Abstract
We have carried out a functional analysis of LysR family transcriptional regulators in Bacillus subtilis. The cell density of cultures of a yofA insertion mutant declined sharply after the end of exponential growth, as measured by optical density at 600 nm. Complementation in trans and analysis of isopropyl-β-d-thiogalactopyranoside (IPTG)-dependent growth of an inducible yofA strain confirmed that YofA contributes to the cell density of a culture after the end of exponential growth. Microscopic observation suggested that cell division is inhibited or delayed in the yofA mutant during entry into stationary phase. Analysis of the transcription of cell division genes revealed that the expression of ftsW is inhibited in yofA mutants, and overexpression of yofA, driven by a multiple-copy plasmid, enhances the induction of ftsW expression. These results suggest that YofA is required for the final round of cell division before entry into stationary phase and that YofA positively regulates ftsW expression. The defects caused by mutation of yofA were suppressed in strains carrying Pspac-ftsW in the presence of IPTG. Furthermore, maximal expression of yofA was observed at the onset of stationary phase, which coincided with the maximal ftsW expression. Our data indicate that YofA is involved in cell division through positive regulation of the expression of ftsW in B. subtilis.
Cell division and growth phase (i.e., vegetative or stationary-phase growth) are coordinated processes in bacteria. In liquid culture, the gram-positive soil bacterium Bacillus subtilis enters a nondividing state after the end of exponential growth. Cells terminate cell division and enter into stationary phase to ensure survival under starvation conditions. Under certain conditions, the bacterium may receive a complex series of internal and external signals for sporulation and initiate a developmental program for spore formation (9, 19, 39) rather than complete cell division and enter into stationary phase (31). The mechanism of initiation of sporulation is well understood (9, 19, 39, 43). In contrast, the mechanism that regulates commitment to a final round of cell division and entry into stationary phase has not been well established.
The division of rod-shaped bacterial cells has been studied primarily in the model organisms Escherichia coli and B. subtilis. These bacteria grow by elongation of the long axis to form a rod, followed by cell division, which occurs at the midpoint of the rod. It has been demonstrated that septum assembly is mediated by a number of proteins that localize to the division site. Among the proteins known to localize to this site in E. coli are FtsZ, FtsA, FtsQ, FtsL, YqbQ, FtsW, FtsI (penicillin-binding protein B [PBPB]), FtsN, and ZipA (13, 44). In B. subtilis, eight cell division proteins have been shown to localize to the division site. They are FtsZ, FtsA, YtpT, FtsQ (DivIB), FtsL, DivIC, FtsW (YlaO), and PBPB (8, 10). The best-characterized protein component of the division site is FtsZ, which forms a cytoskeletal structure called the FtsZ ring. FtsZ is a highly conserved protein that appears to be widely present in prokaryotic cells (6, 16). The FtsZ ring and its associated proteins at the division site are called the divisome. Overall, the mechanisms of cell division are similar in E. coli and B. subtilis, although some aspects of the assembly of the protein complex at the division site differ. In E. coli, for example, FtsW is an integral membrane protein that is required for subsequent recruitment of its cognate transpeptidase, FtsI (32, 36). In contrast, B. subtilis FtsW is thought to be involved in the function of the PBP proteins, but the mechanism by which it does so is unknown.
Our lab is in the process of carrying out a functional analysis of LysR-type regulators in B. subtilis (30, 41). LysR family members typically contain an N-terminal helix-turn-helix motif and function as positive regulators of target promoters and negative autoregulators (41). In B. subtilis seven members of this family of regulators have been well investigated, whereas the functions of an additional 12 LysR-type regulators are unknown (Table 1) (30).
TABLE 1.
LysR-type regulator genes in B. subtilis
| Gene | Function | Reference(s) |
|---|---|---|
| alsR | Activation of acetoin production genes (alsSD) | 37 |
| ccpC | Repression of the aconitase gene (citB) | 28, 29 |
| citR | Repression of the citrate synthase gene (citA) | 26, 27 |
| cysL | Activation of cysteine biosynthesis genes (cysJI) | 15 |
| gltC | Activation of glutamate biosynthesis genes (gltAB) | 1, 3 |
| gltR | Activation of glutamate biosynthesis genes (gltAB) | 2 |
| ytlI | Activation of the sulfur metabolism genes (ytmI operon) | 5 |
| ycgK | Unknown | |
| yclA | Unknown | |
| yoaU | Unknown | |
| yofA | This study | |
| yraN | Unknown | |
| yrdQ | Unknown | |
| yusT | Unknown | |
| yvbU | Unknown | |
| ywbI | Unknown | |
| ywqM | Unknown | |
| yxJO | Unknown | |
| yybE | Unknown |
We report here the identification of a novel cell division-associated function for the LysR-type protein YofA, 1 of the 12 LysR-type regulators whose function was unknown. We show that YofA is essential for cell viability during stationary-phase growth of B. subtilis. We also show that maximal expression of ftsW at the transition from exponential growth to stationary phase is regulated by YofA and that the final round of cell division before entry into stationary phase is a prerequisite for cellular survival during stationary phase.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Most of the bacterial strains and plasmids used in this study are listed in Table 2. The exceptions are the ycgK, yclA, yoaU, yraN, yrdQ, yusT, yvbU, ywbI, ywqM, yxjO, and yybE mutant strains. These strains were constructed as part of European and Japanese projects for functional characterization of the B. subtilis genome and are listed on the following websites: http://locus.jouy.inra.fr/cgibin/genmic/madbase/progs/madbase.operl and http://bacillus.genome.ad.jp.
TABLE 2.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype, phenotype, and relevant characteristics | Source, reference, or constructiona |
|---|---|---|
| E. coli JM105 | supE endA sbcB15 hsdR4 rpsL thi Δ(lac-proAB) F′ [traD36 proAB+lacIqlacZΔM15] | 45 |
| B. subtilis strains | ||
| 168 | trpC2 | Laboratory stock |
| ZL001 | trpC2 yofA::pUCN192 | This study |
| ZL002 | trpC2 yofA::pMUTinyofA (Pspac-yofA) | This study |
| ZL003 | trpC2 thrC:: pTCCyofA (PyofA-yofA) | This study |
| ZL004 | trpC2 yofA:: pUCN192 thrC::pTCCyofA | ZL001 → ZL003 |
| ZL005 | trpC2 thrC::pTCCZ1 (ftsAZ-lacZ) | This study |
| ZL006 | trpC2 thrC::pTCCZ2 (divIB-lacZ) | This study |
| ZL007 | trpC2 thrC::pTCCZ3 (divIC-lacZ) | This study |
| ZL008 | trpC2 thrC::pTCCZ4 (ftsL-pbpB-lacZ) | This study |
| ZL009 | trpC2 thrC::pTCCZ5 (rodA-lacZ) | This study |
| ZL010 | trpC2 thrC::pTCCZ6 (ftsW-lacZ) | This study |
| ZL011 | trpC2 yofA::pUCN192 thrC::pTCCZ6 | ZL001 → ZL010 |
| ZL012 | trpC2 Pxyl-yofA (multicopy yofA) | This study |
| ZL013 | trpC2 Pxyl-yofA thrC::pTCCZ6 | ZL012 → ZL010 |
| ZL014 | trpC2 ftsW::pMUTinftsW (Pspac-ftsW) | This study |
| ZL015 | trpC2 yofA::pUCN192 ftsW::pMUTinftsW | ZL001 → ZL014 |
| ZL016 | trpC2 yofA::pUCN192 thrC::pTCCZ1 | ZL001 → ZL005 |
| ZL017 | trpC2 yofA::pUCN192 thrC::pTCCZ2 | ZL001 → ZL006 |
| ZL018 | trpC2 yofA::pUCN192 thrC::pTCCZ3 | ZL001 → ZL007 |
| ZL019 | trpC2 yofA::pUCN192 thrC::pTCCZ4 | ZL001 → ZL008 |
| ZL020 | trpC2 yofA::pUCN192 thrC::pTCCZ5 | ZL001 → ZL009 |
| Plasmids | ||
| pMUTinT3 | Vector carrying bla erm | 35 |
| pMUTinyofA | pMUTinT3 carrying Pspac-yofA | This study |
| pMUTinftsW | pMUTinT3 carrying Pspac-ftsW | This study |
| pUCN192 | Vector carrying bla neo | 20 |
| pUCNyofA | pUCN192 carrying an internal region of yofA gene | This study |
| pTCC1 | Integration vector at thrC carrying bla cat | 24 |
| pMF20 | Vector carrying bla cat | 34 |
| pHY300PLK | Vector carrying bla tet | 25 |
| pTCCZ | pTCC1 carrying the coding region of lacZ gene | This study |
| pTCCZ1 | pTCCZ carrying the upstream region of ftsA (ftsAZ-lacZ) | This study |
| pTCCZ2 | pTCCZ carrying the upstream region of divIB (divIB-lacZ) | This study |
| pTCCZ3 | pTCCZ carrying the upstream region of divIC (divIC-lacZ) | This study |
| pTCCZ4 | pTCCZ carrying the upstream region of ftsL (ftsL-pbpB-lacZ) | This study |
| pTCCZ5 | pTCCZ carrying the upstream region of rodA (rodA-lacZ) | This study |
| pTCCZ6 | pTCCZ carrying the upstream region of ftsW (ftsW-lacZ) | This study |
| pTCCyofA | pTCC1 carrying native yofA | This study |
| pHYXY1 | Vector carrying bla tet | This study |
| pHYXYyofA | pHYXY1 carrying the entire yofA open reading frame | This study |
Arrows indicate transformation from donor DNA to recipient strain.
The oligonucleotide primers used for PCR amplification are listed in Table 3. E. coli JM105 was the host for all plasmid construction. B. subtilis strain 168 (wild type) served as the host for all strain construction. Transformation of B. subtilis was performed according to standard procedures (7).
TABLE 3.
Oligonucleotide primers used in this study
| Primer | Sequence (5′ to 3′)a | Description | Locationb | Restriction site |
|---|---|---|---|---|
| yofAF | CCCAAGCTTGGAAGCATAACGAAAGCA | yofA sense sequence | 45 | HindIII |
| yofAR | CGCGGATCCCACTGATAAATCCACTTC | yofA antisense sequence | 369 | BamHI |
| yofAF1 | CCCAAGCTTCATAGCAAGCAGGTGAG | yofA sense sequence | −22 | HindIII |
| yofAR1 | CGCGGATCCCAGATGCGTTACCGCCAT | yofA antisense sequence | 315 | BamHI |
| yofAfor | AAAACTGCAGAATCCCAGTTCAATGTCGG | yofA sense sequence | −403 | PstI |
| yofArev | TGCTCTAGATACTCGCTTCAAATGAG | yofA antisense sequence | 948 | XbaI |
| yofABamHI | CGCGGATCCCATAGCAAGCAGGTGAG | yofA sense sequence | −22 | BamHI |
| yofAHindIII | CCCAAGCTTTACTCGCTTCAAATGAG | yofA antisense sequence | 948 | HindIII |
| ftsAZF | AAAACTGCAGGTTTCCGGTTTCTTTTTT | ftsA sense sequence | −365 | PstI |
| ftsAZR | TGCTCTAGATTTCTATTCTATTATTTG | ftsA antisense sequence | −18 | XbaI |
| divIBF | AAAACTGCAGGACAGTTATGGTCGGAAC | divIB sense sequence | −2883 | PstI |
| divIBR | TGCTCTAGACTGTTAAAAGTCTGTCTA | divIB antisense sequence | −30 | XbaI |
| divICF | AAAACTGCAGCGACACAATTCTATACAA | divIC sense sequence | −643 | PstI |
| divICR | TGCTCTAGAATCTCTTCAAAACCGT | divIC antisense sequence | −20 | XbaI |
| ftsLpbpBF | AAAACTGCAGCTTTTTATGGGTAAACA | ftsL sense sequence | −1564 | PstI |
| ftsLpbpBR | TGCTCTAGAATTTTGCGTTCGCGTTTC | ftsL antisense sequence | −21 | XbaI |
| rodAF | AAAACTGCAGTGCGGCGATATGAGCGAC | rodA sense sequence | −711 | PstI |
| rodAR | TGCTCTAGATCACTAATGTTTATTATA | rodA antisense sequence | −31 | XbaI |
| ftsWF | AAAACTGCAGTGGACGCTGAGAAGATTT | ftsW sense sequence | −432 | PstI |
| ftsWR | TGCTCTAGAATTATCTATGGTTTTTAT | ftsW antisense sequence | −28 | XbaI |
| ftsWF1 | CCCAAGCTTAGAAGCAGGGAAGAGGATG | ftsW sense sequence | −21 | HindIII |
| ftsWR1 | CGCGGATCCGACTACCGGAGGGCTACT | ftsW antisense sequence | 453 | BamHI |
| PXYP2 | CGCGGATCCCCGGCATTCAAATACAG | xylR sense sequence | −322 | BamHI |
| PXYR | CCGGAATTCTGCCATGTCACTATTGC | xylR antisense sequence | 1133 | EcoRI |
| yofARTF | GGAAGCATAACGAAAGCA | yofA sense sequence | 46 | |
| yofARTR | GCAGATGCGTTACCGCC | yofA antisense sequence | 299 | |
| ftsWRTF | CTCACTGATATTCGCAAT | ftsW sense sequence | 32 | |
| ftsWRTR | ACTACCGGAGGGGCTACT | ftsW antisense sequence | 452 | |
| rpsRRTF | GCAGAGGCGGTCGTGCGAAA | rpsR sense sequence | 14 | |
| rpsRRTR | ACGTGCGCGTTTGATCGCTGCA | rpsR antisense sequence | 183 |
Additional sequences and restriction sites that do not correspond to the sequences of genes are indicated by boldface type and underlining, respectively.
The locations are the 3′-end positions of the primers corresponding to the number of nucleotides from the initiation codons of the genes.
Construction of plasmids and bacterial strains.
pUCNyofA carries an internal fragment of yofA, which was amplified by PCR using primers yofAF and yofAR (Table 3). The amplified PCR product was digested with BamHI and HindIII and ligated into the corresponding sites of pUCN192 (20).
To construct the conditional yofA and ftsW mutants, DNA fragments that corresponded to nucleotides (nt) −22 to 315 of yofA and −21 to 435 of ftsW relative to the transcriptional start site at nt 1 were amplified by PCR with primers yofAF1 and yofAR1 and primers ftsWF1 and ftsWR1, respectively. After digestion with BamHI and HindIII, the PCR fragments were inserted into the corresponding sites of the Pspac integrational vector pMUTinT3 (35) to create pMUTinyofA and pMUTinftsW. Wild-type B. subtilis was transformed with pMUTinyofA and pMUTinftsW to generate the fusion strains ZL002 (Pspac-yofA) and ZL014 (Pspac-ftsW), in which expression of yofA and ftsW, respectively, was driven by the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible Pspac promoter.
To construct pTCCZ, the coding region of the lacZ gene from pMUTinT3 was subcloned into the thrC integration vector pTCC1 (24) using the PstI and XbaI restriction sites. To construct pTCCyofA, a DNA fragment containing the open reading frame and the promoter region of yofA was amplified with primers yofAfor and yofArev. The amplified PCR product was digested with PstI and XbaI and ligated into the corresponding sites of pTCC1. Before transformation of B. subtilis, the plasmid was linearized by digestion with ScaI.
To construct pTCCZ1, pTCCZ2, pTCCZ3, pTCCZ4, pTCCZ5, and pTCCZ6, DNA fragments of ftsA (nt −365 to −18), divIB (nt −2883 to −30), divIC (nt −643 to −20), ftsL (nt −1564 to −21), rodA (nt −711 to −31), and ftsW (nt −432 to −28) were amplified by PCR with primer pairs ftsAZF/ftsAZR, divIBF/divIBR, divICF/divICR, ftsLpbpBF/ftsLpbpBR, rodAF/rodAR, and ftsWF/ftsWR, respectively. Amplified PCR products were digested with PstI and XbaI and then ligated into the corresponding sites of pTCCZ. Before transformation into B. subtilis, the plasmids were linearized by digestion with ScaI.
To construct pHYXY1, the coding regions of Pxyl and xylR from pMF20 (34) were amplified with primers PXYP2 and PXYR. The amplified PCR product was completely digested with BamHI and EcoRI and ligated into the corresponding sites of pHY300PLK (25).
For overexpression of yofA in B. subtilis, the entire yofA open reading frame was amplified using primers yofABamHI and yofAHindIII. The amplified PCR product was digested with BamHI and HindIII and ligated into the corresponding sites of pHYXY1 to generate the multicopy plasmid pHYXYyofA, in which expression of yofA was driven by the xylose-inducible Pxyl promoter.
β-Galactosidase assay.
Bacteria were grown in LB medium at 37°C and harvested at the indicated times by centrifugation. β-Galactosidase activity was assayed as described previously (33), using o-nitrophenyl-β-d-galactopyranoside as the substrate. Specific activity was calculated relative to the optical density at 600 nm (OD600) of each sample and is expressed below in nanomoles of substrate (o-nitrophenyl-β-d-galactopyranoside) hydrolyzed per milligram of protein per minute.
Fluorescence microscopy.
Cells were grown at 37°C in LB medium supplemented with FM4-64 (final concentration, 0.5 μg/ml; Molecular Probes) for labeling cell membranes and with SYTO16 (final concentration, 5 μg/ml; Molecular Probes) for labeling of cell nucleotides. A portion (2 μl) of each sample was mounted on a glass slide coated with 0.1% poly-l-lysine (Sigma), and slides were viewed by microscopy using an Olympus BX50 microscope with a 100× UplanApo objective lens. Images were captured using a SenSys charge-coupled camera device (Photometrics). FM4-64 and SYTO16 were visualized using a fluorescence isothiocyanate filter set (Olympus) and a wide interference green filter set (Olympus), respectively. Photos were viewed and analyzed using the Metamorph, version 6.1, software (Universal Image) and Adobe Photoshop, version 7.0.
RT-PCR experiments.
Wild-type bacterial cells were grown in LB medium at 37°C, and samples were removed for analysis 2 and 1 h before the end of log phase and 1, 2, 3, 4, and 5 h after the end of log phase. Total RNA was extracted from the cells at the indicated time points, as described previously (22), and used as the template for reverse transcription (RT)-PCR analysis. Primer pairs yofARTF-yofARTR, ftsWRTF-ftsWRTR, and rpsRRTF-rpsRRTR were used to amplify yofA, ftsW, and rpsR, respectively (Table 3). Prior to RT-PCR, RNA was treated with DNase I (TAKARA) to remove any residual DNA. RT-PCR was performed using 0.5 μg of total RNA and an RNA PCR kit (TAKARA) according to the manufacturer's instructions. cDNA was amplified using an Ex Taq PCR kit (TAKARA). To obtain semiquantitative RT-PCR results, the number of PCR cycles was limited to 27, which is in the log-linear range of amplification. mRNA encoding ribosomal protein S18 (rpsR) was used as an internal standard to control for variations in the amount of total RNA used as the starting material. We assumed that the levels of rpsR mRNA were unaffected by the time of incubation of cells. The level of rpsR mRNA was also used to rule out the presence of contaminating chromosomal DNA (data not shown).
RESULTS
YofA is a regulator of cell growth in B. subtilis.
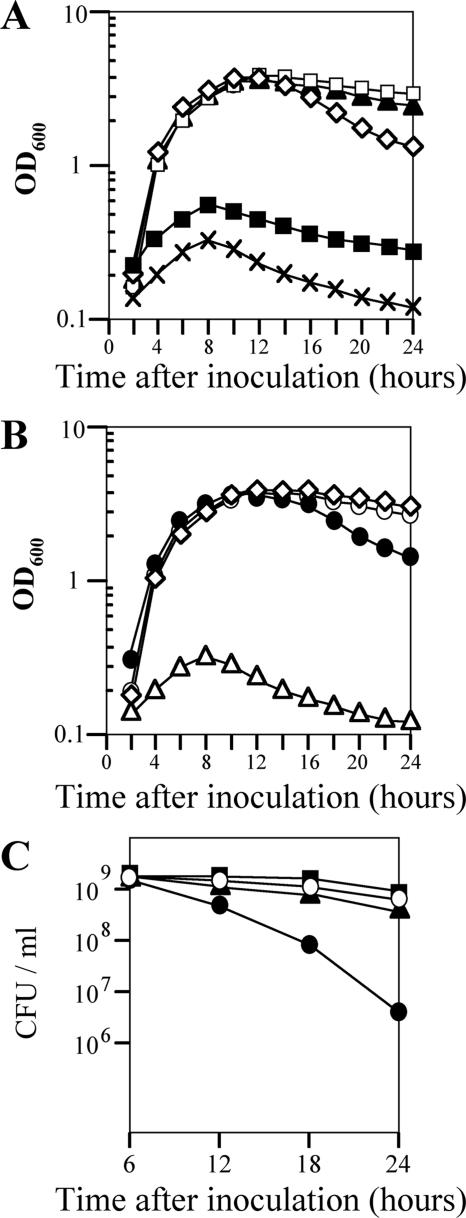
As an initial step in the characterization of the 12 LysR-type regulators with unknown functions, we first examined the growth of 12 mutant strains of B. subtilis in LB medium at 37°C under nonsporulating conditions. These strains carried mutations in ycgK, yclA, yoaU, yofA, yraN, yrdQ, yusT, yvbU, ywbI, ywqM, yxjO, and yybE (30), and the growth rate was determined by monitoring the rate of increase of the OD600 of the cultures. Wild-type B. subtilis and all of the mutant strains except the yofA strain exhibited similar growth patterns (data not shown). The cell density of cultures of the yofA mutant strain declined sharply 14 to 24 h after inoculation (Fig. 1A). This decline continued up to 3 days after inoculation (OD600 at 3 days, ∼0.8) (data not shown). The growth phenotype of the yofA mutant was confirmed by complementation experiments, in which yofA was supplied in trans, and in an inducible expression system, in which the gene was placed under the control of the Pspac promoter. An approximately 1.5-kb segment of DNA (nt −403 to 948) that contained the yofA locus was inserted into the B. subtilis thrC gene to generate the expression vector pTCCyofA. When pTCCyofA was introduced into yofA mutant cells (strain ZL004), normal growth was restored (Fig. 1B). We next examined the growth of a yofA conditional mutant. The yofA gene is located between yogA and ggt and is transcribed in a different direction than these two genes. Thus, it appears that yofA is monocistronic. We fused the ribosome binding site and the first 105 codons of the yofA gene to the inducible Pspac promoter, generating pMUTinyofA. Introduction of pMUTinyofA into wild-type strain 168 resulted in chromosomal integration of a full-length copy of yofA under the control of the Pspac promoter, which is repressed by the LacI repressor (strain ZL002). We then examined the growth of strain ZL002 (carrying Pspac-yofA) in LB medium in the absence and presence of various concentrations of inducer. An overnight culture grown in the presence of 0.05 mM IPTG was used to inoculate liquid cultures, and the growth rate was observed in the presence of 0.05 or 1 mM IPTG (Fig. 1C). The results showed that YofA is essential for cell viability during stationary phase and that a certain threshold level of YofA is required for cell survival during stationary phase.
FIG. 1.
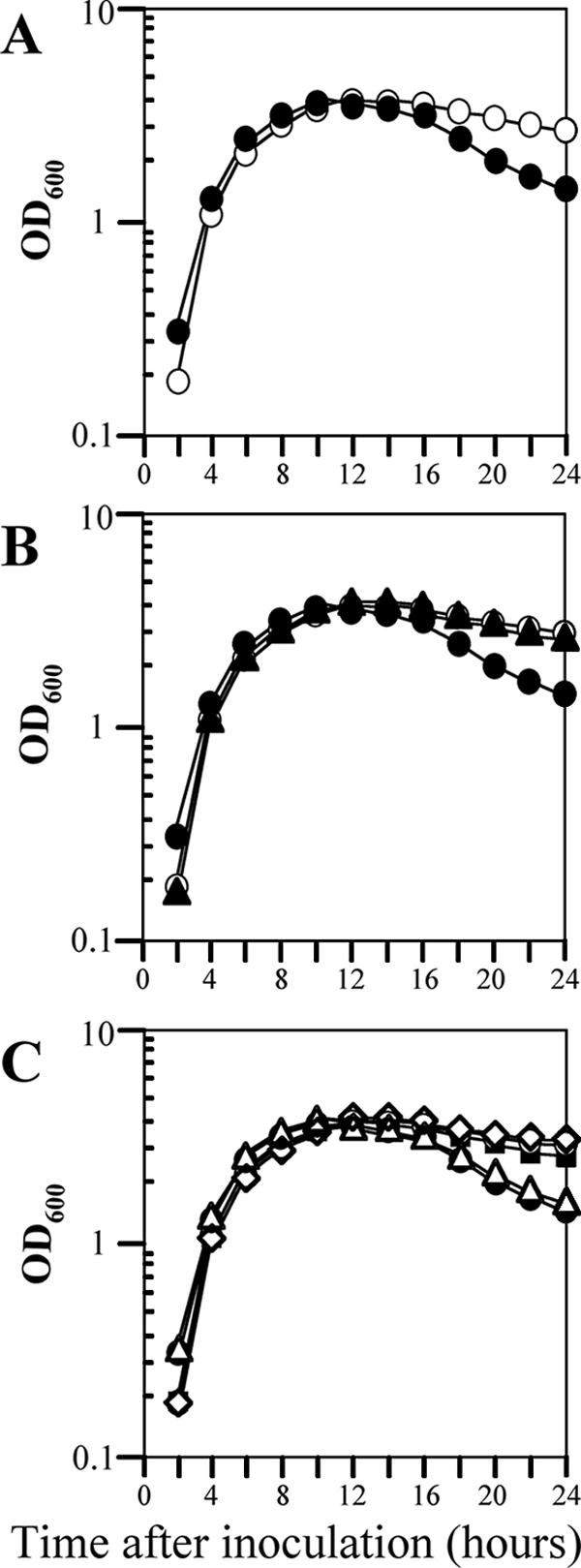
Effect of yofA expression on cell growth. Cells were grown in LB broth at 37°C to stationary phase. (A) Growth assay of wild-type 168 (○) and ZL001 (yofA::neo) (•) cells. (B) Complementation of yofA deficiency by introduction of thrC::yofA in trans: growth assay of wild-type 168 (○), ZL001 (yofA::neo) (•), and ZL004 (yofA::neo thrC::yofA) (▴) cells. (C) Growth of wild-type 168 (○), ZL001 (yofA::neo) (•), and ZL002 (Pspac-yofA) cells in liquid LB medium in the absence of IPTG (▵) or in the presence of the following concentrations of IPTG: 0.05 mM (▪) and 1 mM (⋄). Growth was determined by measuring the OD600, and the data represent the means of three independent experiments.
To investigate whether YofA was required for sporulation, we examined the growth of the yofA mutant strain in DS medium (sporulating conditions) at 37°C. However, there was no difference in the growth curve during sporulation between wild-type and yofA mutant cells (data not shown). We also examined the effect of yofA mutation on the activation of the sporulation-specific sigma factors, σF and σE. We found no differences in the expression patterns of the σF-regulated gene spoIIQ and the σE-regulated gene spoIID in DS medium between wild-type and yofA mutant cells (data not shown). Thus, the effect of yofA appeared to be specific for the transition from log phase to stationary phase for cultures incubated in LB medium.
YofA controls cell viability and the formation of constrictions during cell division.
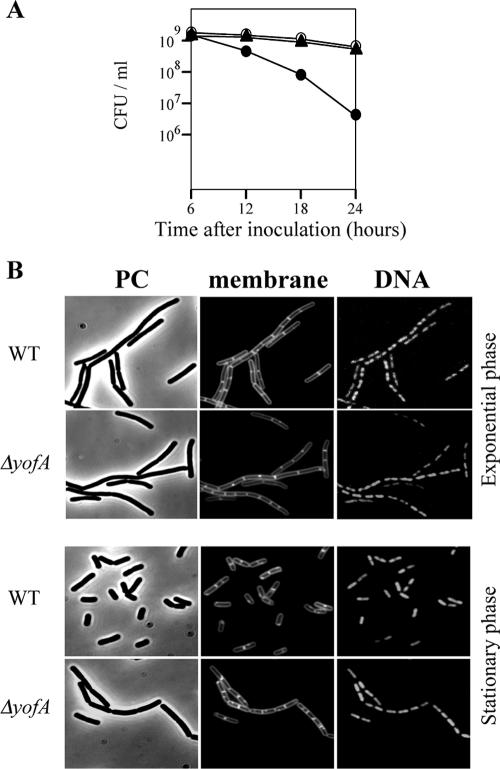
To characterize the underlying defect in yofA mutant cells, we first performed a colony formation assay. During vegetative phase, wild-type and yofA mutant cells displayed similar numbers of viable cells (data not shown). However, as the yofA mutant strain entered stationary phase (12, 18, or 24 h after inoculation), there was a sharp decline in the number of CFU/ml, resulting in a 100-fold decrease 24 h after inoculation compared to wild-type or ZL004 cells (Fig. 2A). Thus, the yofA mutant exhibited a decreased ability to grow in culture, most likely due to a loss of viability.
FIG. 2.
Effect of deletion of yofA on the viability and morphology of B. subtilis. (A) Colony formation assay of wild-type 168 (○), ZL001 (yofA::neo) (•), and ZL004 (yofA::neo thrC::yofA) (▴) cells. The data represent the means of three independent experiments. (B) Fluorescence microscopy of ZL001 (ΔyofA) and wild-type 168 (WT) cells during exponential growth and at 4 h after the end of exponential phase. Cells were treated with FM4-64 and SYTO16 to visualize the membranes and DNA, respectively. PC, phase contrast.
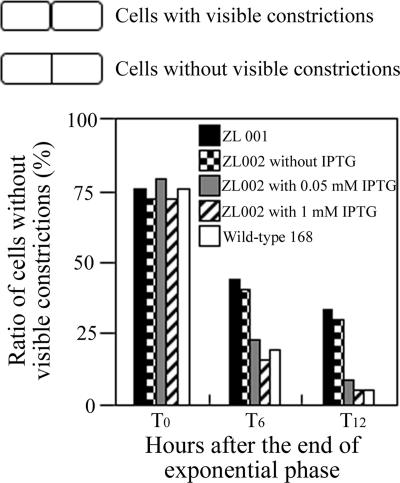
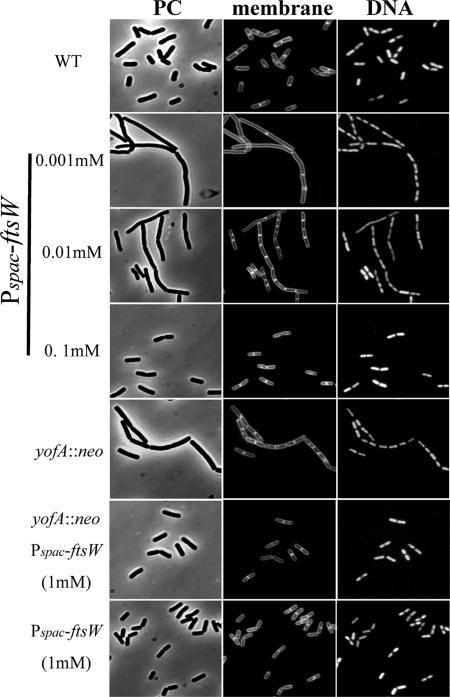
We next examined the cytological features of the yofA mutants. Samples were taken from early-stationary-phase cultures, and cells were double stained with SYTO16 and FM4-64 to visualize cytoplasmic nucleic acids and the cell membrane, respectively. Phase-contrast and fluorescence microscopy revealed that vegetative cells of the wild-type and yofA mutant strains were similar in shape and that the percentages of dividing cells were similar (Fig. 2B). Upon incubation for 4 h after entry into stationary phase, wild-type cells exhibited a dispersed distribution pattern, while yofA mutant cells displayed a highly filamentous morphology (Fig. 2B). The mutant cells at this time point were elongated with segments, and ghost cells were occasionally observed. In addition, the elongated mutant cells contained no visible constrictions of the cell wall and cell membrane at their septa, which suggested that yofA functions during cell division (38, 42). In wild-type B. subtilis, dividing cells can be identified by a visible constriction at the site of the septum, which is involved in the generation of two newborn daughter cells. We quantified the number of cells with constrictions in wild-type cells and yofA mutants harvested 0 to 12 h after the end of the exponential growth phase (Fig. 3). At the end of exponential phase, approximately 78% of wild-type and yofA mutant cells had no visible constrictions. However, 6 and 12 h after the end of log phase, the fraction of yofA mutant cells that had no visible membrane constrictions was larger than that of wild-type cells (45% compared to 20% at 6 h after the end of log phase and 33% compared to 5% at 12 h after the end of log phase) (Fig. 3). We also examined the population of dividing cells in the yofA conditional mutant strain ZL002. In the presence of 1 mM IPTG, the fraction of dividing cells without visible constrictions for strain ZL002 was similar to that for wild-type cells, whereas in the absence of IPTG, it was similar to that for the yofA mutant (Fig. 3). These results indicated that cell division is inhibited or delayed in a yofA mutant during entry into stationary phase. Thus, YofA appeared to be essential in the regulation of the final round of division before entry into stationary phase.
FIG. 3.
Effect of yofA mutation on cell division: ratio of the number of cells lacking visible constrictions to the total number of cells in ZL001 (yofA::neo), wild-type 168, and ZL002 (Pspac-yofA) cultures in the absence of IPTG or in the presence of 0.05 and 1 mM IPTG. The data represent the means of three independent experiments.
YofA controls the expression of ftsW, a cell division protein.
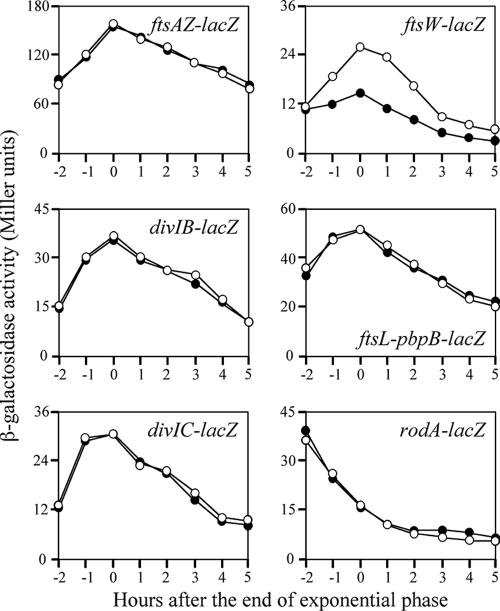
The initiation of cell division involves the formation of a ring of FtsZ protein around the inner membrane of the cell at the midcell division site. Other division proteins, such as FtsA, FtsW (YlaO), FtsL, DivIB, DivIC, and PBPB, are believed to act after the formation of the FtsZ ring (10). RodA is a transmembrane protein that is involved in cell elongation and is required for the synthesis of peptidoglycan (18). To investigate whether depletion of YofA affected the expression of these division or elongation genes, we constructed several additional yofA mutant strains. The coding regions of ftsA, ftsW, divIB, divIC, pbpB, and rodA-lacZ were inserted into the thrC locus of the yofA mutant strain, which generated transcriptional lacZ fusions of each division or elongation gene. As a control, the gene fusions were also generated in a wild-type background. Strains were grown in LB medium at 37°C, and β-galactosidase activity was measured. As shown in Fig. 4, the expression of ftsAZ, divIB, divIC, ftsL-pbpB, and rodA in the yofA mutant background was similar to that in the wild-type cells. However, the transcription of ftsW was partially blocked in the yofA mutant. In wild-type cells, β-galactosidase activity of the ftsW gene fusion gradually increased during exponential phase and then declined during stationary phase. In the yofA mutant background, ftsW expression reached a peak level at the end of exponential phase and declined thereafter, but it was significantly lower at all time points than the expression in wild-type cells. These results suggested that the expression of ftsW is dependent on YofA. Of note, we observed a high level of expression of rodA in both wild-type and yofA mutant cells during vegetative phase, whereas transcription of the other five genes, including ftsW, reached a maximum level upon entry into stationary phase.
FIG. 4.
Effect of yofA mutation on the expression of cell division genes. Wild-type (○) and yofA::neo (•) cells were grown in LB medium, and the β-galactosidase specific activities of the following reporter genes were examined: ftsAZ-lacZ, divIB-lacZ, divIC-lacZ, ftsW-lacZ, ftsL-pbpB-lacZ, and rodA-lacZ. The data represent the means of three independent experiments.
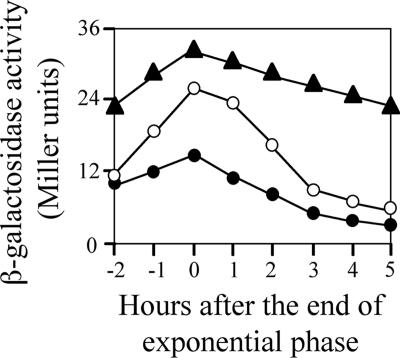
We next examined whether overexpression of YofA elevated the expression of ftsW. The wild type carrying the ftsW-lacZ fusion gene (strain ZL010, thrC::ftsW-lacZ) was transformed with a xylose-inducible yofA expression vector (pHYXYyofA). As shown in Fig. 5, induction of overexpression of yofA by incubation of the cells in 10 mM xylose resulted in enhanced expression of ftsW in both early stationary phase and late stationary phase. These results suggested that YofA regulates the transcription of ftsW.
FIG. 5.
Effect of yofA overexpression on the activity of an ftsW-lacZ reporter gene: β-galactosidase activity of ZL010 (wild-type) (○) and ZL011 (yofA::neo) (•) cells and of ZL013 (Pxyl-yofA ftsW-lacZ) cells supplemented with 10 mM xylose (▴). The data represent the means of three independent experiments.
Induction of FtsW overcomes the final cell division defect caused by yofA mutation during entry into stationary phase.
We next examined whether the final cell division defect caused by yofA mutation at entry into stationary phase could be suppressed by induction of ftsW. The FtsW protein is an integral membrane protein, and its function in B. subtilis is poorly characterized. Therefore, we first examined the growth phenotype of an inducible ftsW strain of B. subtilis. We fused the ribosome binding site and the first 151 codons of ftsW to the Pspac promoter, generating the expression vector pMUTinftsW. Integration of pMUTinftsW into the chromosome of wild-type 168 cells created a strain in which ftsW was under the control of the LacI-repressible, IPTG-inducible Pspac promoter (strain ZL014, Pspac-ftsW). The growth rate of strain ZL014 in the absence or presence of different concentrations of IPTG was determined by monitoring the rate of increase in the OD600. As shown in Fig. 6A, there was a severe growth defect of the Pspac-ftsW cells in liquid medium in the absence of IPTG, suggesting that ftsW is essential for cell growth. We also examined ZL014 cells by microscopy after treatment with different concentrations of IPTG. Cells were double labeled with SYTO16 and FM4-64 to visualize cytoplasmic nucleic acids and the cell membrane, respectively. As shown in Fig. 7, ZL014 cells were extremely filamentous in the presence of low concentrations of IPTG (0.001 to ∼0.01 mM), indicating that there was a block in cell division under these conditions. In contrast, in the presence of 0.1 mM IPTG, the shape of the cells was similar to the shape of wild-type cells. These results suggested that ftsW is required for cell division in B. subtilis.
FIG. 6.
Suppression of growth and viability defects of the yofA mutant upon induction of FtsW. (A) Effect of FtsW induction on growth. The growth of ZL014 (Pspac-ftsW) cells in liquid LB medium in the absence of IPTG (×) or in the presence of the following concentrations of IPTG was determined: 0.001 mM (▪), 0.01 mM (⋄), 0.1 mM (▴), and 1 mM (□). (B) Effect of FtsW induction on growth of yofA mutants: growth of wild-type 168 (○) and ZL001 (yofA::neo) (•) cells and of ZL015 (yofA::neo Pspac-ftsW) cells in liquid LB medium in the absence (▵) or presence of 1 mM IPTG (⋄). (C) Effect of FtsW induction on colony formation of yofA mutants: CFU/ml for wild-type 168 (○), ZL001 (yofA::neo) (•), ZL014 (Pspac-ftsW) in the presence of 1 mM IPTG (▪), and ZL015 (yofA::neo Pspac-ftsW) supplemented with 1 mM IPTG (▴). The data represent the means of three independent experiments, and mean OD600 values are shown.
FIG. 7.
Suppression of the cell division defect of yofA mutants by induction of FtsW: fluorescence micrography of wild-type 168 (WT), ZL001 (yofA::neo), ZL015 (yofA::neo Pspac-ftsW) in the presence of 1 mM IPTG, and ZL014 (Pspac-ftsW) cells in the presence of 0.001 mM, 0.01 mM, 0.1 mM, and 1 mM IPTG. Cells incubated until 4 h after the end of exponential phase were stained with FM4-64 and SYTO16 to visualize cell membranes and DNA, respectively. PC, phase contrast.
To determine whether induction of FtsW overcomes the cell division defect caused by yofA mutation, we analyzed the cell growth (OD600) and colony formation (CFU/ml) of strain ZL015 (Pspac-ftsW yofA::neo) in the presence or absence of IPTG. As shown in Fig. 6B, under inducing conditions (1 mM IPTG), the rate of increase of the OD600 of strain ZL015 was similar to that of wild-type cells. In the presence of 0.01 mM IPTG, there was a sharp decline in the OD600 of strain ZL014 compared to that of the wild type during stationary phase. The yofA mutant exhibited a similar pattern of growth. Under inducing conditions (1 mM IPTG), ZL015 cells also displayed a number of CFU/ml similar to that of wild-type cells (Fig. 6C), suggesting that the growth phenotype of the yofA mutant during stationary phase was completely suppressed by the induction of FtsW. We also examined the morphology of ZL015 cells in the presence of IPTG by fluorescence microscopy. Induction of FtsW resulted in suppression of the final cell division defect caused by yofA mutation during entry into stationary phase (Fig. 7). These results indicated that cellular survival during stationary phase requires maximal expression of ftsW, which is controlled by YofA.
Transcription of yofA increased to the maximum level at entry into stationary phase.
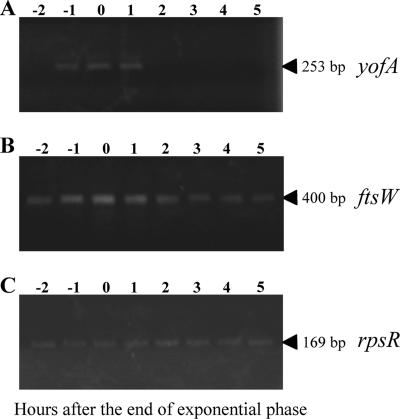
To examine the expression profile of yofA, we generated a strain carrying a lacZ gene fusion of yofA, in which the lacZ gene was integrated into the yofA locus. When we measured β-galactosidase activity in the cells, we observed very low or no activity (data not shown). Thus, we performed RT-PCR to detect yofA and ftsW transcription. As shown in Fig. 8, the transcription of yofA increased over time to a maximum level at entry into stationary phase, which coincided with the transcriptional profile of ftsW.
FIG. 8.
RT-PCR analysis of expression of yofA and ftsW: time course of expression of yofA (A), ftsW (B), and rpsR (C). RNA (0.5 μg) was isolated at the indicated time points from LB medium cultures of wild-type strain 168 and then analyzed by RT-PCR as described in Material and Methods. The rpsR transcript encoding ribosomal protein S18 served as a control for fluctuations in total RNA amounts. The arrowheads indicate the positions of the expected RT-PCR products.
DISCUSSION
Simple unicellular organisms must undergo cell division in order to generate progeny. This is one of the most critical processes in biology. In the current study, we uncovered a novel role for yofA in septum formation during cell division in B. subtilis. We showed that yofA has a role in maintaining cell density after the end of exponential growth. Our analysis further indicated that YofA plays an important role in cell division through the regulation of expression of ftsW, which is an essential component of the cell division machinery in B. subtilis.
YofA is composed of 285 amino acids and shares sequence similarity with members of the LysR family of transcriptional regulators. Among the LysR family members in B. subtilis with known functions, AlsR (37), GltC (1, 3), and YtlI (5) act as positive regulators of target genes located close to them, whereas CysL (15) and CitR (26, 27) act as negative regulators that inhibit the transcription of neighboring genes. Other LysR-type regulators, such as GltR (2) and CcpC (28, 29), act in trans to regulate the expression of target genes, such as gltA and citB, respectively. yofA is a monocistronic operon, located 128 bp from the yogA gene, which encodes a putative alcohol dehydrogenase, and 107 bp from the ggt gene, which encodes gamma-glutamyltranspeptidase. The direction of transcription of yofA is opposite that of yogA and ggt. By analogy with the mechanisms of gene regulation of other LysR family members, YofA was thought to regulate the expression of neighboring genes, such as yogA and/or ggt. However, we found that YofA had no effect on the expression of these genes (data not shown). Thus, YofA is most likely to be a trans-acting regulatory protein, similar to GltR and CcpC. In the current study, we showed that the cell division gene ftsW is a putative target of YofA in B. subtilis.
While the role of cell division in B. subtilis has been clarified, the transcriptional regulatory proteins involved in cell division are not well understood. The only known transcriptional regulator involved in cell division is YycGF, a two-component regulatory protein that is essential for cell growth (12, 21). YycG and YycF function as a sensor kinase and response regulator, respectively (11). YycFG has been shown to play a role in cell division and in cell membrane and cell wall homeostasis. YycF binds to the P1 promoter of the ftsAZ operon, which is involved in cell division (12). Howell et al. (21) identified putative YycF binding sites in 14 genes, including ykvT, which encodes a putative cell wall hydrolase, and the tagAB/tagDEF divergon, which encodes essential components of the teichoic acid biosynthesis pathway. In the current study, we obtained evidence that mutation in a novel transcriptional regulator gene, yofA, inhibits the final round of cell division prior to entry into stationary phase.
Following exponential growth, most cells experience a significant reduction in the rate of cell division as they enter stationary phase. It has been proposed that this phenomenon is the result of the ability of the cells to detect certain signals (e.g., nutrient depletion and cell density) in the extracellular environment. The reduction in the division rate results in completion of the division process, a process that may be required for the accumulation of excess cell division proteins. For example, expression of ftsAZ reaches a maximum at the transition from exponential growth to stationary phase (14). One of the three ftsAZ promoters, P2, is recognized by σH-associated RNA polymerase, which is an alternative RNA polymerase sigma factor that directs the transcription of many genes that function at the transition state (4, 14). In a similar manner, we showed that expression of the division genes ftsW, divIB, divIC, and pbpB, which includes ftsAZ, was also maximal at the transition state (Fig. 4). These data imply that the expression of division genes increases until the end of log phase in order to increase the rate of division. The expression pattern of rodA, which is involved in cell elongation, was opposite that of the other division genes. Although we observed peak expression of ftsW at the transition state in the yofA mutant, the magnitude of expression appeared to be significantly enhanced by YofA, and the transcription profile of yofA correlated with that of ftsW. Thus, it appears that expression of yofA is modulated by nutritional status and that YofA in turn regulates the expression of ftsW.
FtsW is essential for septum formation; however, the function of ftsW has not been elucidated in B. subtilis. FtsW is a paralog of B. subtilis RodA and SpoVE (23, 30). RodA (18) and SpoVE (17, 40) are required for the maintenance of normal cell shape and the synthesis of spore cortex peptidoglycan, respectively. They are members of the SEDS family of proteins (shape, elongation, division, and sporulation) and contain 10 transmembrane-spanning segments (18). In E. coli FtsW is an essential gene for cell division and appears to be involved in the translocation of the lipid-linked peptidoglycan precursor through the cytoplasmic membrane. FtsW plays a role in the stabilization of the FtsZ ring and recruitment of the FtsW cognate transpeptidase FtsI (PBP3) to the division site during cell division (32). Based on these reports, the level of FtsW appears to be important for stabilization of the division machinery. We demonstrated that strain ZL014, which contained the inducible Pspac-ftsW expression construct, undergoes IPTG-dependent cell growth and division (Fig. 6 and 7). In addition, we showed that maximal expression of ftsW is dependent on YofA in B. subtilis (Fig. 5). Interestingly, the growth pattern of the yofA mutant was similar to that of strain ZL014 in the presence of 0.01 mM IPTG (Fig. 6). The reduced level of FtsW in the yofA mutant correlated with the level in ZL014 in the presence of 0.01 mM IPTG, which suggests that there is a threshold level of FtsW that is required for the final round of cell division during entry into stationary phase. Of note, the OD600 and the number of CFU/ml declined in both the yofA mutant and strain ZL014 in the presence of 0.01 mM IPTG during stationary phase. This is may be due to a failure of chains of cells to separate, which may present a growth disadvantage under starvation conditions and eventually leads to cell lysis.
Taken together, our findings indicate that FtsW is specifically required for the formation of the division septum and that maximal expression of ftsW is required for cellular survival during stationary phase. In fact, we observed that decreased ftsW expression caused by yofA mutation led to a defect in septum formation and a growth defect after the end of exponential phase (Fig. 6 and Fig. 7).
To begin to characterize the phenotype of the yofA mutant during entry into stationary phase, we examined the effect of yofA mutation on the transition from logarithmic to stationary phase. We demonstrated that the effect of yofA mutation was specific for the stationary phase in cultures growing in LB medium (nonsporulating conditions). We speculate that the high rate of growth in LB medium requires cells to “change gears” at the transition, which requires YofA, whereas under sporulation conditions, the partial deficiency of FtsW caused by yofA mutation may be overcome during the early stage of sporulation.
The mechanism of activation of transcription of ftsW by YofA may involve binding of YofA to the ftsW promoter region. However, we have not yet determined if YofA binds to the promoter of ftsW or whether other ligands are involved. Another possibility is that YofA is involved in controlling the transcription of other genes, which affect the expression of ftsW. Thus, further studies, such as DNA microarray analysis, are needed to distinguish among these and other possibilities. Furthermore, investigation of growth state- and cell cycle-dependent fluctuations in the expression of ftsW will be essential for understanding the molecular mechanisms of cell division. The identification of yofA as a gene involved in cell division during entry into stationary phase provides a significant piece of information concerning the complex process of cell division. Further work on YofA should lead to a better understanding of the regulation of cell division in B. subtilis.
Acknowledgments
This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
Published ahead of print on 25 May 2007.
REFERENCES
- 1.Belitsky, B. R., and A. L. Sonenshein. 1995. Mutations in GltC that increase Bacillus subtilis gltA expression. J. Bacteriol. 177:5696-5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belitsky, B. R., and A. L. Sonenshein. 1997. Altered transcription activation specificity of a mutant form of Bacillus subtilis GltR, a LysR family member. J. Bacteriol. 179:1035-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bohannon, D. E., and A. L. Sonenshein. 1989. Positive regulation of glutamate biosynthesis in Bacillus subtilis. J. Bacteriol. 171:4718-4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Britton, R. A., P. Eichenberger, J. E. Gonzalez-Pastor, P. Fawcett, R. Monson, R. Losick, and A. D. Grossman. 2002. Genome-wide analysis of the stationary-phase sigma factor (sigma-H) regulon of Bacillus subtilis. J. Bacteriol. 184:4881-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burguiere, P., J. Fert, I. Guillouard, S. Auger, A. Danchin, and I. Martin-Verstraete. 2005. Regulation of the Bacillus subtilis ytmI operon, involved in sulfur metabolism. J. Bacteriol. 187:6019-6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dajkovic, A., and J. Lutkenhaus. 2006. Z ring as executor of bacterial cell division. J. Mol. Microbiol. Biotechnol. 11:140-151. [DOI] [PubMed] [Google Scholar]
- 7.Dubnau, D., and R. Davidoff-Abelson. 1971. Fate of transforming DNA following uptake by competent Bacillus subtilis. Formation and properties of the donor-recipient complex. J. Mol. Biol. 56:209-221. [DOI] [PubMed] [Google Scholar]
- 8.Errington, J., and R. A. Daniel. 2002. Cell division during growth and sporulation, p. 97-109. In A. L. Sonenshein, J. H. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria. American Society for Microbiology, Washington, DC.
- 9.Errington, J. 2003. Regulation of endospore formation in Bacillus subtilis. Nat. Rev. Microbiol. 1:117-126. [DOI] [PubMed] [Google Scholar]
- 10.Errington, J., R. A. Daniel, and D. J. Scheffers. 2003. Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 67:52-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fabret, C., and J. A. Hoch. 1998. A two-component signal transduction system essential for growth of Bacillus subtilis: implications for anti-infective therapy. J. Bacteriol. 180:6375-6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fukuchi, K., Y. Kasahara, K. Asai, K. Kobayashi, S. Moriya, and N. Ogasawara. 2000. The essential two-component regulatory system encoded by yycF and yycG modulates expression of the ftsAZ operon in Bacillus subtilis. Microbiology 146:1573-1583. [DOI] [PubMed] [Google Scholar]
- 13.Goehring, N. W., and J. Beckwith. 2005. Diverse paths to midcell: assembly of the bacterial cell division machinery. Curr. Biol. 15:514-526. [DOI] [PubMed] [Google Scholar]
- 14.Gonzy-Treboul, G., C. Karmazyn-Campelli, and P. Stragier. 1992. Developmental regulation of transcription of the Bacillus subtilis ftsAZ operon. J. Mol. Biol. 224:967-979. [DOI] [PubMed] [Google Scholar]
- 15.Guillouard, I., S. Auger, M. F. Hullo, F. Chetouani, A. Danchin, and I. Martin-Verstraete. 2002. Identification of Bacillus subtilis CysL, a regulator of the cysJI operon, which encodes sulfite reductase. J. Bacteriol. 184:4681-4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harry, E., L. Monahan, and L. Thompson. 2006. Bacterial cell division: the mechanism and its precision. Int. Rev. Cytol. 253:27-94. [DOI] [PubMed] [Google Scholar]
- 17.Henriques, A. O., H. de Lencastre, and P. J. Piggot. 1992. A Bacillus subtilis morphogene cluster that includes spoVE is homologous to the mra region of Escherichia coli. Biochimie 74:735-748. [DOI] [PubMed] [Google Scholar]
- 18.Henriques, A. O., P. Glaser, P. J. Piggot, and C. P. Moran, Jr. 1998. Control of cell shape and elongation by the rodA gene in Bacillus subtilis. Mol. Microbiol. 28:235-247. [DOI] [PubMed] [Google Scholar]
- 19.Hilbert, D. W., and P. J. Piggot. 2004. Compartmentalization of gene expression during Bacillus subtilis spore formation. Microbiol. Mol. Biol. Rev. 68:234-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hosoya, S., Z. Lu, Y. Ozaki, M. Takeuchi, and T. Sato. 2007. Cytological analysis of the mother cell death process during sporulation in Bacillus subtilis. J. Bacteriol. 189:2561-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Howell, A., S. Dubrac, K. K. Anderson, D. Noone, J. Fert, T. Msadek, and K. Devine. 2003. Genes controlled by the essential YycG/YycF two-component system of Bacillus subtilis revealed through a novel hybrid regulator approach. Mol. Microbiol. 49:1639-1655. [DOI] [PubMed] [Google Scholar]
- 22.Igo, M. M., and R. Losick. 1986. Regulation of a promoter that is utilized by minor forms of RNA polymerase holoenzymes in Bacillus subtilis. J. Mol. Biol. 191:615-624. [DOI] [PubMed] [Google Scholar]
- 23.Ikeda, M., T. Sato, M. Wachi, H. K. Jung, F. Ishino, Y. Kobayashi, and M. Matsuhashi. 1989. Structural similarity among Escherichia coli FtsW and RodA proteins and Bacillus subtilis SpoVE protein, which function in cell division, cell elongation, and spore formation, respectively. J. Bacteriol. 171:6375-6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imamura, D., K. Kobayashi, J. Sekiguchi, N. Ogasawara, M. Takeuchi, and T. Sato. 2004. spoIVH (ykvV), a requisite cortex formation gene, is expressed in both sporulating compartments of Bacillus subtilis. J. Bacteriol. 186:5450-5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishiwa, H., and H. Shibahara. 1985. New shuttle vectors from Escherichia coli and Bacillus subtilis. II. Plasmid pHY300PLK, a multipurpose cloning vector with a polylinker, derived from pHY460. Jpn. J. Genet. 60:235-243. [Google Scholar]
- 26.Jin, S., and A. L. Sonenshein. 1994. Identification of two distinct Bacillus subtilis citrate synthase genes. J. Bacteriol. 176:4669-4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin, S., and A. L. Sonenshein. 1994. Transcriptional regulation of Bacillus subtilis citrate synthase genes. J. Bacteriol. 176:4680-4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jourlin-Castelli, C., N. Mani, M. M. Nakano, and A. L. Sonenshein. 2000. CcpC, a novel regulator of the LysR family required for glucose repression of the citB gene in Bacillus subtilis. J. Mol. Biol. 295:865-878. [DOI] [PubMed] [Google Scholar]
- 29.Kim, S. I., C. Jourlin-Castelli, S. R. Wellington, and A. L. Sonenshein. 2003. Mechanism of repression by Bacillus subtilis CcpC, a LysR family regulator. J. Mol. Biol. 334:609-624. [DOI] [PubMed] [Google Scholar]
- 30.Kunst, F., N. Ogasawara, I. Moszer, et al. 1997. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 31.Mandelstam, J., and S. A. Higgs. 1974. Induction of sporulation during synchronized chromosome replication in Bacillus subtilis. J. Bacteriol. 120:38-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mercer, K. L., and D. S. Weiss. 2002. The Escherichia coli cell division protein FtsW is required to recruit its cognate transpeptidase, FtsI (PBP3), to the division site. J. Bacteriol. 184:904-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller, J. H. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 34.Murakami, T., K. Haga, M. Takeuchi, and T. Sato. 2002. Analysis of the Bacillus subtilis spoIIIJ gene and its paralog gene, yqjG. J. Bacteriol. 184:1998-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ogasawara, N. 2000. Systematic function analysis of Bacillus subtilis genes. Res. Microbiol. 151:129-134. [DOI] [PubMed] [Google Scholar]
- 36.Pastoret, S., C. Fraipont, T. den Blaauwen, B. Wolf, M. E. G. Aarsman, A. Piette, A. Thomas, R. Brasseur, and M. Nguyen-Disteche. 2004. Functional analysis of the cell division protein FtsW of Escherichia coli. J. Bacteriol. 186:8370-8379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Renna, M. C., N. Najimudin, L. R. Winik, and S. A. Zahler. 1993. Regulation of the Bacillus subtilis alsS, alsD, and alsR genes involved in post-exponential-phase production of acetoin. J. Bacteriol. 175:3863-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rothfield, L. I., and S. S. Justice. 1997. Bacterial cell division: the cycle of the ring. Cell 88:581-584. [DOI] [PubMed] [Google Scholar]
- 39.Rudner, D. Z., and R. Losick. 2001. Morphological coupling in development: lessons from prokaryotes. Dev. Cell 1:733-742. [DOI] [PubMed] [Google Scholar]
- 40.Sato, T., G. Theeragool, T. Yamamoto, M. Okamoto, and Y. Kobayashi. 1990. Revised nucleotide sequence of the sporulation gene spoVE from Bacillus subtilis. Nucleic Acids Res. 18:4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597-626. [DOI] [PubMed] [Google Scholar]
- 42.Sievers, J., and J. Erringtion. 2000. The Bacillus subtilis cell division protein FtsL localizes to sites of septation and interacts with DivIC. Mol. Microbiol. 36:846-855. [DOI] [PubMed] [Google Scholar]
- 43.Strauch, M. A., and J. A. Hoch. 1993. Signal transduction in Bacillus subtilis sporulation. Curr. Opin. Genet. Dev. 3:203-212. [DOI] [PubMed] [Google Scholar]
- 44.Vicente, M., A. I. Rico, R. Martinez-Arteaga, and J. Mingorance. 2006. Septum enlightenment: assembly of bacterial division proteins. J. Bacteriol. 188:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]