Abstract
In budding yeast, the spindle position checkpoint (SPC) delays mitotic exit until the mitotic spindle moves into the neck between the mother and bud. This checkpoint works by inhibiting the mitotic exit network (MEN), a signaling cascade initiated and controlled by Tem1, a small GTPase. Tem1 is regulated by a putative guanine exchange factor, Lte1, but the function and regulation of Lte1 remains poorly understood. Here, we identify novel components of the checkpoint that operate upstream of Lte1. We present genetic evidence in agreement with existing biochemical evidence for the molecular mechanism of a pathway that links microtubule-cortex interactions with Lte1 and mitotic exit. Each component of this pathway is required for the spindle position checkpoint to delay mitotic exit until the spindle is positioned correctly.
INTRODUCTION
Chromosomes segregate into daughter cells upon cell division. To accomplish this task in budding yeast, the mitotic spindle must intersect the plane of cell cleavage, which is predetermined by the position of the mother/bud neck. The spindle position checkpoint (SPC) prevents chromosome mis-segregation by delaying mitotic exit until the spindle is correctly positioned (Muhua et al., 1998). The mitotic spindle is pulled into the neck by the combined forces of the dynein and Kar9 pathways acting on cytoplasmic microtubules (Adames and Cooper, 2000). The small GTPase Tem1 then activates the mitotic exit network (MEN), which causes spindle breakdown, degradation of mitotic cyclins, cytokinesis, and cell separation (Bardin et al., 2000). Tem1 activation can be regulated by several mechanisms, including putative GTPase-activating protein (GAP; Bfa1/Bub1) and guanine exchange factor (GEF; Lte1) proteins (see Figure 1A; Bardin et al., 2000; Molk et al., 2004; Yoshida et al., 2005) as well as localization and dynamics of Tem1 at the spindle pole body (SPB; Molk et al., 2004).
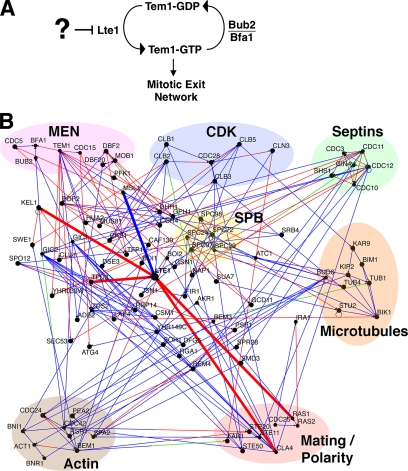
Figure 1.
Potential regulators of the mitotic exit network (MEN). (A) Schematic for regulation of the MEN. Lte1 activates Tem1, and Bub2/Bfa1 inactivates Tem1. Sensors upstream of Lte1 may inhibit its activity until correct spindle position has been achieved. (B) Diagram of physical interactions among key proteins in mitosis including the MEN, cyclin-dependent kinase (CDK), septins, the spindle pole body (SPB), the microtubule and actin cytoskeletons, and mating polarity proteins. Key proteins in this study are circled in red. Interactions of Lte1 are thick lines. Arrowheads on lines point to prey. Open black circles indicate that a protein interacts with itself. The color of the line indicates the experimental method used to determine the physical interaction; red, affinity capture; blue, yeast two-hybrid; and green, affinity chromatography. Osprey 1.2.0 and the yeast GRID database were used (see Materials and Methods).
Lte1 and Bub2/Bfa1 can be powerful controllers of Tem1. Either the overexpression of Lte1 or the loss of Bub2/Bfa1 can be sufficient to cause a complete failure of the checkpoint, with 100% of cells undergoing premature mitotic exit with the spindle in the mother (Bardin et al., 2000). In addition, Lte1 is necessary for cells to exit mitosis at cold temperature or in other conditions where mitotic exit is partially defective, such as in cdc14 early anaphase release (FEAR) network mutants (Stegmeier et al., 2002). Whether Lte1 activates Tem1 by catalyzing GTP exchange has been called into question, because the Lte1 GEF domain appears to be dispensable for Tem1 activation, although it is required for localization of Lte1 to the bud cortex (Yoshida et al., 2003).
The localization of Lte1 appears to be critical to checkpoint function. Lte1 is normally restricted to the bud, where it has been proposed to activate Tem1 that is carried into the bud on the daughter-bound SPB. The arrival of the SPB into the bud implies that the spindle has entered the neck, so this model provides an appealing mechanism for sensing spindle position and signaling mitotic exit (Bardin et al., 2000; Pereira et al., 2001). In support of this model, mis-localizing Lte1 to the mother cell, by overexpression of Lte1 or by disrupting the diffusion barrier for Lte1 at the neck, results in premature Tem1 activation and mitotic exit (Bardin et al., 2000). On the other hand, mitotic exit can occur prematurely when Lte1 localization is normal, in mutants with altered microtubule dynamics, and mitotic exit can occur at the appropriate time in the complete absence of Lte1 (Adames et al., 2001). In addition, the daughter-bound SPB, which accumulates Tem1, does not need to physically contact the bud cortex, which accumulates Lte1, in order to trigger mitotic exit (Molk et al., 2004). Thus, the precise role of Lte1 localization in the regulation of mitotic exit is unclear.
At normal temperatures, Tem1 exchanges and hydrolyzes nucleotide rapidly, which may be sufficient to drive mitotic exit without the aid of its putative GAP and GEF (Geymonat et al., 2002). The presumed functions of the GAP Bub2/Bfa1 and GEF Lte1 may be to make mitotic exit more robust at lower temperatures and to delay mitotic exit in response to a mispositioned spindle. In particular, the spindle position checkpoint appears to inhibit Tem1, and this may occur by inhibition of Lte1 or activation of Bub2/Bfa1.
Here, we investigate how Lte1 is regulated and how the Lte1-dependent pathway recognizes mis-aligned spindles to inhibit mitotic exit. Using databases of protein–protein interactions, we built a physical interaction map of proteins in the mitotic exit network and the cytoskeleton to identify proteins that interact with Lte1. Mutants lacking several of these proteins showed defects in the spindle position checkpoint, and genetic and cell biological analyses of these proteins is consistent with a linear signaling pathway connecting spindle position to the MEN via Lte1.
MATERIALS AND METHODS
Chemicals and Reagents
Chemicals were from Sigma Chemical Co. (St. Louis, MO) or Fisher Scientific (Hanover Park, IL) unless otherwise specified. Yeast medium was from Bio101 (Carlsbad, CA).
Yeast Strains and Plasmids
Yeast strains (Supplementary Table S2) were isogenic with the Research Genetics (Invitrogen, Carlsbad, CA) parental strain BY4741 (MATa his3Δ leu2Δ met15Δ ura3Δ) unless otherwise noted. Deletion strains were created using a PCR-based deletion method using pBJ1153 and pBJ1155. Strains expressing GFP-TUB1 were transformed as described with pBJ1333 (Bloom et al., 1999) or pBJ1351 (Song and Lee, 2001; for plasmids see Supplementary Table S3). Strains expressing 3GFP-LTE1 were transformed with pBJ1368 (Castillon et al., 2003).
Proteomic Map Construction
Osprey 1.2.0 and the yeast GRID database (Breitkreutz et al., 2003a,b) were used to identify protein–protein interaction networks using data from different high and low throughput-binding assays. All components of the MEN pathway, the septins, the microtubule, and the actin cytoskeletons were placed on the map in functional clusters. The program was then used to add all proteins known to physically interact with these components. Proteins that interacted with only one other protein were filtered out.
Fluorescence Microscopy
Time-lapse movies of living yeast cells were collected as described (Castillon et al., 2003). Images were acquired using QED Imaging software (Pittsburgh, PA) and analyzed using ImageJ (Wayne Rasband, NIH, http://rsb.info.nih.gov/ij/). Still images were collected on an Olympus IX70 fluorescence microscope (Melville, NY) with a 100×/1.4 NA oil immersion objective lens and a Coolsnap HQ camera (Roper Scientific, Duluth, GA). For Bud6-GFP/Tub1-CFP colocalization, green and cyan fluorescent protein (GFP and CFP, respectively) signals were observed simultaneously using a HiQ fluorescein isothiocyanate (FITC) or CFP filter cube (41001 707, Chroma, Rockingham, VT). Rhodamine-phalloidin was observed using a TRITC filter cube (U-MNC, Olympus).
Assays for the Spindle Position Checkpoint
Analysis of spindle-position checkpoint used time-lapse microscopy to observe the timing of mitotic exit in individual cells as described (Castillon et al., 2003). Cells with anaphase spindles in the mother were followed for the mean plus 2 SD of normal mitotic exit (generally 25–35 min). Spindles that broke down during that time were considered checkpoint minus. To test the temperature dependence of the checkpoint, cells were grown in liquid culture for 24 h at 12° or 30°. Where movies were not feasible, such as in the cold, we assayed checkpoint integrity by counting interphase spindle pole bodies labeled with GFP-tubulin. A cell was considered multinucleate if two or more interphase spindle pole bodies were observed in the mother cell irrespective of bud status.
Screen for Checkpoint Mutants
We screened a set of ∼150 haploid null mutants from the genome collection (Invitrogen, Carlsbad, Ca) chosen based on a connection to the mother/bud neck, such as localization of the protein, mutant phenotype, or physical or genetic interaction with a neck protein or gene. The set was begun with genes listed in a review by Gladfelter et al. (2001). The initial screen was for the presence of multinucleate cell bodies in asynchronous cultures grown at 30° in liquid YPD, assessed by fluorescence imaging of DAPI-stained cells. No multinucleate phenotype was seen for 100 mutants, including abm1, adk1, apg17, ars137, aut2, aut7, axl2, bem1, bik1, bna3, boi1, boi2, bud3, bud4, bud5, bud7, but1, cin2, cla4, clb2, cpr3, crn1, ctf4, ctf8, dbf2, ddi1, dog1, dog2, elm1, gic1, gic2, gin4, gpg1, hof1, hsl1, hsl7, ilm1, imd4, kar3, kcc4, kel1, kex2, kin1, kin2, kin3, mcr1, mlc2, msb2, msl1, nap1, nif3, nis1, pst2, rax2, rga1, rgd1, rps25a, rrd1, rvs161, sap185, sap190, sap4, she3, sho1, sic1, sit4, siz1, skm1, slt2, spt10, srl2, std1, ste20, ste23, swe1, syp1, tpd3, uba4, ubp15, ybr137w, yck2, ycr087c, ycr087w, ydr465c, ydr532c, yel015w, yfl054c, ygr058w, ygr066c, ygr112c, ygr113w, ygr153w, ygr268c, yhr033w, yhr198c, yip3, yjl218w, yjr056c, ykl056c, ykl063c, ymr071c, ymr181c, ymr196w, ymr226c, ynl101w, ynl116w, ynl187w, ynl335w, yor033w, and ypl070w. A mild phenotype was seen for 18 mutants, including bni4, bni5, bnr1, bud14, bud2, chs3, chs4, cln3, cyk3, hom2, hua2, nfi1, pyc1, vma22, ydr229w, yhr115c, ynl092w, and ypl157w. A moderate phenotype was seen for 13 mutants, including bem2, bem4, bud6, bud9, caf17, cdc10, eng1, shs1, tos10, ufd4, ybr281c, ycr076c, and zds2. A strong phenotype was observed for bub2, serving as a positive control.
Next, 25 of the mutants with phenotypes were assayed for integrity of the spindle position checkpoint by movie analysis, as described above, with GFP-Tub1 fluorescence. Those mutants and their values for checkpoint integrity, calculated as a percentage, were as follows: bem2, 100%; bem4, 70%; bni4, 100%; bni5, 92%; bnr1, 91%; bud14, 35%; bud2, 55%; bud6, 62%; cdc10, 3%; chs3, 93%; chs4, 60%; cln3, 96%; eng1, 86%; hom2, 59%; nfi1, 80%; pyc1, 100%; shs1/sep7, 49%; tos10, 90%; ybr281c, 94%; ycr076c, 89%; ydr229, 71%; yhr115c, 93%; ynl152, 93%; ypl152w, 75%; and zds2, 71%.
Fluorescence Loss in Photobleaching
To detect the possibility of Lte1 diffusion from the bud into the mother, cells expressing Lte1-3GFP at endogenous levels were subjected to fluorescence photobleaching on a wide-field inverted microscope (Olympus IX81) with an Hg lamp and an Olympus 100×/NA 1.35 UPlanApo infinity-corrected oil objective lens with its iris fully open. The fluorescence aperture was minimized, and a small-budded cell was positioned such that a portion of the mother cell opposite the bud was within the circle of illumination. Next, the aperture was opened, and a 300-ms fluorescence image of the entire field was collected, with an EM-CCD video camera (C9100, Hamamatsu, Bridgewater, NJ). The aperture was minimized, and the excitation light was turned on for 10 s at nearly full intensity. The aperture was opened and the process repeated, for 10 cycles. The fluorescence intensity in the bud after each round of photobleaching was quantified. The entire field was subject to a small amount of photobleaching during image collection, which was assessed from the fluorescence intensity of buds of cells that were near but clearly outside the aperture-photobleached area. A slight decrease in fluorescence intensity was observed during the course of the experiment, similar to the amount seen for wild-type and bud6 cells in the figure.
Assessment of DNA Damage and Morphogenesis Checkpoints
To assess the DNA damage checkpoint, liquid cultures of exponentially growing cells were diluted to an OD600 = 0.6 and plated as 10-fold serial dilutions onto a YPD plate containing 100 mM hydroxyurea. The morphogenesis checkpoint was assayed as described (McMillan et al., 1998).
RESULTS
Identification of Potential Regulators of Lte1
To identify candidate regulators of Lte1, we constructed a physical interaction map for Lte1 using the yeast GRID database and Osprey software (Breitkreutz et al., 2003a,b). Protein interactions from comprehensive two-hybrid and affinity purification screens were included. Known MEN proteins were placed on the map, followed by structural, motor, and regulatory proteins of the actin, tubulin, and septin cytoskeletons. Signaling and structural proteins known to interact with these cytoskeletal components were added, such as components of the polarisome, the SPB, and Cdc42 signaling. Proteins with only one binding partner, i.e., “dead ends,” were removed. The result of the analysis is shown in Figure 1B. Lte1 is seen to interact with Kel1, Tpd3, Cla4, Ras2, and Msl1. First, we analyzed the functional role of these proximate interactors. Later, we used the map to make more distant connections.
Function of Lte1-interacting Proteins in the Spindle Position Checkpoint
Null mutants lacking the map's proximate Lte1-interacting proteins were tested for loss of integrity of the spindle position checkpoint. Time-lapse fluorescence movies of the time course of mitosis in single living cells expressing GFP-tubulin were performed, as described (Castillon et al., 2003; Figure 2A). Live movie assays allow one to follow individual cells, rather than observe the average behavior of a synchronized population, providing definitive information as to whether and when mitotic exit occurs in cells with abnormal or delayed positioning of the mitotic spindle. To increase the fraction of cells that delay movement of the spindle into the mother-bud neck, we inactivated dynein by deleting the gene for the dynactin component Arp1. To analyze the data, an observer viewed the movie and identified all cells with late-anaphase (long) spindles in the mother. The observer followed each cell forward in time and recorded whether the cell exited mitosis and the time of mitotic exit, if it occurred. Mitotic exit was marked by breakdown of the fluorescent spindle. The spindle buckles, breaks in the middle, and then spindle microtubules disassemble. Many cells were followed through the end of and into the next cell cycle, and in every case, breakdown of the spindle was followed by a full course of events, including completion of cell division, new bud formation and SPB duplication. A number of other cellular events compose the processes of mitotic exit and the ensuing cell division (Juang et al., 1997; Adames et al., 2001).
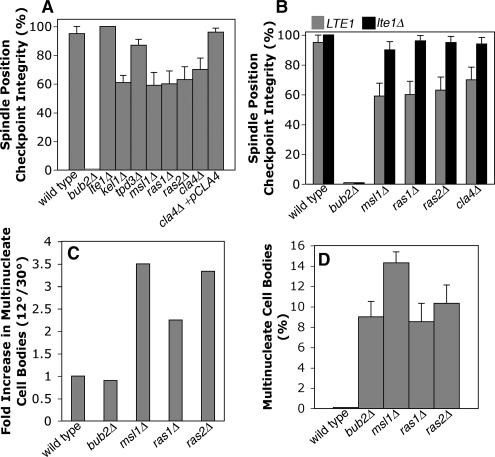
Figure 2.
Spindle position checkpoint in cells lacking Lte1-interacting proteins. (A) Lte1-interacting proteins are required for checkpoint integrity. arp1Δ GFP-TUB1 cells with the indicated additional mutation were assayed for checkpoint integrity. Cells with mispositioned anaphase spindles in the mother were followed by time-lapse movie analysis of GFP-labeled microtubules. The spindle position checkpoint integrity value is the percentage of such cells that remain arrested as opposed to undergoing mitotic exit, revealed by breakdown of the spindle. Mutants lacking Kel1, Msl1, Ras1, Ras2, and Cla4 had defects compared with wild type (p = 0.003, 0.002, 0.003, 0.001, and 0.015, respectively). Error bars, SE of proportion. n ≥ 20 cells for each sample. (B) Deleting LTE1 suppressed the checkpoint integrity phenotype of msl1, ras1, ras2, and cla4 mutants, assayed as in A. In each case, checkpoint integrity was significantly increased by deleting LTE1 (p = 0.003, 0.0002, 0.002, and 0.01, respectively), to levels similar to that of wild type. (C) Checkpoint defects of ras1, ras2, and msl1 mutants were enhanced in the cold. Multinucleate cell bodies were counted, as a percentage of all cells, in asynchronous cultures at 12° and 30°C. The fold-increase at 12 versus 30°C is plotted. n ≥ 281 cells. (D) Multinucleate cell bodies as a percentage of all cells in an asynchronous culture at 12°C. ras1, ras2, msl1, and bub2mutants are similar, suggesting a similar severity in loss of checkpoint.
Otherwise wild-type arp1Δ cells with late-anaphase spindles in the mother remained arrested indefinitely, whereas mitotic exit occurred promptly in mutants lacking spindle position checkpoint components (Figure 2A), as seen in previous studies (Adames et al., 2001; Castillon et al., 2003). Checkpoint integrity was calculated as the number of cells remaining arrested divided by the sum of the number of arrested cells plus the number of cells undergoing mitotic exit with the spindle in the mother. In some cells, the late-anaphase spindle was able to enter the neck. When this happened, mitotic exit ensued promptly, creating two cells with single nuclei. The frequency of this correction of spindle position was 6–8%, with similar values in wt, bub2Δ, bud6Δ, and msl1Δ strains. These cells were not included in the checkpoint integrity calculation.
As a positive control in this assay, GAP-deficient bub2Δ arp1Δ cells displayed 0% checkpoint integrity, meaning that every late-anaphase spindle in the mother underwent mitotic exit in the mother. In addition, mitotic exit occurred promptly, which was defined as a time less than the mean plus 2 SDs of the time for normal mitotic exit.
One Lte1 interactor, Msl1, identified by a high throughput two-hybrid screen, was not previously implicated in the cell cycle (Fromont-Racine et al., 1997). Deletion of MSL1 in an arp1Δ background caused a substantial decrease of checkpoint integrity, to a value of 59%. The tpd3Δ mutant was normal. We also found loss of SPC integrity in null mutants lacking Ras1, Ras2, Cla4, or Kel1, each of which have been implicated in Lte1 function in some way in previous studies (Hofken and Schiebel, 2002; Seshan et al., 2002; Yoshida et al., 2003; Seshan and Amon, 2005).
Msl1 has been characterized as the U2B″ subunit of the U2 small nuclear ribonucleoprotein (snRNP; Tang and Cai, 1996). To assess the potential role of RNA splicing or snRNPs in the checkpoint, we assayed lea1 null mutants, which lack the U2A′ subunit of the U2 snRNP. Lea1 interacts biochemically with Msl1 (also known as Yib9), and msl1 and lea1 null mutants have similar phenotypes with respect to U2 assembly, splicing activity, and growth (Caspary and Seraphin, 1998). A lea1Δ mutant had full integrity of the spindle position checkpoint (Supplementary Figure 1), as did a mud1Δ mutant, which lacks the U1-A component of the U1 snRNP (Liao et al., 1993; Neubauer et al., 1997).
Our hypothesis was that the checkpoint works through upstream regulators to inhibit Lte1 when the spindle has not yet entered the neck. In null mutants lacking such regulators, Lte1 would then be more active than normal, which would cause mitotic exit to fail to wait. If the checkpoint functions of these Lte1-interacting proteins are upstream of Lte1, then the loss of checkpoint in the null mutants should be suppressed by loss of Lte1. We combined the null mutations with an lte1Δ mutation. For the msl1Δ mutant, the checkpoint phenotype was completely suppressed by the lte1Δ mutation. The checkpoint defects of mutants lacking Ras1, Ras2, and Cla4, also depended on Lte1 (Figure 2B). Bub2/Bfa1, in its role as a putative GAP for Tem1, might not be expected to depend on Lte1 in this assay. Indeed, the checkpoint phenotype of a bub2Δ mutant was not suppressed by the addition of an lte1Δ mutation (Figure 2B). Thus, Msl1 appears to function upstream of Lte1 with respect to checkpoint function.
To further characterize the role of Msl1 and other Lte1-interactors, we tested checkpoint integrity at low temperature. During the normal course of mitosis at 30°C, Tem1 can be activated and mitotic exit can occur even in the absence of Lte1, probably because the intrinsic GTP exchange activity of Tem1 is sufficiently high at this temperature (Geymonat et al., 2002). In the absence of Lte1, inhibition of Bub2/Bfa1 GAP activity is presumed to be the regulatory event needed to activate Tem1 and drive mitotic exit. However, at low temperature, where the intrinsic GTP exchange rate of Tem1 is lower, Lte1 is required for mitotic exit, and lte1Δ mutants fail to grow because they arrest in anaphase (Shirayama et al., 1994b).
If Msl1 inhibits Lte1, then an msl1Δ mutant should have increased Lte1 activity, and thus, in the cold, msl1Δ cells might exit from mitosis instead of arrest in mitosis. To test this hypothesis, we counted multinucleated cell bodies, as an indicator of mitotic exit and checkpoint failure, in arp1Δ cells at 12 or 30°C and (Figure 2C). For msl1Δ arp1Δ, the value was 2.5-fold higher at 12 than at 30°C, and similar results were seen for ras1Δ arp1Δ and ras2Δ arp1Δ mutants. A bub2Δ arp1Δ mutant was also included, as a control (Figure 2D), and its value for multinucleate cell bodies was similar to that of msl1Δarp1Δ. At 30°C, the bub2Δ mutation is known to cause a complete loss of the checkpoint, by movie analysis (Castillon et al., 2003; Figure 2B). Movie analysis is not practical at 12°C, but the similarity of phenotypes for msl1Δ arp1Δ and bub2Δ arp1Δ in this assay, in the cold, suggests that Msl1, working through Lte1, may have a critical role in delaying mitotic exit.
In these assays, loss of Cla4, Ras1, or Ras2 also promoted mitotic exit in an arp1Δ background, and this phenotype was suppressed by loss of Lte1 in each case (Figure 2, A and B). The simplest interpretation of these results is that Cla4, Ras1, and Ras2 inhibit Lte1. However, this seems paradoxical because, in previous studies, cla4Δ and lte1Δ mutants had a similar phenotype of poor growth in the cold (Hofken and Schiebel, 2002; Seshan et al., 2002), and cla4, ras1, ras2, and lte1 show synthetic lethality with FEAR mutations (Stegmeier et al., 2002; Goehring et al., 2003; Yoshida et al., 2003). These results suggest that Lte1 and the three other proteins function in the same direction, not opposing directions (Hofken and Schiebel, 2002; Seshan et al., 2002). We confirmed that a cla4Δ single mutant, which is otherwise wild type, grows poorly in the cold in our strain background (data not shown), and we confirmed the cla4Δ checkpoint phenotype by rescue with CLA4 on a plasmid (Figure 2A). Several considerations may account for the apparent discrepancy—the movie assay for mitotic exit is quite different and far more specific than the assay for growth on plates in the cold, the assay for mitotic exit is done in a background without dynein function, and Cla4 is a kinase known to have functions in other pathways unrelated to mitosis (Gulli et al., 2000; Gladfelter et al., 2004).
In previous studies, mutants with unstable cytoplasmic microtubules suffered a loss of the spindle position checkpoint, and movies suggested that loss of cytoplasmic microtubule contact with the mother/bud neck was the common event that preceded mitotic exit (Adames et al., 2001). These observations suggested that microtubule/neck interaction might activate the checkpoint and delay mitotic exit (Adames et al., 2001). To determine whether the premature or “inappropriate” mitotic exit in msl1Δ cells was due to a primary loss of checkpoint activity or was instead secondary to a defect in microtubule dynamics, we followed the dynamics of cytoplasmic microtubules during mitotic exit. This experiment required image collection at short time intervals over a long time duration, which is challenging (Figure 3A, Supplementary Movie 1).
Figure 3.
Loss of spindle position checkpoint integrity in an msl1 mutant is not coupled with defects in microtubule dynamics. (A) Frames at 2-min intervals from a time-lapse movie of a representative msl1Δ arp1Δ GFP-TUB1 cell (Supplementary Movie 1). The cytoplasmic microtubule does not withdraw from the bud before mitotic exit, and the microtubule remains through the bud neck after mitotic exit. Pseudocolor is based on the “Fire” lookup table in ImageJ. Each panel is 8.7 μm wide. (B) Microtubule behavior during mitotic exit in 13 msl1 mutant cells. Cytoplasmic microtubules were observed to leave the neck after, during, or before (top, middle, and bottom, respectively) breakdown of the mitotic spindle.
In 6 of 13 cases of spindle breakdown in the mother cell in an msl1 mutant, we saw that cytoplasmic microtubules were present in the neck when the spindle broke down (Figure 3B). In several cases, the microtubule appeared to be cleaved in two at the neck by the process of cytokinesis. In the other 7 of 13 cases, the cytoplasmic microtubules withdrew from the neck at the same time as the spindle broke down. These results are consistent with msl1Δ cells having a primary defect in the checkpoint, as opposed to a defect secondary to microtubules.
Only about half of the mutant cells with a mispositioned spindle proceeded to mitotic exit with the spindle in the mother; in the other half, the cell cycle was arrested. To understand the difference between these populations, we asked how rapidly mitotic exit occurred. In the movie analysis above, cells with mispositioned anaphase spindles were chosen for observation, raising the possibility that the cells had been arrested in mitosis for a significant period of time before the movie began. To test this possibility, we repeated the experiment, choosing cells in early anaphase allowing one to follow the entire time course of mitosis (Supplementary Figure 2). As usual, a dynein-deficient background (arp1Δ) was used, so that movement of the spindle into the neck was delayed in about one-third of cells. In an otherwise wt cell in which the spindle entered the neck normally, mitosis took an average of 25 min (timed from the start of spindle elongation to spindle breakdown). For msl1Δ, ras1Δ, ras2Δ, and cla4Δ cells in which the spindle entered the neck normally, the time was similar. However, when spindles did not enter the neck and mitosis occurred in the mother cell, mitosis in wild-type cells was significantly longer. In contrast, msl1Δ, ras1Δ, and ras2Δ mutants did not significantly delay mitosis when the spindle stayed in the mother. The cla4Δ mutant had a small but significant delay compared with wild type. In other words, in mutant cells in which the checkpoint failed, the timing of mitosis was normal. Thus, these cells did not display any delay before the checkpoint failed, indicating a complete loss in the checkpoint in those individual cells.
To further investigate the timing of mitosis, we returned to the datasets of movies in which cells with mispositioned spindles in the mother were selected for analysis. We measured the time required for breakdown of the spindle in the mother. These data are shown as histograms in Supplementary Figure 3. Otherwise wild-type arp1Δcells, with mispositioned spindles in the mother, usually remained arrested throughout the movie, usually ∼90 min. When spindles in the mother of otherwise wild-type cells did break down, they did so after being arrested for a highly variable length of time, and this generally followed loss of cytoplasmic microtubules from the neck or abortive entry of the spindle into the neck. When spindles in the mother of msl1Δ, ras1Δ, ras2Δ, and cla4Δ cells broke down, the majority of cells did so in 20–30 min, similar to the time for mitosis of a spindle entering the neck of an otherwise wild-type cell. Only a minority of cells in each mutant significantly delayed mitosis before spindle breakdown. These data are also consistent with mitotic exit occurring with normal rapidity when the checkpoint is lost in mutant cells. The results do not contradict the observation that the checkpoint mechanism fails in only ∼40% of cells. The mechanism is apparently intact in the other ∼60% of cells, which do arrest. Therefore, the mechanism may involve a level of cooperativity or positive feedback.
Linking Lte1 Regulation with Microtubule-Cortex Interactions
To further investigate the role of Msl1 activating the checkpoint, we returned to the proteomic map and looked for connections of Msl1 with proteins of the bud neck and the cytoskeleton (Figure 1B). The map shows that Msl1 binds Atc1 according to a high-throughput two-hybrid assay, which is named Aip Three Complex 1 for its ability to interact, by two-hybrid assay, with the protein Aip3, also known as Bud6 (Freedman et al., 2000; Amberg and Haarer, SUNY Upstate, personal communication, 2007). We will refer to Aip3/Bud6 as Bud6 for simplicity. Bud6 is located in a ring at the bud neck and as puncta on the bud cortex (Huisman et al., 2004; Huisman and Segal, 2005). In both locations, Bud6 has been observed to interact with cytoplasmic microtubules, based on two-color movies (Huisman and Segal, 2005).
A previous study of Bud6 found multinucleated cells in asynchronous cultures of bud6 null mutants and observed mitotic exit in some cells with late-anaphase spindles in the mother (Huisman et al., 2004). We uncovered bud6Δ mutants in a screen for spindle position checkpoint mutants (Heil-Chapdelaine et al., 2002; unpublished data), as described in Materials and Methods.
We analyzed atc1 and bud6 null mutants as described above for msl1Δ. Based on movie analysis in an arp1 background, atc1Δ and bud6Δ mutants had decreased values for checkpoint integrity, with values similar to that of the msl1Δ mutant (Figure 4A). Deletion of LTE1 strongly suppressed the checkpoint phenotypes of the atc1Δ and bud6Δ mutants, as seen for msl1 (Figure 4A). Placing cells in the cold, we found an increase in the number of multinucleate cell bodies in bud6 and atc1Δ mutants, at a level similar to that seen for msl1Δ and bub2Δ (Figure 4, C and D).
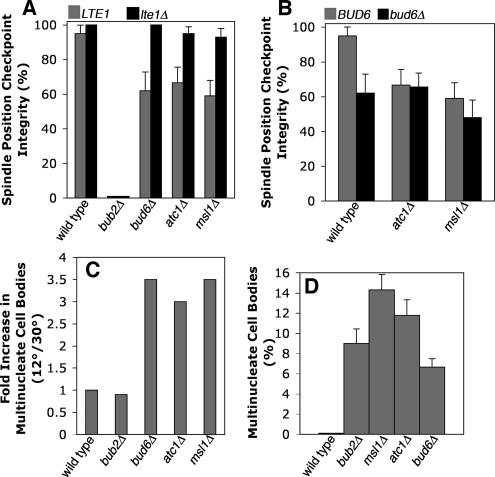
Figure 4.
Genetic analysis of MSL1, ATC1, and BUD6 in the spindle position checkpoint. (A) bud6 and atc1 mutants display decreased checkpoint integrity, with values similar to that of msl1 (p = 0.004, 0.009, and 0.002, respectively, with respect to wild type). Deleting LTE1 restores SPC integrity in bud6 and atc1 mutants, as seen for msl1 mutants (p = 2.3 × 10−6, 0.005, and 7.16 × 10−4, respectively). Cells with the indicated mutations were assayed for checkpoint integrity as in Figure 2A. (B) Deleting BUD6 does not enhance the checkpoint defects of atc1 and msl1 mutants (p = 0.293 and 0.467, respectively). (C) Checkpoint defects of bud6, atc1, and msl1 mutants are enhanced in the cold. The fold-increase in multinucleate cell bodies at 12 versus 30°C is plotted, as in Figure 2C. n ≥ 281 cells. (D) The checkpoint phenotypes of bud6, atc1, and msl1 mutants at 12°C are as severe as that of bub2, based on the percentage of multinucleate cell bodies.
To extend the genetic analysis, we analyzed double null mutants (Figure 4B). Deleting BUD6 did not enhance or suppress the checkpoint phenotype of atc1Δ or msl1Δ mutants, suggesting that ATC1, MSL1, and BUD6 lie in the same genetic pathway with respect to checkpoint function. Additionally, like msl1, ras1, and ras2 mutants, mitotic exit in the mother occurred at the same rate as mitotic exit in the mother-bud neck (Supplementary Figures 2 and 3).
As a further test of the existence of a pathway, we asked whether overexpression of one gene was able to suppress the checkpoint phenotype in a null mutant lacking another gene (Table 1). Based on the physical interactions in the database, one might hypothesize that Bud6 lies upstream of Atc1, Atc1 is upstream of Msl1, and Msl1 inhibits Lte1. To test this simple model, we overexpressed each gene in a strain lacking the other genes, in pair wise combinations. The most straightforward result would be that overexpression of a gene that is downstream of a second gene suppresses the phenotype of a mutant lacking the second gene. The assay was counting multinucleate cell bodies in asynchronous cultures of strains with an arp1Δ background. The checkpoint defect of bud6Δ cells was completely suppressed by overexpression of ATC1 or MSL1 (Table 2). Deletion of LTE1 also suppressed the checkpoint phenotype of bud6Δ, atc1Δ, and msl1Δ mutants, confirming the results above with movie analysis. The checkpoint defect of atc1Δ cells was completely suppressed by overexpression of MSL1 and partially suppressed by overexpression of BUD6. The checkpoint defect of msl1Δ cells was not suppressed by overexpression of BUD6 or ATC1. No overexpression allele caused a loss of checkpoint in lte1Δ cells (Table 2). Furthermore, no gene deletion was able to suppress the checkpoint defect of cells overexpressing LTE1. All the results, with the sole exception of partial, not full, suppression of atc1Δ by BUD6, are consistent with Bud6, Atc1, and Msl1 acting in an ordered pathway to inhibit Lte1 function. This order revealed by this assay is consistent with the order of the protein interactions in the two-hybrid analysis (Figure 1B). The results do not exclude the possible existence of simultaneous multisubunit interactions.
Table 1.
Predicted results of overexpression suppression analysis of Lte1 regulators in the checkpoint pathway
| Deletion | Overexpression |
|||||
|---|---|---|---|---|---|---|
| KIP2 | BUD6 | ATC1 | MSL1 | LTE1 | Control | |
| kip2Δ | NA | + | + | + | − | − |
| bud6Δ | − | NA | + | + | − | − |
| atc1Δ | − | − | NA | + | − | − |
| msl1Δ | − | − | − | NA | − | − |
| lte1Δ | + | + | + | + | NA | + |
Results shown are for checkpoint integrity, based on a linear pathway. In an arp1Δ GFP-TUB1 background, the indicated genes in the first row were overexpressed by integrating the GAL1 promoter at the endogenous locus and adding galactose to the medium. Control is a strain without an integrated GAL1 promoter. The first column lists the null mutant haploid strains tested. +, suppression of the loss-of-checkpoint phenotype; −, no suppression.
Table 2.
Observed results of overexpression suppression analysis of Lte1 regulators in the checkpoint pathway
| Deletion | Overexpression |
|||||
|---|---|---|---|---|---|---|
| KIP2 | BUD6 | ATC1 | MSL1 | LTE1 | Control | |
| kip2Δ | NA | 2.1 | 3.1 | 2.5 | 11.0 | 8.2 |
| bud6Δ | 12.0 | NA | 1.4 | 2.3 | 11.1 | 8.6 |
| atc1Δ | 9.7 | 6.1 | NA | 2.2 | 11.6 | 9.6 |
| msl1Δ | 15.8 | 13.7 | 10.2 | NA | 15.5 | 10.7 |
| lte1Δ | 1.8 | 0.0 | 0.0 | 3.0 | NA | 2.7 |
Checkpoint integrity was assayed by counting cells with multiple GFP-TUB1—labeled spindle pole bodies, expressed here as a percentage of all multinucleate cells in asynchronous culture. n ≥ 242 in each case.
Atc1-GFP and Msl1-GFP were both found throughout the cytoplasm in the mother and bud, with a uniform distribution (Supplementary Figure 4). Msl1-GFP was slightly concentrated in the nucleus, and it appeared to be excluded from the vacuole. The fluorescence signal of Atc1-GFP and Msl1-GFP was significantly higher than the background fluorescence of a cell not expressing GFP. The uniform cytoplasmic distribution of these proteins was similar to that of plain GFP, which was excluded from the nucleus and not concentrated anywhere. The localization of Atc1 and Msl1 was similar in cells at different stages of the cell cycle.
The Cellular Role of Bud6 in Spindle Position Checkpoint Integrity
Bud6 forms a ring at the neck and foci in the bud cortex in late mitosis, where it captures microtubule ends (Huisman and Segal, 2005). We asked whether loss of microtubule capture might be responsible for the checkpoint defect in bud6Δ mutants. First, we colocalized microtubules and Bud6 in anaphase cells in which the checkpoint was activated due to the presence of a late-anaphase spindle in the mother. Microtubule ends, seen by CFP-Tub1, were often colocalized with cortical foci of GFP-Bud6 (Figure 5A), as predicted.
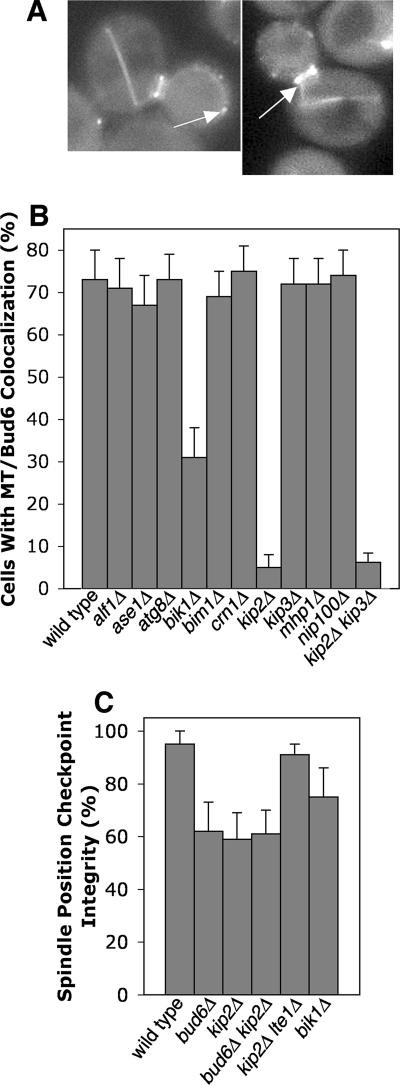
Figure 5.
Microtubule capture and the spindle position checkpoint. (A) Microtubule plus ends colocalize with Bud6 foci at the bud cortex (left) and at the mother-bud neck (right) when cells have anaphase spindles in the mother and the checkpoint is active. Representative examples of arp1Δ cells expressing GFP-BUD6 and CFP-TUB1. The first panel is 11.1 μm wide. The second panel is 9.6 μm wide. (B) Bud6-Mt colocalization requires Kip2. Cells expressing GFP-BUD6 and CFP-TUB1 with the indicated null mutation were viewed using wide-field fluorescence microscopy. Percentage of cells lacking colocalization between Bud6 foci and microtubule plus ends are shown. n ≥ 100 cells. (C) Mutants lacking Kip2 have an Lte1-dependent SPC defect that is not enhanced by loss of Bud6. Cells with the indicated mutations were assayed for SPC integrity as in Figure 2A.
To test the significance of the Bud6-microtubule interaction and to identify other molecular components involved in the interaction, we examined the colocalization of microtubule ends with Bud6 at cortical foci or neck rings in mutants lacking cytoplasmic microtubule-binding proteins (Figure 5B). In 73% of wild-type cells, microtubule ends were observed to be colocalized with Bud6 at the neck or at cortical foci. This value was reduced to 5% in a kinesin/kip2Δ mutant, and 31% in a CLIP170/bik1Δ mutant. All other mutants tested had normal values, including tubulin-folding cofactor B/alf1Δ, EB1/bim1Δ, coronin/crn1Δ, kinesin/kip3Δ, p150/Glued/nip100Δ, ase1Δ, atg8Δ, and mhp1Δ. Thus, the kinesin Kip2 and, to a lesser extent, CLIP170/Bik1, are specifically required for Bud6-mediated microtubule capture.
To test whether microtubule/Bud6 interactions are required for the checkpoint, we assayed checkpoint integrity in kip2Δ and bik1Δ strains because these mutants have poor microtubule/Bud6 interactions. The kip2Δ mutant displayed decreased checkpoint integrity, at a level similar to that of a bud6Δ mutant (Figure 5C). A kip2Δ bud6Δ double mutant had the same value that the single mutants did, suggesting that the two proteins participate in the same process. bik1Δ strains displayed an intermediate value for checkpoint integrity, consistent with the intermediate loss of Bud6/microtubule colocalization in this mutant. Therefore, microtubule capture by the neck or cortex, involving the participation of Bik1, Kip2, and Bud6, correlates with and appears to be necessary for activation of the checkpoint.
Mutants lacking Bik1 and Kip2 have short cytoplasmic microtubules(Berlin et al., 1990; Carvalho et al., 2004), suggesting that this trait might account for the observed decrease in cortical Bud6/microtubule interactions. To test this possibility, we quantified the frequency of colocalization of microtubules with the bud cortex at sites away from Bud6 foci in wild-type, bik1Δ, and kip2Δ cells (Supplementary Figure 5). No significant differences were found, indicating that microtubules in the mutants are able to touch the cortex and thus have the opportunity to make productive capture interactions. We also addressed this issue by deleting the kinesin kip3 in the kip2 mutant, which has been shown to restore microtubule length (Cottingham and Hoyt, 1997). In our strain background, the kip3Δ kip2Δ mutant had the same low value of microtubule/Bud6 interactions as seen in the single kip2 mutant (Figure 5B), despite having qualitatively longer cytoplasmic microtubules. Therefore, kip2Δ and bik1Δ mutations appear to have a direct effect on microtubule interactions with the cortex at Bud6 foci.
Because Kip2 appears to connect microtubules to Bud6 in the cell, we hypothesized that the KIP2 gene might lie upstream of the BUD6 gene in the genetic pathway for regulation of LTE1. To test this hypothesis, we performed suppression analysis, as above. The kip2Δ checkpoint phenotype was suppressed by overexpression of BUD6, ATC1, or MSL1 (Table 2). Conversely, overexpression of KIP2 in bud6Δ, atc1Δ, msl1Δ, and lte1Δ mutants did not suppress the checkpoint phenotype of any of these mutants (Table 2). These results place KIP2 upstream of BUD6 in the checkpoint pathway, as defined by genetic interactions.
To test the specificity of the role of Bud6 in the spindle position checkpoint, as opposed to other checkpoints, we assayed bud6Δ mutants for defects in the bud morphogenesis and DNA damage checkpoints by assaying growth in latrunculin A and hydroxyurea, respectively (Supplementary Figure 6). Both checkpoints were intact.
In addition to roles in microtubule capture, BUD6 is also known to be important for bud-site selection and actin cable formation (Amberg et al., 1997; Moseley et al., 2004). We found that bud1 and bud5 mutants, which are completely defective in bud-site selection, showed no loss of spindle position checkpoint integrity by movie analysis (Supplementary Figure 7A). In addition, loss of actin cables, caused by mutations in genes encoding formins, fimbrin, or tropomyosin, did not cause a significant loss of checkpoint integrity (Supplementary Figure 7B). These results suggest that the role of Bud6 in the spindle position checkpoint is based on its role in microtubule capture.
Lte1 Localization in Checkpoint Mutants
In the events leading up to mitotic exit, Tem1 accumulates at the daughter-bound SPB, and Lte1 localizes to the bud cortex (Bardin et al., 2000; Molk et al., 2004). These observations led to a model in which movement of the SPB into the bud allows interaction of Tem1 with Lte1 (Bardin et al., 2000). In support of this model, spindle position checkpoint failure has been observed to result from septin mutations that allow Lte1 to cross the neck from the bud into the mother (Castillon et al., 2003). In the mother, this ectopic Lte1 is presumed to activate Tem1 and thus the MEN (Shirayama et al., 1994b).
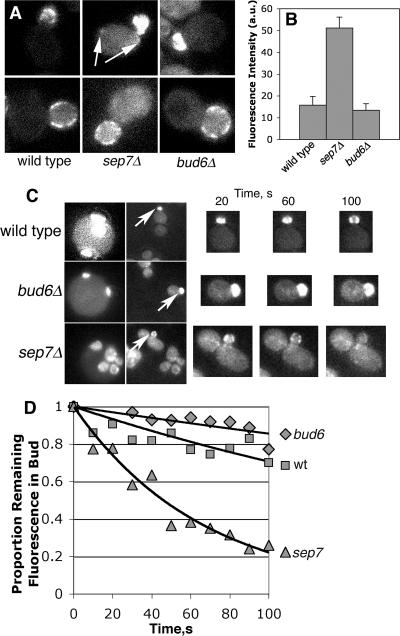
Bud6 is a component of the mother-bud neck, so we considered whether the checkpoint defect of a bud6Δ mutant might be due a defective neck leading to the presence of ectopic Lte1 in the mother. In a previous study, Lte1 was observed to be localized normally in a bud6 mutant (Jensen et al., 2002). We confirmed this result with Lte1-3GFP and found that Lte1 localization appeared normal in atc1Δ, msl1Δ, and kip2Δ cells as well (data not shown). To address the issue of Lte1 in the mother directly, we digitally imaged and quantified the fluorescence of Lte1-3GFP in the mother cytoplasm. In a wild-type cell, the level of this fluorescence is not greater than the fluorescence associated with a control cell that does not express any GFP (Castillon et al., 2003). We found that bud6Δ cells have no more Lte1-3GFP fluorescence in the mother than do wild-type cells. As a positive control, a sep7Δ mutant had significantly increased levels, as seen previously (Castillon et al., 2003).
As a more stringent test for the diffusion of a small amount of Lte1 from the bud into the mother of bud6Δ cells, at a level undetectable by steady-state fluorescence imaging, we used fluorescence loss in photobleaching (FLIP; Figure 6, C and D). We photobleached a portion of a mother cell and quantified the fluorescence in the bud. Sequential rounds of photobleaching and imaging were performed. If Lte1-3GFP was present in the mother, it should have been bleached, leading to a loss of fluorescence intensity in the bud over time. Wild-type and bud6Δ cells gave similar results, with only a small loss of fluorescence after 10 rounds of photobleaching, an amount consistent with the degree of photobleaching over the entire field. As a positive control, in sep7Δ cells, the bud fluorescence fell to background levels after three rounds of photobleaching.
Figure 6.
Lte1 localization and dynamics. (A) Lte1-3GFP is excluded from the mother in bud6 mutants. Representative images of Lte1-3GFP small- and large-budded cells are shown. The mother cell cytoplasm of sep7Δ cells is brighter than that of wt or bud6 cells, which have very low levels of fluorescence, similar to the autofluorescence of cells not expressing GFP (Castillon et al., 2003). Arrows indicate abnormal foci of Lte1 in the mother. Each image is 7.5 μm wide. (B) Quantification of Lte1-3GFP fluorescence in the mother. bud6 mutants have a level of Lte1 in the mother similar to that of wt cells, which is similar to that of cells not expressing GFP (Castillon et al., 2003). (C) Fluorescence loss in photobleaching (FLIP) analysis of Lte1-3GFP cells. The first column shows a field of cells, with arrows indicating the bud fluorescence that was followed during the experiment. The second column shows the region of the field that was bleached, produced by narrowing the fluorescence aperture. The first two images in each series are 15 μm wide. Note that the aperture is positioned so that the mother portion of the cell in question will be bleached. During each 10-s time interval, one round of photobleaching was performed, and a fluorescence image was collected. To the right are representative fluorescence images collected at the indicated times after photobleaching, shown at higher magnification. (D) Quantification of bud fluorescence during the course of photobleaching. The mean remaining proportion of the initial fluorescence intensity is plotted over time. The FLIP results show that concentration of Lte1 that diffuses from the bud into the mother is little to none in wild-type or bud6 mutant cells, in contrast to the sep7Δ positive control cells. n = 8 cells.
These results suggest that the checkpoint phenotype of bud6Δ cells is not due to mislocalization of Lte1 in the mother. Furthermore, they suggest that, in the bud6 mutant, Tem1 activation can occur with Lte1 confined to the bud and with the Tem1-rich daughter-bound SPB still in the mother. A possible resolution of this apparent paradox is provided by the observation that Tem1 at the SPB rapidly exchanges with a cytoplasmic pool of Tem1 (Molk et al., 2004). Based on this and other findings, Molk and colleagues proposed a model in which the cytoplasmic pool of Tem1 is activated by Lte1 in the bud (Molk et al., 2004).
The relationship of the cortical localization of Lte1 to the activity state of Lte1 in the cell is not well understood. Lte1 is lost from the bud cortex at the end of the cell cycle, assuming a diffuse distribution in the cytoplasm, which suggests that cortical localization may influence Lte1 activity (Jensen et al., 2002; Seshan et al., 2002). The cortical localization of Lte1 requires Ras2 (Yoshida et al., 2003), Cla4 (Hofken and Schiebel, 2002), and Kel1 (Seshan et al., 2002). Here, ras1, ras2, and cla4 mutants had decreased checkpoint integrity, in the dynein-deficient background (Figure 2). Their phenotypes were suppressed by the loss of Lte1, and they were not enhanced by the loss of Bud6 (data not shown), suggesting that these proteins influence mitotic exit via Lte1.
DISCUSSION
The spindle position checkpoint delays exit from mitosis until the spindle is positioned correctly. Cytoplasmic microtubules position the mitotic spindle through the mother-bud neck in anticipation of cell division (Adames et al., 2001). The presence or absence of a cytoplasmic microtubule in the bud neck is sufficient to determine whether or not an anaphase cell with its spindle in the mother will remain arrested by the checkpoint or proceed to exit mitosis inappropriately (Adames et al., 2001). Therefore, the existence of a signaling pathway linking microtubule-cortex interactions with mitotic exit makes intuitive sense. In this study, we discovered such a pathway, which prevents mitotic exit by inhibiting the Tem1-activating protein Lte1. Of note, a microtubule-cortex interaction appears to be a necessary component of this pathway. Finally, we identified the molecular components and their order in this pathway, revealing unexpected new roles for proteins not previously implicated in cell cycle control.
We set out to understand how Lte1 is regulated, because Lte1 is known to be a strong activator of mitotic exit via the MEN. Overexpression of Lte1 can completely override the spindle position checkpoint, and Lte1 is necessary for mitotic exit when cells are grown in the cold or when the FEAR network is disrupted (Shirayama et al., 1994b; Bardin et al., 2000; Stegmeier et al., 2002). Genetic evidence indicates that Lte1 drives mitotic exit in a Tem1-dependent manner (Shirayama et al., 1994a; Bardin et al., 2000; Molk et al., 2004).
Lte1 localization and phosphorylation are known to depend on the Ras GTPases Ras1 and Ras2, the PAK kinase Cla4, and the kelch repeat-containing Kel1 (Seshan et al., 2002; Seshan and Amon, 2005). Because ras1Δ, ras2Δ, and cla4Δ mutants are sensitive to the cold, as lte1Δ mutants are, the genes are presumed to be required for normal LTE1 function (Seshan et al., 2002; Seshan and Amon, 2005). Whether and how Lte1 is regulated by the spindle position checkpoint to coordinate spindle position with mitotic exit was unknown. To investigate this question, we searched for new modes of Lte1 regulation.
Using a map of protein–protein interactions, we identified proteins that interact with Lte1. We tested null mutants lacking each of these proteins for loss of checkpoint integrity. A checkpoint mutant lacking a component critical for inhibition of Lte1 should have the following characteristics: 1) loss of checkpoint integrity, namely exit from mitosis by a spindle in the mother, 2) suppression of the checkpoint phenotype by loss of Lte1, and 3) exacerbation of the checkpoint phenotype at low temperatures, where the requirement of Tem1 for activation by Lte1 is greater.
Using the protein-interaction map, we were able to trace a potential pathway linking Lte1 to the cortical microtubule capture protein Bud6. Capture of microtubules by cortical Bud6 helps to orient the spindle parallel to the mother-bud axis in anaphase (Huisman et al., 2004). In addition, a role for Bud6 in cell polarity has been well characterized. Here, we found a novel function for Bud6, as required for the spindle position checkpoint.
We showed that Bud6 can be connected to Lte1 via two unexpected proteins, Msl1 and Atc1. The BUD6, MSL1, and ATC1 genes meet the requirements for LTE1-based regulation of the spindle position checkpoint outlined above. First, bud6Δ, atc1Δ, and msl1Δ mutants had checkpoint defects of the same penetrance, and combinations of these alleles did not enhance the phenotype. Second, deletion of LTE1 suppressed the checkpoint defect of bud6Δ, atc1Δ, and msl1Δ mutants. Third, the checkpoint phenotypes of bud6Δ, atc1Δ, and msl1Δ mutants were enhanced in the cold, where Tem1 has greater need for activation by Lte1.
Having met these criteria, BUD6, ATC1, and MSL1 were subjected to further genetic analysis to investigate the mechanism of their checkpoint activity. Because combinations of alleles did not result in a further decrease in checkpoint integrity, and given the order of the protein–protein interactions (Bud6 with Atc1, Atc1 with Msl1, and Msl1 with Lte1), we hypothesized the existence of an ordered linear pathway. Overexpression suppression analysis placed BUD6 upstream of ATC1, and ATC1 upstream of MSL1. These genetic results are consistent with a linear pathway but they do not rule out the possibility that the proteins act together in a complex.
One interesting aspect of these results is that the checkpoint fails in about half of null mutant cells with mis-positioned spindles. Single and double null mutations of pathway genes consistently gave this result. In the other half of the cells with mis-positioned spindles, the cell cycle halted normally, suggesting that the checkpoint was intact. Furthermore, when the checkpoint was lost, mitotic exit occurred promptly, similar to the timing for normal mitosis. One explanation for these results is that activation of the checkpoint has an element of cooperativity or positive feedback, which is lost in the mutants. This idea is consistent with the existence of other pathways to control the activity of Tem1, such as the Bub2/Bfa1 GAP.
The cell uses cytoplasmic microtubules to position the spindle in the cell, making it logical for the cell to derive information from cytoplasmic microtubules to detect spindle position. We found that Bud6-mediated microtubule attachment to the cell cortex appears to be necessary for the spindle position checkpoint. Bud6 is known to mediate microtubule-cortex interactions, from previous studies (Huisman and Segal, 2005). Here, we discovered that the capture of microtubules by cortical Bud6 required the kinesin Kip2, a cytoplasmic microtubule protein, and that cortical capture correlated with integrity of the checkpoint. A need for Bud6/microtubule associations in checkpoint activation is consistent with and extends a previous model in which a microtubule-interacting spindle position sensor resides at the mother/bud neck (Adames et al., 2001). This sensor was hypothesized to inhibit mitotic exit until microtubule contact with the neck was lost, which would normally occur when the spindle entered the neck (Adames et al., 2001). Our results here suggest that the proposed sensing mechanism requires Bud6 and Kip2, but they do not identify Bud6 or Kip2 as the source of the signal.
In addition, we found that mutants lacking RAS1, RAS2, and CLA4, which are important for normal Lte1 function by mediating Lte1 phosphorylation and localization to the bud cortex, also had Lte1-dependent checkpoint defects. These proteins appear to act on Lte1 outside of the Bud6 checkpoint pathway because these proteins affect Lte1 localization, whereas Kip2, Bud6, Atc1, and Msl1 do not. These data suggest that Msl1, Ras1, Ras2, and Cla4 all inhibit Lte1 function when the checkpoint is active. These data seem inconsistent with previous studies, which suggested that these proteins activate Lte1 based on observations that mutations in their genes exhibit synthetic genetic interactions similar to those of lte1 (Hofken and Schiebel, 2002; Yoshida et al., 2003). The previous studies assayed growth under conditions where lte1 mutants grow poorly. These conditions would not be expected to trigger the spindle position checkpoint, and the assays were not designed to assay checkpoint integrity. Therefore, comparing these results is difficult. The potential discrepancy could be explained by the existence of multiple functions for Cla4 and Ras, which is likely.
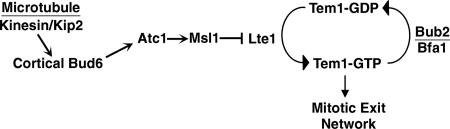
We propose a working model for a molecular pathway that couples monitoring of spindle position to activation of the MEN through Lte1 (Figure 7). Kip2 at the plus end of a microtubule interacts with Bud6 at the bud neck or cortex. This interaction is necessary for a signal that senses the absence of the spindle from the neck. This signal is inactivated when the spindle moves through the neck, because microtubule-bud cortex interaction or tension is lost. The signal passes to Atc1, which in turn signals to Msl1, causing it to cease its inhibition of Lte1. Finally, loss of inhibition allows Lte1 to activate Tem1, and therefore the MEN.
Figure 7.
Model for Lte1 regulation. Bud6 at the neck or bud cortex interacts with Kip2 at the plus end of a cytoplasmic microtubule, effecting capture of that end. This interactions generates a signal that inhibits Lte1, transmitted through Atc1 and Msl1.
An interesting question is where Lte1 activates Tem1. In our mutants, mitotic exit occurred with the daughter-bound SPB in the mother, apparently not in contact with Lte1, which is restricted to the bud. This result is consistent with the prior observation that the Tem1-rich daughter-bound SPB does not make contact with foci of Lte1 at the bud cortex before mitotic exit in wild-type cells (Molk et al., 2004). Tem1 at the SPB exchanges rapidly, revealing the existence of a cytoplasmic pool (Molk et al., 2004). This cytoplasmic pool of Tem1 may interact with Lte1 in the bud, which is either diffuse in the cytoplasm or concentrated at the cortex.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Drs. Rick Heil-Chapdelaine and Ben Leacock for performing the initial assays in the checkpoint screen and for extensive advice and assistance throughout the course of the work. We thank Dr. Mark Longtine for help with fluorescence photobleaching, as well as advice and assistance at many points. This work was supported by National Institutes of Health (NIH) Grants GM 47337 and GM 69895. S.N. was supported by NIH Training Grant T32 GM07067 and the Lucille P. Markey Special Emphasis Pathway in Human Pathobiology.
Abbreviations used:
- SPC
spindle position checkpoint
- MEN
mitotic exit network
- GAP
GTPase-activating protein
- GEF
guanine exchange factor
- SPB
spindle pole body
- FEAR
cdc14 early anaphase release
- snRNP
small nuclear ribonucleoprotein
- FLIP
fluorescence loss in photobleaching.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-03-0242) on July 5, 2007.

 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Adames N. R., Cooper J. A. Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J. Cell Biol. 2000;4:863–874. doi: 10.1083/jcb.149.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adames N. R., Oberle J. R., Cooper J. A. The surveillance mechanism of the spindle position checkpoint in yeast. J. Cell Biol. 2001;1:159–168. doi: 10.1083/jcb.153.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg D. C., Zahner J. E., Mulholland J. W., Pringle J. R., Botstein D. Aip3p/Bud6p, a yeast actin-interacting protein that is involved in morphogenesis and the selection of bipolar budding sites. Mol. Biol. Cell. 1997;4:729–753. doi: 10.1091/mbc.8.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardin A. J., Visintin R., Amon A. A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell. 2000;1:21–31. doi: 10.1016/s0092-8674(00)00007-6. [DOI] [PubMed] [Google Scholar]
- Berlin V., Styles C. A., Fink G. R. BIK1, a protein required for microtubule function during mating and mitosis in Saccharomyces cerevisiae, colocalizes with tubulin. J. Cell Biol. 1990;6(Pt 1):2573–2586. doi: 10.1083/jcb.111.6.2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom K. S., Beach D. L., Maddox P., Shaw S. L., Yeh E., Salmon E. D. Using green fluorescent protein fusion proteins to quantitate microtubule and spindle dynamics in budding yeast. Methods Cell Biol. 1999;61:369–383. doi: 10.1016/s0091-679x(08)61990-1. [DOI] [PubMed] [Google Scholar]
- Breitkreutz B. J., Stark C., Tyers M. Osprey: a network visualization system. Genome Biol. 2003a;4:R22. doi: 10.1186/gb-2003-4-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitkreutz B. J., Stark C., Tyers M. The GRID: the General Repository for Interaction Datasets. Genome Biol. 2003b;4:R23. doi: 10.1186/gb-2003-4-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P., Gupta M. L., Jr, Hoyt M. A., Pellman D. Cell cycle control of kinesin-mediated transport of Bik1 (CLIP-170) regulates microtubule stability and dynein activation. Dev. Cell. 2004;6:815–829. doi: 10.1016/j.devcel.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Caspary F., Seraphin B. The yeast U2A′/U2B complex is required for pre-spliceosome formation. EMBO J. 1998;17:6348–6358. doi: 10.1093/emboj/17.21.6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillon G. A., Adames N. R., Rosello C. H., Seidel H. S., Longtine M. S., Cooper J. A., Heil-Chapdelaine R. A. Septins have a dual role in controlling mitotic exit in budding yeast. Curr. Biol. 2003;8:654–658. doi: 10.1016/s0960-9822(03)00247-1. [DOI] [PubMed] [Google Scholar]
- Cottingham F. R., Hoyt M. A. Mitotic spindle positioning in Saccharomyces cerevisiae is accomplished by antagonistically acting microtubule motor proteins. J. Cell Biol. 1997;138:1041–1053. doi: 10.1083/jcb.138.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman T., Porter A., Haarer B. Mutational and hyperexpression-induced disruption of bipolar budding in yeast. Microbiology. 2000;146:2833–2843. doi: 10.1099/00221287-146-11-2833. [DOI] [PubMed] [Google Scholar]
- Fromont-Racine M., Rain J. C., Legrain P. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat. Genet. 1997;16:277–282. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- Geymonat M., Spanos A., Smith S. J., Wheatley E., Rittinger K., Johnston L. H., Sedgwick S. G. Control of mitotic exit in budding yeast. In vitro regulation of Tem1 GTPase by Bub2 and Bfa1. J. Biol. Chem. 2002;32:28439–28445. doi: 10.1074/jbc.M202540200. [DOI] [PubMed] [Google Scholar]
- Gladfelter A. S., Pringle J. R., Lew D. J. The septin cortex at the yeast mother-bud neck. Curr. Opin. Microbiol. 2001;6:681–689. doi: 10.1016/s1369-5274(01)00269-7. [DOI] [PubMed] [Google Scholar]
- Gladfelter A. S., Zyla T. R., Lew D. J. Genetic interactions among regulators of septin organization. Eukaryot. Cell. 2004;3:847–854. doi: 10.1128/EC.3.4.847-854.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goehring A. S., Mitchell D. A., Tong A. H., Keniry M. E., Boone C., Sprague G.F.J. Synthetic lethal analysis implicates Ste20p, a p21-activated protein kinase, in polarisome activation. Mol. Biol. Cell. 2003;14:1501–1516. doi: 10.1091/mbc.E02-06-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulli M. P., Jaquenoud M., Shimada Y., Niederhauser G., Wiget P., Peter M. Phosphorylation of the Cdc42 exchange factor Cdc24 by the PAK-like kinase Cla4 may regulate polarized growth in yeast. Mol. Cell. 2000;6:1155–1167. doi: 10.1016/s1097-2765(00)00113-1. [DOI] [PubMed] [Google Scholar]
- Hofken T., Schiebel E. A role for cell polarity proteins in mitotic exit. EMBO J. 2002;18:4851–4862. doi: 10.1093/emboj/cdf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman S. M., Bales O. A., Bertrand M., Smeets M. F., Reed S. I., Segal M. Differential contribution of Bud6p and Kar9p to microtubule capture and spindle orientation in S. cerevisiae. J. Cell Biol. 2004;167:231–244. doi: 10.1083/jcb.200407167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman S. M., Segal M. Cortical capture of microtubules and spindle polarity in budding yeast—where's the catch? J. Cell Sci. 2005;118:463–471. doi: 10.1242/jcs.01650. [DOI] [PubMed] [Google Scholar]
- Jensen S., Geymonat M., Johnson A. L., Segal M., Johnston L. H. Spatial regulation of the guanine nucleotide exchange factor Lte1 in Saccharomyces cerevisiae. J. Cell Sci. 2002;115:4977–4991. doi: 10.1242/jcs.00189. [DOI] [PubMed] [Google Scholar]
- Juang Y. L., Huang J., Peters J. M., McLaughlin M. E., Tai C. Y., Pellman D. APC-mediated proteolysis of Ase1 and the morphogenesis of the mitotic spindle. Science. 1997;5304:1311–1314. doi: 10.1126/science.275.5304.1311. [DOI] [PubMed] [Google Scholar]
- Liao X. C., Tang J., Rosbash M. An enhancer screen identifies a gene that encodes the yeast U1 snRNP A protein: implications for snRNP protein function in pre-mRNA splicing. Genes Dev. 1993;7:419–428. doi: 10.1101/gad.7.3.419. [DOI] [PubMed] [Google Scholar]
- McMillan J. N., Sia R.A.L., Lew D. J. A morphogenesis checkpoint monitors the actin cytoskeleton in yeast. J. Cell Biol. 1998;6:1487–1499. doi: 10.1083/jcb.142.6.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molk J. N., Schuyler S. C., Liu J. Y., Evans J. G., Salmon E. D., Pellman D., Bloom K. The differential roles of budding yeast Tem1p, Cdc15p, and Bub2p protein dynamics in mitotic exit. Mol. Biol. Cell. 2004;4:1519–1532. doi: 10.1091/mbc.E03-09-0708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley J. B., Sagot I., Manning A. L., Xu Y., Eck M. J., Pellman D., Goode B. L. A conserved mechanism for Bni1- and mDia1-induced actin assembly and dual regulation of Bni1 by Bud6 and profilin. Mol. Biol. Cell. 2004;15:896–907. doi: 10.1091/mbc.E03-08-0621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhua L., Adames N. R., Murphy M. D., Shields C. R., Cooper J. A. A cytokinesis checkpoint requiring the yeast homolog of an APC-binding protein. Nature. 1998;393:487–491. doi: 10.1038/31014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubauer G., Gottschalk A., Fabrizio P., Seraphin B., Luhrmann R., Mann M. Identification of the proteins of the yeast U1 small nuclear ribonucleoprotein complex by mass spectrometry. Proc. Natl. Acad. Sci. USA. 1997;94:385–390. doi: 10.1073/pnas.94.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G., Tanaka T. U., Nasmyth K., Schiebel E. Modes of spindle pole body inheritance and segregation of the Bfa1p-Bub2p checkpoint protein complex. EMBO J. 2001;22:6359–6370. doi: 10.1093/emboj/20.22.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshan A., Amon A. Ras and the Rho effector Cla4 collaborate to target and anchor Lte1 at the bud cortex. Cell Cycle. 2005;4:940–946. doi: 10.4161/cc.4.7.1785. [DOI] [PubMed] [Google Scholar]
- Seshan A., Bardin A. J., Amon A. Control of Lte1 localization by cell polarity determinants and Cdc14. Curr. Biol. 2002;24:2098–2110. doi: 10.1016/s0960-9822(02)01388-x. [DOI] [PubMed] [Google Scholar]
- Shirayama M., Matsui Y., Tanaka K., Toh-e A. Isolation of a CDC25 family gene, MSI2/LTE1, as a multicopy suppressor of ira1. Yeast. 1994a;4:451–461. doi: 10.1002/yea.320100404. [DOI] [PubMed] [Google Scholar]
- Shirayama M., Matsui Y., Toh E. A. The yeast TEM1 gene, which encodes a GTP-binding protein, is involved in termination of M phase Mol. Cell. Biol. 1994b;11:7476–7482. doi: 10.1128/mcb.14.11.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Lee K. S. A novel function of Saccharomyces cerevisiae CDC5 in cytokinesis J. Cell Biol. 2001;152:451–469. doi: 10.1083/jcb.152.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F., Visintin R., Amon A. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell. 2002;2:207–220. doi: 10.1016/s0092-8674(02)00618-9. [DOI] [PubMed] [Google Scholar]
- Tang H. Y., Cai M. The EH-domain-containing protein Pan1 is required for normal organization of the actin cytoskeleton in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;9:4897–4914. doi: 10.1128/mcb.16.9.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S., Guillet M., Pellman D. MEN signaling: daughter bound pole must escape her mother to be fully active Dev. Cell. 2005;9:168–170. doi: 10.1016/j.devcel.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Yoshida S., Ichihashi R., Toh-e A. Ras recruits mitotic exit regulator Lte1 to the bud cortex in budding yeast. J. Cell Biol. 2003;5:889–897. doi: 10.1083/jcb.200301128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.