Abstract
Proliferating cell nuclear antigen (PCNA) is the sliding clamp that is essential for the high processivity of DNA synthesis during DNA replication. Pyrococcus furiosus, a hyperthermophilic archaeon, has at least two DNA polymerases, polymerase BI (PolBI) and PolD. Both of the two DNA polymerases interact with the archaeal P. furiosus PCNA (PfuPCNA) and perform processive DNA synthesis in vitro. This phenomenon, in addition to the fact that both enzymes display 3′-5′ exonuclease activity, suggests that both DNA polymerases work in replication fork progression. We demonstrated here that both PolBI and PolD functionally interact with PfuPCNA at their C-terminal PIP boxes. The mutant PolBI and PolD enzymes lacking the PIP-box sequence do not respond to the PfuPCNA at all in an in vitro primer extension reaction. This is the first experimental evidence that the PIP-box motif, located at the C termini of the archaeal DNA polymerases, is actually critical for PCNA binding to form a processive DNA-synthesizing complex.
DNA replication is a highly coordinated process that ensures accurate and efficient genome duplication. Many protein factors involved in DNA replication have already been identified, and their functions have been deduced. Biochemical research efforts have suggested that the principle of the DNA replication mechanism seems to be the same in the three biological domains, Bacteria, Eukarya, and Archaea (6, 10, 11, 19), although the protein factors with conserved functions are predicted to have evolved from different origins based on the distinct diversity of the amino acid sequences (23). Multiple DNA polymerases have been identified in each living organism, and these enzymes are thought to share the functions in the cells to process DNA transactions and maintain genome integrity. The DNA polymerases have been classified by amino acid sequence similarity, and seven classes, the A, B, C, D, E, X, and Y families, are now widely recognized (1, 6, 24, 34). Among them, the cells belonging to the Crenarchaeota, a subdomain of the Archaea, have at least two family B DNA polymerases (4, 38) and, in some strains, also have family Y DNA polymerases (22). On the other hand, there is only one family B DNA polymerase and one family D DNA polymerase in the Euryarchaeota, the other subdomain of the Archaea (7, 15, 16, 17). Notably, the family D DNA polymerase is an enzyme specific for euryarchaea, and this enzyme is mysterious from an evolutional point of view, since we discovered polymerase D (PolD) as a new DNA polymerase consisting of two subunits from the hyperthermophilic archaeon Pyrococcus furiosus (37).
Replication factor C (RFC) and proliferating cell nuclear antigen (PCNA) work as the clamp and clamp loader, respectively, and they are essential for DNA polymerase to perform processive DNA synthesis (Fig. 1). RFC loads PCNA onto the DNA strand in an ATP-dependent manner. The central hole of the PCNA homotrimer ring encircles double-stranded DNA so that DNA polymerases can catalyze DNA synthesis with PCNA without falling off the DNA strand (2, 13). P. furiosus has one homolog of eukaryotic PCNA (3) and one homolog of RFC (5). The one-to-four complex of the large subunit (RFCL) and the small subunits (RFCS) of P. furiosus RFC (PfuRFC) loads P. furiosus PCNA (PfuPCNA) onto the DNA strand. We have been investigating the structural and functional aspects of the clamp-loading mechanism (14, 18, 25, 26, 27, 28, 29, 32, 35) and finally detected an intermediate complex in which the PCNA ring is opened by single-particle analysis of electron microscopic images. These results contributed to our understanding of the molecular mechanism of the clamp-loading process (29).
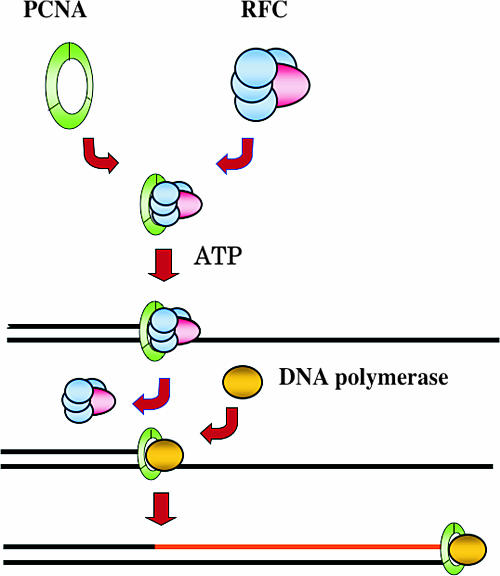
FIG. 1.
Molecular mechanism of processive DNA synthesis. The clamp loader (RFC) opens and recruits the clamp (PCNA) ring onto the primer terminus of the DNA strand in an ATP-dependent manner. The clamp loader then changes off with DNA polymerase, which can synthesize the DNA strand processively without falling off the DNA strand.
After clamp loading, DNA polymerase accesses the PCNA, and the polymerase-clamp complex catalyzes processive DNA synthesis. Therefore, structural and functional studies of the DNA polymerase-PCNA interaction are the next target to elucidate the overall mechanisms of replication fork progression. Extensive studies of the PCNA-interacting proteins revealed that the proteins contain a small conserved sequence motif, called the PCNA-interacting protein box (PIP box), which binds to a common site on PCNA (41). The PIP box consists of the sequence “Qxxhxxaa,” where “x” represents any amino acid, “h” represents a hydrophobic residue (e.g., L, I, or M), and “a” represents aromatic residues (e.g., F, Y, or W). Archaeal DNA polymerases have PIP-box-like motifs in their sequences (reviewed in reference 40). However, few studies have experimentally investigated the function of the motifs. The C-terminal 50 amino acid fragments of the family B DNA polymerases from Archaeoglobus fulgidus and Pyrococcus horikoshii, including the putative PIP-box-like sequence, generated positive signals in two-hybrid analyses and thus interacted with PCNA (31). In the case of Sulfolobus solfataricus, a family B DNA polymerase (PolBI) was unable to interact with PCNA after the deletion of the PIP-box-like motif located at the N terminus (8).
We previously proposed that PIP-box-like sequences exist in the C termini of PolBI and DP2 of PolD from P. furiosus (3). In this study, we created mutant PolBI and PolD, which lack their putative PIP-box motifs, and investigated whether the motifs are actually involved in the functional interaction with P. furiosus PCNA.
MATERIALS AND METHODS
Materials.
The various pET vectors used in this study were obtained from Novagen. Escherichia coli strain JM109, used for plasmid amplification, was obtained from Takara Bio. Epicurian coli BL21-CodonPlus(DE3)-RIL cells for protein expression were obtained from Stratagene. Oligonucleotides used as primers for PCR were synthesized by Hokkaido System Science. The M13mp18 single-stranded circular DNA, used as the template for the DNA polymerase assay, was obtained from Takara Bio.
Construction of an expression system for the PolBI and PolD deletion mutants.
The preparation of the Δ1 mutant PolBI, lacking 29 amino acids from the C terminus, was described in our previous report (21). The gene encoding PolBIΔPIP, with a deletion of the C-terminal 14 amino acids, was amplified by PCR using oligonucleotides 5′-GGGCCCCATATGATTTTAGATGTGGAT-3′ (the NdeI recognition sequence is underlined) and 5′-GAGGGCGCGGCCGCCTATGTCTTTTGGTA-3′ (containing the NotI recognition sequence [underlined]) as primers from the P. furiosus genome. The amplified gene was cloned into the pGEM-T Easy vector (Promega), and the entire nucleotide sequence was confirmed. The cloned gene was digested by NdeI-NotI and was inserted into the corresponding sites of pET21a. The constructed plasmid was designated pPOLBΔPIP. To construct an expression plasmid for DP2 using the pET system, the DP2 gene was amplified by PCR as two fragments by using a nested PCR method. The N-terminal fragment was amplified by PCR using oligonucleotides 5′-GTGGTGCTGATGGAGCTTCC-3′ and 5′-GCCTTTACGAACTCTTGGATCC-3′ as primers for the first PCR and then 5′-GGCATATGGAGCTTCCAAAGGAAATTGAGG-3′ (the NdeI recognition sequence is underlined) and 5′-CCGGATCCACCACTCCTCTACATAG-3′ (the BamHI recognition sequence is underlined) for the second PCR. The C-terminal fragment was amplified by PCR using oligonucleotides 5′-CTATGTAGAGGAGTGGTGGATCC-3′ and 5′-CTTAAAAGTTGTGGTCAGCGTTTGG-3′ as primers for the first PCR and then 5′-CCGGATCCAAGAGTTCGTAAAGGCCGTTAATGAGGCCTATG-3′ (the BamHI recognition sequence is underlined) and 5′-CCGCGGCCGCTTAGCGTTTGGAGAAGAAGTCGTCC (the NotI recognition sequence is underlined) for the second PCR. The C-terminal deletion mutant of the DP2 gene was also amplified by PCR as two fragments using oligonucleotides 5′-CCGGATCCAAGAGTTCGTAAAGGCCGTTAATGAGGCCTATG-3′ (the BamHI recognition sequence is underlined) and 5′-CCCGCGGCCGCTCACTTCTTTTTAGGC-3′ (containing the NotI recognition sequence [underlined]) as primers from the P. furiosus genome. The amplified gene was cloned into the pGEM-T Easy vector, and the entire nucleotide sequence was confirmed. The two cloned gene fragments were connected at the BamHI site, and the fragment was then inserted into the NdeI-NotI sites of pET21a (Ampr). The constructed plasmids were designated pWTDP2 and pDP2ΔPIP. The gene encoding DP1 was amplified and inserted into the NdeI-BamHI sites of pET28a (Kmr) to construct the expression plasmid pWTDP1.
Overproduction and purification of the DNA polymerases, PCNA, and RFC.
To obtain recombinant PolBI, PolBIΔPIP, PolD, and PolDΔPIP, the host E. coli cells (Epicurian coli BL21-CodonPlus(DE3)-RIL) carrying the corresponding plasmids were grown in 1 liter of LB medium containing 50 μg/ml ampicillin and 34 μg/ml chloramphenicol (in addition, kanamycin was added to 50 μg/ml in the cultivation for PolD) at 37°C. For the preparation of PolD, recombinant E. coli producing PolD was obtained by cotransformation of the resultant plasmids pWTDP1 and pWTDP2. Both of the pET vectors have the ColE1 ori and were incompatible for cotransformation in general. However, transformants containing both pWTDP1 and pWTDP2 were obtained by selection for Ampr and Kmr colonies, although the efficiency was quite low. The cells were cultured to an A600 of 0.5, the expression of the pol genes was then induced by adding isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 1 mM, and cultivation was continued for a further 5 h at 37°C. The cells were harvested and disrupted by sonication in buffer A containing 50 mM Tris-HCl (pH 8.0), 0.5 mM dithiothreitol, 0.1 mM EDTA, and 10% glycerol. The soluble cell extracts, obtained by centrifugation (12,000 × g, 20 min), were heated at 80°C for 15 min. The heat-resistant fractions obtained by centrifugation were treated with 0.15% polyethylenimine to remove the nucleic acids. The soluble proteins were precipitated by 80% saturation with ammonium sulfate. The precipitate was resuspended in buffer B containing 50 mM Tris-HCl (pH 8.0), 1 M (NH4)2SO4, 0.5 mM dithiothreitol, 0.1 mM EDTA, and 10% glycerol and was subjected to chromatography on a HiTrap Phenyl column (GE Healthcare Bioscience). The PolBI, PolBIΔPIP, PolD, and PolDΔPIP proteins were eluted with ammonium sulfate with a 1.0 to 0 M concentration gradient. The eluted proteins were dialyzed against buffer A containing 0.04 M NaCl, and the dialysate was subjected to chromatography on a HiTrap Q column (GE Healthcare Bioscience). PolBI and PolBIΔPIP were obtained in the unbound fraction. The purification procedure for PolD and PolDΔPIP was the same as that for PolBI. However, these enzymes were bound to the HiTrap Q matrix. The fractions eluted at the 0.35 to 0.4 M NaCl concentration were dialyzed against buffer A containing 0.1 M NaCl and were fractionated further on a HiTrap Heparin HP column (GE Healthcare Bioscience). The column was developed with a linear gradient of 0 to 1 M NaCl, and PolD and PolDΔPIP were eluted at about 0.4 to 0.5 M NaCl. The purified fractions were pooled and stored at 4°C. Preparation of PfuPCNA and PfuRFC was performed as described in our previous reports (3, 5).
Primer extension reactions by PolBI and PolD.
An in vitro primer elongation reaction in the absence and presence of various combinations of PCNA and RFC, using M13mp18 single-stranded DNA annealed with a 32P-labeled oligonucleotide primer, was performed basically as described previously (5). Briefly, the reaction mixture, containing 20 mM Tris HCl (pH 8.8), 100 mM NaCl, 5 mM MgCl2, 10 mM KCl, 0.1% Triton X-100, 250 μM deoxynucleoside triphosphate, and 0.2 μg of template-primer DNA (30 μl), was preheated at 70°C for 1 min, and the reaction was then started by adding DNA polymerases to 1 nM. Aliquots (8 μl) were obtained after 2.5, 5, and 10 min, and a 2-μl aliquot of the solution (98% deionized formamide, 1 mM EDTA, 0.1% xylene cyanol, 0.1% bromphenol blue) was added. For the reactions with PfuPCNA and PfuRFC, these accessory factors were added to the reaction mixture at an equal molar ratio to the DNA polymerases. The reaction products were analyzed by electrophoresis on a polyacrylamide gel (10%) containing 8 M urea or an alkaline agarose gel (1%) containing 50 mM NaOH and 1 mM EDTA and were visualized by autoradiography.
Surface plasmon resonance analysis.
The Biacore system was used to study the physical interactions between DNA polymerases and PCNA. To monitor the interactions, highly purified PfuPCNA was fixed on a Sensor Chip CM5 (Biacore) according to the manufacturer's recommendations. To measure the kinetic parameters, various concentrations of purified PolBI, PolBIΔPIP, PolD, and PolDΔPIP were applied to the PCNA-immobilized sensor chips. All measurements were performed at a continuous flow rate of 30 μl/min in a buffer containing 10 mM HEPES (pH 7.4), 150 mM NaCl, and 0.005% Tween 20. At the end of each cycle, the bound protein was removed by washing with 2 M NaCl. The association and dissociation phase data were fit simultaneously using a data analysis program, BIAevaluation 3.2 (Biacore).
RESULTS
The C-terminal region of PolB is required for the functional interaction with PCNA.
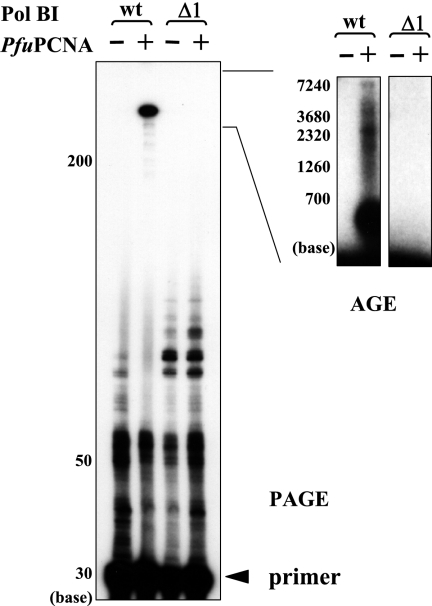
We previously made a series of deletion mutants from the C-terminal end of PolBI and analyzed their DNA-polymerizing and 3′-5′ exonucleolytic activities (21). We used one of these mutant PolBIs, Δ1, lacking 29 amino acids from the C terminus, to investigate the response from PfuPCNA for primer extension abilities during a constant period (5 min), with the ratio of DNA/DNA polymerase as 10:1 in the reaction. As shown in Fig. 2, there was no difference in the product sizes by the Δ1 mutant with and without PCNA, while in contrast, the extension rate of wild-type PolBI was dramatically stimulated by PfuPCNA. This result indicated that the C-terminal region containing 29 amino acids of PolBI is critical for the functional interaction with PCNA.
FIG. 2.
Effects of the C-terminal deletion on PfuPCNA-dependent DNA synthesis by PolBI. The primer extension reaction mixtures, with the indicated proteins in each lane, were analyzed by electrophoresis on a 10% denaturing polyacrylamide gel (PAGE) (left) and a 1% alkaline agarose gel (AGE) (right). wt, wild type.
Preparation of the PIP-box deletion mutant of PolBI and PolD.
We found the PIP-box-like sequences in the C-terminal regions of PolBI (763-QVGLTSWL-770) and DP2, the large subunit of PolD (1253-VISLDDFF-1260). In order to investigate whether these sequences in PolBI and PolD are actually involved in the interactions with PfuPCNA, we made a deletion mutant for each DNA polymerase. Since these PIP-box motifs are located in the C terminus, we deleted amino acids 762 to 775 from the total of 775 amino acids of PolBI and from amino acids 1252 to 1263 from the total of 1,263 amino acids of DP2. Using recombinant expression plasmids containing these mutant genes, the PolBI and PolD mutant proteins were purified by using the same procedures as those for wild-type DNA polymerases, as described in Materials and Methods. The recombinant E. coli cells were cultivated, and PolD (DP1-DP2 complex) was successfully overproduced by IPTG induction. PolBI, PolBIΔPIP, PolD, and PolDΔPIP were purified almost to homogeneity by sequential column chromatographies. About 0.5 mg of each purified protein was obtained from 1 liter of culture.
Analyses of physical interactions between PCNA and DNA polymerases.
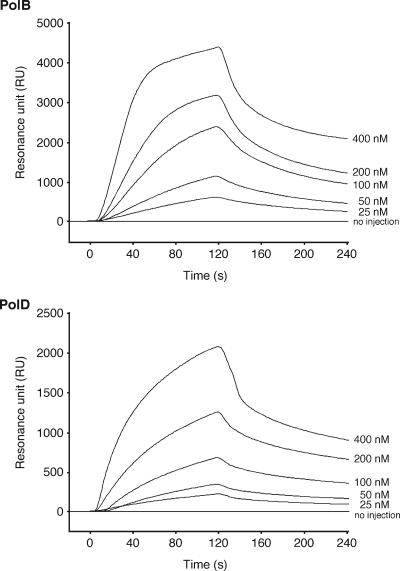
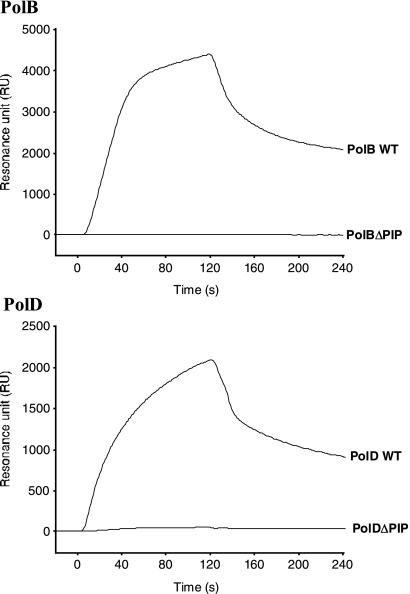
To determine the physical interactions between PCNA and DNA polymerases, we previously reported the binding of PfuPCNA to PolBI and PolD (DP2 but not DP1) by immunoprecipitations and pull-down assays (3). To analyze these interactions more quantitatively, surface plasmon resonance (SPR) experiments were performed. PfuPCNA was immobilized on a Biacore CM5 sensor chip, and subsequently, wild-type PolBI and PolD were injected at different concentrations. The physical interactions between the two proteins were identified by SPR sensorgrams (Fig. 3). When the DP1 and DP2 proteins were injected separately onto the chip, DP2, but not DP1, bound to PfuPCNA on the chip, supporting the interactions observed in our previous immunoprecipitation and pull-down assays described above (data not shown). To measure the binding affinities between the PCNA and DNA polymerases quantitatively, the KD values were obtained from the association and dissociation curves of the sensorgrams (Fig. 3). The association (ka) and dissociation (kd) rate constants were evaluated to be 9.50 × 104 M−1 s−1 and 9.38 × 10−3 s−1, respectively, for PolBI, and 9.61 × 104 M−1 s−1 and 7.34 × 10−3 s−1, respectively, for PolD from the nonlinear curve fitting of the sensorgrams. The binding affinities of PolB and PolD molecules to the PfuPCNA anchored on the sensor chip were calculated from the equilibrium dissociation constant values (KD = kd/ka) to be 98.7 nM and 76.4 nM, respectively. To determine whether our predicted PIP boxes in PolBI and DP2 of PolD are involved in PfuPCNA binding, the PIP-box deletion mutants PolBIΔPIP and PolDΔPIP were subjected to SPR analysis under the same conditions as those used for wild-type DNA polymerases. As shown in Fig. 4, neither PolBIΔPIP nor PolDΔPIP bound to PfuPCNA, in contrast to the cases of wild-type DNA polymerases. These results clearly showed that the C-terminal PIP-box-like sequences are critically important at least for forming stable complexes for both PolBI-PCNA and PolD-PCNA, although other regions of the DNA polymerases may also contribute to the PCNA interactions.
FIG. 3.
Physical interactions between DNA polymerases and PCNA. SPR analyses were performed using a Biacore system to detect the physical interactions between PolBI and PfuPCNA and between PolD and PfuPCNA. Purified PfuPCNA was immobilized on a Biacore sensor chip, and purified PolBI or PolD was analyzed at five different concentrations, as indicated on the right side of each sensorgram. We checked separately that proteins not related to PfuPCNA do not change their sensorgrams from the baseline when this PfuPCNA-immobilized sensor chip is used. The equilibrium constant (KD) was calculated from the obtained sensorgrams.
FIG. 4.
The PIP-box-like motifs of PolBI and PolD are essential for PfuPCNA binding. Physical interactions with PfuPCNA were compared among the wild type (WT) and the PIP-box-truncated mutant (ΔPIP) for both PolBI and PolD by using the Biacore system. Purified PfuPCNA was immobilized on a Biacore sensor chip, and purified DNA polymerases were analyzed.
PfuPCNA does not stimulate the DNA synthesis activities of either PolBIΔPIP or PolDΔPIP.
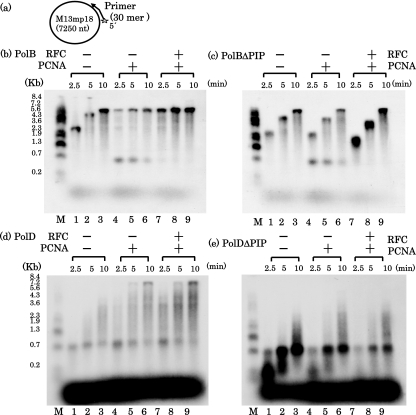
We investigated the effect of PfuPCNA on the activities of PolBIΔPIP and PolDΔPIP by a primer extension reaction assay. Consistent with the data described above, an increase in the rate of extension was enhanced by the addition of PfuPCNA, and this enhancement was more clearly observed with the addition of PfuRFC (Fig. 5b). However, no increase in the rate of extension was observed with PfuPCNA and also by the further addition of PfuRFC in the case of PolBIΔPIP (Fig. 5c). The same result was obtained in the case of PolDΔPIP reactions, in which no effect was observed with PfuPCNA and PfuPCNA/PfuRFC. One pausing site was observed in both the PolBI and PolD reactions at around 700 bases from the primer, when this template-primer combination was used for the primer extension assay. The priming site corresponds to the M13mp18 sequence from positions 6234 to 6205 (GenBank accession no X02513), and therefore, the primer extension reaction from this primer pauses at one of the stable stem-loop structures based on the many palindromic sequences in the intergenic region (39). This pausing was more obvious in the PolD reactions. In the PolBI reaction, pausing was observed only in the presence of PfuPCNA. This pausing was alleviated by the addition of PfuRFC, probably because PfuRFC can unload the PCNA stacked at the pause site. It is interesting that this pausing was also observed in the reaction with PolBIΔPIP and was relieved by the addition of PfuRFC. The amount of the extended products was increased, but the lengths of the products were the same as those from the reaction without PfuPCNA. This observation can be explained by the idea that PfuPCNA by itself loads onto DNA and prohibits the independent PolBI from proceeding along the template DNA. When PfuRFC unloads the stacked PfuPCNA from the DNA, PolBI can move ahead without pausing.
FIG. 5.
Requirement of PCNA and RFC for the primer elongation activities of PolBI and PolD. M13mp18 single-stranded circular DNA, annealed with a 32P-labeled oligonucleotide, was used as the template-primer (a), and primer elongation reactions by PolBI (b), PolBIΔPIP (c), PolD (d), and PolDΔPIP (e) were performed in the presence and absence of PfuPCNA and PfuRFC. The reaction products were analyzed by 1% alkaline agarose gel electrophoresis, and the products were visualized by autoradiography. The sizes indicated on the left were from BstI-digested λ phage DNA, labeled with 32P at each 5′ end.
DISCUSSION
The interaction of the replicative DNA polymerases with the sliding clamp is important for processive DNA synthesis. The clamp binding is also important for the repair DNA polymerases to access the problem site to be repaired. The roles of each DNA polymerase in archaeal cells have not been elucidated. In this study, we made deletion mutants of PolBI and PolD in which only the PIP-box-like motifs were missing and found that these PIP-box-like motifs are likely to be directly involved in the polymerase-PCNA interactions. This result suggests that the binding mode, in which PolBI and PolD interact with the PCNA ring via their C-terminal tails, allows flexible movement of the DNA polymerases on the surface of the PCNA ring to attach to and detach from the DNA strand. The stable binding to PCNA for processive DNA synthesis supports the proposal that both DNA polymerases are involved in the DNA replication process. We previously reported that both PolBI and PolD possess an evident 3′-5′ exonuclease activity (21, 37), and therefore, these enzymes are probably replicative DNA polymerases. It is important to determine how these two DNA polymerases contribute to replication fork processing.
The binding affinities of PolBI and PolD for PfuPCNA are almost the same, as judged by their equilibrium constant (KD) values, suggesting that both DNA polymerases can access the replication fork equally after clamp loading. It is also interesting that the two DNA polymerases showed equal KD values for the PCNA, even though the PIP-box sequences differ somewhat between the proteins (QVGLTSWL for PolBI and VISLDDFF for DP2). Crystal structure analyses will reveal the distinct interaction modes and specificities of the two PIP boxes with regard to PCNA. We are currently analyzing the cocrystal of PolBI and PfuPCNA (33). The cocrystal structures of PolBI-PfuPCNA and DP2-PfuPCNA have yet to be solved.
The KD values for the DNA polymerases are slightly smaller than those for other PCNA binding proteins from P. furiosus, 110 nM for DNA ligase (20), 220 nM for Hjm helicase (9), and 500 nM for Hjc endonuclease (S. Matsumiya et al., unpublished data), determined by previous studies. We also determined the binding affinities of PfuPCNA and PfuRFC by SPR analyses (32). In the case of PfuRFC, even the PIP-box deletion mutant can bind tightly to PfuPCNA, with an equilibrium constant (about 8 nM) that is much lower than those of the two DNA polymerases and other binding proteins. In addition, no difference was observed between wild-type and mutant PfuRFCs, at least in the PCNA-dependent primer extension reactions in vitro (32). This result indicates that PfuRFC has a binding site other than the C-terminal PIP-box sequence, as supported by the three-dimensional structural model based on electron microscopic observations of the clamp-loading complex (30). However, we confirmed that the C-terminal PIP-box-like sequence, QATLFDF, in RFCL can specifically bind to the PfuPCNA and determined the cocrystal structure of a peptide containing this sequence with PfuPCNA (25). Therefore, the C-terminal PIP box of RFCL also contributes to PfuPCNA binding, and the equilibrium constant between PfuPCNA and wild-type PfuRFC should be lower than that of mutant PfuRFC, although the binding constant of wild-type RFC-PCNA could not be obtained, because wild-type PfuRFC binds to the sensor chip nonspecifically. According to the current model of the clamp-loading process, RFC first catches PCNA and opens the clamp ring in an ATP-dependent manner, and the DNA strand then enters the PCNA ring from the opened site. This event should occur before the binding of DNA polymerase to PCNA, and therefore, the stronger affinity between PCNA and RFC may be important to regulate the clamp-loading process. RFC is also required for clamp unloading. It would be interesting to determine how the change from RFC to DNA polymerase and also from DNA polymerase to RFC occurs on the PCNA ring to maintain the appropriate order of the clamp-loading and -unloading processes.
There are conflicting reports describing the interactions of PolD and PCNA from Pyrococcus horikoshii and Pyrococcus abyssi. In the case of P. horikoshii PolD, DP1, but not DP2, can bind to P. horikoshii PCNA, as determined by two-hybrid and SPR analyses (36). On the other hand, P. abyssi PolBI and PolD bind to P. abyssi PCNA in a DNA-dependent manner. Their SPR analyses showed that both of the DNA polymerases bind to the immobilized PCNA only in the presence of DNA and not in the absence of DNA (12). It is not clear at this stage why different SPR analysis results were obtained with DNA polymerases from three different Pyrococcus species. Further investigations are required to understand this issue.
Acknowledgments
We thank Shinichi Kiyonari for help with SPR analyses and making the figures.
Y.I. was supported by a research grant from the Human Frontier Science Program. This work was also supported in part by grants-in-aid from the Ministry of Education, Culture, Sports, Science, and Technology, Japan, to Y.I. We thank the Hou-ansha Foundation for their support of our work.
Footnotes
Published ahead of print on 11 May 2007.
REFERENCES
- 1.Braithwaite, D. K., and J. Ito. 1993. Compilation, alignment, and phylogenetic relationships of DNA polymerases. Nucleic Acids Res. 21:787-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bruck, I., and M. O'Donnell. 2001. The ring-type polymerase sliding clamp family. Genome Biol. 2:REVIEWS3001. http://genomebiology.com/2001/2/1/REVIEWS/3001. [DOI] [PMC free article] [PubMed]
- 3.Cann, I. K. O., S. Ishino, I. Hayashi, K. Komori, H. Toh, K. Morikawa, and Y. Ishino. 1999. Functional interactions of a homolog of proliferating cell nuclear antigen with DNA polymerases in Archaea. J. Bacteriol. 181:6591-6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cann, I. K. O., S. Ishino, N. Nomura, Y. Sako, and Y. Ishino. 1999. Two family B DNA polymerases from Aeropyrum pernix, an aerobic hyperthermophilic crenarchaeote. J. Bacteriol. 181:5984-5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cann, I. K. O., S. Ishino, M. Yuasa, H. Daiyasu, H. Toh, and Y. Ishino. 2001. Biochemical analysis of replication factor C from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 183:2614-2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cann, I. K. O., and Y. Ishino. 1999. Archaeal DNA replication: identifying the pieces to solve a puzzle. Genetics 152:1249-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cann, I. K. O., K. Komori, H. Toh, S. Kanai, and Y. Ishino. 1998. A heterodimeric DNA polymerase: evidence that members of Euryarchaeota possess a distinct DNA polymerase. Proc. Natl. Acad. Sci. USA 95:14250-14255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dionne, I., R. K. Nookala, S. P. Jackson, A. J. Doherty, and S. D. Bell. 2003. A heterotrimeric PCNA in the hyperthermophilic archaeon Sulfolobus solfataricus. Mol. Cell 11:275-282. [DOI] [PubMed] [Google Scholar]
- 9.Fujikane, R., H. Shinagawa, and Y. Ishino. 2006. The archaeal Hjm helicase has recQ-like functions, and may be involved in repair of stalled replication fork. Genes Cells 11:99-110. [DOI] [PubMed] [Google Scholar]
- 10.Garg, P., and P. M. Burgers. 2005. DNA polymerases that propagate the eukaryotic DNA replication fork. Crit. Rev. Biochem. Mol. Biol. 40:115-128. [DOI] [PubMed] [Google Scholar]
- 11.Grabowski, B., and Z. Kelman. 2003. Archeal DNA replication: eukaryal proteins in a bacterial context. Annu. Rev. Microbiol. 57:487-516. [DOI] [PubMed] [Google Scholar]
- 12.Henneke, G., D. Flament, U. Hubscher, J. Querellou, and J. P. Raffin. 2005. The hyperthermophilic euryarchaeota Pyrococcus abyssi likely requires the two DNA polymerases D and B for DNA replication. J. Mol. Biol. 350:53-64. [DOI] [PubMed] [Google Scholar]
- 13.Indiani, C., and M. O'Donnell. 2006. The replication clamp-loading machine at work in the three domains of life. Nat. Rev. Mol. Cell Biol. 7:751-761. [DOI] [PubMed] [Google Scholar]
- 14.Ishino, S., T. Oyama, M. Yuasa, K. Morikawa, and Y. Ishino. 2003. Mutational analysis of Pyrococcus furiosus replication factor C based on the three-dimensional structure. Extremophiles 7:169-175. [DOI] [PubMed] [Google Scholar]
- 15.Ishino, Y., and I. K. O. Cann. 1998. The euryarchaeotes, a subdomain of Archaea, survive on a single DNA polymerase: fact or farce? Genes Genet. Syst. 73:323-336. [DOI] [PubMed] [Google Scholar]
- 16.Ishino, Y., K. Komori, I. K. O. Cann, and Y. Koga. 1998. A novel DNA polymerase family found in Archaea. J. Bacteriol. 180:2232-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishino, Y., and S. Ishino. 2001. DNA polymerases from euryarchaeota. Methods Enzymol. 334:249-260. [DOI] [PubMed] [Google Scholar]
- 18.Ishino, Y., T. Tsurimoto, S. Ishino, and I. Cann. 2001. Functional interactions of an archaeal sliding clamp with mammalian clamp loader and DNA polymerase δ. Genes Cells 6:699-706. [DOI] [PubMed] [Google Scholar]
- 19.Johnson, A., and M. O'Donnell. 2005. Cellular DNA replicases: components and dynamics at the replication fork. Annu. Rev. Biochem. 74:283-315. [DOI] [PubMed] [Google Scholar]
- 20.Kiyonari, S., K. Takayama, H. Nishida, and Y. Ishino. 2006. Identification of a novel binding motif in Pyrococcus furiosus DNA ligase for the functional interaction with proliferating cell nuclear antigen. J. Biol. Chem. 281:28023-28032. [DOI] [PubMed] [Google Scholar]
- 21.Komori, K., and Y. Ishino. 2000. Functional interdependence of DNA polymerizing and 3′-5′ exonucleolytic activities in Pyrococcus furiosus DNA polymerase I. Protein Eng. 13:41-47. [DOI] [PubMed] [Google Scholar]
- 22.Kulaeva, O. I., E. V. Koonin, J. P. McDonald, S. K. Randall, N. Rabinovich, J. F. Connaughton, A. S. Levine, and R. Woodgate. 1996. Identification of a DinB/UmuC homolog in the archaeon Sulfolobus solfataricus. Mutat. Res. 357:245-253. [DOI] [PubMed] [Google Scholar]
- 23.Leipe, D. D., L. Aravind, and E. V. Koonin. 1999. Did DNA replication evolve twice independently? Nucleic Acids Res. 27:3389-3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lipps, G., S. Rother, C. Hart, and G. Krauss. 2003. A novel type of replicative enzyme harbouring ATPase, primase and DNA polymerase activity. EMBO J. 22:2516-2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsumiya, S., S. Ishino, Y. Ishino, and K. Morikawa. 2002. Physical interaction between proliferating cell nuclear antigen and replication factor C from Pyrococcus furiosus. Genes Cells 7:911-922. [DOI] [PubMed] [Google Scholar]
- 26.Matsumiya, S., S. Ishino, Y. Ishino, and K. Morikawa. 2003. Intermolecular ion pairs maintain the toroidal structure of Pyrococcus furiosus PCNA. Protein Sci. 12:823-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsumiya, S., Y. Ishino, and K. Morikawa. 2001. Crystal structure of an archaeal DNA sliding clamp: proliferating cell nuclear antigen from Pyrococcus furiosus. Protein Sci. 10:17-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayanagi, K., T. Miyata, T. Oyama, Y. Ishino, and K. Morikawa. 2001. Three-dimensional electron microscopy of the clamp loader small subunit from Pyrococcus furiosus. J. Struct. Biol. 134:35-45. [DOI] [PubMed] [Google Scholar]
- 29.Miyata, T., T. Oyama, K. Mayanagi, S. Ishino, Y. Ishino, and K. Morikawa. 2004. The clamp-loading complex for processive DNA replication. Nat. Struct. Mol. Biol. 11:632-636. [DOI] [PubMed] [Google Scholar]
- 30.Miyata, T., H. Suzuki, T. Oyama, K. Mayanagi, Y. Ishino, and K. Morikawa. 2005. Open clamp structure in the clamp-loading complex visualized by electron microscopic image analysis. Proc. Natl. Acad. Sci. USA 102:13795-13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Motz, M., I. Kober, C. Girardot, E. Loeser, U. Bauer, M. Albers, G. Moeckel, E. Minch, H. Voss, C. Kilger, and M. Koegl. 2002. Elucidation of an archaeal replication protein network to generate enhanced PCR enzymes. J. Biol. Chem. 277:16179-16188. [DOI] [PubMed] [Google Scholar]
- 32.Nishida, H., S. Ishino, T. Miyata, K. Morikawa, and Y. Ishino. 2005. Identification of the critical region in replication factor C from Pyrococcus furiosus for the stable complex formation with proliferating cell nuclear antigen and DNA. Genes Genet. Syst. 80:83-93. [DOI] [PubMed] [Google Scholar]
- 33.Nishida, H., S. Matsumiya, D. Tsuchiya, Y. Ishino, and K. Morikawa. 2006. Stoichiometric complex formation by proliferating cell nuclear antigen (PCNA) and its interacting protein: purification and crystallization of the DNA polymerase and PCNA monomer mutant complex from Pyrococcus furiosus. Acta Crystallogr. F Struct. Biol. Cryst. Commun. 62:253-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohmori, H., E. C. Friedberg, R. P. Fuchs, M. F. Goodman, F. Hanaoka, D. Hinkle, T. Kunkel, C. W. Lawrence, Z. Livneh, T. Nohmi, L. Prakash, S. Prakash, T. Todo, G. C. Walker, Z. Wang, and R. Woodgate. 2001. The Y-family of DNA polymerases. Mol. Cell 8:7-8. [DOI] [PubMed] [Google Scholar]
- 35.Oyama, T., Y. Ishino, I. K. O. Cann, S. Ishino, and K. Morikawa. 2001. Atomic structure of the clamp loader small subunit from Pyrococcus furiosus. Mol. Cell 8:455-463. [DOI] [PubMed] [Google Scholar]
- 36.Tang, X. F., Y. Shen, E. Matsui, and I. Matsui. 2004. Domain topology of the DNA polymerase D complex from a hyperthermophilic archaeon Pyrococcus horikoshii. Biochemistry 43:11818-11827. [DOI] [PubMed] [Google Scholar]
- 37.Uemori, T., Y. Sato, I. Kato, H. Doi, and Y. Ishino. 1997. A novel DNA polymerase in the hyperthermophilic archaeon, Pyrococcus furiosus: gene cloning, expression, and characterization. Genes Cells 2:499-512. [DOI] [PubMed] [Google Scholar]
- 38.Uemori, T., Y. Ishino, H. Doi, and I. Kato. 1995. The hyperthermophilic archaeon Pyrodictium occultum has two α-like DNA polymerases. J. Bacteriol. 177:2164-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vieira, V., and J. Messing. 1987. Production of single-stranded plasmid DNA. Methods Enzymol. 153:3-11. [DOI] [PubMed] [Google Scholar]
- 40.Vivona, J. B., and Z. Kelman. 2003. The diverse spectrum of sliding clamp interacting proteins. FEBS Lett. 546:167-172. [DOI] [PubMed] [Google Scholar]
- 41.Warbrick, E. M. 2000. The puzzle of PCNA's many partners. BioEssays 22:997-1006. [DOI] [PubMed] [Google Scholar]