Abstract
The pathway for the synthesis of di-myo-inositol-phosphate (DIP) was recently elucidated on the basis of the detection of the relevant activities in cell extracts of Archaeoglobus fulgidus and structural characterization of products by nuclear magnetic resonance (NMR) (N. Borges, L. G. Gonçalves, M. V. Rodrigues, F. Siopa, R. Ventura, C. Maycock, P. Lamosa, and H. Santos, J. Bacteriol. 188:8128-8135, 2006). Here, a genomic approach was used to identify the genes involved in the synthesis of DIP. Cloning and expression in Escherichia coli of the putative genes for CTP:l-myo-inositol-1-phosphate cytidylyltransferase and DIPP (di-myo-inositol-1,3′-phosphate-1′-phosphate, a phosphorylated form of DIP) synthase from several (hyper)thermophiles (A. fulgidus, Pyrococcus furiosus, Thermococcus kodakaraensis, Aquifex aeolicus, and Rubrobacter xylanophilus) confirmed the presence of those activities in the gene products. The DIPP synthase activity was part of a bifunctional enzyme that catalyzed the condensation of CTP and l-myo-inositol-1-phosphate into CDP-l-myo-inositol, as well as the synthesis of DIPP from CDP-l-myo-inositol and l-myo-inositol-1-phosphate. The cytidylyltransferase was absolutely specific for CTP and l-myo-inositol-1-P; the DIPP synthase domain used only l-myo-inositol-1-phosphate as an alcohol acceptor, but CDP-glycerol, as well as CDP-l-myo-inositol and CDP-d-myo-inositol, were recognized as alcohol donors. Genome analysis showed homologous genes in all organisms known to accumulate DIP and for which genome sequences were available. In most cases, the two activities (l-myo-inositol-1-P cytidylyltransferase and DIPP synthase) were fused in a single gene product, but separate genes were predicted in Aeropyrum pernix, Thermotoga maritima, and Hyperthermus butylicus. Additionally, using l-myo-inositol-1-phosphate labeled on C-1 with carbon 13, the stereochemical configuration of all the metabolites involved in DIP synthesis was established by NMR analysis. The two inositol moieties in DIP had different stereochemical configurations, in contradiction of previous reports. The use of the designation di-myo-inositol-1,3′-phosphate is recommended to facilitate tracing individual carbon atoms through metabolic pathways.
Di-myo-inositol phosphate (DIP) is the most widespread organic solute in microorganisms adapted to hot environments (27). This compound was first identified by Scholz and coworkers in the archaeon Pyrococcus woesei (28) and was later encountered in many other hyperthermophiles, including members of the genera Methanotorris, Thermococcus, Thermotoga, Aquifex, Pyrodictium, Aeropyrum, Archaeoglobus, Stetteria, and Pyrolobus) (6, 11-13, 15, 16, 27). Additionally, DIP occurs as a minor solute in the thermophilic bacteria Rubrobacter xylanophilus and Persephonella marina (10, 27). Thus far, DIP has never been encountered in organisms with optimal growth temperatures below 60°C, and hence, the assumption that it plays a role in the thermoprotection of cellular components in vivo is often stated (22, 26, 28).
The question concerning the contribution of compatible solutes from hyperthermophiles to the mechanisms of osmo- and thermoadaptation can only be answered once the pathways for the synthesis of these solutes are known in detail, i.e., once the genes, enzymes, substrates, and reaction products are fully characterized. With this goal in mind, our team has elucidated the biosynthetic pathways of mannosylglycerate and characterized the respective genes and enzymes (1, 9, 17). Moreover, we recently reported the routes for the synthesis of DIP, diglycerol-phosphate, and glycerol-phospho-inositol (GPI) in Archaeoglobus fulgidus, but the genetic and biochemical characterization was lacking (2). In this context, the main objective of the present work was to identify the genes responsible for the synthesis of DIP, a canonical solute of hyperthermophiles.
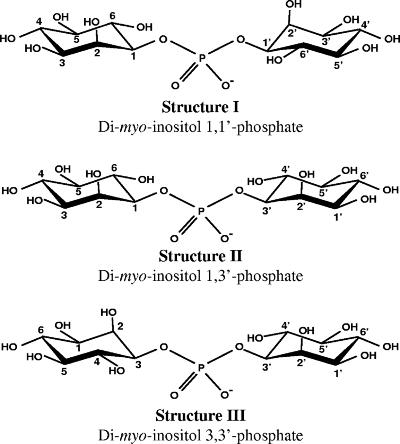
Given that the structural peculiarities of the myo-inositol molecule led to great confusion in the nomenclature of inositol derivatives (20), the three possible configurations of DIP are represented in Fig. 1 for the sake of clarity and easy perception of the structural differences. Throughout this article, stereospecific numbering of the inositol carbon atoms will be used based on the numbering of free myo-inositol. The carbon atom bearing the axial hydroxyl group is designated 2; if the molecule is oriented so that the axial hydroxyl group is above the plane of the molecule, the numbering of carbon atoms goes clockwise (see Fig. S1 in the supplemental material).
FIG. 1.
The structures of the three possible stereoisomers of DIP. Structure I represents the configuration of DIP as reported by van Leeuwen et al. (30) and designated l,l-di-myo-inositol-1,1′-phosphate. Compounds I and III are stereoisomers, while compound II is optically inactive. In these representations, the numbering of the inositol atoms is based on the l configuration according to the “relaxation of lowest-locant rule” as recommended by the Nomenclature Committee of the International Union of Biochemistry in cases where metabolic relationships should be evidenced (20). A short note on myo-inositol nomenclature is given in the supplemental material.
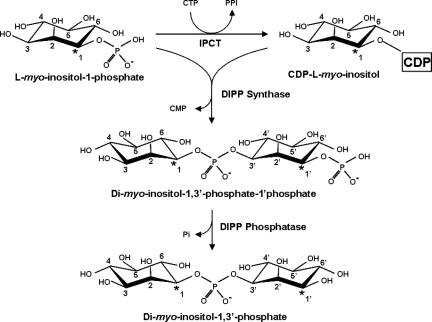
The sequence of reactions leading to DIP synthesis has been established on the basis of the detection of the relevant enzymatic activities in cell extracts of A. fulgidus and of the structural characterization of intermediates and final products by nuclear magnetic resonance (NMR) (2). l-myo-inositol-1-P is activated to CDP-inositol via the activity of CTP:l-myo-inositol-1-P cytidylyltransferase (IPCT); subsequent condensation of CDP-inositol and l-myo-inositol-1-P yields a phosphorylated form of DIP designated DIPP (di-myo-inositol-1,3′-phosphate-1′-phosphate). The extra phosphorylation position was unambiguously assigned. Finally, DIPP is dephosphorylated to yield DIP. Intriguingly, DIP synthesized via this reaction scheme was expected to exhibit the stereochemical configuration of structure II (Fig. 1), in disagreement with the configuration earlier established by van Leeuwen et al. (30) in P. woesei (structure I in Fig. 1). This apparent inconsistency required definite resolution.
Even though DIP was discovered more than 15 years ago, the genes and enzymes for the synthesis of the solute remain elusive. In this work, a genomic approach was used to identify the genes involved in DIP biosynthesis. Genes encoding IPCT and DIPP synthase (DIPPS; equivalent to CDP-inositol:l-myo-inositol-1-phosphate transferase) from several (hyper)thermophiles were uncovered by functional expression in Escherichia coli. Additionally, l-myo-inositol-1-P labeled with carbon 13 was synthesized from [6-13C]glucose in a coupled, two-step reaction catalyzed by Thermoproteus tenax hexokinase and A. fulgidus l-myo-inositol-1-P synthase. This l-myo-[1-13C]inositol-1-P was used as a substrate for recombinant or native enzymes. As a result of this strategy, the configuration of the inositol moieties of DIP was firmly established by tracing the fate of the label through the biosynthetic pathway by NMR analysis of the reaction products.
MATERIALS AND METHODS
Materials.
dl-glycerol-3-P, glucose-6-P, NAD+, CTP, ATP, UTP, GTP, CDP-glycerol, glycerol, and myo-inositol were purchased from Sigma-Aldrich (St. Louis, MO). [6-13C]glucose was obtained from Omicron Biochemicals, Inc. (Indiana). CDP-d-myo-inositol was obtained by chemical synthesis (this work), as well as l-myo-inositol-1-P and d-myo-inositol-1-P (2). CDP-l-myo-inositol and l-[1-13C]myo-inositol-1-P were synthesized enzymatically (this work). DIP from P. woesei was obtained from Bitop AG (Witten, Germany), and 3-(1X-glycerol)-myo-inositol-phosphate, or GPI, was isolated from A. fulgidus (13).
Organisms and growth conditions.
Pyrococcus furiosus strain 3638T, A. fulgidus strain 7324, R. xylanophilus strain 9941T, and T. maritima strain 3109T were obtained from Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany. Thermococcus kodakaraensis strain KOD1 and Aquifex aeolicus strain VF5 biomasses were kindly provided by T. Imanaka (Kyoto University, Japan) and H. Huber (University of Regensburg, Germany), respectively. P. furiosus, A. fulgidus, and R. xylanophilus were cultivated as previously described by Martins and Santos (14), Borges et al., (2), and Empadinhas et al. (10), respectively. T. maritima was grown in medium 343 as described in the Deutsche Sammlung von Mikroorganismen und Zellkulturen.
Enzyme assays.
The activity of DIPPS was determined in cell extracts of E. coli harboring ipct/dipps genes. The reaction mixtures, in a total volume of 0.5 ml containing 20 mM Tris-HCl (pH 7.6), 10 mM MgCl2, 5 mM of the putative substrates, and cell extract (around 12 mg of total protein), were incubated for 1 h at 80°C (or 45°C for the enzyme from R. xylanophilus). The reaction products were analyzed by 31P-NMR spectroscopy. For details, see Borges et al. (2).
Determination of DIP stereochemistry.
The stereochemical configuration of the inositol moieties in intermediate metabolites and end products was determined by 31P-NMR and 13C-NMR spectroscopy using l-[1-13C]myo-inositol-1-P as a substrate for the recombinant enzymes. The stereochemistry of DIP was also investigated in cell extracts of natural producers (A. fulgidus, P. furiosus, and T. maritima). Except for the labeled substrate provided here, the assays were carried out as previously described (2).
Partial purification of native A. fulgidus DIPPS.
Cells of A. fulgidus were harvested during the late exponential phase of growth (optical density at 600 nm = 0.35) and suspended in Tris-HCl (20 mM; pH 7.6) containing 5 mM MgCl2 and DNase I (10 μg/ml). After cell disruption and removal of cell debris by centrifugation, the supernatant was dialyzed against Tris-HCl (20 mM; pH 7.6) prior to fractionation by fast protein liquid chromatography (Amersham Biosciences). The cell extract was applied to a Q-Sepharose column (20 mM Tris-HCl, pH 7.6) and eluted with a linear gradient of NaCl in the same buffer. Activities of cytidylyltransferase and DIPPS were detected in the flowthrough. Active samples were loaded onto a second Q-Sepharose column, equilibrated at pH 8.1, and again, the two activities were not adsorbed. Subsequently, the active fractions were applied to an SP-Sepharose column (20 mM Tris-HCl, pH 7.0), and both activities were detected in the flowthrough. The active fraction was dialyzed against Tris-HCl (50 mM; pH 7.6) containing 1 M ammonium sulfate. The sample was applied to a phenyl-Sepharose column, and elution was carried out with a linear gradient of ammonium sulfate (1 M to 0 M). The two activities eluted at 0.1 M (NH4)2SO4.
Cloning and expression of putative ipct/dipps genes.
Chromosomal DNAs from A. fulgidus, P. furiosus, T. kodakaraensis, R. xylanophilus, and A. aeolicus were isolated according to the method of Ramakrishnan and Adams (21). The ipct/dipps genes from P. furiosus (PF1058), A. fulgidus (AF0263), and A. aeolicus (aq_1367) were amplified by PCR using Pfu DNA polymerase (Fermentas) and cloned in pET23a plasmid following standard protocols (25). The homologous genes from T. kodakaraensis (TK2279) and R. xylanophilus (EF523341) were cloned in pTRC99a and pET30a, respectively. Many sequencing errors were found in the gene Rxyl_1212 (http://www.jgi.doe.gov), with around 10% of the residues incorrectly determined. Therefore, the correct sequence was determined using Pfu DNA polymerase and submitted to GenBank. E. coli BL21(DE3) cells bearing the constructs were grown at 37°C in LB medium with ampicillin (100 μg/ml) to an optical density at 600 nm of 0.5 and induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 4 h. Kanamycin (30 μg/ml) was used for the selection of E. coli BL21(DE3) bearing the R. xylanophilus gene. The cells were harvested, suspended in Tris-HCl (20 mM; pH 7.6) containing 10 mM MgCl2, and disrupted in a French press; cell debris was removed by centrifugation (30,000 × g; 4°C; 1 h). Total protein was estimated by the Bradford method (3).
Characterization of recombinant enzymes.
The substrate specificity of inositol-1-P cytidylyltransferase was studied in assays using CTP and each of the following alcohol donors: myo-inositol, l-myo-inositol-1-P, d-myo-inositol-1-P, glycerol, and dl-glycerol-3-P; additionally, CTP, ATP, GTP, and UTP were examined as putative nucleotidyl donors in combination with l-myo-inositol-1-P. The substrate specificity of DIPPS was investigated using myo-inositol, l-myo-inositol-1-P, d-myo-inositol-1-P, glycerol, and dl-glycerol-3-P as alcohol acceptors and CDP-l-myo-inositol, CDP-d-myo-inositol, and CDP-glycerol as alcohol donors.
Preparation of CDP-l-myo-inositol and [1-13C]myo-inositol-1-P.
CDP-l-myo-inositol was produced by using cell extracts of E. coli BL21(DE3) harboring the gene EF523341 (R. xylanophilus). The cell extract was incubated at 45°C with CTP and l-myo-inositol-1-P. After 1 hour, the reaction mixtures were treated with alkaline phosphatase (30 min at 37°C) to dephosphorylate residual myo-inositol-1-P. The sample was lyophilized, and the residue was dissolved in 1-propanol-25% ammonia (1:1.5 [vol/vol]) and applied to a silica gel S column. The same solvent system was used for elution. CDP-l-myo-inositol eluted first and was followed by myo-inositol. The fractions containing CDP-l-myo-inositol were pooled after analysis by 1H- and 31P-NMR for purity assessment.
l-[1-13C]myo-inositol-1-P was produced from d-[6-13C]glucose by coupling hexokinase from T. tenax and myo-inositol-1-P synthase (IPS) from A. fulgidus. The ips gene was cloned into pET23a; pET-11c:hxk, containing the hk gene, was kindly donated by Bettina Siebers (University Duisburg-Essen, Germany). E. coli BL21(DE3) cells bearing the constructs were grown and induced as described above. Hexokinase and IPS were partially purified by heat treatment: 30 min at 90°C and 15 min at 60°C, respectively. The synthesis of l-[1-13C]myo-inositol-1-P was performed in a reaction mixture containing 6 mM d-[6-13C]glucose, 6 mM ATP, 6 mM NAD+, 10 mM MgCl2, 3 μg of hexokinase, and 3 μg IPS in 20 mM Tris-HCl (pH 7.6). After 1 hour of incubation at 70°C, the labeled compound was purified by anionic-exchange chromatography (2).
NMR spectroscopy.
The identification of metabolites involved in the synthesis of DIP and GPI was accomplished by using 1H-, 31P-, and 13C-NMR on a Bruker DRX500 (Bruker, Rheinstetten, Germany). Proton, carbon, and phosphorus chemical shifts are relative to 3-(trimethylsilyl)propanesulfonic acid (at 0 ppm), methanol (at 49.3 ppm), or 85% H3PO4 (at 0 ppm), respectively. During this work, we found that the assignment of the 13C resonances of myo-inositol reported in the literature is incorrect (4). Therefore, the assignment of the myo-inositol resonances was performed by running heteronuclear multiple quantum coherence and homonuclear correlation spectroscopy spectra. The carbon chemical shift values of myo-inositol are C-1/C-3, 72.54 ppm; C-2, 72.54 ppm; C-4/C6, 71.50 ppm; and C-5, 74.71 ppm.
Chemical synthesis.
d- and l-myo-inositol-1-P were obtained through a significant modification and improvement of the established procedures, as recorded previously (2). Namely, the selective protection of the 1-OH of 3,4,5,6-tetra-O-benzyl-myo-inositol was carried out using tert-butvldimethylsilyl trifluoromethanesulfonate in the presence of diisopropylethylamine in a 90% yield of the desired product. Benzylation of the remaining free 2-OH group, followed by fluorolysis of the silyl ether, afforded racemic 2,3,4,5,6-penta-O-benzyl-myo- inositol (see Fig. 1 in reference 2). Enantiomerically pure d- and l-myo-inositol-1-P were prepared by resolution of racemic 2,3,4,5,6-penta-O-benzyl-myo-inositol via separation of the diastereoisomeric camphanates (8). After hydrolysis, 1d- and 1l-2,3,4,5,6-penta-O-benzyl-myo-inositol were obtained separately {l [a]D20 +5.96 (c = 0.805, CH2Cl2) (8); [a]D20 +9.2 (c = 1, CHCl3); d [a]D20 −5.39 (c = 0.715, CH2Cl2) (8); [a]D20 −9.0 (c = 1, CHCl3)}, and converted into optically pure d- and l-myo-inositol-1-P as for the racemic compound. The coupling to form CDP-d-myo-inositol was accomplished according to established procedures (18, 24).
Nucleotide sequence accession number.
The correct sequence of the gene Rxyl_1212 was submitted to GenBank with the accession number EF523341.
RESULTS
Identification of genes for DIP synthesis in A. fulgidus.
In a first approach, the purification of the DIPPS activity from A. fulgidus cell extracts was attempted in order to obtain amino acid sequence information that would lead to the identification of the gene in the genome sequence of this organism. We failed to reach a satisfactory purification yield but noticed that DIPPS activity was not separated from the IPCT activity even after four chromatographic steps (Q-Sepharose [pH 7.6 and 8.1], SP-Sepharose, and phenyl-Sepharose). The observed copurification of the two activities led to the hint that they could be present in a single polypeptide chain. Further evidence in support of this hypothesis came from the analysis of the A. fulgidus genome (http://www.tigr.org). We found five genes (AF0263, AF1143, AF1744, AF2044, and AF2299) whose predicted products belong to the family of proteins containing a domain characteristic of CDP-alcohol phosphatidyltransferases (http://www.sanger.ac.uk//cgi-bin/Pfam). Three of them (AF1143, AF1744, and AF2044) are annotated as coding for enzymes implicated in the synthesis of phospholipids, leaving two genes (AF0263 and AF2299) as good candidates to encode DIPPS. Gene AF0263, encoding a bifunctional protein with high homology to nucleotidyltransferases (N-terminal domain) and to CDP-alcohol phosphatidyltransferases (C-terminal domain), appeared especially promising. Importantly, genes homologous to AF0263 were found in all the genomes of organisms known to accumulate DIP (P. furiosus, Pyrococcus horikoshi, T. kodakaraensis, A. aeolicus, Aeropyrum pernix, T. maritima, and R. xylanophilus). Furthermore, genes encoding l-myo-inositol-1-P synthase, the first enzyme in DIP synthesis, were found in the immediate flanking region in T. kodakaraensis, R. xylanophylus, T. maritima, and A. pernix, substantiating the suspected connection between AF0263 and DIP synthesis. Therefore, we proceeded with cloning and expression of this gene in E. coli with the objective of validating the predicted functions.
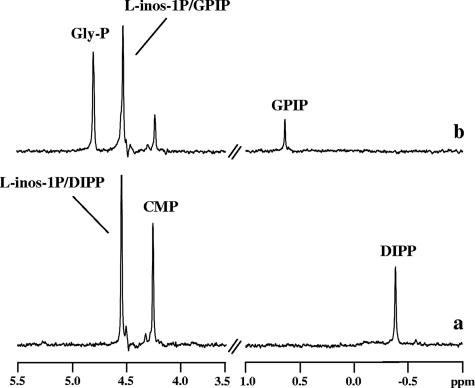
Cell extracts of E. coli BL21(DE3) harboring the gene AF0263 were examined for the presence of IPCT/DIPPS activities using CTP and l-myo-inositol-1-P as substrates. DIPP and vestigial amounts of CDP-inositol were identified by 31P-NMR as products in the reaction mixture (Fig. 2a). We verified that these products were not formed in extracts of E. coli BL21(DE3) cells harboring the empty plasmid. These results showed that AF0263 encodes a bifunctional enzyme able to synthesize DIPP from CTP and l-myo-inositol-1-P via CDP-inositol.
FIG. 2.
31P-NMR spectra of the final products resulting from incubation at 80°C of an E. coli extract (12 mg total protein), harboring the ipct/dipps gene from A. fulgidus, with the following substrates (5 mM): (a) CTP plus l-myo-inositol-1-phosphate and (b) CDP-glycerol plus l-myo-inositol-1-phosphate. The spectra are the differences between the spectra of the reaction mixture with addition of substrates and the spectrum of the control (the reaction mixture with no substrate addition). The spectra were run at a sample temperature of 25°C. Gly-P, dl-glycerol-3-P; L-inos-1P, l-myo-inositol-1-P; CMP, cytidine-5′-monophosphate.
Characterization of recombinant enzymes.
Research effort was directed to purifying the recombinant enzyme from A. fulgidus, but all purification strategies (heat treatment and/or liquid chromatography) applied to extracts of E. coli BL21(DE3) harboring the gene AF0263 led to loss of the DIPPS activity, i.e., only the cytidylyltransferase activity was detected after heat treatment or chromatography. The gene was also expressed in E. coli Rosetta2(DE), which carries a plasmid containing the tRNA genes for codons rarely used in E. coli, with equally poor results. Expression of homologs from T. kodakaraensis, P. furiosus, and A. aeolicus led to similar results. In view of the apparent instability of the second activity, the subsequent characterization of the enzyme activities was performed in cell extracts of E. coli BL21(DE3).
The recombinant enzyme from R. xylanophilus showed distinctive behavior in that the DIPPS activity was never detected; only the cytidylyltransferase activity was observed. The enzyme synthesized CDP-l-myo-inositol from CTP and l-myo-inositol-1-P with high yield, and this property was exploited to produce CDP-l-myo-inositol required for the substrate specificity assays (see below). It was also verified that myo-inositol was not a substrate for this enzyme.
Hereafter, the bifunctional enzyme encoded by gene AF0263 (and homologs) will be designated bifunctional DIPPS and the corresponding fused gene ipct/dipps. The substrate specificity of the bifunctional DIPPS from A. fulgidus was investigated in regard to both cytidylyltransferase and DIPPS activities. The cytidylyltransferase was absolutely specific for CTP and l-myo-inositol-1-P. Other nucleotide donors (ATP, GTP, and UTP) and other alcohol acceptors (myo-inositol, d-myo-inositol-1-P, glycerol, and dl-glycerol-3-P) were not substrates for the enzyme. The DIPPS recognized CDP-l-myo-inositol, CDP-d-myo-inositol, and CDP-glycerol as alcohol donors, but l-myo-inositol-1-P was the only alcohol acceptor used by the enzyme. The other potential substrates examined were myo-inositol, d-myo-inositol-1-P, glycerol, and dl-glycerol 3-P. It was verified by 31P-NMR analysis that GPI-phosphate (GPIP) was the final product when CDP-glycerol was provided as the alcohol donor (Fig. 2b). We checked that the recombinant, bifunctional DIPPS from P. furiosus, T. kodakaraensis, and A. aeolicus used l-myo-inositol-1-P and CTP as substrates for the synthesis of CDP-l-myo-inositol and DIPP. All recombinant enzymes were also able to catalyze the synthesis of GPIP (phosphorylated GPI) when CDP-glycerol was provided in combination with l-myo-inositol-1-P.
Identification of the stereochemical configuration of DIP.
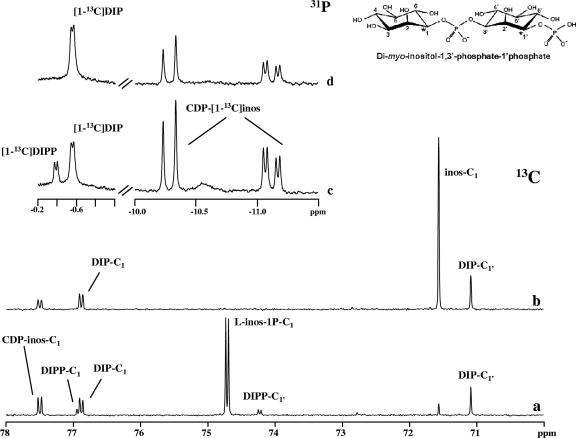
Earlier work by our team established that DIP synthesis occurs via a phosphorylated intermediate, DIPP (2). The proposed pathway would produce DIP with a stereochemical configuration differing from that reported in the literature (30), so we deemed it important to elucidate the stereoconfiguration of the inositol moieties in DIP. For this purpose, l-[1-13C]myo-inositol-1-P was produced from d-[6-13C]glucose, using the hexokinase from T. tenax (7) and l-myo-inositol-1-P synthase from A. fulgidus. After purification, the labeled product was analyzed by 1H-NMR, 13C-NMR, and 31P-NMR. The C-1 resonance at 74.70 ppm was split due to coupling with phosphorus (2JC-P = 5.2 Hz) (Fig. 3), and as expected, the same coupling constant was apparent in the phosphorus spectrum of the same sample. These results, together with the information that only the l stereoisomer is produced by the A. fulgidus inositol-1-P synthase (5), led to the firm identification of the product as l-[1-13C]-myo-inositol-1-P. The use of this labeled compound as a substrate for DIP synthesis in cell extracts of the natural producers allowed us to determine the stereochemical configurations of inositol moieties in intermediates and end products simply by observing 31P-13C coupling patterns (Fig. 4). If DIP were produced in the configuration of the inositol moieties displayed in structure I (Fig. 1), its phosphorus signal (in a spectrum with proton decoupling) should be a triplet, whereas structures II and III would give rise to a doublet and a singlet, respectively.
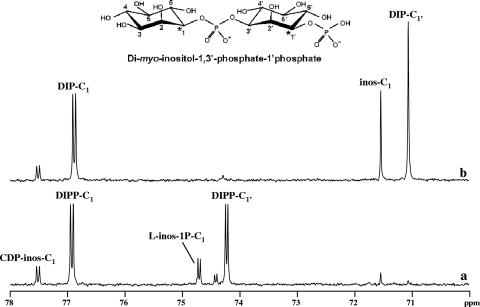
FIG. 3.
13C-NMR and 31P-NMR spectra showing the formation of 13C-labeled products in a cell extract of A. fulgidus after incubation at 80°C. Traces a and c represent spectra of the reaction mixture to which CTP and l-[1-13C]myo-inositol-1-phosphate were added; traces b and d represent spectra of the same sample after treatment with alkaline phosphatase. A schematic representation of DIPP is also shown. Spectra were run at a temperature of 25°C. inos-C1, l-inos-1P-C1, DIP-C1, DIPP-C1, and CDP-inos-C1 designate resonances due to carbon 1 of the inositol moiety in myo-inositol, l-myo-inositol-1-P, DIP, DIPP, and CDP-l-myo-inositol. The splitting of the carbon (or phosphorus) resonances denotes two-bond couplings with phosphorus (or carbon) nuclei.
FIG. 4.
Pathway for DIP biosynthesis using l-[1-13C]myo-inositol-1-phosphate as a labeled precursor. The asterisks highlight the positions of labeled carbon atoms in the several intermediate metabolites and end products as established in this work from 13C- and 31P-NMR analyses of reaction mixtures containing the bifunctional enzyme IPCT/DIPPS (see the text for details). Dashes are used to designate the carbon atoms in the phosphorylated inositol moiety.
The enzymatic assays were performed in A. fulgidus cell extract using CTP and l-[1-13C]myo-inositol-1-P and analyzed by NMR. The 31P-NMR spectrum showed two doublets at −0.38 ppm (JP-C = 6.0 Hz) and at −0.56 ppm (JP-C = 5.6 Hz), corresponding to the phosphodiester resonances of DIPP and DIP, respectively (Fig. 3c and d), meaning that both molecules have configurations identical to structure II in Fig. 1. This conclusion is further supported by the 13C-NMR spectra (Fig. 3a), where the resonances due to the two labeled carbons of DIPP (C-1 and C-1′) are clearly visible at 76.92 ppm and 74.22 ppm, respectively. Treatment of this sample with alkaline phosphatase led to the disappearance of these resonances and a concomitant increase in the intensity of the resonances due to carbons 1 and 1′ of DIP (Fig. 3b). The identity of DIP was also confirmed by spiking with the pure natural compound.
The same strategy allowed us to determine the configuration of the inositol moiety in GPI, whose synthesis also proceeds via a phosphorylated intermediate, GPIP (2). Incubation of A. fulgidus extract with CDP-glycerol and l-[1-13C]myo-inositol-1-P (the substrates for GPI synthesis) resulted in no splitting of the respective phosphorus resonances in the signals due to GPI or GPIP, thus establishing that the phosphate group is linked to carbon 3 of the inositol moiety in both these compounds (data not shown).
The stereochemical configurations of DIPP and GPIP were also confirmed using crude extracts of E. coli producing the recombinant enzymes. The results with the bifunctional, recombinant enzyme of P. furiosus are illustrated in Fig. 5, where the doublets assigned to C-1 and C-1′ in DIPP are clearly seen. Similar results were obtained in E. coli extracts producing the recombinant enzymes from A. fulgidus, T. kodakaraensis, and A. aeolicus. Therefore, in all cases examined, the correct structure of DIP as established in this work is represented by structure II in Fig. 1.
FIG. 5.
13C-NMR spectra showing the formation of 13C-labeled products resulting from incubation at 80°C of an extract of E. coli, bearing the ipct/dipps gene from P. furiosus, with CTP and l-[1-13C]myo-inositol-1-phosphate, before (a) and after (b) treatment of the final reaction mixture with alkaline phosphatase. The inset shows a schematic representation of di-myo-inositol-1,3′-phosphate-1′-phosphate. The spectra were run at a temperature of 25°C. Symbols are as in Fig. 2.
DISCUSSION
Recently, we unequivocally established the biosynthetic pathway of DIP in A. fulgidus and showed that CDP-inositol and the phosphorylated form of DIP are intermediate metabolites in DIP synthesis. Furthermore, the presence of IPCT, DIPPS, and DIPP phosphatase activities was demonstrated in extracts of A. fulgidus and P. furiosus (2). As a logical extension of this work, we set about characterizing the genes and enzymes ascribed to this biosynthetic pathway. A genomic approach combined with hints originating from our attempts to purify the relevant activities led to the proposal of gene AF0263 as a strong candidate to encode the bifunctional IPCT/DIPPS. The final proof came from the functional expression of the gene in E. coli: the recombinant protein in cell extracts catalyzed the synthesis of DIPP from l-myo-inositol-1-P and CTP via CDP-inositol. Moreover, cloning and expression of the homologous genes from P. furiosus, A. aeolicus, and T. kodakaraensis revealed identical functions.
A BLAST search in public databases with the bifunctional IPCT/DIPPS from A. fulgidus resulted in the identification of homologous proteins in several cultured organisms, all of them known to thrive in hot environments (see Fig. S2 in the supplemental material). The two activities, cytidylyltransferase and DIPPS, are encoded in a single polypeptide chain in Pyrococcus spp., T. kodakaraensis, R. xylanophilus, P. marina, and A. aeolicus, while separate genes were found in Hyperthermus butylicus, A. pernix, and T. maritima. Shortly before the manuscript was submitted for publication, the demonstration of this function for the genes of T. maritima appeared in the literature (23).
The N-terminal domain of the bifunctional IPCT/DIPPS showed high similarity to the nucleotidyltransferase family (pfam00483), comprising enzymes that catalyze the transfer of nucleotides onto phosphosugars, like UTP:glucose-1-P uridylyltransferase, CTP:glucose-1-P cytidylyltransferase, or GTP:mannose-1-P guanylyltransferase. The C-terminal domain of IPCT/DIPPS contains the conserved motif (D-G-2[X]-A-R-8[X]-G-3[X]-D-3[X]-D) that is characteristic of the CDP-alcohol phosphatidyltransferase family (pfam01066). These enzymes catalyze the displacement of CMP from CDP-alcohol (diacylglycerol is the most common alcohol) to a second alcohol with formation of a phosphodiester bond and concomitant hydrolysis of pyrophosphate. The phosphatidylinositol synthase from yeast is a well-characterized representative of this family, having 23% identity with the amino acid sequence of A. fulgidus DIPPS (19). Hydropathic profiles predict the presence of three transmembrane segments in the yeast enzyme (19), and similar features are found in the DIPPS domains identified in our study: the stretches comprising residues 272 to 294, 334 to 356, and 393 to 435 have sufficient length and hydrophobicity to span the membrane. The predicted membrane association of the DIPPS domain in the bifunctional IPCT/DIPPS provides a plausible explanation for our failure to purify the enzyme.
The reaction scheme established for DIP synthesis points to a configuration of the inositol units as displayed in structure II (Fig. 1), in disagreement with the stereochemical configuration earlier determined in DIP isolated from Pyrococcus spp. (30). This intriguing inconsistency was definitely resolved in the present work. By using labeled l-[1-13C]myo-inositol-1-P as a substrate for the bifunctional, recombinant enzyme or for the enzymes in cell extracts of the natural producers, the 13C label was traced through the biosynthetic pathway, and we showed that the two inositol moieties in DIP have the configuration represented by structure II (Fig. 1) in all the organisms examined (A. fulgidus, P. furiosus, A. aeolicus, T. kodakaraensis, and T. maritima). We propose that henceforth the designation di-myo-inositol-1,3′-phosphate should be used to highlight the fact that the phosphate group is linked to stereochemically distinct carbons of the two inositol groups and to facilitate spotting individual carbon atoms through metabolic pathways.
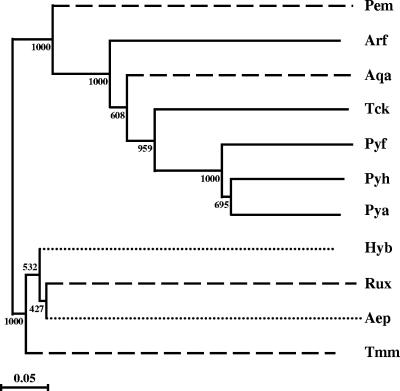
An unrooted phylogenetic tree based on the amino acid sequences of the bifunctional IPCT/DIPPS showed significant topological differences compared with the 16S rRNA tree (31). Although the limited number of sequences available hampers a reliable comparison, it is interesting that only two clusters emerged in the protein-based tree (Fig. 6). In the predominantly euryarchaeote cluster, the Thermococcales form the expected tight cluster and the position of A. fulgidus suggests an early separation from the common ancestor of the genera Thermococcus and Pyrococcus. Curiously, the bacterial species A. aeolicus and P. marina also fall in this “euryarchaeote” cluster. The second cluster comprises the crenarchaeote members, A. pernix and H. butylicus, but also the bacteria T. maritima and R. xylanophilus. It is tempting to speculate that bacterial IPCT/DIPPS either have evolved from distinct ancestors or the respective genes were acquired from archaea by lateral gene transfer. However, analysis of the GC contents and the codon compositions of bacterial ipct/dipps genes did not provide evidence for lateral gene transfer, since their GC contents and codon usages were in line with the properties found in the bulk genes of the respective genomes (data not shown). It is noteworthy that the lack of unusual codon composition or GC content does not rule out the occurrence of lateral gene transfer, since it is conceivable that genes acquired by this means will adapt to the background genomic codon usage during evolution.
FIG. 6.
Unrooted phylogenetic tree based on available amino acid sequences of IPCT/DIPPS. The ClustalX program (29) was used for sequence alignments and to generate the phylogenetic tree by the neighbor-joining tree construction method. The significance of the branching order was evaluated by bootstrap analysis of 1,000 computer-generated trees. The bootstrap values are indicated. Bar = 0.05 change/site. The species and GenBank accession numbers are as follows: A. fulgidus (Arf; NP_069101), P. furiosus (Pyf; NP_578787); P. horikoshii (Pyh, NP_143114); Pyrococcus abyssi (Pya; NP_126714); R. xylanophilus (EF523341); A. aeolicus (NP_213943); T. kodakaraensis (Tck; YP_184692); A. pernix (Aep; NP_147991 for IPCT and NP_147993 for DIPPS), H. butylicus (Hyb; YP_001012367 for IPCT and YP_001012368 for DIPPS); T. maritima (AE000512 for IPCT and DIPPS); and P. marina (Pem; preliminary sequence data were obtained from The Institute for Genomic Research through the website at http://www.tigr.org). Bacteria, euryarchaeotes, and crenarchaeotes are distinguished by using dashed, solid, and dotted lines, respectively.
The cytidylyltransferase domain (C-terminal) of recombinant IPCT/DIPPS was highly specific for CTP and l-myo-inositol-1-P, while the DIPPS domain showed some plasticity in regard to the alcohol-activated donor. All the DIPPS examined (i.e., from A. fulgidus, P. furiosus, A. aeolicus, and T. kodakaraensis) used both CDP-inositol and CDP-glycerol in combination with l-myo-inositol-1-P. Therefore, either DIPP or GPIP can be end products of this activity, depending on the specific CDP-alcohol provided (CDP-inositol or CDP-glycerol, respectively). Thus far, the compatible solute GPI has been found only in species of the genera Archaeoglobus and Aquifex (13). Thus, the observation that all DIPPS examined in this work are also able to catalyze GPI synthesis makes us wonder why GPI does not occur in members of other hyperthermophilic genera, namely, Pyrococcus or Thermococcus. A plausible explanation could be related to insufficient levels of CDP-glycerol, the substrate, besides l-myo-inositol-1-P, that is needed for the synthesis of GPI (2). In A. aeolicus, one of the two organisms known to accumulate GPI, the genes encoding glycerol-3-P cytidylyltransferase and DIPPS are organized in the same operon-like structure and probably under the control of the same promoter. In the other GPI-accumulating organism, A. fulgidus, this close spatial organization is not found, with the gene encoding glycerol-3-P cytidylyltransferase located elsewhere. Nevertheless, CDP-glycerol is expected to be highly available in the cytoplasm of this organism, since CDP-glycerol is also the precursor for the synthesis of diglycerol phosphate, a prominent component of the solute pool of A. fulgidus (11).
In conclusion, this work characterized the key genes and enzymes for the synthesis of DIP, the most widespread compatible solute in hyperthermophiles. Furthermore, it was shown that the same genes are used in the synthesis of GPI, a solute chemically related to DIP and also confined to (hyper)thermophiles. The gene encoding the phosphatase activity that dephosphorylates the phosphorylated intermediates DIPP and GPIP was not characterized; however, the sequence information available for the uncultured organism GZfos13E1 revealed a gene encoding three remarkably expressive activities: cytidylyltransferase, DIPPS, and phosphatase. The hypothesis that the last domain bears the activity that dephosphorylates DIPP appears highly plausible and deserves further investigation. The results disclosed in the present work represent an important milestone in the elucidation of the regulation of DIP biosynthesis and its role in the thermoadaptation of hyperthermophiles.
Supplementary Material
Acknowledgments
This work was funded by Fundação para a Ciência e a Tecnologia, POCTI Portugal, and FEDER Projects A004/2005 Action V.5.1., POCI/BIA-PRO/57263/2004 and POCI/BIA-MIC/56511/2004. M.V.R. (SFRH/BD/25539/2005), N.B. (SFRH/BPD/14841/2003), P.L. (SFRH/BPD/26606/2006), C.F. (SFRH/BD/19599/2004), and N.E. (SFRH/BPD/14828/2003) held fellowships from FCT. Sequencing of P. marina was accomplished with support from NSF.
We thank Vera Lopes and Carla P. Almeida for technical support.
Footnotes
Published ahead of print on 25 May 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Borges, N., J. D. Marugg, N. Empadinhas, M. S. da Costa, and H. Santos. 2004. Specialized roles of the two pathways for the synthesis of mannosylglycerate in osmoadaptation and thermoadaptation of Rhodothermus marinus. J. Biol. Chem. 279:9892-9898. [DOI] [PubMed] [Google Scholar]
- 2.Borges, N., L. G. Gonçalves, M. V. Rodrigues, F. Siopa, R. Ventura, C. Maycock, P. Lamosa, and H. Santos. 2006. Biosynthetic pathways of inositol and glycerol phosphodiesters used by the hyperthermophile Archaeoglobus fulgidus in stress adaptation. J. Bacteriol. 188:8128-8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Breitmaier, E., and W. Voelter. 1989. Carbon-13 NMR spectroscopy: high-resolution methods and applications in organic chemistry and biochemistry, 3rd ed., p. 401. VCH, Weinheim, Germany.
- 5.Chen, L., C. Zhou, H. Yang, and M. F. Roberts. 2000. Inositol-1-phosphate synthase from Archaeoglobus fulgidus is a class II aldolase. Biochemistry 39:12415-12423. [DOI] [PubMed] [Google Scholar]
- 6.Ciulla, R. A., S. Burggraf, K. O. Stetter, and M. F. Roberts. 1994. Occurrence and role of di-myo-inositol-1,1′-phosphate in Methanococcus igneus. Appl. Environ. Microbiol. 60:3660-3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dorr, C., M. Zaparty, B. Tjaden, H. Brinkmann, and B. Siebers. 2003. The hexokinase of the hyperthermophile Thermoproteus tenax. ATP-dependent hexokinases and ADP-dependent glucokinases, two alternatives for glucose phosphorylation in Archaea. J. Biol. Chem. 278:18744-18753. [DOI] [PubMed] [Google Scholar]
- 8.Dreef, C. E., M. Douwes, C. J. Elie, G. A. Marel, and J. H. Boom. 1991. Application of the bifunctional phosphorylating agent bis[6-(trifluoromethyl)benzotriazol-1-yl] methylphosphonate towards the preparation of isosteric di-myo-inositol phospholipid and phosphate analogues. Synthesis 1991:443-447. [Google Scholar]
- 9.Empadinhas, N., J. D. Marugg, N. Borges, H. Santos, and M. S. da Costa. 2001. Pathway for the synthesis of mannosylglycerate in the hyperthermophilic archaeon Pyrococcus horikoshii—biochemical and genetic characterization of key enzymes. J. Biol. Chem. 276:43580-43588. [DOI] [PubMed] [Google Scholar]
- 10.Empadinhas, N., V. Mendes, C. Simões, M. S. Santos, A. Mingote, P. Lamosa, H. Santos, and M. S. da Costa. 18 May 2007. Organic solutes in Rubrobacter xylanophilus: the first example of di-myo-inositol-phosphate in a thermophile. Extremophiles. doi: 10.1007/s00792-007-0084-z. [DOI] [PubMed]
- 11.Gonçalves, L. G., R. Huber, M. S. da Costa, and H. Santos. 2003. A variant of the hyperthermophile Archaeoglobus fulgidus adapted to grow at high salinity. FEMS Microbiol. Lett. 218:239-244. [DOI] [PubMed] [Google Scholar]
- 12.Lamosa, P., L. O. Martins, M. S. da Costa, and H. Santos. 1998. Effects of temperature, salinity, and medium composition on compatible solute accumulation by Thermococcus spp. Appl. Environ. Microbiol. 64:3591-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamosa, P., L. G. Gonçalves, M. V. Rodrigues, L. O. Martins, N. D. H. Raven, and H. Santos. 2006. Occurrence of 1-glyceryl-1-myo-inosityl-phosphate in hyperthermophiles. Appl. Environ. Microbiol. 72:6169-6173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martins, L. O., and H. Santos. 1995. Accumulation of mannosylglycerate and di-myo-inositol-phosphate by Pyrococcus furiosus in response to salinity and temperature. Appl. Environ. Microbiol. 61:3299-3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martins, L. O., L. S. Carreto, M. S. da Costa, and H. Santos. 1996. New compatible solutes related to di-myo-inositol-phosphate in members of the order Thermotogales. J. Bacteriol. 178:5644-5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martins, L. O., R. Huber, H. Huber, K. O. Stetter, M. S. da Costa, and H. Santos. 1997. Organic solutes in hyperthermophilic Archaea. Appl. Environ. Microbiol. 63:896-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martins, L. O., N. Empadinhas, J. D. Marugg, C. Miguel, C. Ferreira, M. S. da Costa, and H. Santos. 1999. Biosynthesis of mannosylglycerate in the thermophilic bacterium Rhodothermus marinus. Biochemical and genetic characterization of a mannosylglycerate synthase. J. Biol. Chem. 274:35407-35414. [DOI] [PubMed] [Google Scholar]
- 18.Moffatt, J. G., and H. G. Khorana. 1961. Nucleoside polyphosphates. X. The synthesis and some reactions of nucleoside-5′ phosphoromorpholidates and related compounds. Improved methods for the preparation of nucleoside-5′ polyphosphates J. Am. Chem. Soc. 83:649-658. [Google Scholar]
- 19.Nikawa, J., T. Kodaki, and S. Yamashita. 1987. Primary structure and disruption of the phosphatidylinositol synthase gene of Saccharomyces cerevisiae. J. Biol. Chem. 262:4876-4881. [PubMed] [Google Scholar]
- 20.Nomenclature Committee of the International Union of Biochemistry. 1989. Numbering of atoms in myo-inositol. Biochem. J. 258:1-2. [PMC free article] [PubMed] [Google Scholar]
- 21.Ramakrishnan, V., and M. W. W. Adams. 1995. Preparation of genomic DNA from sulfur-dependent hyperthermophilic Archaea, p. 95-96. In F. T. Robb and A. R. Place (ed.) Archaea: a laboratory manual—thermophiles. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 22.Ramakrishnan, V., M. F. J. M. Verhagen, and M. W. W. Adams. 1997. Characterization of di-myo-inositol-1,1′-phosphate in the hyperthermophilic bacterium Thermotoga maritima. Appl. Environ. Microbiol. 63:347-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodionov, D. A., O. V. Kurnasov, B. Stec, Y. Wang, M. F. Roberts, and A. L. Osterman. 2007. Genomic identification and in vitro reconstitution of a complete biosynthetic pathway for the osmolyte di-myo-inositol-phosphate. Proc. Natl. Acad. Sci. USA 104:4279-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roseman, S., J. J. Distler, J. G. Moffatt, and H. G. Khorana. 1961. Nucleoside polyphosphates. XI. An improved general method for the synthesis of nucleotide coenzymes. Syntheses of uridine-5′, cytidine-5′ and guanosine-5′ diphosphate derivatives. J. Am. Chem. Soc. 83:659-663. [Google Scholar]
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 26.Santos, H., and M. S. da Costa. 2002. Compatible solutes of organisms that live in hot saline environments. Environ. Microbiol. 4:501-509. [DOI] [PubMed] [Google Scholar]
- 27.Santos, H., P. Lamosa, T. Q. Faria, N. Borges, and C. Neves. 2007. The physiological role, biosynthesis, and mode of action of compatible solutes from (hyper)thermophiles, p. 86-104. In C. Gerday and N. Glandorff (ed.), Physiology and biochemistry of extremophiles. ASM Press, Washington, DC.
- 28.Scholz, S., J. Sonnenbichler, W. Schäfer, and R. Hensel. 1992. Di-myo-inositol-1,1′-phosphate: a new inositol phosphate isolated from Pyrococcus woesei. FEBS Lett. 306:239-242. [DOI] [PubMed] [Google Scholar]
- 29.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Leeuwen, S. H., G. A. van der Marel, R. Hensel, and J. H. van Boom. 1994. Synthesis of l,l di-myo-inositol-1,1′-phosphate: a novel inositol phosphate from Pyrococcus woesei. Recl. Trav. Chim Pays-Bas 113:335-336. [Google Scholar]
- 31.Woese, C. R., O. Kandler, and M. L. Wheelis. 1990. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. USA 87:4576-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.