Abstract
Type IV secretion systems require peptidoglycan lytic transglycosylases for efficient secretion, but the function of these enzymes is not clear. The type IV secretion system gene cluster of Neisseria gonorrhoeae encodes two peptidoglycan transglycosylase homologues. One, LtgX, is similar to peptidoglycan transglycosylases from other type IV secretion systems. The other, AtlA, is similar to endolysins from bacteriophages and is not similar to any described type IV secretion component. We characterized the enzymatic function of AtlA in order to examine its role in the type IV secretion system. Purified AtlA was found to degrade macromolecular peptidoglycan and to produce 1,6-anhydro peptidoglycan monomers, characteristic of lytic transglycosylase activity. We found that AtlA can functionally replace the lambda endolysin to lyse Escherichia coli. In contrast, a sensitive measure of lysis demonstrated that AtlA does not lyse gonococci expressing it or gonococci cocultured with an AtlA-expressing strain. The gonococcal type IV secretion system secretes DNA during growth. A deletion of ltgX or a substitution in the putative active site of AtlA severely decreased DNA secretion. These results indicate that AtlA and LtgX are actively involved in type IV secretion and that AtlA is not involved in lysis of gonococci to release DNA. This is the first demonstration that a type IV secretion peptidoglycanase has lytic transglycosylase activity. These data show that AtlA plays a role in type IV secretion of DNA that requires peptidoglycan breakdown without cell lysis.
Neisseria gonorrhoeae carries a type IV secretion system (T4SS) in a 57-kb region of the gonococcal chromosome known as the gonococcal genetic island (GGI). This secretion system has the unique property of secreting chromosomal DNA in a noncontact-dependent manner (15, 19, 20). The secreted DNA acts in the transformation of other gonococci in the culture, and type IV secretion mutants are deficient in DNA donation for natural transformation, even when cultures are allowed to progress to autolysis (15, 20). Thus, the T4SS is important for DNA donation for natural transformation and may contribute to the spread of antibiotic resistance markers throughout the population. The T4SS may also facilitate evasion of the host immune response by increasing genetic diversity, providing variant alleles of genes for surface molecules (18).
The first gene identified to be involved in type IV secretion in gonococci was atlA. An insertion mutation in atlA eliminated detectable DNA in the culture supernatant and greatly reduced DNA donation for transformation (15). AtlA is similar to the bacteriophage lambda endolysin (lambda R) (14), an enzyme that digests peptidoglycan (PG) to break open Escherichia coli during the lytic stage of lambda infection in order to release phage particles (6, 7). No other secretion system has been found to require an endolysin homologue for function. These data suggested that AtlA might function like lambda R to breakdown PG, but they also suggested the possibility that DNA donation requiring the T4SS might occur by a process involving cell lysis.
Peptidoglycanases have been found to be required for efficient secretion in other secretion systems and are hypothesized to cause a localized break in the cell wall in order to allow for assembly of the secretion apparatus (3, 12, 27). This is an attractive hypothesis, since the openings in the PG of gram-negative bacteria are only large enough to allow the passage of a protein of approximately 50 kDa (9). However, it has not been demonstrated that the PG transglycosylases produce a localized break in the cell wall. Also, the reaction products of T4SS peptidoglycanases have not been characterized. Data supporting the localized-break hypothesis come from Bayer et al., who showed that expression of the putative lytic transglycosylase P19 from the plasmid R1 resulted in localized membrane blebbing from the surface of E. coli (4). Recently, Zahrl et al. showed that multiple secretion system lytic transglycosylase homologues caused zones of clearing on PG zymogram gels and that putative lytic transglycosylase IpgF from the Shigella flexneri type III secretion system degraded PG in vitro (40). In addition to encoding AtlA, the gonococcal T4SS gene cluster encodes a protein, LtgX, that is similar to other secretion system PG transglycosylases. LtgX is similar to the geneX product (Orf169) of the F-plasmid conjugation system (19). The presence of two lytic transglycosylase homologues in the GGI suggests that two separate peptidoglycanase functions are involved in type IV secretion in N. gonorrhoeae.
Here, we investigated the function of AtlA. Using an in vitro reaction with a purified AtlA fusion protein, we show that AtlA degrades macromolecular PG and generates 1,6-anhydro PG monomers. Also, atlA complemented the function of lambda R in E. coli lysis, demonstrating that AtlA can lyse bacteria. A mutation changing the putative active-site residue of AtlA resulted in the loss of activity in the in vitro assay and also the loss of type IV secretion in gonococci, indicating that the lytic transglycosylase activity is the function required for type IV secretion. Deletion of ltgX also eliminated DNA secretion, indicating that both LtgX and AtlA are required for efficient DNA secretion. The function of AtlA in N. gonorrhoeae was found not to be cell lysis, since assays measuring autolysis or lysis of neighboring cells demonstrated no differences between the atlA mutant and the wild-type strain.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains and plasmids are described in Table 1. All gonococcal strains were constructed using N. gonorrhoeae strain MS11 as the parent (36). All gonococci used in experimental procedures were nonpiliated, except for gonococci used for transformation or the DNA secretion assay. Gonococci were grown in GCB liquid medium (GCBL) (Difco) containing Kellogg's supplements (24) and 0.042% NaHCO3 (30) at 37°C with aeration, in Graver-Wade medium (37) at 37°C with aeration, or on GCB agar plates containing Kellogg's supplements at 37°C in 5% CO2. Overexpression of AtlA fusion proteins was carried out in E. coli BL21 Star cells (Invitrogen). All other E. coli strains were constructed using TAM1 cells (Active Motif). E. coli strains were grown in liquid Luria-Bertani (LB) broth (Difco) or in minimal medium with M9 salts (Difco), 2% Casamino Acids, and 0.4% glycerol. E. coli strains were also grown on LB agar plates (33). Ampicillin was used at 100 μg/ml for E. coli. Erythromycin was used at 500 μg/ml for E. coli. Chloramphenicol was used at 25 μg/ml for E. coli and at 10 μg/ml for N. gonorrhoeae.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Property(ies) | Source or reference |
|---|---|---|
| Plasmids | ||
| pBAD30 | Arabinose induction vector (Apr) | 17 |
| pGCC6 | Complementation plasmid | 28 |
| pIDN1 | Insertion duplication mutagenesis vector (Emr) | 20 |
| pIDN2 | Insertion duplication mutagenesis vector (Emr) | 20 |
| pIDN3 | Insertion duplication mutagenesis vector (Emr) | 20 |
| pJD1103 | atlA-ych region in pHSS6 | 14 |
| pJD1141 | Fragment of atlA in pUC19 | 15 |
| pJD1195 | atlA(E48A) as an EcoRI-ClaI fragment in pIDN2 | This work |
| pKH9 | Complementation vector (Cmr) | 19 |
| pKH35 | Complementation vector (Cmr) | 19 |
| pKH37 | Complementation vector (Cmr) | This work |
| pMAL-c2x | MBP fusion vector (Apr) | New England Biolabs |
| pPK20 | Lambda lysis genes in pBAD30 | 26 |
| pPK21 | atlA in place of lambda R in pPK20 | This work |
| pPK22 | atlA(E48A) in place of lambda R in pPK20 | This work |
| pPK53 | atlA in pKH35 for complementation | This work |
| pPK61 | atlA in pMAL-c2X | This work |
| pPK63 | atlA(E48A) in pMAL-c2X | This work |
| pPK82 | Vector for deletion of ltgX | This work |
| pPK83 | ltgX in pKH37 | This work |
| pPK87 | atlA and 1.4 kb of upstream DNA in pIDN1 | This work |
| pPK89 | Vector for internal deletion in atlA | This work |
| N. gonorrhoeae strains | ||
| MS11 | Clinical isolate of N. gonorrhoeae (Srr) | 36 |
| JD1510 | Insertion mutation in atlA | 14 |
| JD1616 | atlA(E48A) mutation in MS11 | This work |
| PK112 | JD1616 complemented with atlA (Cmr) | This work |
| PK120 | Deletion of ltgX in MS11 | This work |
| PK124 | PK120 complemented with ltgX (Cmr) | This work |
| PK127 | Internal deletion in atlA in MS11 | This work |
| ND500 | Deletion of the GGI in MS11 | 19 |
Cloning of atlA and overexpression of MBP-AtlA for PG degradation assays.
To create a plasmid to overexpress maltose-binding protein (MBP)-AtlA (pPK61), atlA was amplified by PCR from the MS11 chromosome using primers XmnatlAF (5′-CGAGGAAGGATTTCAATGTGGCGTGGAATATCAAGTG-3′) and EcoRIatlAR (5′-GCGGAATTCTTAAAATCCTCTCTGCCTAAAGAAATT-3′). The PCR product was digested with XmnI and EcoRI and ligated to pMAL-c2x, which was digested with the same enzymes. The MBP-AtlA(E48A) overexpression plasmid (pPK63) was created the same way, except that atlA(E48A) was amplified by PCR from strain JD1616 and DNA sequencing was used to verify the point mutation. MBP-AtlA and MBP-AtlA(E48A) were overexpressed by growing cells in LB broth containing 0.6 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h at 37°C. We found that it was necessary to express AtlA as a fusion protein with MBP in order to enhance solubility. The overexpressed proteins were purified using amylose resin (New England Biolabs) according to the manufacturer's instructions for purifying cytoplasmic proteins.
PG degradation assay and analysis of PG monomers.
Gonococcal PG was radiolabeled by growing cells with [6-3H]glucosamine as described previously (8). The PG was purified by resuspending cells from a 6-ml culture in 500 μl of 50 mM sodium acetate (pH 5.0), adding an equal volume of 8% sodium dodecyl sulfate (SDS), and boiling the mixture for 30 min. The insoluble PG was then collected by centrifugation at 41,000 × g for 30 min. The insoluble PG pellet was resuspended in water and centrifuged four times at 41,000 × g. The washed PG was resuspended in water and assayed for radioactivity. A volume of resuspended PG that contained approximately 20,000 cpm (approximately 4 × 106 gonococcal sacculi) was added to 1 μg of MBP-AtlA or MBP-AtlA(E48A), which was dissolved in buffer from the protein purification process. The volume of PG used was brought up to 400 μl if necessary. The total reaction mixture volume was brought up to 900 μl by adding the same buffer in which the purified protein was dissolved. At 0, 1, 2.5, and 5 h, a 200-μl aliquot was taken from each reaction and added to 300 μl of water. Trichloroacetic acid (TCA) was added to a concentration of 13%, and the reaction was incubated on ice for 30 min. The precipitated material was centrifuged at 41,000 × g for 30 min at 4°C, and the supernatant was assayed for radioactivity. The fragments generated by digestion with MBP-AtlA were separated as described above for the PG fragment release assay. PG fragments generated by digestion with hen egg white lysozyme and PG fragments liberated by growing gonococci were separated in an identical manner. The monomer fractions were collected and desalted by size-exclusion chromatography. Reversed-phase high-performance liquid chromatography (HPLC) using a C18 column (4.6 by 250 mm; Alltec) was used to characterize the PG monomers as described by Rosenthal and Dziarski (32). The monomers were separated for 30 min by using a 4 to 13% gradient of acetonitrile in water with 0.5% trifluoroacetic acid. Fractions were collected every 30 s, and radioactivity was measured by liquid scintillation counting. The retention times of PG monomers were compared to those of mass spectroscopy-characterized PG standards provided by R. Rosenthal.
Cloning of atlA and overexpression of AtlA for zymogram analysis.
atlA was amplified by PCR with primers AtlANsi3F (5′-GTAATGCATCCTCATATGGAGGC-3′) and AtlABcl-His8R (5′-AGCACTAGGGCCGGCCGTGATCAATGGTGGTGATGATGGTGGTGATGAAATCCTCTCTGCCTAAAG-3′). The PCR product was digested with NsiI and PmeI and cloned into the corresponding sites in pGCC6 (28). atlA-his was subcloned from the resulting plasmid as a BamHI-ScaI fragment into the BamHI and Ecl136II sites in pIDN2, yielding plasmid pJD1186. pPK45 was created to optimize the expression of AtlA-His by changing the ribosome binding site of atlA-his to make it closer to the consensus ribosome binding site for E. coli. pJD1186 was amplified by PCR using primers fullatlARBS1 (5′-CACCGGTACCTCCTTAATGAGGATGCATAGTCT-3′) and fullatlARBS2 (5′-GCACCGGTATGTGGCGTGGAATATCAAGTGGA-3′). The resulting PCR product was digested with AgeI and self ligated, and the resultant plasmid was designated pPK45. pPK48 was created by PCR amplifying atlA(E48A) from gonococcal strain JD1616 with primers closeatlAF and sacIcloseatlaR and using a BbsI-BsrGI fragment from this PCR product to replace the BbsI-BsrGI fragment of atlA-his in pPK45.
AtlA-His and AtlA(E48A)-His were overexpressed by growing E. coli overnight in LB broth at 37°C without induction. Both proteins were insoluble and were purified using nickel-nitrilotriacetic acid agarose beads (QIAGEN) under denaturing conditions according to the manufacturer's instructions.
Zymogram gel analysis.
Zymogram gel analysis was performed as described by others, with modifications (5, 11, 31, 40). Purified AtlA-His and AtlA(E48A)-His were subjected to electrophoresis in an SDS-14% polyacrylamide gel containing 0.2% lyophilized Micrococcus lysodeikticus cells (Sigma) or 0.2% purified gonococcal PG. Gonococcal PG was purified as described previously (13). The denatured proteins were allowed to renature by washing the gel in 25 mM Tris-HCl (pH 7.5), 1% Triton X-100 for 48 h. The buffer was changed frequently, especially in the first few hours, to remove the SDS. The gel was stained with 1% methylene blue in 0.1% KOH and was destained in deionized water to improve the visibility of bands of clearance in the PG. The gel was then stained with Coomassie brilliant blue to allow visualization of the purified protein in the gel.
E. coli lysis assay and construction of strains.
atlA and atlA(E48A) were amplified by PCR from the MS11 and JD1616 chromosomes, respectively, using primers closeatlAF (5′-CGGGTACCATGTGGCGTGGAATATCAAG-3′) and sacIcloseatlaR. pPK20 (26) was amplified by PCR to eliminate lambda R using primers sacIlysisrevF (5′-CGTGCGAGCTCATGAGCAGAGTCACCGCG −3′) and lysisrevR (5′-CGCGGTACCTTATTGATTTCTACCATCTTCTACTCC-3′). Both the amplified atlA and atlA(E48A) PCR products and the pPK20 PCR product were digested using SacI and KpnI and ligated together. The resulting plasmids were designated pPK21(atlA) and pPK22[atlA(E48A)]. The lysis assays were carried out as described previously (26).
Strain construction for DNA secretion, autolysis, and allolysis assays.
For creation of the atlA point mutation, the 5′ two-thirds of atlA were cloned from the JD1510 (14) chromosome as a 0.5-kb EcoRI, partial HindIII fragment into pUC19, generating pJD1141 (15). The putative active-site residue of AtlA was replaced by mutagenic PCR of pJD1141 with primers atlA-Pst1 (5′-CGGCATCTGCAGGTGTAAA-3′) and atlA-Pst2 (5′-ACCTGCAGATGCCGCAATAA-3′), followed by digestion with PstI and religation to generate pJD1185. The mutated atlA was subcloned as an EcoRI-ClaI fragment into pIDN2, generating plasmid pJD1195. This plasmid was used to transform MS11 as described by Gunn and Stein (16), generating strain JD1616. For the complementation of strain JD1616 [atlA(E48A)], a wild-type copy of atlA was amplified with PCR from MS11 DNA using primers Nsi3F (5′-GTAATGCATCCTCATATGGAGGC-3′) and SacIcloseatlAR (5′-CGTCGAGCTCTTAAAATCCTCTCTGCCTAAAGA-3′). The PCR product was cut with NsiI and SacI and inserted into the NsiI and SacI sites of the vector pKH35 (19). The resultant plasmid, pPK53, was used to transform strain JD1616. Chloramphenicol-resistant gonococci were screened for insertion of the complementation construct into the desired site between lctP and aspC on the gonococcal chromosome. The resulting gonococcal strain was designated PK112.
The plasmid for the internal deletion in atlA was created by site-directed mutagenesis using a plasmid (pPK87) containing atlA and approximately 1.4 kb of upstream DNA in pIDN1. The plasmid was amplified with PCR using the mutagenic primers atlANdelF (5′-GGCAACGGGGTTATGGAATTGGAGCGATATGTTCATAATCC-3′) and atlANdelR (5′-AACATATCGCTCCAATTCCATAACCCCGTTGCCTCCATAT-3′). The PCR was treated with DpnI to digest the original template plasmid. The DpnI-treated PCR products were used to transform E. coli, and colonies were screened by PCR to confirm the internal deletion in atlA. The plasmid containing the deletion, pPK89, was used to transform MS11, generating strain PK127.
The plasmid for creating an in-frame deletion of ltgX, pPK82, was created from a plasmid containing ltgX and surrounding regions in pIDN3 (20). The plasmid was amplified by PCR, excluding the coding region of ltgX except for the start and stop codons and two internal codons, using the primers ltgXdelF (5′-GCGCATATGCATGTCTTTACTGTCGTTGTATGCATG-3′) and ltgXdelR (5′-GCGCATATGTAGCCTCCGCAATGATTCTGAC-3′). The PCR product was cut with NdeI and self ligated. The resulting plasmid, pPK82, was transformed into strain MS11. Gonococcal colonies were screened by PCR for the loss of ltgX, and the resulting strain containing a deletion of ltgX, PK120, was sequenced to confirm the loss of ltgX. A plasmid for complementing PK120 with a wild-type copy of ltgX was created by amplifying ltgX with PCR from the MS11 chromosome using primers 109F (5′-TCCCTTGAACCCTTCCTTTA-3′) and PstIltgXR (5′-GCGCTGCAGCGGAGGCTACGAAGAGCGTG-3′). The vector for complementation, pKH37, was created from pKH9 (19) by filling in the HindIII site, deleting the BseRI-Ecl136II fragment, and inserting the multiple cloning site from pIDN1 in the BseRI (blunted with T4 DNA polymerase) and AgeI sites. The resulting PCR product and the vector, pKH37, were cut with NsiI and PstI and ligated together. Plasmids were restriction mapped for the correct orientation of ltgX, and the resulting plasmid, pPK83, was transformed into PK120, generating PK124.
Autolysis and allolysis RNA release assays.
To label gonococcal RNA, we used a modified method based on that of Kingsbury and Duncan (25). Gonococcal strains were grown on GCB agar plates overnight and then were grown in GCBL with supplements for 3 h. Cells were then diluted to an optical density at 540 nm of 0.2 in 4 ml of defined medium (20). [3H]adenine was added to the cultures at 2 μCi/ml, and the cultures were grown for 30 min. After being labeled, the cells were centrifuged for 1 min at 9,000 × g and washed with 1 ml GCBL. The cells were centrifuged again for 1 min at 9,000 × g, resuspended in 4 ml GCBL, and grown for 5 h. Culture (0.5 ml) was harvested and centrifuged at 11,000 × g at 0, 2.5, and 5 h. The radioactivity in the supernatants was measured by scintillation counting. In addition, the radioactivity in the cell pellet at 0 h was determined by scintillation counting. To correct for radioactivity present in the supernatant at 0 h, the counts per minute (cpm) in the supernatants at 0 h were subtracted from the counts at 2.5 and 5 h, and the fraction of RNA released at 2.5 and 5 h was calculated by dividing the corrected cpm in the supernatants by the cpm in the cell pellet at 0 h. In order to confirm that [3H]adenine was effectively labeling the RNA, we treated supernatants of the wild-type culture with RNase A (Sigma) at 10 μg/ml for 30 min at 37°C and precipitated the remaining macromolecular material with TCA. Any radioactive material present in RNA would be liberated into small fragments by RNase A, and any radioactive material incorporated into other macromolecules would be precipitated. The radioactivity in the supernatant of the TCA precipitation reaction was measured by scintillation counting. We also treated culture supernatants with TCA in the absence of RNase A in order to determine if nonmacromolecular cellular components were being labeled and released. The supernatants of these TCA precipitation reactions were assayed for radioactivity by scintillation counting. We found that about 16% of the released labeled material was not TCA precipitable and was likely made up of AMP, ADP, and ATP (23). Release of this material also would occur by lysis of gonococci.
For the allolysis assays, the RNA of strain JD1616 [atlA(E48A)] was labeled as described above, and strains JD1616 and MS11 were grown as described above for the labeling process, but no radiolabel was added to the cultures at any time. After the labeling period, 4-ml cultures in GCBL were made such that half of the cells were labeled JD1616 cells and half of the cells were unlabeled JD1616 or MS11 cells. These mixed cultures were grown for 5 h. Samples of each culture were collected at 0, 2.5, and 5 h, and the amount of released RNA was determined as described above.
DNA secretion assay.
DNA secretion assays were carried out as described previously (19, 20), with modifications. Gonococci were grown overnight on GCB agar plates and swabbed into 3 ml of Graver-Wade medium to an optical density at 540 nm of 0.18. After 2 h of growth, 0.5 ml of the culture was diluted into 3 ml of fresh Graver-Wade medium, and cultures were grown for 2 to 2.5 h. Culture supernatants were collected at 0 h and at the end of the growth period. One hundred microliters of a 1:200 dilution of PicoGreen fluorescent dye (Invitrogen) was added to 100 μl of culture supernatant, and DNA secretion was measured in a fluorometer at an excitation wavelength of 485 nm and an emission wavelength of 535 nm. DNA secretion was determined by using a standard curve of known DNA concentrations. The amount of DNA in culture supernatants was normalized to the total protein in the cell pellet, as determined by the Bio-Rad protein assay. Only cultures that reached between 15 and 30 μg of cellular protein per ml of culture were used for the assay. IPTG was added to strain PK124 to a concentration of 0.33 mM during the secretion period.
PG fragment release assay.
PG fragment release assays were carried out as described previously (8). Gonococcal PG was labeled metabolically by growth of the bacteria in medium containing [6-3H]glucosamine. PG fragments released into the culture medium were analyzed by size-exclusion chromatography and scintillation counting.
RESULTS
AtlA has lytic PG transglycosylase activity in vitro.
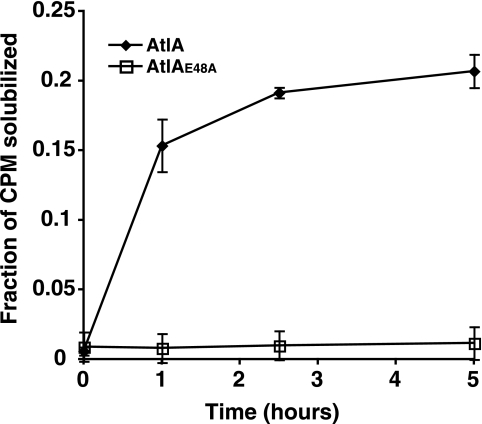
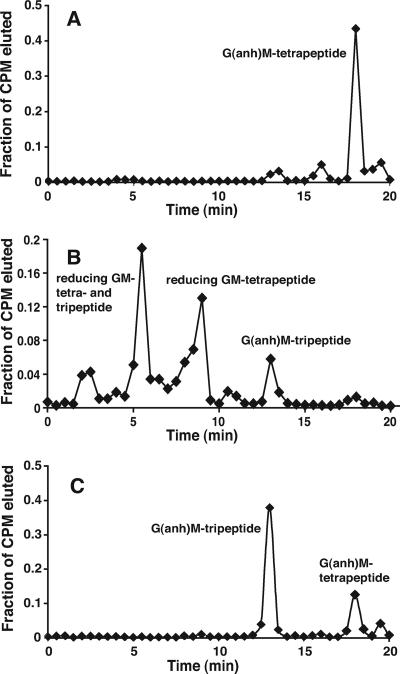
T4SSs include homologues of PG lytic transglycosylases, but in no case has it been demonstrated that a T4SS lytic transglycosylase homologue has this specific biochemical function. To determine if AtlA has lytic transglycosylase activity, we expressed AtlA as a fusion with E. coli MBP and assayed the activity of the fusion protein. Purified MBP-AtlA was found to degrade gonococcal PG, converting radiolabeled insoluble gonococcal sacculi to soluble PG fragments (Fig. 1). However, an equivalent MBP-AtlA fusion protein with an amino acid substitution in the predicted active-site residue [AtlA(E48A)] showed no PG solubilization activity. Using reversed-phase HPLC, we compared AtlA-generated PG monomers (Fig. 2A) to reducing PG monomers generated by hen egg white lysozyme (Fig. 2B) and 1,6-anhydro PG monomers released by growing gonococci (Fig. 2C). Gonococci are known to release 1,6-anhydro disaccharide tripeptide PG monomers and 1,6-anhydro disaccharide tetrapeptide PG monomers (32, 34). PG monomers liberated by AtlA activity consisted primarily of 1,6-anhydro disaccharide tetrapeptide PG monomers, and a small portion of the released monomers consisted of the 1,6-anhydro disaccharide tripeptide. These data demonstrate that AtlA functions as a lytic transglycosylase.
FIG. 1.
Degradation of radiolabeled PG by MBP-AtlA. MBP-AtlA and MBP-AtlA(E48A) were purified and incubated with insoluble, radiolabeled, macromolecular PG. Samples of the reactions were collected periodically, and the insoluble, macromolecular PG was precipitated by addition of TCA and collected by centrifugation. The soluble fragments were quantified by scintillation counting. The fraction of total radioactivity released into the soluble fraction was plotted over time. Values reported are the means of three separate experiments, and the error values are the standard deviations.
FIG. 2.
HPLC analysis of PG monomers generated by MBP-AtlA. Radiolabeled PG monomers generated by MBP-AtlA (A), generated by hen egg white lysozyme (B), and released from growing gonococci (C) were purified and analyzed by reversed-phase HPLC. Fractions were collected and analyzed for radioactivity. The fraction of radioactivity in each fraction per total radioactivity eluted was plotted over time. G, N-acetylglucosamine; M, N-acetylmuramic acid; (anh)M, 1,6-anhydro N-acetylmuramic acid.
We also assayed AtlA activity using a PG zymogram gel. Zymogram gels have often been used to identify peptidoglycanases in bacterial lysates and to characterize peptidoglycanase activity in suspected PG-degrading enzymes (5, 11, 31, 40). We purified AtlA with a polyhistidine tag on the C terminus and found that this protein (AtlA-His) showed a zone of clearing on a PG zymogram gel containing Micrococcus lysodeikticus cells (data not shown). However, the AtlA(E48A)-His protein showed an identical zone of clearing. Similar results were obtained using zymogram gels containing purified gonococcal PG. We diluted AtlA-His until a band of clearance in the gel was barely visible and found that the same amount of AtlA(E48A)-His showed an identical band of clearance (data not shown). It was previously noted by A. Dijkstra that two proteins from Haemophilus influenzae produced zones of clearing by zymogram analysis, even though the proteins do not degrade PG (10). We hypothesize that AtlA(E48A)-His is binding PG and preventing dye from binding to the PG at that location. This result may indicate that AtlA binds more tightly to PG than do some other previously characterized peptidoglycanases.
Cell lysis.
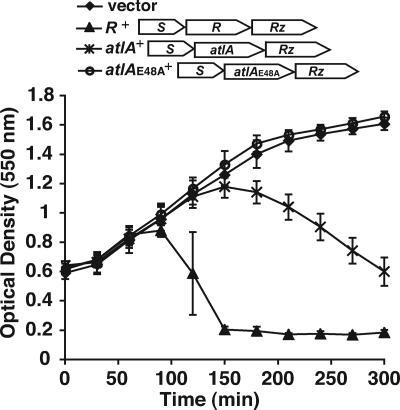
AtlA shows significant similarity to lambda R, the endolysin that breaks open E. coli during the lytic stage of lambda infection (6, 14). This similarity suggested that AtlA might also be able to lyse bacterial cells. We tested the ability of AtlA to cause cell lysis by replacing lambda R with atlA in a construct expressing the lambda lysis genes S, R, and Rz. Lambda S is a holin that creates a disruption in the bacterial inner membrane, allowing lambda R access to the bacterial cell wall (39), while lambda Rz performs an accessory function necessary for lysis under certain conditions (41). Once inside the periplasm, lambda R degrades PG, leading to cell lysis (6). Like lambda R, AtlA was able to lyse E. coli when it was coexpressed with lambda S and lambda Rz. However, AtlA did not lyse E. coli as rapidly as lambda R (Fig. 3). The atlA(E48A) mutant allele was not able to complement the loss of lambda R, confirming that lytic transglycosylase activity was necessary for cell lysis. It should also be noted that expression of AtlA alone does not cause lysis, since we were able to express atlA for protein purification without significant lysis of E. coli.
FIG. 3.
Lysis of E. coli using the lambda lysis system. An inducible promoter was used to drive expression of the bacteriophage lambda lysis genes. The lytic transglycosylase gene from lambda, R, was replaced with atlA or atlA(E48A). The optical density of cultures was plotted over time. The lysis genes were induced by the addition of 0.2% l-arabinose at t = 0. E. coli expressing the lambda lysis genes (PK20; R+) was lysed shortly after induction. Expression of atlA compensated for the loss of R (PK21; atlA+), while substituting alanine for glutamic acid at amino acid 48 of AtlA eliminated lysis [PK22; atlA(E48A)+]. Shown next to the genotype of each strain in the legend is a map of the lambda lysis cassette expressed by that strain. The values reported are the means of three separate experiments, and the error values are the standard deviations.
The ability of AtlA to lyse E. coli suggested an alternative model for the function of AtlA in DNA donation. AtlA might release DNA into the supernatant by lysing the cells that were producing it (autolysis) or by lysing other cells in the culture (allolysis). Since other T4SS genes also are required for DNA donation during growth of gonococci (19), for this hypothesis to be correct the T4SS would have to be necessary for allowing AtlA to access the periplasm, or AtlA would have to be secreted by the T4SS to allow it to harvest DNA from other bacteria in the culture.
In order to investigate the possible role of AtlA in autolysis, we developed a sensitive assay for monitoring the release of RNA by gonococci in culture. The RNA of gonococcal strains was metabolically labeled by growth in medium containing [3H]adenine. It was previously shown that the majority of label incorporated into Neisseria using this method is incorporated into RNA (25). Labeled cells were grown in fresh liquid medium, and culture supernatants were collected periodically to determine the amount of RNA released by the cells. Treatment of the medium from the wild-type strain with RNase A confirmed that most of the released material was RNA (data not shown). In contrast to the differences in DNA secretion observed between the wild-type and atlA strains (15, 20), levels of RNA release for these strains were similar during the log phase (Fig. 4A). These results indicate that these strains undergo autolysis at similar levels and that AtlA is not significantly involved in autolysis in growing cultures. These data also demonstrate that the observed release of DNA by the wild-type strain is not due to autolysis caused by AtlA.
FIG. 4.
Lysis of gonococci. The RNA of strains was metabolically labeled, and the amount of RNA released into the culture supernatant over time was used to measure lysis. Radioactivity in culture supernatants was measured, and the fraction of the total radioactivity released into the supernatant was plotted over time. (A) Labeled wild-type gonococci (MS11) and gonococci containing a point mutation in atlA (JD1616) were grown in pure culture to examine autolysis. (B) In order to examine allolysis, cells with radiolabeled RNA and containing a point mutation in atlA (JD1616) were grown in coculture with unlabeled, wild-type gonococci (MS11) or gonococci containing a point mutation in atlA (JD1616). The values reported are the means of three separate experiments, and the error values are the standard deviations.
While AtlA does not act as an autolysin during log-phase growth, we wanted to examine its potential role as an allolysin. Streptococcus pneumoniae has been shown to lyse neighboring pneumococcal cells in order to harvest DNA during the development of competence (35). It was possible that AtlA similarly lysed gonococcal cells from the outside, causing the release of DNA. In a homogenous population of cells, it was possible that we would not be able to detect allolysis by using the described RNA release assay, so we developed a variation of the radiolabeled-RNA release assay to directly examine lysis from without. The RNA of gonococci containing atlA(E48A) was metabolically labeled, and these cells were grown in mixed culture with wild-type, unlabeled cells or with unlabeled cells containing atlA(E48A). If AtlA acted as an allolysin, then the mixed culture containing the wild-type strain should show increased release of labeled RNA. The point mutation in atlA did not affect how much RNA was liberated into the culture supernatant during coculture with a labeled strain (Fig. 4B), indicating that AtlA does not lyse gonococci from the outside to cause the release of cellular components, including DNA.
Lytic transglycosylase requirements for type IV secretion.
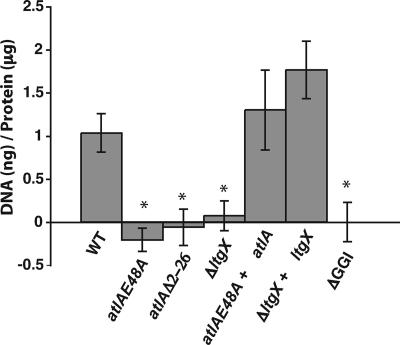
It was previously shown that a gonococcal strain containing an insertion mutation in atlA is deficient for DNA secretion. The level of fluorescence detected in the culture supernatants of this strain in the fluorometric DNA detection assay was the same as the background levels of fluorescence for the assay (15, 20), indicating that little or no DNA was secreted. To examine the necessity of the lytic transglycosylase activity of AtlA for DNA secretion and to rule out a polar effect as the cause of the defect in the original insertion mutant, we tested a strain containing a point mutation in the putative active site of atlA. A strain encoding AtlA(E48A), JD1616, did not secrete detectable DNA into the culture medium. Complementation with a wild-type copy of atlA rescued the ability of gonococci to secrete DNA (Fig. 5). These data indicate that the lytic PG transglycosylase activity of AtlA is required for type IV secretion.
FIG. 5.
Fluorometric detection of secreted DNA during log-phase growth of gonococci. DNA secretion was measured by treating culture supernatants with a fluorescent, DNA binding dye. Secreted DNA was normalized to the total protein in the cell pellet. The background fluorescence was determined by carrying out the assay using gonococci with the GGI deleted (ND500; ΔGGI). This background was subtracted from the fluorescence level of each culture. Strains containing a point mutation in atlA (JD1616; atlAE48A), an internal deletion of atlA (PK127; atlAΔ2-26), or a deletion of ltgX (PK120; ΔltgX) were deficient in DNA secretion. Complementation of the atlA point mutant (PK112; atlAE48A + atlA) or the ltgX deletion mutant (PK124; ΔltgX + ltgX) restored DNA secretion. Each reported value is the result of at least three separate experiments, and the errors are the standard deviations. *, P < 0.005 compared to results for the wild type (WT).
The main difference between AtlA and the related bacteriophage lytic transglycosylases is the presence of a 28-amino-acid extension on the N terminus of AtlA (14). We hypothesize that this region of the protein may be necessary for localization of AtlA to the T4SS or interaction with other T4SS proteins. An in-frame deletion of atlA removing the coding region for amino acids 2 to 26 was found to eliminate DNA secretion (Fig. 5), suggesting that this region of AtlA is necessary for its function in type IV secretion.
In addition to AtlA, there is another putative lytic transglycosylase, LtgX, encoded by the GGI (19). An in-frame deletion of ltgX eliminated DNA secretion by N. gonorrhoeae, suggesting that LtgX also acts in type IV secretion. Complementation with ltgX restored DNA secretion to the levels observed for wild-type gonococci (Fig. 5). These results indicate that the gonococcal T4SS requires two peptidoglycanases for type IV secretion. The requirement of two peptidoglycanases has not previously been described for a T4SS.
Mutation of atlA does not affect PG fragment release during growth.
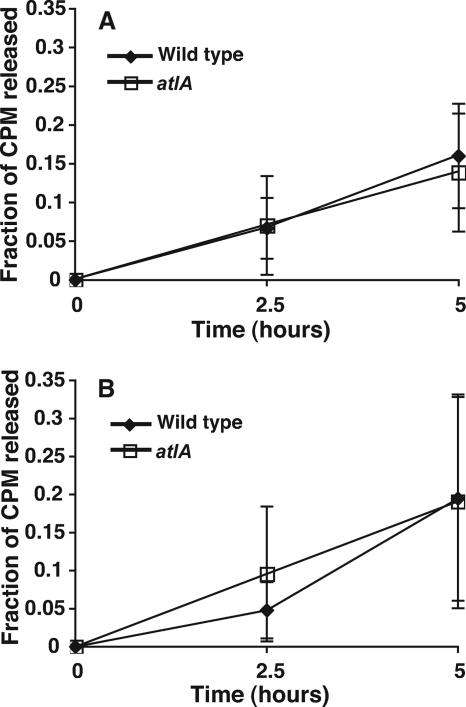
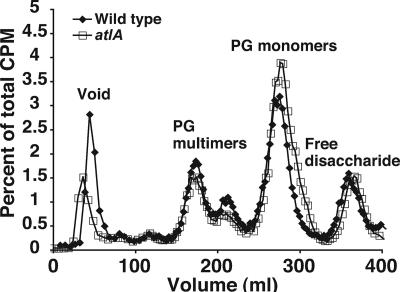
Gonococci are known to release soluble PG fragments as multimers, monomers, and free disaccharide into the culture supernatant during growth (34). Organ culture studies using human fallopian tubes have shown that PG monomers released from N. gonorrhoeae cause the death of ciliated epithelial cells (29). Our laboratory has identified a putative PG transglycosylase, LtgA, that is involved in the release of cytotoxic PG monomers (8). ltgA mutants release about 50% as much PG monomer as the wild-type parent. Because atlA encodes a PG transglycosylase, we considered the possibility that it might have a role in the release of PG fragments. The PG of wild-type and atlA(E48A) gonococcal strains was radioactively labeled by growing cells with [6-3H]glucosamine. After a period of growth without label, supernatants were collected from the cultures, and the released PG fragments were separated by gel filtration. The point mutation in atlA did not reduce PG fragment release (Fig. 6), indicating that the lytic transglycosylase activity of AtlA is not involved in the release of PG monomers from gonococci.
FIG. 6.
Profile of PG fragments released from gonococci during growth. The PG of strains JD1616 [atlA(E48A)] and MS11 (wild type) was 3H labeled, and then strains were grown without label. Culture supernatants were collected, and released 3H-labeled PG fragments were separated by gel filtration. The percentage of radioactivity in each fraction per total radioactivity eluted was plotted over time. Wild-type gonococci and gonococci with an alteration in the putative active site of AtlA did not differ in their profiles of released PG fragments during growth.
DISCUSSION
The original studies with AtlA suggested that it might be an autolysin (14), but later studies identified a requirement of AtlA for DNA release during the growth phase. atlA insertion mutants had the same phenotype as other T4SS mutants, failing to donate DNA during coculture transformation and showing reduced release of DNA during growth (15, 20). These results suggested that AtlA might act for assembly of the T4SS, as has been proposed for other secretion system peptidoglycanases (3, 12, 27). However, the DNA sequence of the GGI revealed the presence of a second peptidoglycanase homologue in the gene cluster encoding the T4SS. This second peptidoglycanase homologue (LtgX) is more similar to secretion system peptidoglycanases than is AtlA (19). These results raised questions about the function of AtlA in DNA release that we have addressed in this study. Is AtlA a lytic transglycosylase? Is it lytic transglycosylase activity or some other function of AtlA that is required for DNA release? Does AtlA function to lyse cells in order to release DNA? Are both AtlA and LtgX required for DNA release?
Using an in vitro reaction, we found that an MBP-AtlA fusion protein had lytic transglycosylase activity, breaking down gonococcal PG and producing 1,6-anhydro PG monomers (Fig. 1 and 2). The monomers were mostly disaccharide tetrapeptides, with a small fraction being disaccharide tripeptides. This result may reflect a larger amount of tetrapeptide in the macromolecular PG used for the assay or may indicate that AtlA has a specificity for PG strands carrying tetrapeptide side chains. These data demonstrate that AtlA has lytic transglycosylase activity in vitro. Many secretion systems use a lytic transglycosylase homologue, and it has long been predicted that these enzymes carry out this function to create a space in the cell wall for the secretion system machinery (2, 3, 12, 27). However, this study is the first demonstration of lytic transglycosylase activity for a T4SS peptidoglycanase homologue.
We carried out PG zymogram analyses to evaluate PG degradation activity of AtlA, but we found that this technique was not useful for this purpose. AtlA produced a zone of clearing on the PG-containing gel. However, the AtlA(E48A) mutant form of the protein and wild-type AtlA produced identical zones of clearing, even when we diluted the proteins to the point that we could barely detect the zones of clearing. The in vitro PG degradation assay demonstrated that the E48A substitution eliminated PG degradation activity. We suspect that AtlA(E48A) is able to bind PG and prevents dye from binding, thereby producing the zone of clearing. Similar results have been reported by A. Dijkstra for proteins from H. influenzae that bind PG but do not degrade it (10) and by G. Koraimann for unspecified proteins (27). PG zymogram analysis has proven its usefulness for identification of PG binding proteins and PG-degrading proteins (5, 11, 31, 40), but we conclude that it cannot be used alone to demonstrate PG degradation activity.
Mutations in atlA demonstrated the importance of the lytic transglycosylase domain as well as the nonconserved N-terminal region of the protein for type IV secretion. A mutation to change the putative active-site residue of AtlA resulted in the loss of PG degradation activity in the in vitro assay and the loss of DNA secretion by N. gonorrhoeae (Fig. 1 and 5). These results indicate that the lytic transglycosylase activity of AtlA is required for DNA secretion. Similarly, an internal deletion removing most of the unique N-terminal coding sequence reduced DNA secretion. The function of this region is not clear, but it does not have the attributes of a Sec-dependent or twin-arginine transport-dependent signal sequence. We hypothesize that this region may be involved in the interaction of AtlA with other T4SS proteins.
Since atlA was able to substitute for lambda R in the bacteriophage lambda lysis system, we considered the possibility that AtlA could be involved in the lysis of gonococci. We developed a sensitive assay of gonococcal lysis in order to determine if AtlA acted in autolysis or allolysis in growing cultures. Using this assay, we found that the lysis of gonococci occurred in growing cultures, but the presence or absence of an active form of AtlA did not affect lysis (Fig. 4). This result is in agreement with previous findings that indicated that T4SS mutants are not reduced in the release of cytoplasmic protein or the degree of cell death in growing cultures (15, 19).
In addition to requiring atlA, gonococci also require ltgX for type IV secretion of DNA. This is the first T4SS that has been found to require two lytic transglycosylase homologues. It is unclear why two PG degradation enzymes should be required. If the purpose of peptidoglycanases for type IV secretion is to create an opening in the cell wall to allow assembly of the T4SS, then is one opening not enough? A possible clue to aid in understanding this problem is provided by variants of the GGI. Some gonococcal strains lack atlA and, in its place, carry another putative peptidoglycanase gene fused in frame to the preceding gene, traG (GenBank accession no. DQ835990) (our unpublished observation). TraG is a predicted membrane protein with a large domain predicted to reside in the periplasm. Perhaps AtlA or the putative peptidoglycanase domain on the variant TraG protein produces a localized break in the PG to allow the periplasmic portion of TraG to interact with other parts of the T4SS. Similarly, ltgX is adjacent to a gene for a putative PG-associated protein, Yag (19). LtgX could be required to allow Yag to bind to PG. One of the peptidoglycanases, AtlA or LtgX, could act to make an initial break in the PG to allow assembly to begin, and the other enzyme might act to allow completion of T4SS assembly and T4SS protein interactions. AtlA or LtgX may interact with other T4SS proteins in order to facilitate the function or assembly of the T4SS apparatus. Interactions of T4SS components with putative PG transglycosylases have been described for the Agrobacterium tumefaciens and Brucella suis T4SSs (1, 21, 38). Cross-species complementation studies by Höppner et al. suggested that interactions of T4SS peptidoglycanases with other T4SS components are necessary for type IV secretion (22). Further experiments will be necessary to understand the order of assembly and T4SS protein interactions in N. gonorrhoeae.
In conclusion, we have shown that AtlA acts as a PG-lytic transglycosylase. Although it is able to lyse E. coli in the lambda lysis system, AtlA does not affect lysis of gonococci during growth, suggesting that its function is specific to the T4SS. Both lytic transglycosylase activity and a unique N-terminal domain of the protein are required for type IV secretion. Both AtlA and LtgX are required for type IV secretion, suggesting that these enzymes act at different times or in different places in the apparatus to allow assembly of a functional T4SS.
Acknowledgments
This work was supported by NIH grant AI47958 to J.P.D. H.L.H. was supported by NIH grant T32 AI055397. P.L.K. was supported by NIH grant T32 GM007215.
We thank R. S. Rosenthal for the kind gift of PG standards and T. Gaal for providing the wild-type bacteriophage lambda. We also thank W. Salgado-Pabón for helpful technical advice and K. Hackett for construction of pKH37.
Footnotes
Published ahead of print on 25 May 2007.
REFERENCES
- 1.Baron, C., M. Llosa, S. Zhou, and P. C. Zambryski. 1997. VirB1, a component of the T-complex transfer machinery of Agrobacterium tumefaciens, is processed to a C-terminal secreted product, VirB1. J. Bacteriol. 179:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bayer, M., K. Bischof, R. Noiges, and G. Koraimann. 2000. Subcellular localization and processing of the lytic transglycosylase of the conjugative plasmid R1. FEBS Lett. 466:389-393. [DOI] [PubMed] [Google Scholar]
- 3.Bayer, M., R. Eferl, G. Zellnig, K. Teferle, A. Dijkstra, G. Koraimann, and G. Hogenauer. 1995. Gene 19 of plasmid R1 is required for both efficient conjugative DNA transfer and bacteriophage R17 infection. J. Bacteriol. 177:4279-4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bayer, M., R. Iberer, K. Bischof, E. Rassi, E. Stabentheiner, G. Zellnig, and G. Koraimann. 2001. Functional and mutational analysis of p19, a DNA transfer protein with muramidase activity. J. Bacteriol. 183:3176-3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernadsky, G., T. J. Beveridge, and A. J. Clarke. 1994. Analysis of the sodium dodecyl sulfate-stable peptidoglycan autolysins of select gram-negative pathogens by using renaturing polyacrylamide gel electrophoresis. J. Bacteriol. 176:5225-5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bienkowska-Szewczyk, K., B. Lipinska, and A. Taylor. 1981. The R gene product of bacteriophage lambda is the murein transglycosylase. Mol. Gen. Genet. 184:111-114. [DOI] [PubMed] [Google Scholar]
- 7.Bienkowska-Szewczyk, K., and A. Taylor. 1980. Murein transglycosylase from phage lambda lysate. Purification and properties. Biochim. Biophys. Acta 615:489-496. [DOI] [PubMed] [Google Scholar]
- 8.Cloud, K. A., and J. P. Dillard. 2002. A lytic transglycosylase of Neisseria gonorrhoeae is involved in peptidoglycan-derived cytotoxin production. Infect. Immun. 70:2752-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Demchick, P., and A. L. Koch. 1996. The permeability of the wall fabric of Escherichia coli and Bacillus subtilis. J. Bacteriol. 178:768-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dijkstra, A. J. 1997. The soluble lytic transglycosylase family of Escherichia coli. Ph.D. thesis. University of Groningen, Groningen, The Netherlands.
- 11.Dijkstra, A. J., and W. Keck. 1996. Identification of new members of the lytic transglycosylase family in Haemophilus influenzae and Escherichia coli. Microb. Drug Resist. 2:141-145. [DOI] [PubMed] [Google Scholar]
- 12.Dijkstra, A. J., and W. Keck. 1996. Peptidoglycan as a barrier to transenvelope transport. J. Bacteriol. 178:5555-5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dillard, J. P., and K. T. Hackett. 2005. Mutations affecting peptidoglycan acetylation in Neisseria gonorrhoeae and Neisseria meningitidis. Infect. Immun. 73:5697-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dillard, J. P., and H. S. Seifert. 1997. A peptidoglycan hydrolase similar to bacteriophage endolysins acts as an autolysin in Neisseria gonorrhoeae. Mol. Microbiol. 25:893-901. [DOI] [PubMed] [Google Scholar]
- 15.Dillard, J. P., and H. S. Seifert. 2001. A variable genetic island specific for Neisseria gonorrhoeae is involved in providing DNA for natural transformation and is found more often in disseminated infection isolates. Mol. Microbiol. 41:263-277. [DOI] [PubMed] [Google Scholar]
- 16.Gunn, J. S., and D. C. Stein. 1996. Use of a non-selective transformation technique to construct a multiply restriction/modification-deficient mutant of Neisseria gonorrhoeae. Mol. Gen. Genet. 251:509-517. [DOI] [PubMed] [Google Scholar]
- 17.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamilton, H. L., and J. P. Dillard. 2006. Natural transformation of Neisseria gonorrhoeae: from DNA donation to homologous recombination. Mol. Microbiol. 59:376-385. [DOI] [PubMed] [Google Scholar]
- 19.Hamilton, H. L., N. M. Domínguez, K. J. Schwartz, K. T. Hackett, and J. P. Dillard. 2005. Neisseria gonorrhoeae secretes chromosomal DNA via a novel type IV secretion system. Mol. Microbiol. 55:1704-1721. [DOI] [PubMed] [Google Scholar]
- 20.Hamilton, H. L., K. J. Schwartz, and J. P. Dillard. 2001. Insertion-duplication mutagenesis of Neisseria: use in characterization of DNA transfer genes in the gonococcal genetic island. J. Bacteriol. 183:4718-4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Höppner, C., A. Carle, D. Sivanesan, S. Hoeppner, and C. Baron. 2005. The putative lytic transglycosylase VirB1 from Brucella suis interacts with the type IV secretion system core components VirB8, VirB9 and VirB11. Microbiology 151:3469-3482. [DOI] [PubMed] [Google Scholar]
- 22.Höppner, C., Z. Liu, N. Domke, A. N. Binns, and C. Baron. 2004. VirB1 orthologs from Brucella suis and pKM101 complement defects of the lytic transglycosylase required for efficient type IV secretion from Agrobacterium tumefaciens. J. Bacteriol. 186:1415-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jyssum, S. 1971. Utilization of thymine, thymidine and TMP by Neisseria meningitidis. 2. Lack of enzymes for specific incorporation of exogenous thymine, thymidine and TMP into DNA. Acta Pathol. Microbiol. Scand. Sect. B Microbiol. Immunol. 79:778-788. [DOI] [PubMed] [Google Scholar]
- 24.Kellogg, D. S., Jr., W. L. Peacock, Jr., W. E. Deacon, L. Brown, and D. I. Pirkle. 1963. Neisseria gonorrhoeae. I. Virulence genetically linked to clonal variation. J. Bacteriol. 85:1274-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kingsbury, D. T., and J. F. Duncan. 1967. Use of exogenous adenine to label the nucleic acids of wild-type Neisseria meningitidis. J. Bacteriol. 94:1262-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohler, P. L., K. A. Cloud, K. T. Hackett, E. T. Beck, and J. P. Dillard. 2005. Characterization of the role of LtgB, a putative lytic transglycosylase in Neisseria gonorrhoeae. Microbiology 151:3081-3088. [DOI] [PubMed] [Google Scholar]
- 27.Koraimann, G. 2003. Lytic transglycosylases in macromolecular transport systems of gram-negative bacteria. Cell. Mol. Life Sci. 60:2371-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mehr, I. J., C. D. Long, C. D. Serkin, and H. S. Seifert. 2000. A homologue of the recombination-dependent growth gene, rdgC, is involved in gonococcal pilin antigenic variation. Genetics 154:523-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melly, M. A., Z. A. McGee, and R. S. Rosenthal. 1984. Ability of monomeric peptidoglycan fragments from Neisseria gonorrhoeae to damage human fallopian-tube mucosa. J. Infect. Dis. 149:378-386. [DOI] [PubMed] [Google Scholar]
- 30.Morse, S. A., and L. Bartenstein. 1974. Factors affecting autolysis of Neisseria gonorrhoeae. Proc. Soc. Exp. Biol. Med. 145:1418-1421. [DOI] [PubMed] [Google Scholar]
- 31.Rambow-Larsen, A. A., and A. A. Weiss. 2002. The PtlE protein of Bordetella pertussis has peptidoglycanase activity required for Ptl-mediated pertussis toxin secretion. J. Bacteriol. 184:2863-2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosenthal, R. S., and R. Dziarski. 1994. Isolation of peptidoglycan and soluble peptidoglycan fragments. Methods Enzymol. 235:253-285. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 34.Sinha, R. K., and R. S. Rosenthal. 1980. Release of soluble peptidoglycan from growing gonococci: demonstration of anhydro-muramyl-containing fragments. Infect. Immun. 29:914-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steinmoen, H., E. Knutsen, and L. S. Havarstein. 2002. Induction of natural competence in Streptococcus pneumoniae triggers lysis and DNA release from a subfraction of the cell population. Proc. Natl. Acad. Sci. USA 99:7681-7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swanson, J., S. J. Kraus, and E. C. Gotschlich. 1971. Studies on gonococcus infection. I. Pili and zones of adhesion: their relation to gonococcal growth patterns. J. Exp. Med. 134:886-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wade, J. J., and M. A. Graver. A fully defined, clear and protein-free liquid medium permitting dense growth of Neisseria gonorrhoeae from very low inocula. FEMS Microbiol. Lett. [Epub ahead of print.] doi: 10.1111/j.1574-6968.2007.00776.x. [DOI] [PubMed]
- 38.Ward, D. V., O. Draper, J. R. Zupan, and P. C. Zambryski. 2002. Peptide linkage mapping of the Agrobacterium tumefaciens vir-encoded type IV secretion system reveals protein subassemblies. Proc. Natl. Acad. Sci. USA 99:11493-11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Young, R. 1992. Bacteriophage lysis: mechanism and regulation. Microbiol. Rev. 56:430-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zahrl, D., M. Wagner, K. Bischof, M. Bayer, B. Zavecz, A. Beranek, C. Ruckenstuhl, G. E. Zarfel, and G. Koraimann. 2005. Peptidoglycan degradation by specialized lytic transglycosylases associated with type III and type IV secretion systems. Microbiology 151:3455-3467. [DOI] [PubMed] [Google Scholar]
- 41.Zhang, N., and R. Young. 1999. Complementation and characterization of the nested Rz and Rz1 reading frames in the genome of bacteriophage lambda. Mol. Gen. Genet. 262:659-667. [DOI] [PubMed] [Google Scholar]