Abstract
The current increase in the incidence and severity of infectious diseases mandates improved understanding of the basic biology and DNA repair profiles of virulent microbes. In our studies of the major pathogen and model organism Neisseria meningitidis, we constructed a panel of mutants inactivating genes involved in base excision repair, mismatch repair, nucleotide excision repair (NER), translesion synthesis, and recombinational repair pathways. The highest spontaneous mutation frequency among the N. meningitidis single mutants was found in the MutY-deficient strain as opposed to mutS mutants in Escherichia coli, indicating a role for meningococcal MutY in antibiotic resistance development. Recombinational repair was recognized as a major pathway counteracting methyl methanesulfonate-induced alkylation damage in the N. meningitidis. In contrast to what has been shown in other species, meningococcal NER did not contribute significantly to repair of alkylation-induced DNA damage, and meningococcal recombinational repair may thus be one of the main pathways for removal of abasic (apurinic/apyrimidinic) sites and strand breaks in DNA. Conversely, NER was identified as the main meningococcal defense pathway against UV-induced DNA damage. N. meningitidis RecA single mutants exhibited only a moderate decrease in survival after UV exposure as opposed to E. coli recA strains, which are extremely UV sensitive, possibly reflecting the lack of a meningococcal SOS response. In conclusion, distinct differences between N. meningitidis and established DNA repair characteristics in E. coli and other species were identified.
Neisseria meningitidis, or the meningococcus (MC), is a gram-negative inhabitant of the human oropharynx that may disseminate into the bloodstream and traverse the blood-brain barrier to cause septicemia and/or meningitis. MC cells residing on mucosal surfaces are exposed to DNA damaging agents, in particular, reactive oxygen species (ROS) generated from normal cellular metabolism or a highly effective immune system through the oxidative burst. ROS from exogenous and endogenous sources can induce a vast number of different types of DNA damage, including single- and double-strand breaks, abasic (apurinic/apyrimidinic, or AP) sites, and base damages, among which the oxidation product of guanine, 7,8-dihydro-8-oxo-2′-deoxyguanosine (8oxoG), is one of the most frequent (13). Oxidative DNA damage is primarily processed by the base excision repair (BER) pathway (52). In Escherichia coli, BER is initiated by DNA glycosylases that nick the N-glycosylic bond and remove the damaged base by a flipping mechanism. The remaining sugar and phosphate moieties are subsequently excised either by an AP-lyase activity inherent in many DNA glycosylases or by an AP endonuclease, leaving the processing of the 3′ or 5′ terminus, respectively, to a deoxyribose phosphodiesterase (52). A triplet of enzymes referred to as the GO system composed of the DNA glycosylases MutY and Fpg, as well as the nucleotide hydrolase MutT, is involved in limiting the mutagenic effects of 8oxoG (38).
In contrast to the endogenous origin of BER lesions, nucleotide excision repair (NER) generally repairs bulky lesions due to stress from exogenous sources interfering with normal base-pairing and impairing transcription and replication (12). In E. coli NER is executed by the UvrABC complex which removes the stretch of nucleotides including the lesion (12, 51). The third excision repair pathway, mismatch repair (MMR), recognizes base-base mismatches and insertion/deletion loops, including those introduced by DNA polymerases. In E. coli MMR, MutS mismatch recognition (33, 43) is followed by MutL recruitment (2). Together, these enzymes activate MutH, an endonuclease that preferentially cleaves the unmethylated strand at hemimethylated GATC sites (3). Additional E. coli DNA repair pathways manage other classes of DNA lesions: double-stranded breaks are primarily repaired by E. coli recombinational repair by the RecABCD pathway (32). A key component, RecA, has the ability to homologously pair, via strand exchange, a damaged duplex with the intact sister duplex (32). Moreover, the binding of E. coli RecA to single-strand regions up-regulates the expression of the more than 40 genes of an inducible SOS system involved in repairing DNA damage and restoring replication (18). Damage tolerance in E. coli is further provided by daughter strand gap repair, as well as translesion synthesis (TLS) constituted by the three TLS DNA polymerases Pol II (8), DinB (58), and UmuDC′ (56) that allow replication past blocking lesions at the cost of mutations (40).
The data available on MC DNA repair components have been derived primarily from genome sequences and experiments conducted in the close relative, Neisseria gonorrhoeae (the gonococcus) (11, 29). The only fully characterized MC DNA repair component to date is the BER DNA glycosylase MutY (10). MC homologues to other components of the BER pathway have been identified; however, the genes encoding DNA glycosylase Nei and endonuclease Nfo are missing (29). An apparent functional MC NER pathway has also been identified since MC genomes harbor homologues of uvrA, uvrB, and uvrC genes (46, 57), and gonococcus NER activity has been experimentally confirmed (4). Additionally, MC genomes reveal the presence of MMR mutS (class I) and mutL homologues, while mutH and dam homologues are missing (46, 49, 57). Due to frequently occurring recombination, transformation, and antigenic variation, recombinational repair proteins, including RecA, are most important in the pathogenic Neisseria (11, 31). Although a recF homologue is missing, both the RecBCD and RecF-like pathways are implicated in DNA repair (37), and gonococcus RecA acts in both pathways (30, 37). In contrast, neither a LexA homologue nor SOS boxes have been detected in the MC or gonococcus genomes, and the lack of an inducible SOS response in the gonococcus has been experimentally confirmed (5). Additionally, homology searches have shown that MC genomes harbor only a single TLS DNA polymerase gene encoding the DinB homologue (11, 35).
Infections caused by MC are associated with significant morbidity and mortality worldwide. However, very limited information exists on MC DNA repair and its influence on genome instability, strain variability, and pathogenesis (11). In this study, we investigated the potential interactions of BER with other DNA repair pathways. We constructed a panel of MC mutants to disrupt the expression of selected BER components and activity of MMR, NER, recombinational repair, and TLS, alone or in combination with BER. Single and double MC DNA repair mutants were assessed with regard to their spontaneous mutation and transformation rates, as well as survival after exposure to various DNA damaging agents. Our results indicate that the interactions between MC DNA repair pathways significantly differ from what has previously been documented in E. coli and other species.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
MC strains included in the study are listed in Table 1. The gonococcus strain N400 containing a recA6 inducible allele was kindly provided by M. Koomey (University of Oslo, Oslo, Norway) (17, 30). MC strains were propagated on 5% blood agar plates, GC plates, or GC plates containing rifampin (Chemical Abstract Service [CAS] 13292-46-1) or nalidixic acid (CAS 389-08-2) to final concentrations of 3 μg/ml or 1 μg/ml, respectively, when appropriate. All incubations were performed in 5% CO2 at 34°C.
TABLE 1.
Relevant characteristics and source of strains used in this study
| Strain | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Escherichia coli ER2566 | Expression host with a chromosomal copy of the T7 RNA polymerase gene | New England Biolabs |
| N. meningitidis strains | ||
| H44/76 mutYK | H44/76 strain with mutY::Kanr | 10 |
| H44/76 mutYE | H44/76 strain with mutY::Ermr | This study |
| H44/76 fpg | H44/76 strain with fpg::Kanr | This study |
| H44/76 mutS | H44/76 strain with mutS::Kanr | This study |
| H44/76 uvrAK | H44/76 strain with uvrA::Kanr | This study |
| H44/76 uvrAE | H44/76 strain with uvrA::Ermr | This study |
| H44/76 dinB | H44/76 strain with dinB::Ermr | This study |
| H44/76 recA6 | H44/76 strain with recA6 allele (Tetr) | This study |
| H44/76 mutY fpg | H44/76 strain with mutY::Ermrfpg::Kanr | This study |
| H44/76 mutY mutS | H44/76 strain with mutY::ErmrmutS::Kanr | This study |
| H44/76 mutY uvrA | H44/76 strain with mutY::ErmruvrA::Kanr | This study |
| H44/76 mutY dinB | H44/76 strain with mutY::KanrdinB::Ermr | This study |
| H44/76 mutY recA6 | H44/76 strain with mutY::KanrrecA6 (Tetr) | This study |
| H44/76 mutS uvrA | H44/76 strain with mutS::KanruvrA::Ermr | This study |
| H44/76 mutS dinB | H44/76 strain with mutS::KanrdinB::Ermr | This study |
| H44/76 mutS recA6 | H44/76 strain with mutS::KanrrecA6 (Tetr) | This study |
| H44/76 fpg uvrA | H44/76 strain with fpg::KanruvrA::Ermr | This study |
| H44/76 recA6 uvrA | H44/76 strain with recA6 (Tetr) uvrA::Kanr | This study |
| H44/76 dinB uvrA | H44/76 strain with dinB::ErmruvrA::Kanr | This study |
| H44/76 dinB recA6 | H44/76 strain with dinB::ErmrrecA6 (Tetr) | This study |
| H44/76 | Serogroup B, ET-5 complex, isolated in Norway | 25 |
| M1080 | Serogroup B, B:1:P1.1,7, isolated in the United States | 16 |
| MC58 | Serogroup B, ET-5 complex, isolated in England | 36 |
| N. gonorrhoeae N400 | MS11mk strain with recA6 allele (Tetr) | 30 |
Construction of MC DNA repair mutants.
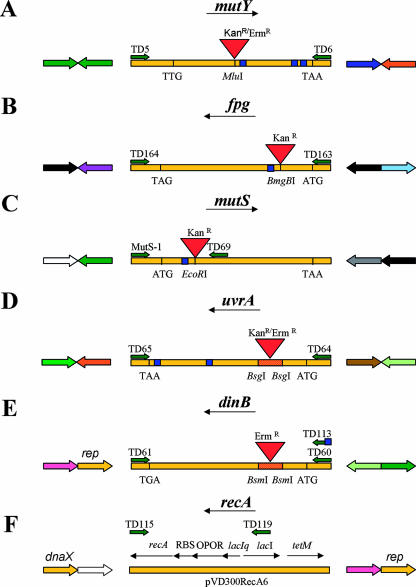
The partial mutS gene (mutS1, 5′-GCTTACGGTCAGTCTCATTCCG; TD69, 5′-GCATCGATGGTAGCGCAAAGGTCGAGCG-3′) as well as the full-length fpg (TD163, 5′-TTCAGAGCTCGTTTCGATATTGAATTTGGG-3′; TD164, 5′-GACAAAGCTTGAAACCGTTTTCAGTCCTAT-3′), dinB (TD60, 5′-CGTATATTTGGAATTCGCCCG-3′; TD61, 5′-GCCGATATCGATAAGGCGG-3′), and uvrA (TD64, 5′-CGCGGCTTCAGACGGGATCCGAG-3′; TD65, 5′-CGGAATTCAAATACTTCCCAGTATAACTCCCC-3′) genes were amplified by PCR using primers as described previously. The DNA fragments were cloned into pBluescript SK+ (pBSK+) (Stratagene), creating plasmids pH 44/76fpg, pM1080mutS, pM1080uvrA, and pMC58dinB. The plasmids pM1080mutY and pM1080mutY-kanr have been described previously (10). The mutY::Ermr, fpg::Kanr, mutS::Kanr, dinB::Ermr, uvrA::Kanr, and uvrA::Ermr alleles were constructed by inserting a kanamycin or erythromycin resistance gene cassette obtained from pUC4K (GE Healthcare, United Kingdom) or pSAPE5A (48) into the respective genes in pBSK+ (Fig. 1). In the mutY, fpg, and mutS genes this involved simple insertions, as opposed to the dinB and uvrA genes, where 125-bp and 524-bp fragments, respectively, were deleted upon cassette insertion. All plasmids were propagated in E. coli ER2566 (New England Biolabs). The DNA repair genes containing antibiotic resistance cassettes were transformed into the MC strain H44/76 as previously described (10). Since dinB does not contain the DNA uptake sequence required for MC transformation, a primer containing the DNA uptake sequence (TD113, 5′-GCGCCGTCTGAAGAATTCGCCCGATGCCGGCAG-3′) was employed to generate the pMC58dinB-Ermr template that was transformed into H44/76. The MC H44/76 recA6 strain was obtained by transforming H44/76 with DNA from the gonococcus strain N400 and selecting for tetracycline resistance. This recA-deficient strain is not a loss-of-function mutant but contains a regulatable recA allele which is not expressed unless induced with isopropyl-d-thiogalactopyranoside (17, 30). Promoter prediction by using the VIMSS Operon Prediction program (www.microbesonline.org) (47) indicated that the antibiotic cassette introduced for gene inactivation of the selected DNA repair genes did not have a downstream effect on gene expression. The MC H44/76 double mutants were made by transforming kanamycin-resistant DNA into erythromycin- or tetracycline-resistant strains. All H44/76 mutants were verified by PCR. The recA6 alleles were assessed by using one recA6-specific primer (TD119, 5′-GCATACTCTGCGACATCGTAT-3′) and one recA-specific primer (TD115, 5′-GCAGCAGAAGTACCGTTTATC-3′).
FIG. 1.
Cloning of N. meningitidis DNA repair genes and construction of the MC H44/76 mutant strains (A to E). Experimental details are outlined in Materials and Methods. The primers used for PCR amplification of open reading frames are denoted TD. The restriction sites used for insertion of antibiotic resistance markers are depicted together with the DNA uptake sequences (blue squares) required for MC transformation of exogenous DNA. The two adjacent genes upstream and downstream of mutY, fpg, mutS, uvrA, dinB, and recA are illustrated according to the following color scheme: yellow, genes involved in DNA metabolism (rep, ATP-dependent DNA helicase; dnaX, DNA polymerase III, gamma and tau subunits); orange, mobile and extrachromosomal element function; dark green, energy metabolism; light green, transport and binding proteins; white, conserved hypothetical proteins; black, hypothetical proteins; dark blue, cellular processes and detoxification; light blue, cell envelope; purple, fatty acid and phospholipids biosynthesis; gray, central intermediary metabolism; pink, amino acid biosynthesis; brown, unknown function.
Spontaneous mutation frequencies of MC DNA repair-deficient strains.
The MC H44/76 wild type and DNA repair mutants (Table 1) were propagated and assessed for spontaneous mutation frequencies by rifampin resistance selection as described previously (10). Rifampin inhibits RNA synthesis by binding specifically to the RNA polymerase β-subunit and thus prevents transcriptional initiation. The ratio of rifampin-resistant cells to the total number of cells yielded the mutation frequency. The assay was repeated at least eight times for each strain. Differences in spontaneous mutation frequencies between strains were monitored by comparing the number of mutant cells giving rise to colonies by using the Wilcoxon test with continuity-adjusted P values. All strains were compared against the wild-type strain, and the double mutants were compared against their corresponding single mutants. This assay does not allow for the measurement of mutation rates as do Luria-Delbrück-like fluctuation experiments (34), and the assay is not sensitive enough to ascertain the significance of slight variations in mutagenicity. However, this relatively quick method of determining mutation frequencies is widely used among researchers and allows for possible comparisons of results among different laboratories (22).
Mutation spectra of rpoB conferring rifampin resistance.
Rifampin-resistant single colonies were propagated overnight in 5% CO2 at 34°C. Three individual rifampin-resistant colonies (each) of the MC H44/76 wild type and H44/76 DNA repair-deficient mutants listed in Table 1 were analyzed. A 230-bp region of the MC rpoB gene spanning the area known to harbor mutations conferring rifampin resistance was PCR amplified and sequenced using primers described by Nolte and coworkers (41).
MC DNA repair mutants and sensitivity to oxidizing agents and alkylation.
The MC H44/76 wild type and DNA repair mutants grown overnight were resuspended to even turbidity in liquid GC medium and then inoculated on GC plates. Nonimpregnated paper disks (Becton Dickinson Microbiology Systems) were saturated with 10, 20, or 30 mM hydrogen peroxide (H2O2) (CAS 7722-84-1); 1, 2, or 3 mM menadione (CAS 58-27-5); 50, 100, or 150 mM paraquat (CAS 1910-42-5); or 0.1, 0.2, or 0.3% methyl methansulfonate (MMS) (CAS 66-27-3). H2O2, paraquat, and menadione are powerful oxidizing agents that create ROS. MMS is an SN2 alkylating agent. The paper disks were placed on top of the agar plates inoculated with the MC cells, preincubated for 10 min at room temperature, and incubated in 5% CO2 at 34°C for 20 h. The diameter of the inhibition zone was measured. The assays were repeated at least twice for each condition. Differences in survival between mutants and wild type and between double mutants and their corresponding single mutants were monitored by analyzing the diameter using a mixed analysis of variance (Proc Mixed procedure in SAS/STAT software; SAS Institute, Cary, NC) assuming a fixed effect of DNA damaging agents at each concentration and random error term per tray and per observation.
MC DNA repair mutants and sensitivity to UV irradiation.
The MC strains were grown on 5% blood agar plates in 5% CO2 at 34°C overnight. Serial dilutions of the MC H44/76 wild type and DNA repair mutants were spread on 5% blood agar plates and exposed to either 10 or 20 J/m2 of UV irradiation (CL-1000 UV Cross-Linker; UVP, United Kingdom) or zero irradiation and incubated in 5% CO2 at 34°C for 20 h. The ratio of UV-irradiated survivors to the total number of cells yielded the survival rate. The assay was repeated three times for each strain. Pairwise comparisons were made by using analysis of variance on the log-transformed survival rates, i.e., assuming the same variance in all groups. Pairwise tests were applied to the 2,000 μJ/cm2 survival rates unless these were both less than 10−3, in which case 1,000 μJ/cm2 survival rates were compared. Tukey P values were used to assess the results.
Quantitative MC genetic transformation.
Colonies of the MC H44/76 wild type and DNA repair mutants were harvested in CO2-saturated liquid GC medium containing 7 mM MgCl2 and 1× Isovitalex (Becton Dickinson Diagnostic Systems) and exposed to either the plasmid pSY6 (donor DNA) or MilliQ-H2O (negative control) for 30 min at 34°C. The plasmid pSY6 contains a point-mutated copy of a DNA gyrase gene conferring resistance to nalidixic acid when incorporated into the chromosome (54). The addition of DNase I (Sigma) to a final concentration of 0.1 mg/ml assured degradation of extracellular DNA before 10 volumes of liquid GC medium was added, and the cell solutions were incubated with tumbling at 37°C for 4.5 h. The solutions were plated on GC medium containing nalidixic acid to a final concentration of 1 μg/ml as well as plain GC medium. The ratio of transformants to the total number of cells yielded the transformation rate. The assay was repeated three times for each strain. To rule out that variation in pilus expression would cause differences in transformation efficiency, the MC colony morphology was assessed by microscopy.
RESULTS
The MC mutS mutants exhibit lower spontaneous mutation frequencies than mutY mutants.
We have recently analyzed the nucleotide and amino acid sequences of selected DNA repair genes in a number of disease-associated MC isolates (9). The MC strain H44/76 showed no frameshift, insertion, or deletion mutations in the mutY, fpg, mutS, uvrA, dinB, and recA genes. Moreover, multilocus sequence typing analysis showed that it was closely related to the fully sequenced MC strain MC58 (9). In addition, the baseline mutation frequency for H44/76 is low (<0.1 per 108 CFU) (9), and this reference strain was therefore employed for the investigation of the in vivo significance of defects in MC DNA repair. The MC DNA repair mutants were assessed for their spontaneous mutation frequencies by rifampin resistance screening. The fpg and mutS mutants displayed only slight increases in mutagenicity (five- and sevenfold; P = 0.008 and 0.00007, respectively), while uvrA-, dinB-, and recA-deficient strains had spontaneous mutation frequencies comparable to the wild-type strain (P > 0.3) (Table 2). Recently, Martin and coworkers measured the spontaneous mutation frequency of MC58 mutS and dinB mutants (35), and our data were in concordance with their findings. We have previously shown that the MC mutY mutant exhibited a significant increase in spontaneous mutation frequency (63-fold increase; P < 0.00001) (10). Compared to the wild-type and mutY single mutant, the mutY fpg double mutant revealed a striking increase in mutation frequency (615-fold increase; P < 0.00004). This synergy is explained by the complete loss of the MC capability to remove chromosomally incorporated 8oxoG, both directly through the action of Fpg on 8oxoG mispaired with C and indirectly through the action of MutY on A mispaired with 8oxoG. 8oxoG lesions present in the genome will lead to GC → TA transversions and, hence, a high mutation frequency. The mutY dinB, mutY uvrA, mutY mutS, and mutY recA mutants retained mutY-level mutator phenotypes (P > 0.2) (Table 2). In summary, this suggests that TLS, NER, MMR, and recombinational repair components do not contribute to the prevention of spontaneous mutations together with MutY. Moreover, no significant differences in the mutagenicity rates between the single mutants and the respective double mutants were observed, except for a 3.8-fold increase in the mutS recA mutant compared to mutS (P = 0.04), indicating that the MMR and recombinational pathways might overlap in the repair of mutagenic lesions (Table 2). However, Bonferroni correction of these P values to account for multiple testing would render the findings on mutS versus mutS recA null mutants together with the wild type versus the fpg mutant statistically insignificant (P > 0.05). It should be noted that the dinB recA mutant exhibited an initial growth defect more pronounced than that of the other recA mutants; however, after 7 to 8 h this defect could no longer be observed.
TABLE 2.
Spontaneous mutation frequencies of N. meningitidis H44/76 wild-type and mutant strains assessed by rifampin resistance
| MC strain genotype | Mutation frequency (no. of Rifr cells/total no. of cells) per 108 CFUa
|
||
|---|---|---|---|
| Median | Q1/Q3 | Increase (n-fold)b | |
| Wild typec | <0.1 | <0.1/0.3 | 1 |
| mutc | 25.0 | 18.1/35.0 | 63 |
| fpg | 2.1 | 0.7/2.9 | 5 |
| mutSc | 2.7 | 2.2/3.4 | 7 |
| uvrA | <0.1 | <0.1/1.0 | 1 |
| dinB | <0.1 | <0.1/<0.1 | 1 |
| recA6 | <0.1 | <0.1/0.2 | 1 |
| mutY fpg | 245.8 | 224.0/390.2 | 615 |
| mutY mutS | 26.2 | 17.3/31.3 | 66 |
| mutY dinB | 12.0 | 10.7/24.4 | 30 |
| mutY uvrA | 36.4 | 21.7/84.0 | 91 |
| mutY recA6 | 24.0 | 14.2/33.9 | 60 |
| mutS uvrA | 1.8 | 0.5/2.9 | 5 |
| mutS dinB | 5.0 | 2.4/6.1 | 13 |
| mutS recA6 | 10.3 | 3.8/11.9 | 26 |
| fpg uvrA | 0.8 | <0.1/1.7 | 2 |
| recA6 uvrA | <0.1 | <0.1/1.1 | 1 |
| dinB uvrA | <0.1 | <0.1/0.8 | 1 |
| dinB recA6 | <0.1 | <0.1/0.6 | 1 |
The results are given as the median of 10 independent measurements. Q, quartile.
The relative increase was calculated from the median using a wild-type frequency of 0.4 (average of 15 independent measurements), since the assay cannot detect mutation frequencies below 0.1.
rpoB mutation spectra.
We have previously assessed the rpoB mutation spectra of the MC and gonococcus wild-type and mutY mutant strains showing that the lack of the MC or gonococcus mutY induces GC-to-TA transversions (10). All MC double mutants containing a mutY defect as well as the fpg uvrA and dinB recA strains exhibited this transversion (Table 3). In general, GC-to-TA transversions are consequences of the preferential mispairing of 8oxoG to adenine (28). The remaining DNA repair mutant strains listed in Table 1 predominantly exhibited GC-to-AT transitions, as did the wild type. This transition may be due to the conversion of C to, e.g., 5-hydroxy-C that will preferentially mispair with A (15). DNA repair mutants also exhibited occasional GC-to-CG transversions (mutS, mutS dinB, mutS recA, and dinB recA strains) (Table 3). Although GC-to-CG transversions are common during oxidative stress, the molecular basis for this transversion was just recently suggested to be caused by oxidative lesion products such as imidazolone, guanidinohydantoin, and spiroiminodihydantoin (28). The positions of the nucleotide changes were identical in all DNA repair mutants and wild-type strains of MC or gonococcus (nucleotide numbers 92, 103, and 119) (10), except for one GC-to-AT transversion and the GC-to-CG transversions appearing at unique locations (nucleotide numbers 64 and 104, respectively) in the mutant strains (Table 3). The results indicate that the mutation spectra obtained for the DNA repair mutants, with the exception of the mutY-induced GC-to-AT transversions and mutS-, recA-, and dinB-induced GC-to-CG transversions, are reflections of the mutagenic processes commonly occurring in neisserial wild-type strains.
TABLE 3.
rpoB mutation spectra of N. meningitidis H44/76 wild-type and DNA repair-deficient strainsa
| H44/76 strain genotype | Nucleotide change(s) |
|---|---|
| Wild typeb | GC → AT(103 × 3) |
| fpg | GC → AT(92 × 3) |
| mutS | GC → AT(103 × 2); GC → CG(104 × 1) |
| dinB | GC → AT(92 × 1, 103 × 1, 119 × 1) |
| uvrA | GC → AT(92 × 1, 103 × 1, 119 × 1) |
| recA6c | GC → AT(92 × 2) |
| mutS dinB | GC → AT(92 × 1, 103 × 1); GC → CG(104 × 1) |
| mutS recA6 | GC → AT(92 × 1, 103 × 1); GC → CG(104 × 1) |
| mutS uvrA | GC → AT(92 × 1, 103 × 2) |
| fpg uvrA | GC → AT(92 × 2); GC → TA(103 × 1) |
| dinB uvrA | GC → AT(103 × 3) |
| dinB recA6 | GC → AT(119 × 1); GC → CG(104 × 1); GC → TA(103 × 1) |
| recA6 uvrA | GC → AT(103 × 3) |
| mutYb | GC → TA(92 × 1, 103 × 1, 119 × 1) |
| mutY fpg | GC → TA(92 × 1, 103 × 2) |
| mutY mutS | GC → TA(103 × 3) |
| mutY dinB | GC → TA(103 × 3) |
| mutY uvrA | GC → TA(92 × 1, 103 × 2) |
| mutY recA6 | GC → TA(64 × 1, 103 × 2) |
Three independent rifampin-resistant strains of the MC H44/76 wild-type and DNA repair-deficient strains listed in Table 1 were analyzed for nucleotide changes conferring rifampin resistance. Subscripts indicate the nucleotide being changed (where nucleotide 1 corresponds to the first nucleotide in the sequencing primer) and the number of strains (× n) with the given transition/transversion.
Previously shown by Davidsen et al. (10).
One strain showed no nucleotide changes inside the sequenced area.
MC DNA repair mutants and sensitivity to oxidizing and alkylating agents.
The MC DNA repair mutants in Table 1 were challenged with H2O2, paraquat, menadione, and MMS in a survival assay. None of the MC DNA repair single and double mutants was particularly resistant or sensitive to H2O2, paraquat, or menadione exposure compared to the wild-type strain (data not shown). However, this finding was expected since MC cells harbor particularly strong defense mechanisms against oxygen radicals, constituted by catalase, SodB, and SodC, as well as other components (53). The gonococcus recA mutants have recently been shown to be sensitive to H2O2 treatment, but the antioxidant system in the gonococcus is slightly different from that in the MC (53). In order to reveal any effect of oxidizing agents in MC DNA repair mutants, selected antioxidative enzymes may need to be inactivated. Additionally, the DNA repair pathways tested may have overlapping substrate specificities. This remains to be examined in triple or quadruple mutants exposed to oxidizing agents.
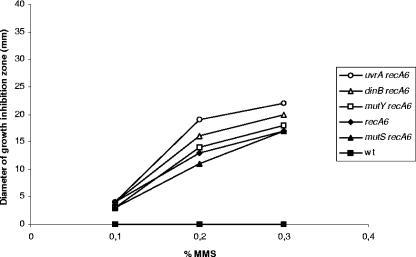
The methylating agent MMS induces predominantly cytotoxic DNA lesions that are removed by a number of different enzymes in E. coli. The genomic complement of the MC contains a single putative gene sequence displaying homology to the E. coli 3-methyladenine DNA glycosylase Tag; however, no homologues to the inducible alkylation/adaptive response constituted by ada, aidB, ogt, alkA, and alkB have been found (11). The products of these genes reverse alkylation damage or initiate pathways for the repair of alkylation damage in many organisms (14). The presence of only a single gene product putatively protecting against methylation damage indicates either that Tag is sufficient for the removal of aberrantly methylated DNA in the meningococcus, since it is likely that the MC does not encounter much alkylation damage in vivo in its habitat, or that other DNA repair pathways may contribute to perform this function. It has previously been shown that both recombinational repair and NER are involved in the repair of MMS-induced DNA damage in E. coli and Schizosaccharomyces pombe (20, 26, 42). However, neither the MC BER, MMR, NER, nor TLS components assessed exhibited MMS sensitivity, and therefore they do not participate in the repair of alkylated bases; alternatively, their contribution is masked by Tag activity (Fig. 2). In contrast, the MC recA single and double mutants were highly sensitive to this agent (Fig. 2), consistent with previous findings showing that the gonococcus RecA participates in the repair of alkylating DNA damage (24). However, no synergy or additive effect of MMS treatment was observed for the double mutants under the conditions tested (mutY recA exhibited a 1.1-fold increase in sensitivity compared to recA when exposed to 0.3% MMS; the increase in sensitivity compared to recA was 1.0-fold for mutS recA, 1.2-fold for dinB recA, and 1.3-fold for uvrA recA), again indicating that BER, MMR, NER, and TLS do not play a role in the repair of alkylated DNA in the MC. The results obtained here indicate a major role of MC recombination repair pathways in the excision of aberrantly methylated DNA; alternatively, MC recombinational repair allows for DNA damage tolerance under the conditions tested.
FIG. 2.
Survival of the MC DNA repair-deficient strains to alkylating stress as measured by growth inhibition zones (mm) surrounding paper diffusion disks after exposure to different concentrations of MMS. Repeated experiments (three to five repetitions) demonstrated that the diameter of the growth inhibition zone of each strain fluctuated; however, the strains always displayed the same MMS sensitivity relative to each other. One representative experiment is shown. The strains are as indicated on the figure; the remaining DNA repair-deficient strains listed in Table 1 showed MMS sensitivity comparable to the wild-type (wt) strain.
MC NER and recombinational repair mutants exhibit sensitivity to UV irradiation.
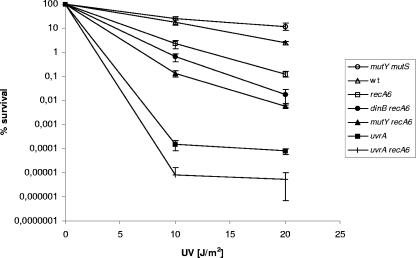
To evaluate the nucleotide excision repair capabilities of the MC DNA repair mutants listed in Table 1, wild-type, single, and double mutant strains were exposed to different doses of UV light. As determined by analysis of variance and Tukey-corrected P values, the strains split distinctly into seven groups, denoted 1 to 7 (Fig. 3), with Tukey P values larger than 0.05 within each group: group 1, mutY mutS, mutY fpg, mutS dinB, and mutY dinB mutants; group 2, wild type, fpg, mutY, mutS, and dinB mutants; group 3, recA6; group 4, dinB recA6; group 5, mutY recA6 and mutS recA6 mutants; group 6, uvrA, mutS uvrA, fpg uvrA, dinB uvrA, and mutY uvrA mutants; and group 7, uvrA recA6. However, groups 1 and 2 could not be totally distinguished from each other since the survival of the fpg strain was not statistically different from the survival of the mutY mutS (P = 0.06) and mutY dinB (P = 0.06) strains. In general, all single mutants of BER, MMR, and TLS survived at wild-type levels, while the double mutants combining BER, MMR, or TLS deficiencies exhibited a slight increase in survival (Fig. 3). There are two possible explanations for this minor increase in UV resistance: either BER, MMR, and TLS do not initiate repair of mutagenic UV-induced DNA lesions that are transformed to cytotoxic intermediates during the repair process, or there are compensatory mutations that provide (temporarily) improved fitness in these mutants. In contrast, the recA and uvrA single mutants exhibited a ∼20- and ∼30,000-fold decrease in survival, respectively, compared to the wild type at a UV dose of 20 J/m2. Moreover, combining the MC recA and uvrA mutants resulted in a ∼15-fold reduction in survival rate compared to the uvrA single mutant (∼440,000-fold decrease compared to wild type with a UV dose of 20 J/m2), providing an additive effect of the recA mutation when there is exposure to UV light. The results indicate that mainly NER and also to a certain extent recombinational repair contribute to the protection against UV-induced damage in the MC. UV light mainly gives rise to cyclobutane pyrimidine dimers and photoproducts, but strand breaks and segregation of strands containing unrepaired lesions may also contribute to lethality (18) and, hence, the sensitivity of recA mutations. Interestingly, the MC RecA-deficient strain was much less sensitive to UV exposure than E. coli recA mutants (18, 32), indicating significant differences in the repair of UV-induced lesions in these two organisms. In contrast, the MC recA dinB double mutant exhibited a fivefold decrease in survival compared to the recA single mutant (UV dose, 20 J/m2). Combining mutY or mutS deficiencies with the recA mutant, a more marked reduction in survival (10-fold) compared to the recA single mutant was observed (UV dose, 20 J/m2). This indicated that when recombinational repair was absent, both BER and MMR sustain more important repair functions in the cell than does TLS. However, combining uvrA deficiency with BER, MMR, and TLS mutations did not result in an increase or decrease in survival after exposure to UV light, arguing that there is no substrate specificity overlap between these repair pathways. In conclusion, MC recombinational repair contributes to the repair of UV-induced DNA damage; however, NER appears to be the major pathway for the repair of such lesions.
FIG. 3.
Survival of the MC DNA repair-deficient strains after exposure to different doses of UV light. The results are given as the averages of three experiments, with error bars showing the standard deviations. The 19 MC strains tested split statistically into seven groups, represented in the figure by one strain each. Group 1 is represented by mutY mutS; mutY fpg, mutS dinB, and mutY dinB mutants showed similar phenotypes. Group 2 is represented by the wild type (wt); fpg, mutY, mutS, and dinB mutants showed similar phenotypes. Group 3 is represented by recA6, and group 4 is represented by dinB recA6. Group 5 is represented by mutY recA6; the mutS recA6 mutant showed a similar phenotype. Group 6 is represented by uvrA; mutS uvrA, fpg uvrA, dinB uvrA, and mutY uvrA mutants showed similar phenotypes. Group 7 is represented by uvrA recA6.
Quantitative genetic transformation in MC DNA repair mutants.
Transformation is the main source of new genetic information integrated into the MC chromosome through horizontal DNA transfer. The MC DNA repair-deficient strains were therefore assessed for their abilities to transform exogenous DNA. Transformation rates in RecA-competent MC mutants ranged from 1.7 × 10−3 to 1.1 × 10−2 transformants/cell, while transformation rates in the recA mutants were below the detection level (data not shown). Thus, none of the MC DNA repair mutants was found to exhibit defective or increased transformation of exogenous DNA, except for the recA mutants, which were noncompetent, as expected.
DISCUSSION
In this study we have assessed the ability of MC DNA repair mutants to counteract DNA damage arising spontaneously or induced by DNA damaging agents. The results demonstrate that mutS mutants exhibit much lower spontaneous mutation frequencies than those of mutY mutants and that, in the MC, MutY and Fpg are particularly important for the repair of spontaneous lesions. Moreover, RecA is critical for MC survival after exposure to alkylating agents, and nucleotide excision repair is the major pathway for the removal of UV-induced DNA damage.
E. coli mutS mutants typically exhibit particularly high mutation frequencies when selected by rifampin resistance, establishing them as the classical mutator phenotype (11). In contrast, the MC mutY mutants exhibited a more pronounced mutator phenotype than the MC mutS-deficient strain (Table 2) (10, 11). MutS null mutants in other mucosal pathogens, such as Streptococcus pneumoniae and Haemophilus influenzae, have also been reported to exhibit low mutation frequencies compared to wild-type strains (22, 39, 60). These results indicate that a different strategy for preventing spontaneous mutations may have developed in the MC and other mucosal pathogens compared to E. coli. Mutator strains exhibit increased spontaneous mutation frequencies compared to those commonly found in the corresponding wild-type strains. Hypermutable strains in bacterial populations might have potential advantages or disadvantages, depending on the nature, rate, and magnitude of environmental change (19). Importantly, mutator strains have been associated with increased survival rates (19), overexpression of virulence factors (23), outbreaks of epidemic disease (50), and increased occurrence of antibiotic resistance (44). Mutators have generally been linked to defects in MMR (19, 60), a notion which to some extent has been verified in the MC (49, 50). However, our findings indicate that mutY mutants potentially may contribute more substantially to the MC mutator pool than mutS deficiencies, as opposed to mutS defects in E. coli. The importance of mutY in promoting pathogenic mutator strains is also emphasized by the finding of four rifampin-resistant Pseudomonas aeruginosa mutator isolates from cystic fibrosis patients exhibiting defects in MutY (44).
A panel of MC DNA repair mutants was constructed to reveal putative interactions of MC BER with NER, MMR, TLS, or recombinational repair pathways. The rifampin resistance assay revealed a synergistic effect of MC MutY and Fpg, demonstrating that the GO system is a major pathway for correcting MC spontaneous DNA damage (Table 2). This is in agreement with previous findings in E. coli (38). Moreover, exposure to the methylating agent MMS showed that only the MC RecA strains exhibited reduced survival rates (Fig. 2). This is consistent with earlier findings showing that the gonococcus RecA participates in the defense against alkylation DNA damage (5, 24), and the results indicate a major role of MC recombinational repair pathways in the excision of aberrantly methylated DNA or, alternatively, in damage tolerance of such lesions. Experiments in E. coli show also that NER participates in the removal of DNA damage by alkylating agents such as adozelesin and nitrogen mustard (26). In addition, when only Tag is present in the cells, UvrABC binds to 7-methylguanine in DNA and starts repair (20). In S. pombe it has been demonstrated that NER is highly important for the repair of alkylated bases, either by direct processing of such lesions or by the repair of AP sites (1). Interestingly, NER was not found to contribute to the defense against alkylation damage in the MC under the conditions tested (Fig. 2). We therefore hypothesize that MC recombinational repair is one of the main pathways for removal of AP sites. Alternatively, MC RecA may be directly involved in the elimination or bypass of MMS-induced alkylation damage. The contribution of MC Tag in the excision of such lesions remains to be elucidated together with the possible roles of endonuclease III (Nth) or exodeoxyribonuclease III (Xth) in the removal of AP sites.
Previously, NER and recombinational repair have been shown to increase gonococcal survival after UV-induced DNA damage (6, 24); however, the relative contribution of each pathway has not been determined. The UV survival assay identified NER as the major repair pathway of UV-induced DNA damage in the MC (Fig. 3). In E. coli, recA strains are extremely UV sensitive, a phenotype which can be attributed to their inability to induce NER proteins through the SOS response. In contrast, although inactivation of the MC RecA affected survival after UV exposure, the MC RecA single mutant showed only a moderate decrease in survival compared to the wild-type strain (Fig. 3). The gonococcus has previously been shown to be devoid of an SOS response (5), and, accordingly, MC strains should be less dependent on RecA for the repair of UV-induced DNA damage compared to E. coli. The importance of protection against UV damage for bacterial mucosal species that seldom will be exposed to light is debatable. However, a fully functional NER pathway argues a role for such DNA repair in the life cycle of both the MC and the gonococcus.
The MC processing of spontaneous, alkylated, and UV-induced DNA damage described here highlights important differences between the MC and E. coli, even though MC DNA repair in other respects resembles that of the established E. coli paradigm (Fig. 4). Although only single and double mutants were assessed, under the conditions tested, MC MutY, MMR, NER, TLS, and recombinational repair did not generally interact with each other to prevent DNA damage (Table 2 and Fig. 2 and 3). The apparent lack of overlap in the processing of DNA lesions indicates that the MC dedicates each pathway to the repair of specific DNA lesions. This is unexpected since an increasing amount of experimental data in other systems indicate a high degree of interactions among different DNA repair pathways: MutY has, for instance, been shown to interact with MutS homologues in humans (21) and to compete with MutS in initiating repair of A·C mismatches in E. coli (27). MutS has also been coupled to the protection of oxidative stress, specifically by binding 8oxoG (59). Furthermore, BER, recombinational repair, and TLS have been shown to be involved in the processing of spontaneous DNA damage in E. coli and yeast by competing for AP sites (45, 55). Additionally, genetic studies in S. pombe (1) have demonstrated a role for NER in the repair of AP sites. NER has also previously been shown to exert activity on untraditional substrates, such as the release of 8oxoG in S. cerevisiae (7).
FIG. 4.
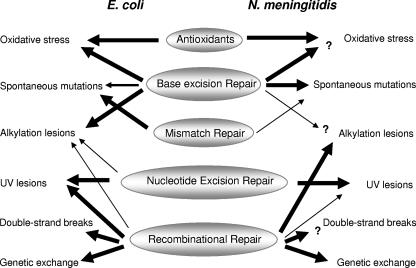
Differences in the contribution of DNA repair pathways in E. coli versus N. meningitidis. Major features discriminating MC repair from E. coli under the conditions tested in this study are (i) the involvement of the MC MutY of the base excision repair pathway in the repair of spontaneous mutations; (ii) the significant contribution of MC recombinational repair and seemingly absent MC nucleotide excision repair for the prevention of alkylation damage; and (iii) the lesser influence of MC recombinational repair in the defense against UV-induced DNA damage. Arrows indicate the relative contribution of each repair pathway in the repair of specific DNA damages. The MC data are based on analysis of single and double DNA repair mutants conducted in this study (Table 1). Interesting questions remaining to be answered concern the involvement of MC BER in the defense against externally induced oxidative stress since at least five MC antioxidants probably mask their contribution (53), the putative participation of the MC Tag in the defense against alkylation damage, and the contribution of Nth and/or Xth in the removal of AP sites. MC RecA is involved in genetic exchange, but it remains to be elucidated whether recombinational repair is the only pathway for the repair of double-strand breaks.
As a basis for characterizing MC DNA repair, our results suggest that this prokaryotic model organism, which also is an important human pathogen, exhibits important differences compared to E. coli. Whether the reduced number of DNA repair genes and the apparent lack of genetic interactions of MC DNA repair pathways identified may be a reflection of the lifestyle of this bacterium is a question to be answered (11). The MC is an exclusive human inhabitant, colonizing only a few sites within its host: the oro- and nasopharynx when commensal, and the blood and/or meninges during disease. MC adaptation is mainly dominated by spontaneous phase and antigenic variation, repeat-associated recombination events, and transformation, while only few regulatory proteins are present (46, 57). In this context, the question of antagonism between adaptive mechanisms and DNA repair in the MC may be raised. However, this and other issues regarding MC genome dynamics remain to be elucidated (11). Studies of DNA metabolism in virulent microorganisms and their relationships to genome (in)stability and strain variation are still in their infancy. Therefore, we need a better understanding of the entire DNA metabolism machinery in bacterial organisms. This new insight is a prerequisite to combating the increasing occurrence of infectious diseases and also to understanding the role of genome dynamics in disease development in general. In this context the MC is a most relevant model organism.
Acknowledgments
This work was supported by grants from The Research Council of Norway to T.T.
We thank the late E. C. Seeberg for his inspiration and unique expertise and K. I. Kristiansen for constructive discussions. E. K. Amundsen, L. Davidsen, and K. L. Tibballs are acknowledged for their technical assistance.
Footnotes
Published ahead of print on 18 May 2007.
REFERENCES
- 1.Alseth, I., F. Osman, H. Korvald, I. Tsaneva, M. C. Whitby, E. Seeberg, and M. Bjoras. 2005. Biochemical characterization and DNA repair pathway interactions of Mag1-mediated base excision repair in Schizosaccharomyces pombe. Nucleic Acids Res. 33:1123-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ban, C., M. Junop, and W. Yang. 1999. Transformation of MutL by ATP binding and hydrolysis: a switch in DNA mismatch repair. Cell 97:85-97. [DOI] [PubMed] [Google Scholar]
- 3.Ban, C., and W. Yang. 1998. Structural basis for MutH activation in E. coli mismatch repair and relationship of MutH to restriction endonucleases. EMBO J. 17:1526-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black, C. G., J. A. Fyfe, and J. K. Davies. 1997. Cloning, nucleotide sequence and transcriptional analysis of the uvrA gene from Neisseria gonorrhoeae. Mol. Gen. Genet. 254:479-485. [DOI] [PubMed] [Google Scholar]
- 5.Black, C. G., J. A. Fyfe, and J. K. Davies. 1998. Absence of an SOS-like system in Neisseria gonorrhoeae. Gene. 208:61-66. [DOI] [PubMed] [Google Scholar]
- 6.Black, C. G., J. A. Fyfe, and J. K. Davies. 1995. A promoter associated with the neisserial repeat can be used to transcribe the uvrB gene from Neisseria gonorrhoeae. J. Bacteriol. 177:1952-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boiteux, S., L. Gellon, and N. Guibourt. 2002. Repair of 8-oxoguanine in Saccharomyces cerevisiae: interplay of DNA repair and replication mechanisms. Free Radic. Biol. Med. 32:1244-1253. [DOI] [PubMed] [Google Scholar]
- 8.Bonner, C. A., S. K. Randall, C. Rayssiguier, M. Radman, R. Eritja, B. E. Kaplan, K. McEntee, and M. F. Goodman. 1988. Purification and characterization of an inducible Escherichia coli DNA polymerase capable of insertion and bypass at abasic lesions in DNA. J. Biol. Chem. 263:18946-18952. [PubMed] [Google Scholar]
- 9.Davidsen, T., E. K. Amundsen, E. A. Rodland, and T. Tonjum. 2007. DNA repair profiles of disease-associated isolates of Neisseria meningitidis. FEMS Immunol. Med. Microbiol. 49:243-251. [DOI] [PubMed] [Google Scholar]
- 10.Davidsen, T., M. Bjoras, E. C. Seeberg, and T. Tonjum. 2005. Antimutator role of DNA glycosylase MutY in pathogenic Neisseria species. J. Bacteriol. 187:2801-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davidsen, T., and T. Tonjum. 2006. Meningococcal genome dynamics. Nat. Rev. Microbiol. 4:11-22. [DOI] [PubMed] [Google Scholar]
- 12.de Laat, W. L., N. G. Jaspers, and J. H. Hoeijmakers. 1999. Molecular mechanism of nucleotide excision repair. Genes Dev. 13:768-785. [DOI] [PubMed] [Google Scholar]
- 13.Demple, B., and L. Harrison. 1994. Repair of oxidative damage to DNA: enzymology and biology. Annu. Rev. Biochem. 63:915-948. [DOI] [PubMed] [Google Scholar]
- 14.Falnes, P. O., and T. Rognes. 2003. DNA repair by bacterial AlkB proteins. Res. Microbiol. 154:531-538. [DOI] [PubMed] [Google Scholar]
- 15.Feig, D. I., L. C. Sowers, and L. A. Loeb. 1994. Reverse chemical mutagenesis: identification of the mutagenic lesions resulting from reactive oxygen species-mediated damage to DNA. Proc. Natl. Acad. Sci. USA 91:6609-6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frasch, C. E., and S. S. Chapman. 1972. Classification of Neisseria meningitidis group B into distinct serotypes. I. Serological typing by a microbactericidal method. Infect. Immun. 5:98-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freitag, N. E., H. S. Seifert, and M. Koomey. 1995. Characterization of the pilF-pilD pilus-assembly locus of Neisseria gonorrhoeae. Mol. Microbiol. 16:575-586. [DOI] [PubMed] [Google Scholar]
- 18.Friedberg, E. C., G. C. Walker, W. Siede, R. D. Wood, R. A. Schultz, and T. Ellenberger. 2006. DNA repair and mutagenesis, 2nd ed. ASM press, Washington, DC.
- 19.Giraud, A., I. Matic, O. Tenaillon, A. Clara, M. Radman, M. Fons, and F. Taddei. 2001. Costs and benefits of high mutation rates: adaptive evolution of bacteria in the mouse gut. Science 291:2606-2608. [DOI] [PubMed] [Google Scholar]
- 20.Grzesiuk, E., A. Gozdek, and B. Tudek. 2001. Contribution of E. coli AlkA, TagA glycosylases and UvrABC-excinuclease in MMS mutagenesis. Mutat. Res. 480-481:77-84. [DOI] [PubMed] [Google Scholar]
- 21.Gu, Y., A. Parker, T. M. Wilson, H. Bai, D. Y. Chang, and A. L. Lu. 2002. Human MutY homolog, a DNA glycosylase involved in base excision repair, physically and functionally interacts with mismatch repair proteins human MutS homolog 2/human MutS homolog 6. J. Biol. Chem. 277:11135-11142. [DOI] [PubMed] [Google Scholar]
- 22.Hall, L. M., and S. K. Henderson-Begg. 2006. Hypermutable bacteria isolated from humans-a critical analysis. Microbiology 152:2505-2514. [DOI] [PubMed] [Google Scholar]
- 23.Harris, S. L., D. A. Elliott, M. C. Blake, L. M. Must, M. Messenger, and P. E. Orndorff. 1990. Isolation and characterization of mutants with lesions affecting pellicle formation and erythrocyte agglutination by type 1 piliated Escherichia coli. J. Bacteriol. 172:6411-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hill, S. A. 2000. Neisseria gonorrhoeae recJ mutants show defects in recombinational repair of alkylated bases and UV-induced pyrimidine dimers. Mol. Gen. Genet. 264:268-275. [DOI] [PubMed] [Google Scholar]
- 25.Holten, E. 1979. Serotypes of Neisseria meningitidis isolated from patients in Norway during the first six months of 1978. J. Clin. Microbiol. 9:186-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin, S. G., J. H. Choi, B. Ahn, T. R. O'Connor, W. Mar, and C. S. Lee. 2001. Excision repair of adozelesin-N3 adenine adduct by 3-methyladenine-DNA glycosylases and UvrABC nuclease. Mol. Cell 11:41-47. [PubMed] [Google Scholar]
- 27.Kim, M., T. Huang, and J. H. Miller. 2003. Competition between MutY and mismatch repair at A·C mispairs in vivo. J. Bacteriol. 185:4626-4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kino, K., and H. Sugiyama. 2005. UVR-induced G-C to C-G transversions from oxidative DNA damage. Mutat. Res. 571:33-42. [DOI] [PubMed] [Google Scholar]
- 29.Kline, K. A., E. V. Sechman, E. P. Skaar, and H. S. Seifert. 2003. Recombination, repair and replication in the pathogenic Neisseriae: the 3 R's of molecular genetics of two human-specific bacterial pathogens. Mol. Microbiol. 50:3-13. [DOI] [PubMed] [Google Scholar]
- 30.Koomey, J. M., and S. Falkow. 1987. Cloning of the recA gene of Neisseria gonorrhoeae and construction of gonococcal recA mutants. J. Bacteriol. 169:790-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koomey, M., E. C. Gotschlich, K. Robbins, S. Bergstrom, and J. Swanson. 1987. Effects of recA mutations on pilus antigenic variation and phase transitions in Neisseria gonorrhoeae. Genetics 117:391-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuzminov, A. 1999. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol. Mol. Biol. Rev. 63:751-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lamers, M. H., A. Perrakis, J. H. Enzlin, H. H. Winterwerp, N. de Wind, and T. K. Sixma. 2000. The crystal structure of DNA mismatch repair protein MutS binding to a G × T mismatch. Nature 407:711-717. [DOI] [PubMed] [Google Scholar]
- 34.Luria, S. E., and M. Delbruck. 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28:491-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin, P., L. Sun, D. W. Hood, and E. R. Moxon. 2004. Involvement of genes of genome maintenance in the regulation of phase variation frequencies in Neisseria meningitidis. Microbiology 150:3001-3012. [DOI] [PubMed] [Google Scholar]
- 36.McGuinness, B. T., I. N. Clarke, P. R. Lambden, A. K. Barlow, J. T. Poolman, D. M. Jones, and J. E. Heckels. 1991. Point mutation in meningococcal porA gene associated with increased endemic disease. Lancet 337:514-517. [DOI] [PubMed] [Google Scholar]
- 37.Mehr, I. J., and H. S. Seifert. 1998. Differential roles of homologous recombination pathways in Neisseria gonorrhoeae pilin antigenic variation, DNA transformation and DNA repair. Mol. Microbiol. 30:697-710. [DOI] [PubMed] [Google Scholar]
- 38.Michaels, M. L., and J. H. Miller. 1992. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine). J. Bacteriol. 174:6321-6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morosini, M. I., M. R. Baquero, J. M. Sanchez-Romero, M. C. Negri, J. C. Galan, R. del Campo, J. C. Perez-Diaz, and F. Baquero. 2003. Frequency of mutation to rifampin resistance in Streptococcus pneumoniae clinical strains: hexA and hexB polymorphisms do not account for hypermutation. Antimicrob. Agents Chemother. 47:1464-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Napolitano, R., R. Janel-Bintz, J. Wagner, and R. P. Fuchs. 2000. All three SOS-inducible DNA polymerases (Pol II, Pol IV and Pol V) are involved in induced mutagenesis. EMBO J. 19:6259-6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nolte, O., M. Muller, S. Reitz, S. Ledig, I. Ehrhard, and H. G. Sonntag. 2003. Description of new mutations in the rpoB gene in rifampin-resistant Neisseria meningitidis selected in vitro in a stepwise manner. J. Med. Microbiol. 52:1077-1081. [DOI] [PubMed] [Google Scholar]
- 42.Nowosielska, A., S. A. Smith, B. P. Engelward, and M. G. Marinus. 2006. Homologous recombination prevents methylation-induced toxicity in Escherichia coli. Nucleic Acids Res. 34:2258-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Obmolova, G., C. Ban, P. Hsieh, and W. Yang. 2000. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature 407:703-710. [DOI] [PubMed] [Google Scholar]
- 44.Oliver, A., R. Canton, P. Campo, F. Baquero, and J. Blazquez. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251-1254. [DOI] [PubMed] [Google Scholar]
- 45.Otterlei, M., B. Kavli, R. Standal, C. Skjelbred, S. Bharati, and H. E. Krokan. 2000. Repair of chromosomal abasic sites in vivo involves at least three different repair pathways. EMBO J. 19:5542-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parkhill, J., M. Achtman, K. D. James, S. D. Bentley, C. Churcher, S. R. Klee, G. Morelli, D. Basham, D. Brown, T. Chillingworth, R. M. Davies, P. Davis, K. Devlin, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Leather, S. Moule, K. Mungall, M. A. Quail, M. A. Rajandream, K. M. Rutherford, M. Simmonds, J. Skelton, S. Whitehead, B. G. Spratt, and B. G. Barrell. 2000. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature 404:502-506. [DOI] [PubMed] [Google Scholar]
- 47.Price, M. N., K. H. Huang, E. J. Alm, and A. P. Arkin. 2005. A novel method for accurate operon predictions in all sequenced prokaryotes. Nucleic Acids Res. 33:880-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Projan, S. J., M. Monod, C. S. Narayanan, and D. Dubnau. 1987. Replication properties of pIM13, a naturally occurring plasmid found in Bacillus subtilis, and of its close relative pE5, a plasmid native to Staphylococcus aureus. J. Bacteriol. 169:5131-5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Richardson, A. R., and I. Stojiljkovic. 2001. Mismatch repair and the regulation of phase variation in Neisseria meningitidis. Mol. Microbiol. 40:645-655. [DOI] [PubMed] [Google Scholar]
- 50.Richardson, A. R., Z. Yu, T. Popovic, and I. Stojiljkovic. 2002. Mutator clones of Neisseria meningitidis in epidemic serogroup A disease. Proc. Natl. Acad. Sci. USA 99:6103-6107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seeberg, E. 1978. Reconstitution of an Escherichia coli repair endonuclease activity from the separated uvrA+ and uvrB+/uvrC+ gene products. Proc. Natl. Acad. Sci. USA 75:2569-2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seeberg, E., L. Eide, and M. Bjoras. 1995. The base excision repair pathway. Trends Biochem. Sci. 20:391-397. [DOI] [PubMed] [Google Scholar]
- 53.Seib, K. L., H. J. Tseng, A. G. McEwan, M. A. Apicella, and M. P. Jennings. 2004. Defenses against oxidative stress in Neisseria gonorrhoeae and Neisseria meningitidis: distinctive systems for different lifestyles. J. Infect. Dis. 190:136-147. [DOI] [PubMed] [Google Scholar]
- 54.Stein, D. C., R. J. Danaher, and T. M. Cook. 1991. Characterization of a gyrB mutation responsible for low-level nalidixic acid resistance in Neisseria gonorrhoeae. Antimicrob. Agents Chemother. 35:622-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Swanson, R. L., N. J. Morey, P. W. Doetsch, and S. Jinks-Robertson. 1999. Overlapping specificities of base excision repair, nucleotide excision repair, recombination, and translesion synthesis pathways for DNA base damage in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:2929-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tang, M., X. Shen, E. G. Frank, M. O'Donnell, R. Woodgate, and M. F. Goodman. 1999. UmuD′(2)C is an error-prone DNA polymerase, Escherichia coli Pol V. Proc. Natl. Acad. Sci. USA 96:8919-8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 58.Wagner, J., P. Gruz, S. R. Kim, M. Yamada, K. Matsui, R. P. Fuchs, and T. Nohmi. 1999. The dinB gene encodes a novel E. coli DNA polymerase, DNA pol IV, involved in mutagenesis. Mol. Cell 4:281-286. [DOI] [PubMed] [Google Scholar]
- 59.Wang, G., P. Alamuri, M. Z. Humayun, D. E. Taylor, and R. J. Maier. 2005. The Helicobacter pylori MutS protein confers protection from oxidative DNA damage. Mol. Microbiol. 58:166-176. [DOI] [PubMed] [Google Scholar]
- 60.Watson, M. E., Jr., J. L. Burns, and A. L. Smith. 2004. Hypermutable Haemophilus influenzae with mutations in mutS are found in cystic fibrosis sputum. Microbiology 150:2947-2958. [DOI] [PubMed] [Google Scholar]