Abstract
Phosphoinositide-3 kinase (PI3K) plays an important role in signal transduction in response to a wide range of cellular stimuli involved in cellular processes that promote cell proliferation and survival. Phosphorylation of the α subunit of the eukaryotic translation initiation factor eIF2 at Ser51 takes place in response to various types of environmental stress and is essential for regulation of translation initiation. Herein, we show that a conditionally active form of the eIF2α kinase PKR acts upstream of PI3K and turns on the Akt/PKB-FRAP/mTOR pathway leading to S6 and 4E-BP1 phosphorylation. Also, induction of PI3K signaling antagonizes the apoptotic and protein synthesis inhibitory effects of the conditionally active PKR. Furthermore, induction of the PI3K pathway is impaired in PKR−/− or PERK−/− mouse embryonic fibroblasts (MEFs) in response to various stimuli that activate each eIF2α kinase. Mechanistically, PI3K signaling activation is indirect and requires the inhibition of protein synthesis by eIF2α phosphorylation as demonstrated by the inactivation of endogenous eIF2α by small interfering RNA or utilization of MEFs bearing the eIF2α Ser51Ala mutation. Our data reveal a novel property of eIF2α kinases as activators of PI3K signaling and cell survival.
INTRODUCTION
The phosphoinositide-3 kinase (PI3K) pathway plays a central role in the transduction of signals from extracellular stimuli such as growth factors, hormones, mitogens, and cytokines to cellular pathways controlling cell growth, proliferation, and survival. The PI3Ks are lipid kinases that generate second messengers by phosphorylating the phosphatidyl group at the 3′ position of the inositol ring (Vivanco and Sawyers, 2002; Engelman et al., 2006). Phosphorylated lipids subsequently recruit proteins containing a pleckstrin homology (PH) domain to the inner leaflet of the cell membrane. An important effector of the PI3K pathway, the serine/threonine kinase Akt/PKB, interacts with phosphorylated lipids through its PH domain. Upon recruitment to the membrane, Akt/PKB is phosphorylated at threonine (Thr) 308 by PDK1 and at serine (Ser) 473 by the FKBP and Rapamycin-associated protein (FRAP)/mTOR-Rictor complex (Scheid and Woodgett, 2003; Hresko and Mueckler, 2005; Sarbassov et al., 2005). The resultant activation of Akt/PKB leads to the induction of cell growth and survival by modulating the function of proteins involved in transcription and translation. Active Akt/PKB dissociates from the membrane and localizes to the cytoplasm and nucleus where it phosphorylates downstream targets such as glycogen synthase kinase-3 (GSK3), procaspase-9, forkhead transcription factors (FKHR, FKHRL1, AFX), IKKα, Ask1, BAD, CREB, and Mdm2. By modulating the activity of these proteins, Akt plays a major role in regulating cellular processes such as proliferation and apoptosis (Franke et al., 2003).
The mammalian target of rapamycin (mTOR) also known as FRAP, belongs to the PI3K-related kinases (PIKK) family of kinases (Abraham, 2004), and its activation in response to growth factors is regulated by Akt/PKB. The best characterized downstream targets of FRAP/mTOR are proteins involved in translation. Activation of translation initiation factors eIF4E, eIF4G, eIF4A, and eIF4B is regulated directly or indirectly by FRAP/mTOR (Gingras et al., 2001b; Shahbazian et al., 2006; Dorrello et al., 2006). Moreover, the S6 Kinase (S6K) and its substrate S6 ribosomal protein, as well as the elongation factor 2 (eEF2), are also targets of this pathway (Hay and Sonenberg, 2004). FRAP/mTOR-mediated regulation of eIF4E is exerted through the phosphorylation of 4E-binding proteins (4E-BPs). Activation of FRAP/mTOR leads to the phosphorylation of 4E-BP1 at its two priming sites: Thr37 and Thr46. These phosphorylations are required for subsequent phosphorylation of 4E-BP1 on Thr 70 and Ser65, which ultimately results in its dissociation from eIF4E (Gingras et al., 2001a).
Metazoans respond to stress signals in part by inhibiting cellular protein synthesis to provide cells with the means to restore a healthy state or by inducing apoptosis if the damage is beyond repair. An important pathway involved in this response is the eIF2α phosphorylation pathway. Phosphorylation of eIF2α at Ser51 by members of the eIF2α kinase family results in inhibition of translation initiation by reducing the levels of functional eIF2B (Dever, 2002). This affects the global rate of protein synthesis, but it can also selectively inhibit translation of specific mRNAs that have greater requirement for active eIF2. To date, there are four distinct eIF2α kinases with unique ability to respond to various stress conditions (Dever, 2002). These kinases share a catalytic domain containing conserved subdomains characteristic of all Ser/Thr protein kinases, but possess highly divergent regulatory domains. Specifically, these kinases show significant homology in subdomain V of the catalytic domain, which may serve as the substrate binding domain (Clemens and Elia, 1997). These kinases include the heme-regulated inhibitor (HRI; Chen, 2000), the general control nonderepressible-2 (GCN2; Kimball and Jefferson, 2000), the protein kinase activated by double-stranded RNA (PKR; Kaufman, 2000), and the endoplasmic reticulum (ER) resident protein kinase (PERK; Ron and Harding, 2000).
Herein, we demonstrate the ability of eIF2α kinases to induce the PI3K pathway, which leads to the activation of FRAP/mTOR and phosphorylation of S6 and 4E-BP1. Our findings reveal a novel role of the eIF2α kinases as regulators of PI3K signaling with implications in translational control and apoptosis.
MATERIALS AND METHODS
Plasmids and Viruses
The construction of GyrB.PKR cDNAs in the pSG5 vector (Stratagene, La Jolla, CA) was previously described (Ung et al., 2001). pCMV-FLAG wild-type PTEN construct was kindly provided by Dr. Georgescu (University of Texas) (Georgescu et al., 1999). Adenoviruses expressing a dominant negative mutant of the p85 subunit of PI3K were kindly provided by Dr. K. Mossman (McMaster University).
Cell Culture, Transfections, and Treatments
Isogenic mouse embryonic fibroblasts (MEFs) from PKR+/+ and PKR−/− mice (Abraham et al., 1999) in 129SvEv background (Durbin et al., 2002) were immortalized based on the standard NIH3T3 protocol. Isogenic PERK+/+ and PERK−/− MEFs (Harding et al., 2000) were immortalized by SV40 infection. Immortalized eIF2 A/A and S/S MEFs were generated as described (Scheuner et al., 2001). Generation and characterization of GyrB.PKR and GyrB.PKRK296H-expressing cells was described previously (Kazemi et al., 2004). GyrB.PKRT487D was generated by site-directed mutagenesis, and HT1080 cells expressing the mutant protein were established as described for GyrB.PKR and GyrB.PKRK296H (Kazemi et al., 2004). HT1080 cells and MEFs were maintained in DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% heat-inactivated calf serum and antibiotics (penicillin-streptomycin, 100 U/ml; ICN Biomedicals, Costa Mesa, CA). For the eIF2α S/S and A/A MEFs, the medium was supplemented with 10% non-heat-inactivated calf serum, antibiotics, 1× essential (Invitrogen) and 1× nonessential amino acids (Invitrogen; Scheuner et al., 2001). Treatment with coumermycin (Biomol, Plymouth Meeting, PA) was performed at a concentration of 100 ng/ml. Rapamycin (Bioshop, Burlington, ON, Canada), LY294002 (Sigma, St. Louis, MO), Wortmannin (Bioshop) and Sal003 (Robert et al., 2006) were used at concentrations of 20 nM, 20 μM, 100 nM, and 75 μM, respectively. Cycloheximide (CHX) and thapsigargin (TG) treatment were performed at a concentration of 20 μg/ml and 1 μM, respectively. Interferon (IFN) γ (Cedarlane, Hornby, ON, Canada) treatment was performed at a concentration of 100 IU/ml. To minimize possible side-effects of serum, the majority of the experiments examining PI3K pathway were performed in cells deprived from serum for 16 h as indicated in the figure legends.
Protein Extraction and Immunoblotting
Cells were washed twice with ice-cold phosphate-buffered saline (PBS) solution (140 mM NaCl, 15 mM KH2PO4, pH 7.2, 2.7 mM KCl), and proteins were extracted in ice-cold lysis buffer containing 10 mM Tris-HCl (pH 7.5), 50 mM KCl, 2 mM MgCl2, 1% Triton X-100, 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF), 0.1 mM Na3VO4, 3 μg/ml aprotinin, 1 μg/ml leupeptin, and 1 μg/ml pepstatin. After incubation on ice for 20 min, lysates were centrifuged at 16,000 × g for 10 min at 4°C. Supernatants were transferred to a fresh tube, and the protein concentration was measured by Bradford assay (Bio-Rad, Richmond, CA). Samples were stored at −80°C.
For immunoblotting, 50 μg of protein extracts were subjected to SDS-PAGE and transferred to polyvinylidene difluoride membranes (PVDF, Immobilon-P; Millipore, Bedford, MA). Immunoblotting was then performed according to the standard protocol (Sambrook et al., 1989). The primary antibodies were as follows: phosphospecific antibodies against 4E-BP1-pThr37/46, -pSer65, and -pThr70 (Cell Signaling, Beverly, MA; 9459, 9451, 9455; 1 μg/ml), anti-4E-BP1 rabbit polyclonal antibody (clone 11209; 1 μg/ml; Gingras et al., 1999a), anti-S6-pSer235/236 rabbit polyclonal antibody (Cell Signaling; 2211; 1 μg/ml), anti-S6 rabbit polyclonal antibody (Cell Signaling; 2212; 1 μg/ml), rabbit serum to phosphoserine 51 of eIF2α (Biosource, Carlsbad, CA; 44-728; 1 μg/ml), anti-eIF2α rabbit polyclonal antibody (FL-315, Santa Cruz Biotechnology, Santa Cruz, CA; sc-11386; 1 μg/ml), anti-Akt-pSer473 rabbit polyclonal antibody (Cell Signaling; 9271; 1 μg/ml), anti-Akt-pThr308 rabbit polyclonal antibody (Cell Signaling; 4056; 1 μg/ml), anti-Akt rabbit polyclonal antibody (Cell Signaling; 9272; 1 μg/ml), anti-GSK3α/β-pSer21/9 rabbit polyclonal antibody (Cell Signaling; 9331; 1 μg/ml), anti-GSK3β rabbit polyclonal antibody (Cell Signaling; 9332; 1 μg/ml), anti-PI3K p85 antibody (Upstate Biotechnology, Lake Placid, NY; 06-195, 1 μg/ml), anti-FLAG antibody (Sigma; F3165; 1 μg/ml), anti-GyrB antibody (7D3, John Innes Centre, Cloney, Norwich, United Kingdom) and anti-actin mouse monoclonal IgG (ICN Biomedicals, Solon, OH; 69100; 0.1 μg/ml). The secondary antibodies were horseradish peroxidase (HRP)-conjugated anti-mouse IgG antibody or HRP-conjugated anti-rabbit IgG antibody (Amersham Pharmacia Biotech, Piscataway, NJ; dilution 1:1000). Proteins were visualized using the enhanced chemiluminescence (ECL) detection system according to the manufacturer's instruction (Perkin Elmer Life Sciences, Boston, MA). Quantification of the bands was done using Scion Image 4.0.3.2 software (Frederick, MD). We quantified the ratio of phosphorylated to total protein. The basal level for each cell line is considered as 1.
PI3K Lipid Kinase Assay
The PI3K assay was performed as described (Naga Prasad et al., 2002) with some modifications. Briefly, cells were subjected to the indicated treatments and proteins were extracted in lysis buffer containing 1% Nonidet P-40, 10% glycerol, 137 mM NaCl, 20 mM Tris-HCl pH 7.4, 1 mM sodium orthovanadate, 1 mM PMSF, 20 mM NaF, 1 mM sodium pyrophosphate, and 2 μg/ml leupeptin and aprotinin. Protein extracts (500 μg) were subjected to immunoprecipitation with anti-PI3K p85 (Upstate Biotechnology, Charlottesville, VA) in the presence of protein A-agarose beads. The samples were centrifuged at 13,000 rpm, and sedimented beads were washed once with lysis buffer and twice with PBS containing 1% NP-40 and 100 μM sodium orthovanadate, three times with 100 mM Tris-HCl, pH 7.4, containing 5 mM LiCl and 100 μM sodium orthovanadate, twice with TNE (10 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA, and 100 μM sodium orthovanadate). The last traces of buffer were completely removed and the pelleted beads were resuspended in 50 μl fresh TNE. To the resuspended pellet, we added 10 μl of 100 mM MgCl2 and 20 μg of l-α-phosphatidylinositol (PI; Jena Biosciences, Jena, Germany) that was previously sonicated in a water bath sonicator for 1 h. The reactions were initiated by adding 10 μCi [γ-32P]ATP and were incubated at 23°C for 15 min with continuous agitation. The reaction was stopped with 20 μl 6 N HCl. Extraction of the lipids was performed by adding 160 μl of chloroform:methanol (1:1), and the samples were vortexed and centrifuged at room temperature to separate the phases. The lower organic phase (30 μl) was spotted onto silica-coated glass thin-layer chromatography (TLC) plates (Sigma Aldrich) precoated with 1% potassium oxalate. The spots were allowed to dry and resolved chromatographically with 2 M acetic acid/isopropanol (1:2). The plates were dried and exposed to film, and the autoradiographic signals were quantified using Scion Image 4.0.3.2 software. The lipid standards were run as a separate lane on the TLC plate to identify the migration of phosphatidylinositol-4,5-phosphate (PIP; Echelon Biosciences, Salt Lake City, UT). TLC plates were stained with iodine to identify the formation of phosphorylated lipid products.
RNA Interference
For small interfering RNA (siRNA) transfection, 1 × 105 cells were seeded in 6-cm plates. The following day, cells were transfected with 200 pmol siRNA for human PIK3R1 (Dharmacon, Boulder, CO), eIF2α (Dharmacon), luciferase reporter gene (Dharmacon), or scrambled RNA (SCR) using 4 μl LipofectAmine 2000 (Invitrogen, Carlsbad, CA) in medium lacking serum. Six hours after transfection, the plates were washed with serum-free DMEM and replenished with medium containing 10% serum. Cells were incubated at 37°C for an additional 72 h before being treated with coumermycin.
Transient Transfections
Cells (6 × 105) seeded in 6-cm plates were incubated with 4 μl Lipofectamine (Invitrogen) and 5 μg of vector DNA in serum-free medium at 37°C for 5 h. The medium was then replenished with 10% serum, and cells were incubated for an additional 24 h before coumermycin treatment.
Adenovirus Infection
Cells (2 × 105) seeded in 6-cm were infected with control adenovirus (Ad BHGdelE1,E3) or dominant negative p85 expressing adenovirus (Ad5dnp85; MOI: 500) in serum-free medium. Cells were incubated at 37°C for 24 h before being treated with coumermycin.
[35S]Methionine Labeling of Cells
Metabolic [35S]methionine labeling, trichloroacetic acid (TCA) precipitation, and counting were performed as described (Kazemi et al., 2004).
Cell Staining and Flow Cytometry Analysis
Cells were subjected to propidium iodide (PI) staining and flow cytometry analysis as previously described (Kazemi et al., 2004).
Double-Strand RNA Transfection
Transfection with double-strand RNA (dsRNA) was performed as described (Baltzis et al., 2002). Briefly, cells (8 × 105) seeded in 10-cm plates were incubated with 10 μg/ml poly(I)-poly(C) RNA, 12 μl Plus reagent, and 20 μl Lipofectamine (Invitrogen) in 2.7 ml serum-free medium at 37°C for 1 h. The medium was then replenished with 10% serum, and cells were incubated for the indicated time points.
RESULTS
A Conditional Form of PKR Induces the Phosphorylation of Components of the PI3K Pathway
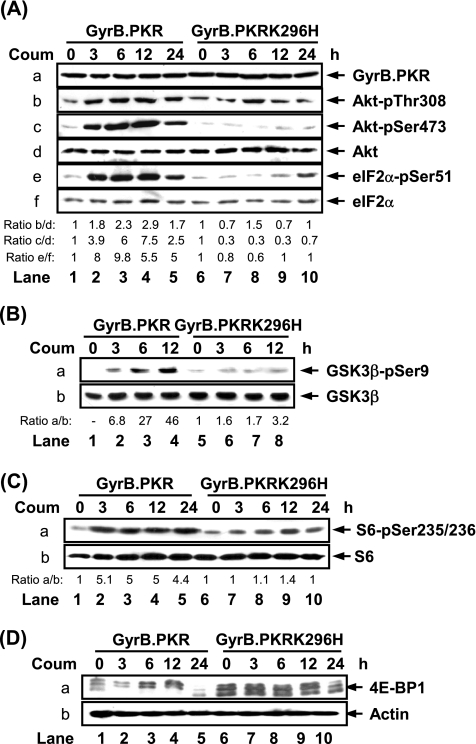
We previously reported the function of a conditionally active form of PKR consisting of the first 220 amino acids (aa) of the bacterial GyrB protein fused to the catalytic domain of the human kinase (GyrB.PKR; Ung et al., 2001). We showed that conditional activation of GyrB.PKR by the antibiotic coumermycin in human fibrosarcoma HT1080 cells leads to eIF2α phosphorylation, inhibition of global protein synthesis, and induction of apoptosis (Kazemi et al., 2004). While investigating the signaling properties of GyrB.PKR expressing cells, we observed that coumermycin treatment resulted in Akt/PKB phosphorylation at Thr308 and Ser473 (Figure 1A, b and c, lanes 1–5). This was consistent with the activation of GyrB.PKR because phosphorylation of eIF2α at Ser51 was induced at 3 h, reaching its peak at 6 h after coumermycin treatment (Figure 1A, e, lanes 1–5). Treatment of cells expressing the catalytically inactive GyrB.PKRK296H (Ung et al., 2001), which is expressed at equal levels to GyrB. PKR (Figure 1A, a, compare lanes 6–10 to 1–5; Kazemi et al., 2004), with coumermycin did not induce Akt/PKB or eIF2α phosphorylation (Figure 1A, b, c, and e, lanes 6–10). Consistent with the phosphorylation of Akt/PKB, activation of GyrB.PKR by coumermycin resulted in the phosphorylation of GSK3β at Ser9 (Figure 1B, a), a downstream target of active Akt/PKB.
Figure 1.
Conditional activation of GyrB.PKR induces the phosphorylation and activation of Akt/PKB. (A–D) Protein extracts (50 μg) from serum-deprived HT1080 cells expressing either GyrB.PKR or GyrB.PKRK296H, untreated or treated with coumermycin (Coum; 100 ng/ml) for up to 12 or 24 h, were subjected to immunoblotting with antibodies against the indicated proteins. The ratio of phosphorylated to total protein is indicated. The ratio was set to 1 for each cell line in the absence of coumermycin treatment. The data represent one of four reproducible experiments.
Two downstream targets of the PI3K pathway are the S6 ribosomal protein and 4E-BP1. Phosphorylation of S6 at Ser235/236 was induced by activated GyrB.PKR (Figure 1C, a). Induction of both Akt/PKB and S6 phosphorylation in GyrB.PKR cells occurred in a time-dependent manner and was reduced after 24 h of coumermycin treatment (lane 5 in Figure 1A, b and c, and C, a). Furthermore, activation of GyrB.PKR by coumermycin also led to the induction of 4E-BP1 phosphorylation as documented by the shift of the low-molecular-weight hypophosphorylated form to the higher molecular weight hyperphosphorylated forms of the protein (Figure 1D, a, lanes 1–5). This shift of hyperphosphorylated 4E-BP1 was not observed in cells containing the catalytically inactive mutant GyrB.PKRK296H (Figure 1D, a, lanes 6–10). Collectively, these data show that the catalytic activity of PKR is capable of inducing the phosphorylation of components of the PI3K pathway such as Akt/PKB, S6, and 4E-BP1.
The above experiments were performed in the absence of serum to diminish the background effects of growth factors on PI3K signaling in response to GyrB.PKR activation. Nevertheless, induction of Akt/PKB, S6 and 4E-BP1 phosphorylation was also observed in serum-repleted GyrB.PKR cells (Supplementary Figure 1A), whereas treatment of parental HT1080 cells with the antibiotic coumermycin did not induce the phosphorylation of Akt/PKB, S6, eIF2α, or 4E-BP1 (Supplementary Figure 1B). These data demonstrate the specificity of GyrB.PKR in mediating the phosphorylation of the above-mentioned proteins.
PKR Acts Upstream of PI3K To Mediate the Phosphorylation of 4E-BP1 and S6
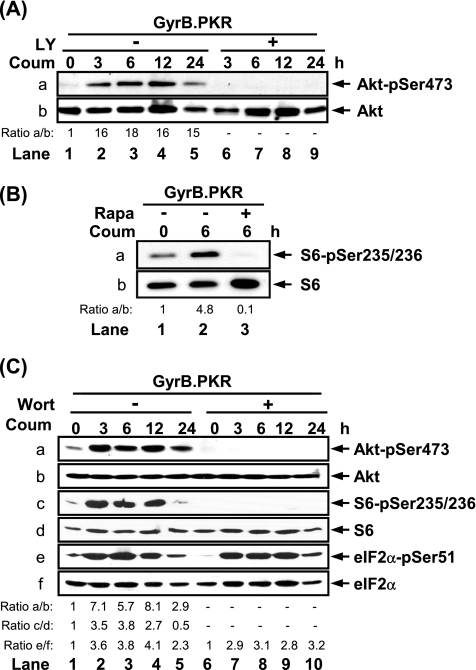
Next, we wanted to determine whether activated GyrB.PKR acts upstream of PI3K. To this end, we examined the effects of LY294002, an inhibitor of PI3K, and rapamycin, an inhibitor of FRAP/mTOR, on Akt/PKB and S6 phosphorylation, respectively. Induction of Akt/PKB phosphorylation at Ser473 in cells with activated GyrB.PKR was undetectable in the presence of LY294002 (Figure 2A, a), indicating that PKR functions upstream of PI3K. Similarly, induction of S6 phosphorylation by activated GyrB.PKR was not possible in the presence of rapamycin (Figure 2B, a), indicating that PKR acts upstream of the FRAP/mTOR kinase. To confirm these observations, we used wortmannin at a concentration of 100 nM to inhibit PI3K without affecting FRAP/mTOR activity. We observed that wortmannin prevented the induction of Akt/PKB and S6 phosphorylation in coumermycin-treated GyrB.PKR cells (Figure 2C, a and c) without affecting the induction of eIF2α phosphorylation by activated GyrB.PKR (Figure 2C, e). This data indicate that catalytically active PKR acts upstream to PI3K.
Figure 2.
GyrB.PKR acts upstream of PI3K. (A–C) Serum-starved HT1080 cells expressing GyrB.PKR were left untreated or treated with coumermycin (100 ng/ml) in the absence or presence of (A) LY294002 (LY; 20 μM), (B) rapamycin (Rapa; 20 nM), or (C) wortmannin (Wort; 100 nM) for the indicated times. Protein extracts (50 μg) were subjected to immunoblotting with antibodies against the indicated proteins. The data represent one of three reproducible experiments.
To further verify the above observations, we tested the lipid kinase activity of PI3K in GyrB.PKR cells. We observed that immunoprecipitated PI3K induced the phosphorylation of PIs, yielding a significant amount of PI(3)P product after GyrB.PKR activation compared with immunoprecipitated PI3K from GyrB.PKRK296H cells (Figure 3A, a). To verify whether PI3K activation was indeed responsible for Akt/PKB phosphorylation in GyrB.PKR cells, we performed several key experiments and found the following: First, that induction of Akt/PKB phosphorylation at Ser473 by GyrB.PKR was not possible after down-regulation of the p85 regulatory subunit of PI3K by siRNA (Figure 3B, a). We further observed that overexpression of a FLAG-tagged form of wild-type PTEN, a phosphatase that antagonizes the PI3K function by dephosphorylating PIP3 to PIP2, impaired the induction of Akt/PKB phosphorylation by activated GyrB. PKR (Figure 3C, a). Moreover, we observed that overexpression of a dominant negative mutant of the p85 subunit of PI3K prevented the induction of Akt/PKB phosphorylation at Ser473 by activated GyrB.PKR (Figure 3D, a). Blockage of PI3K activity by the above means did not interfere with GyrB.PKR activity as indicated by the induction of eIF2α phosphorylation in response to coumermycin treatment (Figure 3, C and D). Collectively, these data demonstrate that the catalytic activity of PKR acts as an activator of PI3K.
Figure 3.
Activation of PI3K by GyrB.PKR. (A) Serum-starved GyrB.PKR or GyrB.PKRK296H-expressing cells were left untreated or treated with coumermycin (100 ng/ml) for 6 h. Protein extracts (500 μg) were subjected to immunoprecipitation with an anti-PI3K p85 antibody followed by an in vitro lipid kinase assay in the presence of [32P-γ]ATP and phosphatidylinositol (PI) as a substrate. Radioactive PIP was visualized by TLC and autoradiography (a). P85 levels in the immunoprecipitates were detected by immunoblotting (b). The data represent one of two reproducible experiments. (B) GyrB.PKR-expressing cells were transiently transfected with scrambled control siRNA (SCR) or siRNA targeting the p85 subunit of PI3K for 72 h. (C) GyrB.PKR cells were transiently transfected with pCMV plasmid lacking or containing FLAG-tagged PTEN for 24 h. (D) GyrB.PKR cells were infected with adenovirus expressing a dominant negative mutant of the p85 subunit (dnp85) of PI3K or a control adenovirus for 24 h. (B–D) Transfected or infected cells were left untreated or treated with coumermycin (100 ng/ml) for 6 h, and protein extracts (30 μg) were subjected to immunoblotting for the indicated proteins. The data represent one of three reproducible experiments.
Induction of 4E-BP1 Phosphorylation by GyrB.PKR Cannot Rescue Protein Synthesis Inhibition by eIF2α Phosphorylation
Next, we were interested in determining whether induction of 4E-BP1 phosphorylation by GyrB.PKR was solely dependent on the activation of PI3K signaling. When GyrB.PKR cells were treated with LY294002 or rapamycin in the absence of coumermycin, we observed a reduction in the basal levels of phosphorylated 4E-BP1 detected by immunoblotting with phosphospecific antibodies (Figure 4A, a–c, lanes 3 and 5). Basal levels of 4E-BP1 phosphorylation were further reduced when both inhibitors were used in concert (Figure 4A, a–c, lane 7). The additive effects of both inhibitors indicated that PI3K-Akt/PKB and FRAP/mTOR form a branched rather than a linear pathway leading to 4E-BP1 phosphorylation in these cells. Also, phosphorylation of Thr70 and Ser65 was much more sensitive to inhibition by rapamycin than Thr37/46 phosphorylation as shown in previous studies (Mothe-Satney et al., 2000; Gingras et al., 2001a; Wang et al., 2005). When GyrB.PKR cells were treated with coumermycin in the presence of rapamycin, we observed that activated GyrB.PKR was partially capable of inducing the phosphorylation of 4E-BP1 at Thr37/46 and Ser65 (Figure 4, a and b, lane 4). This indicates that GyrB.PKR can affect the phosphorylation of these sites either independently of FRAP/mTOR or through a rapamycin-insensitive function of FRAP/mTOR as recently reported (Sarbassov et al., 2005; Wang et al., 2005). When the PI3K inhibitor LY294002 was used, we found that induction of 4E-BP1 phosphorylation at Thr37/46 by activated GyrB.PKR was not possible (Figure 4A, a, compare lanes 5 and 6), whereas phosphorylation of 4E-BP1 at Ser65 and Thr70 fell below detectable levels (Figure 4A, b and c, lanes 5 and 6). Similar results were obtained when both LY294002 and rapamycin were used in conjunction (Figure 4A, a–c, lanes 7 and 8). Immunoblot analysis with a pan-specific antibody against 4E-BP1 verified the lack of hyperphosphorylated forms of the protein in the presence of the inhibitors (Figure 4A, d) confirming the phosphorylation pattern of 4E-BP1 obtained with the phosphospecific antibodies. Significantly, the presence of LY294002 and/or rapamycin did not affect GyrB.PKR activity, because induction of eIF2α phosphorylation took place efficiently in cells treated with the inhibitors (Figure 4A, e, lanes 2, 4, 6, and 8). These data show that the ability of GyrB.PKR to induce the phosphorylation of 4E-BP1 mediated through PI3K and the downstream effector FRAP/mTOR.
Figure 4.
Regulation of 4E-BP1 phosphorylation and its implication in cap-dependent translation in GyrB.PKR cells. (A and B) Serum-deprived GyrB.PKR cells were left untreated or treated with coumermycin (100 ng/ml) for 6 h, in the absence or presence of rapamycin (20 nM) and/or LY294002 (20 μM), as indicated. (A) Protein extracts (50 μg) were subjected to immunoblotting with antibodies against the indicated proteins. The data represent one of three reproducible experiments. (B) Cells expressing either GyrB.PKR (□) or GyrB.PKRK296H (■) were incubated in media lacking methionine and supplemented with 10% dialyzed fetal bovine serum for 1 h. Cells were then treated with LY294002 or rapamycin for 1 h followed by the addition of coumermycin for 4 h. Subsequently, [35S]methionine was added to cells for a further 2 h followed by the quantification of radioactive TCA precipitates. (C, coumermycin; R, rapamycin; LY, LY294002). One hundred percent (%) protein synthesis represents the average value of 35S-labeled proteins in untreated GyrB.PKR or GyrB.PKRK296H-expressing cells. Values represent the average of three separate experiments performed in triplicate.
Given the important role of 4E-BP1 phosphorylation in the stimulation of cap-dependent translation (Gingras et al., 1999b), we wanted to examine whether induction of 4E-BP1 phosphorylation is capable of bypassing the inhibitory effects of eIF2α phosphorylation. To do so, we assessed the levels of global protein synthesis by [35S]methionine labeling of GyrB.PKR-expressing cells treated with LY294002 and/or rapamycin in the absence or presence of coumermycin (Figure 4B). We found that in cells with latent GyrB.PKR (i.e., without coumermycin treatment), inhibition of PI3K by LY294002 resulted in an approximate 40% decrease of global protein synthesis. In cells with activated GyrB.PKR (i.e., coumermycin-treated cells), cellular protein synthesis was inhibited by ∼50%, and was further reduced by an additional 40% upon inhibition of PI3K by LY294002. Treatment with LY294002 resulted in the same degree of protein synthesis inhibition in cells expressing GyrB.PKRK296H either in the absence or presence of coumermycin. Given that PI3K inhibition by LY294002 did not further enhance eIF2α phosphorylation by GyrB.PKR (Figure 4A), this indicates that the PI3K pathway counterbalances translation inhibition by the eIF2α kinases without interfering with its ability to phosphorylate eIF2α. Conversely, rapamycin treatment did not decrease the global inhibition of protein synthesis by activated GyrB.PKR (Figure 4B). Interestingly, we noticed that rapamycin treatment did not inhibit the overall levels of protein synthesis in cells with latent GyrB.PKR (Figure 4B) despite the significant decrease of phosphorylated 4E-BP1 (Figure 4A, a–c, compare lane 1 with 3). This indicates that inhibition of FRAP/mTOR pathway does not always exert global effects on protein synthesis. This is consistent with previous observations (Grolleau et al., 2002) and our data (not shown) that in some cells rapamycin treatment induces qualitative rather than quantitative effects on protein synthesis. Collectively, these data indicate that the antagonistic effects of the PI3K pathway on the inhibition of protein synthesis by activated GyrB.PKR proceeds independently of 4E-BP1 phosphorylation. Also, 4E-BP1 phosphorylation cannot bypass the inhibitory effects of eIF2α phosphorylation on translation initiation consistent with the notion that induction of eIF2α phosphorylation is dominant over the stimulatory activity of eIF4F in translation initiation.
The PI3K Pathway Antagonizes PKR-mediated Cell Death
We previously demonstrated that activation of GyrB.PKR leads to the induction of cell death as a result of inhibition of protein synthesis (Kazemi et al., 2004). Given the ability of GyrB.PKR to induce PI3K activity, we wanted to examine the role of the anti-apoptotic PI3K pathway in PKR-mediated cell death. To this end, we assessed the apoptotic function of GyrB.PKR in the presence of LY294002 or rapamycin (Figure 5). In the absence of coumermycin, treatment of GyrB.PKR cells with either rapamycin or LY294002 did not induce cell death. When cells were treated with coumermycin, activation of GyrB.PKR led to a significant induction of cell death as previously described (Kazemi et al., 2004). The presence of rapamycin, however, did not significantly affect cell death induced by GyrB.PKR as opposed to treatment with LY294002, which resulted in a considerable (∼50%) increase in GyrB.PKR-mediated cell death (Figure 5). These results demonstrate that activation of PI3K pathway antagonizes cell death induced by activated PKR independently of FRAP/mTOR activation and 4E-BP1 phosphorylation.
Figure 5.
Control of PKR-mediated apoptosis by PI3K pathway. HT1080 cells expressing GyrB.PKR were left untreated or treated with coumermycin (100 ng/ml) in the absence or presence of either LY294002 (20 μM) or rapamycin (20 nM) for 24 h. Cells were harvested, fixed in ethanol, stained with PI, and subjected to flow cytometry analysis. The percentage (%) of apoptotic cells or cells in various phases of the cell cycle is indicated. Data represent one of four reproducible experiments.
Biological Relevance of PI3K Activation by PKR
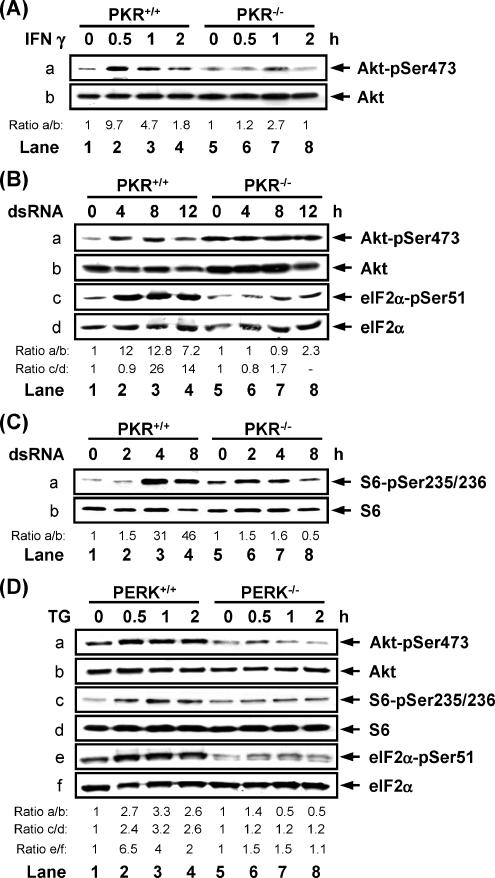
The role of eIF2α kinase activity in PI3K signaling was further addressed under more physiological settings. Previous findings provided evidence for the ability of IFNs to induce the phosphorylation of 4E-BP1 through activation of the PI3K pathway (Lekmine et al., 2003, 2004). Given that PKR is downstream of IFN signaling (Wong et al., 1997, 2001; Kumar et al., 1997; Wang et al., 2006), we addressed its possible role in the induction of PI3K signaling by IFNs using PKR+/+ and PKR−/− MEFs. We observed that treatment with IFN-γ resulted in higher levels of phosphorylated Akt/PKB in PKR+/+ MEFs than in PKR−/− MEFs (Figure 6A). When the same MEFs were subjected to dsRNA treatment, we found that a higher amount of Akt/PKB was phosphorylated at Ser473 in PKR+/+ MEFs than in PKR−/− MEFs (Figure 6B, a). We also observed induction of S6 phosphorylation in PKR+/+ MEFs that was not seen in PKR−/− MEFs (Figure 6C, a). Furthermore, treatment of the MEFs with wortmannin or rapamycin prevented Akt/PKB or S6 phosphorylation respectively upon dsRNA transfection (Supplementary Figure 2, A and B). These results clearly implicate PKR in the activation of PI3K under a physiological stimulation.
Figure 6.
PKR or PERK mediates the induction of PI3K pathway in response to IFN, dsRNA or ER stress respectively. (A) PKR+/+ and PKR−/− MEFs were treated with mouse IFN-γ (100 IU/ml) for the indicated time points. Protein extracts (50 μg) were subjected to immunoblotting against the indicated proteins. (B and C) PKR+/+ and PKR−/− MEFs were transfected with dsRNA (10 μg/ml) for the indicated time points. Protein extracts (50 μg) were subjected to immunoblotting against the indicated proteins. (D) PERK+/+ and PERK−/− MEFs were treated with thapsigargin (TG; 1 μM) in the presence of serum for the indicated time points. Protein extracts (50 μg) were subjected to immunoblotting with phosphospecific antibodies against the indicated proteins. (A–D) Data represent one of three reproducible experiments.
Next we sought evidence for a role of other eIF2α kinases in the induction of PI3K signaling. Accordingly, induction of ER stress by thapsigargin treatment resulted in the induction of both Akt/PKB and eIF2α phosphorylation in PERK+/+ MEFs but not in PERK−/− MEFs (Figure 6D, a and e). The higher levels of Akt/PKB phosphorylation were consistent with the higher induction of S6 phosphorylation in PERK+/+ cells than in PERK−/− cells in response to ER stress (Figure 6D, c). Moreover, infection of MEFs with adenovirus expressing dnp85 inhibited the induction of Akt/PKB phosphorylation in response to either dsRNA transfection or TG treatment (Supplementary Figure 2C). These data further confirm the role PKR and PERK in PI3K activation and provide evidence for rather a general role of eIF2α kinases in the induction of PI3K signaling.
Induction of PI3K Signaling by eIF2α Kinases Requires eIF2α Phosphorylation
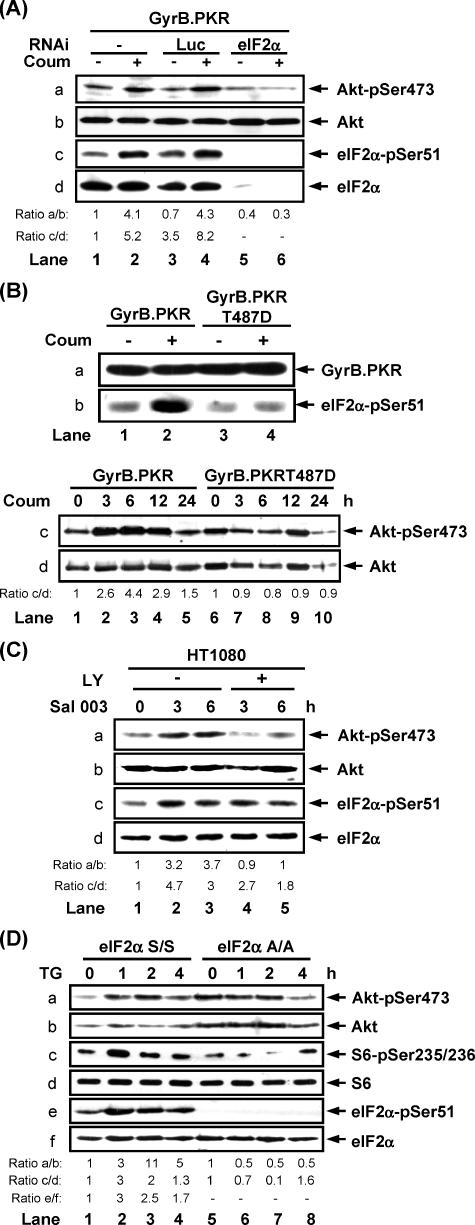
We next addressed the role of translational inhibition by eIF2α phosphorylation in the induction of PI3K signaling. When endogenous eIF2α in GyrB.PKR cells was targeted by siRNA, we observed that induction of Akt/PKB phosphorylation at Ser473 by coumermycin treatment was not possible as opposed to cells treated with an irrelevant control siRNA (Figure 7A). To further test the role of eIF2α phosphorylation, we expressed the GyrB.PKRT487D mutant in HT1080 cells. It was recently shown that introduction of T487D mutation in PKR abolishes its eIF2α kinase activity without affecting its capacity to autophosphorylate (Dey et al., 2005). Using cells expressing equal amounts of GyrB.PKR WT and GyB.PKRT487D (Figure 7B, a), we found that treatment with coumermycin resulted in the induction of Akt/PKB phosphorylation at Ser473 in GyrB.PKR cells but not in GyrB.PKRT487D cells (Figure 7B, c), consistent with the inability of the mutant kinase to phosphorylate eIF2α (Figure 7B, b). Furthermore, induction of eIF2α phosphorylation in cells treated with Sal003, which is a potent inhibitor of eIF2α dephosphorylation (Boyce et al., 2005; Robert et al., 2006), resulted in the induction of both Akt/PKB and eIF2α phosphorylation (Figure 7C, a and c, lanes 1–3). However, when cells were pretreated with LY294002 to inhibit the PI3K pathway, the induction of Akt/PKB phosphorylation by Sal003 was compromised (Figure 7C, a, lanes 4 and 5), showing that induction of Akt/PKB phosphorylation as a result of eIF2α phosphorylation requires PI3K activity. Finally, Akt/PKB phosphorylation was induced in eIF2α S/S MEFs treated with thapsigargin but not in eIF2α A/A MEFs that received the same treatment (Figure 7D, a). The higher background phosphorylation of Akt/PKB in untreated eIF2α A/A cells was reproducibly observed in these cells for reasons that are not immediately clear. In the same experiment we observed induction in S6 phosphorylation in eIF2α S/S MEFs, which was not observed in eIF2α A/A cells (Figure 7D, c). Moreover, dsRNA transfection resulted in a higher induction of Akt/PKB and S6 phosphorylation in eIF2α S/S than in eIF2α A/A MEFs (Supplementary Figure 3A). Furthermore, treatment of the same cells with either wortmannin or rapamycin prevented the induction of Akt/PKB and S6 phosphorylation upon TG treatment (Supplementary Figure 3, B and C). Collectively, these data substantiate the involvement of eIF2α phosphorylation in the induction of PI3K signaling.
Figure 7.
Induction of PI3K signaling by eIF2α kinases requires eIF2α phosphorylation. (A) GyrB.PKR cells were left untransfected or transfected with siRNA for the luciferase reporter gene (Luc; negative control) or siRNA for eIF2α for 72 h followed by treatment with 100 ng/ml coumermycin for 6 h. (B) GyrB.PKR and GyrB.PKRT487D cells were left untreated or treated with coumermycin for the indicated times. (C) HT1080 cells were left untreated or pretreated with 20 μM LY294002 for 1 h before treatment with 75 μM Sal003 for the indicated times. (D) eIF2α S/S and eIF2α A/A MEFs were left untreated or treated with thapsigargin (TG; 1 μM) for indicated times. (A–D) Protein extracts (50 μg) were subjected to immunoblotting against the indicated proteins. The data represents one out of three reproducible experiments.
DISCUSSION
In this study we demonstrate that activation of the eIF2α kinases results in induction of the PI3K signaling pathway. We have used the conditionally active GyrB.PKR to demonstrate that the catalytic activity of eIF2α kinases acts to induce PI3K activity, both in vivo and in vitro (Figures 2 and 3). Our data indicates that activation of GyrB.PKR results in induction of PI3K lipid kinase activity (Figure 3A) and protein kinase activity (data not shown). We further demonstrate that induction of PI3K activity by eIF2α kinases is responsible for the subsequent activation of Akt/PKB and FRAP/mTOR and phosphorylation of their downstream targets such as GSK3β, S6, and 4E-BP1 in response to GyrB.PKR activation (Figure 1). Induction of PI3K signaling by GyrB.PKR is an intracellular effect and does not require the secretion of either a growth factor or a cytokine that functions in an autocrine manner, as demonstrated by the inability of conditional media from GyrB.PKR cells to induce the PI3K pathway in parental HT1080 cells (data not shown).
Our data indicate that eIF2α phosphorylation is essential for activation of the PI3K pathway by eIF2α kinases (Figure 7 and Supplementary Figure 3). Because phosphorylation of eIF2α results in inhibition of translation, through inactivation of eIF2B, the most conceivable interpretation is that eIF2α kinases function indirectly through the translational regulation of a factor(s), which is involved in PI3K activation. In fact, treatment of GyrB.PKR cells with the protein synthesis inhibitor CHX caused the induction of Akt/PKB phosphorylation at Ser473 as previously reported (Beugnet et al., 2003) at the same levels as the activation of GyrB.PKR (Supplementary Figure 4). Given that concomitant CHX treatment and conditional activation of GyrB.PKR did not further enhance Akt/PKB phosphorylation (Supplementary Figure 4), the most conceivable interpretation is that inhibition of protein synthesis by eIF2α phosphorylation blocks the synthesis of a protein(s) that negatively regulates PI3K activity (for a model see Figure 8). PTEN is an example of an inhibitor of PI3K pathway that is regulated at the translational level (Han et al., 2003). However, in our studies we did not observe any changes in expression levels of PTEN upon activation of PKR (data not shown). Ruk, also known as CIN85 or SETA, is an adaptor-type protein belonging to the CD2AP/CMS family, which functions as an inhibitor of PI3K (Gout et al., 2000; Verdier et al., 2002). Ruk interaction with the p85 subunit of PI3K requires the proline-rich domain of Ruk and the SH3 domain of P85 (6732). Therefore, the possibility remains that Ruk itself or a Ruk cofactor (Schmidt et al., 2003) is sensitive to translational inhibition by eIF2α kinases. It is also possible that the negative regulator(s) act at a level upstream of PI3K by affecting the activity of Src, Ras, or other small GTP-binding proteins that are known to induce PI3K signalings (Cuevas et al., 2001; Chan et al., 2002). Previous data provided evidence that HT1080 cells contain an active form of N-Ras (Gupta et al., 2001). However, the ability of GyrB.PKR to induce PI3K signaling proceeds independently of N-Ras as indicated by binding assays of the activated forms of Ras to c-Raf1 (data not shown). Further analyses are required to identify the target of eIF2α phosphorylation pathway, which activates the PI3K signaling pathway.
Figure 8.
Model of PI3K activation by eIF2α kinases. Activation of the eIF2α kinases (namely PERK or PKR) leads to the translational inhibition of a protein (X) that negatively regulates PI3K activity. This leads to the activation of the downstream components of PI3K signaling as documented in this article. As explained in the Discussion, the negative regulator may act either directly on PI3K or indirectly by suppressing the activity of upstream activators of PI3K.
With regard to mRNA translation, our work shows that eIF2α phosphorylation exerts a dominant inhibitory effect on global protein synthesis regardless of 4E-BP1 phosphorylation. Specifically, inhibition of 4E-BP1 phosphorylation by rapamycin did not further decrease the translation inhibition induced by activated GyrB.PKR (Figure 4B). On the other hand, overexpression of eIF4E in GyrB.PKR cells was unable to overcome the translational inhibitory effects of activated GyrB.PKR (data not shown). Unlike rapamycin, treatment of cells with LY294002 further decreased the overall levels of protein synthesis in response to GyrB.PKR activation (Figure 4B), indicating that PI3K signaling counterbalances the translation inhibitory effects of active PKR independently of 4E-BP1 phosphorylation. Significantly, inhibition of the PI3K pathway by LY294002 treatment did not affect eIF2α phosphorylation by activated GyrB.PKR (Figure 4A), providing evidence that the antagonistic effect of PI3K signaling on GyrB.PKR-mediated translation inhibition is eIF2α phosphorylation independent.
Concerning the biological relevance of our findings, activation of the PI3K pathway serves as a negative feedback mechanism impeding GyrB.PKR-mediated apoptosis (Figure 5). This anti-apoptotic function of PI3K is not exerted at the translational level through 4E-BP1 phosphorylation because treatment with rapamycin, which blocks the phosphorylation of 4E-BP1 (Figure 4A), did not increase GyrB.PKR-mediated apoptosis (Figure 5). Consistent with this, overexpression of eIF4E was not able to rescue GyrB.PKR-dependent apoptosis (data not shown). Thus, inhibition of PKR-dependent apoptosis by PI3K may involve a novel translational pathway that proceeds independently of 4E-BP1 and eIF2α phosphorylation. An alternative but not mutually exclusive possibility is that posttranslational regulation of anti-apoptotic or proapoptotic proteins through Akt/PKB-mediated phosphorylation results in their activation or inactivation, respectively, and thus exerts an antagonistic effect on PKR-mediated apoptosis (Franke et al., 2003; Downward, 2004).
Our findings also provide strong evidence for a physiologically relevant role of PKR in PI3K activation. Specifically, we show that PKR is required for the induction of Akt/PKB phosphorylation in response to either IFN-γ (Figure 6A) or dsRNA treatment (Figure 6B), both of which induce the PI3K pathway (Platanias, 2005; Sen and Sarkar, 2005). At first glance, the ability of PKR to mediate the induction of PI3K signaling in IFN- or dsRNA-treated cells does not reconcile with its well-established anti-proliferative and proapoptotic properties resulting from global inhibition of protein synthesis (Su et al., 2006). However, induction of PI3K signaling by PKR may provide a window for the efficient expression of genes required for mounting an antiviral response before the general shut-off of protein synthesis resulting from eIF2α phosphorylation. It is interesting to note that induction of PI3K signaling is not specific for PKR but appears to be a property shared by other eIF2α kinases. That is, inhibition of protein synthesis by activation of the ER-resident eIF2α kinase PERK also leads to the induction of the PI3K pathway as observed by the increased Akt/PKB phosphorylation in PERK+/+ MEFs but not in PERK−/− MEFs (Figure 6D). Induction of PI3K signaling by ER stress may be a cytoprotective mechanism that mediates the cellular adaptive response to stress (Schroder and Kaufman, 2005) with possible important implications in regulation of protein synthesis as a result of PERK activation. It is of interest that although both PERK and PKR can induce PI3K signaling, each kinase does so only in response to treatment that is specific for their activation (data not shown).
In conclusion, we show that induction of eIF2α kinase activity results in a functional interplay between PI3K and translational control via eIF2α phosphorylation. This is in line with other findings that demonstrate the ability of PKR to act as a molecular clock by temporally inducing cell survival by activating the anti-apoptotic NF-κB (nuclear factor κB) before the induction of cell death by eIF2α phosphorylation (Donze et al., 2004).
Supplementary Material
ACKNOWLEDGMENTS
We thank Ana Maria Rivas-Estillas, Like Qu, Qiaozhu Su, S. Huang, and S. Papadopoulou for help with some experiments; N. Sonenberg for helpful discussions; H. Harding (New York University School of Medicine) and D. Ron (New York University School of Medicine) for PERK+/+ and PERK−/− MEFs; R. Kaufman (University of Michigan Medical School) for providing the eIF2α S/S and A/A MEFs; M. Georgescu for PTEN cDNA; K. Mossman for adenoviruses containing the p85 cDNA; and Russell and Joan Durbin (The Ohio State University) for PKR+/+ and PKR−/− MEFs. This work has been supported by a grant from the Canadian Institutes of Health Research (CIHR) to A.E.K. and J.P. S.K. and D.B. are research students of the Terry Fox Foundation through awards from the National Cancer Institute of Canada (NCIC). Z.M. is the recipient of a Canada Graduate Doctoral Award administered by CIHR and a CIHR predoctoral award. J.F.R. is an awardee of a U.S. Army Breast Cancer Research Predoctoral Traineeship. O.P. is a postdoctoral researcher of the Terry Fox Foundation through an award from the NCIC.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-01-0053) on June 27, 2007.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Abraham N., et al. Characterization of transgenic mice with targeted disruption of the catalytic domain of the double-stranded RNA-dependent protein kinase, PKR. J. Biol. Chem. 1999;274:5953–5962. doi: 10.1074/jbc.274.9.5953. [DOI] [PubMed] [Google Scholar]
- Abraham R. T. PI 3-kinase related kinases: ‘big’ players in stress-induced signaling pathways. DNA Repair (Amst.) 2004;3:883–887. doi: 10.1016/j.dnarep.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Baltzis D., Li S., Koromilas A. E. Functional characterization of pkr gene products expressed in cells from mice with a targeted deletion of the N terminus or C terminus domain of PKR. J. Biol. Chem. 2002;277:38364–38372. doi: 10.1074/jbc.M203564200. [DOI] [PubMed] [Google Scholar]
- Beugnet A., Tee A. R., Taylor P. M., Proud C. G. Regulation of targets of mTOR (mammalian target of rapamycin) signalling by intracellular amino acid availability. Biochem. J. 2003;372:555–566. doi: 10.1042/BJ20021266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce M., et al. A selective inhibitor of eIF2alpha dephosphorylation protects cells from ER stress. Science. 2005;307:935–939. doi: 10.1126/science.1101902. [DOI] [PubMed] [Google Scholar]
- Chan T. O., Rodeck U., Chan A. M., Kimmelman A. C., Rittenhouse S. E., Panayotou G., Tsichlis P. N. Small GTPases and tyrosine kinases coregulate a molecular switch in the phosphoinositide 3-kinase regulatory subunit. Cancer Cell. 2002;1:181–191. doi: 10.1016/s1535-6108(02)00033-8. [DOI] [PubMed] [Google Scholar]
- Chen J. J. Heme-regulated EIF2alpha kinase. In: Sonenberg N., Hershey J.W.B., Mathews M. B., editors. Translational Control of Gene Expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. pp. 529–546. [Google Scholar]
- Clemens M. J., Elia A. The double-stranded RNA-dependent protein kinase PKR: structure and function. [Review] J. Interf. Cytokine Res. 1997;17:503–524. doi: 10.1089/jir.1997.17.503. [DOI] [PubMed] [Google Scholar]
- Cuevas B. D., Lu Y., Mao M., Zhang J., LaPushin R., Siminovitch K., Mills G. B. Tyrosine phosphorylation of p85 relieves its inhibitory activity on phosphatidylinositol 3-kinase. J. Biol. Chem. 2001;276:27455–27461. doi: 10.1074/jbc.M100556200. [DOI] [PubMed] [Google Scholar]
- Dever T. E. Gene-specific regulation by general translation factors. Cell. 2002;108:545–556. doi: 10.1016/s0092-8674(02)00642-6. [DOI] [PubMed] [Google Scholar]
- Dey M., Cao C., Dar A. C., Tamura T., Ozato K., Sicheri F., Dever T. E. Mechanistic link between PKR dimerization, autophosphorylation, and eIF2alpha substrate recognition. Cell. 2005;122:901–913. doi: 10.1016/j.cell.2005.06.041. [DOI] [PubMed] [Google Scholar]
- Donze O., Deng J., Curran J., Sladek R., Picard D., Sonenberg N. The protein kinase PKR: a molecular clock that sequentially activates survival and death programs. EMBO J. 2004;23:564–571. doi: 10.1038/sj.emboj.7600078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrello N. V., Peschiaroli A., Guardavaccaro D., Colburn N. H., Sherman N. E., Pagano M. S6K1- and beta-TRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science. 2006;314:467–471. doi: 10.1126/science.1130276. [DOI] [PubMed] [Google Scholar]
- Downward J. PI 3-kinase, Akt and cell survival. Semin. Cell Dev. Biol. 2004;15:177–182. doi: 10.1016/j.semcdb.2004.01.002. [DOI] [PubMed] [Google Scholar]
- Durbin R. K., Mertz S. E., Koromilas A. E., Durbin J. E. PKR protection against intranasal vesicular stomatitis virus infection is mouse strain dependent. Viral Immunol. 2002;15:41–51. doi: 10.1089/088282402317340224. [DOI] [PubMed] [Google Scholar]
- Engelman J. A., Luo J., Cantley L. C. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat. Rev. Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- Franke T. F., Hornik C. P., Segev L., Shostak G. A., Sugimoto C. PI3K/Akt and apoptosis: size matters. Oncogene. 2003;22:8983–8998. doi: 10.1038/sj.onc.1207115. [DOI] [PubMed] [Google Scholar]
- Georgescu M. M., Kirsch K. H., Akagi T., Shishido T., Hanafusa H. The tumor-suppressor activity of PTEN is regulated by its carboxyl-terminal region. Proc. Natl. Acad. Sci. USA. 1999;96:10182–10187. doi: 10.1073/pnas.96.18.10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras A. C., Gygi S. P., Raught B., Polakiewicz R. D., Abraham R. T., Hoekstra M. F., Aebersold R., Sonenberg N. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 1999a;13:1422–1437. doi: 10.1101/gad.13.11.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras A. C., Raught B., Gygi S. P., Niedzwiecka A., Miron M., Burley S. K., Polakiewicz R. D., Wyslouch-Cieszynska A., Aebersold R., Sonenberg N. Hierarchical phosphorylation of the translation inhibitor 4E–BP1. Genes Dev. 2001a;15:2852–2864. doi: 10.1101/gad.912401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras A. C., Raught B., Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 1999b;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- Gingras A. C., Raught B., Sonenberg N. Regulation of translation initiation by FRAP/mTOR. Genes Dev. 2001b;15:807–826. doi: 10.1101/gad.887201. [DOI] [PubMed] [Google Scholar]
- Gout I., Middleton G., Adu J., Ninkina N. N., Drobot L. B., Filonenko V., Matsuka G., Davies A. M., Waterfield M., Buchman V. L. Negative regulation of PI 3-kinase by Ruk, a novel adaptor protein. EMBO J. 2000;19:4015–4025. doi: 10.1093/emboj/19.15.4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grolleau A., Bowman J., Pradet-Balade B., Puravs E., Hanash S., Garcia-Sanz J. A., Beretta L. Global and specific translational control by rapamycin in T cells uncovered by microarrays and proteomics. J. Biol. Chem. 2002;277:22175–22184. doi: 10.1074/jbc.M202014200. [DOI] [PubMed] [Google Scholar]
- Gupta S., Stuffrein S., Plattner R., Tencati M., Gray C., Whang Y. E., Stanbridge E. J. Role of phosphoinositide 3-kinase in the aggressive tumor growth of HT1080 human fibrosarcoma cells. Mol. Cell. Biol. 2001;21:5846–5856. doi: 10.1128/MCB.21.17.5846-5856.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B., Dong Z., Liu Y., Chen Q., Hashimoto K., Zhang J. T. Regulation of constitutive expression of mouse PTEN by the 5′-untranslated region. Oncogene. 2003;22:5325–5337. doi: 10.1038/sj.onc.1206783. [DOI] [PubMed] [Google Scholar]
- Harding H. P., Zhang Y., Bertolotti A., Zeng H., Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol. Cell. 2000;5:897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- Hay N., Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- Hresko R. C., Mueckler M. mTOR.RICTOR is the Ser473 kinase for Akt/protein kinase B in 3T3–L1 adipocytes. J. Biol. Chem. 2005;280:40406–40416. doi: 10.1074/jbc.M508361200. [DOI] [PubMed] [Google Scholar]
- Kaufman R. J. Double-stranded RNA-activated protein kinase PKR. In: Sonenberg N., Hershey J.W.B., Mathews M. B., editors. Translational Control of Gene Expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. pp. 503–527. [Google Scholar]
- Kazemi S., Papadopoulou S., Li S., Su Q., Wang S., Yoshimura A., Matlashewski G., Dever T. E., Koromilas A. E. Control of alpha subunit of eukaryotic translation initiation factor 2 (eIF2 alpha) phosphorylation by the human papillomavirus type 18 E6 oncoprotein: implications for eIF2 alpha-dependent gene expression and cell death. Mol. Cell. Biol. 2004;24:3415–3429. doi: 10.1128/MCB.24.8.3415-3429.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimball S. R., Jefferson L. S. Regulation of translation initiation in mammalian cells by amino acids. In: Sonenberg N., Hershey J.W.B., Mathews M. B., editors. Translational Control of Gene Expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. pp. 561–579. [Google Scholar]
- Kumar A., Yang Y. L., Flati V., Der S., Kadereit S., Deb A., Haque J., Reis L., Weissmann C., Williams B. R. Deficient cytokine signaling in mouse embryo fibroblasts with a targeted deletion in the PKR gene: role of IRF-1 and NF-kappaB. EMBO J. 1997;16:406–416. doi: 10.1093/emboj/16.2.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lekmine F., Sassano A., Uddin S., Smith J., Majchrzak B., Brachmann S. M., Hay N., Fish E. N., Platanias L. C. Interferon-gamma engages the p70 S6 kinase to regulate phosphorylation of the 40S S6 ribosomal protein. Exp. Cell Res. 2004;295:173–182. doi: 10.1016/j.yexcr.2003.12.021. [DOI] [PubMed] [Google Scholar]
- Lekmine F., Uddin S., Sassano A., Parmar S., Brachmann S. M., Majchrzak B., Sonenberg N., Hay N., Fish E. N., Platanias L. C. Activation of the p70 S6 kinase and phosphorylation of the 4E-BP1 repressor of mRNA translation by type I interferons. J. Biol. Chem. 2003;278:27772–27780. doi: 10.1074/jbc.M301364200. [DOI] [PubMed] [Google Scholar]
- Mothe-Satney I., Brunn G. J., McMahon L. P., Capaldo C. T., Abraham R. T., Lawrence J. C., Jr Mammalian target of rapamycin-dependent phosphorylation of PHAS-I in four (S/T)P sites detected by phospho-specific antibodies. J. Biol. Chem. 2000;275:33836–33843. doi: 10.1074/jbc.M006005200. [DOI] [PubMed] [Google Scholar]
- Naga Prasad S. V., Laporte S. A., Chamberlain D., Caron M. G., Barak L., Rockman H. A. Phosphoinositide 3-kinase regulates beta2-adrenergic receptor endocytosis by AP-2 recruitment to the receptor/beta-arrestin complex. J. Cell Biol. 2002;158:563–575. doi: 10.1083/jcb.200202113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platanias L. C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- Robert F., et al. Initiation of protein synthesis by hepatitis C virus is refractory to reduced eIF2·GTP·Met-tRNAiMet ternary complex availability. Mol. Biol. Cell. 2006;17:4632–4644. doi: 10.1091/mbc.E06-06-0478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D., Harding H. P. PERK and translational control by stress in the endoplasmic reticulum. In: Sonenberg N., Hershey J.W.B., Mathews M. B., editors. Translational Control of Gene Expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. pp. 547–560. [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. pp. 60–74. [Google Scholar]
- Sarbassov D. D., Guertin D. A., Ali S. M., Sabatini D. M. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- Scheid M. P., Woodgett J. R. Unravelling the activation mechanisms of protein kinase B/Akt. FEBS Lett. 2003;546:108–112. doi: 10.1016/s0014-5793(03)00562-3. [DOI] [PubMed] [Google Scholar]
- Scheuner D., Song B., McEwen E., Liu C., Laybutt R., Gillespie P., Saunders T., Bonner-Weir S., Kaufman R. J. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol. Cell. 2001;7:1165–1176. doi: 10.1016/s1097-2765(01)00265-9. [DOI] [PubMed] [Google Scholar]
- Schmidt M. H., Chen B., Randazzo L. M., Bogler O. SETA/CIN85/Ruk and its binding partner AIP1 associate with diverse cytoskeletal elements, including FAKs, and modulate cell adhesion. J. Cell Sci. 2003;116:2845–2855. doi: 10.1242/jcs.00522. [DOI] [PubMed] [Google Scholar]
- Schroder M., Kaufman R. J. The mammalian unfolded protein response. Annu. Rev. Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- Sen G. C., Sarkar S. N. Transcriptional signaling by double-stranded RNA: role of TLR3. Cytokine Growth Factor Rev. 2005;16:1–14. doi: 10.1016/j.cytogfr.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Shahbazian D., Roux P. P., Mieulet V., Cohen M. S., Raught B., Taunton J., Hershey J. W., Blenis J., Pende M., Sonenberg N. The mTOR/PI3K and MAPK pathways converge on eIF4B to control its phosphorylation and activity. EMBO J. 2006;25:2781–2791. doi: 10.1038/sj.emboj.7601166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Q., Wang S., Baltzis D., Qu L. K., Wong A. H., Koromilas A. E. Tyrosine phosphorylation acts as a molecular switch to full-scale activation of the eIF2{alpha} RNA-dependent protein kinase. Proc. Natl. Acad. Sci. USA. 2006;103:63–68. doi: 10.1073/pnas.0508207103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ung T. L., Cao C., Lu J., Ozato K., Dever T. E. Heterologous dimerization domains functionally substitute for the double-stranded RNA binding domains of the kinase PKR. EMBO J. 2001;20:3728–3737. doi: 10.1093/emboj/20.14.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdier F., Valovka T., Zhyvoloup A., Drobot L. B., Buchman V., Waterfield M., Gout I. Ruk is ubiquitinated but not degraded by the proteasome. Eur. J. Biochem. 2002;269:3402–3408. doi: 10.1046/j.1432-1033.2002.03031.x. [DOI] [PubMed] [Google Scholar]
- Vivanco I., Sawyers C. L. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat. Rev. Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- Wang S., Raven J. F., Baltzis D., Kazemi S., Brunet D. V., Hatzoglou M., Tremblay M. L., Koromilas A. E. The catalytic activity of the eukaryotic initiation factor-2alpha kinase PKR is required to negatively regulate Stat1 and Stat3 via activation of the T-cell protein-tyrosine phosphatase. J. Biol. Chem. 2006;281:9439–9449. doi: 10.1074/jbc.M504977200. [DOI] [PubMed] [Google Scholar]
- Wang X., Beugnet A., Murakami M., Yamanaka S., Proud C. G. Distinct signaling events downstream of mTOR cooperate to mediate the effects of amino acids and insulin on initiation factor 4E-binding proteins. Mol. Cell. Biol. 2005;25:2558–2572. doi: 10.1128/MCB.25.7.2558-2572.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A. H., Durbin J. E., Li S., Dever T. E., Decker T., Koromilas A. E. Enhanced antiviral and antiproliferative properties of a STAT1 mutant unable to interact with the protein kinase PKR. J. Biol. Chem. 2001;276:13727–13737. doi: 10.1074/jbc.M011240200. [DOI] [PubMed] [Google Scholar]
- Wong A. H., Tam N. W., Yang Y. L., Cuddihy A. R., Li S., Kirchhoff S., Hauser H., Decker T., Koromilas A. E. Physical association between STAT1 and the interferon-inducible protein kinase PKR and implications for interferon and double-stranded RNA signaling pathways. EMBO J. 1997;16:1291–1304. doi: 10.1093/emboj/16.6.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.