Abstract
Pseudomonas syringae translocates effector proteins into plant cells via an Hrp1 type III secretion system (T3SS). T3SS components HrpB, HrpD, HrpF, and HrpP were shown to be pathway substrates and to contribute to elicitation of the plant hypersensitive response and to translocation and secretion of the model effector AvrPto1.
Pseudomonas syringae is a phytopathogenic proteobacterium whose host-specific pathovars collectively attack a wide variety of crop plants (21). P. syringae has a type III secretion system (T3SS), which is encoded by hrp and hrc genes (3). The former are so named because they are required (or in operons required) for P. syringae to elicit the defense-associated hypersensitive response (HR) in nonhost plants or to be pathogenic in host plants; the latter encode a subset of nine proteins that are required for the HR and are highly conserved components of the T3SS of both plant and animal pathogens. The Hrp T3SS is required to inject effectors, known as Hop (Hrp outer protein) or Avr (avirulence) proteins, into plant cells, which is an essential process in P. syringae pathogenesis (4).
P. syringae pv. syringae strain 61 is a weak pathogen of bean whose Hrp system has been extensively characterized because cosmid pHIR11, which expresses the system, enables nonpathogens such as Pseudomonas fluorescens to secrete harpin proteins in culture and inject test effectors in planta, which facilitates study of the T3SS and the action of individual effectors in activating or suppressing HR and basal defenses (5, 25, 30, 42). Although the tomato and Arabidopsis pathogen P. syringae pv. tomato DC3000 now has emerged as the primary model for studying P. syringae T3SS-related virulence mechanisms (7, 46), much early work on the Hrp system was done with P. syringae pv. syringae 61 (5, 8, 10, 11, 20, 22, 24, 26, 35, 55, 58). (As part of this study we have collected the strain 61 hrp-hrc sequences carried on pHIR11 into a single file with GenBank accession number EF514224, and we have also made available the complete sequence, along with a list of corrections [http://pseudomonas-syringae.org].) The Hrp systems of these two strains are functionally similar, and the P. syringae pv. syringae 61 hrp-hrc gene cluster can restore pathogenicity on tomato (but not Arabidopsis) to a DC3000 Δhrp-hrc mutant (14). The P. syringae pv. syringae 61 Hrp system is also representative of Hrp1 T3SSs, which are carried by phytopathogens in the Pseudomonadaceae and Enterobacteriaceae and differ in many ways from the Hrp2 T3SSs of phytopathogenic Ralstonia and Xanthomonas spp. (3, 9).
Components of the Hrp1 T3SS machinery that are themselves substrates for the pathway are of particular interest in exploring adaptations for plant cell wall penetration. In addition to forming the channel through which effectors travel, T3SS component-substrates are involved in breaching host cell barriers and regulating protein traffic through the pathway upon host contact (9). Here we address the potential of 11 of the 17 P. syringae pv. syringae 61 Hrp proteins to be T3SS substrates. We excluded from our analysis HrpL, an alternative sigma factor (57), HrpR and HrpS, which are σ54 enhancer-binding proteins (17), and three previously demonstrated T3SS substrates: the HrpA2 pilin protein (47), the HrpK putative translocator (44), and the HrpZ1 harpin (19). Regarding HrpJ, our results largely corroborate extensive analyses of the HrpJ proteins of P. syringae pv. tomato DC3000 and Erwinia amylovora, which appeared while the present study was in preparation (16, 41).
The locations of the 11 hrp genes we investigated are shown in Fig. 1. To test the ability of the encoded proteins to be substrates for the T3SS pathway, each gene was cloned using PCR primers into the Gateway pENTR/SD/D-TOPO vector and then transferred into pCPP3234, which enables the tac promoter to drive expression of a C-terminal fusion of the test protein with a Cya (Bordetella pertussis adenylate cyclase) translocation reporter (49). Strains and plasmids and the PCR primers we used are presented in Tables S1 and S2 in the supplemental material, respectively. Culture conditions were as previously described (49). All pENTR/SD/D-TOPO hrp clones were sequence confirmed. Clones were conjugated from Escherichia coli DH5α into P. fluorescens strain 55 carrying pLN18 or pCPP3297 with the aid of E. coli HB101 harboring the helper plasmid pRK2013 (13, 14). Plasmid pLN18 harbors the complete P. syringae pv. syringae 61 hrp-hrc cluster but lacks genes for the HopA1 effector and its chaperone, and pCPP3297 is a ΔhrcC derivative of pLN18 (30). Production of the predicted Hrp-Cya hybrid proteins in P. fluorescens carrying pLN18 or pCPP3297 was confirmed by immunoblotting with anti-Cya antibodies as described previously (49) (see Fig. S1 in the supplemental material). An advantage of this translocation assay system for identifying T3SS substrates is that it queries the T3SS when it is interacting with plant cells. As shown in Table 1, HrpB-Cya, HrpD-Cya, HrpF-Cya, HrpJ-Cya, and HrpP-Cya were translocated into Nicotiana benthamiana cells in a T3SS-dependent manner. Thus, five of the eleven P. syringae pv. syringae 61 Hrp proteins tested are capable of being T3SS substrates. It should be noted that we also tested for the secretion of the eleven Hrp proteins from P. syringae cells grown in Hrp-inducing minimal medium (28), by using C-terminal Cya and hemagglutinin tags and immunoblot analysis of culture fluids (49). We detected secretion in culture only for HrpJ (data not shown). The secretion and translocation of HrpJ has recently been reported with P. syringae pv. tomato DC3000 (16).
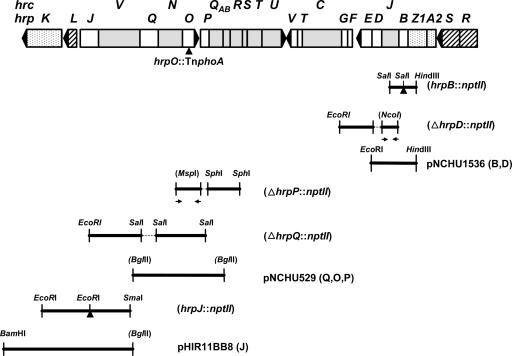
FIG. 1.
The T3SS gene cluster of P. syringae pv. syringae and newly constructed hrp mutations. The figure shows the hrpK-hrpR gene cluster of P. syringae pv. syringae 61, which is carried on pLN18 and encodes a functional T3SS enabling P. fluorescens to translocate T3SS substrates. The hrc genes are shaded, genes encoding positive regulators are denoted with diagonal lines, genes encoding Hrp proteins previously shown to be T3SS substrates are stippled, and the 11 hrp genes analyzed in this report are white (including hrpJ, whose product in P. syringae pv. tomato DC3000 was recently shown to travel the Hrp pathway [16]). The cloned DNA fragments used to construct or complement mutations are also shown (the parenthetical letters after each complementing plasmid denote the mutations complemented by that plasmid), and their origins are described in Table S1 in the supplemental material (25, 32). The ΔhrpQ mutation was also complemented with pCPP2082 (hrcN::TnphoA) (23). Fragments generated by PCR are indicated with opposing arrows, and restriction sites that were lost during mutant construction are given in parentheses. Note that designations such as hrpA2 and hrpZ1 reflect a new, unified nomenclature for P. syringae T3SS substrates (36).
TABLE 1.
Hrp-dependent translocation of P. syringae pv. syringae Hrp proteins
| Proteina | Translocation by P. fluorescens carrying the indicated plasmid (mean pmol of cAMP/μg of protein ± SD)b
|
|
|---|---|---|
| pLN18 (Hrp+) | pCPP3297 (Hrp−) | |
| HrpB-Cya | 172.9 ± 17.4 | 0.5 ± 0.2 |
| HrpD-Cya | 81.8 ± 8.8 | 0.2 ± 0.1 |
| HrpE-Cya | 4.2 ± 1 | 0.9 ± 0.2 |
| HrpF-Cya | 315.3 ± 34.6 | 1.4 ± 0.5 |
| HrpG-Cya | 0.33 ± 0 | 0.5 ± 0 |
| HrpJ-Cya | 310.9 ± 36.9 | 1.6 ± 0.3 |
| HrpO-Cya | 0.2 ± 0.1 | 0.3 ± 0.2 |
| HrpP-Cya | 384.8 ± 5.8 | 1.5 ± 0.7 |
| HrpQ-Cya | 2.1 ± 1.9 | 0.4 ± 0.1 |
| HrpT-Cya | 0.5 ± 0.3 | 0.4 ± 0.2 |
| HrpV-Cya | 2.2 ± 0 | 0.3 ± 0 |
Proteins were expressed from pCPP3234 derivatives (described in Table S1 in the supplemental material), which were carried in P. fluorescens cells also carrying either pLN18 or pCPP3297, as indicated.
cAMP was quantified in N. benthamiana leaf samples 7 h after infiltration with 108 CFU/ml. Values are the means for two samples for each treatment. The experiment was repeated three times with similar results.
We next used functionally nonpolar mutations in P. syringae pv. syringae 61 to determine the role of each of the eleven Hrp proteins in HR elicitation. Strain 61 elicits the HR in N. tabacum (tobacco), N. benthamiana, and other solanaceous plants (6, 25, 54). Elicitation of the HR by P. syringae in nonhost plants is thought to result from R-gene-mediated recognition of one or more translocated effectors (34, 40, 54). The construction of nonpolar hrp mutations marked with a 1.5-kb nptII cassette lacking a rho-independent transcription terminator was previously reported for hrpF, hrpG, hrpT, and hrpV (11). The same methods were used here to construct mutations in hrpB, hrpD, hrpJ, hrpP, and hrpQ, as depicted in Fig. 1. Mutations were initially constructed in subclones derived from pHIR11 and then marker-exchanged into P. syringae pv. syringae 61 as previously described (11). All mutations were confirmed by Southern blot or PCR analysis, and the ability of each mutant to elicit a wild-type HR was restored by the complementing DNA fragments shown in Fig. 1. The location of hrpE and hrpO at the end of their respective operons obviated construction of nonpolar mutations, and previously isolated TnphoA insertions were used (23, 24). The HR tests were performed as previously described, using bacteria inoculated at a titer of 3 × 108 CFU/ml and then rated for visual tissue collapse 48 h later (25).
As summarized in Table 2, the hrpB, hrpD, hrpE, hrpF, hrpO, and hrpQ mutations abolished the ability of P. syringae pv. syringae 61 to elicit the HR in both N. tabacum and N. benthamiana, whereas the hrpG, hrpJ, hrpP, and hrpT mutations had weaker phenotypes in N. benthamiana. Among the hrp mutants with weaker phenotypes, we were particularly interested in hrpP and hrpJ because they involved Hrp proteins that could be translocated. The hrpP gene was completely deleted. The hrpJ mutant produced only the first 115 amino acids of a 345-amino-acid protein. The differing sensitivities of the two Nicotiana spp. was particularly noticeable with the hrpP mutation, which completely abolished HR elicitation in N. tabacum but permitted an HR that was only slightly reduced from wild-type in N. benthamiana. As expected, the mutation affecting HrpV, a negative regulator of the Hrp regulon (45), functioned as a positive control and showed no reduction in HR elicitation in either plant, whereas the hrcC mutant provided a T3SS-deficient negative control that completely abolished HR elicitation. Thus, the use of N. benthamiana reveals that five of the eleven Hrp proteins are not absolutely required for HR elicitation, including two, HrpJ and HrpP, which are themselves substrates for the pathway.
TABLE 2.
Ability of P. syringae pv. syringae hrp mutants to elicit the HR in test plants
| P. syringae pv. syringae strain | Genotype | Phenotype classa
|
|
|---|---|---|---|
| N. tabacum | N. benthamiana | ||
| 61 | Wild type | I | I |
| 61-N540 | hrpB::nptII | IV | IV |
| 61-N363 | ΔhrpD::nptII | IV | IV |
| 61-N19 | hrpE::TnphoA | IV* | IV |
| 61-N491 | ΔhrpF::nptII | IV*† | IV |
| 61-N492 | hrpG::nptII | II*† | I |
| 61-N460 | hrpJ::nptII | II‡ | I |
| 61-2083 | hrpO::TnphoA | IV | IV |
| 61-N534 | ΔhrpP::nptII | IV | II |
| 61-N526 | ΔhrpQ::nptII | IV | IV |
| 61-N402 | ΔhrpT::nptII | III*† | II |
| 61-N407 | ΔhrpV::nptII | I† | I |
| 61-N393 | ΔhrcC::nptII | IV* | IV |
Phenotype classes: I, wild type HR; II, HR slightly reduced and/or delayed; III, HR weak and spotty, some leaves with no response; IV, no HR. Mutants deficient in hrpB, hrpD, hrpJ, hrpO, hrpP, and hrpQ were restored to wild-type ability to elicit the HR in N. tabacum by the complementing clones shown in Fig. 1. Results are based on at least four independent tests for each strain. *, The ability of the mutant to secrete HrpZ1 in culture was previously tested (24); †, similar HR phenotypes in N. tabacum were previously reported for these mutants (11); ‡, a similar HR phenotype in N. tabacum was reported for a P. syringae pv. tomato DC3000 hrpJ mutant (16).
Each of the eleven P. syringae pv. syringae 61 hrp mutants was then tested for its ability to translocate AvrPto1-Cya into N. benthamiana cells (49). Plasmid pCPP3221, which carries avrPto1 fused to cya, was electroporated as described above into the P. syringae pv. syringae 61 mutants. As shown in Table 3, only the hrpG and hrpV mutants translocated significant amounts of AvrPto1-Cya, based on increased levels of cyclic AMP (cAMP) in inoculated tissues. The ΔhrcC mutant provided a negative control for these assays. Interestingly, the hrpG and hrpV mutants translocated ca. two times more AvrPto1-Cya than wild-type P. syringae pv. syringae 61 in repeated experiments. Importantly, all five of the Hrp proteins newly identified to be T3SS substrates in P. syringae pv. syringae 61 are required for translocation of AvrPto1-Cya.
TABLE 3.
Translocation of AvrPto1-Cya into N. benthamiana cells by P. syringae pv. syringae hrp mutants
| P. syringae pv. syringae straina | Genotype | Translocationb (mean pmol of cAMP/μg of protein ± SD) |
|---|---|---|
| 61 | Wild type | 284.7 ± 111.1 |
| 61-N540 | hrpB::nptII | 2.4 ± 0.1 |
| 61-N363 | ΔhrpD::nptII | 2.2 ± 0.5 |
| 61-N19 | hrpE::TnphoA | 1.9 ± 0.1 |
| 61-N491 | ΔhrpF::nptII | 3.1 ± 0.4 |
| 61-N492 | hrpG::nptII | 627.0 ± 83.2 |
| 61-N460 | hrpJ::nptII | 3.9 ± 0.1 |
| 61-2083 | hrpO::TnphoA | 5.4 ± 1.5 |
| 61-N534 | ΔhrpP::nptII | 5.8 ± 2.2 |
| 61-N526 | ΔhrpQ::nptII | 2.5 ± 0.1 |
| 61-N402 | ΔhrpT::nptII | 4.9 ± 3.7 |
| 61-N407 | ΔhrpV::nptII | 739.2 ± 120.8 |
| 61-N393 | ΔhrcC::nptII | 14.3 ± 12.1 |
All strains carried pCPP3221, which expresses AvrPto1-Cya from a vector tac promoter (49).
Levels of cAMP were determined in N. benthamiana leaf samples 7 h after infiltration with 108 CFU/ml (49). Values represent the mean of three samples for each treatment. The experiment was repeated three times with similar results.
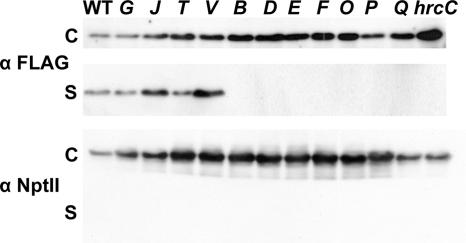
Mutants lacking extracellular Hrp translocation factors would be expected to be deficient in translocation of AvrPto1 into plant cells but not in secretion of AvrPto1 to culture medium. We used anti-FLAG antibodies in immunoblots to follow the distribution of AvrPto1-FLAG between cell-bound and culture supernatant fractions of P. syringae pv. syringae 61 mutants growing in Hrp-inducing minimal medium (28) (Fig. 2). AvrPto1-FLAG was used here because a higher background level of cell lysis was observed with AvrPto1-Cya. We used a P. syringae pv. syringae 61 hrcC mutant and NptII as controls to detect Hrp-independent release of proteins to the culture medium. Plasmid pCPP3026, which carries avrPto1 tagged with a FLAG epitope (18), was electroporated into each of the 11 P. syringae pv. syringae 61 mutant strains. Assays to detect the secretion of AvrPto1-FLAG were done as previously described (52). As shown in Fig. 2, only the hrpG, hrpJ, hrpT, and hrpV mutants secreted AvrPto1. Furthermore, we repeatedly observed that the hrpJ and hrpV mutants secreted higher levels of AvrPto1 than wild-type or hrpG or hrpT mutants. In contrast, Fu et al. (16) observed that AvrPto1 secretion was substantially reduced in the hrpJ mutant of P. syringae pv. tomato DC3000.
FIG. 2.
AvrPto1-FLAG is secreted by hrpG, hrpJ, hrpV, and hrpT mutants in P. syringae pv. syringae 61. The indicated wild-type, hrp mutant, or ΔhrcC strains were grown in Hrp-inducing minimal medium to an optical density at 600 nm of 0.3. Cultures were centrifuged to separate cell pellet (C) and supernatant (S) fractions. Fractions were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to a polyvinylidene difluoride membrane. AvrPto1-FLAG and NptII were detected by immunoblotting with either anti-FLAG or anti-NPTII antibodies, followed by secondary antibodies conjugated to alkaline phosphatase. NptII is a cytoplasmic protein used as a control for cell lysis.
Four of the five Hrp1 T3SS pathway substrates that we have identified here are similar to components of the well-studied T3SSs of Yersinia and Salmonella spp.: HrpB (YscI, PrgJ), HrpD (YscK), HrpJ (YopN, InvE), and HrpP (YscP, InvJ/SpaN) (27, 43). Only HrpF has no obvious homolog or analog among the animal pathogens. The Yersinia YscI and Salmonella PrgJ proteins are thought to form the inner rod of the T3SS needle and are also secreted by the T3SS (33, 37). YscK has been shown by yeast two-hybrid experiments to be part of a peripheral membrane/cytoplasmic ATPase complex that also involves YscL, YscN, and YscQ (29). YscK has a homolog, PscK, in the P. aeruginosa T3SS (15), but the localization of neither YscK nor PscK has been investigated and little else is known about the function of these proteins. Much more is known about the Yersinia YopN and YscP proteins, both of which are translocated and have T3SS regulatory functions (9). The translocation of the HrpP and HrpF proteins is particularly noteworthy because this is the first report that they travel the Hrp1 T3SS, and they may have interesting roles in T3SS regulation during host cell contact. The Yersinia YscP protein functions as a molecular ruler that determines the length of the YscF needle (31). The YscP protein also has a type III secretion substrate specificity switch domain (T3S4), which acts in concert with YscU to switch pathway traffic from channel components to effectors (1). The length of YscP controls the length of the needle so that the needle can extend beyond bacterial surface structures (notably the YadA adhesin), thus enabling needle contact with the host cell cytoplasmic membrane (39). Importantly, YscP is secreted and this secretion is required to control needle length but not substrate specificity (2, 50).
HrpP has a C-terminal T3S4 like that of the Yersinia YscP, but HrpP and its homologs in other plant pathogens are much smaller (1). This observation suggests that HrpP has a role in substrate switching but is not a molecular ruler controlling pilus length in P. syringae. Indeed, the needs for pilus/needle length control are fundamentally different in plant pathogens because a variably thick plant cell wall of 100 to 200 nm must be penetrated. Thus, the plant pathogen pilus must be much longer than the animal pathogen needle, and pilus length must be indeterminate. Studying the conditions and consequences associated with HrpP release may yield important clues to the regulated penetration of plant cell walls.
HrpF was of interest because it is strongly translocated, absolutely required for AvrPto1 secretion and translocation, as well as for bacterial HR elicitation, and it has no obvious homolog or analog outside of Hrp1 T3SSs, where it appears to be ubiquitous (38, 48). It is also noteworthy that HrpF is the second most variable Hrp protein produced by the Hrp systems of P. syringae pv. syringae 61 (after HrpA2) and P. syringae pv. tomato DC3000 (11), which suggests that this protein interacts with the host during natural infections. The P. syringae pv. syringae 61 HrpF also shows similarity in sequence and general properties to the DC3000 HrpA1 pilin protein (11). The hrpF operon also encodes the HrpV-negative regulator and HrpG, which is a chaperone-like suppressor of HrpV (45, 55). HrpF, HrpG, and HrpV are unique to Hrp1 T3SSs (3), which raises the possibility that HrpF has a regulatory function in concert with HrpG and HrpV. The DC3000 HrpA1 protein already has been shown to have a role in regulation: a ΔhrpA1 mutant is strongly reduced in expression of hrp/hrc genes (56).
These observations prompted us to use real-time PCR to test the effects of deleting hrpF and hrpP on the expression of three hrp genes: hrpA2, hrpP, and hrpL, the last of which encodes the alternative sigma factor that activates transcription of the Hrp regulon (51). Real-time PCR using SYBR green I technology and gap1 as a constitutively expressed control was performed as described previously (12). The ΔhrpF mutation strongly reduced expression of all three test genes, which further demonstrates the intriguing similarities between HrpA2 and HrpF (Table 4). In contrast, the ΔhrpP mutation had little, if any, effect on the expression of the hrp genes tested, which is consistent with its proposed role as a substrate specificity switch.
TABLE 4.
Expression of hrp genes in P. s. syringae ΔhrpF and ΔhrpP mutants relative to wild-type strain 61
| Mutant | Expression relative to wild-type P. syringae pv. syringae 61 (mean OD600 ± SD)a
|
|||
|---|---|---|---|---|
| gap1b | hrpA2 | hrpL | hrpP | |
| 61-N491 (ΔhrpF::nptII) | 1 | 0.66 ± 0.14 | 0.54 ± 0.04 | 0.57 ± 0.07 |
| 61-N534 (ΔhrpP::nptII) | 1 | 1.09 ± 0.1 | 0.85 ± 0.06 | ND |
RNA levels were determined by real-time PCR from cultures grown in Hrp minimal medium to an optical density at 600 nm of 0.4. Values represent the mean of three biological replicates. ND, not detectable.
Several intriguing observations should be useful for future studies. First, it is interesting that each of the four operons encoding structural components of the P. syringae T3SS begins with a gene encoding a protein that can travel the pathway and has a demonstrated or potential role in regulating T3SS gene expression or traffic switching (HrpA2, HrpF, HrpJ, and HrpP). Second, the observation that the hrpJ and hrpP mutants are unable to translocate AvrPto1-Cya but still retain an ability to elicit the HR in N. benthamiana suggests that some effectors can still be translocated by these mutants, at least in levels sufficient to elicit the HR. This concept is consistent with the finding that a P. syringae pv. tomato DC3000 hrpJ mutant can still translocate AvrB weakly (16), and it suggests that HR assays may be more sensitive than the Cya translocation assay. Third, it is noteworthy that HrpF, HrpJ, and HrpP are the most abundantly translocated of the five Hrp proteins we have studied here and that previous studies have shown that HrpA and HrpZ1 are secreted in relative abundance in culture (19, 59). These five abundantly trafficked Hrp proteins are likely the major players in the adaptation of the Hrp1 T3SS to the special needs of delivering effectors across plant cell walls, and understanding those adaptations is the next challenge.
Supplementary Material
Acknowledgments
This study was supported by NSF grant MCB-0544066.
We thank Lisa M. Schechter and Kathy R. Munkvold for constructing pCPP3235.
Footnotes
Published ahead of print on 25 May 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Agrain, C., I. Callebaut, L. Journet, I. Sorg, C. Paroz, L. J. Mota, and G. R. Cornelis. 2005. Characterization of a type III secretion substrate specificity switch (T3S4) domain in YscP from Yersinia enterocolitica. Mol. Microbiol. 56:54-67. [DOI] [PubMed] [Google Scholar]
- 2.Agrain, C., I. Sorg, C. Paroz, and G. R. Cornelis. 2005. Secretion of YscP from Yersinia enterocolitica is essential to control the length of the injectisome needle but not to change the type III secretion substrate specificity. Mol. Microbiol. 57:1415-1427. [DOI] [PubMed] [Google Scholar]
- 3.Alfano, J. R., and A. Collmer. 1997. The type III (Hrp) secretion pathway of plant pathogenic bacteria: trafficking harpins, Avr proteins, and death. J. Bacteriol. 179:5655-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alfano, J. R., and A. Collmer. 2004. Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42:385-414. [DOI] [PubMed] [Google Scholar]
- 5.Alfano, J. R., H.-S. Kim, T. P. Delaney, and A. Collmer. 1997. Evidence that the Pseudomonas syringae pv. syringae hrp-linked hrmA gene encodes an Avr-like protein that acts in a hrp-dependent manner within tobacco cells. Mol. Plant-Microbe Interact. 10:580-588. [DOI] [PubMed] [Google Scholar]
- 6.Baker, C. J., M. M. Atkinson, and A. Collmer. 1987. Concurrent loss in Tn5 mutants of Pseudomonas syringae pv. syringae of the ability to induce the hypersensitive response and host plasma membrane K+/H+ exchange in tobacco. Phytopathology 77:1268-1272. [Google Scholar]
- 7.Buell, C. R., V. Joardar, M. Lindeberg, J. Selengut, I. T. Paulsen, M. L. Gwinn, R. J. Dodson, R. T. Deboy, A. S. Durkin, J. F. Kolonay, R. Madupu, S. Daugherty, L. Brinkac, M. J. Beanan, D. H. Haft, W. C. Nelson, T. Davidsen, J. Liu, Q. Yuan, H. Khouri, N. Fedorova, B. Tran, D. Russell, K. Berry, T. Utterback, S. E. Vanaken, T. V. Feldblyum, M. D'Ascenzo, W.-L. Deng, A. R. Ramos, J. R. Alfano, S. Cartinhour, A. K. Chatterjee, T. P. Delaney, S. G. Lazarowitz, G. B. Martin, D. J. Schneider, X. Tang, C. L. Bender, O. White, C. M. Fraser, and A. Collmer. 2003. The complete sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 100:10181-10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collmer, A., J. L. Badel, A. O. Charkowski, W.-L. Deng, D. E. Fouts, A. R. Ramos, A. H. Rehm, D. M. Anderson, O. Schneewind, K. van Dijk, and J. R. Alfano. 2000. Pseudomonas syringae Hrp type III secretion system and effector proteins. Proc. Natl. Acad. Sci. USA 97:8770-8777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cornelis, G. R. 2006. The type III secretion injectisome. Nat. Rev. Microbiol. 4:811-825. [DOI] [PubMed] [Google Scholar]
- 10.Deng, W.-L., and H.-C. Huang. 1998. Cellular locations of Pseudomonas syringae pv. syringae HrcC and HrcJ proteins, required for harpin secretion via the type III pathway. J. Bacteriol. 181:2298-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng, W.-L., G. Preston, A. Collmer, C.-J. Chang, and H.-C. Huang. 1998. Characterization of the hrpC and hrpRS operons of Pseudomonas syringae pathovars syringae, tomato, and glycinea and analysis of the ability of hrpF, hrpG, hrcC, hrpT, and hrpV mutants to elicit the hypersensitive response and disease in plants. J. Bacteriol. 180:4523-4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferreira, A. O., C. R. Myers, J. S. Gordon, G. B. Martin, M. Vencato, A. Collmer, M. D. Wehling, J. R. Alfano, G. Moreno-Hagelsieb, W. F. Lamboy, G. DeClerck, D. J. Schneider, and S. W. Cartinhour. 2006. Whole-genome expression profiling defines the HrpL regulon of Pseudomonas syringae pv. tomato DC3000, allows de novo reconstruction of the Hrp cis element, and identifies novel co-regulated gene. Mol. Plant-Microbe Interact. 19:1167-1179. [DOI] [PubMed] [Google Scholar]
- 13.Figurski, D., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fouts, D. E., J. L. Badel, A. R. Ramos, R. A. Rapp, and A. Collmer. 2003. A Pseudomonas syringae pv. tomato DC3000 Hrp (type III secretion) deletion mutant expressing the Hrp system of bean pathogen P. syringae pv. syringae 61 retains normal host specificity for tomato. Mol. Plant-Microbe Interact. 16:43-52. [DOI] [PubMed] [Google Scholar]
- 15.Frank, D. W. 1997. The exoenzyme S regulon of Pseudomonas aeruginosa. Mol. Microbiol. 26:621-629. [DOI] [PubMed] [Google Scholar]
- 16.Fu, Z. Q., M. Guo, and J. R. Alfano. 2006. Pseudomonas syringae HrpJ is a type III secreted protein that is required for plant pathogenesis, injection of effectors, and secretion of the HrpZ1 Harpin. J. Bacteriol. 188:6060-6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grimm, C., and N. J. Panopoulos. 1989. The predicted protein product of a pathogenicity locus from Pseudomonas syringae pv. phaseolicola is homologous to a highly conserved domain of several procaryotic regulatory proteins. J. Bacteriol. 171:5031-5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ham, J. H., D. W. Bauer, D. E. Fouts, and A. Collmer. 1998. A cloned Erwinia chrysanthemi Hrp (type III protein secretion) system functions in Escherichia coli to deliver Pseudomonas syringae Avr signals to plant cells and to secrete Avr proteins in culture. Proc. Natl. Acad. Sci. USA 95:10206-10211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He, S. Y., H.-C. Huang, and A. Collmer. 1993. Pseudomonas syringae pv. syringae harpinPss: a protein that is secreted via the Hrp pathway and elicits the hypersensitive response in plants. Cell 73:1255-1266. [DOI] [PubMed] [Google Scholar]
- 20.Heu, S., and S. W. Hutcheson. 1993. Nucleotide sequence and properties of the hrmA locus associated with the Pseudomonas syringae pv. syringae 61 hrp gene cluster. Mol. Plant-Microbe Interact. 6:553-564. [DOI] [PubMed] [Google Scholar]
- 21.Hirano, S. S., and C. D. Upper. 2000. Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae: a pathogen, ice nucleus, and epiphyte. Microbiol. Mol. Biol. Rev. 64:624-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang, H.-C., S. Y. He, D. W. Bauer, and A. Collmer. 1992. The Pseudomonas syringae pv. syringae 61 hrpH product: an envelope protein required for elicitation of the hypersensitive response in plants. J. Bacteriol. 174:6878-6885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang, H.-C., S. W. Hutcheson, and A. Collmer. 1991. Characterization of the hrp cluster from Pseudomonas syringae pv. syringae 61 and TnphoA tagging of genes encoding exported or membrane-spanning Hrp proteins. Mol. Plant-Microbe Interact. 4:469-476. [Google Scholar]
- 24.Huang, H.-C., R.-W. Lin, C.-J. Chang, A. Collmer, and W.-L. Deng. 1995. The complete hrp gene cluster of Pseudomonas syringae pv. syringae 61 includes two blocks of genes required for harpinPss secretion that are arranged colinearly with Yersinia ysc homologs. Mol. Plant-Microbe Interact. 8:733-746. [DOI] [PubMed] [Google Scholar]
- 25.Huang, H.-C., R. Schuurink, T. P. Denny, M. M. Atkinson, C. J. Baker, I. Yucel, S. W. Hutcheson, and A. Collmer. 1988. Molecular cloning of a Pseudomonas syringae pv. syringae gene cluster that enables Pseudomonas fluorescens to elicit the hypersensitive response in tobacco plants. J. Bacteriol. 170:4748-4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang, H.-C., Y. Xiao, R.-H. Lin, Y. Lu, S. W. Hutcheson, and A. Collmer. 1993. Characterization of the Pseudomonas syringae pv. syringae hrpJ and hrpI genes: homology of HrpI to a superfamily of proteins associated with protein translocation. Mol. Plant-Microbe Interact. 6:515-520. [DOI] [PubMed] [Google Scholar]
- 27.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huynh, T. V., D. Dahlbeck, and B. J. Staskawicz. 1989. Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science 245:1374-1377. [DOI] [PubMed] [Google Scholar]
- 29.Jackson, M. W., and G. V. Plano. 2000. Interactions between type III secretion apparatus components from Yersinia pestis detected using the yeast two-hybrid system. FEMS Microbiol. Lett. 186:85-90. [DOI] [PubMed] [Google Scholar]
- 30.Jamir, Y., M. Guo, H.-S. Oh, T. Petnicki-Ocwieja, S. Chen, X. Tang, M. B. Dickman, A. Collmer, and J. R. Alfano. 2004. Identification of Pseudomonas syringae type III secreted effectors that suppress programmed cell death in plants and yeast. Plant J. 37:554-565. [DOI] [PubMed] [Google Scholar]
- 31.Journet, L., C. Agrain, P. Broz, and G. R. Cornelis. 2003. The needle length of bacterial injectisomes is determined by a molecular ruler. Science 302:1757-1760. [DOI] [PubMed] [Google Scholar]
- 32.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 33.Kimbrough, T. G., and S. I. Miller. 2000. Contribution of Salmonella typhimurium type III secretion components to needle complex formation. Proc. Natl. Acad. Sci. USA 97:11008-11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kobayashi, D. Y., S. J. Tamaki, and N. T. Keen. 1989. Cloned avirulence genes from the tomato pathogen Pseudomonas syringae pv. tomato confer cultivar specificity on soybean. Proc. Natl. Acad. Sci. USA 86:157-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lidell, M. C., and S. W. Hutcheson. 1994. Characterization of the hrpJ and hrpU operons of Pseudomonas syringae pv. syringae Pss61: similarity with components of enteric bacteria involved in flagellar biogenesis and demonstration of their role in harpinPss secretion. Mol. Plant-Microbe Interact. 7:488-497. [DOI] [PubMed] [Google Scholar]
- 36.Lindeberg, M., J. Stavrinides, J. H. Chang, J. R. Alfano, A. Collmer, J. L. Dangl, J. T. Greenberg, J. W. Mansfield, and D. S. Guttman. 2005. Proposed guidelines for a unified nomenclature and phylogenetic analysis of type III Hop effector proteins in the plant pathogen Pseudomonas syringae. Mol. Plant-Microbe Interact. 18:275-282. [DOI] [PubMed] [Google Scholar]
- 37.Marlovits, T. C., T. Kubori, A. Sukhan, D. R. Thomas, J. E. Galan, and V. M. Unger. 2004. Structural insights into the assembly of the type III secretion needle complex. Science 306:1040-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mor, H., S. Manulis, M. Zuck, R. Nizan, D. L. Coplin, and I. Barash. 2001. Genetic organization of the hrp gene cluster and dspAE/BF operon in Erwinia herbicola pv. gypsophilae. Mol. Plant-Microbe Interact. 14:431-436. [DOI] [PubMed] [Google Scholar]
- 39.Mota, L. J., L. Journet, I. Sorg, C. Agrain, and G. R. Cornelis. 2005. Bacterial injectisomes: needle length does matter. Science 307:1278. [DOI] [PubMed] [Google Scholar]
- 40.Mysore, K. S., and C.-M. Ryu. 2004. Nonhost resistance: how much do we know? Trends Plant Sci. 9:97-104. [DOI] [PubMed] [Google Scholar]
- 41.Nissinen, R. M., A. J. Ytterberg, A. J. Bogdanove, K. J. Van Wijk, and S. V. Beer. 2007. Analysis of the secretomes of Erwinia amylovora and selected hrp mutants reveal novel type III secreted proteins and an effect of HrpJ on extracellular harpin levels. Mol. Plant Pathol. 8:55-67. [DOI] [PubMed] [Google Scholar]
- 42.Oh, H.-S., and A. Collmer. 2005. Basal resistance against bacteria in Nicotiana benthamiana leaves is accompanied by reduced vascular staining and suppressed by multiple Pseudomonas syringae type III secretion system effector proteins. Plant J. 44:348-359. [DOI] [PubMed] [Google Scholar]
- 43.Pallen, M. J., S. A. Beatson, and C. M. Bailey. 2005. Bioinformatics, genomics and evolution of non-flagellar type-III secretion systems: a Darwinian perspective. FEMS Microbiol. Rev. 29:201-229. [DOI] [PubMed] [Google Scholar]
- 44.Petnicki-Ocwiega, T., K. van Dijk, and J. R. Alfano. 2005. The hrpK operon of Pseudomonas syringae pv. tomato DC3000 encodes two proteins secreted by the type III (Hrp) protein secretion system: HopB1 and HrpK, a putative type III translocator. J. Bacteriol. 187:649-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Preston, G., W.-L. Deng, H.-C. Huang, and A. Collmer. 1998. Negative regulation of hrp genes in Pseudomonas syringae by HrpV. J. Bacteriol. 180:4532-4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Preston, G. M. 2000. Pseudomonas syringae pv. tomato: the right pathogen, of the right plant, at the right time. Mol. Plant Pathol. 1:263-275. [DOI] [PubMed] [Google Scholar]
- 47.Roine, E., W. Wei, J. Yuan, E.-L. Nurmiaho-Lassila, N. Kalkkinen, M. Romantschuk, and S. Y. He. 1997. Hrp pilus: an hrp-dependent bacterial surface appendage produced by Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 94:3459-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rojas, C. M., J. H. Ham, J. F. Kim, S. V. Beer, and A. Collmer. 2004. The Erwinia chrysanthemi EC16 hrp/hrc gene cluster encodes an active Hrp type III secretion system that is flanked by virulence genes functionally unrelated to the Hrp system. Mol. Plant-Microbe Interact. 17:644-653. [DOI] [PubMed] [Google Scholar]
- 49.Schechter, L. M., K. A. Roberts, Y. Jamir, J. R. Alfano, and A. Collmer. 2004. Pseudomonas syringae type III secretion system targeting signals and novel effectors studied with a Cya translocation reporter. J. Bacteriol. 186:543-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stainier, I., S. Bleves, C. Josenhans, L. Karmani, C. Kerbourch, I. Lambermont, S. Totemeyer, A. Boyd, and G. R. Cornelis. 2000. YscP, a Yersinia protein required for Yop secretion that is surface exposed, and released in low Ca2+. Mol. Microbiol. 37:1005-1018. [DOI] [PubMed] [Google Scholar]
- 51.Tang, X., Y. Xiao, and J.-M. Zhou. 2006. Regulation of the type III secretion system in phytopathogenic bacteria. Mol. Plant-Microbe Interact. 19:1159-1166. [DOI] [PubMed] [Google Scholar]
- 52.van Dijk, K., V. C. Tam, A. R. Records, T. Petnicki-Ocwieja, and J. R. Alfano. 2002. The ShcA protein is a molecular chaperone that assists in the secretion of the HopPsyA effector from the type III (Hrp) protein secretion system of Pseudomonas syringae. Mol. Microbiol. 44:1469-1481. [DOI] [PubMed] [Google Scholar]
- 53.Vencato, M., T. Tian, J. R. Alfano, C. R. Buell, S. Cartinhour, G. A. DeClerck, D. S. Guttman, J. Stavrinides, V. Joardar, M. Lindeberg, P. A. Bronstein, J. W. Mansfield, C. R. Myers, A. Collmer, and D. J. Schneider. 2006. Bioinformatics-enabled identification of the HrpL regulon and type III secretion system effector proteins of Pseudomonas syringae pv. phaseolicola 1448A. Mol. Plant-Microbe Interact. 19:1193-1206. [DOI] [PubMed] [Google Scholar]
- 54.Wei, C.-F., B. H. Kvitko, R. Shimizu, E. Crabill, J. R. Alfano, N.-C. Lin, G. B. Martin, H.-C. Huang, and A. Collmer. A Pseudomonas syringae pv. tomato DC3000 mutant lacking the type III effector HopQ1-1 is able to cause disease in the model plant Nicotiana benthamiana. Plant J., in press. [DOI] [PubMed]
- 55.Wei, C. F., W. L. Deng, and H. C. Huang. 2005. A chaperone-like HrpG protein acts as a suppressor of HrpV in regulation of the Pseudomonas syringae pv. syringae type III secretion system. Mol. Microbiol. 57:520-536. [DOI] [PubMed] [Google Scholar]
- 56.Wei, W., A. Plovanich-Jones, W.-L. Deng, A. Collmer, H.-C. Huang, and S. Y. He. 2000. The gene coding for the Hrp pilus structural protein is required for type III secretion of Hrp and Avr proteins in Pseudomonas syringae pv. tomato. Proc. Natl. Acad. Sci. USA 97:2247-2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiao, Y., S. Heu, J. Yi, Y. Lu, and S. W. Hutcheson. 1994. Identification of a putative alternate sigma factor and characterization of a multicomponent regulatory cascade controlling the expression of Pseudomonas syringae pv. syringae Pss61 hrp and hrmA genes. J. Bacteriol. 176:1025-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiao, Y., Y. Lu, S. Heu, and S. W. Hutcheson. 1992. Organization and environmental regulation of the Pseudomonas syringae pv. syringae 61 hrp cluster. J. Bacteriol. 174:1734-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yuan, J., and S. Y. He. 1996. The Pseudomonas syringae Hrp regulation and secretion system controls the production and secretion of multiple extracellular proteins. J. Bacteriol. 178:6399-6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.