Abstract
The essential Bacillus anthracis nrdE gene carries a self-splicing group I intron with a putative homing endonuclease belonging to the GIY-YIG family. Here, we show that the nrdE pre-mRNA is spliced and that the homing endonuclease cleaves an intronless nrdE gene 5 nucleotides (nt) upstream of the intron insertion site, producing 2-nt 3′ extensions. We also show that the sequence required for efficient cleavage spans at least 4 bp upstream and 31 bp downstream of the cleaved coding strand. The position of the recognition sequence in relation to the cleavage position is as expected for a GIY-YIG homing endonuclease. Interestingly, nrdE genes from several other Bacillaceae were also susceptible to cleavage, with those of Bacillus cereus, Staphylococcus epidermidis (nrdE1), B. anthracis, and Bacillus thuringiensis serovar konkukian being better substrates than those of Bacillus subtilis, Bacillus lichenformis, and S. epidermidis (nrdE2). On the other hand, nrdE genes from Lactococcus lactis, Escherichia coli, Salmonella enterica serovar Typhimurium, and Corynebacterium ammoniagenes were not cleaved. Intervening sequences (IVSs) residing in protein-coding genes are often found in enzymes involved in DNA metabolism, and the ribonucleotide reductase nrdE gene is a frequent target for self-splicing IVSs. A comparison of nrdE genes from seven gram-positive low-G+C bacteria, two bacteriophages, and Nocardia farcinica showed five different insertion sites for self-splicing IVSs within the coding region of the nrdE gene.
Group I introns are self-splicing RNA sequences that in the splicing event merge the two flanking sequences that they interrupt into a contiguous mRNA (2). They are widespread and can be found in eukaryotes and viruses, as well as bacteria and bacteriophages (2, 15, 18, 30). Bacterial group I introns are mostly found in tRNA genes and rarely interrupt protein-coding genes, except in some prophage sequences (32). The annotated Bacillus anthracis chromosome has two genes that are interrupted by group I intron sequences, the nrdE and recA genes (17, 27). The closely related Bacillus thuringiensis serovar konkukian contains a shorter group I intron sequence in the nrdE gene and another group I intron in the recA gene that is identical to the group I intron in the B. anthracis recA gene (36). The group I introns in the B. anthracis and B. thuringiensis nrdE genes are unique among bacterial chromosomes, as they interrupt an essential protein-coding gene, the large component (α polypeptide) of class Ib ribonucleotide reductase. This enzyme catalyzes de novo production of deoxyribonucleotides that are required for aerobic DNA synthesis and bacterial proliferation (24, 35).
The nrdE intron in B. anthracis, but not the recA intron, also encodes a newly reported homing endonuclease gene (HEG). Homing endonucleases (HEases) cleave non-HEG-containing DNA close to sites that represent their own locations, leaving a single-strand break (nick) or a double-strand break (DSB) (1). The nick or DSB leads to a recombination/repair between the allele carrying the HEG and the other allele lacking the HEG. The recombination/repair results in copying of the HEG, together with parts of the flanking genomic regions, to the non-HEG allele, and HEGs can therefore be described as selfish genetic elements.
HEGs have been found within intervening self-splicing intron sequences and self-splicing intein sequences (here collectively called IVSs) and at intergenic regions (1). A HEase encoded in a self-splicing intron or intein usually has a recognition site that spans the IVS insertion site and is interrupted by the IVS harboring the HEG. The IVS-associated HEase almost exclusively cleaves alleles lacking the IVS, since the IVS with few exceptions interrupts the recognition site, preventing the HEase from cleaving the IVS-containing allele. The freestanding IVSs can also cleave a HEG-containing allele, since the HEase target site resides in the neighboring gene. Both types of HEases promote a non-Mendelian inheritance of the HEG-containing allele.
Based on their active-site motifs, HEases are divided into four families, LAGLIDADG, HNH, His box, and GIY-YIG (1, 33). The HEases also contain a DNA binding motif(s). The specific target sites recognized by the HEases span 14 to 35 bp (1). This means that cleavage by HEase is rare, occurring typically once per genome. Although HEases have long target recognition sites, they display a rather low stringency and allow some variation within the recognition sequence. The low stringency in turn allows spreading to other alleles with a nonidentical recognition site. Once a HEG is established at a new location, it has a good chance to spread efficiently at that site in the population (11, 12, 14, 29). Phage genes are ideal hosts for HEGs, as a double infection event is an opportunity for the HEG to spread to a HEG-less allele.
In this paper, we show that the nrdE intron in B. anthracis is spliced and encodes a fully functional HEase (denoted I-BanI) of the GIY-YIG family. We map the cleavage site and the recognition site of I-BanI and show that it has a variable capacity to spread among several closely and distantly related Bacillaceae, even though it has been found only in B. anthracis and B. thuringiensis serovar konkukian so far.
MATERIALS AND METHODS
Bacterial strains and general methods.
Wild-type B. anthracis Sterne 7700 pXO1−/pXO2− (lacking both virulence plasmids) was obtained from the Swedish Defense Research Agency. Strains of Bacillus subtilis 168, Bacillus cereus ATCC 14579, Bacillus licheniformis ATCC 14580, Staphylococcus epidermidis ATCC 12228, S. epidermidis RP62A, Salmonella enterica serovar Typhimurium LT2, Corynebacterium ammoniagenes ATCC 6872, and Escherichia coli K-12 were obtained from the Spanish Type Culture Collection. Lactococcus lactis MG1363 was kindly provided by Jan Kok. All strains were grown in brain heart infusion medium (Becton Dickinson) at 30°C, with the exceptions of B. anthracis, S. enterica serovar Typhimurium, and E. coli, which were grown at 37°C. Genomic DNA was extracted using the Genomic DNA extraction kit (QIAGEN) according to the manufacturer's recommendations. All primers mentioned below are specified in the supplemental material. All target and template PCR products were amplified from genomic DNA using Taq DNA polymerase (Fermentas) according to the manufacturer's recommendations and purified using a QIAquick PCR purification kit (QIAGEN).
In vivo splicing analysis.
RNA isolation from B. anthracis Sterne cells harvested at an A640 of 1.0 was carried out using the RNeasy kit for total-RNA isolation (QIAGEN) according to the manufacturer's recommendations, including the on-column DNase I treatment. Reverse transcriptase (RT)-PCRs were carried out using the ThermoScript RT-PCR system (Invitrogen) according to the manufacturer's recommendations. The RNA concentration used in the RT step was 1.37 μg/ml. After the RT step, the presence of spliced and unspliced products was analyzed by PCR. The primers used were ex1, int1, and ex2. PCRs were performed using standard conditions with Taq DNA polymerase (Fermentas) according to the manufacturer's recommendations. Products were separated on agarose gels and visualized with ethidium bromide and UV light. The PCR product of the spliced RNA was sequenced (by MWG Biotech AG) with primers B.ant forw and B.ant rev.
Prediction of intron secondary structure.
Prediction of the intron secondary structure was performed using Mfold default settings (http://www.bioinfo.rpi.edu/applications/mfold/rna/form1.cgi) and corrected by hand.
In vitro translation of endonuclease constructs.
Templates for in vitro translation were amplified using primers I-BanI TNT start and I-BanI end. To make the I-BthI variant of the endonuclease primers, I-BthI del and I-BthI del inv were used in conjunction with the start and end primers. The deletion primers also included the single amino acid residue change (F24L) at the 24th position that differs between I-BthI and I-BanI. Templates were amplified from genomic DNA from B. anthracis Sterne and were used for in vitro translation using the kit TNT T7 Quick for PCR DNA (Promega) according to the manufacturer's recommendations. Radiolabeled [35S]Met was included in the reactions, and the products were separated on a denaturing 15% polyacrylamide gel and analyzed with a phosphorimager (Fujifilm FLA-3000).
In vitro endonuclase activity assays.
Targets for HEase cleavage were amplified using genomic DNA as a template and one of the two primers labeled with fluorescein according to the method described previously (29). Cleavage reactions were performed using in vitro-translated HEases directly in the reaction according to the method described previously (29). The reaction mixtures contained 500 fmol target DNA, 3 μl in vitro translation reaction mixture, and 0.1 μg RNaseA in 50 μl of 10 mM Tris-HCl (pH 8.5), 10 mM MgCl2, 100 mM KCl, and 0.1 mg/ml bovine serum albumin and were incubated at 37°C for 30 min. Incubations with mock in vitro translation without template DNA served as negative controls. All cleavage products were separated on agarose gels except for one experiment (see Fig. 4), in which the products were separated on a 5% polyacrylamide gel with 7 M urea, analyzed by excitation of fluorescein-labeled targets and products at 473 nm, and filtered at 520 nm (FujiFilm FLA-3000). Note that only one cleavage product was visualized, as only one strand was fluorescein labeled. The cleavage efficiencies of the different targets were analyzed with ImageGauge v3.45 (Fujifilm). The cleavage efficiency was calculated by dividing the fluorescence of the product by the total fluorescence of the substrate and product. The most efficiently cleaved target was set to 100% cleavage. All cleavage efficiencies were adjusted accordingly to give the relative efficiency of cleavage.
FIG. 4.
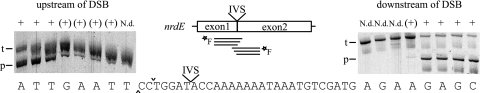
Mapping of minimum sequence required for I-BanI cleavage. Polyacrylamide gels show products from cleavage assays with fluorescein-labeled targets with short flanking sequences upstream and downstream of the cut on the coding strand, −10 bp and + 35 bp, respectively. The target (t) and product (p) are indicated for each gel. Note that only one cleavage product was visualized, as only one strand was fluorescein labeled. The left and right gels show cleavage of the targets with successively shorter sequence upstream and downstream. The end positions are indicated below the lanes. Cleavage is indicated with + for efficient cleavage, (+) for low or nearly no cleavage, and N.d. for no cleavage detected. DSB is indicated with ^ and ∨ for the cuts on the template and coding strands, respectively.
Cleavage site mapping.
For mapping of the DSB, one 32P-radiolabeled and one unlabeled primer were used in the target PCR for labeling of each strand separately. Primers [32P]-nrdE seq forw and B.ant rev were used for identifying the cut on the coding strand. Primers [32P]-nrdE seq rev and B.ant forw were used for identifying the cut on the template strand. The targets were then cleaved with I-BanI. The cleavage products were purified and separated on 10% polyacrylamide gels with 7 M urea, together with sequencing ladders produced with the fmol DNA Cycle Sequencing System (Promega) according to the method described previously (29). The sequencing ladders were labeled using [γ-35S]dATP incorporation and the unlabeled primers nrdE seq forw and nrdE seq rev. Template sequences for both cleaving and sequencing were IVS-less nrdE variants constructed from B. anthracis Sterne. The gel was visualized and analyzed with a phosphorimager (Fujifilm FLA-3000).
Mapping of the recognition site.
Targets used in mapping of the required site for cleavage of I-BanI were amplified from a IVS-less nrdE PCR product using fluorescein-labeled primers and primers successively shortening the target sequence. The targets for the downstream part of the site were amplified using primers B.ant forw 2 (F) and Ltd bind +28 to +35. The targets for the upstream part of the site were amplified using primers B.ant rev (F) and Ltd bind −3 to −10. (F) denotes fluorescein labeling.
Cleavage of nrdE from related Bacillaceae.
The targets used in the activity assays were amplified from genomic DNAs from respective strains by PCR using one fluorescein-labeled primer and one unlabeled primer. For a full list of the primers and templates used, see the supplemental material.
Sequence data were obtained from the GenBank genome data (accession numbers NC_003997 for B. anthracis Ames (NP_843829 for the HEG), NC_005945 for B. anthracis Sterne (YP_027537 for the HEG), NC_004722 for B. cereus, NC_006270 for B. licheniformis, NC_000964 for B. subtilis 168 and B. subtilis 168 prophage SPβ, NC_005957 for B. thuringiensis serovar konkukian strain 97-27, Y09572 for C. ammoniagenes, NC_000913 for E. coli, NZ_AAGO01000069 for L. lactis, NC_003197 for S. enterica serovar Typhimurium, NC_004461 for S. epidermidis ATCC 12228, and NC_002976 for S. epidermidis RP62A).
Sequence alignments and phylogenetic analysis.
Sequence alignments of target sites were done using Clustalw (34). Phylogenetic analyses were done as previously described (35) and visualized with TreeView (25).
RESULTS
In vivo splicing and secondary structure of the group I intron in B. anthracis nrdE.
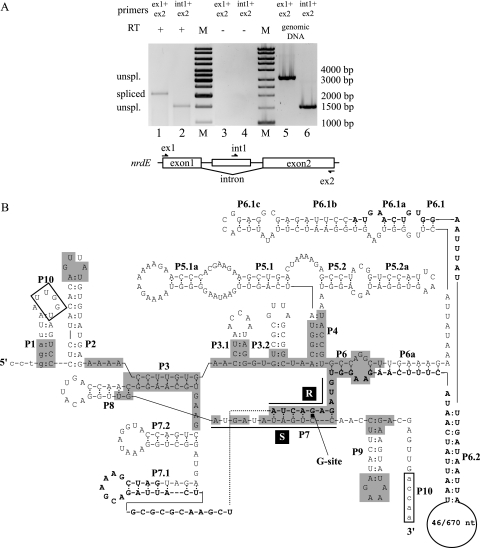
The intron of the B. anthracis nrdE gene was tested for in vivo splicing activity by nonquantitative RT-PCR (Fig. 1A). RNA isolated from cells in the late log phase showed the presence of both unspliced and spliced nrdE mRNAs. Exon1- and exon2-specific primers gave a product of 2.1 kbp from RT-PCR, corresponding to the size of the spliced product, and no unspliced products could be detected in this way (Fig. 1A, lane 1). As a control, intron- and exon2-specific primers gave a product of 1.5 kbp from RT-PCR, corresponding to unspliced mRNA (Fig. 1A, lane 2). In addition, these primer pairs were used in PCR with genomic DNA, giving products of 3.2 kbp and 1.5 kbp, respectively, corresponding to the length of unspliced mRNA (Fig. 1A, lanes 5 and 6). The primer pairs were also used as controls in an RT-PCR without RT, giving no observed product (Fig. 1A, lanes 3 and 4), indicating that genomic contamination did not occur. Sequencing of the spliced product showed the spliced site to be as predicted (see the sequence in Fig. 4). These results show that the group I intron of B. anthracis nrdE is efficiently spliced in vivo.
FIG. 1.
Efficient splicing of the group I self-splicing intron in B. anthracis nrdE. (A) Agarose gel showing RT-PCR products of unspliced and spliced nrdE mRNA in total-RNA extraction from B. anthracis Sterne 7700 pXO1−/pXO2−. Exon1- and exon2-specific primers (ex1 and ex2, respectively) in RT-PCR gave a product of 2.1 kbp, corresponding to spliced nrdE mRNA (lane 1). The intron-specific primer int1, in combination with ex2, gave an RT-PCR product of 1.5 kbp, corresponding to unspliced nrdE mRNA (lane 2). Primer pairs were used in control RT-PCR without RT and showed no DNA contamination in the RNA extraction (lanes 3 and 4). Primer pairs were also used in PCRs with genomic DNA as primer controls and showed products of 3.2 kbp and 1.5 kbp, respectively (lanes 5 and 6). A molecular size marker (M) was run for reference, where bands 4 to 6, 8, and 10 from the bottom correspond to 1,000 bp, 1,500 bp, 2,000 bp, 3,000 bp, and 4,000 bp, respectively. (B) Predicted secondary structure of the nrdE intron. The lowercase letters indicate the coding sequence of the nrdE gene. The uppercase letters indicate the intronic sequence. Boldface uppercase letters indicate the ORF in the intron, with the numbers in the loop in P6.2 representing the sequences of B. thuringiensis serovar konkukian and B. anthracis Sterne. Conserved sequence elements (R and S), conserved base-paired regions (P1 to P9), and additional pairings (P3.1, P3.2, P5.1, P5.1a, P5.2, p5.2a, P6a, P6.1a to P6.1c, P6.2, P7.1, and P7.2) are shown. Alignment between the 5′ and 3′ splice sites can be promoted by the boxed nucleotides, UUGGU, in the P1 loop, and ACCAA, near P9, making pairing P10. The shaded boxes represent nucleotides identical with the group I intron in the recA gene in B. anthracis (17).
Prediction of the secondary structure of the intron showed that it contained all conserved regions of pairing (P1 to P10) and the conserved sequence elements R and S and shared characteristics with the subgroup IA2 introns (23). The open reading frame (ORF) is predicted to start in the P6.1b region and to span the P7 region with the conserved R element (Fig. 1B). The intron also showed high similarity to the predicted secondary structure of the group I intron in the recA gene of B. anthracis (17) over the conserved regions of the intron. The group I intron in the nrdE gene of the B. subtilis BSG40 prophage (20) also shared similar secondary structure with the nrdE intron in B. anthracis and was found at the same insertion site in their respective nrdE genes, suggesting a distant relationship between the introns. In addition, the ORF found in the group I intron in the nrdE gene of the B. subtilis BSG40 prophage (20) shared 50% identity with the ORF encoding I-BanI.
A GIY-YIG endonuclease in the B. anthracis nrdE intron and a remnant HEG in B. thuringiensis serovar konkukian.
The annotated 1,102-bp B. anthracis nrdE intron, located between Asp255 and Thr256, contains an ORF encoding a 253-amino-acid-residue putative HEase of the GIY-YIG family (Fig. 2A). We named the ORF I-BanI, according to the suggested nomenclature for HEases (28). I-BanI contains two major domains found in the GIY-YIG endonuclease family, the N-terminal GIY-YIG domain, containing the GIY-YIG motif, and the C-terminal GIY-YIG domain, containing a minor-groove DNA binding α-helix motif and a helix-turn-helix motif (8, 22). The closely related B. thuringiensis serovar konkukian includes a much shorter ORF (encoding 45 amino acid residues) in its nrdE intron (478 bp). The ORF here called I-BthI was found to be a deletion variant of I-BanI. The B. thuringiensis serovar konkukian nrdE intron has a deletion of 624 bp compared to the B. anthracis nrdE intron (36). This deletion gives rise to an in-frame deletion of major parts of both the N-terminal and C-terminal domains of the ORF of the HEase (Fig. 2A and 1B) plus a 1-amino-acid substitution, but it still retains the GIY-YIG motif. A structure prediction experiment using the SwissModel First Approach Mode at http://swissmodel.expasy.org/ (13, 26, 31) showed the I-BanI to have high similarity to the I-TevI GIY-YIG catalytic N-terminal domain (36), while no similarities were found for I-BthI. These findings indicate a loss of function for I-BthI (data not shown).
FIG. 2.
The B. anthracis nrdE intron encodes a GIY-YIG HEase. (A) I-BanI, identified as a GIY-YIG endonuclease, contains an N-terminal GIY-YIG domain (red box) with the GIY-YIG endonuclease motif, a C-terminal GIY-YIG domain (blue box) with a minor-groove DNA binding α-helix motif, and a helix-turn-helix motif (yellow box). A large deletion in I-BthI spans most of both the N-terminal and C-terminal domains. (B) [35S]Met-radiolabeled in vitro translation of I-BthI and I-BanI showed products of 5 kDa and 30 kDa, respectively.
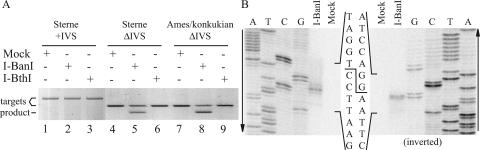
Several attempts at cloning and overexpressing I-BanI in E. coli failed, presumably due to its toxicity to E. coli. Also, with I-BanI expressed in and secreted from Drosophila sp. strain Schneider S2 cells, no specific cleavage was detected (data not shown). On the other hand, cleavage activity was observed with in vitro-translated I-BanI (see below). In vitro translation of I-BanI and I-BthI gave rise to ∼30-kDa and ∼5-kDa products, respectively (Fig. 2B), which are in agreement with the predicted molecular masses for these ORFs. The products were tested for DSB cleavage activity on target sequences amplified from B. anthracis Sterne nrdE. No cleavage activity was detected when the wild-type nrdE sequence containing the intron sequence was incubated with I-BanI or I-BthI in vitro translations or with mock translation (no DNA template in the in vitro translation reaction) (Fig. 3A, lanes 1 to 3). As IVSs are known to divide the target sites of most HEases residing within them, two IVS-less target variants were constructed on the assumption that the target site would be recreated if the intron sequence was deleted. The Sterne and Ames/serovar konkukian IVS-less targets both showed cleavage products when incubated with I-BanI (Fig. 3A, lanes 5 and 8, respectively), indicating that the target sites were recreated by deletion of the intron. No significant difference in I-BanI cleavage was seen for the Sterne and Ames/serovar konkukian nrdE sequences that differ by a T-to-C transition 17 bp downstream and an A-to-G transition 334 bp upstream of the intron insertion site. No cleavage activity was found when the IVS-less targets were incubated with either I-BthI or mock translations (Fig. 3A, lanes 6 and 9 and lanes 4 and 7, respectively). Note that only one cleavage product was visualized, as only one strand was fluorescein labeled.
FIG. 3.
Mapping of the cleavage site of the I-BanI HEase. (A) Agarose gel showing products from a cleavage assay with fluorescein-labeled targets, wild-type Sterne (2,276 bp; lanes 1 to 3), Sterne IVS-less (1,174 bp; lanes 4 to 6), and Ames/serovar konkukian (1,174 bp; lanes 7 to9) incubated with mock translation, I-BanI, and I-BthI. No cleavage products were found for targets incubated with mock translation or I-BthI (lanes 1, 4, and 7 and lanes 3, 6, and 9, respectively). Cleavage products corresponding to the predicted 722 bp were found for IVS-less targets incubated with I-BanI (lanes 5 and 8). No cleavage product was found for wild-type Sterne incubated with I-BanI (lane 2). Note that only one cleavage product was visualized, as only one strand was fluorescein labeled. (B) Polyacrylamide gel showing sequencing reactions run alongside 32P single-strand-labeled cleavage reactions to identify the I-BanI cleavage site and overhang. The second gel is inverted to more clearly show the cleavage sites on both strands.
Mapping of the I-BanI cleavage site.
Based on fragment sizes from cleavage assays on IVS-less target variants (Fig. 3A), a DSB was estimated to occur close to the intron insertion site. In separate experiments, the DSB could be identified by the isotopic labeling of one or the other strand of the IVS-less target incubated with I-BanI. The cleavage products were run alongside sequencing reactions using the same primers as for target construction. Figure 3B shows that the DSB occurs 5 and 7 nucleotides (nt) upstream of the intron insertion site, with 2-nt 3′ extensions, i.e., 760 nt and 758 nt from the start of nrdE on the coding and template strands, respectively.
Mapping the length of the I-BanI recognition site.
Sterne IVS-less nrdE was used as a template for producing targets flanking the cleavage site and with successively shorter upstream or downstream sequences. Figure 4 shows that we could map the approximate length of the I-BanI recognition site with this approach. When the target sequence was limited to 8 bp upstream (and 452 bp downstream) of the cut on the coding strand, it was readily cleaved by I-BanI. However, targets with shorter upstream sequences showed a gradual decrease in cleavage efficiency, and cleavage was abolished when the target was limited to only 3 bp upstream of the cleavage site. Limiting the target sequence to 30 bp downstream (and 431 bp upstream) of the cut on the coding strand abolished cleavage of I-BanI. A very small amount of cleavage product was detected with 31 bp downstream, whereas a target sequence limited to 32 bp downstream showed full activity of I-BanI cleavage. These experiments suggest that the I-BanI recognition site is 35 to 40 bp and covers the cleavage site with a bias toward the downstream region including the IVS insertion site.
I-BanI cleaves nrdE sequences from related strains of Bacillaceae.
PCR-amplified target sequences from several related Bacillaceae strains were tested for I-BanI cleavage. Sequences of nrdE were amplified from B. subtilis 168 prophage SPβ, B. subtilis 168, B. licheniformis, S. epidermidis RP62A nrdE1 (identical to S. epidermidis ATCC 12228 nrdE) and nrdE2, and B. cereus ATCC 14579. I-BanI was shown to produce DSBs with varying efficiencies of cleavage for all Bacillaceae targets tested (Fig. 5A, lanes 1 to 6). Surprisingly, the most efficiently cleaved targets were B. cereus and S. epidermidis nrdE1 (100 to 95% cleavage) compared to the IVS-less targets from the strains Ames/serovar konkukian and Sterne (77 to 80% cleavage), where we originally found the HEG variants. The least efficiently cleaved target was S. epidermidis nrdE2 (12% cleavage). However, this target differs less in sequence from S. epidermidis nrdE1 (4 bp substitutions between position −8 and position +31 of the cut on the coding strand) than the other susceptible targets (10 to 15 bp substitutions compared to S. epidermidis nrdE1) (Fig. 5B). To further investigate the stringency of I-BanI, target nrdEs from L. lactis, C. ammoniagenes, E. coli, and S. enterica serovar Typhimurium were also tested for I-BanI cleavage with no cleavage products detected (Fig. 5A, lanes 7 to 10).
FIG. 5.
I-BanI can cleave several closely related Bacillaceae nrdE genes. (A) Agarose gel showing products (p) from a cleavage assay with fluorescein-labeled targets (t) from the B. subtilis 168 prophage SPβ (2,106 bp), B. subtilis 168 (2,103 bp), B. licheniformis (2,103 bp), S. epidermidis nrdE1 (2,103 bp) and nrdE2 (2,103 bp), B. cereus (1,174 bp), C. ammoniagenes (2,106 bp), E. coli (2,106 bp), S. enterica serovar Typhimurium (2,106 bp), and L. lactis (2,106 bp) incubated with I-BanI. Cleavage products corresponding to the predicted sizes were found for targets B. subtilis 168 prophage SPβ (1,073 bp), B. subtilis 168 (784 bp), B. licheniformis (784 bp), S. epidermidis nrdE1 (787 bp) and nrdE2 (766 bp), and B. cereus (722 bp) (lanes 1 to 6). No cleavage products were detected for the targets C. ammoniagenes, E. coli, S. enterica serovar Typhimurium, and L. lactis (lanes 7 to 10). Note that only one cleavage product was visualized, as only one strand was fluorescein labeled. (B) Target site alignments showing a region from positions −12 to +37 flanking the cuts on the coding strands of all target sequences tested. For each target, the measured relative activity is indicated, with the most efficiently cleaved target set as 100%. N.d. indicates no cleavage detected. Means and standard deviations (SD) were calculated from measurements from three experiments. DSB is indicated with ^ and ∨ for the cuts on the template and coding strands, respectively. Uppercase letters in the consensus sequence and gray boxes indicate fully conserved nucleotides within all cleaved targets. Lowercase letters in the consensus sequence indicate conserved nucleotides in target sequences cleaved 50% or more.
Abundant occurrence of IVSs in the nrdE gene in bacteria, prophages, and phages.
The previously sequenced nrdE genes available in public databases were observed to encode IVSs with five different insertion sites, IVS1 to IVS5 (Fig. 6A) (7, 19, 20, 32); the IVS in B. anthracis is at IVS3. B. cereus E33L (GenBank accession number NC_006274) has an intron at IVS4. One of the two nrdE genes in S. epidermidis RP62A (10) has an intein at IVS4 instead and an intron with a HEG at IVS5. The closely related S. epidermidis ATCC 12228 has no IVSs. The B. subtilis 168 prophage SPβ has an intron at IVS2 and an intein at IVS4 (32), and the Staphylococcus aureus phage Twort has an intron each at IVS2 and IVS5 (19). In addition, the actinobacterium Nocardia farcinica has an intein at IVS1 (16).
FIG. 6.
The nrdE gene in Bacillaceae is a refuge for self-splicing IVSs. (A) Alignments of sequences flanking the insertion sites of IVSs in the nrdE gene (IVS1 to -5 in different colors) found within the family Bacillaceae, S. epidermidis, Staphylococcus phage Twort, and N. farcinica. The insertion sites of the IVSs and fully conserved nucleotides are indicated with light gray and dark gray, respectively. The numbers indicate the distances in nucleotides between the shown aligned sequence locations in the B. anthracis Sterne nrdE. Symbols denote group I intron with HEG (filled circles), group I intron without HEG (open circles), and intein with HEG (filled triangles). Inteins IVS1 and IVS4 have also been called RIR1-j and RIR1-b, respectively (http://bioinfo.weizmann.ac.il/∼pietro/inteins/), and introns IVS2, IVS3, and IVS4 have also been called RIR1-I, RIR1-II, and RIR1-III, respectively (20). (B) Phylogenetic tree of bacterial NrdEs with a selected set of Bacillaceae. The low-G+C gram-positive bacteria are shown with a shaded background. Vertical lines indicate subgroups within the B. cereus group, underlining denotes phage sequences, and dashed underlining denotes prophage. The occurrence of IVS1 to IVS5 is indicated with symbols and colors as shown in panel A, and relative cleavage efficiencies by I-BanI according to Fig. 4 are indicated. Only bootstrap values below 950 are shown. The distance bar in the lower left corner indicates 0.1 changes per site.
The presence of IVSs and HEases in the nrdE gene in Bacillaceae is scattered, and a sequence similarity tree of the NrdE protein shows that the Bacillaceae NrdEs are distributed in three different branches (Fig. 6B). As expected, the B. anthracis Sterne NrdE protein clusters together with NrdEs from the B. cereus group of bacteria (B. cereus, B. anthracis, and B. thuringiensis), whereas close relatives of the B. cereus group, like the other Bacillus species (e.g., B. subtilis), form an independent cluster and the B. thuringiensis subsp. israelensis NrdE2 is on a branch diverging from the B. cereus branch (Fig. 6B). NrdEs from other low-G+C gram-positive members are found in three additional branches, Streptococcus/Lactococcus in one branch and Staphylococcus in two branches, one together with NrdEs in S. aureus phages. NrdEs from other bacteria, including all proteobacteria, gram-positive high-G+C bacteria, spirochetes, and mollicutes (e.g., Mycoplasmas), are clustered together and independently of the NrdE branches of the low-G+C gram-positive bacteria. The diversity in the NrdEs from species of the low-G+C gram-positive bacteria was previously also observed for the class Ib ribonucleotide reductase protein NrdF (β polypeptide) (35), whereas this group of bacteria contains close family members, based on a 16S rRNA tree (6).
There are subtle differences in the branch clustering within the B. cereus group, in most cases related to whether the gene contains an IVS. For example, B. thuringiensis subsp. israelensis NrdE1 and B. cereus ATCC 14597 without an IVS cluster apart from other IVS-containing B. thuringiensis serovar konkukian and B. cereus strains (Fig. 6B). On the other hand, the IVS2-containing Bacillus weihenstephanensis KBAB4 and B. cereus subsp. cytotoxis are in different subclusters of the B. cereus branch. As shown in Fig. 5, S. epidermidis nrdE1 and nrdE2, the B. subtilis 168 prophage SPβ, B. subtilis 168, and B. lichenformis lacking IVS3 are susceptible to I-BanI cleavage, whereas low-G+C gram-positive L. lactis, the gammaproteobacteria E. coli and S. enterica serovar Typhimurium, and the high-G+C gram-positive C. ammoniagenes are not. Thus, the lack of IVSs at the third site (IVS3) in some species of the low-G+C gram-positive group is not due to a lack of DNA sequences that are susceptible to cleavage by the HEase.
DISCUSSION
Self-splicing group I introns are rarely found in protein-coding genes in bacteria (2, 9, 15). In this study, we show that the B. anthracis nrdE gene, an essential component of the aerobic ribonucleotide reductase in this bacterium, carries a group I intron that is efficiently spliced in vivo and that the intron encodes a functional GIY-YIG HEase. From the results presented in Fig. 4, we suggest that the target site covers 4 to 8 bp upstream and 31 to 32 bp downstream of the cut on the coding strand. This correlates with what is expected of HEases of the GIY-YIG family.
Interestingly, the B. anthracis endonuclease denoted I-BanI has the potential to cleave several nrdE genes in the family Bacillaceae. As was found for other HEases (1, 33), the target site has relatively low stringency. It would be tempting to deduce a preliminary optimal cleavage sequence based on our data, but an extensive mutagenesis study would be needed to characterize the target site. Despite the wide range of cleaved targets, it seems that the spread of the IVS encoding the I-BanI HEase has been limited, since it has been found only in B. anthracis and B. thuringiensis serovar konkukian so far. Only with more sequence data from many strains of the same species might the spread of this IVS be traced and reveal clues to how non-prophage-associated HEases might spread in a bacterial host.
HEases may hop into new sites quite frequently (11, 12, 29), but they will be detected in bacterial genomes only if they are fixed. The homing event is more likely to occur if the recognition site of the HEase is in a conserved sequence, e.g., an essential gene. Such a recognition site will be encountered more frequently, and therefore the homing event has a higher probability to lead to fixation. In addition, an essential gene will not be lost from the genome despite a higher cost that an IVS may inflict on the organism. Essential genes, such as nrdE, may therefore act as a haven for IVSs and HEases. The ribonucleotide reductase class Ib NrdE protein has highly conserved sequences, especially in proximity to the active-site regions and the allosteric regulatory sites (24), and it is essential for B. anthracis DNA replication and proliferation during aerobiosis (35). Among the more than 100 sequenced nrdE genes (see http://rnrdb.molbio.su.se/ for a full account of RNR occurrences in organisms), 16 contain one or more IVSs in this gene (and several B. subtilis prophages have at least one IVS in the nrdF gene) (20, 21, 32). Interestingly, the nrdE IVSs are found at five different insertion sites, IVS1 to IVS5 (Fig. 6A), and they are found exclusively in some members of the Bacillaceae (including B. subtilis prophages), Staphylococcus and S. aureus phages, Streptococcus/Lactococcus groups, and N. farcinica (Fig. 6B).
Group I self-splicing IVSs have been found in all types of organisms and frequently in rRNA genes in mitochondria and chloroplasts of unicellular eukaryotes, in tRNA genes in bacteria, and in genes involved in DNA metabolism in bacteriophages (2, 15, 18, 30). In bacteria, IVSs have also been observed to interrupt protein-coding genes in prophages (32), but interestingly, the nrdE genes in B. anthracis and B. thuringiensis do not appear to be associated with prophage sequences, even though members of the B. anthracis family carry annotated prophage sequences elsewhere in their genomes (27). In addition, bacterial group I introns in protein-coding genes have been reported only for the family Bacillaceae, S. epidermidis, and N. farcinica (Fig. 6). It has been argued that the occurrence of HEG-containing IVSs in phages is an effective source of horizontal gene transfer (HGT) (9, 12, 19, 20, 29, 30, 32, 33), as phages multiply to several copies in an infected bacterium and bacteria may be infected by more than one phage, promoting horizontal spread among phages, as well as between phages and host cell genomes. The potential group I intron-encoded GIY-YIG HEG in the B. subtilis BSG40 prophage nrdE gene (20) could indicate that the nrdE intron in B. anthracis has been transmitted by phage. In addition, natural competence is widespread among soil organisms and may contribute to HGT among the Bacillaceae (3, 5). Several authors suggest that HGT is the major force for incorporating different and new metabolic functions in a given bacterium (4). As more examples of IVSs and HEGs within the nrdE gene in the family Bacillaceae are found, the different possibilities for inheritance can be tested statistically. Here, we have shown that not only phage genes are prone to harbor IVSs with or without HEGs, but host cells may also encounter IVS copies. A similar observation has been made for the recA gene (17). Despite being widespread, the distribution of group I introns is irregular, and they may be found in one gene of an organism but not in the same gene of a closely related species.
Supplementary Material
Acknowledgments
We thank Solveig Hahne for technical assistance, Daniel X. Johansson for expression of I-BanI in Drosophila sp. strain Schneider S2 cells, and Linus Sandegren and Patrick Young for constructive criticism.
This work was supported by a grant from the Swedish Research Council.
Footnotes
Published ahead of print on 11 May 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Belfort, M., and R. J. Roberts. 1997. Homing endonucleases: keeping the house in order. Nucleic Acids Res. 25:3379-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cech, T. R. 1990. Self-splicing of group I introns. Annu. Rev. Biochem. 59:543-568. [DOI] [PubMed] [Google Scholar]
- 3.Chen, I., P. J. Christie, and D. Dubnau. 2005. The ins and outs of DNA transfer in bacteria. Science 310:1456-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, I., and D. Dubnau. 2004. DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2:241-249. [DOI] [PubMed] [Google Scholar]
- 5.Claverys, J. P., M. Prudhomme, and B. Martin. 2006. Induction of competence regulons as a general response to stress in gram-positive bacteria. Annu. Rev. Microbiol. 60:451-475. [DOI] [PubMed] [Google Scholar]
- 6.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, S. A. Kulam, D. M. McGarrell, G. M. Garrity, and J. M. Tiedje. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33:D294-D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derbyshire, V., and M. Belfort. 1998. Lightning strikes twice: intron-intein coincidence. Proc. Natl. Acad. Sci. USA 95:1356-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dodd, I. B., and J. B. Egan. 1990. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 18:5019-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edgell, D. R., M. Belfort, and D. A. Shub. 2000. Barriers to intron promiscuity in bacteria. J. Bacteriol. 182:5281-5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gill, S. R., D. E. Fouts, G. L. Archer, E. F. Mongodin, R. T. Deboy, J. Ravel, I. T. Paulsen, J. F. Kolonay, L. Brinkac, M. Beanan, R. J. Dodson, S. C. Daugherty, R. Madupu, S. V. Angiuoli, A. S. Durkin, D. H. Haft, J. Vamathevan, H. Khouri, T. Utterback, C. Lee, G. Dimitrov, L. Jiang, H. Qin, J. Weidman, K. Tran, K. Kang, I. R. Hance, K. E. Nelson, and C. M. Fraser. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187:2426-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goddard, M. R., and A. Burt. 1999. Recurrent invasion and extinction of a selfish gene. Proc. Natl. Acad. Sci. USA 96:13880-13885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gogarten, J. P., and E. Hilario. 2006. Inteins, introns, and homing endonucleases: recent revelations about the life cycle of parasitic genetic elements. BMC Evol. Biol. 6:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 14.Haugen, P., V. A. Huss, H. Nielsen, and S. Johansen. 1999. Complex group-I introns in nuclear SSU rDNA of red and green algae: evidence of homing-endonuclease pseudogenes in the Bangiophyceae. Curr. Genet. 36:345-353. [DOI] [PubMed] [Google Scholar]
- 15.Haugen, P., D. M. Simon, and D. Bhattacharya. 2005. The natural history of group I introns. Trends Genet. 21:111-119. [DOI] [PubMed] [Google Scholar]
- 16.Ishikawa, J., A. Yamashita, Y. Mikami, Y. Hoshino, H. Kurita, K. Hotta, T. Shiba, and M. Hattori. 2004. The complete genomic sequence of Nocardia farcinica IFM 10152. Proc. Natl. Acad. Sci. USA 101:14925-14930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ko, M., H. Choi, and C. Park. 2002. Group I self-splicing intron in the recA gene of Bacillus anthracis. J. Bacteriol. 184:3917-3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lambowitz, A. M., and M. Belfort. 1993. Introns as mobile genetic elements. Annu. Rev. Biochem. 62:587-622. [DOI] [PubMed] [Google Scholar]
- 19.Landthaler, M., U. Begley, N. C. Lau, and D. A. Shub. 2002. Two self-splicing group I introns in the ribonucleotide reductase large subunit gene of Staphylococcus aureus phage Twort. Nucleic Acids Res. 30:1935-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lazarevic, V. 2001. Ribonucleotide reductase genes of Bacillus prophages: a refuge to introns and intein coding sequences. Nucleic Acids Res. 29:3212-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lazarevic, V., B. Soldo, A. Dusterhoft, H. Hilbert, C. Mauel, and D. Karamata. 1998. Introns and intein coding sequence in the ribonucleotide reductase genes of Bacillus subtilis temperate bacteriophage SPbeta. Proc. Natl. Acad. Sci. USA 95:1692-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marchler-Bauer, A., J. B. Anderson, P. F. Cherukuri, C. DeWeese-Scott, L. Y. Geer, M. Gwadz, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, G. H. Marchler, M. Mullokandov, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, R. A. Yamashita, J. J. Yin, D. Zhang, and S. H. Bryant. 2005. CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res. 33:D192-D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michel, F., and E. Westhof. 1990. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J. Mol. Biol. 216:585-610. [DOI] [PubMed] [Google Scholar]
- 24.Nordlund, P., and P. Reichard. 2006. Ribonucleotide reductases. Annu. Rev. Biochem. 75:681-706. [DOI] [PubMed] [Google Scholar]
- 25.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 26.Peitsch, M. C., T. N. Wells, D. R. Stampf, and J. L. Sussman. 1995. The Swiss-3DImage collection and PDB-Browser on the World-Wide Web. Trends Biochem. Sci. 20:82-84. [DOI] [PubMed] [Google Scholar]
- 27.Read, T. D., S. N. Peterson, N. Tourasse, L. W. Baillie, I. T. Paulsen, K. E. Nelson, H. Tettelin, D. E. Fouts, J. A. Eisen, S. R. Gill, E. K. Holtzapple, O. A. Okstad, E. Helgason, J. Rilstone, M. Wu, J. F. Kolonay, M. J. Beanan, R. J. Dodson, L. M. Brinkac, M. Gwinn, R. T. DeBoy, R. Madpu, S. C. Daugherty, A. S. Durkin, D. H. Haft, W. C. Nelson, J. D. Peterson, M. Pop, H. M. Khouri, D. Radune, J. L. Benton, Y. Mahamoud, L. Jiang, I. R. Hance, J. F. Weidman, K. J. Berry, R. D. Plaut, A. M. Wolf, K. L. Watkins, W. C. Nierman, A. Hazen, R. Cline, C. Redmond, J. E. Thwaite, O. White, S. L. Salzberg, B. Thomason, A. M. Friedlander, T. M. Koehler, P. C. Hanna, A. B. Kolsto, and C. M. Fraser. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 423:81-86. [DOI] [PubMed] [Google Scholar]
- 28.Roberts, R. J., M. Belfort, T. Bestor, A. S. Bhagwat, T. A. Bickle, J. Bitinaite, R. M. Blumenthal, S. Degtyarev, D. T. Dryden, K. Dybvig, K. Firman, E. S. Gromova, R. I. Gumport, S. E. Halford, S. Hattman, J. Heitman, D. P. Hornby, A. Janulaitis, A. Jeltsch, J. Josephsen, A. Kiss, T. R. Klaenhammer, I. Kobayashi, H. Kong, D. H. Kruger, S. Lacks, M. G. Marinus, M. Miyahara, R. D. Morgan, N. E. Murray, V. Nagaraja, A. Piekarowicz, A. Pingoud, E. Raleigh, D. N. Rao, N. Reich, V. E. Repin, E. U. Selker, P. C. Shaw, D. C. Stein, B. L. Stoddard, W. Szybalski, T. A. Trautner, J. L. Van Etten, J. M. Vitor, G. G. Wilson, and S. Y. Xu. 2003. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 31:1805-1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sandegren, L., D. Nord, and B.-M. Sjöberg. 2005. SegH and Hef: two novel homing endonucleases whose genes replace the mobC and mobE genes in several T4-related phages. Nucleic Acids Res. 33:6203-6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandegren, L., and B.-M. Sjöberg. 2004. Distribution, sequence homology, and homing of group I introns among T-even-like bacteriophages: evidence for recent transfer of old introns. J. Biol. Chem. 279:22218-22227. [DOI] [PubMed] [Google Scholar]
- 31.Schwede, T., J. Kopp, N. Guex, and M. C. Peitsch. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31:3381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stankovic, S., B. Soldo, T. Beric-Bjedov, J. Knezevic-Vukcevic, D. Simic, and V. Lazarevic. 2007. Subspecies-specific distribution of intervening sequences in the Bacillus subtilis prophage ribonucleotide reductase genes. Syst. Appl. Microbiol. 30:8-15. [DOI] [PubMed] [Google Scholar]
- 33.Stoddard, B. L. 2005. Homing endonuclease structure and function. Q. Rev. Biophys. 38:49-95. [DOI] [PubMed] [Google Scholar]
- 34.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Torrents, E., M. Sahlin, D. Biglino, A. Gräslund, and B.-M. Sjöberg. 2005. Efficient growth inhibition of Bacillus anthracis by knocking out the ribonucleotide reductase tyrosyl radical. Proc. Natl. Acad. Sci. USA 102:17946-17951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tourasse, N. J., E. Helgason, O. A. Økstad, I. K. Hegna, and A. B. Kolstø. 2006. The Bacillus cereus group: novel aspects of population structure and genome dynamics. J. Appl. Microbiol. 101:579-593. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.