Abstract
It is becoming clear that in vivo phage DNA ejection is not a mere passive process. In most cases, both phage and host proteins seem to be involved in pulling at least part of the viral DNA inside the cell. The DNA ejection mechanism of Bacillus subtilis bacteriophage φ29 is a two-step process where the linear DNA penetrates the cell with a right-left polarity. In the first step ∼65% of the DNA is pushed into the cell. In the second step, the remaining DNA is actively pulled into the cytoplasm. This step requires protein p17, which is encoded by the right-side early operon that is ejected during the first push step. The membrane protein p16.7, also encoded by the right-side early operon, is known to play an important role in membrane-associated phage DNA replication. In this work we show that, in addition, p16.7 is required for efficient execution of the second pull step of DNA ejection.
Despite being a key step in the phage life cycle, the process whereby phages eject their genome into the cell cytoplasm during the early steps of infection has been one of the major unsolved problems. The current understanding of the phage DNA ejection process has recently been reviewed by Molineux (29). The ejection process of phages that infect gram-negative bacteria is now beginning to be understood at a detailed molecular level (24, 30, 33). This process is far from being a passive mechanism, just driven by the release of the pressure built inside the capsid. The cell cytoplasm has a pressure of several atmospheres with respect to the external medium. Therefore, the pressure-based mechanism would be responsible for ejection of only part of the phage genome; i.e., ejection of phage DNA by pressure will stop when the decreasing packing forces in the mature virion equal those inside the cytoplasm. Thus, internalization of the remaining part of the phage genome requires additional mechanisms, and diverse strategies have been described for different phages. For instance, some Escherichia coli phages eject their genomes into the cell largely through enzyme-catalyzed, energy-requiring processes. Phage T5 DNA ejection takes place in two steps (23). In the first step, 8% of the genome enters the cytoplasm. Then, there is a pause of about 4 min during which two proteins encoded by this DNA fragment, A1 and A2, are synthesized. The transfer of the remaining DNA (92%) takes place in a second step only if these proteins are synthesized. A2 is a DNA binding protein, thought to pull DNA into the cell (39). Phage T7 is the only one for which it has been clearly demonstrated that DNA internalization is coupled to transcription (reviewed in reference 29). About 850 bp of the left end of the T7 genome are ejected into the host (11). A molecular motor formed by viral proteins gp16 and gp15 has been proposed to control the amount of DNA that enters the cell (21). Transcription from promoters located on this short DNA sequence by the host RNA polymerase provides the force to internalize about 20% of the genome into the cell. T7 RNA polymerase is then synthesized and is responsible for transcription-driven internalization of the remaining part of the phage genome.
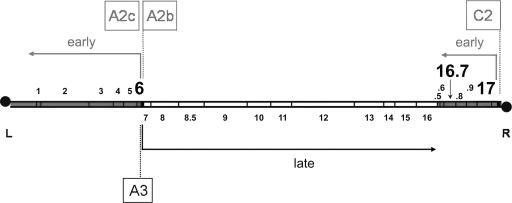
In contrast with the well-characterized internalization of the DNA of some phages of gram-negative bacteria, little is known about the mechanisms of DNA ejection by phages infecting gram-positive bacteria. Bacillus subtilis phage φ29 has a linear double-stranded DNA with a terminal protein covalently linked at each 5′ end (35) (see Fig. 1 for a genetic and transcriptional map). Phage φ29 DNA transcription is time controlled into an early and a late stage (reviewed in reference 26). All late genes are clustered in a single, centrally located operon that is transcribed by promoter A3. The early expressed genes are present in two operons flanking the late operon. The one located at the left side encodes all essential phage DNA replication proteins as well as the transcriptional regulator protein p4 and is expressed from the promoters A2b and A2c organized in tandem. The other early operon, located at the right side of the φ29 genome, is under the control of the C2 promoter. This operon contains genes 17 and 16.7 and four additional open reading frames. The abundantly synthesized φ29 protein p6, encoded by the left-side early operon, is required for activation of the initiation step of viral DNA replication and is involved in transcriptional control as it represses early promoter C2 and cooperates with phage protein p4 in the repression of the early promoters A2b and A2c and in the activation of the late promoter A3 (reviewed in reference 12). Protein p6 binds DNA with little sequence specificity, compacting and organizing the whole viral genome (15). We took advantage of the DNA binding properties of p6 to monitor the entry of φ29 DNA into the cell and showed that the viral DNA penetrates the cell with a right-to-left polarity (16), in agreement with in vitro studies (4, 22). Furthermore, it was found that the viral genome is ejected by a two-step process, described as a “push-pull” mechanism. During the first push step, ∼65% of the genome enters the cell, most likely at the expense of the packing pressure of DNA in the capsid. Entrance of the remaining left part of the φ29 genome requires viral protein(s), as this second pull step is prevented in the presence of chloramphenicol (16). Protein(s) involved in the pull step of ejection must be encoded by the right-side early operon (Fig. 1) because this is the only operon present in the cell after the push step. Protein p17, encoded by the first gene of the right-side early operon, has been demonstrated to be important, but not essential, for the second pull step of ejection (16). Thus, although internalization is strongly impaired in the absence of p17, some full genome entry can still be detected under these conditions. In contrast, internalization is fully blocked when protein synthesis is inhibited by chloramphenicol. This supports the idea that other phage protein(s) synthesized de novo may be required for the efficient pull step of φ29 DNA ejection.
FIG. 1.
Genetic and transcriptional map of the 19.3-kbp linear phage φ29 DNA (adapted from reference 13). The main early promoters A2b, A2c, and C2 and late promoter A3 are boxed. The directions of transcription and lengths of the transcripts are indicated by arrows. The positions of genes are indicated with numbers. The early genes 6, 16.7, and 17, relevant for this work, are shown in bold. Note that whereas gene 6 is present in the early left-side operon, genes 17 and 16.7 are located in the early right-side operon. Black circles represent the terminal protein attached to the 5′ DNA ends. L and R indicate the left and right end of the φ29 genome, respectively.
Early reports showed that cells infected with sus17 mutant phages produced reduced numbers of phage progeny and appeared to be affected in their viral DNA synthesis (7, 20, 31). As mentioned above, analysis of the φ29 ejection process revealed that p17 is important for the second pull step of this process (16), which could account for the earlier observed results. However, the absence of p17 was also shown to drastically affect the level of φ29 DNA replication once the entire sus17 genome was internalized (14), demonstrating that p17 also is important for in vivo φ29 DNA replication.
In addition to gene 17, gene 16.7, also located in the right-side early operon, is highly conserved in all φ29-related phages (26). Protein p16.7 (130 residues) is a membrane protein that plays an important role in membrane-associated in vivo φ29 DNA replication (27, 28). Its first 20 residues constitute a membrane anchor that is responsible for membrane localization (28). Immunofluorescence studies showed that p16.7 is required for efficient spreading of φ29 DNA replication from its initial to additional sites at the membrane of the infected cell (27). In vitro analyses of a soluble p16.7 derivative lacking the membrane anchor revealed that (i) it is a dimer with unspecific DNA binding activity, (ii) it has affinity for the φ29 terminal protein, and (iii) it is able to form higher-order multimers upon DNA binding (36, 37). The solution and crystal structures of the dimeric functional C-terminal half of p16.7, p16.7C (residues 63 to 130) (32), as well as the crystal structure of p16.7C complexed with double-stranded DNA have been recently solved (1, 2). Based on its properties we considered the possibility that p16.7 has a role in the pull step of DNA ejection. Here, we show that p16.7 is required for efficient implementation of the second pull step of φ29 DNA ejection. Thus, at least two proteins, p16.7 and p17, play a role in the pull step of φ29 DNA ejection. Interestingly, both p16.7 and p17 also play a role in in vivo φ29 DNA replication.
MATERIALS AND METHODS
Bacterial strains, plasmids, and phages.
B. subtilis 110NA strain (trpC2 spo0A3 su−) (31) harboring plasmid pPR55w6 or pPR55ow6 was used. Both plasmids are pUB110 derivatives containing bacteriophage φ29 gene 6 inserted in its functional (pPR55w6) or in its opposite (pPR55ow6) orientation behind the phage λ PR promoter (5). Infections were carried out at multiplicity of 3 with either of the following φ29 phages: sus14(1242), a delayed lysis mutant with otherwise wild-type phenotype (20); sus3(91), a replication null mutant (31); sus17(112) (31); or sus16.7(48)/sus14(1242) (28). The genomes of these mutant φ29 phages contain a suppressible nonsense mutation in the gene(s) indicated. For simplicity, these phages will be referred to as sus14, sus3, sus17, and sus16.7/sus14, respectively. Besides the indicated exception, analyses were limited to 30-min postinfection times in the experiments presented to avoid possible interference of phage-induced cell lysis.
Medium, enzymes, drugs, and reactives.
Bacteria were grown in LB medium supplemented with 5 mM MgSO4. Phleomycin (Cayla S.A.R.L.) was added at a final concentration of 0.8 μg/ml. Micrococcal nuclease was from Amersham Pharmacia Biotech, and proteinase K was from Boehringer Mannheim. Protein A-Sepharose CL-4B, lysozyme, RNase A, chloramphenicol, and novobiocin were from Sigma-Aldrich. Formaldehyde at 37% was from Calbiochem.
DNAs and oligonucleotides.
Proteinase K-digested φ29 DNA was obtained as described previously (18). The sequences of the oligonucleotides used for PCR (Isogen) are given, together with the coordinates of the DNA sequence they amplify (U, upper strand; L, lower strand). φ29 DNA region L, comprising the 259-bp left terminal region, was amplified with oligonucleotides (U-1) 5′-AAAGTAAGCCCCCACCCTCACATG and (L-259) 5′-GCCCACATACTTTGTTGATTGG. φ29 DNA region R, comprising the 298-bp right terminal region, was amplified with oligonucleotides (U-18988) 5′-AAAGTAGGGTACAGCGACAACATAC and (L-19285) 5′-AAATAGATTTTCTTTCTTGGCTAC.
Cross-linking, immunoprecipitation, and DNA amplification.
Bacteria were grown at 30°C up to 108 cells/ml and infected at a multiplicity of 3. Drugs (34 μg/ml chloramphenicol and 500 μg/ml novobiocin) were added at the indicated times, and cross-linking was performed 10 min later. Chloramphenicol was added to prevent expression of p6, and novobiocin was used to enhance protein p6 binding (16). Cross-linking and chromatin immunoprecipitation (X-ChIP) were carried out essentially as described previously (25). Culture samples, 20 ml each, were treated with 1% formaldehyde, together with 10 mM sodium phosphate, pH 7.2. After 5 min at room temperature without shaking, reactions were stopped by the addition of 125 mM glycine. Cells were harvested by centrifugation, washed twice with phosphate-buffered saline buffer, and resuspended in 1 ml of buffer A (10 mM Tris-HCl, pH 8.0, 50 mM NaCl, 10 mM EDTA) containing 3 mg/ml lysozyme. Samples were incubated for 30 min at 37°C prior to lysis by the addition of 1 ml of 2× immunoprecipitation buffer (100 mM Tris-HCl, pH 7.0, 300 mM NaCl, 2% Triton X-100) and 0.1% sodium dodecyl sulfate (SDS). Then, samples were subjected to mild digestion with micrococcal nuclease (0.05 U in the presence of 13 mM CaCl2 for 10 min at 37°C), and the reactions were stopped with 20 mM EDTA. DNA was sheared by sonication using a 150-W Ultrasonic Disintegrator (MSE, United Kingdom). Fragments with an average size of about 750 bp were obtained by sonicating samples (2 ml) during three separate pulses of 10 s at an amplitude of 15 μm. To avoid heating, samples were packed with crushed ice during the entire sonication procedure, and sonication pulses were separated by at least 20-s intervals. Next, cell debris was eliminated by centrifugation. Part (1/20) of each sample was kept for total DNA analysis, and the remainder was split to immunoprecipitate overnight at 4°C with either anti-p6 polyclonal antibodies (αp6 sample) or preimmune serum (20 μl each), followed by incubation for 2.5 h at 4°C with 120 μl of a 25% protein A-Sepharose slurry. Complexes were collected by centrifugation and washed twice with 1× immunoprecipitation-0.1% SDS buffer, three times with 1× immunoprecipitation buffer, and twice with TE buffer (10 mM Tris-HCl, pH 7.5, 1 mM EDTA). The slurry was resuspended in 150 μl of TE buffer containing 1% SDS to disrupt immune complexes. Total DNA samples were also brought to a total volume of 150 μl of TE containing 1% SDS. All samples were incubated overnight at 65°C with shaking to reverse cross-links. Slurry was removed by centrifugation, and the supernatant was transferred to a fresh tube. DNA was purified by phenol and chloroform extraction, ethanol-precipitated, and finally resuspended in water. Analysis of DNA samples L and R from left and right φ29 ends, respectively, was performed by real-time PCR in a Light-Cycler instrument using a Light Cycler-FastStart DNA Master SYBR Green I hot-start reaction mixture (Roche). Amplification conditions included a preheating step of 20 min at 95°C to activate the polymerase, followed by 30 cycles comprising a denaturation step of 15 s at 95°C for both regions, a hybridization step of 5 s at 53°C for region L and 50°C for R, and an elongation step at 72°C lasting 15 s for L and 40 s for R. Finally, a melting analysis was performed by continuous fluorescence measurement from 65°C to 95°C to check that a single product was amplified. Protein p6 binding values were expressed as the immunoprecipitation coefficient, or IC: [(amount of αp6 DNA − amount of PI DNA)/amount of T DNA] × 106, where T stands for total DNA and αp6 and PI values are for DNA immunoprecipitated with serum against p6 and preimmune serum, respectively.
Western blot analysis.
Cells were grown at 30°C up to 108 cells/ml and infected at a multiplicity of 3. At the times indicated in Fig. 2 and 4, 1.5-ml aliquots were transferred to ice-cold tubes, concentrated 7.5-fold in loading buffer (60 mM Tris-HCl, pH 6.8, 2% SDS, 5% β-mercaptoethanol, 30% glycerol), and disrupted by sonication. Samples were subjected to SDS-polyacrylamide gel electrophoresis on a 15% polyacrylamide gel, and proteins were transferred using a Mini Trans Blot apparatus (Bio-Rad) at 100 mA and 4°C for 60 min. Immobilon-P membranes (Millipore) were probed with αp6 or αp16.7 polyclonal antibodies diluted 1:2,000 and αp17 polyclonal antibodies diluted 1:4,000 for 70 min. Then, membranes were washed twice and incubated with anti-rabbit horseradish peroxidase-conjugated antibodies (dilution, 1:4,000) for another 70 min, and the immune complexes were detected by ECL detection reagents (Amersham).
FIG. 2.
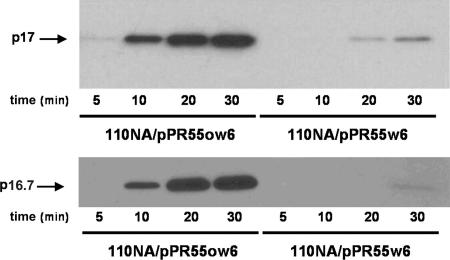
Time course synthesis of early proteins p17 and p16.7 in p6-producing (110NA/pPR55w6) and nonproducing (110NA/pPR55ow6) cells infected with phage sus14 at a multiplicity of infection of 10. Samples taken at the indicated postinfection times were analyzed by Western blotting using antibodies against p17 or p16.7.
DNA replication.
B. subtilis cells were grown and infected as described above. At the times indicated in Fig. 5, cells corresponding to 1-ml aliquots were sedimented, washed, and lysed as previously described (5). Samples were treated with proteinase K (50 μg/ml), and DNA was extracted with phenol. The amount of DNA from L sequence was quantified by real-time PCR, using the amplification protocol described above.
FIG. 5.
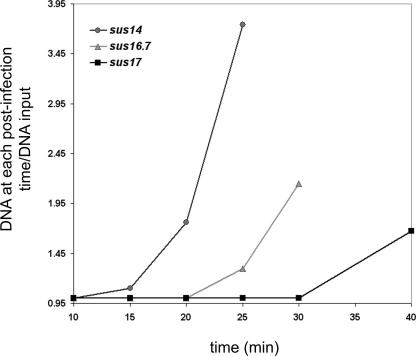
Determination of the onset of DNA replication of mutant phages sus14, sus16.7/sus14, and sus17. Non-p6-producing cells (110NA/pPR55ow6) were infected with the indicated mutants. Aliquots were taken at the indicated times after infection, and the DNA was purified by phenol extraction and ethanol precipitation as described in Materials and Methods. The amount of φ29 DNA at each postinfection time was calculated by real-time PCR of the left terminal sequence (L), and divided by the input amount of infecting DNA (determined at minute 10 postinfection).
RESULTS
Ectopically expressed protein p6 decreases synthesis of proteins under the control of promoter C2.
Previous studies on φ29 DNA ejection were based on the φ29 DNA binding properties of protein p6. Thus, entry of φ29 DNA into the cell was monitored by measuring binding of p6, constitutively expressed from plasmid pPR55w6, to various φ29 DNA regions by X-ChIP and real-time PCR (16, 17). Under these conditions, protein p16.7, in contrast to p17, did not seem to play a significant role in the pull step of ejection. However, protein p6 represses the φ29 promoter C2 that drives expression of the right-side early operon containing genes 17 and 16.7 (3, 40), which is ejected during the first step of φ29 DNA ejection. The ectopically expressed protein p6 may therefore impair transcription of the promoter C2-driven operon immediately upon ejection of the right half of the φ29 genome. This might result in levels of p16.7 that are insufficient to exert a function in the pull step of DNA ejection. In addition, it has been reported that the presence of p6 before infection causes a delay of at least 20 min in the ejection process (16).
To study if the plasmid-encoded protein p6 affects expression of the φ29 right-side operon, the kinetics of p16.7 and p17 synthesis were compared in cells producing or not producing protein p6. Thus, cells harboring the p6-producing plasmid pPR55w6 or a derivative containing gene 6 in the opposite orientation, pPR55ow6, were infected with φ29, and samples withdrawn at different times after infection were processed and subjected to Western blot analysis using antibodies against p16.7 and p17. Phage φ29 sus14, containing a suppressor mutation in gene 14 encoding the holin protein (20), was used in these experiments. As shown in Fig. 2, in the absence of ectopically synthesized protein p6, proteins p17 and p16.7 were detected at 10 min after infection, their levels increased greatly during the next 10 min (time = 20), and their further increase leveled off during the next 10 min (time = 30) (Fig. 2, 110NA/pPR55ow6). However, the amounts of proteins p17 and p16.7 were drastically reduced in the presence of ectopically synthesized protein p6. Thus, faint and barely detectable signals were obtained for p17 and p16.7, respectively, at 30 min after infection (Fig. 2, 110NA/pPR55w6). These results demonstrate that plasmid-encoded p6 protein strongly represses expression of these proteins.
Protein p16.7 is required for efficient implementation of the pull step of φ29 DNA ejection.
The results presented above show that the experimental setup in which protein p6 is present in the cell before infection causes a strong and immediate repression of the right-side early operon, which may mask a possible involvement of protein p16.7 in the pull step of DNA ejection. To test this possibility, the φ29 ejection process was studied in non-p6-producing B. subtilis cells. Whereas the major aim was to study whether p16.7 plays a role in the second pull step of φ29 ejection, we also analyzed the effects of the absence of p17 under these conditions. This allows comparison to the situation in which wild-type levels of p17 are synthesized as well as possible differential effects on the ejection process in the absence of either p16.7 or p17.
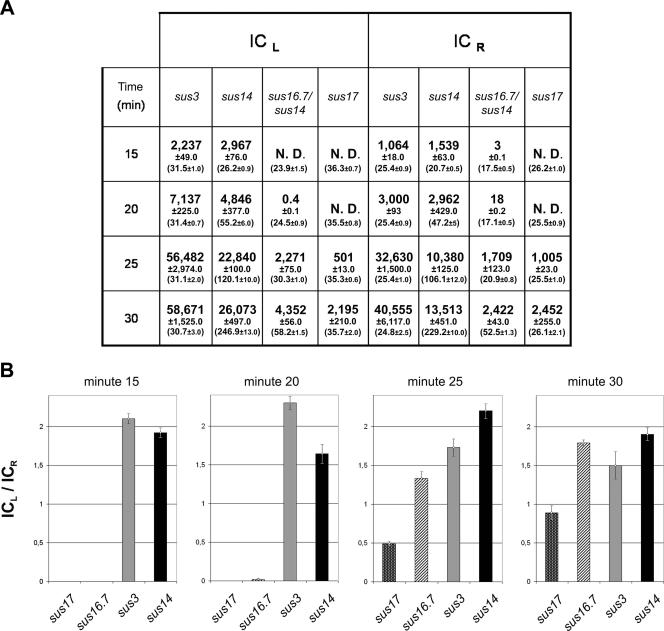
As a first approach we monitored the entrance of the φ29 genome in non-p6-producing cells infected with φ29 mutant phage sus16.7/sus14 or sus17 using X-ChIP (see Materials and Methods). Infections with phage sus14 or sus3 served as wild-type and replication-deficient controls, respectively. A logical consequence of this experimental setup is that p6-DNA complexes can only be formed after φ29-encoded p6 is synthesized. Binding of protein p6 to the φ29 left (L) and right (R) DNA end regions was measured by quantitative PCR on the immunoprecipitated DNA using appropriate primers and expressed as the IC (see Materials and Methods). The ratio of the IC values, ICL/ICR, indicates the degree of internalization of the left end. Since the intrinsic affinity of p6 for the left-end φ29 DNA region is about twofold higher than that for the right-end φ29 DNA region (13), completion of φ29 DNA ejection is reflected by ICL/ICR ratios of about 2. The calculated IC values corresponding to the L and R regions at different times after infection for each phage analyzed are given in Fig. 3A. To visualize internalization efficiencies, the ICL/ICR ratios were plotted at different postinfection times for each mutant (Fig. 3B). ICL/ICR ratios of about 2 were observed 15 min after infection for phages sus14 and sus3. This indicates that, in the absence of promoter C2 repression, internalization of the entire genome of these phages takes less than 15 min. The situation was very different though for φ29 phages sus16.7/sus14 and sus17. Immunoprecipitation of substantial amounts of the left genome end in mutant sus16.7/sus14 was not detected until minute 25. Complete ejection, an ICL/ICR ratio close to 2, was not reached until minute 30 postinfection, indicating a delay in the pull step of the ejection process of this mutant of at least 15 min with respect to the wild-type situation. In the case of phage sus17, DNA fragments corresponding to the left genome end region were first detected by immunoprecipitation 25 min after infection. However, the ICL values obtained for the 25- and 30-min postinfection times were lower than the corresponding ones for sus16.7/sus14 (Fig. 3A). Moreover, the maximum ICL/ICR ratio obtained at 30 min postinfection was still less than 1, indicating that only about 50% of the sus17 phage genomes had been fully internalized at this time. These results corroborate earlier findings that protein p17 is required for efficient ejection of the left half of the φ29 genome (16, 17). Importantly, these results strongly indicate that protein p16.7 is also required for efficient execution of the pull step of DNA ejection, although its absence affects this step less severely than the absence of protein p17.
FIG. 3.
Efficiency of φ29 DNA ejection in cells infected with φ29 mutants sus3, sus14, sus16.7/sus14, or sus17. A culture of non-p6-producing cells (110NA/pPR55ow6) grown at 30°C was split into four when a density of 108 cells/ml was reached, and cells were infected with phage sus3, sus14, sus16.7/sus14, or sus17. At the indicated times after infection, aliquots of 20 ml, incubated with chloramphenicol and novobiocin during 10 min, were cross-linked with formaldehyde, and ChIP was performed as described in Materials and Methods. (A) Protein p6 binding to DNA fragments corresponding to the left and right φ29 genome end are expressed as the IC. The value of the total DNA, T (in ng/ml), is shown within brackets below each IC together with the standard deviation. Besides internalized DNA, the T value also includes DNA from adsorbed phages that have not (yet) ejected their DNA. ND, not detected. (B) Graphic representation of the ICL/ICR ratios for phages sus3, sus14, sus16.7/sus14, and sus17 at different postinfection times; data are taken from panel A. Values are the average of (at least) three independent experiments, which among themselves differed less than 10%. The mean values of the IC ratios together with the standard deviation are presented.
The absence of p16.7 leads to a delay in protein p6 synthesis and onset of phage DNA replication.
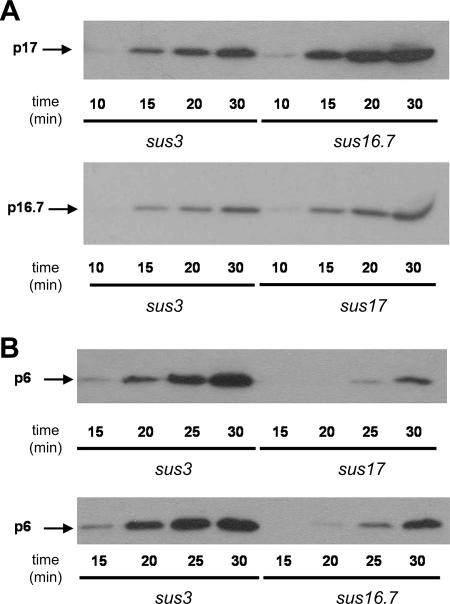
Genes 16.7 and 17 are located at the right-side early operon that is injected during the first push step. Gene 6 is located in the left-side early operon that is ejected during the second pull step of infection, implying that protein p6 synthesis will not occur until the DNA region containing gene 6 is present in the cell. Hence, impairment of the pull step of DNA ejection will cause a delay in the synthesis of protein p6. Analysis of the kinetics of protein p6 synthesis serves, therefore, as an additional method to study the efficiency of the pull step of ejection. In addition, the analysis of the pull step by X-ChIP described above is an indirect method with the limitation that the efficiency of the immunoprecipitation may be affected by different p6/φ29 DNA ratios that are likely to occur during the progress of the infection cycle. Thus, the kinetics of protein p6 synthesis in cells infected with mutant phage sus3, sus16.7/sus14, or sus17 were studied by Western blot analysis. The same samples were also used to analyze the kinetics of synthesis of proteins encoded by genes present in the right-side early operon (i.e., p17 or p16.7). The results of these experiments are presented in Fig. 4. As expected, proteins encoded by the right-side operon (p17 or p16.7) were detected for all three mutant phages as soon as 10 to 15 min after infection (Fig. 4A), indicating that the mutations do not affect the push step. However, protein p6 encoded by the left-side operon was first detected at 15, 20, and 25 min after infection with the mutant phages sus3, sus16.7/sus14, and sus17, respectively (Fig. 4B). In addition, although for all three phages the amount of p6 increased with time, clear differences were observed even at 30 min after infection. At this postinfection time the highest amount of p6 was detected in cells infected with phage sus3. Furthermore, the amount of p6 in sus16.7/sus14-infected cells was significantly higher than that detected in sus17-infected cells. It is worth mentioning that the amount of protein p6 detected at 25 and 30 min postinfection may be overestimated in cells infected with phage sus16.7/sus14, as the amount of sus16.7/sus14 DNA at 25 and 30 min postinfection is 1.3- and 2-fold higher, respectively, than that of sus3 DNA due to replication of sus16.7/sus14 DNA (not shown). These results confirm that (i) both p16.7 and p17 are required for efficient internalization of the left early operon (pull step) and (ii) the absence of p16.7 affects the pull step less drastically than the absence of p17.
FIG. 4.
Synthesis of φ29 early proteins encoded by the left- and right-side early operon in sus3-, sus16.7/sus14-, or sus17-infected cells. Time course synthesis of right-side operon-encoded proteins p17 and p16.7 (A) and left-side operon-encoded protein p6 (B) in non-p6-producing cells (110NA/pPR55ow6), infected with φ29 mutants sus3, sus16.7/sus14, or sus17. Samples taken at the indicated postinfection times were analyzed by Western blotting using antibodies against proteins p17 and p16.7 (A) or p6 (B). Note that higher amounts of p16.7 and p17 were detected at 20 and 30 min after infection in sus17- and sus16.7/sus14-infected cells with respect to sus3-infected cells. Most probably this is due to a delayed synthesis of protein p6 in the sus16.7/sus14- and sus17-infected cells, causing a delayed and/or less efficient repression of the C2 promoter.
Protein p6, together with other proteins encoded by the left-side early operon, is essential for φ29 DNA replication. Therefore, it is expected that impairment of the pull step will also delay the onset of phage DNA replication. To test this prediction, we used real-time PCR to quantify the amount of accumulated φ29 DNA at different times after infection with phage sus14, sus16.7/sus14, or sus17. As shown in Fig. 5, while the onset of DNA replication is observed 15 min after infection for phage sus14, it is delayed for phage sus16.7/sus14 and sus17 to at least 20 and 30 min after infection, respectively.
DISCUSSION
A critical step during the early stages of phage infection is the DNA ejection from the virion capsid into the cytosol of the host. In the case of B. subtilis phage φ29, the linear genome penetrates with a right-to-left polarity by a two-step push-pull mechanism (16). During the first push step, about 65% of the right-side φ29 genome is ejected into the host. This step does not require an external energy source and is probably driven by the high packing pressure of the DNA inside the phage head (16, 17, 38). The φ29 right-side early operon, which is under the control of the strong C2 promoter, is ejected during this push step. The finding that the DNA ejection process is completely stalled after the push step when cells are infected in the presence of chloramphenicol indicated that one or more proteins encoded by the right-side early operon is required to pull the remaining part of the φ29 genome into the host (16). The capacity of protein p6 to bind φ29 DNA was used to develop an X-ChIP assay combined with real-time PCR to monitor the φ29 ejection process. This approach provided conclusive evidence that protein p17 plays an important role in the pull step of ejection (16). In those experiments, φ29 protein p6 was produced ectopically in B. subtilis cells before infection upon constitutive expression from a high-copy-number plasmid. Under these conditions, protein p16.7 did not seem to play a significant role in the pull step of DNA ejection.
In the work presented here we show that the plasmid-expressed p6 protein strongly represses the C2 promoter, causing a drastic reduction in the synthesis of proteins encoded by the right-side early operon. This result, together with the fact that whereas the pull step was fully blocked by chloramphenicol, it was only impaired in sus17-infected cells (16), prompted us to reassess a possible role of p16.7 in the pull step. The efficiency of the pull step was therefore determined using non-p6-producing B. subtilis cells in which the right-side early operon becomes expressed at wild-type levels. As a first approach, the entrance of viral DNA of various mutant phages was monitored by the X-ChIP assay combined with real-time PCR by measuring binding of phage-encoded protein p6 to the left and right φ29 DNA ends at different times after infection. These results showed that protein p16.7 is required for efficient execution of the pull step (Fig. 3). This conclusion was confirmed by another approach in which the kinetics of protein p6 synthesis was studied in cells infected with different φ29 mutant phages. Thus, synthesis of protein p6, whose gene is located in the left-side early operon that is internalized during the pull step, is delayed in the absence of protein p16.7 (Fig. 4B). Finally, the fact that the onset of φ29 DNA replication was delayed in the absence of p16.7 (Fig. 5) is in agreement with the conclusion that p16.7 is required for efficient execution of the pull step.
In order to compare the effects of p16.7 and p17 in the pull step, these experiments were performed in parallel using mutant phages sus17 and sus16.7/sus14. The results obtained by different approaches congruently showed that the absence of p17 affected the efficiency of the pull step more severely than the absence of p16.7. Altogether, the results presented in this work demonstrate that at least two proteins encoded by the right-side operon, p17 and p16.7, are required for an efficient pull step of DNA ejection, although the absence of p17 affects this step more drastically than the absence of p16.7.
Available data (see introduction) indicate that the native dimeric DNA binding protein p16.7 is responsible for attaching φ29 DNA to the membrane of the infected cell. Interestingly, 16.7 appears to be the only gene present in the right-side operon to encode a membrane protein (10, 28; also our unpublished results). In prokaryotes, transcription, translation, and membrane insertion of membrane proteins are coupled processes, referred to as transertion (8, 9, 34, 41). The transertion process of p16.7 will position the right half of the φ29 DNA, ejected during the push step, at or near the membrane where it is subsequently bound by the p16.7 protein. We envision that p16.7-mediated association of this part of the φ29 genome at the membrane contributes to the formation and/or organization of a functional complex, including at least protein p17, that is responsible for internalization of the remaining part of the φ29 genome via an active process, known to require energy (17). This view implies that the pull step of φ29 DNA ejection is a membrane-associated process, which is supported by the following observations. Although linear, the φ29 DNA is topologically constrained in vivo (13), and alteration of the topological conformation of the φ29 DNA upon the addition of novobiocin abruptly blocks the pull step (17). In vivo φ29 DNA replication occurs at the membrane of the infected cell (6, 19, 27). Interestingly, both p16.7 and p17 are required for efficient membrane-associated φ29 DNA replication (14, 27, 28). Thus, proteins p16.7 and p17 are involved in (i) active internalization of the left part of the φ29 DNA during ejection and (ii) membrane-associated in vivo DNA replication. To our knowledge, this is the first example for which it has been shown that phage-encoded proteins are involved in these two different aspects of the phage life cycle.
Acknowledgments
We are grateful to A. Bravo for providing us B. subtilis strain 110NA/pPR55w6.
This work was supported by research grant BFU2005-00733 from the Spanish Ministry of Education and Science to M.S. and by an institutional grant from the Fundación Ramón Areces to the Centro de Biología Molecular Severo Ochoa. M.A. is a predoctoral fellow of the Ministry of Education and Science.
Footnotes
Published ahead of print on 25 May 2007.
REFERENCES
- 1.Albert, A., D. Muñoz-Espín, M. Jiménez, J. L. Asensio, J. A. Hermoso, M. Salas, and W. J. J. Meijer. 2005. Structural basis for membrane anchorage of viral φ29 DNA during replication. J. Biol. Chem. 280:42486-42488. [DOI] [PubMed] [Google Scholar]
- 2.Asensio, J. L., A. Albert, D. Muñoz-Espín, C. González, J. Hermoso, L. Villar, J. Jiménez-Barbero, M. Salas, and W. J. J. Meijer. 2005. Structure of the functional domain of φ29 replication organizer: insights into oligomerization and DNA binding. J. Biol. Chem. 280:20730-20739. [DOI] [PubMed] [Google Scholar]
- 3.Barthelemy, I., R. P. Mellado, and M. Salas. 1989. In vitro transcription of bacteriophage φ29 DNA: inhibition of early promoters by the viral replication protein p6. J. Virol. 63:460-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bjornsti, M. A., B. E. Reilly, and D. L. Anderson. 1983. Morphogenesis of bacteriophage φ29 of Bacillus subtilis: oriented and quantized in vitro packaging of DNA protein gp3. J. Virol. 45:383-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bravo, A., J. M. Hermoso, and M. Salas. 1994. A genetic approach to the identification of functional amino acids in protein p6 of Bacillus subtilis phage φ29. Mol. Gen. Genet. 245:529-536. [DOI] [PubMed] [Google Scholar]
- 6.Bravo, A., and M. Salas. 1997. Initiation of bacteriophage φ29 DNA replication in vivo: assembly of a membrane-associated multiprotein complex. J. Mol. Biol. 269:102-112. [DOI] [PubMed] [Google Scholar]
- 7.Carrascosa, J. L., A. Camacho, F. Moreno, F. Jiménez, R. P. Mellado, E. Viñuela, and M. Salas. 1976. Bacillus subtilis phage φ29: characterization of gene products and functions. Eur. J. Biochem. 66:229-241. [DOI] [PubMed] [Google Scholar]
- 8.Chang, C. N., P. Model, and G. Blobel. 1979. Membrane biogenesis: cotranslational integration of the bacteriophage f1 coat protein into an Escherichia coli membrane fraction. Proc. Natl. Acad. Sci. USA 76:1251-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cordes, C., R. Meima, B. Twiest, B. Kazemier, G. Venema, J. M. van Dijl, and S. Bron. 1996. The expression of a plasmid-specified exported protein causes structural plasmid instability in Bacillus subtilis. J. Bacteriol. 178:5235-5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crucitti, P., J. M. Lázaro, V. Beneš, and M. Salas. 1998. Bacteriophage φ29 early protein p17 is conditionally required for the first rounds of viral DNA replication. Gene 223:135-142. [DOI] [PubMed] [Google Scholar]
- 11.García, L. R., and I. J. Molineux. 1996. Transcription-independent DNA translocation of bacteriophage T7 DNA into Escherichia coli. J. Bacteriol. 178:6921-6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.González-Huici, V., M. Alcorlo, M. Salas, and J. M. Hermoso. 2004. Bacteriophage φ29 protein p6: an architectural protein involved in genome organization, replication and control of transcription. J. Mol. Recognit. 17:390-396. [DOI] [PubMed] [Google Scholar]
- 13.González-Huici, V., M. Alcorlo, M. Salas, and J. M. Hermoso. 2004. Binding of phage φ29 architectural protein p6 to the viral genome: evidence for topological restriction of the phage linear DNA. Nucleic Acids Res. 32:3493-3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.González-Huici, V., M. Alcorlo, M. Salas, and J. M. Hermoso. 2004. Phage φ29 proteins p1 and p17 are required for efficient binding of architectural protein p6 to viral DNA in vivo. J. Bacteriol. 186:8401-8406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.González-Huici, V., M. Salas, and J. M. Hermoso. 2004. Genome wide, supercoiling-dependent, in vivo binding of a viral protein involved in DNA replication and transcriptional control. Nucleic Acids Res. 32:2306-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.González-Huici, V., M. Salas, and J. M. Hermoso. 2004. The push-pull mechanism of bacteriophage φ29 DNA injection. Mol. Microbiol. 52:529-540. [DOI] [PubMed] [Google Scholar]
- 17.González-Huici, V., M. Salas, and J. M. Hermoso. 2006. Requirements for Bacillus subtilis bacteriophage φ29 DNA ejection. Gene 374:19-25. [DOI] [PubMed] [Google Scholar]
- 18.Inciarte, M. R., J. M. Lázaro, M. Salas, and E. Viñuela. 1976. Physical map of bacteriophage φ29 DNA. Virology 74:314-323. [PubMed] [Google Scholar]
- 19.Ivarie, R. D., and J. J. Pène. 1973. DNA replication in bacteriophage φ29: the requirement of a viral-specific product for association of φ29 DNA with the cell membrane of Bacillus amyloliquefaciens. Virology 52:351-362. [DOI] [PubMed] [Google Scholar]
- 20.Jiménez, F., A. Camacho, J. De la Torre, E. Viñuela, and M. Salas. 1977. Assembly of Bacillus subtilis phage φ29. 2. Mutants in the cistrons coding for the non-structural proteins. Eur. J. Biochem. 73:57-72. [DOI] [PubMed] [Google Scholar]
- 21.Kemp, P., M. Gupta, and I. J. Molineux. 2004. Bacteriophage T7 DNA ejection into cells is initiated by an enzyme-like mechanism. Mol. Microbiol. 53:1251-1265. [DOI] [PubMed] [Google Scholar]
- 22.Krawiec, S., F. Jiménez, J. A. García, N. Villanueva, J. M. Sogo, and M. Salas. 1981. The orderly, in vitro emergence of DNA from bacteriophage φ29 particles. Virology 111:440-454. [DOI] [PubMed] [Google Scholar]
- 23.Lanni, Y. T. 1968. First-step-transfer deoxyribonucleic acid of bacteriophage T5. Bacteriol. Rev. 32:227-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Letellier, L., P. Boulanger, M. de Frutos, and P. Jacquot. 2003. Channeling phage DNA through membranes: from in vivo to in vitro. Res. Microbiol. 154:283-287. [DOI] [PubMed] [Google Scholar]
- 25.Lin, D. C., and A. D. Grossman. 1998. Identification and characterization of a bacterial chromosome partitioning site. Cell 92:675-685. [DOI] [PubMed] [Google Scholar]
- 26.Meijer, W. J. J., J. A. Horcajadas, and M. Salas. 2001. φ29 family of phages. Microbiol. Mol. Biol Rev. 65:261-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meijer, W. J. J., P. J. Lewis, J. Errington, and M. Salas. 2000. Dynamic relocalization of phage φ29 DNA during replication and the role of the viral protein p16.7. EMBO J. 19:4182-4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meijer, W. J. J., A. Serna-Rico, and M. Salas. 2001. Characterization of the bacteriophage φ29-encoded protein p16.7: a membrane protein involved in phage DNA replication. Mol. Microbiol. 39:731-746. [DOI] [PubMed] [Google Scholar]
- 29.Molineux, I. J. 2006. Fifty-three years since Hershey and Chase: much ado about pressure but which pressure is it? Virology 344:221-229. [DOI] [PubMed] [Google Scholar]
- 30.Molineux, I. J. 2001. No syringes please, ejection of phage T7 DNA from the virion is enzyme driven. Mol. Microbiol. 40:1-8. [DOI] [PubMed] [Google Scholar]
- 31.Moreno, F., A. Camacho, E. Viñuela, and M. Salas. 1974. Supressor-sensitive mutants and genetic map of Bacillus subtilis bacteriophage φ29. Virology 62:1-16. [DOI] [PubMed] [Google Scholar]
- 32.Muñoz-Espín, D., M. G. Mateu, L. Villar, A. Marina, M. Salas, and W. J. J. Meijer. 2004. Phage φ29 DNA replication organizer membrane protein p16.7 contains a coiled-coil and a dimeric, homeodomain-related, functional domain. J. Biol. Chem. 279:50437-50445. [DOI] [PubMed] [Google Scholar]
- 33.Rossmann, M. G., V. V. Mesyanzhinov, F. Arisaka, and P. G. Leiman. 2004. The bacteriophage T4 DNA injection machine. Curr. Opin. Struct. Biol. 14:171-180. [DOI] [PubMed] [Google Scholar]
- 34.Ryter, A., and A. Chang. 1975. Localization of transcribing genes in the bacterial cell by means of high resolution autoradiography. J. Mol. Biol. 98:797-810. [DOI] [PubMed] [Google Scholar]
- 35.Salas, M. 1991. Protein-priming of DNA replication. Annu. Rev. Biochem. 60:37-71. [DOI] [PubMed] [Google Scholar]
- 36.Serna-Rico, A., D. Muñoz-Espín, L. Villar, M. Salas, and W. J. Meijer. 2003. The integral membrane protein p16.7 organizes in vivo φ29 DNA replication through interaction with both the terminal protein and ssDNA. EMBO J. 22:2297-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Serna-Rico, A., M. Salas, and W. J. J. Meijer. 2002. The Bacillus subtilis phage φ29 protein p16.7, involved in φ29 DNA replication, is a membrane-localized single-stranded DNA-binding protein. J. Biol. Chem. 277:6733-6742. [DOI] [PubMed] [Google Scholar]
- 38.Smith, D. E., S. J. Tans, S. B. Smith, S. Grimes, D. L. Anderson, and C. Bustamante. 2001. The bacteriophage φ29 portal motor can package DNA against a large internal force. Nature 413:748-752. [DOI] [PubMed] [Google Scholar]
- 39.Snyder, C. E., Jr., and R. H. Benzinger. 1981. Second-step transfer of bacteriophage T5 DNA: purification and characterization of the T5 gene A2 protein. J. Virol. 40:248-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whiteley, H. R., W. D. Ramey, G. B. Spiegelman, and R. D. Holder. 1986. Modulation of in vivo and in vitro transcription of bacteriophage φ29 early genes. Virology 155:392-401. [DOI] [PubMed] [Google Scholar]
- 41.Woldringh, C. L., P. R. Jensen, and H. V. Westerhoff. 1995. Structure and partitioning of bacterial DNA: determined by a balance of compaction and expansion forces? FEMS Microbiol. Lett. 131:235-242. [DOI] [PubMed] [Google Scholar]