Abstract
Phosphate import is required for the growth of mycobacteria and is regulated by environmental inorganic phosphate (Pi) concentrations, although the mechanism of this regulation has not been characterized. The expression of genes involved in Pi acquisition is frequently regulated by two-component regulatory systems (2CRs) consisting of a sensor histidine kinase and a DNA-binding response regulator. In this work, we have identified the senX3-regX3 2CR as a Pi-dependent regulator of genes involved in phosphate acquisition in Mycobacterium smegmatis. Characterization of senX3 mutants with different PhoA phenotypes suggests a dual role for SenX3 as a phosphatase or a phosphodonor for the response regulator RegX3, depending upon Pi availability. Expression of PhoA activity required phosphorylation of RegX3, consistent with a role for phosphorylated RegX3 (RegX3∼P) as a transcriptional activator of phoA. Furthermore, purified RegX3∼P bound to promoter sequences from phoA, senX3, and the high-affinity phosphate transporter component pstS, demonstrating direct transcriptional control of all three genes. DNase I footprinting and primer extension analyses have further defined the DNA-binding region and transcriptional start site within the phoA promoter. A DNA motif consisting of an inverted repeat was identified in each of the promoters bound by RegX3∼P. Based upon our findings, we propose a model for Pi-regulated gene expression mediated by SenX3-RegX3 in mycobacteria.
Bacterial pathogens must adapt to numerous harsh conditions in order to persist within their hosts. The expression of genes required for survival under specific environmental conditions is often controlled by two-component regulatory systems (2CRs) consisting of a sensor histidine kinase (HK) and a response regulator (RR) (17, 34). Commonly, HKs are membrane-associated proteins which autophosphorylate upon detection of an environmental stimulus and mediate transfer of the phosphoryl group to a cognate RR at a highly conserved aspartic acid (Asp) residue. Phosphorylation of the RR leads to activation of its effector function, which usually involves DNA binding and transcriptional regulation. In the absence of an environmental stimulus, the HK can often catalyze the reverse reaction, resulting in dephosphorylation and inactivation of the RR effector function (7, 31, 44).
Recent data have emerged regarding the 2CRs of Mycobacterium tuberculosis, and important roles have been demonstrated for several, including DevRS, MprAB, PhoPR, PrrAB, and SenX3-RegX3 (14, 22, 24, 25, 27, 29, 43). Parish et al. have performed microarray analyses of M. tuberculosis H37Rv with a deletion in the regX3 RR and identified approximately 100 differentially regulated genes involved in diverse cellular pathways (25). However, a consensus DNA-binding motif within the promoters of deregulated genes was not apparent. In addition, no activation of senX3-regX3 expression was observed in response to a variety of stress conditions, including pH extremes, nutrient deprivation, antibiotics, sodium dodecyl sulfate (SDS), and H2O2 (25). Thus, the genes that directly require senX3-regX3 for their regulation and the governing environmental stimuli remain to be determined.
Inorganic phosphate (Pi) is an essential nutrient for living organisms and must be acquired by pathogens during the course of infection. M. tuberculosis carries multiple copies of genes for components of a high-affinity, phosphate-specific transporter (Pst) (8, 21). Mutations within any of several Pst genes result in reduced phosphate import, and such strains are significantly attenuated for growth within macrophages and in animal models of infection (10, 26, 28). Furthermore, expression of pstS genes increases when mycobacteria are grown under Pi-limiting conditions, demonstrating Pi-regulated expression of the Pst transporters (2, 13, 21). However, none of the M. tuberculosis 2CRs have been shown to regulate gene expression in response to limiting available Pi.
Analysis of Pi-regulated gene expression is often facilitated by the detection of alkaline phosphatase (encoded by phoA), a monoesterase which liberates Pi from macromolecules. Although a low level of nonspecific phosphatase activity is present on the cell surface, pathogenic mycobacteria do not carry a phoA homolog (5, 8, 9, 15). We recently reported the unexpected presence of a Pi-regulated phoA gene in the saprophytic organism Mycobacterium smegmatis, providing a convenient means to study Pi-regulated gene expression in mycobacteria (19). Here, the senX3-regX3 2CR is shown to respond to limiting environmental phosphate concentrations and to regulate phoA expression in M. smegmatis. This work provides key information for further analysis of Pi-regulated gene expression in pathogenic mycobacterial species.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Escherichia coli strains used for cloning, protein expression, and cosmid propagation were grown in LB. Wild-type M. smegmatis mc2155 (33) was grown in LB broth supplemented with 0.02% tyloxapol for those experiments where Pi was not a limiting nutrient. Phosphate-limiting growth assays were performed using a MOPS (morpholinepropanesulfonic acid)-glucose minimal medium (25 mM MOPS, pH 7.2, 25 mM KCl, 10 mM Na2SO4, 20 mM NH4Cl, 10 μM FeCl3, 2 mM MgSO4, 0.1 mM CaCl2, 0.4% glucose, 0.02% tyloxapol) with K2HPO4 added as a source of Pi.
Plasmid construction.
Plasmid construction and molecular biology techniques were performed according to methods described by Sambrook et al. (32). A list of plasmids used in this work is provided in Table 1. All cloned DNA alleles were verified by DNA sequencing. The complete senX3-regX3 operon (including 300 bp of upstream promoter sequence) from M. smegmatis and M. tuberculosis was PCR amplified and cloned into pMV306 (35) to generate pYUB1113 and pYUB1114, respectively.
TABLE 1.
Plasmids used in this work
| Plasmid name | Relevant characteristics | Reference |
|---|---|---|
| p0004.SacB | Suicide vector, Hygr, SacB counterselection | Gift from T. Hsu |
| pYUB76 | pMV306; (promoterless) E. coli lacZ | 4 |
| pYUB1106 | pYUB76; PphoA-lacZ fusion | This work |
| pMV306 | Integrative, single copy | 35 |
| pYUB1113 | pMV306; PsenX3-senX3-regX3 from M. smegmatis | This work |
| pYUB1114 | pMV306; PsenX3-senX3-regX3 from M. tuberculosis | This work |
| pET45b | E. coli expression vector | Novagen |
| pTIC6 | Integrative, single-copy, tetracycline-inducible promoter | This work |
| pYUB1122 | pTIC6; His6RegX3 | This work |
| pYUB1123 | pTIC6; His6RegX3(D52N) | This work |
N-terminal His6 fusion protein constructs were generated as follows. regX3 was PCR amplified using the oligonucleotides (s)regX3fwd (ATAAGATCTAACCAGCGTGTTGATCGTTG) and (s)regX3rvr2 (GCAAGCTTTCAGCCCTCCAGCTTGTATC), where introduced BglII and HindIII sites are underlined. A truncated fragment of senX3 (lacking the N-terminal 393-bp transmembrane domain) was amplified using (s)senX3sol (TAACGGTACCCGGCTGCTGACCGACGAC) and (s)senX3rvr (GCAAGCTTCGCTGGTCATCGTTCTCGTTG), where KpnI and HindIII sites are underlined. PCR products were digested and ligated into pET45b vector DNA (Novagen, Madison, WI). A missense mutation (underlined) at codon 52 (GAT to AAT) was introduced into regX3 using the Quick-Change Site-directed mutagenesis kit (Stratagene, La Jolla, CA) and oligonucleotides Rmut1 (GCCGACATCGTGCTGCTGAATCTCATGCTCCCCGGCA; the mutagenized nucleotide is in italics) and Rmut2 (TGCCGGGGAGCATGAGATTCAGCAGCACGATGTCGGC). The resulting product, His6regX3(D52N), contains an Asn substitution at position Asp52, which corresponds to the conserved site of RegX3 phosphorylation (16).
To assemble the integrative tetracycline-inducible vector pTIC6, a fragment containing the M. smegmatis rrnT2 transcription terminator and the Psmyc1tetO promoter/operator (12) was amplified by PCR using pUV15tetORs as a template. This fragment was inserted into pMP118 (a gift from Martin Pavelka), a pMV261 derivative containing the Tn10 tetR gene fused to the Mycobacterium bovis hsp60 promoter. A synthetic multiple cloning site was inserted downstream of Psmyc1tetO. Finally, the pMV261 backbone was replaced with that of pMV361 to generate pTIC6 (http://www.aecom.yu.edu/tbresearch/Resources/Vectors/6.html). The His6regX3 and His6regX3(D52N) genes were amplified from the pET45b-derived vectors described above (including the vector-derived ribosome binding site [RBS]) and cloned into the multiple cloning site of pTIC6 to generate pYUB1122 and pYUB1123, respectively.
Plasmid pYUB1106 was constructed by PCR amplification of the phoA promoter with BglII sites (underlined) introduced using primers Pph1 (AAAGATCTCGGCCACCCGGACGCAGG) and Pph2 (GTCGGATCCACGGCTTCTCCTAGACTC). The PCR product was digested and ligated with pYUB76 (4). The final construct consisted of approximately 300 bp of promoter sequence with the phoA initiation codon in frame with the promoterless E. coli lacZ.
Transposon mutagenesis.
Transposon mutagenesis was performed using phAE181(Kanr), a temperature-sensitive mycobacteriophage containing the mariner-based transposon Tn5371 (19, 30). Preparation of high-titer phages and transductions were performed as previously described (20). Transposon mutants that constitutively express PhoA activity were isolated on LB agar plates containing 50 μg/ml kanamycin and 60 μg/ml 5-bromo-4-chloro-3-inlolylphosphate (BCIP). Genetic sites of transposon insertion were identified as previously described (3, 11).
Specialized and generalized transductions.
DNA regions of approximately 1 kb flanking senX3 or regX3 were amplified by PCR and cloned into the counterselectable vector p0004.SacB (a gift from T. Hsu) (unpublished data). Sequenced plasmids were linearized with PacI and ligated to PacI-digested phAE159 DNA. Ligation reaction products were packaged using Gigapack Gold III (Stratagene, La Jolla, CA) and grown as cosmids in E. coli HB101. Cosmid DNA containing the deletion constructs was electroporated into M. smegmatis mc2155, and high-titer phage was prepared as previously described (20). Following transduction, strains with successful allelic exchange were selected on LB agar plates containing 50 μg/ml hygromycin at 37°C. Generalized transductions using mycobacteriophage Bxz1 were performed as previously described (20).
Alkaline phosphatase assays.
Determination of alkaline phosphatase activity on whole cells was performed as described previously, with minor modifications (6). Bacterial cultures were diluted to an optical density at 600 nm (OD600) of approximately 0.05 and grown at 37°C with gentle shaking for 40 h. One-milliliter aliquots were removed and washed twice in an equal volume of TT buffer (1 M Tris, pH 8.0, 0.1% Tween 80). The cells were resuspended in 1 ml TT buffer, and 100-μl aliquots were divided into individual tubes. One milliliter of substrate solution (10 mM p-nitrophenyl phosphate, 10 mM MgCl2) was added, and the tubes were incubated at 37°C and protected from light until a pale yellow color developed; 0.2 ml of 1 M K2HPO4 was added to stop the reaction, and cellular debris was removed by centrifugation. The OD of each sample was read at 410 nm, and alkaline phosphatase activity was determined using the following formula: 1,000 × OD410 × (min of reaction)−1 × OD600−1 × (0.1 ml volume of cells)−1.
β-Galactosidase assays.
M. smegmatis was grown in MOPS-glucose medium with K2HPO4 added as a source of Pi, as described above. At the indicated time points, aliquots were removed; washed in 1 M Tris, pH 8.0; lysed by sonication; and centrifuged for 10 min to remove insoluble material. Total protein in the soluble fraction was quantified using the Bio-Rad Protein Assay Kit (Bio-Rad, Hercules, CA). β-Galactosidase activity was determined as previously described (23). Units of activity were defined as follows: 1,000 × OD420 × (min of reaction)−1 × (mg protein)−1.
Northern blot analysis.
RNA was isolated from 10 ml bacteria using the FastRNA Pro Blue kit and FastPrep Instrument (Qbiogene, Carlsbad, CA) for 40 s at a speed setting of 6. Five hundred nanograms of RNA was run per lane on a 1% denaturing agarose gel using NorthernMax Northern blot reagents (Ambion, Austin, TX). Following electrophoresis, the separated RNA was transferred to a cellulose membrane using capillary transfer and cross-linked by UV irradiation. Single-stranded RNA probes were prepared by amplifying intragenic target DNA with PCR primers introducing a 3′ T7 sequence. PCR products were used as transcription templates and labeled with [α-32P]CTP using Ambion's Maxiscript Kit. Membranes were incubated with 1 × 106 cpm of labeled probe for 1 h at 80°C in UltraHyb buffer and washed six times in 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) buffer. The membranes were air dried and exposed on a phosphorimager screen overnight.
Nickel affinity purification of recombinant proteins.
Recombinant E. coli Rosetta(DE3)/pLysS (Novagen, Madison, WI) was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h at 37°C. The bacteria were washed in phosphate-buffered saline, resuspended in 1/50 volume of lysis buffer (50 mM Tris, pH 8.0, 60 mM NaCl, 1 mg/ml fresh lysozyme) supplemented with protease inhibitor cocktail (Roche, Mannheim, Germany), lysed by sonication, and centrifuged for 30 min at 28,000 × g at 4°C. NaCl and imidazole were then added to the cleared lysate to 500 mM and 5 mM, respectively. Protein purification was performed using Amersham HiTrap chelating columns charged with NiSO4, in accordance with the manufacturer's instructions (Amersham Biosciences, Piscataway, NJ). Briefly, the columns were equilibrated with buffer A (50 mM Tris, pH 8.0, 500 mM NaCl, 5 mM imidazole), loaded with lysate, and washed with 5 volumes of buffer A to remove nonspecific protein binding. Stepwise elutions were performed using 5 ml buffer A plus 15 mM, 60 mM, and 300 mM imidazole, with collection volumes of 1 ml each. Fractions with positive OD280 values were checked for protein content by standard SDS-polyacrylamide gel electrophoresis and stained with Coomassie blue.
Samples containing protein of the predicted molecular weight were subjected to buffer exchange using an Amersham HiTrap desalting column in buffer B (50 mM sodium phosphate, pH 7.0, 25 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol) and were concentrated using Amicon Centriprep YM-3 columns. The purified proteins were >95% pure, as determined by Coomassie blue staining on 12% SDS-polyacrylamide gel electrophoresis gels. Aliquots were mixed with an equal volume of glycerol and stored at −80°C for long-term storage.
EMSAs.
Target DNA for use in electrophoretic mobility shift assays (EMSAs) was amplified by PCR and labeled with [γ-32P]ATP using T4 polynucleotide kinase. Ten micromolar His6RegX3 was phosphorylated by 2 μM His6SenX3 in phosphorylation buffer (10 mM MgCl2, 50 mM KCl, 25 mM Tris-HCl, pH 8.0, 0.5 mM EDTA, 10 mM ATP, 5% glycerol) for 20 to 30 min at room temperature. As a control, the same reaction was performed in the absence of ATP or with His6RegX3(D52N). Binding reactions consisted of 4 μM His6RegX3 and 100 pM of target DNA in phosphorylation buffer supplemented with 6 × 10−4 U poly(dI-dC) and 0.3 mg/ml bovine serum albumin for 30 min at room temperature. Samples were run on 5% nondenaturing polyacrylamide gels (45 mM Tris-borate, 1.25 mM EDTA, 10 mM MgCl2), vacuum dried, and exposed overnight on a phosphorimager screen.
DNase I footprint analysis.
The PCR fragment of the phoA promoter was digested with BamHI, and the recessed 3′ end was labeled using [α32P]TTP and Klenow exo enzyme (Epicenter, Madison, WI). His6RegX3 was phosphorylated as described above and incubated with 5 fmol/μl of radiolabeled DNA in phosphorylation buffer supplemented with 1 mM CaCl2 and 0.3 mg/ml bovine serum albumin for 45 min at room temperature; 0.1 U DNase I was added to each tube, and the reactions were stopped after 30 seconds with stop solution (5 mM EDTA, 5 mM EGTA, 1% SDS, 0.3 M sodium acetate, pH 5.2). Following phenol-chloroform extraction and ethanol precipitation, samples were run on a 5% acrylamide sequencing gel, dried, and exposed to a phosphorimager screen overnight.
Primer extensions.
Complementary-strand oligonucleotides PhPEA1 (GCCACCGTTGGTCACGATGT) and SsPEA1 (GCAGTAACGCCGACACCAAG) were end labeled with [γ-32P]ATP using T4 polynucleotide kinase. Reverse transcription was performed using avian myeloblastosis virus reverse transcriptase (Roche, Mannheim, Germany) on RNA extracted from mc2155 grown for 1 h in Pi-limiting MOPS-glucose medium. To make a single-nucleotide marker, a plasmid carrying phoA was sequenced with the Sequitherm II kit (Epicenter, Madison, WI), using the same primers as for the reverse transcription reactions. Samples were run on a 4% polyacrylamide-urea gel, dried, and exposed to a phosphorimager screen overnight.
RESULTS
Transposon insertions within senX3 result in PhoA-constitutive mutant strains.
In order to identify genes involved in the regulation of phoA expression, M. smegmatis transposon insertion mutant libraries were plated on rich medium supplemented with the chromogenic PhoA substrate BCIP. From approximately 5,000 transductants, 24 mutants with constitutive PhoA activity were isolated. Twenty-one of the mutants had transposon insertions within genes of the phosphate- specific transporter encoded by pstSCAB, consistent with our previous observation that phoA repression requires an intact Pst transporter during growth in excess Pi (19). Three novel, independent transposon insertions mapped within the senX3 HK, with each transposon inserted in the same orientation (Fig. 1). To confirm that the senX3::Tn5371 transposon insertions mediated the PhoA-constitutive phenotype and did not result from unrelated secondary mutations, generalized transductions were performed on each mutant strain using the mycobacteriophage Bxz1 (20). Greater than 99.5% of the resulting kanamycin-resistant transductants cleaved BCIP to yield “blue” colonies, demonstrating tight linkage between the senX3::Tn5371 insertions and the PhoA-constitutive phenotype.
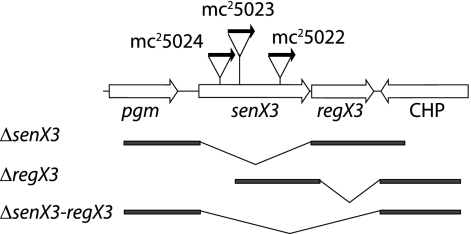
FIG. 1.
The senX3-regX3 region of M. smegmatis. senX3, regX3, and flanking genes are shown as open arrows. Vertical lines indicate sites of transposon insertion within senX3. A horizontal arrow indicates the orientation of the kanamycin-selectable marker within each transposon, with strain designations indicated above each arrow. The DNA fragments used for homologous integration (bold lines) and targeted deletions (slanted lines) are shown below the gene map.
M. smegmatis and M. tuberculosis senX3-regX3 complement senX3 Tn5371 mutants.
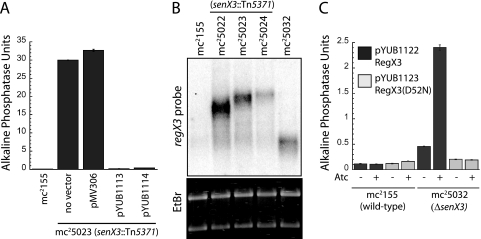
The previous result suggested that SenX3 may be required for the repression of phoA in the presence of excess Pi. To test this prediction, each of the senX3::Tn5371 mutant strains was transformed with an integrative vector carrying the senX3-regX3 operon cloned from either M. smegmatis or M. tuberculosis H37Rv. For all three mutants, 100% of transformants reverted to a PhoA-repressed phenotype, appearing as “white” colonies on agar plates containing BCIP. The senX3::Tn5371 mutant mc25023 was further used in a quantitative alkaline phosphatase assay to measure the degree of complementation. As shown in Fig. 2A, the presence of the senX3-regX3 operon cloned from either M. smegmatis (pYUB1113) or M. tuberculosis (pYUB1114) reduced alkaline phosphatase activity in mc25023 to levels comparable to that of the parental mc2155, while the vector control (pMV306) had no significant effect. The ability of M. tuberculosis senX3-regX3 to complement the senX3::Tn5371 PhoA-constitutive M. smegmatis mutants further suggests a shared regulatory role between these species.
FIG. 2.
Anaylsis of gene expression in senX3 mutant strains. (A) The PhoA-constitutive senX3::Tn5371 mutant mc25023 was transformed with a vector containing the complete senX3-regX3 operon cloned from M. smegmatis (pYUB1113) or M. tuberculosis (pYUB1114). Recombinant bacteria were grown in Pi-rich media and assayed for alkaline phosphatase activity as described in Materials and Methods. (B) Total RNA was extracted from the indicated strains, and Northern blot analysis was performed using a regX3-specific RNA probe. Ethidium bromide (EtBr) staining was used to confirm equal loading. (C) Wild-type mc2155 and the ΔsenX3 strain mc25032 were transformed with pYUB1122 (black bars) and pYUB1123 (gray bars). pYUB1122 encodes His6RegX3, while pYUB1123 encodes the phosphorylation-defective mutant His6RegX3(D52N), each under the transcriptional control of a tetracycline-inducible promoter. Protein expression was induced in the indicated cultures by the addition of 50 ng/ml anhydrotetracycline and assayed for alkaline phosphatase activity. The error bars indicate standard deviations.
senX3 and regX3 deletion analysis.
To further analyze the individual roles of SenX3 and RegX3 in phoA regulation, deletions of the coding region of each gene (Fig. 1) were made using specialized transduction (3). Numerous attempts to delete regX3, either alone or together with senX3, were unsuccessful, suggesting that regX3 is essential in M. smegmatis. In support of this prediction, disruptions of regX3 were obtained in merodiploid strains containing an extra copy of the senX3-regX3 operon cloned from either M. smegmatis or M. tuberculosis. Allelic exchange of senX3 was successful in mc2155, and the integrated construct was resolved to generate an unmarked ΔsenX3 mutant strain, designated mc25032. Surprisingly, unlike the transposon insertion mutants, the ΔsenX3 strain did not exhibit detectable PhoA activity, even under growth conditions where Pi was the limiting nutrient. The PhoA-uninducible phenotype observed in mc25032 suggests that SenX3 is required for the induction, rather than the repression, of PhoA activity. Because senX3 precedes regX3 in an operon, the different PhoA phenotypes most likely result either from residual SenX3 activity (in the transposon insertion mutants) or from deregulated transcription/translation of downstream regX3. Because the sites of transposon insertion are broadly distributed within senX3, residual SenX3 activity conserved in each mutant seems unlikely. The latter possibility is reasonable, as the transposon used in this study lacks a transcriptional terminator and all three transposons are inserted in the same orientation (Fig. 1).
To determine whether transcription of regX3 is constitutive among the mutant strains, Northern blot analysis was performed. As shown in Fig. 2B, mRNA corresponding to regX3 is more abundant in each of the transposon mutant strains than in wild-type M. smegmatis. In addition, the sizes of regX3-specific mRNAs are increased relative to each site of transposon insertion, consistent with regX3 transcription from the Kanr promoter. Because regX3 is essential in M. smegmatis, we predicted that some level of regX3 transcription must occur in mc25032, and a regX3-specific mRNA species of approximately 1 kb was detected in this strain.
RegX3 is required for PhoA activity.
The senX3 deletion strategy included the removal a putative RBS within the coding region. We therefore predicted that regX3 translation in mc25032 may be impaired. In order to precisely determine the effect of RegX3 expression, regX3 bearing an N-terminal His6 tag was cloned into a tetracycline-inducible vector to generate pYUB1122. An additional construct, pYUB1123, was created using site-directed mutagenesis for the expression of His6RegX3(D52N), a mutant that cannot be phosphorylated at the conserved Asp52 residue due to an Asn substitution. As expected, mc2155 transformed with either pYUB1122 or pYUB1123 showed no change in PhoA activity following promoter induction with anhydrotetracycline (Fig. 2C). In contrast, expression of His6RegX3 in mc25032 resulted in >5-fold induction of PhoA activity compared to the uninduced control, an induction level consistent with the predicted promoter activity using the tetracycline-inducible promoter in mycobacteria (12). These data are consistent with the transcription of a stable, regX3-encoding mRNA species in mc25032 which cannot be efficiently translated into functional RegX3 protein. Importantly, PhoA activity under these conditions also requires the expression of wild-type RegX3, as expression of the phosphorylation-defective mutant RegX3(D52N) had no effect on PhoA activity. Together, these observations suggest a requirement for phosphorylated RegX3 (RegX3∼P), as well as the absence of functional SenX3, in the positive regulation of PhoA activity.
Both senX3-regX3 and phoA are transcriptionally activated under Pi-limiting growth conditions.
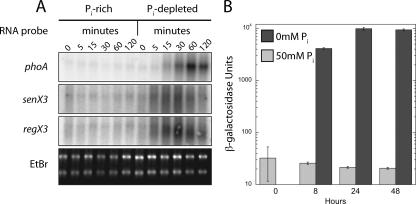
Expression of PhoA activity in M. smegmatis is known to be induced during Pi-limiting growth (19). To determine the kinetics of phoA and senX3-regX3 regulation at the level of transcription, Northern blot analysis was performed on total RNA isolated from wild-type mc2155 grown in MOPS-glucose medium in the presence of excess Pi or without exogenously supplied Pi. mRNAs corresponding to phoA, senX3, and regX3 were rapidly induced when bacteria were grown in media lacking Pi (Fig. 3A). The senX3 and regX3 mRNAs were not well resolved in the denaturing agarose gel and appeared to be between 1.2 and 2 kb and 0.9 and 2 kb, respectively, with the lower range corresponding to the size of each individual gene. Both senX3 and regX3 mRNAs were detectable as early as 5 min following phosphate depletion and preceded detectable phoA transcription. The levels of all three transcripts began to decline by 2 h poststarvation, possibly reflecting a global decrease in transcription as a result of insufficient available Pi.
FIG. 3.
Phosphate-dependent transcriptional regulation of phoA and senX3-regX3. (A) Northern blot. Total RNA was extracted from bacteria grown in medium supplemented with excess Pi (50 mM) or with no added Pi, as shown. The single-stranded RNA probes used for each blot are identified on the left. Ethidium bromide (EtBr) staining of total RNA was used to demonstrate equivalent loading of RNA. (B) Recombinant mc2155 containing the PphoA-lacZ reporter plasmid pYUB1106 was grown in the presence of excess (50 mM) Pi (gray bars) or in Pi-limiting medium (black bars). At the time points indicated, aliquots were removed and assayed for β-galactosidase activity as described in Materials and Methods. The error bars indicate standard deviations.
To analyze the regulation of the phoA promoter independently of PhoA activity, we utilized a reporter plasmid, pYUB1106, containing a PphoA-lacZ translational fusion. Recombinant M. smegmatis was grown in MOPS-glucose medium containing excess Pi (50 mM) or without added Pi and assayed for lacZ activity at various time points. In agreement with the data obtained from Northern blots, translation of lacZ from the phoA promoter was induced only when bacteria were grown under Pi-limiting growth conditions (Fig. 3B). After 24 h, the level of LacZ activity was increased more than 450 times when bacteria were grown in Pi-depleted media. In contrast, lacZ expression from the constitutive M. tuberculosis hsp60 promoter remained constant throughout the assay, regardless of the phosphate concentration (data not shown).
Phosphorylated RegX3 directly binds to promoters of phosphate-regulated genes.
Analysis of M. tuberculosis SenX3-RegX3 has indicated canonical 2CR characteristics, including autophosphorylation of SenX3, transfer of the phosphoryl group to RegX3, and binding of RegX3 to the senX3 promoter (autoregulation) (16). The data described here provide strong evidence for the transcriptional control of phoA by SenX3-RegX3 but cannot exclude an indirect relationship within a more complex regulatory network. In order to determine if RegX3 acts directly on the phoA promoter, EMSAs were performed.
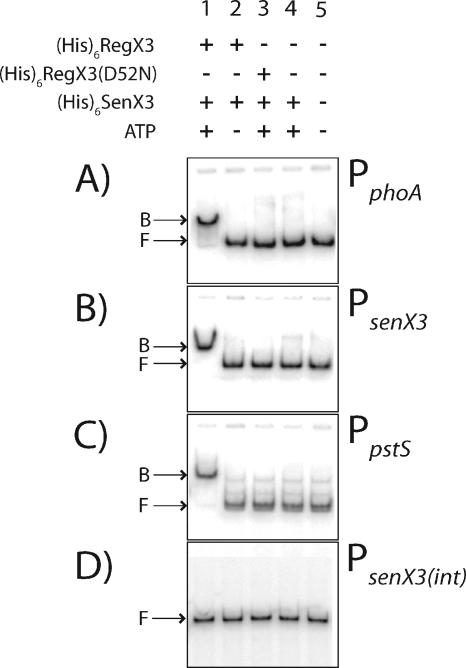
M. smegmatis His6SenX3, His6RegX3, and His6RegX3(D52N) were expressed in E. coli and purified using nickel affinity chromatography. EMSAs were performed using His6RegX3 and PCR-amplified DNA promoter elements of phoA (167 bp), senX3 (199 bp), and the phosphate-regulated gene pstS (139 bp), with the sizes of the respective PCR products in parentheses. His6RegX3 directly bound to each of the regulated promoters only when it was first phosphorylated by preincubation with His6SenX3 (Fig. 4). No binding was observed when the phosphorylation reaction was performed in the absence of ATP, with the mutant His6RegX3(D52N), or with His6SenX3 alone. In addition, RegX3∼P was unable to bind to a 355-bp DNA fragment corresponding to an internal senX3 coding sequence, PsenX3(int). These results demonstrate a specific and direct regulatory role for RegX3∼P in the transcriptional control of phoA, senX3, and pstS.
FIG. 4.
Direct binding of RegX3∼P to Pi-regulated promoter elements using EMSA. Promoter regions were amplified upstream of (A) phoA (157 bp), (B) senX3 (199 bp), and (C) pstS (139 bp) or from (D) an internal coding region of senX3 (355 bp). Radiolabeled PCR products were used in binding reactions with the protein combinations indicated above each lane. Bound (B) and Free (F) DNA is indicated on the left of each panel. Binding reactions consisted of 4 μM His6RegX3 or His6RegX3(D52N), 0.8 μM His6SenX3, and 200 pM [γ-32P]ATP-labeled DNA. The final concentration of phosphorylated His6SenX3 in lane 4 was 4 μM.
Identification of the RegX3∼P DNA-binding region and transcriptional start site (TSS) within the phoA promoter.
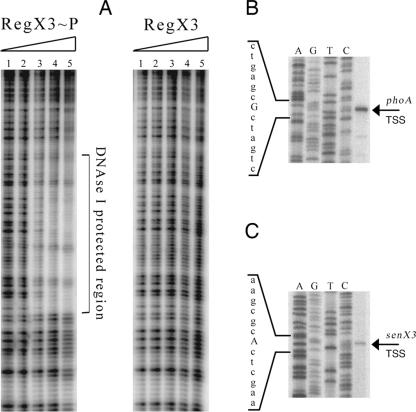
To determine the specific DNA sequence within the phoA promoter recognized by RegX3∼P, DNase I footprinting was performed. As shown in Fig. 5A, incubation with RegX3∼P protected a region of the phoA promoter from DNase I cleavage. The protected region consisted of approximately 50 bp located between positions −110 and −59 upstream of the phoA coding sequence. As predicted by the EMSA results, the same promoter fragment was not protected from DNase I cleavage using purified His6RegX3 that was not phosphorylated.
FIG. 5.
(A) DNase I footprinting of the phoA promoter bound by RegX3∼P. End-labeled phoA promoter DNA was incubated with (lane 1) 1.5 μM, (lane 2) 3 μM, (lane 3) 6 μM, (lane 4) 12 μM, or (lane 5) 24 μM phosphorylated His6RegX3. Following DNase I treatment, samples were run on a 5% acrylamide sequencing gel and visualized on a phosphorimager. The DNA region protected from DNase I cleavage is indicated. As a control, the same experiment was performed with the omission of ATP in the phosphorylation reaction. (B and C) Total RNA isolated from Pi-starved cells was used in a primer extension analysis with antisense oligonucleotide primers located within phoA (B) and senX3 (C), as indicated. Samples were run adjacent to a sequencing reaction performed using the same primer as for the reverse transcription on a 4% polyacrylamide-urea gel and visualized using a phosphorimager. The unique band identified in each primer extension is distinguished from adjacent sequence by a capital letter.
To further characterize the phoA promoter, the TSS was determined by primer extension analysis of total RNA extracted from M. smegmatis grown in Pi-limiting medium. The antisense primer PhPEA1 was used to identify a unique, dominant TSS at a cytosine residue 41 nucleotides upstream of the GTG start codon (Fig. 5B). The same experiment was performed using an additional primer, PhPEA2, and confirmed the location of the phoA TSS (data not shown). Using the same pool of total RNA and the antisense primer SsPEA1, the senX3 TSS was identified at a thymidine residue located at least 7 bp upstream of several potential translational initiation codons (Fig. 5C).
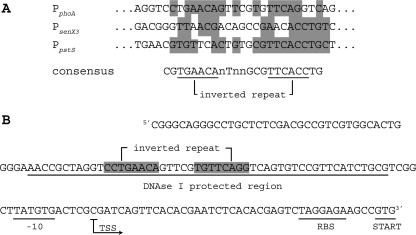
Using Gibbs motif analysis (36, 37) of the sequences of the three promoters bound by RegX3∼P, a putative “pho box” consisting of an inverted repeat (GTGAAC) and separated by seven unconserved nucleotides was identified in each promoter (Fig. 6A). This conserved motif was also identified within a 42-bp region of the M. tuberculosis senX3 promoter previously shown to bind recombinant purified RegX3 from M. tuberculosis (16). The predicted “pho box” within the M. smegmatis senX3 promoter is located 78 bp upstream of the senX3 TSS.
FIG. 6.
Conserved regulatory sequences identified within Pi-regulated promoters. (A) The DNA sequences used in the EMSA assay were compared using Gibbs motif analysis. Sequences were aligned based on the identified motif present in each. Nucleotides conserved between at least two of the three sequences are highlighted. The consensus motif is shown below the alignment with the inverted-repeat sequence underlined. n, unconserved nucleotide. (B) The sequence of the phoA promoter is shown with regulatory elements indicated, including the DNase I protected region, −10 consensus, TSS, RBS, and translation initiation codon.
Based on sequence analysis of the phoA promoter, the DNase I protected region is located approximately 6 bp upstream of a consensus −10 sequence, with the TSS preceding a well-conserved RBS sequence (Fig. 6B). The positions of regulatory elements within the phoA promoter are all consistent with a role for RegX3∼P in the transcriptional activation of phoA. Interestingly, the “pho box” sequence present in the phoA promoter exists as an exact inverted 7-bp repeat, extending 1 bp from the conserved motif at each end. It is unclear if the extended motif or exact repeat plays a role in the activity of phoA regulation by RegX3∼P. At this time, the RegX3∼P recognition sequence we have identified is loosely conserved at specific nucleotides among the three regulated promoters. As such, the binding motif we have identified is insufficient to predict additional RegX3∼P binding sequences in the M. smegmatis genome. The identification of additional genes directly regulated by RegX3∼P and the invariant nucleotides within the RegX3∼P binding motif should help to define the mycobacterial “pho” regulon.
DISCUSSION
M. smegmatis has previously been shown to carry a Pi-regulated alkaline phosphatase (phoA) gene (19). Here, we have identified the senX3-regX3 2CR as a direct regulator of phoA expression. We isolated two classes of senX3 mutants with dichotomous PhoA phenotypes; three independent senX3::Tn5371 mutants were PhoA constitutive, while a ΔsenX3 strain was PhoA null. Because SenX3 is a sensor HK, its role in regulating phoA expression is predicted to involve modification of the phosphorylation state of RegX3. The opposing PhoA phenotypes observed in the different senX3 mutant strains most likely resulted from deregulated expression of regX3, which lies immediately downstream of senX3. Differing levels of RegX3 were expected among the different mutants, as the transposon used in this study does not contain a transcriptional terminator and all three transposon insertions are in the same orientation (Fig. 1). Furthermore, because regX3 is essential in M. smegmatis, some level of RegX3 protein must be present in the transposon insertion mutants, as well as the ΔsenX3 strain.
Under growth conditions where Pi is abundant, we observed PhoA activity when SenX3 was absent and RegX3 was present in a phosphorylatable state. Because PhoA activity requires RegX3∼P and the absence of functional SenX3, we conclude that SenX3 catalyzes the removal of the phosphate moiety from RegX3. In vitro experiments, on the other hand, suggest that SenX3 acts as a phosphodonor for RegX3 and that phosphorylation of RegX3 is required for its DNA-binding activity. The latter result is consistent with a role for SenX3 in facilitating the phosphorylation and consequent positive regulatory activity of RegX3.
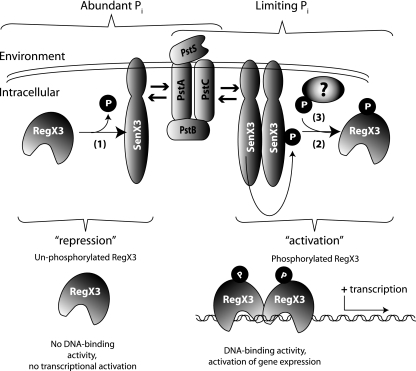
Based on our data and current information regarding other prokaryotic 2CRs (18, 31), we propose a model for phoA regulation by SenX3-RegX3 as outlined in Fig. 7. According to our model, the phosphatase activity of SenX3 maintains RegX3 in an unphosphorylated (and inactive) state when Pi is abundant. When environmental Pi becomes limiting, SenX3 loses its phosphatase activity, probably through dimerization and autophosphorylation, and acts as a preferred phosphodonor for RegX3. The accumulation of RegX3∼P then leads to the transcriptional activation of several genes, including phoA. Our model requires that RegX3 be phosphorylated in the absence of SenX3, although an alternative phosphodonor has not been identified. However, the M. tuberculosis dormancy RR DosR can acquire a phosphor group from its cognate HK, DosS, or by the orphan HK, DosT, demonstrating the presence of alternative phosphodonors in mycobacteria (35). In addition, RR can often use small phosphate-containing metabolic intermediates directly as phosphodonors (34). The apparent similarities between Pi-regulated gene expression in M. smegmatis and E. coli are noteworthy, particularly because these organisms are so genetically distinct.
FIG. 7.
Model of phoA regulation by SenX3-RegX3. When environmental Pi is abundant, SenX3 is required to maintain RegX3 in an unphosphorylated state, where it is unable to bind target DNA and gene transcription is repressed (1). When Pi becomes a limiting nutrient, SenX3 autophosphorylates and becomes a phosphodonor for RegX3 (2). The increased affinity of RegX3∼P for its consensus motif permits DNA binding and activates gene transcription. In the absence of functional SenX3, RegX3 can acquire phosphate from another, undetermined donor molecule (3). Because the extracellular sequence of SenX3 is only a few amino acids in length, the concentration of available Pi is probably not sensed by SenX3 directly. Instead, the Pst transporter is likely to relay this information to SenX3 and to regulate its phosphatase/phosphodonor activity toward RegX3, as indicated by arrows on either side of the Pst transporter.
In E. coli, phoA expression is regulated by a 2CR consisting of the RR PhoB and the HK PhoR (38, 40). Like SenX3, PhoR lacks a significant extracellular domain, and the Pst transporter is predicted to be the sensor of available Pi. Accordingly, Pi sampling is most likely performed by the Pst transporter encoded by pstS-C-A-B, as mutations in any of these genes are sufficient for deregulated phoA expression. PhoR has also been shown to have phosphatase activity toward PhoB (7), demonstrating a similar mechanism for phoA repression in E. coli. Like the senX3::Tn5371 identified in this work, E. coli phoR mutants are PhoA constitutive and PhoB is known to acquire a phospho group from either an unrelated kinase, CreC, or directly from acetyl phosphate, neither of which is regulated by environmental Pi (1, 41, 42).
The essentiality of regX3 that we observed in M. smegmatis was unexpected, because neither senX3 nor regX3 is essential in M. tuberculosis (25, 29). It is possible that a low level of regX3 expression is required in M. smegmatis to permit sufficient Pi uptake for normal growth. Due to the slower growth and/or presence of multiple phosphate transporters (21), this requirement may be reduced in M. tuberculosis. Alternatively, the essential gene(s) that requires RegX3 for its expression in M. smegmatis may be under different or additional regulatory controls in M. tuberculosis.
SenX3-RegX3 comprise one of several complete 2CRs highly conserved in mycobacteria (8, 9, 15, 39), and independent studies have shown attenuated virulence of M. tuberculosis senX3-regX3 mutants in vivo (25, 29). Nevertheless, previous efforts to identify an environmental stimulus for senX3-regX3 have been unsuccessful (25), and the environmental stimuli that govern 2CR responses can rarely be predicted based solely on gene homology. The work described in this paper provides a mechanism for Pi-regulated gene expression in mycobacteria that is mediated by the SenX3-RegX3 2CR and suggests that pathogenic mycobacteria must adapt to Pi-limiting conditions in vivo. Because the M. tuberculosis senX3-regX3 operon was able to complement the M. smegmatis senX3 transposon insertion mutants, it is possible that both organisms utilize SenX3 and RegX3 for Pi-dependent gene regulation. Future research should focus on identifying those genes that are directly regulated by SenX3-RegX3 and determining the significance of the Pi-regulated adaptive response for the virulence of pathogenic mycobacteria.
Acknowledgments
This work was supported by grants from the National Institutes of Health (AI26170, AI57158, AI051519, and AI52816 to W.R.J).
We thank Howard Steinman and Michelle Larsen for scientific discussions and review of the manuscript.
Footnotes
Published ahead of print on 25 May 2007.
REFERENCES
- 1.Amemura, M., K. Makino, H. Shinagawa, and A. Nakata. 1990. Cross talk to the phosphate regulon of Escherichia coli by PhoM protein: PhoM is a histidine protein kinase and catalyzes phosphorylation of PhoB and PhoM open reading frame 2. J. Bacteriol. 172:6300-6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen, A. B., L. Ljungqvist, and M. Olsen. 1990. Evidence that protein antigen b of Mycobacterium tuberculosis is involved in phosphate metabolism. J. Gen. Microbiol. 136:477-480. [DOI] [PubMed] [Google Scholar]
- 3.Bardarov, S., J. Kriakov, C. Carriere, S. Yu, C. Vaamonde, R. A. McAdam, B. R. Bloom, G. F. Hatfull, and W. R. Jacobs, Jr. 1997. Conditionally replicating mycobacteriophages: a system for transposon delivery to Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 94:10961-10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barletta, R. G., D. D. Kim, S. B. Snapper, B. R. Bloom, and W. R. Jacobs, Jr. 1992. Identification of expression signals of the mycobacteriophages Bxb1, L1 and TM4 using the Escherichia-Mycobacterium shuttle plasmids pYUB75 and pYUB76 designed to create translational fusions to the lacZ gene. J. Gen. Microbiol. 138:23-30. [DOI] [PubMed] [Google Scholar]
- 5.Braibant, M., and J. Content. 2001. The cell surface associated phosphatase activity of Mycobacterium bovis BCG is not regulated by environmental inorganic phosphate. FEMS Microbiol. Lett. 195:121-126. [DOI] [PubMed] [Google Scholar]
- 6.Braunstein, M., T. I. Griffin, J. I. Kriakov, S. T. Friedman, N. D. Grindley, and W. R. Jacobs, Jr. 2000. Identification of genes encoding exported Mycobacterium tuberculosis proteins using a Tn552′phoA in vitro transposition system. J. Bacteriol. 182:2732-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carmany, D. O., K. Hollingsworth, and W. R. McCleary. 2003. Genetic and biochemical studies of phosphatase activity of PhoR. J. Bacteriol. 185:1112-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 9.Cole, S. T., K. Eiglmeier, J. Parkhill, K. D. James, N. R. Thomson, P. R. Wheeler, N. Honore, T. Garnier, C. Churcher, D. Harris, K. Mungall, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. M. Davies, K. Devlin, S. Duthoy, T. Feltwell, A. Fraser, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, C. Lacroix, J. Maclean, S. Moule, L. Murphy, K. Oliver, M. A. Quail, M. A. Rajandream, K. M. Rutherford, S. Rutter, K. Seeger, S. Simon, M. Simmonds, J. Skelton, R. Squares, S. Squares, K. Stevens, K. Taylor, S. Whitehead, J. R. Woodward, and B. G. Barrell. 2001. Massive gene decay in the leprosy bacillus. Nature 409:1007-1011. [DOI] [PubMed] [Google Scholar]
- 10.Collins, D. M., R. P. Kawakami, B. M. Buddle, B. J. Wards, and G. W. de Lisle. 2003. Different susceptibility of two animal species infected with isogenic mutants of Mycobacterium bovis identifies phoT as having roles in tuberculosis virulence and phosphate transport. Microbiology 149:3203-3212. [DOI] [PubMed] [Google Scholar]
- 11.Connell, N. D. 1994. Mycobacterium: isolation, maintenance, transformation, and mutant selection. Methods Cell Biol. 45:107-125. [DOI] [PubMed] [Google Scholar]
- 12.Ehrt, S., X. V. Guo, C. M. Hickey, M. Ryou, M. Monteleone, L. W. Riley, and D. Schnappinger. 2005. Controlling gene expression in mycobacteria with anhydrotetracycline and Tet repressor. Nucleic Acids Res. 33:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Espitia, C., M. Elinos, R. Hernandez-Pando, and R. Mancilla. 1992. Phosphate starvation enhances expression of the immunodominant 38-kilodalton protein antigen of Mycobacterium tuberculosis: demonstration by immunogold electron microscopy. Infect. Immun. 60:2998-3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ewann, F., M. Jackson, K. Pethe, A. Cooper, N. Mielcarek, D. Ensergueix, B. Gicquel, C. Locht, and P. Supply. 2002. Transient requirement of the PrrA-PrrB two-component system for early intracellular multiplication of Mycobacterium tuberculosis. Infect. Immun. 70:2256-2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garnier, T., K. Eiglmeier, J. C. Camus, N. Medina, H. Mansoor, M. Pryor, S. Duthoy, S. Grondin, C. Lacroix, C. Monsempe, S. Simon, B. Harris, R. Atkin, J. Doggett, R. Mayes, L. Keating, P. R. Wheeler, J. Parkhill, B. G. Barrell, S. T. Cole, S. V. Gordon, and R. G. Hewinson. 2003. The complete genome sequence of Mycobacterium bovis. Proc. Natl. Acad. Sci. USA 100:7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Himpens, S., C. Locht, and P. Supply. 2000. Molecular characterization of the mycobacterial SenX3-RegX3 two-component system: evidence for autoregulation. Microbiology 146:3091-3098. [DOI] [PubMed] [Google Scholar]
- 17.Hoch, J. A. 2000. Two-component and phosphorelay signal transduction. Curr. Opin. Microbiol. 3:165-170. [DOI] [PubMed] [Google Scholar]
- 18.Hoch, J. A., and T. J. Silhavy. 1995. Two-component signal transduction. ASM Press, Washington, DC.
- 19.Kriakov, J., S. Lee, and W. R. Jacobs, Jr. 2003. Identification of a regulated alkaline phosphatase, a cell surface-associated lipoprotein, in Mycobacterium smegmatis. J. Bacteriol. 185:4983-4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, S., J. Kriakov, C. Vilcheze, Z. Dai, G. F. Hatfull, and W. R. Jacobs, Jr. 2004. Bxz1, a new generalized transducing phage for mycobacteria. FEMS. Microbiol. Lett. 241:271-276. [DOI] [PubMed] [Google Scholar]
- 21.Lefevre, P., M. Braibant, L. de Wit, M. Kalai, D. Roeper, J. Grotzinger, J. P. Delville, P. Peirs, J. Ooms, K. Huygen, and J. Content. 1997. Three different putative phosphate transport receptors are encoded by the Mycobacterium tuberculosis genome and are present at the surface of Mycobacterium bovis BCG. J. Bacteriol. 179:2900-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malhotra, V., D. Sharma, V. D. Ramanathan, H. Shakila, D. K. Saini, S. Chakravorty, T. K. Das, Q. Li, R. F. Silver, P. R. Narayanan, and J. S. Tyagi. 2004. Disruption of response regulator gene, devR, leads to attenuation in virulence of Mycobacterium tuberculosis. FEMS Microbiol. Lett. 231:237-245. [DOI] [PubMed] [Google Scholar]
- 23.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 24.Parish, T., D. A. Smith, S. Kendall, N. Casali, G. J. Bancroft, and N. G. Stoker. 2003. Deletion of two-component regulatory systems increases the virulence of Mycobacterium tuberculosis. Infect. Immun. 71:1134-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parish, T., D. A. Smith, G. Roberts, J. Betts, and N. G. Stoker. 2003. The senX3-regX3 two-component regulatory system of Mycobacterium tuberculosis is required for virulence. Microbiology 149:1423-1435. [DOI] [PubMed] [Google Scholar]
- 26.Peirs, P., P. Lefevre, S. Boarbi, X. M. Wang, O. Denis, M. Braibant, K. Pethe, C. Locht, K. Huygen, and J. Content. 2005. Mycobacterium tuberculosis with disruption in genes encoding the phosphate binding proteins PstS1 and PstS2 is deficient in phosphate uptake and demonstrates reduced in vivo virulence. Infect. Immun. 73:1898-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perez, E., S. Samper, Y. Bordas, C. Guilhot, B. Gicquel, and C. Martin. 2001. An essential role for phoP in Mycobacterium tuberculosis virulence. Mol. Microbiol. 41:179-187. [DOI] [PubMed] [Google Scholar]
- 28.Rengarajan, J., B. R. Bloom, and E. J. Rubin. 2005. From The Cover: Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc. Natl. Acad. Sci. USA 102:8327-8332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rickman, L., J. W. Saldanha, D. M. Hunt, D. N. Hoar, M. J. Colston, J. B. Millar, and R. S. Buxton. 2004. A two-component signal transduction system with a PAS domain-containing sensor is required for virulence of Mycobacterium tuberculosis in mice. Biochem. Biophys. Res. Commun. 314:259-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rubin, E. J., B. J. Akerley, V. N. Novik, D. J. Lampe, R. N. Husson, and J. J. Mekalanos. 1999. In vivo transposition of mariner-based elements in enteric bacteria and mycobacteria. Proc. Natl. Acad. Sci. USA 96:1645-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Russo, F. D., and T. J. Silhavy. 1993. The essential tension: opposed reactions in bacterial two-component regulatory systems. Trends Microbiol. 1:306-310. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 33.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser, and W. R. Jacobs, Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911-1919. [DOI] [PubMed] [Google Scholar]
- 34.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 35.Stover, C. K., V. F. de la Cruz, G. P. Bansal, M. S. Hanson, T. R. Fuerst, W. R. Jacobs, Jr., and B. R. Bloom. 1992. Use of recombinant BCG as a vaccine delivery vehicle. Adv. Exp. Med. Biol 327:175-182. [DOI] [PubMed] [Google Scholar]
- 36.Thompson, W., M. J. Palumbo, W. W. Wasserman, J. S. Liu, and C. E. Lawrence. 2004. Decoding human regulatory circuits. Genome Res. 14:1967-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson, W., E. C. Rouchka, and C. E. Lawrence. 2003. Gibbs recursive sampler: finding transcription factor binding sites. Nucleic Acids Res. 31:3580-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Torriani, A. 1990. From cell membrane to nucleotides: the phosphate regulon in Escherichia coli. Bioessays 12:371-376. [DOI] [PubMed] [Google Scholar]
- 39.Tyagi, J. S., and D. Sharma. 2002. Mycobacterium smegmatis and tuberculosis. Trends Microbiol. 10:68-69. [DOI] [PubMed] [Google Scholar]
- 40.Wanner, B. L. 1993. Gene regulation by phosphate in enteric bacteria. J. Cell Biochem. 51:47-54. [DOI] [PubMed] [Google Scholar]
- 41.Wanner, B. L., and P. Latterell. 1980. Mutants affected in alkaline phosphatase expression: evidence for multiple positive regulators of the phosphate regulon in Escherichia coli. Genetics 96:353-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wanner, B. L., and M. R. Wilmes-Riesenberg. 1992. Involvement of phosphotransacetylase, acetate kinase, and acetyl phosphate synthesis in control of the phosphate regulon in Escherichia coli. J. Bacteriol. 174:2124-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zahrt, T. C., and V. Deretic. 2001. Mycobacterium tuberculosis signal transduction system required for persistent infections. Proc. Natl. Acad. Sci. USA 98:12706-12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zahrt, T. C., C. Wozniak, D. Jones, and A. Trevett. 2003. Functional analysis of the Mycobacterium tuberculosis MprAB two-component signal transduction system. Infect. Immun. 71:6962-6970. [DOI] [PMC free article] [PubMed] [Google Scholar]