Abstract
Despite its notoriety as a human pathogen, Vibrio cholerae is an aquatic microbe suited to live in freshwater, estuarine, and marine environments where biofilm formation may provide a selective advantage. Here we report characterization of biofilms formed on abiotic and biotic surfaces by two non-O1/O139 V. cholerae strains, TP and SIO, and by the O1 V. cholerae strain N16961 in addition to the isolation of 44 transposon mutants of SIO and TP impaired in biofilm formation. During the course of characterizing the mutants, 30 loci which have not previously been associated with V. cholerae biofilms were identified. These loci code for proteins which perform a wide variety of functions, including amino acid metabolism, ion transport, and gene regulation. Also, when the plankton colonization abilities of strains N16961, SIO, and TP were examined, each strain showed increased colonization of dead plankton compared with colonization of live plankton (the dinoflagellate Lingulodinium polyedrum and the copepod Tigriopus californicus). Surprisingly, most of the biofilm mutants were not impaired in plankton colonization. Only mutants impaired in motility or chemotaxis showed reduced colonization. These results indicate the presence of both conserved and variable genes which influence the surface colonization properties of different V. cholerae subspecies.
The ecology of the cholera-causing bacterium, Vibrio cholerae, has been considered since the 1850s, when contaminated water sources were first implicated by John Snow as a key factor associated with its epidemiology (59). However, many of the strategies that this organism employs for survival outside the human host have come to light only recently. Perhaps one of the most important discoveries is the role that plankton have in aiding the survival of V. cholerae within aquatic environments. Seminal work by Huq et al. (25) demonstrated that the presence of copepods improved the survival of V. cholerae populations. Analogous experiments performed by Islam et al. (26, 27) showed that V. cholerae survival can be enhanced via association with cyanobacteria. In both cases, surface colonization by V. cholerae was evident. The propensity of V. cholerae for epibiosis is also evident in its attachment to chironomid egg masses and vascular plants (20, 56).
The significance of V. cholerae higher-order assemblages can be further illustrated in two ways. First, there is a correlation between V. cholerae population levels and phytoplankton and zooplankton blooms (14, 24), as well as the physical factors which drive them (10, 36). The biofilms formed by V. cholerae during these associations may aid in survival, as biofilms have been shown to offer shelter from many different environmental stresses, including protozoan grazing (40), UV light (15), oxidants (13), and toxic metals (60). Second, filtration of drinking water through sari cloth (pore size, >20 μm) is an effective method for removing V. cholerae assemblages and reducing the incidence of cholera (11).
The biofilm characteristics of clinical isolates of V. cholerae have been examined previously (65, 71). Cells of V. cholerae O1 El Tor strain N16961 form biofilms in a stepwise manner, first contacting the surface using flagellar motility and the mannose-sensitive hemagglutinin (MSHA) type IV pilus. After surface contact occurs, a three-dimensional biofilm structure is established by growth of the sessile population and production of an exopolymer matrix composed of Vibrio polysaccharide (VPS). In addition, it has been noted that V. cholerae can exhibit a diverse range of growth phenotypes on surfaces, including rugose and smooth colony variation on agar surfaces (45, 63) or distinct biofilms ranging from thick, mature biofilms to flat single layers of cells colonizing a surface (71).
The MSHA pilus also contributes to the attachment of V. cholerae to the carapace of the common water flea, Daphnia pulex (8). mshA mutations diminish attachment of V. cholerae O1 El Tor strains and O139 strains but have no significant effect on the attachment of O1 classical strains due to the fact that the vast majority of classical strains do not elaborate functional MSHA (17). This suggests that additional factors outside the MSHA pilus mediate attachment to biotic surfaces and that different strains of V. cholerae may employ different strategies to attach to surfaces. In fact, further work has identified an additional colonization factor of V. cholerae, GbpA, which is an adhesin that binds to sugars present on both chitin-containing surfaces and mammalian cell membranes (31).
The current study examined biofilm formation by two environmental, nontoxigenic strains of V. cholerae, SIO and TP. The phenotypic differences between the biofilms formed by each of these strains and biofilms formed by an O1 El Tor clinical strain were examined, and genes important in SIO and TP biofilm formation were identified using a transposon mutagenesis screen. To understand biofilm formation in a more environmentally relevant context, we also examined the abilities of these strains to form biofilms on biotic surfaces, such as dinoflagellates and copepods.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
All strains and plasmids used in this study are listed in Table 1. Wild-type strains SIO and TP of V. cholerae were isolated from Southern California coastal waters. All Escherichia coli and V. cholerae strains were grown in LB broth (43) supplemented with appropriate antibiotics at 37°C, unless stated otherwise. The antibiotics used in this study were kanamycin (Km) (50 μg/ml for E. coli and 200 μg/ml for V. cholerae), chloramphenicol (Cm) (20 μg/ml for E. coli and 5 μg/ml for V. cholerae), and rifampin (Rif) (100 μg/ml).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or description | Reference |
|---|---|---|
| E. coli S17-1λpir | recA pro hsdR RP4-2-Tc::Mu-Km::Tn7 | 57 |
| V. cholerae strains | ||
| SIO | Wild type | 53 |
| TP | Wild type | 53 |
| N16961 | Wild-type O1 El Tor | 22 |
| S2151 | SIO ΔmshA | This study |
| S2153 | SIO ΔvpsL | This study |
| S2163 | SIO ΔpepN | This study |
| S2164 | SIO ΔVC2215 | This study |
| S2101 | SIO ΔtnaA | This study |
| S2123 | SIO ΔpomA | This study |
| S2104 | SIO ΔflaA | This study |
| T2114 | TP Δzur | This study |
| T2151 | TP ΔmshA | This study |
| T2153 | TP ΔvpsL | This study |
| T2127 | TP ΔVC1437 | This study |
| T2112 | TP ΔlacI | This study |
| T2101 | TP ΔtnaA | This study |
| T2130 | TP ΔVCA0859 | This study |
| T2123 | TP ΔpomA | This study |
| T2104 | TP ΔflaA | This study |
| Plasmids | ||
| pRL27 | Tn5-Rl27 oriR6K Kmr | 32 |
| pFL122 | RSF1010 derivative, lacZ MCS mob Smr | 33 |
| pFL122L | Linker containing SacI site ligated into BglII site of pFL122 | This study |
| pBSL181 | oriR6K Apr Cmr | 1 |
| pFLcm | pFL122L containing cat gene from pBSL181 | This study |
| p519ngfp | RSF1010 derivative containing nptII promoter before gfp gene, mob Kmr | 39 |
| pFLgfp | pFLcm122L containing PCR amplicon of nptIIp-gfpmut2 of p519ngfp | This study |
| pGP704sac28 | pGP704 derivative containing sacB gene of B. subtilis oriR6K, mob Apr | 6 |
| pMB2190 | pBR327 derivative, Kmr Apr | 3 |
| pGPKm | pGP704sac28 containing Kmr gene of pMB2190 | This study |
| pGPVX0408 | ΔmshA in pGPKm | This study |
| pGPVX0892 | ΔpomA in pGPKm | This study |
| pGPVX0934 | ΔvpsL in pGPKm | This study |
| pGPVX1494 | ΔpepN in pGPKm | This study |
| pGPVX2188 | ΔflaA in pGPKm | This study |
| pGPVX2215 | ΔVC2215 in pGPKm | This study |
| pGPVCX0161 | ΔtnaA in pGPKm | This study |
| pGPVX0378 | Δzur in pGPKm | This study |
| pGPVX1437 | ΔVC1437 in pGPKm | This study |
| pGPVX2337 | ΔlacI in pGPKm | This study |
| pGPVCX0859 | ΔVCA0859 in pGPKm | This study |
V. cholerae mating.
E. coli strain S17-1λpir (57) was used as a donor strain for all conjugation experiments with strains of V. cholerae. All strains were grown overnight to the stationary phase at 37°C. V. cholerae strains were subcultured at a 1:100 dilution and grown in LB medium at 22°C until the mid-log phase of growth (optical density at 600 nm [OD600], ∼0.7) was reached. E. coli and V. cholerae were washed to remove antibiotics by centrifugation (2 min, 13,000 × g) and concentrated 5- and 20-fold in LB medium, respectively. Fifteen microliters of each culture was transferred to a 0.22-μm polycarbonate filter on an LB agar plate, and the cultures were mixed. The conjugation mixtures were left overnight at 37°C. The filters were then transferred to tubes containing 1 ml of LB medium, and cells were removed by vortexing and plated onto thiosulfate citrate bile salts sucrose (TCBS) (89 g/liter; Difco) or LB medium supplemented with appropriate antibiotics and grown at 37°C overnight.
Transposon mutagenesis library and biofilm screening.
The suicide vector pRL27 (32), containing the hypertransposable mini-Tn5 element, was introduced into V. cholerae strains SIO and TP via conjugation. Transposon mutant libraries were made by selecting individual colonies from the TCBS selective plates containing 200 μg/ml Km. These colonies were then inoculated into 200 μl of LB medium in 96-well microtiter dishes, grown overnight, and subsequently converted into frozen stocks by adding glycerol to a final concentration of 15% to each well.
The method used for an initial screen to search for biofilm mutants was adapted from the method described by O'Toole et al. (50). Individual transposon mutants were arrayed in 96-well polystyrene microtiter dishes and grown overnight in 200 μl of fresh LB medium at 37°C with moderate shaking. Crystal violet was then added to each well to stain the adherent cells. The medium was then removed from each well and replaced with 200 μl of 95% ethanol, and the OD570 of individual wells were recorded. Wells with absorbance values below the wild-type levels were rescreened for biofilm-forming ability using borosilicate glass tubes. Cultures were grown in triplicate in 5 ml of LB medium overnight and were retested using the crystal violet method described above. The biofilm accumulation of mutant strains was normalized to the accumulation of the corresponding parental strain by dividing the OD570 of each mutant strain by the OD570 of the wild type to obtain a percentage. Pairwise comparisons of the data for each mutant strain and wild type were then analyzed for significance using a Student's t test (α = 0.05). Mutants displaying significant reductions in biofilm accumulation were also tested for possible growth differences from the wild-type strain, which may have contributed to the observed biofilm formation deficiencies. In this analysis, growth curves for each mutant and wild-type strain grown in 10 ml of LB medium were recorded over a 24-h period. No gross differences between the growth of the mutant strain and the growth of the wild type were noted for any of the mutants reported in this study (data not shown).
Transposon flanking DNA amplification and sequencing.
The protocol for arbitrary PCR described by O'Toole et al. (50) was used in order to obtain sequence data for the loci disrupted by the transposon insertions in each biofilm mutant (for a description of the primers, see Table S1 in the supplemental material). The PCRs resulted in approximately 500-bp amplicons, which were cleaned and sequenced using an Amersham MegaBace 100 sequencer. Recovered sequence data were analyzed in December 2006 by performing a BLASTX analysis (2) with the GenBank nonredundant database (http://www.ncbi.nlm.nih.gov). In some cases an additional flanking sequence was obtained using a panhandle PCR method adapted from the method of Jones and Winistorfer (28). Several rounds of chromosome walking were used in these instances to obtain additional sequence data for these undetermined regions.
Biofilm formation under continuous-flow conditions.
Biofilms were formed in flow cells covered with glass coverslips. The continuous-flow cell systems were prepared and assembled as previously described (46), with minor modifications. The flow cells were constructed from polycarbonate. Each flow chamber was 1 by 4 by 40 mm and was covered by a glass microscope coverslip, which was glued on top of the flow cell to serve as the substratum for biofilm growth. All tubing and flow cells were autoclaved prior to assembly.
V. cholerae strains were grown overnight in 2M minimal medium (51). Approximately 1 ml of an overnight culture was inoculated into the flow channels, and cells were allowed to attach under no-flow conditions for 1 h, during which time the tubing upstream and downstream of the flow cell was clamped. After attachment, the flow rate was adjusted to approximately 18 ml h−1, and the cells were perfused with 2M minimal medium with a glucose concentration of 0.04% at room temperature supplied by a peristaltic pump for up to 10 days.
Biofilms formed in flow cell chambers were observed by confocal laser scanning microscopy over a 10-day period in order to assess biofilm formation and maturation. At appropriate times, the flow was stopped, flow cells were clamped, and biofilms were stained with a LIVE/DEAD BacLight bacterial viability kit (Molecular Probes Inc., Eugene, OR). This kit contains two fluorescent dyes that stain living cells green (SYTO 9) and dead cells red (propidium iodide). Images were obtained with a Bio-Rad MRC 1024 confocal laser scanning microscope.
In-frame deletion constructs.
To engineer in-frame deletions of specific genes within the V. cholerae genomes, SOEing PCR was performed (23). For each deletion approximately 500 bp at the 5′ and 3′ ends of the desired gene was amplified (for a description of the primers, see Table S1 in the supplemental material). The amplicons included approximately 50 bp of the ends of each gene that, when fused by SOEing PCR, created in-frame deletions of the interior portions of the gene of interest. The resulting products were then cloned into pGPKm (this study) and transformed into S17-1λpir cells. Inserts were verified by PCR using pGPkm-based primers, and each suicide plasmid was conjugated into the appropriate Rif-resistant V. cholerae strain and plated onto LB agar (containing 100 μg/ml Rif and 200 μg/ml Km). Resulting colonies were selected for single crossover events using medium containing Rif, Km, and sucrose and were subsequently grown in LB medium containing no antibiotics, which allowed a second crossover to occur. After growth in LB medium, selection for loss of the genome-integrated sacB-containing plasmid was performed on LB medium plates containing 10% sucrose in place of NaCl. After overnight incubation at 37°C, colonies were picked and screened by PCR for conversion to the deletion construct using primers external to the original amplicons (see Table S1 in the supplemental material). These PCR products were sequenced to verify each in-frame deletion.
Colonization assays.
For copepod experiments, the green fluorescent protein-expressing plasmid pFLgfp (this study) was introduced into various strains of V. cholerae via conjugation. These strains were grown in LB medium (containing 5 μg/ml Cm) overnight, and fluorescence was verified using epifluorescence microscopy. After growth to the stationary phase, the cells were washed two times with autoclaved and filtered seawater and diluted to a concentration of 106 cells/ml in sterile seawater amended with 5 μg/ml Cm. Copepods were then washed on GFF filters as 30 to 50 ml of sterile seawater was filtered over the organisms to remove bacteria from the zooplankton surface and surrounding medium. Two live or dead (heat killed at 65°C for 10 min) copepods were then added to microcosms containing 106 washed V. cholerae cells per ml. Each experiment was performed in triplicate, and the preparations were incubated overnight at room temperature in the dark. Individual copepods were then collected and washed three times with 15 to 20 ml of sterile seawater to remove unattached V. cholerae. The specimens were then observed using epifluorescence microscopy to detect the level of surface colonization of the copepod by the bacteria. At specific time points, colonization of Tigriopus californicus was quantified by homogenizing individual washed copepods in sterile seawater and plating serially diluted homogenates onto TCBS containing 5 μg/μl Cm.
Colonization experiments with axenic Lingulodinium polyedrum were done in a similar manner. Axenic L. polyedrum strain CCMP 1932 (a gift from P. Von Dassow, M. Latz, X. Mayali, and F. Azam) was cultured at 16°C in F/2 medium (18) with a cycle consisting of 12 h of light supplied by cool white fluorescent tubes and 12 h of darkness. V. cholerae strains carrying pFLgfp were grown overnight in LB medium, washed in sterile seawater, and diluted as described above. L. polyedrum was taken from exponentially growing cultures and diluted 1:10 in either 1 ml of sterile seawater or artificial seawater (30 parts per thousand) in 24-well microtiter dishes. V. cholerae was then added to the wells at a concentration of 106 cells/ml. Each experiment was performed in triplicate, and the preparations were incubated overnight. After incubation the 1-ml microcosms were filtered onto 5-μm polycarbonate filters and washed three times with 10 ml of sterile artificial seawater to collect the fraction of V. cholerae attached to L. polyedrum. Epifluorescence microscopy was then used to examine the samples in order to determine the attachment of the bacteria to the dinoflagellate. Colonization was quantified by manually counting individual bacteria attached to washed dinoflagellates. For each sample at least 20 fields were counted, and averages were used for comparison of strains. For each colonization experiment whose results were quantified, pairwise comparisons of the data for each mutant strain and the wild type were analyzed for significance using a Student's t test (α = 0.05). If the data did not meet initial assumptions of normality and homogeneity of variance, they were transformed to perform the analysis.
Colonization experiments were carried out in a manner similar to the manner described above using both types of plankton and gfp-tagged wild-type strains in the presence of 100 μM phenamil, which has been shown to inhibit motility in vibrios by binding to the sodium-driven motor of the polar flagella (49). Loss of motility was examined in wild-type strains throughout each experiment using epifluorecent microscopy, and colonization phenotypes of V. cholerae strains were monitored as described above.
RESULTS
Surface attachment properties of clinical and environmental isolates of V. cholerae.
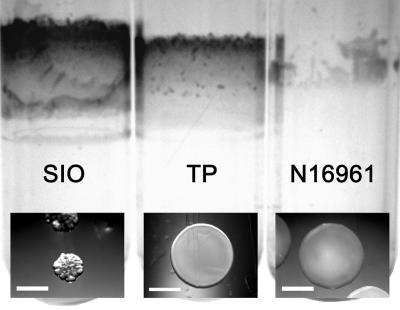
The surface-associated growth characteristics of two Southern California nonpathogenic V. cholerae isolates, SIO and TP (53), were examined and compared to the characteristics of an O1 serotype, El Tor biotype strain, N16961 (22). On LB agar plates, strain TP grew as smooth, opaque colonies, like the smooth variant of toxigenic strain N16961, whereas strain SIO grew as compact, rugose colonies (Fig. 1), presumably due to an increase in the production of VPS (71). The biofilms elaborated by the strains were also very different. When strains were grown in LB medium overnight in borosilicate glass tubes, the biofilms formed by the environmental strains, SIO and TP, were thicker and more robust than those formed by strain N16961 (Fig. 1).
FIG. 1.
Crystal violet staining of biofilms attached to borosilicate tubes of two environmental V. cholerae strains, SIO and TP, and clinical strain N16961. The images at the bottom show the colony morphology of each strain. Scale bars = 1 mm. Magnification, ×60.
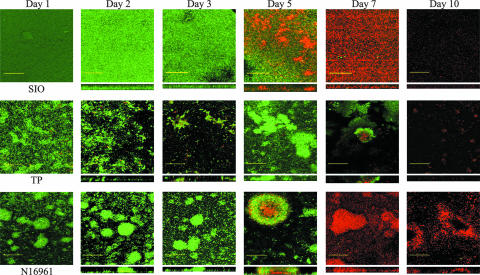
SIO and TP biofilms were examined in finer detail using flow cells and confocal laser scanning microscopy. It was discovered that the morphology and kinetics of biofilm formation of the environmental and clinical strains of V. cholerae also differed. Under these conditions, all three strains rapidly colonized the coverslip surfaces, but strains TP and N16961 appeared to form more differentiated structures than strain SIO, which colonized the entire surface homogeneously (Fig. 2). Within the first 3 days of the experiment, strain TP aggregated into discrete cell clusters on the glass substratum; however, these clusters did not resemble the typical rounded microcolonies found in N16961 biofilms, and the biomass was never as great as the biomass of the N16961 microcolonies. Despite this, the patterns of death of the N16961 and TP biofilms were similar, and the centers of the cell clusters exhibited significant death after days 5 and 7, respectively. After this, death of the remaining biofilm occurred, followed by sloughing of the attached cells from the substratum between days 7 and 10 of the experiment.
FIG. 2.
Steps in the development of V. cholerae SIO, TP, and N16961 biofilms determined using the LIVE/DEAD BacLight staining kit. By the first day, strain SIO rapidly colonizes the entire glass surface as a monolayer of cells (green cells). As the biofilm ages, strain SIO cells begin to die by day 5 (red cells) and completely detach from the glass substratum by day 10. The development of V. cholerae TP proceeds differently, with discrete clusters of cells developing on the substratum between days 2 and 5. On day 7 the centers of these clusters appear to die, and by day 10 no significant attachment to the glass can be seen. Strain N16961 begins forming distinct, rounded microcolonies (∼100 μm) immediately after attachment. Similar to strain TP, the centers of the microcolonies begin to die by day 5, and death of the entire biofilm occurs by day 7. This is followed by dispersal of the biofilm, leaving hollow colonies devoid of cells. Scale bars = 50 μm.
SIO biofilms formed in a very different manner. In contrast to the other strains tested, SIO cells never differentiated into clusters of cells and remained attached to the glass surface in a homogeneous layer. After the initial colonization of the flow cell surface, the undifferentiated SIO biofilm grew until day 5 of the experiment, when significant death of the entire biofilm was seen. Unlike the death of strains TP and N16961, this death did not seem to coincide with a dispersal event; rather, the cells remained attached to the surface until gradual sloughing by day 10.
Biofilm mutant screen.
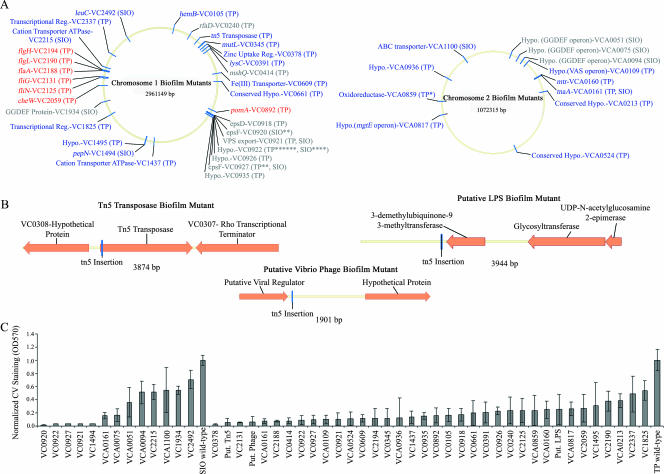
Based on the results described above indicating that both TP and SIO form distinctive biofilms, an inquiry into the genetic basis for the phenotypic differences was conducted. Mini-Tn5 transposon mutagenesis was performed, followed by screening for biofilm-defective phenotypes and identification of transposon insertion sites using arbitrary PCR. Forty-four unique mutations conferring a biofilm formation deficiency were identified for strains TP and SIO, representing ∼1% of the total collection of mutants screened. The nature of the biofilm loci identified based upon TBLASTX analysis (2) is presented in Fig. 3A and Table 2. Figure 3C shows the biofilm formation by each mutant relative to the biofilm formation by the corresponding parental strain after 16 h, as measured by crystal violet staining.
FIG. 3.
Biofilm mutants of V. cholerae SIO and TP. (A) Genomic map of the genes important for biofilm formation recovered in this study. The map includes the TIGR numbers of the mutated loci, the putative gene functions, and the parental V. cholerae strains. Asterisks indicate that there are multiple mutations in the same locus. Hypo., hypothetical; Reg., regulator. (B) Loci of the three biofilm transposon insertions isolated in strain TP whose genome sequence is not present in the N16961 strain. (C) Biofilm formation measured by crystal violet staining of the biofilm mutants. The values are normalized to wild-type biofilm formation. The error bars represent one standard deviation from the mean. CV, crystal violet.
TABLE 2.
Genes important for biofilm formation in V. cholerae strains SIO and TP
| Strain | TIGR gene or genome region | Gene | Common name or conserved domainsa | Predicted function |
|---|---|---|---|---|
| Cell motility | ||||
| T1104 | VC2188 | flaA | Flagellin subunit A | Flagellar motility |
| T1106 | VC2131 | fliH | Flagellar assembly protein | Flagellar motility |
| T1108 | VC2190 | flgL | Flagellar hook-associated protein | Flagellar motility |
| T1109 | VC2194 | flgH | Flagellar L-ring protein precursor | Flagellar motility |
| T1116 | VC2125 | fliN | Flagellar motor switch | Flagellar motility |
| T1123 | VC0892 | pomA | Flagellar motor protein | Flagellar motility |
| T1139 | VC2059 | cheW | Purine-binding chemotaxis protein | Chemotaxis |
| T1141 | VC0414 | mshQ | MshQ | MSHA pilus biosynthesis |
| Amino acid transport and metabolism | ||||
| S1163 | VC1494 | pepN | Aminopeptidase N | Glutathione metabolism |
| S1168 | VC2492 | leuC | 3-Isopropylmalate dehydratase | Leucine metabolism |
| S1165 | VCA1100 | ABC transporter | Peptide/nickel transport | |
| S/T1101 | VCA0161a | tnaA | Tryptophanase | Tryptophan metabolism |
| T1124 | VCA0160 | mtr | Mtr | Tryptophan transport |
| T1133 | VC0391 | lysC | Aspartokinase III | Amino acid metabolism |
| Inorganic ion transport and metabolism | ||||
| S1164 | VC2215 | Cation transporter ATPase, E1-E2 family | Divalent ion transport | |
| T1114 | VC0378 | Zinc uptake regulation protein | Zinc transport | |
| T1127 | VC1437 | Cation transporter ATPase, E1-E2 family | Divalent ion transport | |
| T1132 | VC0609 | ABC transporter | Iron(III) transport | |
| Cell wall/membrane biogenesis | ||||
| S/T1105 | VC0927a | cpsF | UDP-N-acetyl-d-mannosamine transferase | EPS biosynthesis |
| S/T1128 | VC0921a | Polysaccharide export protein | EPS biosynthesis | |
| T1102 | VC0918 | epsD | UDP-N-acetyl-d-mannosaminuronic acid dehydrogenase | EPS biosynthesis |
| T1142 | VC0240 | rfaD | ADP-l-glycero-d-mannoheptose-6-epimerase | LPS biosynthesis |
| General function prediction only | ||||
| S1150 | VC0920a | epsF | Glycosyl transferase | EPS biosynthesis |
| T1130 | VCA0859a | Aldo/keto reductase 2 family | Phosphate starvation | |
| Coenzyme transport and metabolism | ||||
| T1118 | VC0105 | hemB | δ-Aminolevulinic acid dehydratase | Porphyrin metabolism |
| Transcription | ||||
| T1112 | VC2337 | lacI | LacI family transcriptional regulator | Transcription regulation |
| Replication, recombination, and repair | ||||
| T1122 | VC0345 | mutL | DNA mismatch repair protein | Hfq operon |
| Signal transduction mechanisms | ||||
| S1166 | VC1934 | Diguanylate cyclase/phosphodiesterase, GGDEF/EAL family | EPS biosynthesis and signaling | |
| Conserved hypothetical | ||||
| S1162 | VCA0094 | MarR transcriptional regulator domainb | Transcription regulation | |
| T1121 | VC0661 | O-Methyltransferase domainb | General | |
| T1131 | VCA0524 | Tellurite resistance protein domainb | Inorganic ion transport | |
| Hypothetical | ||||
| S1154 | VCA0051-GGDEF operon | TPR transcriptional regulator domainb | ||
| S1160 | VCA0075-GGDEF operon | |||
| S/T1103 | VC0922a-VPS operon | |||
| T1111 | VC1495- pepN operon | |||
| T1115 | VCA0817-mgtE operon | Small mechanosensitive channel domainb | ||
| T1120 | VCA0213 | Na/H transporter domainb | Energy production | |
| T1125 | VC0926-VPS operon | |||
| T1134 | VC0935-VPS operon | |||
| T1144 | VCA0109 | Conserved domain: DUF1316b | Unknown | |
| T1146 | VCA0936 | Deacylase/carboxypeptidase domainb | General | |
| Other | ||||
| T1143 | Tn5 transposase | |||
| T1140 | Vibrio phage VJK locus | |||
| T1138 | Putative LPS locus |
There are multiple mutations in the same locus.
Function assigned using the KEGG database.
Mutations involved in surface attachment and biofilm structure.
Many of the mutations recovered in this screen were mutations in genes important for initial surface attachment, microcolony formation, and subsequent biofilm maturation. Mutations in VPS genes, which occurred in a large segment of the biofilm mutants in this category, were found in both of the VPS biosynthetic operons of the V. cholerae genome, and multiple transposon insertions occurred in four of the seven mutated VPS genes. Microarray studies have shown that in rugose colonies of V. cholerae the expression of both operons is up-regulated compared to the expression in smooth colonies (70). Accordingly, VPS mutations occurring in the SIO parental strain resulted in a switch from rugose colony morphology to smooth colony morphology. In addition to VPS gene insertions, numerous mutations were found in genes responsible for chemotaxis, flagellum production, and biosynthesis of the MSHA pilus. These types of genes have been hypothesized to aid in V. cholerae biofilm formation by facilitating initial attachment of cells to surfaces (47, 65).
Interestingly, while mutations in the MSHA biosynthetic operon were obtained with strain TP, no such insertions were obtained in our biofilm screen of strain SIO mutants. In order to assess the role of the MSHA in SIO biofilms, an in-frame deletion of the mshA gene, encoding a pilin subunit of the pilus, was constructed, and the biofilm-forming ability of the mutant was assessed. Surprisingly, it was discovered that the ΔmshA mutant exhibited increased surface accumulation compared to that of the wild type. Thus, the MSHA pilus is not essential in all strains for the formation of biofilms under the conditions tested.
Finally, two genes were found in loci which appear to be important in lipopolysaccharide (LPS) production. One transposon insertion disrupted the rfaD gene (VC0240), which has been shown in Salmonella to be involved in the biosynthesis of a conserved sugar in the LPS molecule (34). The second mutation was in a region of the TP chromosome not found in the N16961 genome. DNA sequencing of this region revealed that this mutation is in a locus that contains sequences with high similarity to LPS biosynthesis genes from various organisms, including V. cholerae serogroup O37 (for sequence data, see Table S2 in the supplemental material).
Biofilm regulatory mutants.
Many regulators of biofilm formation have recently been discovered in V. cholerae, and most of these regulators directly or indirectly affect the transcription of the VPS genes (6, 21, 69, 72). For example, the modulation of cytosolic cyclic diguanylate (c-di-GMP) levels by a family of proteins containing GGDEF and/or EAL domains appears to be one way in which V. cholerae cells regulate VPS gene expression (61).
In accordance with the importance of this signaling system, two of the genes (VC1934 and VCA0075) disrupted in our screen were in syntenic and paralogous operons encoding enzymes of this protein family. It should be noted that although the arrangements of these operons are similar, the VC1934 product contains both GGDEF and EAL domains, implying that it has a role in both c-di-GMP breakdown and synthesis, while its paralog, the VCA0074 product, contains only the GGDEF domain. Interestingly, microarray studies have shown that genes in both of these operons are up-regulated in rugose V. cholerae colonies compared to the expression in smooth variants, suggesting a possible role in modulating VPS expression (70). Consistent with this regulatory role, an SIO-derived VCA0075 mutant did not have the rugose phenotype of its parental strain, similar to the finding by Lim et al. (35) that noted a similar phenotypic shift in the colony morphology for mutations in the operon containing VCA0075 of V. cholerae strain 92A1552.
Another biofilm mutant contained a transposon insertion within VCA0051, resulting in a hypothetical protein containing a tetratricopeptide repeat motif. The tetratricopeptide domain appears to coordinate protein-protein interactions between the motif-containing protein and a target protein (for a review, see reference 12). VCA0051 is located within a highly conserved operon in members of the Vibrionaceae, and there is a downstream gene coding for another GGDEF/EAL protein.
In addition to these types of regulators, two mutations were found to be located within genes encoding transcriptional regulators. One of the insertions is located in the lacI (VC2337) gene. The second interruption is in a gene containing a conserved PadR transcriptional regulator domain. This family of regulators has been implicated in the response to phenolic acid compounds (19).
Amino acid metabolism mutants.
Various mutations were found within genes responsible for amino acid metabolism, including the leuC and lysC genes. Although the role of these genes with respect to biofilm formation is unknown, their recovery is consistent with other mutagenesis screens which have discovered similar amino acid mutants that are unable to form proper biofilms (54).
Both the TP and SIO strains contained a mutation in the peptidase N operon (VC1494 and VC1495). The peptidase N product is related to proteins in the KEGG database (29) which are involved in glutathione catabolism by catalyzing the breakdown of l-cysteinyl-glycine to cysteine and glycine. Similar to other SIO mutants, the pepN mutant grew as smooth colonies on LB agar plates, indicating a possible deficiency in polysaccharide production, although it is not known how PepN enzymatic activity may govern VPS production.
Another gene which is important for both SIO and TP biofilm formation is tnaA, the gene encoding tryptophanase, which is a pyridoxyl 5′-phosphate-dependent enzyme that catalyzes the breakdown of tryptophan. Recent work has shown that indole, a product of this reaction, is a key factor in E. coli biofilm formation. Addition of this molecule to E. coli tnaA mutants causes significant changes in the proteome of the cells (9), and the molecule can complement biofilm deficiencies in various bacterial strains simply through exogenous addition (38). In the same way, indole was added to liquid cultures of the tnaA transposon mutants of SIO and TP. Crystal violet staining of the biofilms after overnight growth in 5 ml of LB broth at 37°C demonstrated that indole was able to complement the biofilm formation defect in both the SIO and TP mutants at concentrations of ≥100 μM (data not shown), which is in accordance with observed extracellular indole concentrations for E. coli grown in culture (64).
The tryptophanse mutant of SIO also grew as smooth, opaque colonies on LB agar, implying that the tryptophanse gene has a role in VPS production. However, colonies of this mutant were not identical to the smooth, translucent colonies of the VPS mutants of SIO, suggesting that VPS expression may not be completely eliminated in the tnaA mutant. To determine whether indole also affected rugosity in this strain, the SIO tnaA mutant was grown on plates containing indole. After growth overnight at 37°C, rugosity similar to that of the SIO wild type was restored in the mutant. These results indicate that indole does indeed influence the production of VPS in strain SIO.
Transporter mutants.
The biofilm mutant screen also uncovered numerous genes which have been annotated to function in the transport of various organic compounds and ions. Two insertions occurred in genes which have putatively been annotated to be involved in the transport of carbohydrates. The assigned function of the product of the one of these genes, VC1437, is an E1-E2 family transporter which has been annotated by the KEGG database to be an isomaltose transporter. Studies examining the transcriptome of E. coli biofilms have discovered that genes encoding a maltose permease transporter and maltose binding protein are among the genes whose expression is significantly increased in biofilm cells compared to the expression in planktonic cells (55). The second mutation is within the last gene of an operon coding for an ABC carbohydrate transporter, VCA1100. This operon is up-regulated by in the presence of chitin, suggesting its role in a surface-attached lifestyle (41).
A second group includes mutations within genes encoding an E1-E2 family putative Cu2+ transporter (VC2215) and a putative Zn2+ uptake regulation protein (VC0378) and an operon coding for a mechanosensitive channel and an Mg2+ transporter (VCA0817) (22). These loci are noteworthy given the connection between divalent cation transport and biofilm formation uncovered in other bacteria. A magnesium transporter is required for biofilm formation in Aeromonas hydrophila (42), and studies by Loo et al. (37) have shown that various proteins involved in zinc and manganese uptake are important for biofilm formation in Streptococcus gordonii.
The last mutation recovered in this class is in a gene coding for an iron(III) ABC transporter (VC0609). It has recently been reported that low-iron environments inhibit the normal formation of biofilms on surfaces by interfering with normal motility during biofilm development (58).
Unclassified mutants.
Numerous mutations occurred in genes without clear connections to specific functions in biofilm formation. Among these genes were various genes that have been annotated to encode hypothetical proteins, some of which appear to be regulated by RpoN, which has been shown in V. cholerae to regulate many genes involved in biofilm formation, including vpsR and hapR (70). Included in this group are two conserved hypothetical genes, VCA0936 and VCA0109. The latter of these genes has also been shown to be located within a conserved operon essential for the secretion of factors involved in eukaryotic cell death (52). Another gene in this group is VCA0859, which encodes a putative family 2 aldo/keto reductase. While the exact role of this protein in biofilm formation is unclear, it is notable that an oxidoreductase, PA3701, was also found to be essential for biofilm formation in Pseudomonas aeruginosa (16).
Other genes included in this group are hemB (VC0105), various other hypothetical genes, and two putative mobile genetic elements not found in the N16961 genome. One of these elements is closely related to a gene encoding a regulatory protein of the V. cholerae phage VGJΦ (5), and the other is a putative Tn5 transposase gene located in the intragenic region between VC0307 and VC0308 (Fig. 3b) (for sequence data, see Table S2 in the supplemental material). When a mutant with a mutation in the latter element was examined under flow cell conditions, it was noticed that it was unable to form microcolonies on the glass surface, preventing subsequent three-dimensional development of the biofilm (data not shown). It has recently been demonstrated that mobile genetic elements may play a large role in biofilms, as phage-related genes have been shown to be activated in biofilms and, in some cases, dictate phenotypic variation in cells, resulting in differences in biofilm development (66).
Phenotypic verification of genetic disruptions.
Many of the biofilm mutants contained insertions within putative operons. Consequently, the biofilm defect could be derived from the loss of function of the disrupted gene or could be due to a polar effect on downstream gene expression. In order to discriminate between these possibilities, in-frame deletions were made for a select group of SIO and TP mutants, and the biofilm-forming abilities of the strains were measured. This method was chosen instead of standard plasmid complementation because of the difficulty in creating suitable and stable expression vectors for strains SIO and TP (unpublished results).
Table 3 lists the in-frame deletions which were created. Also shown are the relative biofilm-forming abilities of some transposon mutants and their in-frame deletion counterparts. As a control, in-frame deletions were created in the vpsL gene (VC0934) of SIO and TP, which is essential for VPS production and proper biofilm formation (69). As expected, this mutation greatly reduced biofilm formation in these strains. All of the other in-frame deletions tested exhibited biofilm defects similar to those of the strains carrying transposon mutations in the same genes. These results indicate that the biofilm deficiency of these mutants is most likely derived from the loss of function of the disrupted genes.
TABLE 3.
Verification of biofilm defects in selected mutants
| Strain | Genetic disruption | Predicted function | Biofilm formationa
|
|
|---|---|---|---|---|
| Transposon insertion | In-frame deletion | |||
| V. cholerae SIO | ||||
| SIO | Wild type | 1.00 ± 0.192 | 1.00 ± 0.192 | |
| S2151 | VC0408 | MSHA pilin | NRb | 1.17 ± 0.294 |
| S2123 | VC0892 | Motility | NR | 0.919 ± 0.070 |
| S2104 | VC2188 | Flagellin | NR | 0.054 ± 0.033 |
| S1155/S2153 | VC0920/VC0934 | VPS genes | 0.018 ± 0.004 | 0.014 ± 0.015 |
| S1163/S2163 | VC1494 | Aminopeptidase | 0.034 ± 0.005 | 0.045 ± 0.200 |
| S1164/S2164 | VC2215 | Cation transport | 0.598 ± 0.793 | 0.645 ± 0.147 |
| S1101/S2101 | VCA0161 | Tryptophanase | 0.157 ± 0.046 | 0.169 ± 0.097 |
| V. cholerae TP | ||||
| TP | Wild type | 1.00 ± 0.182 | 1.00 ± 0.182 | |
| T1114/T2114 | VC0378 | Zinc transport | 0.025 ± 0.004 | 0.133 ± 0.073 |
| T1141/T2151 | VC0414/VC0408 | MSHA pilus genes | 0.081 ± 0.040 | 0.051 ± 0.064 |
| T1123/T2123 | VC0892 | Motility | 0.161 ± 0.117 | 0.009 ± 0.001 |
| T1102/T2153 | VC0935/VC0934 | VPS genes | 0.154 ± 0.053 | 0.026 ± 0.012 |
| T1127/T2127 | VC1437 | Cation transport | 0.134 ± 0.094 | 0.045 ± 0.029 |
| T1104/T2104 | VC2188 | Flagellin | 0.076 ± 0.012 | 0.013 + 0.001 |
| T1112/T2112 | VC2337 | Transcription | 0.490 ± 0.272 | 0.136 ± 0.059 |
| T1101/T2101 | VCA0161 | Tryptophanase | 0.076 ± 0.035 | 0.057 ± 0.050 |
| T1130/T2130 | VCA0859 | Oxidoreductase | 0.240 ± 0.230 | 0.453 ± 0.558 |
Biofilms were quantified using the crystal violet staining technique described in Materials and Methods. The values are means ± one standard deviation from the mean.
NR, not recovered by transposon mutagenesis screen.
Colonization of plankton by V. cholerae wild-type strains.
To determine whether diminution of attachment to abiotic surfaces translates into impairment of biotic attachment, the abilities of various strains to colonize marine plankton were evaluated. Initial colonization experiments examined the ability of gfp-tagged strains of N16961, SIO, and TP to attach to the marine copepod T. californicus and to the marine dinoflagellate L. polyedrum. For each experiment approximately 106 cells per ml were incubated with copepods or dinoflagellates, and colonization was allowed to proceed overnight at room temperature.
When attachment to copepods was examined, it was noted that all strains of V. cholerae were able to colonize T. californicus but that colonization of heat-killed copepods was greatly enhanced compared with the colonization of live animals (data not shown). On live copepods colonization was greatest in the oral and anal regions and the egg sacs, as previously reported (25). In contrast, after the copepod was killed, the entire carapace was quickly colonized by all three V. cholerae strains tested.
Similar results were obtained when attachment to L. polyedrum was investigated. Each strain exhibited a strong affinity for dead organisms, and there was little attachment to live, exponentially growing phytoplankton. Attachment to both live and dead L. polyedrum was dependent on the phase of growth of the dinoflagellate, as no strains of V. cholerae were able to attach appreciably to live or dead organisms when the L. polyedrum was taken from older cultures in the final stages of growth. Unlike colonization of copepods, there were no distinct differences in the abilities of the strains of V. cholerae to attach to the dinoflagellates.
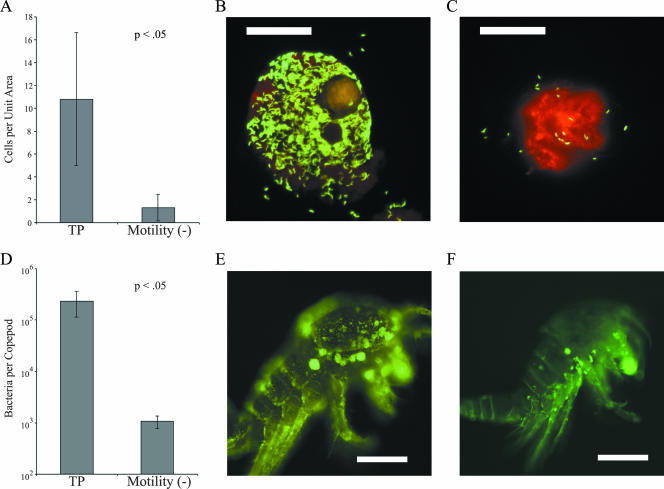
The biofilms formed on dead dinoflagellates and copepods under seawater conditions were different than those formed on glass or plastic surfaces by LB medium cultures. Similar to biofilms formed on glass under artificial seawater conditions (30), the biofilms on biotic substrates consisted mainly of a single layer of cells which eventually covered the entire surface of the organism (Fig. 4B and E). When time course experiments were performed using T. californicus as the substrate for colonization, rapid initial attachment occurred, with significant numbers of V. cholerae attaching after 4 h of incubation at 30°C. At this point the numbers of attached bacteria increased from ∼103 cells per copepod to ∼105 cells per copepod. After 24 h the amounts of SIO and TP cells attached to copepods were ∼5- and ∼10-fold greater than the amounts of N16961 cells, respectively (data not shown).
FIG. 4.
Comparison of the abilities of gfp-tagged wild-type V. cholerae and motility mutants to colonize the dinoflagellate L. polyedrum and T. californicus. (A) Quantification of cells of the TP wild-type strain and a motility mutant of TP attached to the surface of the dinoflagellate. (B and C) Epifluorescent images of the strains on the surface of dead L. polyedrum. Magnification, ×1,000. Scale bars = 20 μm. (D) Quantification of cells of the TP wild-type strain and a motility mutant attached to the surface of copepods. (E and F) Epifluorescent images of TPf and T2123f on the surface of dead T. californicus. Magnification, ×100. Scale bars = 200 μm. The error bars in the graphs represent one standard deviation from the mean.
Colonization of plankton by biofilm mutants.
In order to evaluate the plankton-colonizing abilities of the SIO and TP biofilm mutants, a selection of gfp-tagged biofilm mutants were also tested. In these experiments, most of the biofilm mutants tested were found to be able to colonize both L. polyedrum and T. californicus at levels similar to the levels of the wild-type SIO and TP strains (Table 4). However, when the motility or chemotaxis mutants of strain TP and SIO were tested for colonization of plankton, little or no visible attachment by the bacteria was observed (Fig. 4C and F). In the few instances where attachment was observed with these mutants, only small clusters of cells were found attached to a surface, which differed dramatically from the biofilms formed by the wild-type strains (Fig. 4B and E).
TABLE 4.
Qualitative evaluation of the colonization abilities of gfp-tagged biofilm mutants on dinoflagellate and copepod surfaces
| Straina | Genetic disruption | Predicted function | Colonization of substrateb
|
|
|---|---|---|---|---|
| Dinoflagellate | Copepod | |||
| V. cholerae SIO | ||||
| SIOf | SIO wild type | + | + | |
| S2151f | VC0408 | MSHA pilin | ++ | + |
| S2123f | VC0892 | Flagellar motor | − | − |
| S1155f | VC0920 | VPS gene | + | ++ |
| S1103f | VC0922 | VPS gene | + | + |
| S1166f | VC1934 | GGDEF/EAL | + | + |
| S1154f | VCA0051 | Hypothetical (GGDEF/EAL operon) | + | + |
| S1160f | VCA0075 | Hypothetical (GGDEF operon) | + | + |
| S1162f | VCA0094 | Hypothetical/transcriptional regulator | + | + |
| S1101f | VCA0161 | Tryptophanase | + | + |
| SIOf + phenamil | Flagellar motor inhibitor | NT | − | |
| V. cholerae TP | ||||
| TPf | TP wild type | + | + | |
| T1118f | VC0105 | Porphyrin metabolism | + | + |
| T1142f | VC0240 | LPS biosynthesis | + | + |
| T1114f | VC0378 | Ion transport | + | + |
| T1130f | VC0391 | Lysine metabolism | + | + |
| T2151f | VC0408 | MSHA pilin | ++ | + |
| T1141f | VC0414 | MSHA pilus biosynthesis | + | + |
| T1132f | VC0609 | Ion transport | + | + |
| T1123f | VC0892 | Flagellar motor | − | − |
| T2123f | VC0892 | Flagellar motor | − | − |
| T1102f | VC0918 | VPS gene | ++ | ++ |
| T1103f | VC0922 | Hypothetical (VPS operon) | + | + |
| T1111f | VC1495 | Hypothetical (pepN operon) | + | + |
| T1139f | VC2059 | Chemotaxis | − | − |
| T1116f | VC2125 | Flagellar motor switch | − | − |
| T1106f | VC2131 | Flagellar assembly | − | − |
| T1104f | VC2188 | Flagellin | − | − |
| T1108f | VC2190 | Flagellar hook protein | − | − |
| T1112f | VC2337 | Transcriptional regulator | + | + |
| T1101f | VCA0161 | Tryptophanase | ++ | + |
| T1120f | VCA0213 | Hypothetical/ion transporter | + | + |
| T1131f | VCA0524 | Hypothetical/ion transporter | + | + |
| T1115f | VCA0817 | Hypothetical/mechanosensitive transporter | + | + |
| T1130f | VCA0859 | Oxidoreductase | + | ++ |
| T1138f | Putative LPS loci | + | ||
| T1140f | Vibrio phage loci | + | + | |
| T1143f | Tn5 transposase | + | ++ | |
| TPf + phenamil | Flagellar motor inhibitor | NT | − | |
The strains contain the gfp-expressing plasmid pFLgfp.
+, wild-type colonization level; ++, colonization level greater than the wild-type colonization level; −, greatly reduced colonization compared to the wild-type colonization; NT, not tested.
Figure 4A illustrates quantitatively the inability of the motility mutants of strain TP to attach to L. polyedrum. Compared to the wild type, the motility mutant of TP was impaired approximately 10-fold in colonization of dinoflagellates. Under the conditions used it was estimated that approximately 3.5 × 102 V. cholerae wild-type cells were able to colonize the surface of a single L. polyedrum cell, whereas only 40 cells of the mutants impaired in motility were able to attach. Similar results were obtained when attachment to copepods was quantified. Motility mutants of strain TP were, on average, 100-fold less proficient in colonization after overnight incubation, and only ∼103 cells attached to an entire copepod, compared to the >105 cells of the wild type that attached (Fig. 4D). These results were further verified when the wild-type SIO and TP strains were tested for colonization ability in the presence of phenamil, which poisons the Na+-driven motor of the Vibrio polar flagellum. It was observed that in the presence of 100 μM phenamil wild-type cells of SIO and TP were nonmotile and were not able to significantly colonize the carapaces of copepods after overnight incubation.
DISCUSSION
The ability of V. cholerae to concentrate on surfaces in aquatic environments has been hypothesized to be one of the main factors controlling environmental survival and disease transmission (11). Our results, which revealed many new genes not previously implicated in V. cholerae biofilm formation and demonstrated that there are differences between abiotic biofilm formation and biotic biofilm formation, show that strains of V. cholerae are highly variable in terms of both genotype and phenotype with regard to surface attachment.
A clear illustration of this variability was seen when different strains of V. cholerae were observed to form distinct biofilms even when they were grown in flow cells under the same experimental conditions. In this experiment, the surface colonization profile of TP mirrored that of clinical strain N16961, while SIO formed unique biofilms. These lacked distinct microcolonies late in the experiment and remained attached to the substratum for multiple days after the film had died. Similarly, data from separate biofilm flow cell experiments showed that another rugose variant of V. cholerae (strain 91A1552) displayed the same biofilm phenotype as SIO (D. McDougald, unpublished data). Therefore, it is possible that overproduction of VPS may lead to the observed lack of water channels, which may be essential for metabolite transport and sustained growth of the biofilms, accounting for the phenotypic differences recorded.
In order to identify genes responsible for these biofilm structure differences, we performed transposon mutagenesis screens with the SIO and TP strains of V. cholerae. While these screens were not saturating, the genes found to be responsible for biofilm formation in each strain did not fully overlap, and a distinct bias was found in the types of genes recovered for each strain. Whereas the mutations in strain TP involved a wide variety of genes, including genes for VPS production and flagellum and MSHA pilus biosynthesis, 75% of the mutations recovered for SIO appeared to directly or indirectly influence VPS production. Also, no mutations in genes responsible for flagellum or MSHA biosynthesis in SIO were recovered. In fact, in-frame deletions of the mshA and flaA genes did not significantly diminish abiotic biofilm formation by this strain, and the former actually resulted in greater surface accumulation than that of the wild type. This mechanism is not unique to SIO, as a study by Watnick and Kolter (65) has shown that a V. cholerae O139 clinical isolate also does not rely on flagella or MSHA pili to form biofilms. The dispensable biofilm role of the MSHA pilus is further reflected by the fact that ΔmshA mutants of both SIO and TP are not impaired in the ability to colonize copepods. This is not entirely surprising since Chiavelli et al. (8) showed that while the MSHA pilus is required for attachment of V. cholerae to exoskeletons of Daphnia pulex by the MSHA-expressing O139 strains, it has little relevance in attachment by O1 classical strains, which are not believed to produce MSHA.
In addition to demonstrating the differences in the significance of certain genes in SIO and TP for biofilm formation, the transposon mutagenesis screen identified many new genes not previously described as genes that are important for V. cholerae biofilms. Six transposon insertions which disrupted the genes involved in the transport of carbohydrates or metal ions, such as Zn2+, Mg2+, and Fe3+, were identified. Numerous studies have suggested that in addition to being essential for many fundamental cellular processes, various transporters perform a critical role in microbial biofilm formation (37, 42). It has been shown that divalent cations have a strong affinity for the exopolymer matrix of biofilms, and it has been hypothesized that they support the biofilm by acting as ionic cross-links between molecules of exopolysaccharide (EPS) (7, 62). It is possible that under this scenario, trace metals are sequestered within the EPS matrix and become limiting in the cytosol of the bacterial cells. Therefore, the role of ion transport systems may become more critical within EPS-containing biofilms.
A second important class of biofilm mutants contained disruptions in genes involved in amino acid metabolism. These types of genes have also been found to be important for biofilms of other species of bacteria in studies using microarray and mutagenesis analyses (54, 67). From this work it has been hypothesized that these genes may have an essential role in the physiological shift occurring within cells which accompanies the transition from planktonic to biofilm cells. One gene in particular that was found to be important for biofilm formation in multiple species of bacteria was the tryptophanse gene (38). In these bacteria the tnaA gene has been proposed to regulate biofilm formation through a signaling mechanism involving a by-product of the tryptophanase reaction, indole. Our results are consistent with these studies and further implicate indole as a regulator of VPS expression, since indole is able to complement the loss of rugosity of the SIO tnaA transposon mutant. While other studies have shown that indole can modulate the expression of genes involved in a variety of cellular functions (9), our findings provide the first evidence that indole can function as an extracellular signaling molecule influencing the expression of genes involved in the production of the exopolymeric matrix extruded by cells within a biofilm.
Many genes which have a role in biofilm formation at the level of gene and protein regulation, including genes coding for GGDEF/EAL family proteins, were also found. Research by Tischler eand Camilli (61) has shown that proteins of this family can modulate intracellular concentrations of the second messenger c-di-GMP, which appears to act as a signaling molecule in V. cholerae affecting the transcription of VPS genes. In the work of these researchers, the GGDEF domain-containing VCA0956 protein was discovered to increase intracellular c-di-GMP levels through its diguanylate cyclase activity, which causes subsequent induction of VPS transcription. Observations of reduced biofilm formation and smooth colony morphology for a mutant with a mutation in the VCA0075 operon, which contains another GGDEF domain-containing protein, in this study and another study (35) suggest that there is a similar deficiency in VPS synthesis.
It is clear from these results that SIO and TP, much like other strains of V. cholerae, rely on disparate sets of genes and regulatory mechanisms in order to produce biofilms on both biotic and abiotic surfaces. While some fundamental genetic mechanisms used to form biofilms may be shared between strains of V. cholerae, there appears to be no set of “universal” biofilm genes for all strains. It is also possible that the same strain relies on many different genes to form distinct biofilm morphologies when it is exposed to a new environment.
This was indeed seen in comparisons of the biofilms of the wild-type strains grown on abiotic surfaces in nutrient-replete rich medium (LB medium) and the biofilms grown on biotic surfaces in unenriched seawater microcosms. In these experiments V. cholerae cells formed an almost exclusively monolayer biofilm on plankton in the seawater microcosms and never produced a three-dimensional film characteristic of LB medium-grown biofilms. Therefore, genetic mutants involved in initial surface attachment could be important for these biofilms, whereas genes involved in producing the three-dimensional structure of biofilms (e.g., VPS genes) could have reduced importance. Consistent with this possibility, only mutants affected in motility and chemotaxis, which have previously been shown to be responsible for defects in the monolayer stage of biofilm formation in V. cholerae (48), were impeded in attachment to L. polyedrum and T. californicus, whereas none of the VPS mutants tested demonstrated appreciable reduction in attachment. This result is supported by a previous finding that motility mutants of Silicibacter sp. strain TM1040 are also not able to colonize the marine dinoflagellate Pfesteria piscicida (44).
It should be noted that it is not merely the presence of a flagellum which is required for attachment to plankton; proper rotation of the flagellum is also required. Flagellated strains grown in the presence of phenamil, an inhibitor of the sodium-driven flagellum motor (4), were also unable to attach appreciably to copepods. Therefore, it may be the motive force that a functional flagellum provides which is needed to overcome the electrostatic repulsive forces found on surfaces (65).
Another result of this study is that under the conditions tested all of the strains of V. cholerae were dramatically better at attaching to dead organisms than at attaching to live organisms. This coincides with previous mesocosm experiments which showed that V. cholerae preferentially associates with the detritus rather than the free-living phase during the demise of intense phytoplankton blooms (68). Therefore, in the context of the true environment, quantification of V. cholerae populations associated with detritus and within sediments as a function of plankton bloom cycles would be helpful in assessing the ecological significance of these colonization preferences. With such knowledge we may be better able to identify the exact nature of the environmental reservoir of V. cholerae.
This study provided evidence that different strains of V. cholerae possess overlapping but distinctive sets of genes for biofilm formation. In addition, clear differences in the genetic requirements for biofilm formation on abiotic and biotic surfaces and in the characteristics of the biofilms were noted.
Supplementary Material
Acknowledgments
We thank Bianca Brahamsha for assistance with manuscript preparation, Fiona Tomas and Xavier Mayali for aiding with statistical analysis, Federico Lauro for designing arbitrary PCR primers and insightful discussions, Paul Rohdenburg for help with initial colonization experiments, and Sinem Beyhan, Bentley Lim, and Fitnat Yildiz for help constructing an initial deletion construct in strain SIO.
This work was supported by NIH grant AI46600-02 and by funding from grant CEQI0047 provided by the UC Marine Council Coastal Environmental Quality Initiative.
Footnotes
Published ahead of print on 11 May 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alexeyev, M. F., and I. N. Shokolenko. 1995. Mini-Tn10 transposon derivatives for insertion mutagenesis and gene delivery into the chromosome of Gram-negative bacteria. Gene 160:59-62. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Arps, P. J., and M. E. Winkler. 1987. Structural analysis of the Escherichia coli K-12 hisT operon by using a kanamycin resistance cassette. J. Bacteriol. 169:1061-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atsumi, T., L. McCarter, and Y. Imae. 1992. Polar and lateral flagellar motors of marine Vibrio are driven by different ion-motive forces. Nature 355:182-184. [DOI] [PubMed] [Google Scholar]
- 5.Campos, J., E. Martinez, E. Suzarte, B. L. Rodriguez, K. Marrero, Y. Silva, T. Ledon, R. del Sol, and R. Fando. 2003. VGJΦ, a novel filamentous phage of Vibrio cholerae, integrates into the same chromosomal site as CTXΦ. J. Bacteriol. 185:5685-5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casper-Lindley, C., and F. H. Yildiz. 2004. VpsT is a transcriptional regulator required for the rugose colonial morphology of Vibrio cholerae O1 El Tor. J. Bacteriol. 186:1574-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, X., and P. S. Stewart. 2002. Role of electrostatic interactions in cohesion of bacterial biofilms. Appl. Microbiol. Biotechnol. 59:718-720. [DOI] [PubMed] [Google Scholar]
- 8.Chiavelli, D. A., J. W. Marsh, and R. K. Taylor. 2001. The mannose-sensitive hemagglutinin of Vibrio cholerae promotes adherence to zooplankton. Appl. Environ. Microbiol. 67:3220-3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collet, A., S. Vilain, P. Cosette, G. A. Junter, T. T. Jouenne, R. S. Phillips, and P. D. Martino. 2007. Protein expression in Escherichia coli S17-1 biofilms: impact of indole. Antonie Leeuwenhoek 91:71-85. [DOI] [PubMed] [Google Scholar]
- 10.Colwell, R. R. 1996. Global climate and infectious disease: the cholera paradigm. Science 274:2025-2031. [DOI] [PubMed] [Google Scholar]
- 11.Colwell, R. R., A. Huq, M. S. Islam, K. M. A. Aziz, M. Yunus, N. H. Khan, A. Mahmud, R. B. Sack, G. B. Nair, J. Chakraborty, D. A. Sack, and E. Russek-Cohen. 2003. Reduction of cholera in Bangladeshi villages by simple filtration. Proc. Natl. Acad. Sci. USA 100:1051-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D'Andrea, L. D., and L. Regan. 2003. TPR proteins: the versatile helix. Trends Biochem. Sci. 28:655. [DOI] [PubMed] [Google Scholar]
- 13.Elkins, J. G., D. J. Hassett, P. S. Stewart, H. P. Schweizer, and T. R. McDermott. 1999. Protective role of catalase in Pseudomonas aeruginosa biofilm resistance to hydrogen peroxide. Appl. Environ. Microbiol. 65:4594-4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Epstein, P. 1993. Algal blooms in the spread and persistence of cholera. Biosystems 31:209-221. [DOI] [PubMed] [Google Scholar]
- 15.Espeland, E. M., and R. G. Wetzel. 2001. Complexation, stabilization, and UV photolysis of extracellular and surface-bound glucosidase and alkaline phosphatase: implications for biofilm microbiota. Microb. Ecol. 42:572-585. [DOI] [PubMed] [Google Scholar]
- 16.Finelli, A., C. V. Gallant, K. Jarvi, and L. L. Burrows. 2003. Use of in-biofilm expression technology to identify genes involved in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 185:2700-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finkelstein, R. A., and S. Mukerjee. 1963. Hemagglutination: a rapid method for differentiating Vibrio cholerae and El Tor vibrios. Proc. Soc. Exp. Biol. Med. 112:355-359. [Google Scholar]
- 18.Guillard, R. R. L. 1975. Culture of phytoplankton for feeding marine invertebrates. Plenum Press, New York, NY.
- 19.Gury, J., L. Barthelmebs, N. P. Tran, C. Divies, and J.-F. Cavin. 2004. Cloning, deletion, and characterization of PadR, the transcriptional repressor of the phenolic acid decarboxylase-encoding padA gene of Lactobacillus plantarum. Appl. Environ. Microbiol. 70:2146-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halpern, M., Y. B. Broza, S. Mittler, E. Arakawa, and M. Broza. 2004. Chironomid egg masses as a natural reservoir of Vibrio cholerae non-O1 and non-O139 in freshwater habitats. Microb. Ecol. 47:341-349. [DOI] [PubMed] [Google Scholar]
- 21.Haugo, A. J., and P. I. Watnick. 2002. Vibrio cholerae CytR is a repressor of biofilm development. Mol. Microbiol. 45:471-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. Mcdonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horton, R. 1997. In vitro recombination and mutagenesis of DNA. SOEing together tailor-made genes. Methods Mol. Biol. 67:141-149. [DOI] [PubMed] [Google Scholar]
- 24.Huq, A., R. B. Sack, A. Nizam, I. M. Longini, G. B. Nair, A. Ali, J. G. Morris, Jr., M. N. H. Khan, A. K. Siddique, M. Yunus, M. J. Albert, D. A. Sack, and R. R. Colwell. 2005. Critical factors influencing the occurrence of Vibrio cholerae in the environment of Bangladesh. Appl. Environ. Microbiol. 71:4645-4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huq, A., E. B. Small, P. A. West, M. I. Huq, R. Rahman, and R. R. Colwell. 1983. Ecological relationships between Vibrio cholerae and planktonic crustacean copepods. Appl. Environ. Microbiol. 45:275-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Islam, M. S., B. S. Drasar, and D. J. Bradley. 1990. Long-term persistence of toxigenic Vibrio cholerae 01 in the mucilaginous sheath of a blue-green alga, Anabaena variabilis. J. Trop. Med. Hyg. 93:133-139. [PubMed] [Google Scholar]
- 27.Islam, M. S., S. Mahmuda, M. G. Morshed, H. B. Bakht, M. N. Khan, R. B. Sack, and D. A. Sack. 2004. Role of cyanobacteria in the persistence of Vibrio cholerae O139 in saline microcosms. Can. J. Microbiol. 50:127-131. [DOI] [PubMed] [Google Scholar]
- 28.Jones, D. H., and S. C. Winistorfer. 1992. Sequence specific generation of a DNA panhandle permits PCR amplification of unknown flanking DNA. Nucleic Acids Res. 20:595-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanehisa, M., and S. Goto. 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28:27-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kierek, K., and P. I. Watnick. 2003. Environmental determinants of Vibrio cholerae biofilm development. Appl. Environ. Microbiol. 69:5079-5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirn, T. J., B. A. Jude, and R. K. Taylor. 2005. A colonization factor links Vibrio cholerae environmental survival and human infection. Nature 438:863-866. [DOI] [PubMed] [Google Scholar]
- 32.Larsen, R. A., M. M. Wilson, A. M. Guss, and W. W. Metcalf. 2002. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch. Microbiol. 178:193-201. [DOI] [PubMed] [Google Scholar]
- 33.Lauro, F. M., E. A. Eloe, N. Liverani, G. Bertoloni, and D. H. Bartlett. 2005. Conjugal vectors for cloning, expression, and insertional mutagenesis in gram-negative bacteria. BioTechniques 38:708, 710, 712. [DOI] [PubMed] [Google Scholar]
- 34.Lehmann, V., G. Hammerling, M. Nurminen, I. Minner, E. Ruschmann, O. Luderitz, T.-T. Kuo, and B. A. D. Stocker. 1973. A new class of heptose-defective mutant of Salmonella typhimurium. Eur. J. Biochem. 32:268-275. [DOI] [PubMed] [Google Scholar]
- 35.Lim, B., S. Beyhan, J. Meir, and F. H. Yildiz. 2006. Cyclic-diGMP signal transduction systems in Vibrio cholerae: modulation of rugosity and biofilm formation. Mol. Microbiol. 60:331-348. [DOI] [PubMed] [Google Scholar]
- 36.Lobitz, B., L. Beck, A. Huq, B. Wood, G. Fuchs, A. S. G. Faruque, and R. Colwell. 2000. Climate and infectious disease: use of remote sensing for detection of Vibrio cholerae by indirect measurement. Proc. Natl. Acad. Sci. USA 97:1438-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loo, C. Y., K. Mitrakul, I. B. Voss, C. V. Hughes, and N. Ganeshkumar. 2003. Involvement of the adc operon and manganese homeostasis in Streptococcus gordonii biofilm formation. J. Bacteriol. 185:2887-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martino, P. D., R. Fursy, L. Bret, B. Sundararaju, and R. S. Phillips. 2003. Indole can act as an extracellular signal to regulate biofilm formation of Escherichia coli and other indole-producing bacteria. Can. J. Microbiol. 49:443-449. [DOI] [PubMed] [Google Scholar]
- 39.Matthysse, A. G., S. Stretton, C. Dandie, N. C. McClure, and A. E. Goodman. 1996. Construction of GFP vectors for use in Gram-negative bacteria other than Escherichia coli. FEMS Microbiol. Lett. 145:87-94. [DOI] [PubMed] [Google Scholar]
- 40.Matz, C., D. McDougald, A. M. Moreno, P. Y. Yung, F. H. Yildiz, and S. Kjelleberg. 2005. Biofilm formation and phenotypic variation enhance predation-driven persistence of Vibrio cholerae. Proc. Natl. Acad. Sci. USA 102:16819-16824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meibom, K. L., X. B. Li, A. T. Nielsen, C. Y. Wu, S. Roseman, and G. K. Schoolnik. 2004. The Vibrio cholerae chitin utilization program. Proc. Natl. Acad. Sci. USA 101:2524-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Merino, S., R. Gavin, M. Altarriba, L. Izquierdo, M. E. Maguire, and J. M. Tomas. 2001. The MgtE Mg2+ transport protein is involved in Aeromonas hydrophila adherence. FEMS Microbiol. Lett. 198:189-195. [DOI] [PubMed] [Google Scholar]
- 43.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Press, Cold Spring Harbor, NY.
- 44.Miller, T. R., and R. Belas. 2006. Motility is involved in Silicibacter sp. TM1040 interaction with dinoflagellates. Environ. Microbiol. 8:1648-1659. [DOI] [PubMed] [Google Scholar]
- 45.Mizunoe, Y., S. N. Wai, A. Takade, and S.-I. Yoshida. 1999. Isolation and characterization of rugose form of Vibrio cholerae O139 strain MO10. Infect. Immun. 67:958-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moller, S., C. Sternberg, J. B. Andersen, B. B. Christensen, J. L. Ramos, M. Givskov, and S. Molin. 1998. In situ gene expression in mixed-culture biofilms: evidence of metabolic interactions between community members. Appl. Environ. Microbiol. 64:721-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moorthy, S., and P. I. Watnick. 2004. Genetic evidence that the Vibrio cholerae monolayer is a distinct stage in biofilm development. Mol. Microbiol. 52:573-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moorthy, S., and P. I. Watnick. 2005. Identification of novel stage-specific genetic requirements through whole genome transcription profiling of Vibrio cholerae biofilm development. Mol. Microbiol. 57:1623-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muramoto, K., Y. Magariyama, M. Homma, I. Kawagishi, S. Sugiyama, Y. Imae, and S. Kudo. 1996. Rotational fluctuation of the sodium-driven flagellar motor of Vibrio alginolyticus is induced by binding of inhibitors. J. Mol. Biol. 259:687-695. [DOI] [PubMed] [Google Scholar]
- 50.O'Toole, G. A., L. A. Pratt, P. I. Watnick, D. K. Newman, V. B. Weaver, and R. Kolter. 1999. Genetic approaches to study of biofilms. Methods Enzymol. 310:91-109. [DOI] [PubMed] [Google Scholar]
- 51.Paludan-Muller, C., D. Weichart, D. McDougald, and S. Kjelleberg. 1996. Analysis of starvation conditions that allow for prolonged culturability of Vibrio vulnificus at low temperature. Microbiology 142:1675-1684. [DOI] [PubMed] [Google Scholar]
- 52.Pukatzki, S., A. T. Ma, D. Sturtevant, B. Krastins, D. Sarracino, W. C. Nelson, J. F. Heidelberg, and J. J. Mekalanos. 2006. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. USA 103:1528-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Purdy, A., F. Rohwer, R. Edwards, F. Azam, and D. H. Bartlett. 2005. A glimpse into the expanded genome content of the Vibrio cholerae species through the identification of genes present in environmental strains. J. Bacteriol. 187:2992-3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sauer, K., and A. K. Camper. 2001. Characterization of phenotypic changes in Pseudomonas putida in response to surface-associated growth. J. Bacteriol. 183:6579-6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schembri, M. A., K. Kjaergaard, and P. Klemm. 2003. Global gene expression in Escherichia coli biofilms. Mol. Microbiol. 48:253-267. [DOI] [PubMed] [Google Scholar]
- 56.Shukla, B. N., D. C. Singh, and S. C. Sanyal. 1995. Attachment of non-culturable toxigenic Vibrio cholerae O1 and non-O1 and Aeromonas spp. to the aquatic arthropod Gerris spinolae and plants in the River Ganga, Varanasi. FEMS Immun. Med. Microbiol. 12:113-120. [DOI] [PubMed] [Google Scholar]
- 57.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784. [Google Scholar]
- 58.Singh, P. K., M. R. Parsek, E. P. Greenberg, and M. J. Welsh. 2002. A component of innate immunity prevents bacterial biofilm development. Nature 417:552. [DOI] [PubMed] [Google Scholar]
- 59.Snow, J. 1855. On the mode of communication of cholera, 2nd ed. John Churchill, London, England.
- 60.Teitzel, G. M., and M. R. Parsek. 2003. Heavy metal resistance of biofilm and planktonic Pseudomonas aeruginosa. Appl. Environ. Microbiol. 69:2313-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tischler, A. D., and A. Camilli. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol. Microbiol. 53:857-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Toner, B., A. Manceau, M. A. Marcus, D. B. Millet, and G. Sposito. 2005. Zinc sorption by a bacterial biofilm. Environ. Sci. Technol. 39:8288-8294. [DOI] [PubMed] [Google Scholar]
- 63.Wai, S. N., Y. Mizunoe, A. Takade, S.-I. Kawabata, and S.-I. Yoshida. 1998. Vibrio cholerae O1 strain TSI-4 produces the exopolysaccharide materials that determine colony morphology, stress resistance, and biofilm formation. Appl. Environ. Microbiol. 64:3648-3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang, D., X. Ding, and P. N. Rather. 2001. Indole can act as an extracellular signal in Escherichia coli. J. Bacteriol. 183:4210-4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Watnick, P. I., and R. Kolter. 1999. Steps in the development of a Vibrio cholerae El Tor biofilm. Mol. Microbiol. 34:586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Webb, J. S., M. Lau, and S. Kjelleberg. 2004. Bacteriophage and phenotypic variation in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 186:8066-8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Whiteley, M., M. G. Bangera, R. E. Bumgarner, M. R. Parsek, G. M. Teitzel, S. Lory, and E. P. Greenberg. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860-864. [DOI] [PubMed] [Google Scholar]
- 68.Worden, A. Z., M. Seidel, S. Smriga, A. Wick, F. Malfatti, D. Bartlett, and F. Azam. 2006. Trophic regulation of Vibrio cholerae in coastal marine waters. Environ. Microbiol. 8:21-29. [DOI] [PubMed] [Google Scholar]
- 69.Yildiz, F. H., N. A. Dolganov, and G. K. Schoolnik. 2001. VpsR, a member of the response regulators of the two-component regulatory systems, is required for expression of vps biosynthesis genes and EPSETr-associated phenotypes in Vibrio cholerae O1 El Tor. J. Bacteriol. 183:1716-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yildiz, F. H., X. S. Liu, A. Heydorn, and G. K. Schoolnik. 2004. Molecular analysis of rugosity in a Vibrio cholerae O1 El Tor phase variant. Mol. Microbiol. 53:497-515. [DOI] [PubMed] [Google Scholar]
- 71.Yildiz, F. H., and G. K. Schoolnik. 1999. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc. Natl. Acad. Sci. USA 96:4028-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhu, J., M. B. Miller, R. E. Vance, M. Dziejman, B. L. Bassler, and J. J. Mekalanos. 2002. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 99:3129-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.