Rcr1 regulates chitin deposition in the cell wall of Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:8275–8284. doi: 10.1074/jbc.M409428200. [DOI] [PubMed] [Google Scholar]
- Inadome H., Noda Y., Adachi H., Yoda K. Immunoisolation of the yeast Golgi subcompartments and characterization of a novel membrane protein, Svp26, discovered in the Sed5-containing compartments. Mol. Cell. Biol. 2005;25:7696–7710. doi: 10.1128/MCB.25.17.7696-7710.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J. Y., Hong Y., Ashida H., Shishioh N., Murakami Y., Morita Y. S., Maeda Y., Kinoshita T. PIG-V involved in transferring the second mannose in glycosylphosphatidylinositol. J. Biol. Chem. 2005;280:9489–9497. doi: 10.1074/jbc.M413867200. [DOI] [PubMed] [Google Scholar]
- Kilmartin J. V., Adams A, E. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J. Cell Biol. 1984;98:922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T., Inoue N. Dissecting and manipulating the pathway for glycosylphosphatidylinositol-anchor biogenesis. Curr. Opin. Chem. Biol. 2000;4:632–638. doi: 10.1016/s1367-5931(00)00151-4. [DOI] [PubMed] [Google Scholar]
- Klinosky D. J., Emr S. D. Membrane protein sorting: biosynthesis, transport and processing of yeast vacuolar alkaline phosphatase. EMBO J. 1989;8:2241–2250. doi: 10.1002/j.1460-2075.1989.tb08348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleizen B., Braakman I. Protein folding and quality control in the endoplasmic reticulum. Curr. Opin. Cell Biol. 2004;16:343–349. doi: 10.1016/j.ceb.2004.06.012. [DOI] [PubMed] [Google Scholar]
- Kodukula K., Gerber L. D., Amthauer R., Brink L., Udenfriend S. Biosynthesis of glycosylphosphatidylinositol (GPI)-anchored membrane proteins in intact cells: specific amino acid requirements adjacent to the site of cleavage and GPI attachment. J. Cell Biol. 1993;120:657–664. doi: 10.1083/jcb.120.3.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lewis M. J., Rayner J. C., Pelham H. R. A novel SNARE complex implicated in vesicle fusion with the endoplasmic reticulum. EMBO J. 1997;16:3017–3024. doi: 10.1093/emboj/16.11.3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C. F., Montijn R. C., Brown J. L., Klis F., Kurjan J., Bussey H., Lipke P. N. Glycosyl phosphatidylinositol-dependent cross-linking of α-agglutinin and β 1,6-glucan in the Saccharomyces cerevisiae Cell Wall. J. Cell Biol. 1995;128:333–340. doi: 10.1083/jcb.128.3.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lussier M., Gentzsch M., Sdicu A. M., Bussey H., Tanner W. Protein O-glycosylation in yeast. The PMT2 gene specifies a second protein O-mannosyltransferase that functions in addition to the PMT1-encoded activity. J. Biol. Chem. 1995;270:2770–2775. doi: 10.1074/jbc.270.6.2770. [DOI] [PubMed] [Google Scholar]
- Maeda Y., Watanabe R., Harris C. L., Hong Y., Ohishi K., Kinoshita K., Kinoshita T. PIG-M transfers the first mannose to glycosylphosphatidylinositol on the luminal side of the ER. EMBO J. 2001;20:250–261. doi: 10.1093/emboj/20.1.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman I., Warren G. The road taken: past and future foundations of membrane traffic. Cell. 2000;100:99–112. doi: 10.1016/s0092-8674(00)81687-6. [DOI] [PubMed] [Google Scholar]
- Mnaimneh S., et al. Exploration of essential gene functions via titratable promoter alleles. Cell. 2004;118:31–44. doi: 10.1016/j.cell.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Muhlrad D., Hunter R., Parker R. A rapid method for localized mutagenesis of yeast genes. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- Muniz M., Nuoffer C., Hauri H. P., Riezman H. The Emp24 complex recruits a specific cargo molecule into endoplasmic reticulum-derived vesicles. J. Cell Biol. 2000;148:925–930. doi: 10.1083/jcb.148.5.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng D. T., Spear E. D., Walter P. The unfolded protein response regulates multiple aspects of secretory and membrane protein biogenesis and endoplasmic reticulum quality control. J. Cell Biol. 2000;150:77–88. doi: 10.1083/jcb.150.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P., Field C., Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- Nuoffer C., Horvath A., Riezman H. Analysis of the sequence requirements for glycosylphosphatidylinositol anchoring of Saccharomyces cerevisiae Gas1 protein. J. Biol. Chem. 1993;268:10558–10563. [PubMed] [Google Scholar]
- Palade G. Intracellular aspects of the process of protein synthesis. Science. 1975;189:347–358. doi: 10.1126/science.1096303. [DOI] [PubMed] [Google Scholar]
- Pittet M., Conzelmann A. Biosynthesis and function of GPI proteins in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2006;1771:405–420. doi: 10.1016/j.bbalip.2006.05.015. [DOI] [PubMed] [Google Scholar]
- Popolo L., Vai M. The Gas1 glycoprotein, a putative wall polymer cross-linker. Biochim. Biophys. Acta. 1999;1426:385–400. doi: 10.1016/s0304-4165(98)00138-x. [DOI] [PubMed] [Google Scholar]
- Schuldiner M., et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Sipos G., Fuller R. S. Separation of Golgi and endosomal compartments. Methods Enzymol. 2002;351:351–365. doi: 10.1016/s0076-6879(02)51857-7. [DOI] [PubMed] [Google Scholar]
- Taron B. W., Colussi P. A., Wiedman J. M., Orlean P., Taron C. H. Human Smp3p adds a fourth mannose to yeast and human glycosylphosphatidylinositol precursors in vivo. J. Biol. Chem. 2004;279:36083–36092. doi: 10.1074/jbc.M405081200. [DOI] [PubMed] [Google Scholar]
- Takahashi M., Inoue N., Ohishi K., Maeda Y., Nakamura N., Endo Y., Fujita T., Takeda J., Kinoshita T. PIG-B, a membrane protein of endoplasmic reticulum with a large luminal domain, is involved in transferring the third mannose of the GPI anchor. EMBO J. 1996;15:4254–4261. [PMC free article] [PubMed] [Google Scholar]
- Tsukada M., Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- Van Den Hazel H. B., Kielland-Brandt M. C., Winther J. R. Biosynthesis and function of yeast vacuolar proteases. Yeast. 1996;12:1–16. doi: 10.1002/(sici)1097-0061(199601)12:1<1::aid-yea902>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Vashist S., Frank C. G., Jakob C. A., Ng D. T. Two distinctly localized P-type ATPases collaborate to maintain organelle homeostasis required for glycoprotein processing and quality control. Mol. Biol. Cell. 2002;21:3955–3966. doi: 10.1091/mbc.02-06-0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooding S., Pelham H. R. The dynamics of Golgi protein traffic visualized in living yeast cells. Mol. Biol. Cell. 1998;9:2667–2680. doi: 10.1091/mbc.9.9.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]