Abstract
In gram-negative bacteria, transporters belonging to the resistance-nodulation-cell division (RND) superfamily of proteins are responsible for intrinsic multidrug resistance. Haemophilus influenzae, a gram-negative pathogen causing respiratory diseases in humans and animals, constitutively produces the multidrug efflux transporter AcrB (AcrBHI). Similar to other RND transporters AcrBHI associates with AcrAHI, the periplasmic membrane fusion protein, and the outer membrane channel TolCHI. Here, we report that AcrABHI confers multidrug resistance when expressed in Escherichia coli and requires for its activity the E. coli TolC (TolCEC) protein. To investigate the intracellular dynamics of AcrABHI, single cysteine mutations were constructed in AcrBHI in positions previously identified as important for substrate recognition. The accessibility of these strategically positioned cysteines to the hydrophilic thiol-reactive fluorophore fluorescein-5-maleimide (FM) was studied in vivo in the presence of various substrates of AcrABHI and in the presence or absence of AcrAHI and TolCEC. We report that the reactivity of specific cysteines with FM is affected by the presence of some but not all substrates. Our results suggest that substrates induce conformational changes in AcrBHI.
Multidrug efflux transporters belonging to the resistance-nodulation-cell division (RND) superfamily of proteins are broadly represented in gram-negative bacteria. Previous studies established that these transporters play a major role in the intrinsic and induced resistance of gram-negative bacteria to multiple antimicrobial agents including clinically important antibiotics (reviewed in references 7, 11, 18, and 27). Their contribution to antibiotic resistance in clinical settings made RND-type transporters attractive targets of a search for specific and broad-spectrum inhibitors that could be used to increase the efficacy of antibiotics (8).
RND-type multidrug transporters from various species share a high degree of sequence and, correspondingly, structure conservation. Structural and functional studies showed that RND-type transporters exist as trimers, which are organized into two large domains: the transmembrane hydrophobic domain comprised by 36 transmembrane α-helices (TMS) with each protomer contributing 12 TMSs and the large periplasmic domain exposed to aqueous environment of the periplasm (17). This impressive structure, however, is not sufficient to provide multidrug resistance. RND transporters associate with two accessory proteins. The periplasmic membrane fusion proteins (MFPs) are believed to mediate the interaction between an RND transporter and an outer membrane channel belonging to the outer membrane factor family of proteins (18, 27). Together, these interacting three components span the inner and outer membranes and the periplasm of gram-negative bacteria. Mutations in any of the three proteins completely abolish intrinsic multidrug resistance of gram-negative bacteria.
In the three-component complexes, RND transporters are responsible for drug recognition. Crystallization studies of AcrB from Escherichia coli (AcrBEC) revealed several possible drug-interacting sites (16, 25, 26). In the first report, several unrelated drugs were detected in the central cavity formed by three protomers of AcrBEC on the interface between periplasm and the inner membrane (26). Two other sites are located in the periplasmic domains: one is in a prominent cleft on the surface of the periplasmic domain (25); another is deep inside this domain (16). These periplasmic drug binding sites were largely delineated by mutagenesis studies of RND-type transporters from various gram-negative bacteria even before the crystal structures became available (5, 10, 12, 23). Consistent with a high degree of sequence conservation, positions affecting substrate specificity are conserved among various RND transporters. Furthermore, recombinant RND-type multidrug transporters from various gram-negative bacteria are functional when expressed in E. coli (23).
Haemophilus influenzae is a gram-negative pathogen causing respiratory diseases in humans and animals. Genetic and sequence studies showed that H. influenzae constitutively produces a homolog of AcrBEC (19). Similar to other RND transporters, H. influenzae AcrB (AcrBHI) associates with AcrAHI, the periplasmic MFP, and the outer membrane channel TolCHI (24). Inactivation of any of the three proteins AcrAHI, AcrBHI, or TolCHI increased the susceptibility of H. influenzae to 16 antimicrobial compounds. In all cases, the susceptibility profiles of AcrAHI, AcrBHI, or TolCHI null mutants were identical, suggesting that these proteins are components of a single pump. Mutations in other RND transporters identified by sequences searches of H. influenzae genome did not affect drug susceptibility (24). Therefore, AcrABHI-TolCHI is a major, constitutively expressed multidrug efflux complex of H. influenzae and a potent target for the development of specific inhibitors of this transporter.
The substrate specificities of AcrABEC-TolCEC and AcrABHI-TolCHI are very similar and include both positively charged (dyes and erythromycin) and negatively charged (novobiocin) compounds (19, 24). In contrast to E. coli, AcrABHI-TolCHI does not protect H. influenzae from β-lactams, chloramphenicol, tetracycline, or fluoroquinolones. It is presently unclear whether this reflects differences in substrate specificities of two transporters or differences in the permeability properties of their outer membranes.
In this study, we found that AcrABHI confers multidrug resistance when expressed in E. coli cells and requires for its activity TolCEC protein. We next used the ease of the protein overproduction in E. coli to investigate the intracellular dynamics of AcrABHI in the presence of various substrates and to gain insight into the possible role of the accessory proteins in multidrug recognition. For this purpose, single cysteine mutations were constructed in AcrABHI in positions previously identified in Pseudomonas aeruginosa MexD as important for substrate recognition (10). The accessibility of these strategically positioned cysteines to the hydrophilic thiol-reactive fluorophore fluorescein-5-maleimide (FM) was studied in vivo in the presence of various substrates of AcrABHI and in the presence or absence of AcrAHI and TolCEC. We found that the reactivity of specific cysteines with FM was affected by the presence of some but not all substrates. Both protection and increased susceptibility of cysteines to FM were observed. Our results suggest that substrates induce conformational changes in AcrBHI.
MATERIALS AND METHODS
Construction of AcrABHI expression plasmid and mutagenesis.
The region of the H. influenzae chromosome encoding the HI0893 (AcrRHI), HI0894 (AcrAHI), and HI0895 (AcrBHI) genes (the homologs of AcrREC, AcrAEC, and AcrBEC, respectively) was amplified in two steps by PCR and cloned into the pSE380 expression vector (Invitrogen) to yield the pRAB plasmid. The first PCR-amplified fragment was obtained using the oligonucleotides 5′-CCGAATTCGAGCAATTATTGCTCC (the EcoRI site is underlined) and 5′-TTCGCCTGCAGGATCGGTATT (the PstI site is underlined) and contained the untranslated region upstream of acrRHI (without its own promoter), complete sequences of the acrRHI and acrAHI genes, and the fragment of acrBHI. This PCR fragment was cloned into EcoRI and PstI sites of pSE380 downstream of the Ptrc promoter. The second PCR fragment was generated using the oligonucleotides 5′-ATCCTGCAGGCGCGTTAGCAGAT (the PstI site is underlined) and 5′-GAGAAGCTTTAGTGGTGGTGGTGGTGGTGGGTTGTTTTATTTTC (the Hind III site is underlined) and contained the remaining fragment of acrBHI fused to the six-histidine (His6) tag coding sequence. After subcloning into PstI and Hind III sites, the resulting pRAB plasmid contained all three genes of the acrRABHI operon transcribed in the same direction under the IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible Ptrc promoter. In addition, the produced AcrBHI contained the C-terminal His6 tag to facilitate the purification of this protein using metal affinity chromatography. The presence of the transcriptional repressor AcrRHI appears to be essential for the optimal expression of AcrABHI. The pSE380-based plasmid containing the acrABHI genes without acrRHI caused strong growth inhibition of host cells even without IPTG induction.
To identify the positions of amino acids for mutagenesis, we performed the multiple alignment analysis using the ClustalW program as a part of the InfoMax package (Invitrogen). Twenty-one sequences of RND transporters including AcrBEC and AcrDEC and MexB, MexD, MexF, MexY, and MexK from P. aeruginosa were aligned with AcrBHI. Positions known to affect the substrate specificity of MexD and MexB were selected for mutagenesis of AcrBHI (Table 1). Site-specific mutagenesis was carried out with a QuickChange kit (Invitrogen) using pRAB as a template. DNA primers were designed according to the manufacturers' instructions. All mutations were verified by sequencing of the resulting plasmids.
TABLE 1.
AcrBHI-Cys variants and the corresponding amino acid residues in AcrBEC and mutations in P. aeruginosa MexD
| AcrBHI-Cys residue | Corresponding residue in:
|
Intensity of FM labeling
|
||
|---|---|---|---|---|
| AcrBECa | MexDb | AcrAHI+ TolCEC+c | AcrAHI−/TolCEC−d | |
| E42C | Q34 | Q34K | ++ | LE/+ |
| T96C | V88 | E89K | ++ | +/+ |
| A288C | G290 | A292V | + | −/− |
| L324C | P325 | P328L | ++ | +/+ |
| I601C | F617 | F608S | +++ | LE/+ |
| Q658C | T678 | N673K | +++ | +/+ |
| L396C | M397 | ND | − | ND |
| I397C | V398 | ND | − | ND |
| G401C | G402 | ND | − | ND |
Protein sequences were aligned using the ClustalW program.
(10).
Qualitative intensity of FM-labeled AcrBHI variants from the visual inspection of Fig. 1B, 2A, and 3A.
LE, low expression. In AcrAHI− background, these AcrBHI variants were produced at or below levels detectable by CBB staining. ND, no data.
The DNA sequence encoding AcrAHI was removed (1,060 bp out of 1,146 bp of the coding sequence) by PCR using the oligonucleotides 5′-GCGAATGCTNAGCGGTGCAATAGAGCCTGCAC and 5′-GCGGAAGCTNAGCAAACCGATGACACCGAC (the BlpI sites are underlined) and respective pRAB derivatives as templates. The PCR fragments were digested with BlpI enzyme and ligated by treatment with T4 DNA ligase.
Strains and growth conditions.
The E. coli DH5α strain was used for all cloning purposes. The expression, FM labeling, and MIC determination were carried out in either the AG100AX strain [argE3 thi-1 rpsL xyl mtl galK supE441 Δ(gal-uvrB) λ− ΔacrAB::kan ΔacrEF::spe) (14) or ECM1694 (MC4100 ΔacrAB::kan). ECM2112 (MC4100 ΔacrAB::kan ΔtolC::Tn10) was used to study the effect of TolCEC. Cells were grown at 37°C with shaking at 200 rpm in Luria-Bertani (LB) broth containing 10 g of Bacto tryptone, 10 g of Bacto yeast extract, and 5 g of NaCl per liter. The antibiotics ampicillin (100 μg/ml), kanamycin (34 μg/ml), spectinomycin (50 μg/ml), and tetracycline (25 μg/ml) were used as selection markers.
MICs.
MICs of various antimicrobial agents were measured using the microdilution technique in 96-well microtiter plates. For this purpose, exponentially growing cultures (A600 of ∼1.0 as determined using a Shimadzu UV-1601 spectrophotometer) were inoculated at a density of 104 cells per ml into LB medium in the presence of twofold-increasing concentrations of the drug under investigation. Cell growth was determined visually after overnight incubation at 37°C.
Protein analyses.
Membrane fractions of various strains were analyzed to determine the expression of AcrBHI and AcrAHI. For this purpose, cells were grown in 10 ml of LB broth supplemented with ampicillin (100 μg/ml), and at an A600 of ∼0.6, cells were induced with 0.1 mM IPTG for 2 h. Cells were pelleted and resuspended in 1 ml of buffer containing 10 mM Tris-HCl (pH 8.0), 1 mM EDTA, and 1 mM phenylmethylsulfonyl fluoride. Cells were lysed by sonication, and unbroken cells were removed by centrifugation at 3,220 × g for 15 min. Membranes were collected by centrifugation at 250,000 × g for 60 min. Membrane fractions were solubilized in 2% sodium dodecyl sulfate (SDS) sample buffer at room temperature and subjected to 8% SDS-polyacrylamide gel electrophoresis (PAGE). For analysis of Cys-containing AcrBHI (AcrBHI-Cys), nonreducing SDS-PAGE was used, where boiling and the addition of β-mercaptoethanol were omitted from the sample preparation. The levels of expression of AcrAHI and AcrBHI were monitored by Western immunoblotting analysis using polyclonal anti-AcrA antibody and HisProbe-HRP (SuperSignal West HisProbe kit; Pierce), a nickel-activated derivative of horseradish peroxidase, respectively. The alkaline phosphatase-conjugated secondary antibody and the chromogenic reaction with 5-bromo-4-chloro-3-indolyl phosphate-nitroblue tetrazolium substrates were used to visualize AcrAHI protein bands.
Whole-cell labeling with FM and mini-protein purification.
The His6-tagged AcrBHI Cys-less and Cys mutants were purified from E. coli cells carrying the corresponding plasmids. For this purpose, cells were grown in 200 ml of LB medium supplemented with ampicillin (100 μg/ml) to an A600 of ∼0.6. The expression of AcrBHI was induced by 0.1 mM IPTG for 1 h 45 min. Cells were harvested and washed once with ice-cold phosphate-buffered saline (PBS), pH 7.0. To determine accessibility of AcrBHI-Cys to FM, cells were resuspended in 1 ml of PBS buffer containing 300 μM FM (Invitrogen-Molecular Probes). To study the effect of substrates on accessibility of AcrBHI-Cys to FM, we adapted the protocol previously described for the MdfA transporter (1). Briefly, prior to the addition of FM, cells (120 optical density units at 600 nm in 1 ml of PBS) were preincubated with substrates (erythromycin, novobiocin, cloxacillin, ethidium bromide, Triton X-100, or inhibitor MC-207,110) in final concentrations of 5 mM (1). After incubation with FM for 30 min at room temperature, the reactions were stopped by the addition of dithiothreitol to a final concentration of 10 mM. Cells were pelleted in a microcentrifuge and resuspended in 2 ml of buffer containing 50 mM Tris-HCl (pH 8.0), 1 mM EDTA, and 0.1 mg/ml lysozyme. Cells were broken by sonication, the unbroken cells were removed by centrifugation at 3,220 × g for 15 min, and the supernatant was further centrifuged at 250,000 × g for 60 min. The pellet was resuspended in 0.5 ml of binding buffer containing 50 mM Tris-HCl (pH 8.0), 500 mM NaCl, and 5 mM imidazole (buffer A); then an equal volume of 10% Triton X-100 in buffer A was slowly added to solubilize membrane proteins. After overnight incubation at 4°C, insoluble material was removed by centrifugation at 250,000 × g for 30 min. Supernatants were loaded onto 0.1-ml His-bind resin (Novagen) mini-columns charged with Cu2+ and equilibrated with buffer A containing 0.2% Triton X-100 (buffer B). The column was washed with buffer B, and then bound proteins were eluted with a step gradient of imidazole (5 mM, 50 mM, and 500 mM) in buffer B. The majority of AcrBHI was eluted with 500 mM imidazole. Purified AcrBHI and its variants were separated by 8% SDS-PAGE. A Storm 840 phosphor and fluorescent screen imaging system (Amersham Pharmacia-Molecular Dynamics) was used to assess fluorescein labeling of cysteine mutants. Next, gels were stained with Coomassie brilliant blue (CBB) to visualize protein bands and determine amounts of proteins. Fluorescence intensity and protein amount were analyzed using the ImageQuant TL program (Amersham Pharmacia). For each protein band, the fluorescence intensity was normalized on the protein amount determined from the same gel after staining with CBB.
RESULTS
AcrABHI is expressed in E. coli and functional.
AcrAHI and AcrBHI were expressed from pRAB plasmid under the IPTG-inducible Ptrc promoter. When pRAB was transformed into the E. coli strain ECM1694 (ΔacrAB::kan), cells gained high levels of resistance to multiple antibiotics (Table 2). The increase in MICs was observed even in the absence of IPTG, indicating that both AcrAHI and AcrBHI are expressed as a result of the promoter leakage. Western immunoblotting confirmed that AcrBHI is expressed in E. coli and that the addition of IPTG induced severalfold overproduction of this protein (data not shown). The activity of AcrABHI was completely abolished in the ECM2112 (ΔacrAB ΔtolC) strain. Thus, similar to other recombinant RND transporters produced in E. coli, the function of AcrABHI in E. coli required the outer membrane channel TolCEC.
TABLE 2.
Antimicrobial susceptibility of the E. coli ECM1694 (ΔacrAB) strain carrying plasmid pRAB with AcrABHI and its variants
| Plasmid | MIC (μg/ml)a
|
||||||
|---|---|---|---|---|---|---|---|
| SDS | NOV | EtB | ERY | CHL | TET | NOR | |
| pSE380 (vector) | 40 | 4 | 3.125 | 4 | 0.4 | 0.39 | 0.04 |
| pRAB (AcrABHI) | >10,240 | 256 | 200 | 256 | 3.2 | 1.56 | 0.16 |
| pRAB (C932S) | >10,240 | 64 | 200 | 64 | 1.6 | 0.78 | 0.08 |
| pRAB (C932S E42C) | >10,240 | 32 | 50 | 32 | 1.6 | 0.78 | 0.08 |
| pRAB (C932S T96C) | >10,240 | 32 | 100 | 32 | 1.6 | 0.39 | 0.08 |
| pRAB (C932S A288C) | >10,240 | 32 | 100 | 64 | 1.6 | 0.39 | 0.08 |
| pRAB (C932S L324C) | >10,240 | 32 | 100 | 16 | 0.8 | 0.39 | 0.08 |
| pRAB (C932S L396C) | >10,240 | 32 | 100 | 32 | 1.6 | 0.195 | 0.04 |
| pRAB (C932S I397C) | >10,240 | 32 | 100 | 32 | 1.6 | 0.39 | 0.08 |
| pRAB (C932S G401C) | >10,240 | 32 | 100 | 32 | 1.6 | 0.39 | 0.08 |
| pRAB (C932S I601C) | >10,240 | 32 | 100 | 32 | 1.6 | 0.39 | 0.08 |
| pRAB (C932S Q658C) | >10,240 | 32 | 50 | 8 | 0.4 | 0.195 | 0.08 |
Data are representative of three independent experiments. SDS, sodium dodecyl sulfate; NOV, novobiocin; EtB, ethidium bromide; ERY, erythromycin; CHL, chloramphenicol; TET, tetracycline; NOR, norfloxacin.
Although disruption of AcrAHI and AcrBHI in H. influenzae led to only two- to eightfold decreases in MICs of such antimicrobials as erythromycin, novobiocin, ethidium bromide, and SDS (19), in E. coli the expression of AcrABHI resulted in a 16- to 128-fold increase in MICs of the same drugs (Table 2). In addition, AcrABHI conferred elevated resistance to tetracycline, chloramphenicol, and norfloxacin, the antibiotics previously reported to be unaffected by the activity of this transporter in H. influenzae cells. We estimate that the uninduced level of AcrABHI expression in E. coli is lower or comparable to that of the native AcrABEC (data not shown). Thus, increased amounts of AcrABHI cannot explain the observed changes in MICs and substrate specificity. These data support the previously stated idea (19) that the apparent difference in the substrate specificities of AcrAB in E. coli and H. influenzae could be attributed to the differences in the size of general porins in the outer membrane of these bacteria.
Single cysteine mutants of AcrBHI are functional.
To investigate the intracellular dynamics of AcrBHI, we introduced unique cysteine residues into the putative substrate binding sites of AcrBHI (AcrBHI-Cys). Previously, Mao et al. (10) identified several spontaneous mutations in P. aeruginosa MexD that altered the substrate specificity of this transporter. MexD and AcrBHI share 29% identity and 53% homology with each other. We reasoned that the overall topology of substrate binding sites is conserved in RND transporters. Using sequence alignment and structure analysis, we identified the amino acid residues located in the sites of AcrBHI similar to the sites of MexD (Table 1). Prior to mutagenesis, a single cysteine residue, C932, of the native AcrBHI was replaced with serine [AcrBHI(C932S)]. Western immunoblotting analysis showed that AcrBHI(C932S) is expressed to the level comparable to that of the wild-type AcrBHI (data not shown). As judged by MIC measurements, this cysteineless AcrBHI was functional although MICs were two- to fourfold lower for all tested substrates (Table 2).
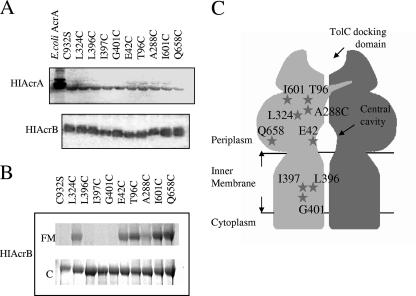
We next introduced nine unique cysteine residues into AcrBHI(C932S) (Fig. 1C and Table 1). Three mutations L396C, I397C, and G401C are located in the TMS4, which is proposed to be the major route for the proton translocation (17). The E42C substitution is exposed to the central cavity of AcrBHI, the site of substrate binding to AcrBEC identified in one of the structural studies (26). Substitutions T96C, A288C, and L324C are located in the major depression of the periplasmic domain, which was recently implicated in substrate binding (16). Finally, I601C and Q658C are located in the external cleft of the periplasmic domain and in proximity to the putative interaction site with the periplasmic AcrAHI (17, 25). All AcrBHI-Cys mutants were expressed at about the same level and retained their functionality (Fig. 1A and Table 2). However, AcrBHI(Q658C) provided only partial levels of resistance to erythromycin and ethidium bromide and lost activity against chloramphenicol (Table 2). The AcrBHI carrying E42C and L324C mutations was only partially active against ethidium bromide and erythromycin, respectively. All Cys mutations with the exception of E42C failed to confer resistance against tetracycline and displayed only a twofold increase in resistance against norfloxacin.
FIG. 1.
Expression and accessibility to FM of AcrBHI and its variants. (A) Western immunoblotting of membrane fractions (15 μg of total protein per lane) isolated from cells carrying pRAB plasmids with the indicated AcrBHI mutants. AcrAHI was visualized using rabbit anti-AcrA antibody, and AcrBHI was visualized with a nickel-activated derivative of horseradish peroxidase. (B) Accessibility of AcrBHI-Cys to FM. Whole cells containing the indicated AcrBHI variants were incubated with 0.3 mM FM for 30 min at 37°C (see Materials and Methods). Purified AcrBHI variants were separated by 8% SDS-PAGE and visualized by fluorescence using a Storm Imager (FM). After fluorescence scanning protein bands were visualized by CBB (C) staining. (C) Schematic representation of AcrBHI and positions of introduced cysteine residues based on the structure of AcrBEC (17). HIAcrA, AcrAHI; HIAcrB, AcrBHI.
Cysteines located in the periplasmic domain of AcrBHI but not in the TMS4 are accessible for labeling with the hydrophilic thiol-reactive probe FM.
Our experimental approach is based on two assumptions: (i) since many substrates of AcrBHI are bulky molecules, a cysteine residue located in the substrate binding site should be accessible to a relatively large thiol-reactive probe; (ii) binding of substrate is expected to modulate the reactivity of a cysteine residue located in proximity to substrate binding sites. Previously, the substrate protection of cysteines from thiol-reactive probes was successfully used to delineate substrate binding sites in various transporters (4, 15).
To investigate whether the constructed cysteine residues of AcrBHI are accessible from the aqueous environment of the periplasm, whole E. coli cells expressing various AcrBHI-Cys mutants were treated with the hydrophilic, thiol-reactive fluorophore FM. The reaction was terminated by the addition of dithiothreitol, and AcrBHI-Cys variants were purified from membrane fractions and analyzed as described in Materials and Methods.
Figure 1B shows that no fluorescent signal was detected in the cysteineless AcrBHI(C932S) and mutants with Cys residues L396C, I397C, and G401C located in TMS4. However, Cys residues located in the periplasmic loops were accessible to FM, albeit to different degrees. As expected, I601C and Q658C exposed to the outer cleft of the periplasmic domain of AcrBHI were the most readily reacting with FM. The A288C mutation was the least accessible to FM (∼10 to 30% of Q658C reactivity). In the crystal structure of the homologous AcrBEC, this A288C residue is located deep in the periplasmic drug binding pocket. The E42C, T96C, and L324C residues were partially reactive, with ∼30 to 50% of accessibility compared to Q658C. In the AcrBEC structure, E42C is accessible from the central cavity formed by three AcrBHI protomers, and T96C and L324C lie in the periplasmic domain (16).
All cysteines are less accessible to FM in TolCEC- and AcrAHI-deficient E. coli.
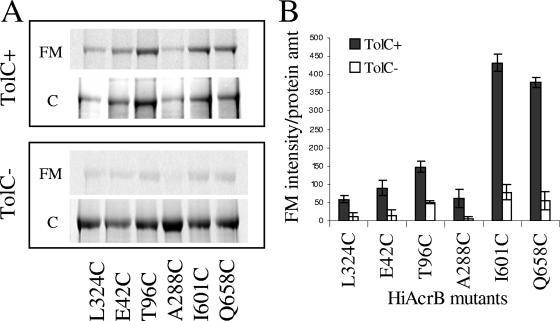
For its function in E. coli AcrABHI requires the outer membrane channel TolCEC. Therefore, we next investigated the accessibility of AcrBHI-Cys mutants to FM in the E. coli strain lacking the chromosomal TolCEC. Consistent with previous observations (6), the high levels of AcrABHI expression were toxic to the tolCEC strain. The levels of AcrABHI mutants varied significantly, and expression was often completely lost after storage of the transformed strains at −80°C. Therefore, for these experiments we used only freshly transformed cells. Surprisingly, although the overall pattern of AcrBHI-Cys accessibility to FM remained the same, the reactivity of all mutants was substantially lower in the tolCEC mutant compared to the wild type (Fig. 2). This result suggested that either the lack of TolCEC dramatically changed the AcrBHI conformation or, more likely, that the outer membrane of the tolCEC mutant is less permeable to FM.
FIG. 2.
FM accessibility of Cys residues in the periplasmic loops of AcrBHI-Cys is reduced in the absence of TolCEC. (A) AcrABHI variants were produced in E. coli strains AG100AX (ΔacrAB ΔacrEF) and ECM2112 (ΔacrAB ΔtolC) and analyzed for FM accessibility as described in the legend of Fig. 1 and in Materials and Methods. (B) Fluorescence intensities of AcrBHI-Cys variants (counts) shown in panel A are normalized based on the amount (amt) of AcrBHI protein present (counts) as determined by CBB staining of SDS-PAGE gels. Error bars are standard deviations (n = 3). HiAcrB, AcrBHI.
The growing body of evidence suggests that MFPs could be directly involved in transport reactions by interacting with substrates or communicating conformational changes taking place in the inner membrane transporter to the outer membrane channel (2, 3, 22). To investigate if AcrAHI affects the reactivity of cysteines in AcrBHI-Cys, DNA sequence encoding AcrAHI was deleted from the plasmids carrying AcrBHI with E42C, T96C, A288C, L324C, I601C, and Q658C substitutions. The AcrAHI-less constructs were then studied for the ability to confer a multidrug resistance phenotype in AG100AX cells, expression of AcrBHI, and accessibility to FM.
No multidrug resistance phenotype was conferred by AcrBHI variants in the absence of AcrAHI, demonstrating that none of the endogenous AG100AX proteins can complement the AcrAHI function (data not shown).
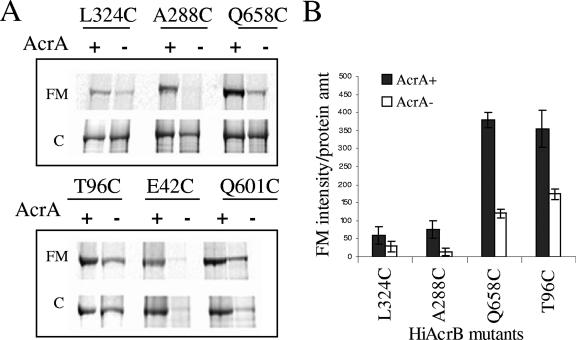
The lack of AcrAHI affected accessibility as well as expression of AcrBHI-Cys variants. Although the expression of all AcrBHI variants was somewhat lower in the absence of AcrAHI, E42C and I601C variants were expressed at the lowest levels (Fig. 3). Surprisingly, similar to the TolCEC-deficient strain, the accessibility of all AcrBHI-Cys variants was reduced in the absence of AcrAHI (Fig. 3). The increase in Cys accessibility of AcrBHI would be expected if any of these residues were in a direct contact with AcrAHI.
FIG. 3.
The lack of AcrAHI negatively affects the expression and FM accessibility of AcrBHI-Cys. (A) AcrBHI-Cys variants were produced in E. coli AG100AX (ΔacrAB ΔacrEF) in the presence and absence of AcrAHI and analyzed for FM accessibility as described in the legend of Fig. 1 and in Materials and Methods. (B) Fluorescence intensities of AcrBHI-Cys variants shown in panel A are normalized as described in the legend of Fig. 2. Error bars are standard deviations (n = 3). HiAcrB, AcrBHI; amt, amount.
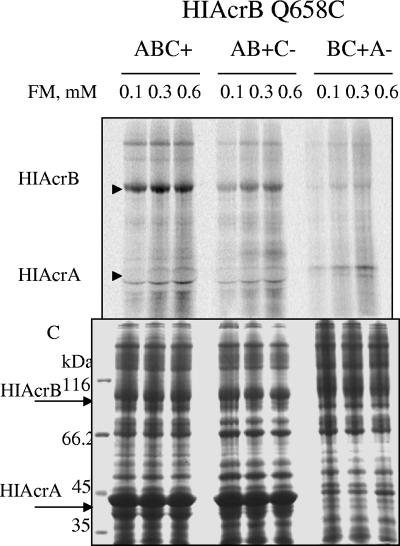
Both TolCEC− and AcrAHI− strains are highly susceptible to various compounds. Possibly the major response of E. coli to the lack of active efflux is to decrease the permeability of the outer membrane. To test whether FM permeability is the major reason for decreased labeling of cysteines in mutants lacking TolCEC or AcrAHI, cells expressing AcrBHI(Q658C) were treated with increasing concentrations of FM; total membrane fractions were then isolated and analyzed by SDS-PAGE (Fig. 4). Even at the lowest concentration of FM (0.1 mM) Q658C was readily labeled with FM in cells expressing all three proteins. The fluorescence intensity slightly increased at 0.3 mM FM and remained constant at 0.6 mM FM. Similarly, the extent of labeling of Q658C increased with increasing concentrations of FM in TolCEC− and AcrAHI− strains but was substantially lower than that in the presence of the functional transporter. In addition to AcrBHI(Q658C), a number of other membrane proteins were labeled with FM. Overall accessibility profiles of total membrane fractions correlated with that of AcrBHI(Q658C), indicating that the labeling of all proteins, not only AcrBHI, decreased in the absence of TolCEC or AcrAHI. We conclude that the lack of either TolCEC or AcrAHI negatively affects accessibility of all proteins to FM, presumably due to the change in outer membrane permeability.
FIG. 4.
Cells lacking either TolCEC or AcrAHI are less permeable for FM. Cells expressing AcrBHI(Q658C) in the presence (+) or absence (−) of TolCEC and AcrAHI (indicated by B, C, and A, respectively, above the lanes) were treated with increasing concentrations of FM for 30 min at room temperature. Total membrane fractions were isolated, and proteins were separated by 12% SDS-PAGE followed by fluorescence imaging (top panel) and then CBB staining (bottom panel; C). Positions of AcrBHI and AcrAHI are indicated by arrows. Total fluorescence intensity of all proteins bands, not only AcrBHI, decreased in the absence of either TolCEC or AcrAHI. HIAcrA, AcrAHI; HIAcrB, AcrBHI.
The reactivity of thiol groups in E42C, T96C, and A288C is modulated by AcrBHI substrates.
All AcrBHI-Cys variants are competent in multidrug recognition, as demonstrated by complementation of multidrug-susceptible phenotypes of acrAB-deficient E. coli (Table 2). The only exception is the Q658C variant, which was found to be partially deficient in efflux of erythromycin and chloramphenicol. To test whether substrates affect the accessibility of Cys in AcrBHI, whole cells of AcrBHI-Cys variants were preincubated with selected substrates: the antibiotics erythromycin, novobiocin, and cloxacillin; the detergent Triton X-100; the dye ethidium bromide; and the broad-spectrum inhibitor of RND-type transporters MC-207,110. Previously, MC-207,110 was shown to be a substrate of these transporters also (9). Next, cells were exposed to FM, and AcrBHI-Cys was analyzed as described above.
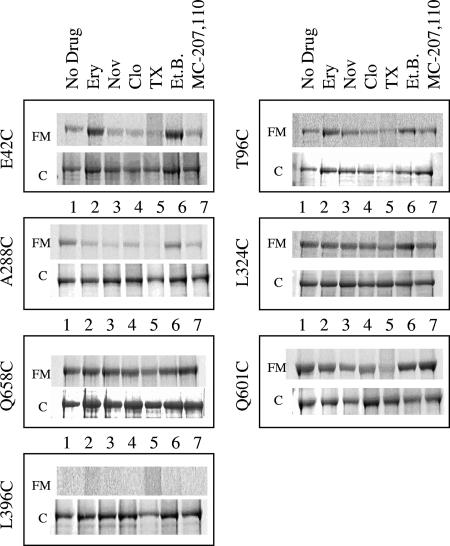
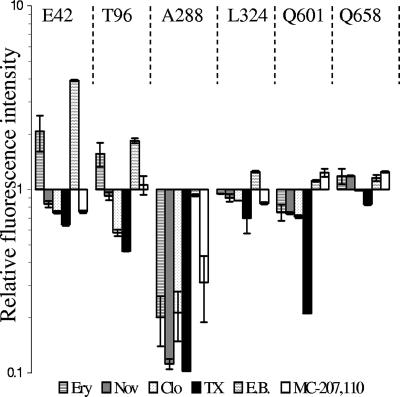
As seen in Fig. 5 and 6, Triton X-100 reduced labeling of cysteines in all positions, albeit to different degrees. It is unlikely that the effect of Triton X-100 is specific to AcrBHI since AG100AX (ΔacrAB ΔacrEF) is resistant to this detergent, and the overproduction of AcrABHI did not affect susceptibility to Triton X-100 (data not shown). Perhaps this detergent interferes with either cysteine or FM reactivity. The accessibility of cysteine residues in the four positions I601C, Q658C, L324C, and L396C was not affected by pretreatment of cells with various compounds. The L396C residue located in the TMS4 remained inaccessible to FM, suggesting that binding of a substrate does not lead to opening of the transmembrane domain. In the presence of all tested substrates, except Triton X-100, the I601C, Q658C, and L324C residues were labeled to the same degree, suggesting that these three residues are not directly involved in binding of substrates or located in the structurally rigid regions of AcrBHI.
FIG. 5.
Substrates modulate the accessibility of AcrBHI-Cys to FM. Whole cells containing the indicated AcrBHI variants were pretreated for 30 min at 37°C with PBS buffer alone (No drug) or containing a 5 mM concentration of the following compounds: erythromycin (Ery), novobiocin (Nov), cloxacillin (Clo), Triton X-100 (TX), ethidium bromide (Et.B.), or inhibitor MC-207,110. After this pretreatment cells were labeled with FM and analyzed as described in the legend of Fig. 1. For each variant, the top panel shows fluorescence intensity (FM) and the bottom panel shows CBB (C) staining.
FIG. 6.
Substrates modulate the accessibility of AcrBHI-Cys to FM. Fluorescence intensities of AcrBHI-Cys variants shown in Fig. 5 were normalized as described in the legend of Fig. 2 and expressed as a F1/F0 ratio, where F1 is the normalized fluorescence intensity of AcrBHI pretreated with drug and F0 is the normalized fluorescence intensity of AcrBHI pretreated with buffer alone (No drug). Error bars are standard deviations (n = 3). Ery, erythromycin; Nov, novobiocin; Clo, cloxacillin; TX, Triton X-100; EB, ethidium bromide.
The three mutations E42C, T96C, and A288C displayed variable accessibility to FM depending on specific substrates (Fig. 5 and 6). The E42C and T96C substitutions showed similar responses. The preincubation with ethidium bromide and erythromycin visibly increased the accessibility of E42C and T96C to FM, whereas other substrates slightly inhibited labeling of these residues. The protection against labeling with FM is expected if these two residues are directly involved in binding of substrates. This result suggests that these two residues are located in the structurally flexible regions of AcrBHI and that substrates promote conformational changes in AcrBHI.
In addition to E42C and T96C, the accessibility of A288C was also affected by substrates. However, all tested substrates with the exception of ethidium bromide protected A288C from FM. This result suggests that substrates restrict access of FM to the A288C residue. In the crystal structure of AcrBEC, the homologous G290 residue is located in the large binding pocket within ∼8 Å distance from the bound substrate (16). Thus, the protection of the A288C residue could arise either from direct substrate interference or from the conformational changes in the protein induced by substrates.
DISCUSSION
In this report, we present evidence that AcrBHI is a dynamic protein undergoing conformational changes in the presence of substrates. We introduced single cysteine substitutions based on the sequence alignment and structure modeling of AcrBHI and MexD from P. aeruginosa (Table 1). Previously Mao et al. (10) showed that mutations in these positions enable MexD transporter to pump out the normally nontransported antibiotic carbenicillin. In addition, these mutations also had a substantial impact, both positive and negative, on the MexD-mediated transport of numerous other substrates, suggesting that these residues play an important role in substrate recognition and/or transport. In the high-resolution structure of AcrBEC, the homologous Cys substitutions could be tentatively assigned to three distinct regions. These three regions map to the three putative substrate binding regions identified in the recent structural studies: the central cavity (E42C) (26), the large pocket in the periplasmic domain (T96C, A288C, and L324C) (16), and the external cavity of the periplasmic domain (Q658C and I601C) (25). In addition, three single cysteines were introduced into TMS4 of AcrBHI in the vicinity of the proposed proton channel (17).
Substitutions with cysteines in all selected positions did not lead to notable changes in AcrBHI activity or substrate specificity (Table 2) with the exception of the E42C, L324C, and Q658C mutants, which were partially defective in the transport of at least one substrate from among erythromycin, ethidium bromide, and chloramphenicol. Thus, none of the mutated amino acid residues is critical in multidrug efflux. However, the reduced activities of the E42C, L324C, and Q658C mutants suggest that these residues could contribute to the binding or transport of drugs.
Consistent with structure predictions, cysteine residues placed into the periplasmic domain were accessible for labeling with the large, hydrophilic probe FM, albeit to different degrees. E42C, T96C, A288C, and L324C residues were labeled only partially, indicating that the reactivity of these residues could be limited due to hindrance from surrounding amino acid residues. Alternatively, partial accessibility could indicate the heterogeneity of AcrBHI conformations. Recently, two independent studies suggested that during transport reaction AcrBEC cycles through three different conformations (16, 21). Interestingly, the access to T96C, A288C, and L324C from the periplasm differs in these three conformations, with the least access in the “tight-binding” conformer. Furthermore, the conformational transitions of AcrBEC also affect the central cavity, the site of E42C. Thus, partial reactivity of these residues could be due to the conformational heterogeneity of AcrBHI.
Located in the external cleft of the periplasmic domain, I601C and Q658C were readily labeled with FM. Interestingly, AcrAHI, which presumably binds to this region of AcrBHI, did not protect these residues from labeling. In contrast, Q658C was visibly less accessible to FM in the absence of AcrAHI. In addition, the accessibilities of T96C, A288C, and L324C also decreased in the absence of AcrAHI. A similar decrease in Cys accessibility was also observed for the TolC-deficient strain of E. coli. This decrease in FM labeling was not restricted to AcrBHI (Fig. 4). FM labeling of other membrane proteins was also reduced in TolCEC− and AcrAHI− backgrounds, suggesting that the change in the FM permeability is the major reason for this effect. Previous studies showed that cells lacking TolCEC replace the outer membrane porin OmpF with OmpC, the porin with the much smaller permeability (13). Such substitution could restrict the access of the hydrophilic FM to the periplasm. Similarly, using N-terminal protein sequencing, we found that ECM2112, the TolCEC− strain used in this study, produces OmpC. Interestingly, in the AG100AX strain, which is hypersusceptible to multiple drugs due to the lack of major efflux pumps, OmpF is also replaced by OmpC (E. B. Tikhonova and H. Zgurskaya, unpublished data). Thus, replacement of OmpF with the less permeable OmpC seems to be a common response of E. coli to the increased multidrug susceptibility due to the lack of active efflux.
In addition, TolCEC itself could be a route for FM to reach the periplasm and the transporter. Defects in TolCEC, for example, led to hyperresistance to the glycopeptide antibiotic vancomycin, which is too large to pass through the general porins (20). In this case, the lack of AcrAHI could keep TolCEC in the closed conformation, leading to the decrease in FM accessibility.
Low fluorescence signals precluded unequivocal determination of the effect of substrates on AcrBHI-Cys reactivity in the absence of TolCEC or AcrAHI (Fig. 2 and 3; also data not shown). The effect of substrates became apparent when all three components of the operational pump were present. The Cys in A288C was protected from labeling with FM by almost all tested compounds (the only exception is ethidium bromide) including MC-207,110 inhibitor. This result is consistent with the recently reported structure of the ligand-bound AcrBEC, in which the homologous G290 residue is in proximity to bound substrates (16). However, this approach cannot distinguish between hindrance effects caused by substrate binding versus conformational changes. Thus, either one of these factors or both could contribute to protection of the A288C residue from FM.
Two other residues, E42C in the central cavity and T96C located in the periplasmic domain, were also differentially affected by substrates. In contrast to A288C, some substrates (erythromycin and ethidium bromide) significantly enhanced the reactivity of E42C and T96C, suggesting that the conformation of AcrBHI changes in response to substrate binding. A similar substrate-dependent enhancement of labeling with thiol-reactive reagents was also reported for cysteines introduced into the multidrug efflux transporter MdfA (1). Interestingly, the modulation of cysteine accessibility of E42C and T96C depended on the specific substrate. Both erythromycin and ethidium bromide are positively charged compounds, and preincubation with these compounds enhanced labeling with FM. In contrast, negatively charged novobiocin and cloxacillin did not affect the labeling or slightly protected the E42C and T96C residues. This result suggests that various compounds could bind to different surfaces on AcrBHI.
None of the other accessible Cys residues of the AcrBHI mutants changed their reactivity in response to the presence of substrates. In contrast to MexD, all cysteine substitutions were well tolerated by AcrBHI transporter and did not lead to significant changes in MICs. Only L324C and Q658C substitutions partially reduced resistance to several tested compounds (Table 2). Therefore, the lack of effect of substrates in the case of Q658C and L324C could be due to defects in substrate binding. However, it is also possible that these residues, similar to Q601C, do not contribute to substrate binding and located in the conformationally rigid regions of AcrBHI.
Acknowledgments
This study was supported by National Institutes of Health Grant 1-RO1-AI052293.
We thank the anonymous reviewers for their helpful and constructive comments.
Footnotes
Published ahead of print on 25 May 2007.
REFERENCES
- 1.Adler, J., and E. Bibi. 2004. Determinants of substrate recognition by the Escherichia coli multidrug transporter MdfA identified on both sides of the membrane. J. Biol. Chem. 279:8957-8965. [DOI] [PubMed] [Google Scholar]
- 2.Aires, J. R., and H. Nikaido. 2005. Aminoglycosides are captured from both periplasm and cytoplasm by the AcrD multidrug efflux transporter of Escherichia coli. J. Bacteriol. 187:1923-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borges-Walmsley, M. I., J. Beauchamp, S. M. Kelly, K. Jumel, D. Candlish, S. E. Harding, N. C. Price, and A. R. Walmsley. 2003. Identification of oligomerization and drug-binding domains of the membrane fusion protein EmrA. J. Biol. Chem. 278:12903-12912. [DOI] [PubMed] [Google Scholar]
- 4.Dodd, J. R., and D. L. Christie. 2005. Substituted cysteine accessibility of the third transmembrane domain of the creatine transporter: defining a transport pathway. J. Biol. Chem. 280:32649-32654. [DOI] [PubMed] [Google Scholar]
- 5.Elkins, C. A., and H. Nikaido. 2002. Substrate Specificity of the RND-type multidrug efflux pumps AcrB and AcrD of Escherichia coli is determined predominately by two large periplasmic loops. J. Bacteriol. 184:6490-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lau, S. Y., and H. I. Zgurskaya. 2005. Cell division defects in Escherichia coli deficient in the multidrug efflux transporter AcrEF-TolC. J. Bacteriol. 187:7815-7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li, X. Z., and H. Nikaido. 2004. Efflux-mediated drug resistance in bacteria. Drugs 64:159-204. [DOI] [PubMed] [Google Scholar]
- 8.Lomovskaya, O., and K. A. Bostian. 2006. Practical applications and feasibility of efflux pump inhibitors in the clinic—a vision for applied use. Biochem. Pharmacol. 71:910-918. [DOI] [PubMed] [Google Scholar]
- 9.Lomovskaya, O., M. S. Warren, A. Lee, J. Galazzo, R. Fronko, M. Lee, J. Blais, D. Cho, S. Chamberland, T. Renau, R. Leger, S. Hecker, W. Watkins, K. Hoshino, H. Ishida, and V. J. Lee. 2001. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob. Agents Chemother. 45:105-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mao, W., M. S. Warren, D. S. Black, T. Satou, T. Murata, T. Nishino, N. Gotoh, and O. Lomovskaya. 2002. On the mechanism of substrate specificity by resistance nodulation division (RND)-type multidrug resistance pumps: the large periplasmic loops of MexD from Pseudomonas aeruginosa are involved in substrate recognition. Mol. Microbiol. 46:889-901. [DOI] [PubMed] [Google Scholar]
- 11.Markham, P. N., and A. A. Neyfakh. 2001. Efflux-mediated drug resistance in gram-positive bacteria. Curr. Opin. Microbiol. 4:509-514. [DOI] [PubMed] [Google Scholar]
- 12.Middlemiss, J. K., and K. Poole. 2004. Differential impact of MexB mutations on substrate selectivity of the MexAB-OprM multidrug efflux pump of Pseudomonas aeruginosa. J. Bacteriol. 186:1258-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Misra, R., and P. R. Reeves. 1987. Role of micF in the tolC-mediated regulation of OmpF, a major outer membrane protein of Escherichia coli K-12. J. Bacteriol. 169:4722-4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyamae, S., O. Ueda, F. Yoshimura, J. Hwang, Y. Tanaka, and H. Nikaido. 2001. A MATE family multidrug efflux transporter pumps out fluoroquinolones in Bacteroides thetaiotaomicron. Antimicrob. Agents Chemother. 45:3341-3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mordoch, S. S., D. Granot, M. Lebendiker, and S. Schuldiner. 1999. Scanning cysteine accessibility of EmrE, an H+-coupled multidrug transporter from Escherichia coli, reveals a hydrophobic pathway for solutes. J. Biol. Chem. 274:19480-19486. [DOI] [PubMed] [Google Scholar]
- 16.Murakami, S., R. Nakashima, E. Yamashita, T. Matsumoto, and A. Yamaguchi. 2006. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature 443:173-179. [DOI] [PubMed] [Google Scholar]
- 17.Murakami, S., R. Nakashima, E. Yamashita, and A. Yamaguchi. 2002. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 419:587-593. [DOI] [PubMed] [Google Scholar]
- 18.Poole, K. 2004. Efflux-mediated multiresistance in gram-negative bacteria. Clin. Microbiol. Infect. 10:12-26. [DOI] [PubMed] [Google Scholar]
- 19.Sanchez, L., W. Pan, M. Vinas, and H. Nikaido. 1997. The acrAB homolog of Haemophilus influenzae codes for a functional multidrug efflux pump. J. Bacteriol. 179:6855-6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schlor, S., A. Schmidt, E. Maier, R. Benz, W. Goebel, and I. Gentschev. 1997. In vivo and in vitro studies on interactions between the components of the hemolysin (HlyA) secretion machinery of Escherichia coli. Mol. Gen. Genet. 256:306-319. [DOI] [PubMed] [Google Scholar]
- 21.Seeger, M. A., A. Schiefner, T. Eicher, F. Verrey, K. Diederichs, and K. M. Pos. 2006. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science 313:1295-1298. [DOI] [PubMed] [Google Scholar]
- 22.Tikhonova, E. B., V. K. Devroy, S. Y. Lau, and H. I. Zgurskaya. 2007. Reconstitution of the Escherichia coli macrolide transporter: the periplasmic membrane fusion protein MacA stimulates the ATPase activity of MacB. Mol. Microbiol. [DOI] [PubMed]
- 23.Tikhonova, E. B., Q. Wang, and H. I. Zgurskaya. 2002. Chimeric analysis of the multicomponent multidrug efflux transporters from gram-negative bacteria. J. Bacteriol. 184:6499-6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trepod, C. M., and J. E. Mott. 2004. Identification of the Haemophilus influenzae tolC gene by susceptibility profiles of insertionally inactivated efflux pump mutants. Antimicrob. Agents Chemother. 48:1416-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu, E. W., J. R. Aires, G. McDermott, and H. Nikaido. 2005. A periplasmic drug-binding site of the AcrB multidrug efflux pump: a crystallographic and site-directed mutagenesis study. J. Bacteriol. 187:6804-6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu, E. W., G. McDermott, H. I. Zgurskaya, H. Nikaido, and D. E. Koshland, Jr. 2003. Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump. Science 300:976-980. [DOI] [PubMed] [Google Scholar]
- 27.Zgurskaya, H. I. 2002. Molecular analysis of efflux pump-based antibiotic resistance. Int. J. Med. Microbiol. 292:95-105. [DOI] [PubMed] [Google Scholar]