Abstract
Packaging DNA into condensed structures is integral to the transmission of genomes. The mammalian mitochondrial genome (mtDNA) is a high copy, maternally inherited genome in which mutations cause a variety of multisystem disorders. In all eukaryotic cells, multiple mtDNAs are packaged with protein into spheroid bodies called nucleoids, which are the fundamental units of mtDNA segregation. The mechanism of nucleoid formation, however, remains unknown. Here, we show that the mitochondrial transcription factor TFAM, an abundant and highly conserved High Mobility Group box protein, binds DNA cooperatively with nanomolar affinity as a homodimer and that it is capable of coordinating and fully compacting several DNA molecules together to form spheroid structures. We use noncontact atomic force microscopy, which achieves near cryo-electron microscope resolution, to reveal the structural details of protein–DNA compaction intermediates. The formation of these complexes involves the bending of the DNA backbone, and DNA loop formation, followed by the filling in of proximal available DNA sites until the DNA is compacted. These results indicate that TFAM alone is sufficient to organize mitochondrial chromatin and provide a mechanism for nucleoid formation.
INTRODUCTION
Mutations affecting mitochondrial oxidative phosphorylation cause a variety of multisystem disorders (DiMauro and Schon, 2003; Taylor and Turnbull, 2005) with an estimated incidence of 1:5000 live births (Thorburn, 2004). The majority of these mutations are found in the mitochondrial genome (mtDNA), which encodes the core hydrophobic proteins involved in oxidative phosphorylation and some of the molecules required for their expression, such as tRNAs and rRNAs. The mutant mtDNA genomes generally coexist alongside wild-type copies in affected patients, a situation referred to as heteroplasmy. The segregation pattern of heteroplasmic DNAs in different tissues is an important determinant of the severity of the clinical phenotype. To develop an animal model of heteroplasmy, our laboratory generated mice containing polymorphisms in otherwise normal mtDNA, and we identified three nuclear quantitative trait loci that affect mtDNA segregation in liver, spleen, and kidney (Jenuth et al., 1997; Battersby et al., 2003). The selection of one variant of the mtDNA over another could be explained by differences in their organization or packaging.
Mammalian mtDNA is packaged in protein–DNA complexes termed nucleoids, which can be visualized as small submitochondrial bodies in the matrix (Nass, 1969). The nucleoid contains two to seven genomes, depending on cell type, and it is present as 450–800 distinct foci in cultured cells (Nass, 1969; Iborra et al., 2004; Legros et al., 2004). Several mammalian mitochondrial nucleoid proteins have been identified after enrichment by sedimentation and immunoprecipitation (Wang and Bogenhagen, 2006; He et al., 2007). However, other than the components of the mitochondrial replisome and transcription apparatus, the only proteins that have been found to bind DNA are the Lon protease (Lu et al., 2003; Liu et al., 2004; Lu et al., 2007), which binds solely to single-stranded DNA in vitro and in vivo, and a truncation of mammalian ATAD3, which binds preferentially though weakly to supercoiled D-loop sequences (He et al., 2007). Because the packaging of the nucleoid is likely a double-stranded (ds) DNA-directed activity, we pursued the identification of mitochondrial proteins that interact directly with dsDNA, and we isolated a single protein, the mitochondrial transcription factor TFAM (data not shown). In addition, our preliminary experiments have identified TFAM and single-stranded DNA binding protein 1 as major components of purified mouse liver nucleoids (data not shown), leading us to focus on understanding the role of TFAM in mtDNA packaging.
TFAM is an essential protein (Larsson et al., 1998) that was initially discovered as a factor that stimulated transcription from the heavy and light strand mtDNA promoters (HSP and LSP, respectively) by mitochondrial RNA polymerase (Fisher and Clayton, 1985). By using DNase I, TFAM was footprinted to the regions upstream of these transcription start sites, revealing a strong preference immediately proximal to the LSP, which creates a specific local pattern or phasing of TFAM molecules (Fisher and Clayton, 1988; Ghivizzani et al., 1994). Additionally, mature human and mouse TFAM proteins, which are 78% similar, both exhibit qualitatively high affinity for nonspecific dsDNA (Ekstrand et al., 2004), which cannot be detected by DNAse I footprinting (Fisher and Clayton, 1988). TFAM has also been localized to the nucleoid by immunocytochemistry (Garrido et al., 2003; Legros et al., 2004).
The exact function of TFAM in the mammalian nucleoid is unclear. Some evidence indicates that mtDNA is coated with TFAM (Takamatsu et al., 2002; Alam et al., 2003; Ohgaki et al., 2007), suggesting a role in packaging (Alam et al., 2003). Any compaction afforded by TFAM binding has been attributed to typical DNA bending caused by high mobility group (HMG) box domains (Fisher et al., 1992; Parisi et al., 1993). Because TFAM expression levels and mtDNA copy number show a strong correlation, TFAM has also been speculated to provide a scaffold upon which the nucleoid is formed (Ekstrand et al., 2004). Recently, evidence has been presented for a low abundance of TFAM in mitochondria that is limiting for transcription initiation, which confines TFAM function to the original designation of transcription factor (Maniura-Weber et al., 2004).
To elucidate the role of TFAM in the mtDNA nucleoid, we developed in vitro systems that model the specific and nonspecific features of TFAM-DNA binding. Here, we show that the protein exists and functions as a homodimer that binds to DNA cooperatively—a characteristic that increases the apparent affinity of TFAM for DNA and drives DNA compaction to completion. In addition, our data corroborate in vivo estimates determined for the stoichiometry between bound TFAM and DNA, further validating our experimental system. Lastly, we demonstrate that TFAM not only has the capacity to compact DNA fully at in vivo stoichiometry but also that it organizes and compacts DNA through loop formation and coordination of multiple DNAs into nucleoid-like structures. Our results indicate that TFAM is a DNA packaging protein and provide a detailed mechanism for nucleoid formation.
MATERIALS AND METHODS
Recombinant TFAM Protein Purification
The mature form of mouse TFAM fused to glutathione S-transferase (GST)-tagged protein was expressed from the pGEX-4T1 plasmid in BL21-AI cells (Invitrogen, Carlsbad, CA) according to the manufacturer's recommendations. Proteins were isolated on a 1-ml Hitrap GST column (GE Healthcare, Piscataway, NJ) at 4°C according to the manufacturer using 300 mM NaCl (TFAM) or 500 mM NaCl (TFAM-high salt [HS]).
Nanoscope Atomic Force Microscopy (AFM) Imaging and Electrophoretic Mobility Shift Assay (EMSA)
Freshly cleaved mica was treated first with 2 μl of poly-l-lysine (0.01%; Sigma-Aldrich, St. Louis, MO) for 2 min, rinsed with deionized water and dried under a flow of nitrogen gas. Protein was incubated with 1 μg/ml DNA in buffer (100 mM NaHCO3, pH 7.6) for 2 min, and 5% of the reaction was diluted 1:1 in buffer and immediately deposited onto the treated mica. Samples were allowed to adsorb for 3–5 min, then they were washed and dried as described above. Experiments conducted with samples prepared on different days yielded similar results. The remainder of the reaction was resolved in 0.5X Tris borate-EDTA and 1.2% agarose, and then it was stained with ethidium bromide.
Images were acquired in air using a Bioscope I with Nanoscope IIIa controller atomic force microscope (Veeco Instruments, Woodbury, NY) operating in tapping mode. The scan rate was set to 2 Hz at 512 × 512 pixels. Broad field scans were 3–5 μm. All images were obtained in air using oxide-sharpened silicon tips (Veeco Instruments) with resonant frequency ∼300 kHz.
Computational Analysis of Atomic Force Height Images
Using the data of all available images of naked DNA on mica, we computed the mean DNA height (h̄DNA) and the SD of the distribution (σDNA) for each protein amount. To identify the pixels that correspond to DNA molecules, a height threshold was set at (h̄DNA − σDNA). Irregularities were removed from the computation by selecting for large objects. The number of objects analyzed in each TFAM sample were 0 μg/ml, 190; 1.7 μg/ml, 50; 3.5 μg/ml, 127; 7 μg/ml, 133; 15 μg/ml, 126; 25 μg/ml, 27; 50 μg/ml, 57; and 100 μg/ml, 43.
To quantify protein occupancy of the DNA molecules, the ratio between the number of pixels above 2h̄DNA and the total number of pixels of the objects was computed, yielding the relative protein occupancy parameter (Figure 2A, inset). Hill transformation of these data were achieved by correcting for maximal binding at 100 μg/ml to θ = 0.99 (Supplemental Figure 1). Proportional corrections were applied to the remaining occupancy values.
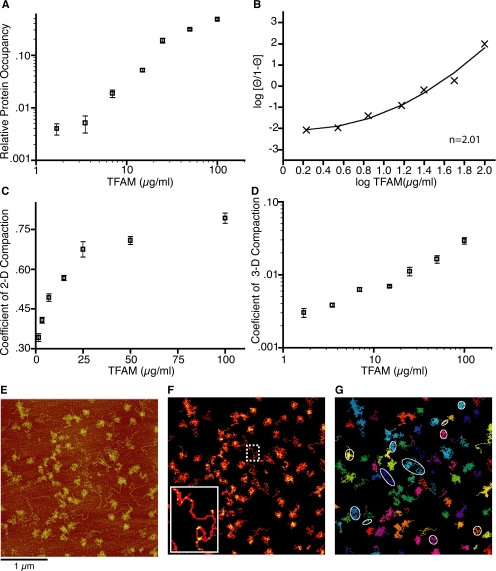
Figure 2.
Quantitation of protein occupancy and compaction derived from AFM images. (A) Relative protein occupancy per structure with SE for each TFAM concentration. (B) Hill plot of adjusted TFAM occupancy. Transformed data points and quadratic best fit are shown with a Hill coefficient (n = 2.01). (C) Coefficient of 2-dimensional compaction per TFAM concentration with SE. (D) Coefficient of 3-dimensional compaction per DNA molecule for each TFAM concentration with SE. (E) Representative AFM image from 7 μg/ml TFAM (see Figure 1C). Bar, is 1 μm. (F) Colorized example of DNA (red) and DNA bound by TFAM (yellow) derived from AFM data shown in E with inset zoom of uncompacted DNA molecule. (G) Colorized individual DNA molecules from E as determined by computational analysis. Examples of ellipses used for calculations used to generate panel C and D are indicated in white.
To quantify horizontal compaction on the substrate plane, we computed the ellipse that best approximates the two-dimensional (2-D) region described by the object. The ratio between the area occupied by an object and the surface area of its bounding ellipse was calculated, yielding the coefficient of 2-D compaction.
To quantify compaction in both the horizontal and vertical planes, we divided the maximum height of each DNA object by the long axis of its corresponding ellipse, yielding the coefficient of three-dimensional (3-D) compaction.
Surface Plasmon Resonance
The binding interaction between TFAM (∼48-kDa dimer) and DNA was examined by surface plasmon resonance (SPR) in real-time by using BIACORE 3000 instrumentation (Biacore AB, Uppsala, Sweden). According to the manufacturer's recommendations, biotinylated ds DNA containing cytochrome c oxidase subunit I (COXI) sequence was immobilized on streptavidin (SA) sensor chips (Biacore AB). Kinetic analyses were performed such that TFAM was diluted in HBS-EP (10 mM HEPES pH. 7.4, 150 mM NaCl, 3.4 mM EDTA, and 0.005% P-20) and injected at 10 μl/min (120-s association + 120-s dissociation) over the reference SA and active oligonucleotide surfaces (∼200 resonance units [RU]). Regeneration was achieved using a 1-min injection of 0.5 M NaCl in HBS-EP, followed by a 1-min running buffer-only wash. All concentrations were analyzed in multiples (2–3) to ensure consistency, and every experiment was tested for the presence of mass transport effects. All binding data were “double-referenced” (Myszka, 1999) and analyzed using BIAevaluation software 4.1 (Biacore AB).
Size Exclusion Chromatography
Samples were injected in 250 μl of buffer (25 mM HEPES, pH 7.6, and 150 mM NaCl) plus 1% deoxycholate (DOC) onto a Tricorn Superdex 200 10/300 GL column (GE Healthcare). Samples were separated at a flow rate 0.4 ml/min in buffer plus 0.1% DOC. The column was calibrated with both high- and low-molecular-weight calibration kits by UV absorption using the Unicorn Analysis Module (GE Healthcare). Recombinant TFAM protein samples were diluted into injection buffer, whereas mitochondrial extracts were generated by lysis in injection buffer and clarified before injection.
Noncontact Atomic Force Microscopy Imaging
High-resolution surface topography measurements were performed by noncontact (NC)-AFM under ultrahigh vacuum conditions (<4 × 10−8 Pa). In this operation mode, the oscillation frequency of the cantilever is used to regulate the tip-sample distance (Albrecht et al., 1991; Morita et al., 2002). The NC-AFM measurements were performed at room temperature using an ultrahigh vacuum JEOL JSPM 4500A AFM (JEOL, Tokyo, Japan), and a Nanosurf phase-locked loop was used for frequency demodulation. Cantilevers were supplied by Nanosensors (Neuchatel, Switzerland), having a typical resonance frequency of 170 kHz and a spring constant of 40–50 N/m, and they were used with an oscillation amplitude of 5 nm.
For additional methods and further details, see Supplemental Material.
RESULTS
TFAM Binds DNA Cooperatively as Determined by Electrophoretic Mobility Shift Assay and Atomic Force Microscopy
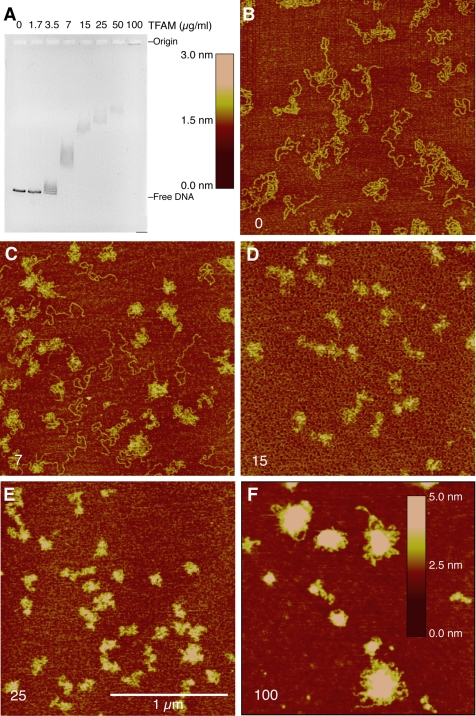
To examine sequence nonspecific association of TFAM with DNA, we incubated linearized pUC19 plasmid DNA with various amounts of purified TFAM and examined each reaction by EMSA and AFM (Figure 1). We observed qualitative indications of positive cooperative protein loading on DNA; specifically, the first binding event affects the probability and location of the second event. By EMSA, we detected a nonlinear response to increasing protein levels as well as a significant shift in mobility between adjacent samples (Figure 1A), which is in agreement with results reported previously (Fisher and Clayton, 1988). The corresponding AFM height images showed that while naked DNAs are similar to pieces of string on a surface (Figure 1B), increasing protein concentrations in the reaction lead to a decrease in the spreading of the DNA on the surface and an increase in the height of these structures (Figure 1, C–F). In addition, heterogeneous compaction was evident at lower protein concentrations where compacted and uncompacted DNA molecules were observed side by side (e.g., Figure 1C), consistent with a cooperative loading mechanism of TFAM onto DNA. These hallmarks of cooperativity were also observed by AFM on circular templates (Supplemental Figure 1).
Figure 1.
Electrophoretic mobility shift assay and atomic force microscopy analyses of TFAM association with pUC19 DNA. (A) Gel shift analysis of TFAM binding to linearized pUC19 DNA. TFAM concentration (micrograms per milliliter) is indicated for each lane, and it correlates with samples analyzed in B–F. Image contrast is inverted. (B–E) AFM height scans of TFAM-DNA complexes at varying concentrations of TFAM (3-nm height scale). (F) AFM height scan of TFAM–DNA complexes at 100 μg/ml TFAM (5-nm height scale). Bar, 1 μm. TFAM concentrations are indicated in the bottom left corner of each panel in micrograms per milliliter. All AFM data are raw flattened images.
We analyzed the data from the AFM images to quantify the association of TFAM with DNA and to determine the amount of compaction in each reaction (Figure 2). Using an algorithm to distinguish unbound DNA from protein-bound DNA by height, we observed that plotting TFAM occupancy as a function protein concentration generated a sigmoidal curve (Figure 2A). A colorized example of this analysis is shown in Figure 2F, where red pixels indicate all DNAs (pixels above the polylysine coating on the mica chip), and yellow pixels indicate the subset of red in which DNA is bound by protein. The yellow-to-red ratio was determined for each structure (unique structures are individually colorized as shown in Figure 2G) and the relative protein occupancy of TFAM per DNA molecule was plotted for each concentration (Figure 2A). Positive cooperativity (n > 1) was indicated by Hill plots calculated using either nonlinear (Figure 2B; n ∼ 2 for all data points) or linear (data not shown; n ∼ 1.9, excluding highest and lowest concentrations) regression analysis.
Analysis of Three-Dimensional DNA Compaction Shows a Linear Relationship to Protein Concentration
The AFM images from the TFAM titrations provide information about the physical intermediates of the compaction process, which prompted us to determine which measurements best described the DNA compaction state. Recently, the worm-chain–like behavior of DNA has been evaluated from AFM measurements by examining the binding of Abf2p, the yeast homologue of TFAM, to a plasmid substrate (Friddle et al., 2004). In that study, a two-dimensional analysis of compaction was calculated by measuring end-to-end distances. Because DNA at low concentrations behaves like a piece of string, with end-to-end distances varying widely, we elected to use the spreading of DNA on the substrate to describe compaction in two dimensions. We calculated the spreading of DNA (coefficient of 2-D compaction) by dividing the area occupied by a DNA molecule by the area of the best-fit ellipse drawn around that molecule (example shown in Figure 2G), which describes how densely the DNA is packed. We observed an asymptote for planar compaction at 25 μg/ml TFAM (Figure 2C). Because we observed further protein loading onto DNA at higher TFAM concentrations, we compared the structure height relative to its spreading to include all three dimensions in the representation of compaction (coefficient of 3-D compaction). To do this, we calculated the maximum height of a structure divided by the length of the long axis of the best-fit ellipse drawn around that object and observed a linear progression of compaction in response to increasing protein concentration by (Figure 2D). Combining these two estimates of compaction suggests that the binding of TFAM initially decreases the spreading of DNA then subsequently loads onto available sites, which increases the rigidity (height) of the structures on the surface. When we consider these characteristics in solution, we conclude that DNA length rapidly decreases early in the process of compaction and the additional TFAM loaded onto proximal sites creates a dense, globular core.
TFAM Binds DNA Cooperatively and with Nanomolar Affinity in Surface Plasmon Resonance Experiments
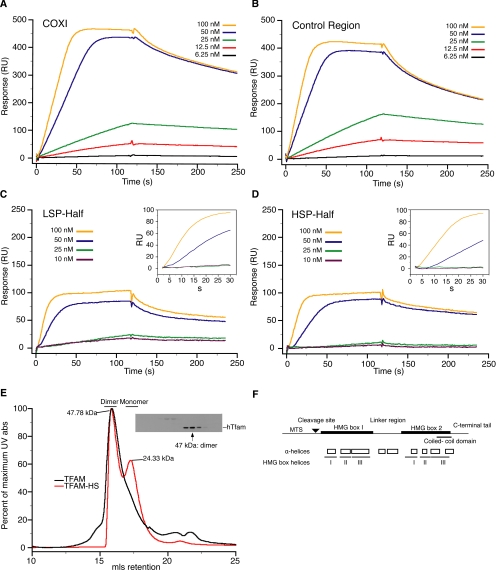
We next used SPR to quantitate the binding kinetics of TFAM for representative mouse mtDNA sequences. Sensorgram profiles monitored specific changes in binding response (RU) as a function of time, and a greater-than-additive binding response to protein concentration was observed for a 99-base pair fragment of the COXI sequence (Figure 3A), especially at TFAM concentrations at 50 nM and above. Titrations using immobilized control region sequence (Figure 3B), higher flow rates, or different TFAM concentrations produced similar results (data not shown). At a low protein concentration, we also observed the described preferential affinity of TFAM (Gaspari et al., 2004) with the 45-base pair sequence immediately upstream of the light strand promoter (Figure 3C, LSP-Half) compared with that of the heavy strand promoter (Figure 3D, HSP-Half) due to slower dissociation. The differences in binding affinities between these two sequences (10-fold) are consistent with differences between sequence-specific and nonspecific binding of other HMG box proteins (10–50-fold) (Steitz, 1990). This sequence specificity is diminished as protein concentration increases.
Figure 3.
Analysis of TFAM-DNA association and TFAM oligomerization state by surface plasmon resonance and size exclusion chromatography. (A and B) SPR sensorgrams for TFAM association and dissociation with immobilized 99-base pair COXI sequence and control region sequence, respectively. (C and D) SPR sensorgrams for the 45-base pair LSP-Half and HSP-Half of control region DNA, respectively. Insets, expanded view of the first 30 s of association phase. The concentration of TFAM is indicated in the legend of each graph. (E) Size exclusion analysis of recombinant TFAM preparations and mitochondrial extracts. UV absorption chromatographs of isolated TFAM and TFAM-HS are shown as the percentage of maximum absorbance (abs) and milliliters retention (mls). Inset, Western blot analysis of even fractions of a mitochondrial extract isolated from HEK293 cells. (F) Schematic representation of mouse TFAM protein. Shown are the major domains, including mitochondrial targeting sequence (MTS), MTS cleavage site, DNA binding domains (HMG box 1 and 2), linker region between HMG boxes, predicted coiled-coil domain, and the C-terminal extension. Also shown are predicted α-helices (Chou and Fasman, 1974) and the HMG box helices inferred from sequence alignment with the crystal structure of HMG-D (Murphy et al., 1999; Matsushima et al., 2003).
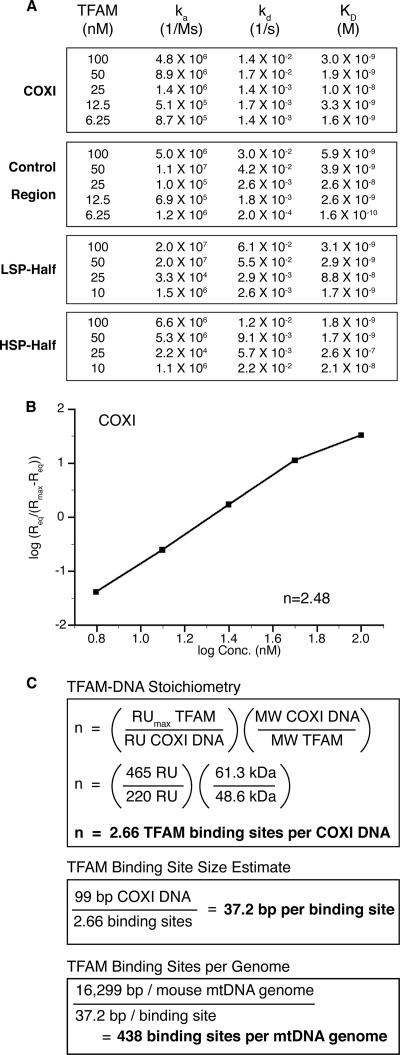
The SPR data sets were initially subjected to global analysis by using the 1:1 binding model, which assumes that the association (ka) and dissociation (kd) rate constants are equivalent for all protein concentrations; however, the model did not fit the data well. A subsequent plot of the observed rate constants (kobs) as a function of concentration was not linear (data not shown), indicating that the ka of TFAM binding to DNA is not constant. Similar issues were encountered for measurements of cooperative interactions of GroEL (Fridmann et al., 2002) and the CD40 ligand (Wieckowski et al., 2007). We therefore analyzed our data locally using a 1:1 binding model, which generates independent association and dissociation rate constants for each protein concentration (Figure 4A). Using the predicted kinetic constants for the TFAM–COXI DNA interactions (i.e., sequence nonspecific), we determined an average affinity (KD) of ∼4 × 10−9 M (Figure 4A). In contrast to TFAM, the yeast homologue Abf2p has an estimated DNA binding affinity of only 0.4 × 10−6 M by EMSA (Cho et al., 2001) or 1.4 × 10−6 M by AFM and circular dichroism (Friddle et al., 2004).
Figure 4.
Predicted kinetic, cooperative, and stoichiometric characteristics of TFAM by SPR. (A) Kinetic analysis of TFAM binding to DNA. DNA ligands are indicated to the left of each box. Data were fitted locally using 1:1 binding model with mass transport correction. (B) Hill plot derived from kinetic parameters of TFAM binding to COXI sequence, yielding a positive Hill constant of 2.48 for TFAM. (C) TFAM–DNA stoichiometry, binding site size estimate and binding sites per genome calculations.
The BIAevaluation software also predicted binding responses at equilibrium (Req) for each TFAM concentration, which was then used to determine the fraction of binding sites occupied at each protein concentration relative to the maximal binding response (Rmax) (Trempe et al., 2005; Majka and Speck, 2007). We used this data to generate a Hill plot, setting the maximal occupancy to the Req value of 100 nM TFAM binding on the COXI substrate (Figure 4B). The calculated Hill constant of ∼2.5 (Figure 4B) from the SPR data is consistent with the Hill constant of ∼2 determined from the analysis of the AFM images (Figure 2).
TFAM Functions In Vivo and In Vitro as a Homodimer
To calculate the binding stoichiometry at saturation from the SPR data, we determined the oligomerization state of both recombinant and endogenous TFAM by size exclusion chromatography (Figure 3E). Using the ionic detergent deoxycholate, which has a low micelle number, endogenous TFAM is a dimer in isolated mitochondria from human embryonic kidney (HEK)293 cells (Figure 3E, inset) as well as from human heart and liver and mouse liver and kidney (data not shown). Higher resolution estimates of protein size can be achieved by analyzing the UV absorption tracing of recombinant mature TFAM. The calculated size of the complex (Figure 3E, black line) from the elution profile matches well with the predicted size of a globular dimer (calibration R2 = 0.97). When the recombinant protein is purified in high salt buffers (500 mM NaCl), a new population is observed at the molecular weight of the monomer (Figure 3E, red line). TFAM dimerization is likely mediated through the predicted coiled-coil domain that resides within the second HMG box (Figure 3F). These results differ from the original estimates by glycerol gradient sedimentation analysis that indicated that TFAM was a monomer (Fisher and Clayton, 1988); however, the significant loss of activity and protein in those gradients suggest the protein was unstable.
Using the SPR data for the COXI ligand, we determined a periodicity of binding of 37.2 base pairs per TFAM dimer (Figure 4C) (Majka and Speck, 2007). This estimate is in close agreement with an in vivo measurement of 17.6 base pairs/TFAM monomer (35.2 base pairs/TFAM dimer), which is based on cosedimentation of TFAM with mtDNA from HeLa cells (Takamatsu et al., 2002). Our data are also corroborated by DNase I sensitivity experiments conducted in organello and in vitro, which detected TFAM footprints of 35 and 36–45 base pairs, respectively (Ghivizzani et al., 1994; Dairaghi et al., 1995). Using our binding site size estimate, we calculate that human mtDNA has up to 445 sites for TFAM dimer binding, which is close to the 470 (Alam et al., 2003) or 450 (Takamatsu et al., 2002) TFAM dimers that associate with mtDNA by cosedimentation. Mouse mtDNA would have a similar number of binding sites per genome, at 438. We conclude that the nonspecific DNA binding characteristic of recombinant TFAM is essentially identical to that of the endogenous protein, and that TFAM binds mtDNA approximately every 35 base pairs as a dimer, independent of sequence; thus, it is the principal contributor to mtDNA organization.
Noncontact Atomic Force Microscopy Reveals Structural Details of TFAM-mediated DNA Compaction Intermediates at Low Nanometer Resolution
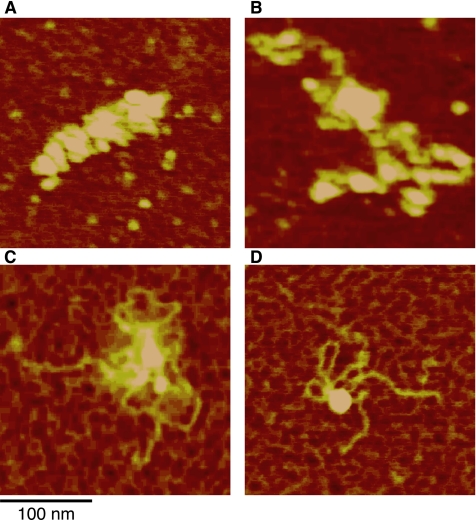
While compiling the AFM height scans, we observed flower-like structures containing loops of DNA exterior to a densely packed center, which were often present in structures with low protein occupancy and compaction (Figure 5). The fine details of the process of forming these structures and promoting compaction, however, were not clear from these height images. To reduce the effects of humidity in the air that occurs in conventional AFM, and thus achieve higher detail images of these structures, we examined the low-protein occupancy DNA molecules by using NC-AFM under high vacuum conditions (Figure 6). NC-AFM provides higher contrast images and reveals more detailed structures of single and double-stranded DNAs than tapping mode AFM (Maeda et al., 1999). In our experiments, NC-AFM height scans of naked DNA revealed the periodicity of the bead structure of polylysine underlying the nucleic acid, but the DNA remained relatively flat unless it crossed over another strand (Figure 6). Thus, protein–DNA associations were clearly evident in a representative NC-AFM scan of DNA with low TFAM occupancy in which we achieved near cryo-electron microscopy (EM) resolution (Figure 6, B and C). By measuring simple binding events, such as those in the top row of Figure 6D, we observed that TFAM binding to DNA induces a bend of 100 ± 20° over a short distance (e.g., Figure 6D, step 1) (both mean and median, n = 25). This measurement differs from the 78° observed for Abf2p by tapping mode AFM (Friddle et al., 2004), although this difference could be attributed to the resolution achieved by NC-AFM. Of particular interest in this image is the protein at the base of two individual omega-shaped (Ω) loop structures (Figure 6B, arrows). The DNA bend angle on both sides of the base of the omega fall within our measured range, suggesting that one binding site does not influence the bend angle of the other. We selected 21 DNA structures at early stages of compaction from the NC-AFM data for estimating protein and loop formation, and we found, on average, 10 ± 3 protein molecules on each, with a range of 6–15, with qualitatively similar levels of compaction. DNA loops contained one to four proteins, one protein being a molecule at the base of the loop. Most loops are small, and four proteins would cover two thirds of a loop, which suggests that a loop is at least 210 base pairs. These data indicate both that TFAM directly mediates loop formation, which decreases the spreading of DNA molecules, and that loops form early in the process of compaction.
Figure 5.
Bioscope atomic force microscopy analysis of TFAM–DNA with decreasing protein occupancy. (A–D) An example titration of TFAM bound on DNA reveals loop structures at low protein occupancy. All images are 250-nm scans. Bar, 100 nm.
Figure 6.
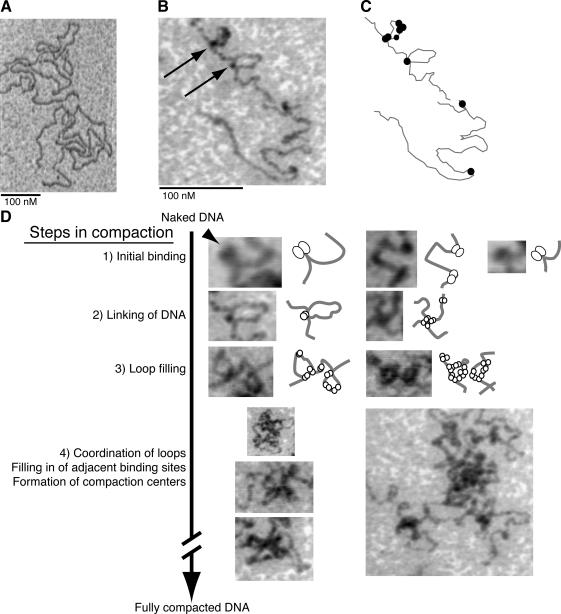
Ultrahigh vacuum noncontact atomic force microscopy of DNA compaction intermediates. (A) NC-AFM scan of naked DNA. (B) NC-AFM scans show TFAM at low occupancy induces the formation of loop structures on DNA. Arrows indicate TFAM protein at the base of two omega (Ω) structures. Steps of compaction are numbered for each row. (C) Outline of protein–DNA complexes observed in B. (D) Model for DNA compaction. The process of compaction progresses from initial binding to fully compacted DNA with numerous stages of intermediate organization between. The arrowhead indicates a possible cleft in a TFAM dimer. Bars (A and B), 100 nm. The raw data are contrast inverted.
The early events in compaction are represented by several NC-AFM scans presented in order of complexity of structure (Figure 6D). The simplest structures occur when TFAM first binds and bends DNA, reminiscent of a light bulb on a string of lights (Figure 6D, step 1). Some protein bound to DNA seems to have a cleft, which is consistent with TFAM binding to DNA as a dimer (Figure 6D, arrowhead). The TFAM dimer can then capture another DNA site, be it on the same or different DNA molecule (i.e., in cis or trans), to form a loop or X-structure, respectively (Figure 6D, step 2). Additional TFAM binds DNA proximal to the initial binding site, which is observed as loop filling or expansion along linear DNA tracts (Figure 6D, step 3). The filled loops are coordinated and organized around a central structure, upon which more protein begins to fill nearby sites (Figure 6D, step 4). In these later stages, we frequently observe multiple DNA molecules coordinated and linked at a single nexus. Loop structures were not described in AFM studies of Abf2p (Friddle et al., 2004) or in EM analysis of xl-TFAM (Antoshechkin et al., 1997).
TFAM Coordinates Multiple DNAs into Single Nucleoid-like Structures
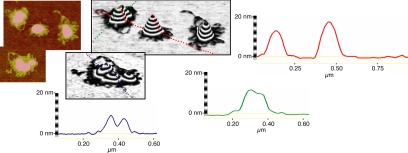
Because the measured amount of TFAM associated with DNA in vivo (Takamatsu et al., 2002) is equivalent to stoichiometry measured at saturation by our in vitro SPR measurements, we focused on the structures observed in tapping mode AFM images at a protein concentration (100 μg/ml) where TFAM binding of DNA approaches saturation (Figure 7). Although the amount of TFAM protein in this reaction is in excess of the predicted number of binding sites, we only observed near saturating amounts of protein loading (Figure 2A) at these volumes when using a buffer compatible with AFM, possibly decreasing the relative binding efficacy of TFAM to DNA. Although height images contain some compacted, presumably single DNA molecules (e.g., Figure 1F), we observed that most structures are significantly larger and contain multiple DNAs with thick, protein-coated loops on the periphery (Figure 7, left). The topology of these structures can be represented as a 3-D projection (Figure 7, center) and as 2-D cross sections (Figure 7, right). These highly organized structures are well compacted and thus quite tall (up to 24.7 nm in height), and they are close to the diameter measured for nucleoids in vivo by fluorescence and electron microscopy (Iborra et al., 2004). We can clearly visualize multiple structures merging, and asymmetric structures that have merged completely (Figure 7, bottom left image).
Figure 7.
Analysis of atomic force microscopy images of higher-order nucleoprotein structures. The left two panels are height images of TFAM–DNA structures at 100 μg/ml protein. The center two panels are 3-D projections of AFM images. Each band in the Z-plane is 2 nm. The right three panels are cross-section traces of the 3-D projections as indicated by color.
DISCUSSION
This study demonstrates that TFAM has the capacity to be the principal organizer of the mtDNA nucleoid, the formation of which requires the organization and compaction of multiple DNAs into a single spheroid structure. We have qualitatively and quantitatively described in detail the process of TFAM-promoted compaction, and we have shown that linear progression of compaction is coincident with the cooperative loading of TFAM on DNA, thereby suggesting that compaction is driven by the nanomolar affinity of TFAM-DNA interactions. We find that TFAM alone is sufficient to coordinate the compaction of DNAs together, with linkages occurring at both low and high protein concentrations. Moreover, TFAM completes the coordinated compaction of DNA in vitro at the same TFAM occupancy observed in vivo (Takamatsu et al., 2002).
Process of Compaction and Coordination of DNAs
The process of TFAM-mediated DNA compaction is seeded by the binding of the TFAM dimer and bending of DNA. Loop or X structures can then form, and multiple DNA molecules may become linked. As additional TFAM loads onto proximal sites, the contour length of the DNA rapidly decreases. This second site bias is observed in the distribution of TFAM on DNA molecules in NC-AFM images, and it is also represented in the cooperative binding characteristics of TFAM as detected by AFM, EMSA, and SPR analysis. Continued loading of TFAM onto the DNA promotes the formation of a globular center, at which DNA–TFAM complexes are densely packed. These compacted structures can merge in solution.
We show that TFAM mediates two types of linkages between DNAs: the formation of X structures at an early point in the compaction process (Figure 6D, step 2, center) and the joining of compacted DNAs (Figure 7). We postulate that the X structures, as well as Ω or loop structures, could occur during replication as nascent nucleotides are exposed, whereas the fusion of compacted molecules might occur as part of nucleoid dynamics (e.g., figure 6 in Garrido et al., 2003). The latter event could be explained by the availability of HMG-boxes in DNA-bound TFAM dimers, and it is supported by the observation that a small percentage of TFAM dimers are fully engaged by loop and X structures (Figure 6, step 4). Together, we do not consider the observed DNA binding cooperativity as a direct predictor of the formation of higher order structures (multiple DNAs associated together); rather, the availability of free HMG-boxes is likely responsible for this observation.
The formation of loop structures rapidly decreases DNA spreading, which may constitute the primary mechanism for DNA compaction, but do loop structures exist in vivo? Similar structures have been observed in nucleoids enriched from rat liver mitochondria (Van Tuyle and McPherson, 1979). In that study, rapidly sedimenting mtDNAs were spread with formamide and visualized using electron microscopy techniques to reveal single genome-length rosettes that contained a dense core (e.g., figure 7 in Van Tuyle and McPherson, 1979). On treatment with SDS and phenol, the same samples retained neither a rosette structure nor the dense core (e.g., figure 8 in Van Tuyle and McPherson, 1979), suggesting that the loop structures were maintained by protein. The authors also observed rosette structures that were consistent with two genome lengths. Although this method was suitable for electron microscopy, the use of detergent and the complexity of isolated material unfortunately render this preparation incompatible with AFM. The strong similarities between the mtDNA rosette structures isolated from rat liver mitochondria and the flower-like structures frequently observed in our AFM images (e.g., Figures 5D and 7) are striking, prompting the speculation that TFAM could form these structures in vivo.
Are these structures unique to TFAM-DNA interactions? Many members of the HMG1/2 family have been shown to affect DNA structure through supercoiling, bending, and looping, and they may recognize altered structures such as four-way junctions and “prebent” DNAs (for review, see Churchill et al., 1999). Some characteristics related to loop formation have been described in vitro for other HMG box proteins, such as DNA inversion by NHP6A and HMG3 (Stros et al., 1994; Paull and Johnson, 1995) and the circularization of short DNA substrates by HMG1 (Paull et al., 1993). These abilities may be common to many HMG proteins, or to subsets of these proteins that contain multiple HMG box domains or that can form dimers, tetramers, and so on. Importantly, most of these proteins function in an in vivo context of nucleosome-loaded DNA and other high-affinity dsDNA binding proteins, which likely limit the influence of HMG box proteins on localized loop structures. This situation contrasts starkly with that of mammalian mitochondria, where TFAM may not be competing with other dsDNA-organizing proteins, such as histones, and, at least in some cell types, TFAM may occupy mtDNA maximally.
Nanomolar Affinity of TFAM–DNA Interactions
We calculated an average affinity for nonspecific mouse TFAM–DNA interactions of ∼4 × 10−9 M. A recent study has determined an eightfold lower KD for TFAM-DNA interactions by SPR (Ohgaki et al., 2007), similar to what we observed for 6.25 nM TFAM on control region sequence. The details of the data fitting in that study are unclear, including whether or not the effects of mass transport were taken into account (Myszka et al., 1998). Furthermore, the increase in RU seen during the dissociation phase suggests some nonspecific binding to the reference flow cell, which would explain the slower off-rate and lower the overall KD. The apparent discrepancy may also be due to their use of an N-terminal six-histidine tag or the difference between human and mouse TFAM affinities for DNA.
The three orders of magnitude difference in affinity for DNA between TFAM and Abf2p may represent distinct differences in their function. Although both proteins have two HMG box domains, TFAM has a C-terminal 29 residue extension that is required for mitochondrial transcription activation in vitro (Dairaghi et al., 1995) and in vivo (Kanki et al., 2004), and it may contribute to the higher affinity for DNA observed for TFAM (Dairaghi et al., 1995; Ohgaki et al., 2007). Both mammalian TFAM and Abf2p contain coiled-coil domains, although in different HMG box domains. Studies of Abf2p association with DNA using AFM and other physical methods conclude that the protein binds DNA weakly (Friddle et al., 2004) due to a fast off-rate and small stabilizing forces (Brewer et al., 2003). Moreover, both of those studies suggest that compaction occurs as a product of a single type of protein–DNA association, without the formation of X or loop structures, consistent with DNA bending mediating the reduction of the contour length of the genome. It has been suggested that the micromolar affinity of Abf2p for DNA would pack DNA loosely, simplifying access to genome by regulatory proteins (Friddle et al., 2004). This notion is consistent with the suggestion that yeast have co-opted other mitochondrial proteins during evolution to contribute to the stabilization of mtDNA (Kaufman et al., 2000; Chen et al., 2005).
Cooperative loading of TFAM on DNA
The ability of TFAM to bind DNA cooperatively is supported by evidence presented here in three different types of experiments. By EMSA, the electrophoretic mobility of the DNA ligand is shifted more than expected for twofold differences in protein concentration (e.g., 3.5 and 7 μg/ml). In AFM images, we observe the persistence of DNA with low protein occupancy next to highly compacted, highly protein-loaded molecules (e.g., Figure 2E). These differences are not explained by protein clumping, because individual TFAM seeding is clearly visualized in the red–yellow colorization of uncompacted TFAM-bound DNA (Figure 2F, inset). The quantification of TFAM-bound DNA in the AFM images generated a sigmoidal curve, indicating that after initial TFAM seeding, protein loading occurs exponentially until saturation is reached. In contrast weak positive cooperativity (Hill coefficient ∼1.35) was also observed for Abf2p by using AFM (Friddle et al., 2004). In SPR experiments, the use of 99-base pair DNA ligands provides multiple TFAM binding sites and reveals positive cooperativity through greater than linear responses to protein concentration, which occurs strongly at and above 50 μM TFAM.
How might TFAM binding be cooperative? Cooperativity indicates that the first binding event influences the kinetics of the second binding event. In positive cooperativity, the effective KD will decrease as protein occupancy on the substrate increases. This higher affinity might be due to interactions between proteins on the DNA, or to changes in proximal DNA structure, such as the local distortion of DNA conformation that would occur adjacent to the kinks observed in TFAM-bound DNA. Although we have observed that TFAM binding and the subsequent formation of loop or X structures creates an obvious bias in second site selection (Figure 6D, bottom two rows), we cannot distinguish between these two modes of cooperativity here.
TFAM Binding DNA Topology
We think that TFAM associates with DNA immediately after the replication or transcription machinery has passed, when the DNA is topologically relaxed. mtDNA has not been observed as a reticular or decondensed structure in cells, indicating that it is largely compacted even during replication and transcription; thus, we surmise that long stretches of DNA do not remain protein free; rather, dimers from the free pool of TFAM would associate as the polymerase exposed DNA. Carefully isolated mtDNA contains mostly relaxed (nicked) circles and some linear molecules, with a limited amount of the total being supercoiled (Pohjoismaki et al., 2006; Kaufman, unpublished data); thus, we expect that the topology of DNA encountered by TFAM in the mitochondria to be relaxed. The use of linearized DNA maintains a relaxed topology when TFAM binds and prevents the compaction of molecules due to protein induced supercoiling.
Moreover, if topological constraints are a factor for TFAM binding in vivo (i.e., proteins have reduced access to mtDNA), then the observed binding stoichiometry in vivo would be lower than what we measure in vitro (in the absence of topological constraints) (Figure 4C). One report conflicts with those cited earlier, and it has suggested that the stoichiometry between TFAM and mtDNA in HEK cells is ∼35 molecules of TFAM per mtDNA (Maniura-Weber et al., 2004), which could be explained by variation among cell types or methodologies used for such a determination. Although our data are in agreement with measurements made in HeLa cells, we cannot exclude that there is a range of TFAM–DNA ratios in different cell and tissue types, which possibly involve differences in DNA topology. Consistent with the proposal that TFAM is abundantly associated with mtDNA is the evidence that TFAM is distributed across the genome by chromatin immunoprecipitation analysis, rather than localized to the control region of transcription (Ohgaki et al., 2007). Finally, in light of the recent discussion in the literature suggesting that oxidation may not be the major source mtDNA-encoded errors (Kujoth et al., 2005), we expect the mitochondrial genome to be fully insulated from its environment in tissues.
The in vivo manipulation of TFAM levels, through overexpression or knockdown, shows a co-dependence of TFAM protein levels and mtDNA copy number. In the TFAM heterozygous knockout mouse, the amount of mtDNA goes down by 35–40% (Larsson et al., 1998), whereas stable overexpression of a single copy of human TFAM in mouse causes an increase in mtDNA of ∼50% in several mouse tissues (Ekstrand et al., 2004). High levels of inducible TFAM overexpression, however, result in mtDNA depletion in cell lines (HEK293) (Pohjoismaki et al., 2006). Our data would suggest that TFAM is required for the stability of mtDNA through its compaction and organization of the nucleoid, whereas high levels of overexpression could drive the formation of complex structures and suppress accessibility of mtDNA to replication and transcription complexes. Consistent with this interpretation, inducing high levels of TFAM was shown to increase the amount of supercoiled mtDNA and cause the formation of replication intermediates similar to those observed during replication stalling caused by dideoxy cytosine treatment (Pohjoismaki et al., 2006). Thus, although the titration model for TFAM and mtDNA seems true to a first approximation, very high levels of TFAM may change the TFAM:DNA ratio, causing aberrant compaction and transmission.
In conclusion, our data provide evidence that TFAM binds mtDNA with an affinity compatible with the idea of the protein coating the genome, shows a strong preference for DNA with protein already bound, and is sufficient to compact and assemble DNAs into multigenomic nucleoid-like structures. How nucleoid dynamics may be regulated, for example during nucleoid division, is not understood. We speculate that mitochondrial chromatin dynamics require the regulation of the high-affinity association of TFAM with DNA to promote mtDNA transcription or replication, similar to those mechanisms involved in nuclear genome dynamics.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Kolesar, K. Hastings, and S. Leary for critical reading of this manuscript. This work was funded by grants from the Canadian Institutes of Health Research (CIHR), Natural Sciences and Engineering Research Council of Canada, Canadian Institute of Advanced Research, and Fonds Québécois de Recherche sur la Nature et les Technologies. B.A.K. holds a postdoctoral fellowship from Muscular Dystrophy Canada/CIHR. The Sheldon Biotechnology Centre at McGill University is supported by a Research Resource Grant from CIHR. N.D. and S.C. are supported by the Neurophysics-CIHR Strategic Training Program grant. E.A.S. is an International Scholar of the Howard Hughes Medical Institute and a Senior Scientist of the CIHR.
Abbreviations used:
- AFM
atomic force microscopy
- EMSA
electrophoretic mobility shift assay
- mtDNA
mitochondrial DNA
- NC-AFM
noncontact atomic force microscopy
- SPR
surface plasmon resonance.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-05-0404) on June 20, 2007.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Alam T. I., Kanki T., Muta T., Ukaji K., Abe Y., Nakayama H., Takio K., Hamasaki N., Kang D. Human mitochondrial DNA is packaged with TFAM. Nucleic Acids Res. 2003;31:1640–1645. doi: 10.1093/nar/gkg251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht T. R., Grutter P., Horne D., Rugar D. Frequency modulation detection using high-Q cantilevers for enhanced force microscope sensitivity. J. Appl. Phys. 1991;69:668–673. [Google Scholar]
- Antoshechkin I., Bogenhagen D. F., Mastrangelo I. A. The HMG-box mitochondrial transcription factor xl-mtTFA binds DNA as a tetramer to activate bidirectional transcription. EMBO J. 1997;16:3198–3206. doi: 10.1093/emboj/16.11.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battersby B. J., Loredo-Osti J. C., Shoubridge E. A. Nuclear genetic control of mitochondrial DNA segregation. Nat. Genet. 2003;33:183–186. doi: 10.1038/ng1073. [DOI] [PubMed] [Google Scholar]
- Brewer L. R., Friddle R., Noy A., Baldwin E., Martin S. S., Corzett M., Balhorn R., Baskin R. J. Packaging of single DNA molecules by the yeast mitochondrial protein Abf2p. Biophys. J. 2003;85:2519–2524. doi: 10.1016/s0006-3495(03)74674-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. J., Wang X., Kaufman B. A., Butow R. A. Aconitase couples metabolic regulation to mitochondrial DNA maintenance. Science. 2005;307:714–717. doi: 10.1126/science.1106391. [DOI] [PubMed] [Google Scholar]
- Cho J. H., Lee Y. K., Chae C. B. The modulation of the biological activities of mitochondrial histone Abf2p by yeast PKA and its possible role in the regulation of mitochondrial DNA content during glucose repression. Biochim. Biophys. Acta. 2001;1522:175–186. doi: 10.1016/s0167-4781(01)00333-5. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Conformational parameters for amino acids in helical, beta-sheet, and random coil regions calculated from proteins. Biochemistry. 1974;13:211–222. doi: 10.1021/bi00699a001. [DOI] [PubMed] [Google Scholar]
- Churchill M. E., Changela A., Dow L. K., Krieg A. J. Interactions of high mobility group box proteins with DNA and chromatin. Methods Enzymol. 1999;304:99–133. doi: 10.1016/s0076-6879(99)04009-4. [DOI] [PubMed] [Google Scholar]
- Dairaghi D. J., Shadel G. S., Clayton D. A. Addition of a 29 residue carboxyl-terminal tail converts a simple HMG box-containing protein into a transcriptional activator. J. Mol. Biol. 1995;249:11–28. doi: 10.1006/jmbi.1995.9889. [DOI] [PubMed] [Google Scholar]
- DiMauro S., Schon E. A. Mitochondrial respiratory-chain diseases. N. Engl. J. Med. 2003;348:2656–2668. doi: 10.1056/NEJMra022567. [DOI] [PubMed] [Google Scholar]
- Ekstrand M. I., Falkenberg M., Rantanen A., Park C. B., Gaspari M., Hultenby K., Rustin P., Gustafsson C. M., Larsson N. G. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum. Mol. Genet. 2004;13:935–944. doi: 10.1093/hmg/ddh109. [DOI] [PubMed] [Google Scholar]
- Fisher R. P., Clayton D. A. A transcription factor required for promoter recognition by human mitochondrial RNA polymerase. Accurate initiation at the heavy- and light-strand promoters dissected and reconstituted in vitro. J. Biol. Chem. 1985;260:11330–11338. [PubMed] [Google Scholar]
- Fisher R. P., Clayton D. A. Purification and characterization of human mitochondrial transcription factor 1. Mol. Cell. Biol. 1988;8:3496–3509. doi: 10.1128/mcb.8.8.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. P., Lisowsky T., Parisi M. A., Clayton D. A. DNA wrapping and bending by a mitochondrial high mobility group-like transcriptional activator protein. J. Biol. Chem. 1992;267:3358–3367. [PubMed] [Google Scholar]
- Friddle R. W., Klare J. E., Martin S. S., Corzett M., Balhorn R., Baldwin E. P., Baskin R. J., Noy A. Mechanism of DNA compaction by yeast mitochondrial protein Abf2p. Biophys. J. 2004;86:1632–1639. doi: 10.1016/S0006-3495(04)74231-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridmann Y., Kafri G., Danziger O., Horovitz A. Dissociation of the GroEL-GroES asymmetric complex is accelerated by increased cooperativity in ATP binding to the GroEL ring distal to GroES. Biochemistry. 2002;41:5938–5944. doi: 10.1021/bi020117v. [DOI] [PubMed] [Google Scholar]
- Garrido N., Griparic L., Jokitalo E., Wartiovaara J., van der Bliek A. M., Spelbrink J. N. Composition and dynamics of human mitochondrial nucleoids. Mol. Biol. Cell. 2003;14:1583–1596. doi: 10.1091/mbc.E02-07-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspari M., Falkenberg M., Larsson N. G., Gustafsson C. M. The mitochondrial RNA polymerase contributes critically to promoter specificity in mammalian cells. EMBO J. 2004;23:4606–4614. doi: 10.1038/sj.emboj.7600465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghivizzani S. C., Madsen C. S., Nelen M. R., Ammini C. V., Hauswirth W. W. In organello footprint analysis of human mitochondrial DNA: human mitochondrial transcription factor A interactions at the origin of replication. Mol. Cell. Biol. 1994;14:7717–7730. doi: 10.1128/mcb.14.12.7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., et al. The AAA+ protein ATAD3 has displacement loop binding properties and is involved in mitochondrial nucleoid organization. J. Cell Biol. 2007;176:141–146. doi: 10.1083/jcb.200609158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iborra F. J., Kimura H., Cook P. R. The functional organization of mitochondrial genomes in human cells. BMC Biol. 2004;2:9. doi: 10.1186/1741-7007-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuth J. P., Peterson A. C., Shoubridge E. A. Tissue-specific selection for different mtDNA genotypes in heteroplasmic mice. Nat. Genet. 1997;16:93–95. doi: 10.1038/ng0597-93. [DOI] [PubMed] [Google Scholar]
- Kanki T., Ohgaki K., Gaspari M., Gustafsson C. M., Fukuoh A., Sasaki N., Hamasaki N., Kang D. Architectural role of mitochondrial transcription factor A in maintenance of human mitochondrial DNA. Mol. Cell. Biol. 2004;24:9823–9834. doi: 10.1128/MCB.24.22.9823-9834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman B. A., Newman S. M., Hallberg R. L., Slaughter C. A., Perlman P. S., Butow R. A. In organello formaldehyde crosslinking of proteins to mtDNA: identification of bifunctional proteins. Proc. Natl. Acad. Sci. USA. 2000;97:7772–7777. doi: 10.1073/pnas.140063197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujoth G. C., et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- Larsson N. G., Wang J., Wilhelmsson H., Oldfors A., Rustin P., Lewandoski M., Barsh G. S., Clayton D. A. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat. Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- Legros F., Malka F., Frachon P., Lombes A., Rojo M. Organization and dynamics of human mitochondrial DNA. J. Cell Sci. 2004;117:2653–2662. doi: 10.1242/jcs.01134. [DOI] [PubMed] [Google Scholar]
- Liu T., Lu B., Lee I., Ondrovicova G., Kutejova E., Suzuki C. K. DNA and RNA binding by the mitochondrial lon protease is regulated by nucleotide and protein substrate. J. Biol. Chem. 2004;279:13902–13910. doi: 10.1074/jbc.M309642200. [DOI] [PubMed] [Google Scholar]
- Lu B., Liu T., Crosby J. A., Thomas-Wohlever J., Lee I., Suzuki C. K. The ATP-dependent Lon protease of Mus musculus is a DNA-binding protein that is functionally conserved between yeast and mammals. Gene. 2003;306:45–55. doi: 10.1016/s0378-1119(03)00403-7. [DOI] [PubMed] [Google Scholar]
- Lu B., et al. Roles for the human ATP-dependent Lon protease in mitochondrial DNA maintenance. J. Biol. Chem. 2007;282:17363–17374. doi: 10.1074/jbc.M611540200. [DOI] [PubMed] [Google Scholar]
- Maeda Y., Matsumoto T., Kawai T. Observation of single- and double-stranded DNA using non-contact atomic force microscopy. Appl. Surf. Sci. 1999;140:400–405. [Google Scholar]
- Majka J., Speck C. Analysis of protein-DNA interactions using surface plasmon resonance. Adv. Biochem. Eng. Biotechnol. 2007;104:13–36. [PubMed] [Google Scholar]
- Maniura-Weber K., Goffart S., Garstka H. L., Montoya J., Wiesner R. J. Transient overexpression of mitochondrial transcription factor A (TFAM) is sufficient to stimulate mitochondrial DNA transcription, but not sufficient to increase mtDNA copy number in cultured cells. Nucleic Acids Res. 2004;32:6015–6027. doi: 10.1093/nar/gkh921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima Y., Matsumura K., Ishii S., Inagaki H., Suzuki T., Matsuda Y., Beck K., Kitagawa Y. Functional domains of chicken mitochondrial transcription factor A for the maintenance of mitochondrial DNA copy number in lymphoma cell line DT40. J. Biol. Chem. 2003;278:31149–31158. doi: 10.1074/jbc.M303842200. [DOI] [PubMed] [Google Scholar]
- Morita S., Wiesendanger R., Meyer E. Noncontact Atomic Force Microscopy. Berlin, Germany: Springer; 2002. [Google Scholar]
- Murphy F., 4th, Sweet R. M., Churchill M. E. The structure of a chromosomal high mobility group protein-DNA complex reveals sequence-neutral mechanisms important for non-sequence-specific DNA recognition. EMBO J. 1999;18:6610–6618. doi: 10.1093/emboj/18.23.6610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myszka D. G. Improving biosensor analysis. J. Mol. Recognit. 1999;12:279–284. doi: 10.1002/(SICI)1099-1352(199909/10)12:5<279::AID-JMR473>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Myszka D. G., He X., Dembo M., Morton T. A., Goldstein B. Extending the range of rate constants available from BIACORE: interpreting mass transport-influenced binding data. Biophys. J. 1998;75:583–594. doi: 10.1016/S0006-3495(98)77549-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nass M. M. Mitochondrial DNA. I. Intramitochondrial distribution and structural relations of single- and double-length circular DNA. J. Mol. Biol. 1969;42:521–528. doi: 10.1016/0022-2836(69)90240-x. [DOI] [PubMed] [Google Scholar]
- Ohgaki K., Kanki T., Fukuoh A., Kurisaki H., Aoki Y., Ikeuchi M., Kim S. H., Hamasaki N., Kang D. The C-terminal tail of mitochondrial transcription factor a markedly strengthens its general binding to DNA. J. Biochem. 2007;141:201–211. doi: 10.1093/jb/mvm020. [DOI] [PubMed] [Google Scholar]
- Parisi M. A., Xu B., Clayton D. A. A human mitochondrial transcriptional activator can functionally replace a yeast mitochondrial HMG-box protein both in vivo and in vitro. Mol. Cell. Biol. 1993;13:1951–1961. doi: 10.1128/mcb.13.3.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull T. T., Haykinson M. J., Johnson R. C. The nonspecific DNA-binding and -bending proteins HMG1 and HMG2 promote the assembly of complex nucleoprotein structures. Genes Dev. 1993;7:1521–1534. doi: 10.1101/gad.7.8.1521. [DOI] [PubMed] [Google Scholar]
- Paull T. T., Johnson R. C. DNA looping by Saccharomyces cerevisiae high mobility group proteins NHP6A/B. Consequences for nucleoprotein complex assembly and chromatin condensation. J. Biol. Chem. 1995;270:8744–8754. doi: 10.1074/jbc.270.15.8744. [DOI] [PubMed] [Google Scholar]
- Pohjoismaki J. L., Wanrooij S., Hyvarinen A. K., Goffart S., Holt I. J., Spelbrink J. N., Jacobs H. T. Alterations to the expression level of mitochondrial transcription factor A, TFAM, modify the mode of mitochondrial DNA replication in cultured human cells. Nucleic Acids Res. 2006;34:5815–5828. doi: 10.1093/nar/gkl703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz T. A. Structural studies of protein-nucleic acid interaction: the sources of sequence-specific binding. Q. Rev. Biophys. 1990;23:205–280. doi: 10.1017/s0033583500005552. [DOI] [PubMed] [Google Scholar]
- Stros M., Stokrova J., Thomas J. O. DNA looping by the HMG-box domains of HMG1 and modulation of DNA binding by the acidic C-terminal domain. Nucleic Acids Res. 1994;22:1044–1051. doi: 10.1093/nar/22.6.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamatsu C., Umeda S., Ohsato T., Ohno T., Abe Y., Fukuoh A., Shinagawa H., Hamasaki N., Kang D. Regulation of mitochondrial D-loops by transcription factor A and single-stranded DNA-binding protein. EMBO Rep. 2002;3:451–456. doi: 10.1093/embo-reports/kvf099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor R. W., Turnbull D. M. Mitochondrial DNA mutations in human disease. Nat. Rev. Genet. 2005;6:389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorburn D. R. Mitochondrial disorders: prevalence, myths and advances. J. Inherit. Metab. Dis. 2004;27:349–362. doi: 10.1023/B:BOLI.0000031098.41409.55. [DOI] [PubMed] [Google Scholar]
- Trempe J. F., Brown N. R., Lowe E. D., Gordon C., Campbell I. D., Noble M. E., Endicott J. A. Mechanism of Lys48-linked polyubiquitin chain recognition by the Mud1 UBA domain. EMBO J. 2005;24:3178–3189. doi: 10.1038/sj.emboj.7600797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Tuyle G. C., McPherson M. L. A compact form of rat liver mitochondrial DNA stabilized by bound proteins. J. Biol. Chem. 1979;254:6044–6053. [PubMed] [Google Scholar]
- Wang Y., Bogenhagen D. F. Human mitochondrial DNA nucleoids are linked to protein folding machinery and metabolic enzymes at the mitochondrial inner membrane. J. Biol. Chem. 2006;281:25791–25802. doi: 10.1074/jbc.M604501200. [DOI] [PubMed] [Google Scholar]
- Wieckowski S., Trouche N., Chaloin O., Guichard G., Fournel S., Hoebeke J. Cooperativity in the interaction of synthetic CD40L mimetics with CD40 and its implication in cell signaling. Biochemistry. 2007;46:3482–3493. doi: 10.1021/bi602434a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.