Abstract
We have previously shown that CbpA, a major pneumococcal virulence factor, is regulated by the two-component signal transduction system RR/HK06 (A. J. Standish, U. H. Stroeher, and J. C. Paton, Proc. Natl. Acad. Sci. USA 102:7701-7706, 2005). However, additional unidentified regulated factors appeared to be responsible for differences in adherence and the ability of Streptococcus pneumoniae to cause disease in a mouse model. Here, we identified a number of other regulated genes by overexpressing the system. cbpA, along with a cotranscribed upstream gene, showed substantial increases in expression when RR06 was overexpressed in S. pneumoniae strains D39 and TIGR4. However, there were no other similarities between these strains. In D39, rr06 overexpression decreased expression of numerous factors, including the major virulence factor gene pspA. Further investigation of cbpA regulation by RR/HK06, using mutants with mutations in both HK06 and RR06, suggested that rather than the norm, cbpA transcription was activated when RR06 was in the nonphosphorylated form. Although other factors, such as pspA and gls24, are regulated by this system, these genes appear to be repressed when RR06 is in its phosphorylated form.
The increasing resistance of Streptococcus pneumoniae to antibiotics, along with the limitations of the current generation of vaccines, has resulted in the pneumococcus remaining a major cause of morbidity and mortality worldwide. During pathogenesis, the pneumococcus encounters a number of different environmental niches in the body, including the nasopharynx, lungs, blood, and brain. Several studies have shown that a large number of pneumococcal genes change expression as the pathogen progresses through the body (19, 30). These changes in expression are likely to be crucial for the ability of the pneumococcus to colonize the host and cause disease. It is likely that two-component signal transduction systems (TCSTSs) are responsible for the regulation of these genes as they respond to changes in external stimuli (for a review, see reference 37).
These systems generally comprise two proteins, a histidine kinase (HK) and a response regulator (RR). The HK is the sensor protein, responding to changes in external stimuli by autophosphorylating a conserved His. The subsequent transfer of the phosphoryl group to the RR leads to conformational changes, enabling the RR to act as a transcription factor by binding to DNA.
Genomic studies identified a total of 13 TCSTSs in the pneumococcus, along with one orphan RR (18, 39). ComDE is the best-characterized TCSTS and is important in the development of competence of the pathogen (5, 31). We have previously shown that 1 of the 13 TCSTSs in S. pneumoniae, RR/HK06, regulates the expression of choline binding protein A (CbpA), a major virulence factor and protective antigen (36). We also saw a number of effects on pathogenesis and adherence, which could not be explained by effects on CbpA alone, suggesting that there are as-yet-uncharacterized RR/HK06-regulated factors that contribute to the observed phenotype. Recently, Ma and Zhang (20) confirmed our previous finding (36) that RR06 does indeed regulate the transcription of cbpA and identified another gene (spd2018) upstream of cbpA which is also regulated by this system. The RR06 binding sites upstream of both spd2018 and cbpA were also identified and shown to consist of an imperfect 19-bp conserved sequence. In the current study, we used microarray analysis to gain a global understanding of the regulation by this system, through overexpression of RR/HK06. We also constructed mutants predicted to abrogate various activities of RR06 and HK06 in an effort to gain a greater understanding of the mechanism behind RR/HK06 regulation. Our results showed that another major virulence factor and protective antigen, pneumococcal surface protein A (PspA), is also regulated by RR/HK06 in the S. pneumoniae serotype 2 strain D39.
MATERIALS AND METHODS
Bacterial strains, plasmids, growth conditions, and transformation.
Bacterial strains and plasmids used in this study are listed in Table 1. S. pneumoniae strains were grown in Todd-Hewitt broth with 1% yeast extract (THY) or on blood agar, as described previously (28). S. pneumoniae strains D39 and TIGR4 were transformed as described previously, using CSP-1 and CSP-2, respectively (2, 3, 44).
TABLE 1.
Bacterial strains and plasmids
| S. pneumoniae strain | Description | Source (reference) |
|---|---|---|
| D39 | Capsular serotype 2 | 1 |
| D39rr06 | Eryr, rr06 insertion-duplication mutant of D39 | 36 |
| D39Δhk06 | hk06 deletion mutant of D39 | 36 |
| D39Δrr06 | rr06 deletion mutant of D39 | 36 |
| D39Δrr/hk06 | rr/hk06 deletion mutant of D39 | This study |
| D39 RR06D51E | Asp51 to Glu51 of RR06 in D39 | This study |
| D39 RR06D51N | Asp51 to Asn51 of RR06 in D39 | This study |
| D39 HK06H242Y | His242 to Tyr242 of HK06 in D39 | This study |
| D39 HK06H242R | His242 to Arg242 of HK06 in D39 | This study |
| D39 HK06S241D | Ser241 to Asp241 of HK06 in D39 | This study |
| R6 | Unencapsultated derivative of D39 | Laboratory strain |
| D39::pControl | pLS1RGFP in D39 | This study |
| D39::phk06 | hk06 overexpressed in D39 | This study |
| D39::prr06 | rr06 overexpressed in D39 | This study |
| D39::prr/hk06 | rr/hk06 overexpressed in D39 | This study |
| D39Δrr/hk06::pControl | pLS1RGFP in D39Δrr/hk06 | This study |
| D39Δrr/hk06::prr06 | rr06 overexpressed in D39Δrr/hk06 | This study |
| D39Δrr/hk06::pRR06D51E | RR06D51E overexpressed in D39Δrr/hk06 | This study |
| D39Δrr/hk06::pRR06D51N | RR06D51N overexpressed in D39Δrr/hk06 | This study |
| TIGR4 | Capsular serotype 4 | 5 |
| TIGR4::pControl | pLS1RGFP in TIGR4 | This study |
| TIGR4::phk06 | hk06 overexpressed in TIGR4 | This study |
| TIGR4::prr06 | rr06 overexpressed in TIGR4 | This study |
| TIGR4::prr/hk06 | rr/hk06 overexpressed in TIGR4 | This study |
Oligonucleotide primers, DNA isolation, and manipulation.
The primers used are listed in Table 2. S. pneumoniae chromosomal DNA was isolated as described previously (24). Amplification of DNA was performed by high-fidelity PCR using the Expand Long Template PCR system (Roche Diagnostics). Overlap extension PCR products were generated from the initial products as described previously (9, 10). Amplification products were sequenced using the Big Dye system (Applied Biosystems). PCR amplification was performed with a Hybaid touchdown thermal cycler, and each 50-μl reaction mixture contained PCR buffer (1.5 mM MgCl2, 10 mM Tris [pH 8.4], 50 mM KCl, 100 μg gelatin per ml), 1.5 U of Taq polymerase, 1 μM of each primer, 100 ng of DNA template, and 200 μM of each of the four deoxynucleoside triphosphates. The program consisted of 25 cycles of denaturation at 95°C for 30 s, annealing at 50°C for 30 s, and elongation at 72°C for 1 to 4 min (1 min for each 1 kb of expected product). A 5-μl sample of the PCR product was analyzed by agarose gel electrophoresis.
TABLE 2.
Oligonucleotides used in study
| Oligonucleotide | Sequence (5′-3′) | Location (nucleotides)a |
|---|---|---|
| AS5 | 5′ GCACCTGATGCTTCAAGATAGTAC 3′ | 1987671-1987694 |
| AS6 | 5′ TGTCGGGAAAACAGCTCTGGC 3′ | 1994684-1994663 |
| 16sF | 5′ GGTGAGTAACGCGTAGGTAA 3′ | 15455-15474 |
| 16sR | 5′ ACGATCCGAAAACCTTCTTC 3′ | 15780-15761 |
| CbpAFNEW | 5′ TCAACTAGATAGAAGAAAACATACCCA 3′ | 1989422-1989396 |
| CbpARedit | 5′ TCCTGGTTTCAATGATCTTTTTTAAACTT 3′ | 1989236-1989265 |
| HKF | 5′ TTTTGCTTTTGTATCTCCCATATAC 3′ | 1991251-1991227 |
| HKR | 5′ TTGCTCATTACGACTTTCCAAC 3′ | 1990993-1991016 |
| RRF | 5′ CTCAGTCACAGGTATTTGATCTC 3′ | 1992066-1992044 |
| RRR | 5′ TATGCTGAATCAACACTTTAGTAC 3′ | 1991833-1991857 |
| AS62 | 5′ GATTCAGAACGAGATAAGGCAAGG 3′ | 2111950-2111867b |
| AS64 | 5′ GTTTAGTGGAAGAGTCTGAACTTG 3′ | 2111608-2111631b |
| AS67 | 5′ ACGGTAGCTAGCAGTAAAGGAGGTAGAGAGATGAACATTTTAG 3′c | 1992326-1992306 |
| AS68 | 5′ ACGGTAGCTAGCTCATAAGCTAATCTTATACCCAACATTTTT 3′c | 1991664-1991695 |
| AS69 | 5′ ACGGTAGCTAGCACCAAAGGAGGTTTAGCTATGATAAAAAATC CTAAATTATTAACC 3′c | 1991675-1991641 |
| AS70 | 5′ ACGGTAGCTAGCCTACAAGCTAATCTTAAATTCCATACC 3′c | 1990338-1990360 |
| PspAF | 5′ CAGAAGAATCTCCCGTAGCCAGTC 3′ | 128447-128471 |
| PspAR | 5′ TCGAACAGTATTAAATTTAGTTTTTGCCTC 3′ | 128794-128764 |
| AS55 | 5′ AGAGAAGGAATTGATTATAGTTTTCTCCCTTATGAACAC 3′ | 1990364-1990391 (end of tail, 1992272-1992283)d |
| AS56 | 5′ CCGAGTTTACTATAATCAATTCCTTCTCTAATCATTTCCTCGTC 3′ | 1992272-1992298 (end of tail, 1990391-1990375)d |
| AS91 | 5′ AAATCATTTAAGATTTTATCTGGTGCC 3′ | 65890-65915 |
| AS92 | 5′ TCATAGTAGCGACCAACTGCC 3′ | 66240-66217 |
| AS93 | 5′ CCAGCTGATTTAGCTACTGCTC 3′ | 3275451-327567 |
| AS94 | 5′ ATCAACAGTTGCATTTCCATTGGC 3′ | 327894-327918 |
| AS97 | 5′ GATTATGATTTAACGTCGCCGCG 3′ | 626055-626078 |
| AS98 | 5′ CATAGCTAATCTAGTACCAGAACC 3′ | 626255-626281 |
| AS99 | 5′ GGTGTTATCATTGCTACTCCTGAGC 3′ | 1312881-1312906 |
| AS100 | 5′ AAGTTATCTAAGCAAGTTTCAAGG 3′ | 1313195-1313171 |
| AS101 | 5′ ATGGGAATCATCGCAAATGTATTCG 3′ | 1598730-1598755 |
| AS102 | 5′ TTACTTTCTACCTGTAAAGAATGAC 3′ | 1598883-1598858 |
| AS103 | 5′ GACCAATCGTGTTTTGGCACGA 3′ | 1640492-1640469 |
| AS104 | 5′ GTAGTCAGATGGAACACGATAAGC 3′ | 1640190-1640216 |
| AS105 | 5′ GAGAATGAGAAACGAATCCTTAGC 3′ | 1990192-1990169 |
| AS106 | 5′ CAACAAGGTCAAGCCAATCAACTC 3′ | 1989854-1989877 |
| AS77 | 5′ CGCGGAGCATCATACGAGTTGAAAACC 3′ | 1990957-1990930 |
| AS78 | 5′ GGTTTTCAAC TCGTATGATG CTCCGCG 3′ | 1990930-1990957 |
| AS85 | 5′ CGCGGAGCATCAGAAGAGTTGAAAACC 3′ | 1990957-1990930 |
| AS86 | 5′ GGTTTTCAACTCTTCTGATGCTCCGCG 3′ | 1990930-1990957 |
| AS87 | 5′ CGCGGAGCAAACCATGAGTTGAAAACC 3′ | 1990957-1990930 |
| AS88 | 5′ GGTTTTCAAC TCATGGTTTG CTCCGCG 3′ | 1990930-1990957 |
| AS75 | 5′ CTCATGGTACTGGAGTTAATGATG 3′ | 1992181-1992157 |
| AS76 | 5′ CATCATTAACTCCAGTACCATGAG 3′ | 1992157-1992181 |
| AS79 | 5′ CTCATGGTACTGAACTTAATGATG 3′ | 1992181-1992157 |
| AS80 | 5′ CATCATTAAGTTCAGTACCATGAG 3′ | 1992157-1992181 |
Unless indicated otherwise, location in the genome sequence of S. pneumoniae R6 strain ADDIN ENRfu (11) (GenBank accession number AE007317).
Location in the genome sequence of S. pneumoniae TIGR4 strain ADDIN ENRfu (4) (GenBank accession number NC_003028).
An NheI site is underlined.
The sequence location in the genome of the end of the “tail” region, which does not bind to DNA in first PCR step of overlap extension PCR, is indicated in parentheses (this region is indicated by boldface type in the sequence).
Construction of S. pneumoniae overexpressing hk06, rr06, and rr/hk06.
The vectors phk06, prr06, and prr/hk06 were constructed as follows. Oligonucleotides AS69 and AS70, AS67 and AS68, and AS67 and AS70 were used to amplify hk06, rr06, and rr/hk06, respectively, using the protocol outlined above. The PCR products were digested with NheI at 37°C for 90 min and cloned into the unique XbaI site of the vector pLS1RGFP (27). The resulting plasmids were transformed into unencapsulated strain R6 and selected on erythromycin. Transformants were then checked for the presence of the correct plasmid insert by sequencing. Plasmids were subsequently transformed into the appropriate S. pneumoniae strain and selected on erythromycin. To induce expression, cells were grown to mid-log phase in THY (A600, ∼0.2) and cells were pelleted by centrifugation at 4,500 × g for 15 min and then resuspended in prewarmed THY containing 0.8% maltose for 30 min.
Construction of D39Δrr/hk06 and amino acid substitutions.
The strains mentioned above were constructed essentially as described previously (36). First-round amplification was undertaken with the following oligonucleotide combinations: for D39Δrr/hk06, AS5/AS55 and AS6/AS56; for D39 RR06D51E, AS5/AS75 and AS6/AS76; for D39 RR06D51N, AS5/AS79 and AS6/AS80; for D39 HK06H242Y, AS5/AS77 and AS6/AS78; for D39 HK06H242R, AS5/AS85 and AS6/AS86; and for D39 HK06S241D, AS5/AS87 and AS6/AS88. Overlap extension PCR was then undertaken using round one PCR products and the external oligonucleotides AS5 and AS6. All mutations were confirmed by sequence analysis.
SDS-PAGE and Western blotting.
Bacterial cell lysates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as described by Laemmli (16). Separated samples were then electroblotted onto nitrocellulose (Pall Life Sciences, United States) as described previously (41) and probed with the appropriate antisera (36). N-terminal sequencing was performed at the Australian Proteome Analysis Facility, Sydney, Australia.
RNA extraction.
RNA was isolated from bacterial pellets with acid-phenol-chloroform/isoamyl alcohol (125:24:1, pH 4·5; Ambion) essentially as described previously (28). RNA was resuspended in nuclease-free water, and contaminating DNA was digested with DNase I (Roche Diagnostics).
Real-time RT-PCR.
Real-time reverse transcription (RT)-PCR was performed with a Rotorgene RG-2000 cycler (Corbett Research, Australia) using the Access RT-PCR system (Promega) according to the manufacturer's instructions, essentially as described previously (33). At least two biological replicate analyses were performed, with each reaction performed in triplicate. Amplification included the following steps: 45 min of RT at 48°C, followed by 2 min of denaturation at 94°C and then 40 cycles of amplification with denaturation at 94°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 45 s. Results were calculated using the comparative cycle threshold (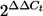 ) method (user bulletin no. 2 [http://docs.appliedbiosystems.com/pebiodocs/04303859.pdf]; Applied Biosystems), in which the amount of target mRNA is normalized relative to an internal control (16S rRNA). Differences in gene expression were analyzed by analysis of variance (ANOVA) with logarithmically transformed data; a P value of ≤0.05 was considered significant. When differences were significant, a Tukey test was performed.
) method (user bulletin no. 2 [http://docs.appliedbiosystems.com/pebiodocs/04303859.pdf]; Applied Biosystems), in which the amount of target mRNA is normalized relative to an internal control (16S rRNA). Differences in gene expression were analyzed by analysis of variance (ANOVA) with logarithmically transformed data; a P value of ≤0.05 was considered significant. When differences were significant, a Tukey test was performed.
RNA labeling and microarray hybridization.
Total bacterial RNA was labeled and hybridized using a Genisphere Array 900 MPX kit (Genisphere, United States) according to the manufacturer's instructions. A total of 3 μg of each RNA preparation was used per microarray slide, and hybridization reactions were carried out at 65°C. S. pneumoniae microarray slides were obtained from the Bacterial Microarray Group, St. Georges's Hospital, University of London, London, United Kingdom. The array was read using a GenePix 4000B (Molecular Devices Sunnyvale, CA), and images were acquired using GenePixPro6.0 (Axon). Results were analyzed using R package and the programs Spot (CSIRO Mathematical and Information Sciences, Image Analysis Group, New South Wales, Australia) and LimmaGUI (Bioinformatics Walter and Eliza Hall Institute, Melbourne, Victoria, Australia). Gene lists were prepared with a twofold cutoff and with a P value of <0.05. The P value was calculated from t tests performed for the intensities of individual spots. The data obtained for D39 were derived from a total of four slides using two biological replicates and two dye reversals; the data for TIGR4 were derived from a total of six slides with three biological replicates and dye reversals.
Microarray data accession numbers.
Microarray data were deposited in the GEO database under accession numbers GSE5322 (D39) and GSE5330 (TIGR4).
RESULTS
Overexpression of RR/HK06.
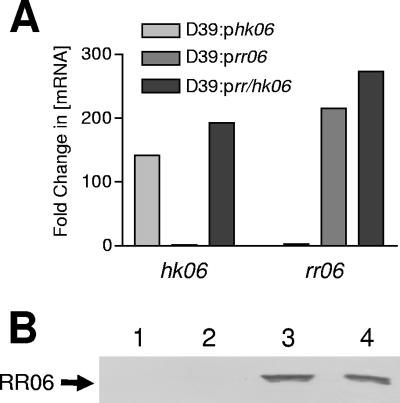
Recently, overexpression of TCSTSs has proven to be a useful tool for identifying regulated genes, particularly when the environmental cue is unknown (29, 43). The maltose-inducible expression vector pLS1RGFP (kindly provided by P. Lopez), which has previously been used to successfully overexpress the essential pneumococcal response regulator YycF (23), was employed to overexpress the RR/HK06 system. We cloned hk06 and rr06, as well as both genes together, into this vector, and the resultant plasmids were designated phk06, prr06, and prr/hk06, respectively. Overexpression of hk06, rr06, and rr/hk06 in S. pneumoniae D39 harboring these plasmids or the control construct pControl (vector alone) was confirmed using real-time RT-PCR with oligonucleotides specific for hk06 and rr06 (HKF plus HKR and RRF plus RRR [Table 2]) (Fig. 1A). 16S rRNA levels (determined in parallel using primers 16sF and 16sR) were used as an internal control. Overexpression of RR06 was also confirmed by Western blot analysis of cell lysates from the strains mentioned above with anti-RR06 (36) (Fig. 1B). The maximal induction, as determined by real-time RT-PCR, was approximately 300-fold and occurred 15 to 20 min after maltose addition (data not shown). We subsequently used 30 min postinduction for all investigations. The growth rates of all strains postinduction were checked and found to be the same, indicating that overexpression alone was unlikely to cause a major stress response (data not shown).
FIG. 1.
Analysis of RR/HK06 overexpression in D39. (A) RNA was isolated from D39::pControl, D39::phk06, D39::prr06, and D39::prr/hk06, and hk06 and rr06 mRNA levels were compared using real-time RT-PCR. The data are the fold changes in the mRNA concentration relative to D39::pControl. (B) Proteins in lysates of (lane 1) D39::pControl, (lane 2) D39::phk06, (lane 3) D39::prr06, and (lane 4) D39::prr/hk06, grown in THY as described in Materials and Methods, were separated by SDS-PAGE, transferred onto nitrocellulose, and probed with polyclonal murine anti-RR06 serum.
Effect of RR/HK06 overexpression on CbpA.
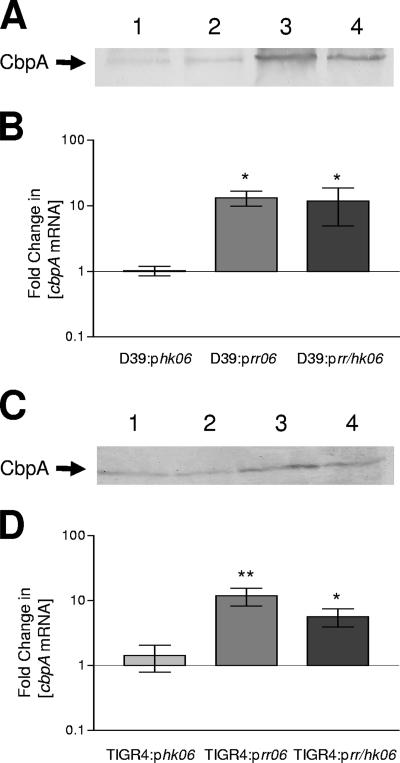
We previously reported that an in-frame chromosomal deletion mutation in hk06 resulted in an increase in cbpA expression, whereas an rr06 deletion mutant exhibited markedly decreased expression of cbpA compared to the wild-type strain S. pneumoniae D39 (36). We therefore investigated the level of cbpA in all three overexpression strains. Western blot analysis using anti-CbpA and real-time RT-PCR using cbpA-specific oligonucleotides (CbpAFNew and CbpARedit) were used to compare CbpA levels in D39::pControl, D39::phk06, D39::prr06, and D39::prr/hk06. Overexpression of rr06 and rr/hk06 led to increases in CbpA at the level of mRNA transcription (approximately 10-fold) and at the protein level (Fig. 2A and 2B). D39::phk06 showed CbpA levels similar to those seen in D39::pControl. These results are consistent with our previous report suggesting that the regulatory effect of RR/HK06 on cbpA occurs at the level of transcription and suggest that RR06 is an activator of cbpA expression. An in-frame deletion mutant of D39 lacking both hk06 and rr06 (D39Δrr/hk06) showed no significant difference in cbpA expression compared to the wild type, as measured by both Western immunoblot analysis and real-time RT-PCR.
FIG. 2.
Analysis of CbpA expression in D39 and TIGR4. (A) Proteins in lysates of (lane 1) D39::pControl, (lane 2) D39::phk06, (lane 3) D39::prr06, and (lane 4) D39::prr/hk06 or (C) proteins in lysates of (lane 1) TIGR4::pControl, (lane 2) TIGR4::phk06, (lane 3) TIGR4::prr06, and (lane 4) TIGR4::prr/hk06, grown in THY as described in Materials and Methods, were separated by SDS-PAGE, transferred onto nitrocellulose, and probed with polyclonal murine anti-CbpA serum. (B) RNA isolated from D39 harboring pControl, phk06, prr06, or prr/hk06 or (D) RNA from TIGR4 harboring pControl, phk06, prr06, or prr/hk06 was analyzed for differences in cbpA mRNA by real-time RT-PCR. The data are fold changes (± standard error) in the cbpA mRNA concentration relative to D39. One asterisk, P < 0.05; two asterisks, P < 0.01, as determined by one-way ANOVA with the post hoc Tukey test).
To determine whether regulation of cbpA by RR/HK06 occurred across serotypes, pControl, phk06, prr06, and prr/hk06 were transformed into the serotype 4 strain TIGR4 (4). Previously we had seen that an erythromycin replacement mutant of hk06 resulted in increased expression of cbpA in TIGR4, whereas the rr06 equivalent mutant resulted in levels similar to the wild-type levels (36). Since in both HK06 and RR06 there is only a single conserved amino acid substitution between D39 and TIGR4 (H217Y and L124I, respectively), we used the original constructs derived from D39 to overexpress the system in TIGR4. Real-time RT-PCR using gene-specific oligonucleotides for TIGR4 cbpA (AS62 and AS64 [Table 2]) and Western immunoblot analysis showed that the presence of prr06 and prr/hk06 resulted in significant increases in CbpA (Fig. 2C and 2D).
RR/HK06 and global regulation.
In order to identify genes other than cbpA which are regulated by RR/HK06, microarray analysis was performed to compare mRNA extracted from D39::pControl and D39::prr06. We used our standard protocol consisting of two biological replicates and performed dye swaps in order to confirm that the differences were not a result of dye bias. A complete list of genes with altered transcription in D39::pControl and D39::prr06 is shown in Table 3. We included only those genes which have an alteration in expression greater than fourfold or those genes which have a P value of <0.01. Increased transcription of both rr06 and cbpA was seen in D39::prr06 relative to D39::pControl. This correlated with results obtained by real-time RT-PCR and Western immunoblotting described above. Transcription of the gene found immediately upstream of cbpA, spd2018, was also increased. In order to determine if cbpA and spd2018 are cotranscribed, we performed an RT-PCR with D39 mRNA using oligonucleotides AS105 and CbpARedit (Table 2), which bind within spd2018 and cbpA, respectively. This resulted in a 956-bp PCR product, indicating that these two genes are in fact cotranscribed (results not shown).
TABLE 3.
Genes differentially regulated with overexpression of rr06
| Genea | Mean fold changeb | P valuec | Annotationd |
|---|---|---|---|
| D39 up-regulated genes | |||
| spd2017 | 7.97 | 1.31E-03 | cbpA |
| spd2018 | 16.68 | 3.43E-03 | Hypothetical protein |
| spd2020 | 57.48 | 1.36E-03 | rr06 |
| D39 down-regulated genes | |||
| spd0126 | 8.57 | 3.87E-02 | pspA |
| spd0065e | 70.03 | 9.80E-04 | β-Galactosidase |
| spd0066 | 62.47 | 1.30E-03 | PTS system, mannose-specific IIB componentg |
| spd0067 | 33.47 | 3.81E-04 | PTS system, mannose-specific IIC component |
| spd0068 | 36.50 | 5.55E-03 | PTS system, mannose-specific IID component |
| spd0069 | 7.24 | 2.47E-01 | PTS system, mannose-specific IIA component |
| spd0070 | 27.00 | 1.20E-02 | Sugar isomerase domain protein AgaS |
| spd0071 | 42.96 | 4.17E-03 | Aldose 1-epimerase |
| spd0335 | 31.89 | 6.82E-03 | Hypothetical protein |
| spd0373 | 2.94 | 8.44E-03 | Macrophage infectivity potentiator protein |
| spd0610e | 4.89 | 1.33E-02 | Hypothetical protein |
| spd0611 | 5.56 | 4.28E-02 | Hypothetical protein |
| spd0612 | 5.90 | 5.09E-03 | Hypothetical protein |
| spd0613 | 5.64 | 8.21E-03 | Hypothetical protein |
| spd0614 | 4.17 | 1.84E-02 | ABC transporter, ATP-binding protein |
| spd0616 | 5.30 | 1.88E-03 | Amino acid ABC transporter, ATP-binding protein |
| spd1300 | 7.41 | 3.41E-02 | Thiamine biosynthesis protein ApbE |
| spd1301 | 6.19 | 8.66E-02 | NADPH-dependent flavin mononucleotide reductase |
| spd1302e | 7.84 | 2.62E-02 | Oxidoreductase |
| spn1472f | 5.70 | 1.49E-02 | Hypothetical protein |
| spn1801e,f | 12.13 | 4.30E-03 | Hypothetical protein |
| spd1588 | 7.67 | 8.61E-03 | Hypothetical protein |
| spd1589 | 13.64 | 4.59E-03 | Lipoprotein, putative |
| spd1590 | 9.42 | 1.91E-02 | General stress protein 24 (gls24), putative |
| spd1591 | 9.61 | 1.57E-02 | Hypothetical protein |
| spd1633 | 9.92 | 2.50E-03 | Galactose-1-phosphate uridylyltransferase |
| spd1634 | 4.35 | 1.21E-01 | Galactokinase |
| spd1635 | 7.57 | 1.52E-03 | Galactose operon repressor |
| TIGR4 up-regulated genes | |||
| spn2190 | 5.16 | 2.08E-04 | cbpA |
| spn2191 | 7.14 | 1.91E-03 | Hypothetical protein |
| spn2193 | 19.31 | 2.63E-03 | rr06 |
Gene designation that matches the microarray open reading frame. The prefix spn indicates the TIGR4 sequence (GenBank accession number NC_003028), and the prefix spd indicates the D39 sequence (GenBank accession number CP000410).
Intensity ratio of prr06 to pControl in microarray experiments in the strains indicated. Genes with >2-fold changes in expression were selected.
Mean P value calculated by t tests for the intensity of the individual spots.
Annotations published for the TIGR4 or D39 genome.
Differences in mRNA levels were confirmed by real-time RT-PCR using gene-specific oligonucleotides AS91 and AS92 (spd0065), AS93 and AS94 (spd0355), AS97 and AS98 (spd0610), AS99 and AS100 (spd1302), AS101 and AS102 (intragenic region corresponding to spn1801), AS103 and AS104 (spd1633), and AS105 and AS106 (spn2191) (Table 2).
For spn1472 and spn806 the corresponding region in D39 sequence is given as the intragenic untranslated sequence.
PTS, phosphotransferase system.
Among the genes with decreased expression in D39::prr06 relative to D39::pControl were the major virulence factor and protective antigen gene pspA and a putative stress protein gene, gls24. Overexpression of rr06 did not appear to affect the expression of the cognate HK gene, hk06, providing further evidence that the system does not autoregulate its own transcription (36). To validate the results from the microarray analysis, real-time RT-PCR carried out using specific oligonucleotides for spd0065, spd0335, spd0610, spd1302, spn1801, spd1633, spd2018, and hk06 (Tables 2 and 3) revealed analogous differences in expression in D39::pControl and D39::prr06 (Fig. 3). These genes were chosen because in many cases they are representative genes in an operon which appears to be regulated and also represent genes which are both highly and weakly regulated. Examination of the upstream regions for the genes mentioned above did not result in identification of a possible RR06 consensus binding site with the exception of spd2018, where we found the RR06 binding site described by Ma and Zhang (20). Considering that binding sites can vary dramatically, as has seen for another pneumococcal RR, YycF (26), this was not surprising. Alternatively, we may have observed indirect effects of RR06 on some of the regulated genes; for example, the regulation of PspA has been reported to also be under the control of another regulator, namely YycF (26).
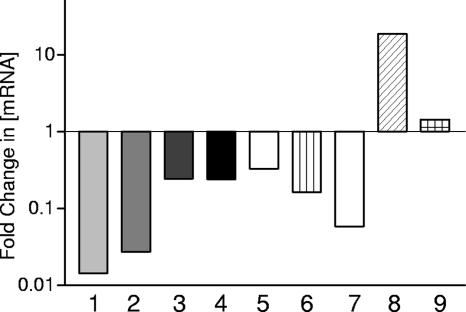
FIG. 3.
Analysis of mRNA levels by real-time RT-PCR. RNA was isolated from induced D39::pControl and D39::prr06 and analyzed for differences in the mRNA levels in D39::prr06 relative to D39::pControl of the following genes using real-time RT-PCR: bar 1, spd0065; bar 2, spd0335; bar 3, spd0610; bar 4, spd1302; bar 5, spd1633; bar 6, spn1801; bar 7, spd1590; bar 8, spd2018; and bar 9, hk06. The data are fold changes in the mRNA concentration relative to D39::pControl.
In order to see if regulation by RR/HK06 is similar across serotypes, a microarray analysis was undertaken in the TIGR4 background. As has been seen in an investigation of pneumococcal TCS04 (21), there was a marked difference in regulation between D39 and TIGR4. While both cbpA and spd2018 still showed a statistically significant increase in expression when rr06 was overexpressed, only three other genes were differentially expressed in TIGR4 (Table 3). Investigation into the upstream regions of a number of genes apparently regulated in D39 but not in TIGR4, such as pspA, did not show any major nucleotide differences. This may indicate that some of the genes that we found to be regulated by TCS06 are indirectly affected by RR06, possibly via another as-yet-unknown regulator.
Effect of RR/HK06 on PspA and Gls24 expression in D39.
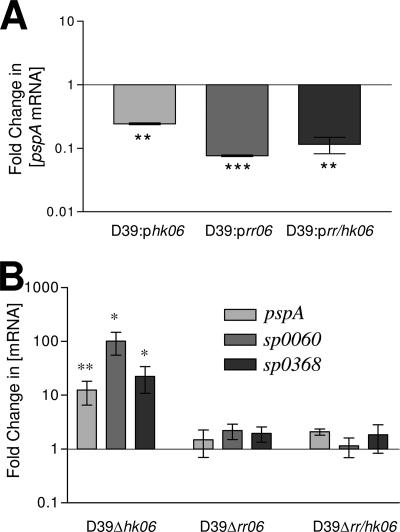
As PspA is a known virulence factor, we chose to further examine its possible regulation by RR/HK06 using real-time RT-PCR with pspA-specific oligonucleotides (PspAF and PspAR [Table 2]). We were able to show statistically significant down-regulation of pspA in D39::phk06, D39::prr06, and D39::prr/hk06 (Fig. 4A), although the down-regulation seen with overexpressed HK06 was significantly lower than that seen with overexpression of RR06 or RR/HK06. These results indicate that RR/HK06 may act as a repressor of pspA. However, since the proposed RR06 binding site is not found upstream of pspA, the regulation of this virulence factor may be indirect or in combination with the regulator Yycf mentioned above (26). To examine whether overexpression of hk06 led to a decrease in pspA via rr06, pControl and phk06 were transformed into an in-frame rr06 deletion mutant of D39 (D39Δrr06). Real-time RT-PCR analysis showed that there was no significant difference in expression of pspA between the two strains (data not shown). This suggests that the overexpression of hk06 affected pspA expression via the chromosomal copy of rr06 rather than through some other intermediate. Overexpression of hk06, rr06, and rr/hk06 in TIGR4 did not alter expression of pspA as measured by real-time RT-PCR (data not shown), confirming the results seen by microarray analysis.
FIG. 4.
Analysis of PspA expression in D39. (A) RNA isolated from D39 harboring pControl, phk06, prr06, and prr/hk06 was analyzed for differences in pspA mRNA by real-time RT-PCR. The data are fold changes (± standard error) in the cbpA mRNA concentration relative to D39::pControl. Two asterisks, P < 0.01; three asterisks, P < 0.001, as determined by one-way ANOVA with the post hoc Tukey test. (B) RNA isolated from D39, D39Δhk06, D39Δrr06, and D39Δrr/hk06 were analyzed for differences in pspA, spd0065, and spd0335 mRNA by real-time RT-PCR. The data are fold changes (± standard error) in the pspA mRNA concentration relative to D39. One asterisk, P < 0.05; two asterisks, P < 0.01, as determined by one-way ANOVA with the post hoc Tukey test.
Real-time RT-PCR, as described above, was then used to compare levels of pspA mRNA in D39 and the in-frame deletion mutants D39Δhk06, D39Δrr06, and D39Δrr/hk06. While both D39Δrr06 and D39Δrr/hk06 showed pspA mRNA levels similar to the wild-type levels, D39Δhk06 showed a significantly increased level of pspA mRNA (Fig. 4B). We also examined mRNA levels in the strains mentioned above for two other genes (spd0065 and spd0335) that had been identified by microarray analysis as genes that are highly repressed by overexpression of rr06. Similar to pspA, both spd0065 and spd0335 showed increased expression in D39Δhk06. However in D39Δrr06 and D39Δrr/hk06 the mRNA levels were similar to the wild-type levels (Fig. 4B). While examining the protein expression profiles of the various D39 mutants, we identified another protein which appeared to show regulation similar to that seen for pspA. The protein was identified by N-terminal sequencing to be Gls24. Since the gls24 and spd0065 genes show expression which is altered in the hk06 mutant without overexpression of either HK or RR06 (Fig. 5D), it seems likely that the differences seen during rr06 overexpression are due to either a direct or indirect effect of RR/HK06 on target genes rather than a general stress response due to the high levels of rr06 expression.
FIG. 5.
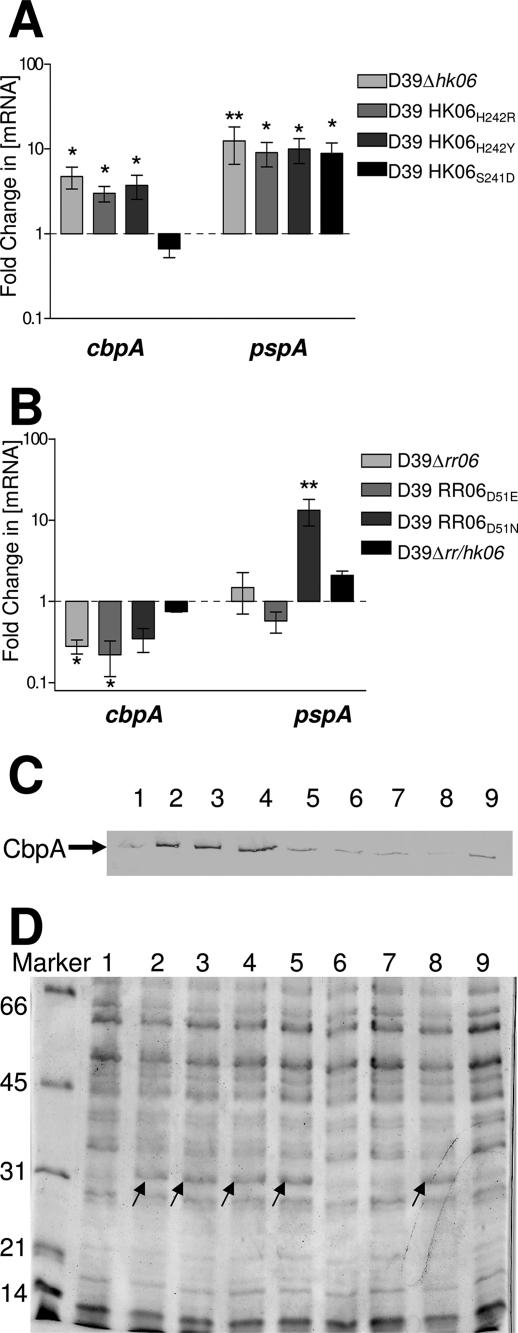
Protein and gene expression analysis of HK06 and RR06 substitutions. (A) RNA isolated from D39, D39Δhk06, D39 HK06H242N, D39 HK06H242Y, and D39 HK06S241D was analyzed for levels of cbpA and pspA mRNA by real-time RT-PCR. The data are fold changes (± standard error) in the mRNA concentration relative to D39. One asterisk, P < 0.05; two asterisks, P < 0.01, as determined by one-way ANOVA with the post hoc Tukey test. (B) RNA isolated from D39, D39Δrr06, D39 RR06D51E, and D39 RR06D51N was analyzed for levels of cbpA and pspA mRNA by real-time RT-PCR. The data are fold changes (± standard error) in the mRNA concentration relative to D39. One asterisk, P < 0.05; two asterisks, P < 0.01, as determined by one-way ANOVA with the post hoc Tukey test. (C and D) Proteins in lysates of (lane 1) D39, (lane 2) D39Δhk06, (lane 3) D39 HK06H242N, (lane 4) D39 HK06H242Y, (lane 5) D39 HK06S241D, (lane 6)D39Δrr06, (lane 7) D39 RR06D51E, (lane 8) D39 RR06D51N, and (lane 9) D39Δrr/hk06 grown in THY as described in Materials and Methods were separated by SDS-PAGE and transferred onto nitrocellulose. Then (C) they were probed with polyclonal murine anti-CbpA serum, or (D) the SDS-PAGE gel was stained with Coomassie blue. The numbers on the left indicate the molecular masses of the markers (in kDa).
Effect of point mutations in RR06 and HK06.
RR/HK06 belongs to the same family of TCSTSs as the EnvZ/OmpR system. Sequence alignments suggested that His242 and Asp51 are likely to be the conserved residues that are phosphorylated in HK06 and RR06, respectively. A large number of HKs, including EnvZ, possess both kinase and phosphatase activities to limit the lifetime of the activated state of the RR. Based on studies of EnvZ, three HK06 point mutations were constructed in D39 in order to try to mutate kinase activity (D39 HK06H242Y), phosphatase activity (D39 HK06S241D), or both (D39 HK06H242R) (12, 13, 40). Real-time RT-PCR and Western immunoblot analysis showed that D39 HK06H242R and D39 HK06H242Y exhibited increased cbpA expression, similar to that seen in D39Δhk06 (Fig. 5A and 5C), indicating that disruption of the proposed kinase sites mimics an HK deletion mutation. Surprisingly, D39 HK06S241D, which has only the putative phosphatase site altered, showed no statistically significant difference in cbpA mRNA compared to the control, again suggesting that it is the lack of HK06 kinase activity which results in the increase in cbpA expression. In comparison, the levels of pspA mRNA (as measured by real-time RT-PCR) (Fig. 5A) and Gls24 (as seen on a Coomassie blue-stained SDS-PAGE gel [Fig. 5D]) were increased in all three strains. Thus, it appears that without a fully functional HK, the level of PspA increases.
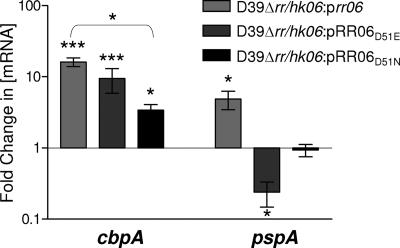
A number of studies have reported that changing the conserved Asp of the RR to Glu mimics the “on” or phosphorylated state (14, 17). Other studies have shown that replacement of the conserved Asp with Asn or Ala blocks activation (6, 7, 14). Accordingly, point mutations in the conserved Asp of RR06 (D39 RR06D51E and D39 RR06D51N) were constructed (see Materials and Methods). The levels of cbpA mRNA and protein for D39 RR06D51E and D39 RR06D51N were determined and compared to the wild-type levels. In both cases, the amount of cbpA mRNA was decreased, although only the data for D39 RR06D51E reached statistical significance (Fig. 5B), indicating that a fully functional RR06 is required for activation of cbpA expression. D39 RR06D51E showed levels of pspA mRNA similar to those of the wild type, while D39 RR06D51N showed an approximately 13-fold increase (Fig. 5B). The RR06D51N mutation has been proposed to mimic the nonphophorylated form of RR06 (14, 17). Similar results were also seen for Gls24 (Fig. 5D). These results suggest that phosphorylation of RR06 is critical for its ability to repress pspA and also Gls24. However, when we overexpressed either the wild-type RR06 or the mutations (D39 RR06D51E and D39 RR06D51N) in an RR/HK06 double-deletion background (similar to what was done by Ma and Zhang [20]), we observed a very different result for both cbpA and pspA mRNA expression. In these circumstances we observed significant up-regulation of the cbpA transcript by the RR06 form which mimics the phosphorylated RR06D51E form and a much smaller increase for the nonphosphorylated form of RR06 (RR06D51N) (Fig. 6). It thus appears that in a double mutant overexpressing the phosphorylated form of RR06 we can reproduce the results seen by other workers (20) regarding the transcriptional regulation of cbpA by a phosphorylated RR06. Examination of the levels of the pspA transcript in this system revealed that the phosphorylated form RR06D51E represses pspA, whereas the nonphosphorylated form RR06D51N has no significant effect (Fig. 6). We saw the same expression profile when examining the levels of the Gls24 protein (data not shown). The results of expression profiles for cbpA and pspA in the RR/HK06 mutant strains are summarized in Table 4.
FIG. 6.
CbpA and PspA mRNA levels. RNA isolated from D39Δrr/hk06::pControl, D39Δrr/hk06::prr06, D39Δrr/hk06::pRR06D51E, and D39Δrr/hk06::pRR06D51N was analyzed for levels of cbpA and pspA mRNA by real-time RT-PCR. The data are fold changes (± standard error) in the mRNA concentration relative to D39Δrr/hk06::pControl. One asterisk, P < 0.05; three asterisks, P < 0.001, as determined by one-way ANOVA with the post hoc Tukey test.
TABLE 4.
Summary of results
| Strain | Expressiona
|
|
|---|---|---|
| cbpA | pspA | |
| D39::pControl | — | — |
| D39::phk06 | — | ↓ |
| D39::prr06 | ↑ | ↓↓ |
| D39::prr/hk06 | ↑ | ↓↓ |
| D39Δrr/hk06::pControl | — | — |
| D39Δrr/hk06::prr06 | ↑↑ | ↑ |
| D39Δrr/hk06::pRR06D51E | ↑↑ | ↓ |
| D39Δrr/hk06::pRR06D51N | ↑ | — |
| D39 | — | — |
| D39Δhk06 | ↑ | ↑ |
| D39 HK06H242Y | ↑ | ↑ |
| D39 HK06S241D | — | ↑ |
| D39 HK06H242R | ↑ | ↑ |
| D39Δrr06 | ↓ | — |
| D39 RR06D51E | ↓ | — |
| D39 RR06D51N | — | — |
| D39Δrr/hk06 | — | — |
↑, up-regulated; ↓, down-regulated; ↓↓, strongly down-regulated; —, no change.
DISCUSSION
Regulation of virulence genes by pneumococcal TCSTSs is likely to be crucial for the ability of S. pneumoniae to colonize and cause disease. Previously, we showed that RR/HK06 regulates the expression of CbpA, a major pneumococcal virulence factor and protective antigen (36). However, regulated factors other than CbpA appeared to be at least partially responsible for the system's effects on epithelial cell adherence and pathogenesis. In the present study, we overexpressed RR/HK06 in order to identify additional regulated genes. Using microarray analysis, we identified 53 genes differentially expressed in D39, while only 5 genes showed differences in TIGR4 when organisms were grown in THY. The only genes commonly regulated were cbpA and the cotranscribed upstream gene spd2018. The genes regulated by another pneumococcal TCSTS, TCS04, have also been shown to differ markedly in these two strains (21). One possible explanation for this difference is that overexpression of RR06 resulted in a serious stress response in D39 but not in TIGR4. However, this seems unlikely as the strain overexpressing RR06 grew like D39::pControl. Additionally, a number of genes repressed during overexpression of RR06 also showed increased expression in an hk06 in-frame deletion mutant, thus further suggesting a direct effect independent of RR06 overexpression.
Another gene found to be repressed in D39 was gls24. This gene encodes a polypeptide with similarity to proteins from Enterococcus faecalis, Lactococcus lactis, and Staphylococcus aureus associated with bile salts resistance and an alkaline shock response (8, 15). Recent studies in E. faecalis have also shown that this protein is a putative protective antigen (38). The level of this protein was also found to be substantially increased in the in-frame deletion mutant D39Δhk06. Investigation of the growth at a range of basic pHs (pH 7.5 to 9.0) illustrated that even with increased levels of Gls24, D39Δhk06 grew at the same rate as the wild type (A. J. Standish, unpublished data).
Microarray analysis also showed that a number of genes potentially involved in sugar metabolism were strongly repressed by overexpression of RR06 (spd0065 to spd0071 and spd1633 to spd1635). Interestingly, expression of the first gene (spd0065) from the possible operon was also shown to be increased in D39Δhk06. This is the same phenomenon that was seen with pspA expression.
Expression of the choline binding protein gene pspA was markedly reduced when rr06 was overexpressed in D39. PspA is an important virulence factor, enabling the pathogen to evade host defenses through its ability to inhibit complement activation and bind lactoferrin (35, 42). In contrast to CbpA, PspA appears to be repressed by RR/HK06. Differential expression of these two proteins has also been reported to occur in the transparent and opaque phase variants (34). However, we previously reported that regulation by RR/HK06 appears to be independent of phase (36). PspA has also been reported to be regulated by another pneumococcal TCSTS, YycFG, in strain R6, an unencapsulated derivative of D39 (26), implicating both of these TCSTSs in the regulation of PspA in D39.
In our previous study of RR/HK06 (36), a deletion in hk06 resulted in increased cbpA expression, whereas overexpression of hk06 led to no change in cbpA expression. Interestingly, a recent report on the TCS06 system found that inactivation of RR06 led to a complete loss of CbpA (20). It should be pointed out that the mutation used by Ma and Zhang (20) to examine the effects of cbpA transcription had strong polar effects, at least on the HK gene and perhaps even on other genes downstream. However, the RR06 deletion replacement and point mutations that we used in this study do not have any polar effects on transcription of the corresponding HK gene. We have previously described a significant reduction in the level of CbpA at both the protein and mRNA levels in our RR06 mutants (36), and we have attempted to reconcile the results of Ma and Zhang for the same S. pneumoniae strain, since these workers also used Western blotting to detect CbpA. We therefore reexamined the levels of CbpA in this study by Western blotting and determined the level of cbpA transcript in our RR06 deletion mutant; we found reduced levels but not complete abolition. Our evidence suggests that RR06 acts to activate cbpA expression in its nonphosphorylated state, unlike the findings of Ma and Zhang, which suggest that cbpA expression is increased when RR06 is phosphorylated (mutations mimicking the nonphosphorylated state were not investigated [20]). Most RRs are active in the phosphorylated state, but there are exceptions, such as the DegU RR from Bacillus subtilis. This RR has been shown to function in either a phosphorylated or nonphosphorylated form, depending on the target gene (for a review, see reference 25). Assuming that HK06 has predominantly kinase activity, overexpression would not result in an increase in nonphosphorylated RR06, and thus there would be no increase in cbpA expression. This would explain why deletion of hk06, removing this kinase activity, results in an increase in cbpA mRNA. Amino acid substitutions in HK06 predicted to mutate kinase and phosphatase or simply kinase activities of HK06 resulted in increased levels of CbpA similar to that seen in D39Δhk06. However, D39 HK06S241D, predicted to retain kinase activity but not phosphatase activity, showed a level of cbpA mRNA comparable to the wild-type level. This suggests that phosphatase activity or loss of kinase activity is critical for the ability of RR/HK06 to increase cbpA expression. In other words, the nonphosphorylated form of RR06 activates cbpA expression. The recent evidence presented by Ma and Zhang (20) suggests that RR06 acts while in its phosphorylated form, since removing the proposed site of phosphorylation leads to a loss of CbpA. An alternative interpretation is that mutating this site prevents the RR from interacting with its DNA binding site but gives the impression that phosphorylation is critical. We subsequently examined cbpA expression (at both the protein and mRNA levels) in a background which still contains a fully functional HK06 but has an altered chromosomal copy of rr06, thereby avoiding any copy number effects potentially present when complementation with plasmid-based mutant or wild-type genes is used in a null background. Our substitutions in the conserved Asp of RR06, which are predicted to mimic either the phosphorylated (D39 RR06D51E) or nonphosphorylated (D39 RR06D51N) form (14, 17), showed that only the D39 RR06D51E mutant exhibited a statistically significant reduction in cpbA expression. Interestingly, this substitution is actually the one that mimics the phosphorylated form of the protein and as such would be expected to increase CpbA levels if this was the active form. Our data suggest that alteration of the phosphorylation site to mimic the phosphorylated form either leads to repression or abolishes the ability of the protein to bind to the DNA. Similarly, it can be argued that alteration to mimic the nonphosphorylated form (D39 RR06D51N) also diminishes the DNA binding ability. However, our data on pspA mRNA expression clearly indicate that these two mutations in RR06 have very different phenotypes. If either the RR06D51E or RR06D51N mutant had lost the ability to bind DNA, then it should have no effect on either cbpA or pspA transcription levels. Nevertheless, there are indeed significant differences in the levels of the transcripts (Table 4), indicating that their ability to bind DNA has not been affected.
Examination of cbpA mRNA levels in an in-frame deletion mutant with deletions in both hk06 and rr06 in D39 showed that this mutant produced cbpA mRNA levels similar to the wild-type levels. This is perhaps not surprising in light of a recent study of S. pneumoniae which showed that a deletion mutant with a mutation in either RR08 or the whole TCS08 resulted in no alteration of the mRNA expression profile, whereas an activating HK08 mutation did alter the profile (22). This suggests that loss-of-function TCSTS mutations do not always influence the expression of regulated genes.
Studies of expression of pspA, and also Gls24, in the strains mentioned above provided results that clearly indicate that the regulation of these factors is different from that of cbpA. Investigation of pspA expression in deletion mutants showed that while D39Δhk06 led to a significant increase in pspA mRNA, both D39Δrr06 and D39Δrr/hk06 resulted in levels similar to the wild-type D39 levels. The levels of Gls24 in these strains mimicked the levels of pspA expression. Robertson et al. (32) saw similar changes when they investigated expression of the vex locus in mutants with mutations in the TCSTS VncRS, and a mutation in the HK resulted in increased expression of vex123, while an RR mutant produced levels of vex123 mRNA similar to the wild-type levels (32). These authors hypothesized that the phosphorylated RR is responsible for the repression of this locus. The overexpression of hk06, rr06, and rr/hk06 in D39 suggests that RR/HK06 represses pspA expression. Studies using the three amino substitutions predicted to mutate various activities of HK06 showed that all three produced increased levels of pspA mRNA and Gls24. Additionally, when prr06 was transformed into D39Δrr/hk06, increased levels of pspA mRNA and Gls24 were evident, which perhaps is not surprising as this is essentially the same genotype as D39Δhk06. This suggests that repression is possible only in the presence of a fully functional copy of hk06. This study illustrates cross-serotype regulation of the major pneumococcal virulence factor and protective antigen CbpA. We have also shown that expression of the virulence factor PspA is also under the regulatory control of RR/HK06. It appears that while CbpA is activated predominantly by nonphosphorylated RR06 and the system acts as an activator, for PspA and another regulated factor, Gls24, regulation occurs by a distinct mechanism whereby the phosphorylated form of RR06 acts as a repressor.
Acknowledgments
This work was funded by program grant 284214 from the National Health and Medical Research Council of Australia.
We thank Paloma Lopez for providing pLS1RGFP, Mark Van der Hoek for assistance with microarray analysis, and the Bacterial Microarray Group at St. George's Hospital, University of London, for the supply of S. pneumoniae microarray chips.
Footnotes
Published ahead of print on 25 May 2007.
REFERENCES
- 1.Avery, O. T., C. M. Macleod, and M. McCarty. 1944. Studies on the chemical nature of the substance inducing transformation of pneumococcal types: induction of transformation by a deoxyribonucleic acid fraction isolated from pneumococcus type III. J. Exp. Med. 79:137-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berry, A. M., A. D. Ogunniyi, D. C. Miller, and J. C. Paton. 1999. Comparative virulence of Streptococcus pneumoniae strains with insertion-duplication, point, and deletion mutations in the pneumolysin gene. Infect. Immun. 67:981-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berry, A. M., J. Yother, D. E. Briles, D. Hansman, and J. C. Paton. 1989. Reduced virulence of a defined pneumolysin-negative mutant of Streptococcus pneumoniae. Infect. Immun. 57:2037-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bricker, A. L., and A. Camilli. 1999. Transformation of a type 4 encapsulated strain of Streptococcus pneumoniae. FEMS Microbiol. Lett. 172:131-135. [DOI] [PubMed] [Google Scholar]
- 5.Cheng, Q., E. A. Campbell, A. M. Naughton, S. Johnson, and H. R. Masure. 1997. The com locus controls genetic transformation in Streptococcus pneumoniae. Mol. Microbiol. 23:683-692. [DOI] [PubMed] [Google Scholar]
- 6.Dahl, M. K., T. Msadek, F. Kunst, and G. Rapoport. 1992. The phosphorylation state of the DegU response regulator acts as a molecular switch allowing either degradative enzyme synthesis or expression of genetic competence in Bacillus subtilis. J. Biol. Chem. 267:14509-14514. [PubMed] [Google Scholar]
- 7.DiGiuseppe, P. A., and T. J. Silhavy. 2003. Signal detection and target gene induction by the CpxRA two-component system. J. Bacteriol. 185:2432-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giard, J. C., A. Rince, H. Capiaux, Y. Auffray, and A. Hartke. 2000. Inactivation of the stress- and starvation-inducible gls24 operon has a pleiotropic effect on cell morphology, stress sensitivity, and gene expression in Enterococcus faecalis. J. Bacteriol. 182:4512-4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 10.Horton, R. M. 1993. In vitro recombination and mutangenesis of DNA. Methods Mol. Biol. 15:251-261. [DOI] [PubMed] [Google Scholar]
- 11.Hoskins, J., W. E. Alborn, Jr., J. Arnold, L. C. Blaszczak, S. Burgett, B. S. DeHoff, S. T. Estrem, L. Fritz, D. J. Fu, W. Fuller, C. Geringer, R. Gilmour, J. S. Glass, H. Khoja, A. R. Kraft, R. E. Lagace, D. J. LeBlanc, L. N. Lee, E. J. Lefkowitz, J. Lu, P. Matsushima, S. M. McAhren, M. McHenney, K. McLeaster, C. W. Mundy, T. I. Nicas, F. H. Norris, M. O'Gara, R. B. Peery, G. T. Robertson, P. Rockey, P. M. Sun, M. E. Winkler, Y. Yang, M. Young-Bellido, G. Zhao, C. A. Zook, R. H. Baltz, S. R. Jaskunas, P. R. Rosteck, Jr., P. L. Skatrud, and J. I. Glass. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 183:5709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsing, W., and T. J. Silhavy. 1997. Function of conserved histidine-243 in phosphatase activity of EnvZ, the sensor for porin osmoregulation in Escherichia coli. J. Bacteriol. 179:3729-3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanamaru, K., H. Aiba, and T. Mizuno. 1990. Transmembrane signal transduction and osmoregulation in Escherichia coli. I. Analysis by site-directed mutagenesis of the amino acid residues involved in phosphotransfer between the two regulatory components, EnvZ and OmpR. J. Biochem. (Tokyo) 108:483-487. [DOI] [PubMed] [Google Scholar]
- 14.Klose, K. E., D. S. Weiss, and S. Kustu. 1993. Glutamate at the site of phosphorylation of nitrogen-regulatory protein NTRC mimics aspartyl-phosphate and activates the protein. J. Mol. Biol. 232:67-78. [DOI] [PubMed] [Google Scholar]
- 15.Kuroda, M., T. Ohta, and H. Hayashi. 1995. Isolation and the gene cloning of an alkaline shock protein in methicillin resistant Staphylococcus aureus. Biochem. Biophys. Res. Commun. 207:978-984. [DOI] [PubMed] [Google Scholar]
- 16.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 17.Lan, C. Y., and M. M. Igo. 1998. Differential expression of the OmpF and OmpC porin proteins in Escherichia coli K-12 depends upon the level of active OmpR. J. Bacteriol. 180:171-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lange, R., C. Wagner, A. de Saizieu, N. Flint, J. Molnos, M. Stieger, P. Caspers, M. Kamber, W. Keck, and K. E. Amrein. 1999. Domain organization and molecular characterization of 13 two-component systems identified by genome sequencing of Streptococcus pneumoniae. Gene 237:223-234. [DOI] [PubMed] [Google Scholar]
- 19.LeMessurier, K. S., A. D. Ogunniyi, and J. C. Paton. 2006. Differential expression of key pneumococcal virulence genes in vivo. Microbiology 152:305-311. [DOI] [PubMed] [Google Scholar]
- 20.Ma, Z., and J.-R. Zhang. 2007. RR06 activates transcription of spr1996 and cbpA in Streptococcus pneumoniae. J. Bacteriol. 189:2497-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCluskey, J., J. Hinds, S. Husain, A. Witney, and T. J. Mitchell. 2004. A two-component system that controls the expression of pneumococcal surface antigen A (PsaA) and regulates virulence and resistance to oxidative stress in Streptococcus pneumoniae. Mol. Microbiol. 51:1661-1675. [DOI] [PubMed] [Google Scholar]
- 22.McKessar, S. J., and R. Hakenbeck. 2007. The two-component regulatory system TCS08 is involved in cellobiose metabolism of Streptococcus pneumoniae R6. J. Bacteriol. 189:1342-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mohedano, M. L., K. Overweg, A. de la Fuente, M. Reuter, S. Altabe, F. Mulholland, D. de Mendoza, P. Lopez, and J. M. Wells. 2005. Evidence that the essential response regulator YycF in Streptococcus pneumoniae modulates expression of fatty acid biosynthesis genes and alters membrane composition. J. Bacteriol. 187:2357-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morona, J. K., R. Morona, and J. C. Paton. 1999. Comparative genetics of capsular polysaccharide biosynthesis in Streptococcus pneumoniae types belonging to serogroup 19. J. Bacteriol. 181:5355-5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Msadek, T., F. Kunst, and G. Rapoport. 1995. A signal transduction network in Bacillus subtilis includes the DegS/DegU and ComP/ComA two-component systems, p. 447-471. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction. ASM Press, Washington, DC.
- 26.Ng, W. L., H. C. Tsui, and M. E. Winkler. 2005. Regulation of the pspA virulence factor and essential pcsB murein biosynthetic genes by the phosphorylated VicR (YycF) response regulator in Streptococcus pneumoniae. J. Bacteriol. 187:7444-7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nieto, C., P. Fernandez de Palencia, P. Lopez, and M. Espinosa. 2000. Construction of a tightly regulated plasmid vector for Streptococcus pneumoniae: controlled expression of the green fluorescent protein. Plasmid 43:205-213. [DOI] [PubMed] [Google Scholar]
- 28.Ogunniyi, A. D., P. Giammarinaro, and J. C. Paton. 2002. The genes encoding virulence-associated proteins and the capsule of Streptococcus pneumoniae are upregulated and differentially expressed in vivo. Microbiology 148:2045-2053. [DOI] [PubMed] [Google Scholar]
- 29.Ogura, M., H. Yamaguchi, K. Yoshida, Y. Fujita, and T. Tanaka. 2001. DNA microarray analysis of Bacillus subtilis DegU, ComA and PhoP regulons: an approach to comprehensive analysis of B. subtilis two-component regulatory systems. Nucleic Acids Res. 29:3804-3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orihuela, C. J., J. N. Radin, J. E. Sublett, G. Gao, D. Kaushal, and E. I. Tuomanen. 2004. Microarray analysis of pneumococcal gene expression during invasive disease. Infect. Immun. 72:5582-5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pestova, E. V., L. S. Havarstein, and D. A. Morrison. 1996. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol. Microbiol. 21:853-862. [DOI] [PubMed] [Google Scholar]
- 32.Robertson, G. T., J. Zhao, B. V. Desai, W. H. Coleman, T. I. Nicas, R. Gilmour, L. Grinius, D. A. Morrison, and M. E. Winkler. 2002. Vancomycin tolerance induced by erythromycin but not by loss of vncRS, vex3, or pep27 function in Streptococcus pneumoniae. J. Bacteriol. 184:6987-7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rogers, T. J., A. W. Paton, S. R. McColl, and J. C. Paton. 2003. Enhanced CXC chemokine responses of human colonic epithelial cells to locus of enterocyte effacement-negative Shiga-toxigenic Escherichia coli. Infect. Immun. 71:5623-5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosenow, C., P. Ryan, J. N. Weiser, S. Johnson, P. Fontan, A. Ortqvist, and H. R. Masure. 1997. Contribution of novel choline-binding proteins to adherence, colonization and immunogenicity of Streptococcus pneumoniae. Mol. Microbiol. 25:819-829. [DOI] [PubMed] [Google Scholar]
- 35.Shaper, M., S. K. Hollingshead, W. H. Benjamin, Jr., and D. E. Briles. 2004. PspA protects Streptococcus pneumoniae from killing by apolactoferrin, and antibody to PspA enhances killing of pneumococci by apolactoferrin. Infect. Immun. 72:5031-5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Standish, A. J., U. H. Stroeher, and J. C. Paton. 2005. The two-component signal transduction system RR06/HK06 regulates expression of cbpA in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 102:7701-7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 38.Teng, F., E. C. Nannini, and B. E. Murray. 2005. Importance of gls24 in virulence and stress response of Enterococcus faecalis and use of the Gls24 protein as a possible immunotherapy target. J. Infect. Dis. 191:472-480. [DOI] [PubMed] [Google Scholar]
- 39.Throup, J. P., K. K. Koretke, A. P. Bryant, K. A. Ingraham, A. F. Chalker, Y. Ge, A. Marra, N. G. Wallis, J. R. Brown, D. J. Holmes, M. Rosenberg, and M. K. Burnham. 2000. A genomic analysis of two-component signal transduction in Streptococcus pneumoniae. Mol. Microbiol. 35:566-576. [DOI] [PubMed] [Google Scholar]
- 40.Tokishita, S., A. Kojima, and T. Mizuno. 1992. Transmembrane signal transduction and osmoregulation in Escherichia coli: functional importance of the transmembrane regions of membrane-located protein kinase, EnvZ. J. Biochem. (Tokyo) 111:707-713. [DOI] [PubMed] [Google Scholar]
- 41.Towbin, H., T. Staehelin, and J. Gordon. 1992. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Bio/Technology 24:145-149. [PubMed] [Google Scholar]
- 42.Tu, A. H., R. L. Fulgham, M. A. McCrory, D. E. Briles, and A. J. Szalai. 1999. Pneumococcal surface protein A inhibits complement activation by Streptococcus pneumoniae. Infect. Immun. 67:4720-4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ulijasz, A. T., D. R. Andes, J. D. Glasner, and B. Weisblum. 2004. Regulation of iron transport in Streptococcus pneumoniae by RitR, an orphan response regulator. J. Bacteriol. 186:8123-8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yother, J., L. S. McDaniel, and D. E. Briles. 1986. Transformation of encapsulated Streptococcus pneumoniae. J. Bacteriol. 168:1463-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]